No new local coronavirus disease 2019 (COVID-19) cases were reported in Beijing for over 56 days before the reemergent COVID-19 cases that were reported on June 11, 2020, and more than 200 confirmed cases have been reported as of June 20 in Beijing. All confirmed COVID-19 cases were associated with Xinfadi Wholesale Market. Here, we reported the phylogenetic characteristics of the COVID-19 viruses (also known as severe acute respiratory syndrome coronavirus 2, SARS-CoV-2, and human coronavirus 2019, HCoV-19) sequenced from the clinical specimens of 2 human cases and 1 environmental sample.
We conducted whole-genome sequencing of COVID-19 viruses using the Illumina NextSeq 550 platform. A total of 3 whole genomes of COVID-19 virus were submitted to GISAID website and these sequences were named as nCoV Beijing IVDC-001 (EPI_ ISL_469254, from the first confirmed case in a 53-year-old male), -002 (EPI_ ISL_469255, from another confirmed case in a 25-year-old male), -003 (EPI_ ISL_469256, from environmental sample), respectively. These genomes (Beijing Xinfadi strains) showed 99.96%–99.97% nucleic acid identity to the reference strain hCoV-19/Wuhan/IVDC-HB-01/2019|EPI_ISL_402119|2019-12-30 that was isolated from Wuhan on January 7, 2020 (1). A total of 7 nucleotide variations were identified among the 3 whole genomes of Xinfadi strains, including C241T, C3037T, C14408T, A23403G, G28881A, G28882A, and G28883C, which were consistent with the characteristics of Branch 1 of the L-lineage from Europe (Figure 1) and different from the previous pandemic COVID-19 virus in Wuhan (Dec-2019) and Beijing (Feb-2020), which belong to the S-lineage (2). Additional genetic variations were also identified among these Xinfadi strains including the C6026T mutation in all 3 genomes of the virus, C12085T and A29694C in nCoV Beijing IVDC-001 and A11910G, A29694C, and G29868A in nCoV Beijing IVDC-003.
Figure 1.
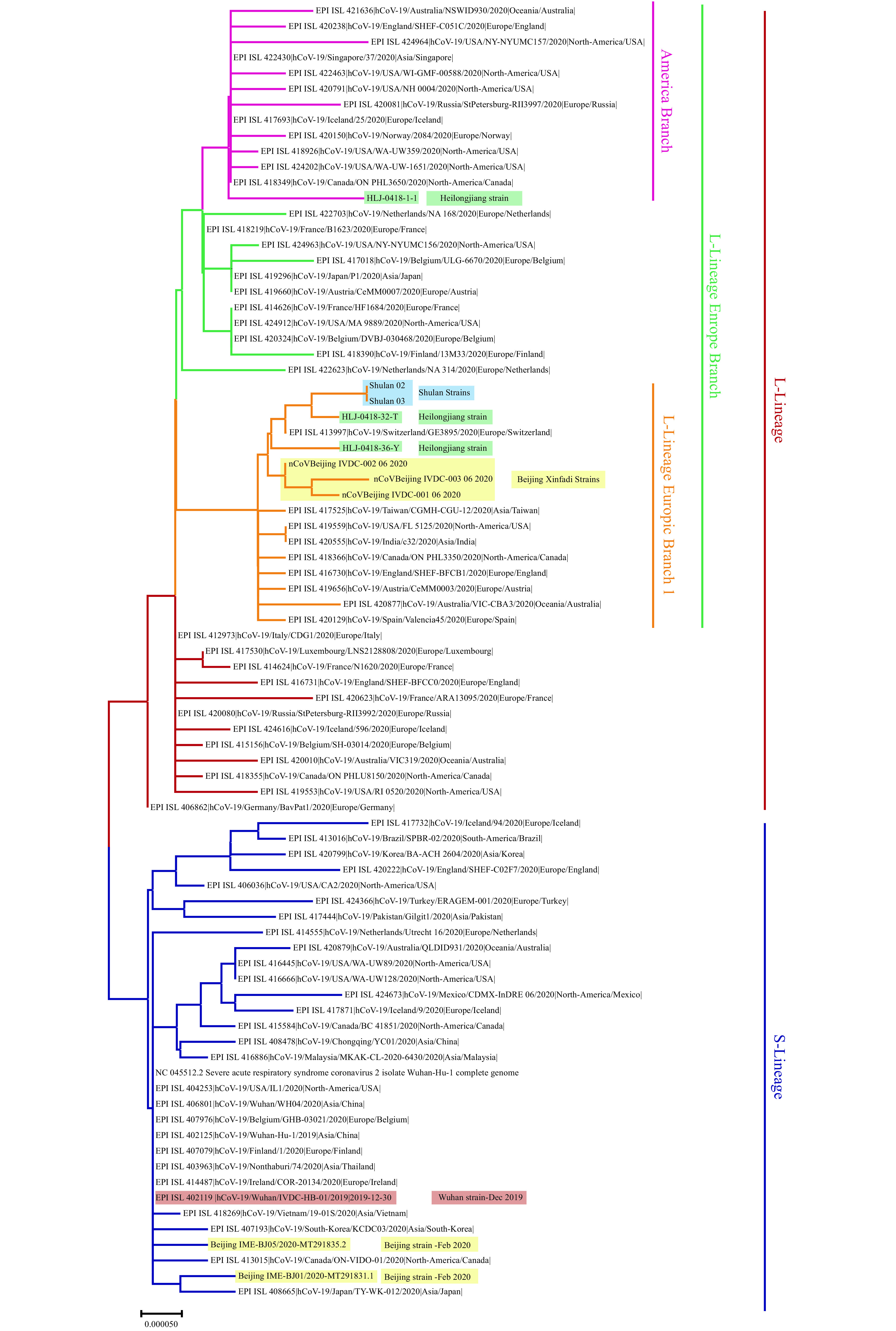
Phylogenetic tree based on the genome sequences of the COVID-19 virus. The genome of the COVID-19 virus from Beijing Municipality and Wuhan City were highlighted in shades of yellow and orange, respectively. The recent reemergence of COVID-19 virus in northeastern China (Shulan and Heilongjiang) that was associated with imported cases was also highlighted in shades of blue and green, respectively. S- or L-lineage of the COVID-19 virus were marked and colored on the right.
Among them, C6026T (P1291S), C14408T (P4715L), A23403G (D614G), and the GGG mutated to AAC at position 28881–28883 (R203K, R203R, G204R), caused amino acid substitutions that were located on ORF 1ab (6026, 14408), S protein (23403), and N protein (28881–28883), respectively. Some amino acid mutations (ORF1ab: P1291S, P4715L; S: D614G; N: R203K, G204R) in the COVID-19 virus may change its biological characteristics significantly. For example, the D614G mutation of the COVID-19 S protein caused by A23403G may increase the ability of the virus to infect cells (3).
The genome sequences of these Xinfadi strains provide strong evidence that the reemergent of COVID-19 cases in Beijing may be caused by an imported source of infections, suggesting that it is not continuous transmission of the local COVID-19 virus strain. It is worth noting that these Xinfadi strains have high homology with several genomes from the recently imported COVID-19 cases in northeastern China (Shulan strain, Heilongjiang strains) (4–5) that also belong to Branch 1 of L-lineage from Europe (Figure 1). Furthermore, the whole genome sequence analysis of the COVID-19 virus further confirmed that the source of the virus in this outbreak did not spread from natural animal hosts or intermediate animal hosts to humans.
COVID-19 cases are associated with the Xinfadi Wholesale Market, the virus could be transmitted from person to person via the respiratory tract or direct contact of the contaminated environment at market. The source of the virus needed to be further determined based on epidemiological, serological, and etiological monitoring.
Funding Statement
This work was supported by the National Key Research and Development Program of China (2016YFD0500301, 2020YFC0840900).
Contributor Information
Wenjie Tan, Email: tanwj@ivdc.chinacdc.cn.
Chun Huang, Email: huangch@bjcdc.org.
Wenbo Xu, Email: xuwb@ivdc.chinacdc.cn.
Guizhen Wu, Email: wugz@ivdc.chinacdc.cn.
References
- 1.Lu RJ, Zhao X, Li J, Niu PH, Yang B, Wu HL, et al Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–74. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang XL, Wu CC, Li X, Song YH, Yao XM, Wu XK, et al On the origin and continuing evolution of SARS-CoV-2. Nat Sci Rev. 2020;7(6):1012–23. doi: 10.1093/nsr/nwaa036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daniloski Z, Guo XY, Sanjana NE The D614G mutation in SARS-CoV-2 Spike increases transduction of multiple human cell types. bioRxiv. 2020 doi: 10.1101/2020.06.14.151357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Zhang Y, Zhao X, Wang DY, Dai WP, Jiao GY, et al. A Reemergent Case of COVID-19 — Harbin City, Heilongjiang Province, China, April 9, 2020. China CDC Weekly, 2020, 2(25):460 − 2. http://weekly.chinacdc.cn/en/article/doi/10.46234/ccdcw2020.127.
- 5.Chen C, Zhao X, Wang DY, Li J, Wang A, Wu DL, et al. The Initial Case of COVID-19 — Shulan City, Jilin Province, China, May 8, 2020. China CDC Weekly, 2020, 2(25):458 − 9. http://weekly.chinacdc.cn/en/article/doi/10.46234/ccdcw2020.115.


