Abstract
Fanconi anemia (FA) is an autosomal recessive cancer susceptibility syndrome with at least eight complementation groups (A to H). Three FA genes, corresponding to complementation groups A, C, and G, have been cloned, but their cellular function remains unknown. We have previously demonstrated that the FANCA and FANCC proteins interact and form a nuclear complex in normal cells, suggesting that the proteins cooperate in a nuclear function. In this report, we demonstrate that the recently cloned FANCG/XRCC9 protein is required for binding of the FANCA and FANCC proteins. Moreover, the FANCG protein is a component of a nuclear protein complex containing FANCA and FANCC. The amino-terminal region of the FANCA protein is required for FANCG binding, FANCC binding, nuclear localization, and functional activity of the complex. Our results demonstrate that the three cloned FA proteins cooperate in a large multisubunit complex. Disruption of this complex results in the specific cellular and clinical phenotype common to most FA complementation groups.
Fanconi anemia (FA) is an autosomal recessive disease characterized by developmental abnormalities, chromosomal instability, cancer susceptibility, and progressive bone marrow failure (1, 6, 23). Somatic cell fusion studies have defined at least eight genetic complementation groups (FA-A through FA-H) (13, 14, 37). The genes corresponding to groups A, C, and G have been cloned (7, 9, 26, 38), and mutations in these genes account for greater than 80% of FA patients (3, 13). FANCA, FANCC, and FANCG have no sequence similarity to each other or to other proteins in the GenBank database, and their biochemical functions remain unknown.
We have recently determined that the FANCA and FANCC proteins interact and form a cytoplasmic and nuclear complex in normal cells (21, 29). FANCA and FANCC fail to bind in vitro (18), suggesting that their interaction requires additional adapter proteins or posttranslational modifications. The FANCA/FANCC protein complex is not detected in cell lines derived from FA patients in groups A, B, C, E, F, G, and H, suggesting that the products of other FA genes are required for its assembly (44). Consistent with this model of regulated assembly, phosphorylation of the FANCA protein correlates with FANCC binding (44). While other proteins have been shown to bind to FANCC (17, 20), the physiological relevance of these binding interactions remains unknown.
The relationship between the newly cloned FANCG/XRCC9 protein and the FANCA/FANCC complex remains unknown. The human XRCC9 cDNA was originally expression cloned by its ability to partially complement the mitomycin C (MMC) sensitivity of the Chinese hamster ovary (CHO) mutant UV40 cell line (25). The cDNA was independently cloned by its ability to complement the MMC sensitivity of an FA-G patient cell line (7). Mutations were found in the human FANCG/XRCC9 gene in cell lines derived from FA-G patients (7). Little is known about the function or expression patterns of the FANCG/XRCC9 protein.
Based on the cellular abnormalities observed in FA, the FA proteins may regulate any one of several biochemical mechanisms. FA cells are hypersensitive to cross-linking agents, such as diepoxybutane and MMC, suggesting a defect in DNA repair. FA cells also exhibit abnormal cell cycle progression (15, 20) and reduced cell survival (5, 28, 33, 34, 41). FA cells are hypersensitive to reactive oxygen radicals as well (12), suggesting a defect in the removal or repair of oxygen-mediated cellular damage.
The selective sensitivity of FA cells to DNA cross-linking agents suggests a specific defect in interstrand DNA cross-link repair. Little is known about the mechanism of DNA cross-link repair in mammalian cells. Studies with Saccharomyces cerevisiae (11, 27) have demonstrated that cross-link repair requires the generation and repair of double-strand breaks (DSBs). In mammalian cells, DSB repair is performed by at least two primary, nonoverlapping mechanisms, homologous recombination (HR) and nonhomologous end joining (NHEJ) (reviewed in reference 4). HR is executed by a family of RAD51 proteins (2, 24). Nonhomologous recombination is performed by a discrete set of proteins, including the Ku, DNA-PK, XRCC4, and DNA ligase IV proteins (reviewed in reference 40). Interestingly, recent studies have demonstrated that FA cells are defective in the fidelity of rejoining of specific DSBs (8, 36), suggesting that the FA proteins interact with the HR or NHEJ pathway.
In the current study, we demonstrated that the FANCG protein is required for the interaction of the FANCA and FANCC proteins. Furthermore, we demonstrated that the FANCG protein is a component of a large nuclear protein complex containing FANCA, FANCG, and FANCC. Increasing evidence suggests that this protein complex plays a direct or indirect role in interstrand DNA cross-link repair and in the maintenance of normal chromosome stability.
MATERIALS AND METHODS
Cell culture.
Epstein-Barr virus-transformed lymphoblasts were maintained in RPMI medium supplemented with 15% heat-inactivated fetal calf serum and grown in a humidified 5% CO2-containing atmosphere at 37°C. FA-G lymphoblast lines EUFA316 and EUFA143 were provided by Hans Joenje (7, 13). Simian virus (SV40)-transformed GM6914 FA-A fibroblasts, expressing various mutant forms of the FANCA polypeptide, have previously been described (29). GM0637 cells are SV40-transformed fibroblasts from a normal adult control.
Retroviral infection of FA cell lines.
The indicated pMMP constructs were transfected by lipofection into 293 producer cells (human embryonic kidney cells) expressing the vesicular stomatitis virus G envelope protein (30). Retroviral supernatants were collected on day 5 following lipofection and contained 4.6 × 106 infectious units/ml, as estimated by Southern blot analysis of infected NIH 3T3 cells (data not shown).
FA lymphoblasts were infected with the various pMMP supernatants by a 4-h incubation in the presence of 8 μg of Polybrene per ml (32). Infected cells were washed free of viral supernatant and resuspended in growth medium. After 48 h, cells were transferred to medium containing puromycin (1 μg/ml). Dead cells were removed over a Ficoll cushion after 5 days, and surviving cells were grown under continuous selection in puromycin.
MMC sensitivity assays.
MMC sensitivity assays for lymphoblasts were performed as previously described (43).
Immunoprecipitation and immunoblotting.
Whole-cell extracts were prepared in lysis buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 1% [vol/vol] Triton X-100) supplemented with protease inhibitors (1 μg each of leupeptin and pepstatin per ml, 2 μg of aprotinin per ml, and 1 mM phenylmethylsulfonyl fluoride) and phosphatase inhibitors (1 mM sodium orthovanadate and 10 mM sodium fluoride). Immunoprecipitation was performed essentially as previously described (21), except that the protein A Sepharose-bound immune complexes were washed with lysis buffer. Immunoblotting was done as previously reported by using anti-FANCA, anti-FANCC, or anti-FANCG antiserum or with anti-β-tubulin antibody (Boehringer Mannheim Biochemicals, Indianapolis, Ind.) or anti-human topoisomerase II antibody (Calbiochem-Novabiochem Corporation, La Jolla, Calif.).
Cell fractionation.
Cell fractions were obtained by hypotonic swelling followed by Dounce homogenization as previously described (21). Prior to immunoprecipitation, both nuclear and cytoplasmic fractions were adjusted to 150 mM NaCl and 1% Triton X-100.
In vitro translation.
For in vitro translation of FANCA, FANCC, and FANCG, the TNT T7 Coupled Reticulocyte System (Promega, Madison, Wis.) was used in accordance with the manufacturer’s instructions. Proteins were independently synthesized in the presence of [35S]methionine, subsequently mixed (where indicated), incubated at 30°C for 30 min, and diluted in lysis buffer prior to immunoprecipitation.
Generation of human FANCG cDNA constructs.
Total RNA (2 μg) prepared from MG63 cells was used for random-primed reverse transcription (RT) in a 50-μl reaction mixture. Four microliters of the RT reaction mixture was subjected to PCR using primers FANCG (5′ primer) (5′GCCGCggatccATGTCCCGCCAGACCACCTCTG3′) and FANCG (3′ primer) (5′GCCGCgaattcCTACAGGTCACAAGACTTTGGC3′). The resulting PCR product of 1,866 bp, encoding the full-length FANCG polypeptide, was subcloned into the NcoI and BglII sites of retroviral vector pMMP. The pMMP vector (32) was modified by ligation with a puromycin resistance cDNA cassette.
Generation of anti-FANCG sera.
The affinity-purified antiantisera specific for FANCA and FANCC have been previously described (43) (21). Rabbit polyclonal antisera against FANCG were generated by using glutathione S-transferase (GST)-FANCG fusion proteins as antigen sources. Initially, we used RT-PCR and the full-length FANCG open reading frame cDNA as the template in order to amplify a cDNA fragment corresponding to the carboxyl-terminal region of the FANCG protein. A 3′ fragment was amplified by using primers 5′GCCGCagatctAGGCTCTATCAGCAACTGGGG3′ and FANCG (3′ primer). The resulting PCR product of 987 bp, encoding the carboxyl-terminal 329 amino acids of the FANCG polypeptide, was digested with BglII/EcoRI and subcloned into the BamHI/EcoRI sites of plasmid pGEX2T (Pharmacia). A GST-FANCG (C-terminal) fusion protein of the expected size (66 kDa) was expressed in Escherichia coli BL21, purified over glutathione-Sepharose, and used to immunize New Zealand White rabbits. FANCG-specific immune antisera were affinity purified over AminoLink Plus columns (Pierce) loaded with the GST-FANCG (C-terminal) fusion protein.
Immunofluorescence microscopy.
Immunofluorescence microscopy of human fibroblasts was performed as previously described (29).
RESULTS
Expression of FANCG restores the binding of FANCA and FANCC.
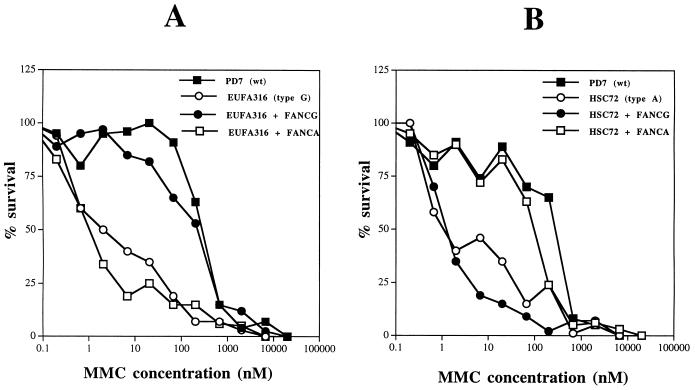
Initially, we isolated the FANCG cDNA by RT-PCR and tested its function in FA lymphoblast lines (Fig. 1). Consistent with previous studies (7), retroviral infection with pMMP-FANCG specifically corrected the MMC sensitivity of FA-G cells (Fig. 1A) but failed to correct the MMC sensitivity of FA-A cells (Fig. 1B).
FIG. 1.
Characterization of the MMC sensitivities of FA-A and FA-G lymphoblast lines. The full-length FANCG cDNA (7, 25) was isolated by RT-PCR of RNA from a human MG63 tumor cell line and subcloned into the murine retroviral vector pMMP(puro) (19). The indicated retroviral supernatants were generated and used to infect FA lymphoblast lines, and puromycin-resistant cells were selected. The cells analyzed included EUFA316 (FA-G) cells, HSC72 (FA-A) cells, and normal (PD7) cells. Consistent with previous studies (7), pMMP-FANCG infection also resulted in functional complementation (correction of MMC sensitivity) of another FA-G cell line, EUFA143 (data not shown). WT, wild type.
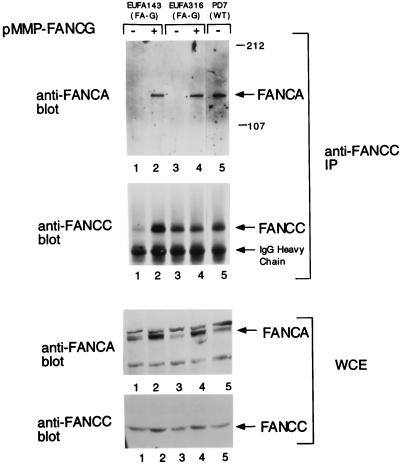
We have previously shown that the FANCA and FANCC proteins bind in normal lymphoblasts but fail to bind in FA-G lymphoblasts (44). We next examined the effect of FANCG expression on the interaction of the FANCA and FANCC proteins in two FA-G cell lines (Fig. 2). For uncorrected FA-G cell lines (lanes 1 and 3), comparatively low levels of the FANCA and FANCC proteins were observed (whole-cell extract immunoblots) and FANCA did not coimmunoprecipitate with FANCC. Interestingly, for FA-G cell lines corrected with pMMP-FANCG (lanes 2 and 4), expression of FANCC, and particularly of FANCA, was increased and FANCA/FANCC binding was restored.
FIG. 2.
Expression of FANCG in FA-G lymphoblasts restores the FANCA/FANCC interaction. Whole-cell extracts (WCE) were generated from lymphoblast lines, including EUFA316, EUFA316-FANCG, EUFA143, EUFA143-FANCG, and PD7 (normal adult control). These protein extracts (100 μg) were probed directly by immunoblotting with either anti-FANCA or anti-FANCC serum. The FANCA protein is indicated by an arrow, and additional bands in the anti-FANCA immunoblot are nonspecific. Alternatively, the same amount of protein from each extract (2 mg) was used for immunoprecipitation with affinity-purified anti-FANCC serum as indicated. Immune complexes were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with anti-FANCA or anti-FANCC serum. WT, wild type; IP, immunoprecipitation. The values to the right of the gel are molecular sizes in kilodaltons.
FANCG is a component of the FANCA/FANCC protein complex.
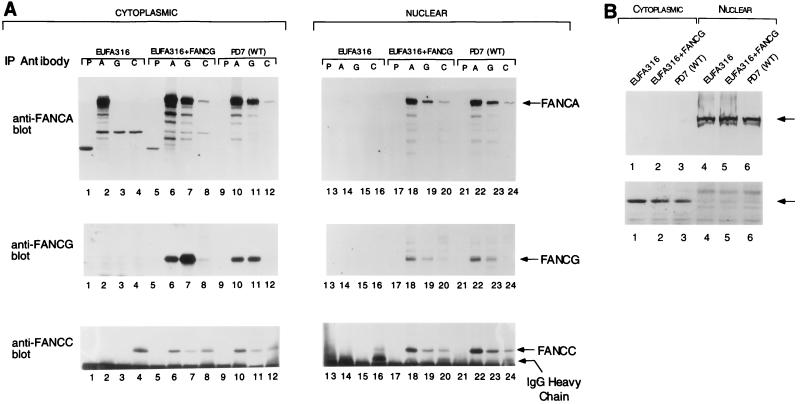
The FANCG protein may restore FANCA/FANCC binding either through a direct physical interaction with the protein complex or through an indirect interaction. To test for a direct physical interaction, we examined the FANCA/FANCC protein complex for the presence of FANCG protein (Fig. 3). For this purpose, we generated an affinity-purified anti-FANCG serum specific for the carboxyl-terminal region of FANCG. For FA-G lymphoblasts complemented with FANCG and for normal lymphoblasts, immunoprecipitation with an anti-FANCA serum resulted in coimmunoprecipitation of the FANCA (163 kDa), FANCG (65 kDa), and FANCC (60 kDa) proteins (Fig. 3A, lanes 6, 10, 18, and 22). Likewise, immunoprecipitation with an anti-FANCG serum (lanes 7, 11, 19, and 23) or an anti-FANCC serum (lanes 8, 12, 20, and 24) resulted in the coimmunoprecipitation of the three proteins. Immunoprecipitation of the complex was less efficient through FANCC, perhaps indicating that the C-terminal epitope of the FANCC protein is concealed in the multimeric complex and less accessible to the antibody.
FIG. 3.
FANCG is a component of the cytoplasmic and nuclear FANCA/FANCC protein complex. (A) The indicated lymphoblast lines, including EUFA316 (FA-G cells), EUFA316 corrected with the FANCG cDNA (FA-G + FANCG), or PD7 (normal adult control) were lysed and fractionated into cytoplasmic and nuclear fractions. Proteins from each fraction were immunoprecipitated with control nonimmune (P), anti-FANCA (A), anti-FANCG (G), or anti-FANCC (C) serum. Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with anti-FANCA (upper panel), anti-FANCG (middle panel) or anti-FANCC (lower panel) serum. To demonstrate that the absence of the FANCA/FANCC interaction in FA-G cells is not simply due to the low expression level of FANCA and FANCC in these cells, three times more cytoplasmic extract (6 mg in 1 ml) was used for EUFA316 (lanes 1 to 4) than for the other two cell lines (lanes 5 to 12). (B) To ensure effective fractionation, samples from the indicated lymphoblast lines described in panel A were analyzed for topoisomerase levels (upper panel) and β-tubulin levels (lower panel). WT, wild type; IgG, immunoglobulin G; IP, immunoprecipitating.
The multisubunit complex of the FANCA, FANCG, and FANCC proteins was observed in cellular fractions derived from the cytoplasm and the nucleus of the respective cell lines (compare lanes 1 to 12 to lanes 13 to 24). Efficient cellular fractionation was confirmed by analyzing the marker proteins topoisomerase II and β-tubulin (Fig. 3B). Consistent with the results in Fig. 2, FANCA failed to coimmunoprecipitate with FANCC or FANCG in the mutant FA-G cell line (Fig. 3A, lane 2). Interestingly, FANCA was only weakly detectable in the nuclear fraction of these cells compared with their corrected counterpart (compare lanes 14 and 18), suggesting that correction with FANCG not only restores FANCA/FANCC binding but also enhances nuclear accumulation of the complex. Whether this observation reflects a real deficiency in the nuclear translocation process or is a direct consequence of the low expression level of FANCA and FANCC in mutant FA-G cells remains to be determined. Taken together, these results demonstrate that the FANCG protein is bound in a protein complex with FANCA and FANCC; absence of the FANCG protein results in absence of FANCA/FANCC binding and appears to decrease FANCA/FANCC nuclear accumulation.
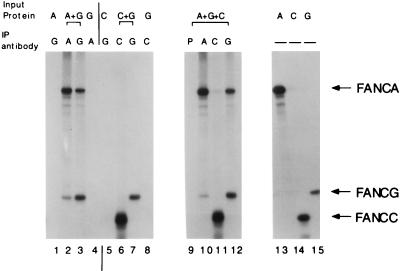
The interaction of FANCA, FANCG, and FANCC was also analyzed by using in vitro-translated proteins (Fig. 4). In vitro-translated FANCA and FANCG proteins coimmunoprecipitated with either anti-FANCA serum (lane 2) or anti-FANCG serum (lane 3). In contrast, in vitro-translated FANCC protein did not coimmunoprecipitate with the FANCA or FANCG protein (lanes 6 and 11), suggesting that the binding of FANCC to the FANCA/FANCG complex is weaker or is regulated by other adapter proteins or posttranslational modifications which are not present in the cell-free in vitro translation mixture. Accordingly, the coimmunoprecipitation of FANCA and FANCG from whole-cell extracts was more efficient than the coimmunoprecipitation of FANCA and FANCC or FANCG and FANCC (Fig. 3).
FIG. 4.
Binding of FANCA and FANCG in vitro. In vitro-translated, [35S]methionine-labeled FANCA (A), FANCG (G), and FANCC (C) were prepared in separate reactions and mixed as indicated (Input Protein). Immunoprecipitation was done with either preimmune (P), anti-FANCA (A), anti-FANCG (G), or anti-FANCC (C) serum. Total reticulocyte lysate (without immunoprecipitation) containing labeled FANCA, FANCG, and FANCC was loaded in lanes 13, 14, and 15, respectively. IP, immunoprecipitating.
The amino-terminal region of FANCA is required for FANCG binding, FANCC binding, and functional activity of the complex.
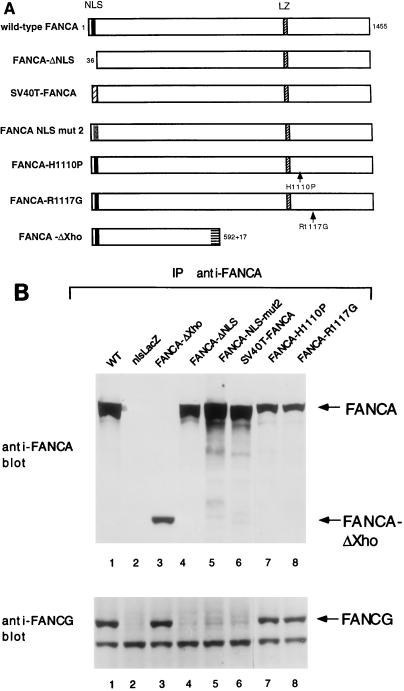
We next determined the region of the FANCA protein required for interaction with FANCG (Fig. 5). By using a mutant series of FANCA polypeptides (Fig. 5A), we have recently determined that the amino-terminal region of FANCA is required for nuclear localization, FANCC binding, and functional activity (29). In the current study, we examined FA-A fibroblasts (GM6914 cells) expressing these mutant FANCA polypeptides (Fig. 5B). Immunoprecipitation of either wild-type FANCA or mutant forms of FANCA containing the intact amino-terminal nuclear localization signal (NLS) sequence resulted in coimmunoprecipitation of FANCG (Fig. 5B, lanes 1, 3, 7, and 8). In contrast, mutant forms of the FANCA protein which lack the N-terminal NLS region of FANCA failed to bind to FANCG (Fig. 5B, lanes 4 to 6), failed to translocate to the cell nucleus and failed to complement MMC sensitivity (Table 1). Taken together, these data demonstrate that the amino-terminal NLS region of the FANCA protein is required for FANCG binding, FANCC binding, nuclear localization, and functional activity. Interestingly, nuclear localization is necessary but not sufficient for functional activity. For instance, the SV40 FANCA protein accumulates in the nucleus but fails to bind FANCG and FANCC and fails to function (Table 1). Whether the amino-terminal NLS region of FANCA directly binds to FANCG or requires additional adapter proteins or posttranslational modifications (indirect binding) remains to be determined.
FIG. 5.
The amino-terminal NLS region of FANCA is required for FANCG binding and functional activity. (A) Schematic representation of wild-type (WT) FANCA and mutant proteins. The wild-type FANCA protein is 1,455 amino acids long and contains a bipartite NLS and a partial leucine zipper (LZ) motif. The FANCA-ΔNLS mutant protein is missing the amino-terminal 35 amino acids. The FANCA-NLS-mut2 protein has all of the basic amino acids of the NLS mutated. The SV40T-FANCA protein contains the 13-amino-acid NLS of the SV40 T antigen in place of the NLS of FANCA. The FANCA(H1110P) and FANCA(R1117G) mutations are based on FANCA mutational screens (22). cDNAs encoding these mutant FANCA proteins were generated and expressed in the FA-A fibroblast line GM6914 as previously described (29). (B) Cell lysates were prepared from GM6914 FA-A fibroblasts expressing the indicated FANCA mutant proteins. The FANCA proteins were immunoprecipitated with an anti-FANCA serum (anti-carboxy-terminal antibody), and the immune complexes were analyzed by anti-FANCA and anti-FANCG immunoblotting. As a negative control, GM6914 cells expressing nlsLacZ (no FANCA protein) were analyzed (lane 2). For the FANCA-ΔXho mutant protein, immunoprecipitation was done with the anti-FANCA (anti-amino-terminal) antibody as previously described (29). IP, immunoprecipitation.
TABLE 1.
Functional analysis of FANCA mutant proteins
| Protein | FA-A cell complementation | FANCC binding | Nuclear localization | FANCG binding |
|---|---|---|---|---|
| Wild-type FANCA | + | + | + | + |
| FANCA-ΔNLS | − | − | − | − |
| SV40T-FANCA | − | − | + | − |
| FANCA-NLS-mut2 | − | − | − | − |
| FANCA-ΔXho | − | − | − | + |
| FANCA(H1110P) | − | − | − | + |
| FANCA(R1117G) | − | − | − | + |
Analysis of the FANCA/FANCG/FANCC complex in other FA complementation groups.
The interaction of the FANCA, FANCG, and FANCC proteins was next examined in various lymphoblast lines derived from normal controls or FA patients (Fig. 6). The FANCA/FANCG/FANCC complex was detected in normal lymphoblasts, in FANCA-corrected FA-A cells, and in FA-D cells (see the anti-FANCC immunoblot, lanes 2, 6, and 16). FANCA/FANCG binding without a FANCC interaction was observed in cells derived from multiple FA complementation groups, including groups A, B, C, E, F, and H (see the anti-FANCG immunoblot, lanes 8, 10, 12, 14, 16, 18, 20, and 24). These results further suggest that the interaction between the FANCA and FANCG proteins is a constitutive (and perhaps direct) interaction which is not regulated by FANCC or by the products of other FA genes. In contrast, the binding of FANCC appears to require FANCA/FANCG binding and the products of other FA genes, as previously described (44).
FIG. 6.
Analysis of the FANCA/FANCG/FANCC protein complex in cell lines from multiple FA complementation groups. (A) Whole-cell extracts were prepared from the indicated lymphoblast lines, and proteins were immunoprecipitated with either preimmune (P) or anti-FANCA (A) serum. Immune complexes were immunoblotted with antiserum to FANCA, FANCG, or FANCC, as indicated. The cell lines analyzed included normal control PD7 cells (lanes 1 and 2) and HSC72 (lanes 3 and 4), HSC72–FA-A (lanes 5 and 6), BD32 (lanes 7 and 8), HSC230 (lanes 9 and 10), PD4 (lanes 11 and 12), HSC536 (lanes 13 and 14), PD20L (lanes 15 and 16), EUFA130 (lanes 17 and 18), EUFA121 (lanes 19 and 20), EUFA316 (lanes 21 and 22), and EUFA173 (lanes 23 and 24) cells. The BD32 (FA-A) cell line expressed a mutant FANCA polypeptide [FANCA(H1110P)]. The HSC536 (FA-C) cell line expressed a mutant FANCC polypeptide [FANCC(L554P)]. All of the cell lines examined expressed comparatively equal levels of FANCC protein as judged by whole-cell lysate immunoblotting (data not shown), except PD4 cells, which expressed no FANCC protein. IP, immunoprecipitating; IgG, immunoglobulin G. WT, wild type.
One FA-A cell line (BD-32) (lanes 7 and 8) expressed a mutant, nonfunctional form of the FANCA protein with a histidine residue replaced with a proline at amino acid 1110 [FANCA(H1110P)] (19). This mutant form of the FANCA protein still coimmunoprecipitated with FANCG (lane 8), although FANCC was not observed in the complex. These results suggest that FANCG binding is necessary but not sufficient for FANCA activity; FANCC binding is also required for functional activity of the complex. Interestingly, FANCA and FANCG levels appeared to be decreased in cells derived from FA groups A, B, E, F, and H (lanes 8, 10, 18, 20, and 24), suggesting that other FA genes may regulate FANCA and FANCG protein expression or stability.
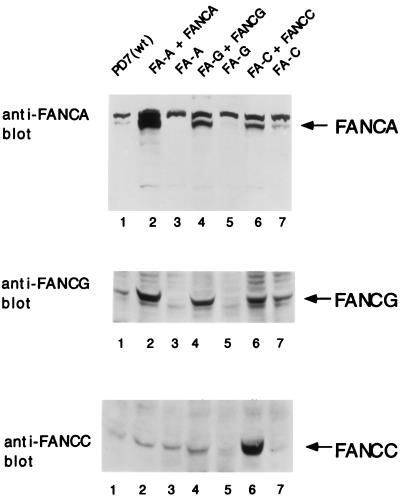
The FANCA, FANCG, and FANCC proteins stabilize the FA protein complex.
In an attempt to establish the effects of individual protein components (FANCA, FANCG, and FANCC) on the stability and expression level of the FA protein complex, we analyzed isogenic pairs of mutant and corrected FA cell lines by whole-cell lysate immunoblots (Fig. 7). For FA-A cells, correction with FANCA resulted in an increase in the expression level of the FANCC and FANCG proteins (lanes 2 and 3). For FA-G cells, correction with FANCG resulted in an increase in the expression level of the FANCA and FANCC proteins (lanes 4 and 5). For FA-C cells, correction with FANCC resulted in an increase in the expression level of the FANCA and FANCG proteins (lanes 6 and 7). Taken together, these results further suggest that the three FA proteins FANCA, FANCG, and FANCC bind in a complex and that the stability of the complex is enhanced by each component.
FIG. 7.
The FANCA, FANCG, and FANCC proteins stabilize the FA protein complex. Whole-cell extracts from the indicated lymphoblast lines were prepared, and the same amount of protein from each was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and immunoblotted with affinity-purified antiserum against the FANCA, FANCC, or FANCG protein, as indicated. WT, wild type.
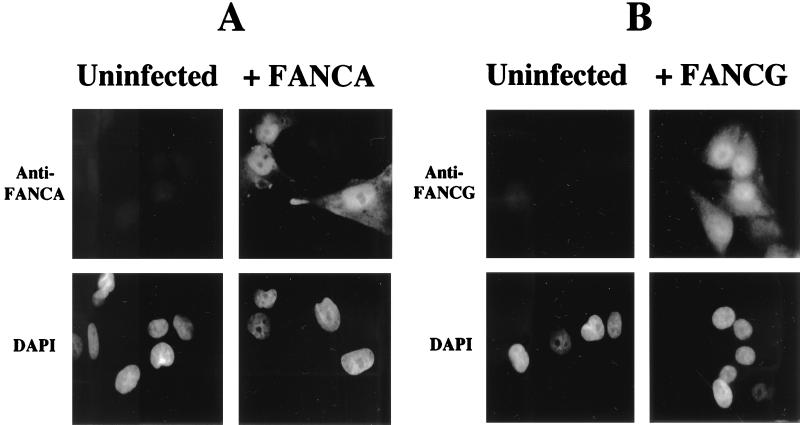
Nuclear localization of the FANCA and FANCG proteins.
To further confirm the cellular localization of the FANCA and FANCG proteins, we performed immunofluorescence on an SV40-transformed normal human fibroblast line (Fig. 8). The FANCA and FANCG proteins were expressed primarily in the nuclei of retrovirus-infected cells, consistent with previous studies (29) and consistent with the functional interaction of the proteins. Some cytoplasmic expression of the FANCA and FANCG proteins was also observed.
FIG. 8.
Nuclear localization of the FANCA and FANCG polypeptides. The human fibroblast line GM0637, derived from a normal (wild-type) control, was infected with either pMMP-FANCA (wild type) (A) or pMMP-FANCG (wild type) (B) as indicated. Pools of infected cells were stained with anti-FANCA or anti-FANCG serum or stained with the DNA-specific dye 4′,6-diamidino-2-phenylindole (DAPI) and analyzed by immunofluorescence assay as previously described (29).
DISCUSSION
Our results demonstrate that the three cloned FA genes encode proteins which interact in a multisubunit complex. The complex was detected in both the cytoplasm and the nucleus, consistent with previous studies which localized FANCC to both cellular compartments (10, 21, 43, 45). Accumulation of the nuclear complex is required for the function of the complex (29). Mutation of any of the three FA genes leads to disruption of the protein complex, resulting in the conserved FA cellular and clinical phenotype.
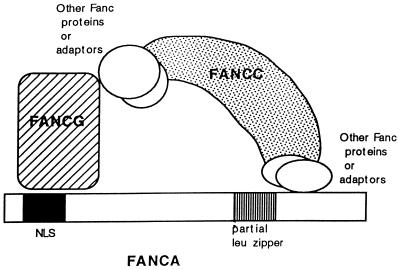
The structural features of the various protein-protein contacts within the multisubunit complex are not fully understood. Based on an analysis of truncated and point mutant forms of the FANCA protein, the amino-terminal NLS of FANCA is required for FANCG binding and FANCC binding (29) (Fig. 9). Based on the FANCA-ΔXho mutant protein, the amino terminus of FANCA is also sufficient for the binding interaction with FANCG. The carboxy-terminal leucine zipper region of FANCA is deleted in the FANCA-ΔXho mutant protein and is therefore not required for FANCG binding. Sequences at the carboxy terminus of FANCA, perhaps including the leucine zipper, are required for FANCC binding. For example, in the patient-derived point mutant proteins FANCA(H1110P) and FANCA(R1117G) (19), the mutations near the leucine zipper region of FANCA disrupt FANCC binding, nuclear localization, and function. Additional posttranslational modifications of FANCA or other adapter proteins may also be required to enable the interaction of FANCC with the complex (44).
FIG. 9.
Schematic model of molecular interactions of the FANCA, FANCG, and FANCC proteins. Based on available data, at least two regions of the FANCA polypeptide are required for the FANCA-FANCG-FANCC interaction. First, the amino-terminal region of the FANCA protein, which includes the bipartite NLS region, is required for interaction with the FANCG protein. Second, a region near the carboxy terminus of FANCA is required for the FANCC interaction. Patient-derived point mutations in this region disrupt the FANCC interaction. The FANCC interaction is a weak or regulated interaction, requiring FANCA/FANCG binding and the products of other FA genes (44). The region of FANCG required for FANCA binding is not known. The region of FANCC required for FANCA binding may be the carboxy terminus of FANCC. This region is highly conserved among human, murine, and bovine FANCC proteins. Also, an FA-C patient-derived point mutation [FANCC(L554P)] ablates FANCA binding (21).
The detailed stoichiometry of the FANCA-FANCG-FANCC interaction is not clear. It is apparent that anti-FANCA antibodies are as efficient as anti-FANCG antibodies in immunoprecipitating FANCA and FANCG. FANCA/FANCG binding is even observed for in vitro-translated proteins, suggesting that the interaction is direct. In contrast, the anti-FANCC antibody is less efficient in immunoprecipitating FANCA and FANCG. These results suggest that there is a large pool of cellular FANCC uncomplexed with FANCA and FANCG. Whether the uncomplexed FANCC protein has a discrete cellular or biochemical function, compared to the complexed form, remains unknown.
There are several possible explanations for the substoichiometric interaction of FANCC with the FANCA/FANCG complex. First, binding of the FANCC protein may be regulated. Consistent with this hypothesis, FANCC binding correlates with the phosphorylation of the FANCA protein (44). The phosphorylation sites of FANCA and the relevant cellular kinase(s) remain unknown. Second, our current immunoprecipitation strategy may be efficient at preserving the FANCA/FANCG interaction but inefficient at preserving the FANCC interaction. Third, the epitope of the FANCC protein recognized by our anti-FANCC antibody may be poorly accessible when FANCC is complexed with FANCA/FANCG.
Analysis of other multisubunit complexes, such as the nucleotide excision repair complex (42) and the RNA polymerase II holoenzyme (16), also reveals some subunits with strong, constitutive interactions and other subunits with weak or regulated interactions. For instance, the ERCC2, ERCC3, and p62 proteins interact within the RNA polymerase transcription factor complex TFIIH but these interactions are dependent on the salt concentration (35).
Our results demonstrate that expression of FANCG not only restores the binding of FANCA and FANCC (Fig. 2) but also increases the expression of FANCA and FANCC (Fig. 2 and 7). Likewise, FANCC correction of an FA-C cell line increases the expression of FANCA and FANCG (Fig. 7), although FANCA/FANCG binding is observed even in the absence of FANCC.
Increased protein expression may result from increased synthesis or decreased turnover. We hypothesize that it probably results from the increased stability conferred by protein-protein interactions in the complex. To support this model, we have performed pulse-chase experiments to determine the half-life of newly synthesized FANCA protein (9a). In uncorrected FA-G lymphoblasts, the FANCA protein was unstable. Following correction of these cells by FANCG protein expression, the FANCA protein half-life was prolonged, suggesting that FANCG binding directly protects FANCA from degradation. Consistent with these results, the stabilization of binding members has been observed for other multisubunit complexes, such as the AP-1 complex and the excision repair complex (31, 39).
Cells from other FA complementation groups, including groups B, E, F, and H, have decreased levels of the FANCA, FANCC, and FANCG proteins. The protein products of the FANCB, FANCE, FANCF, and FANCH genes may therefore regulate the stability of the FA protein complex, perhaps through direct association with the complex. Based on sucrose gradient sedimentation (20a), the nuclear protein complex is large (>400 kDa), suggesting the presence of additional adapter proteins. Purification of the FA complex may therefore allow the identification of other FA gene products.
Finally, the biochemical function of the nuclear FA protein complex remains unknown. Since FA cells have an underlying defect in the fidelity of rejoining of specific DSBs (9, 36), the complex may represent a novel DSB repair pathway or may modulate a known DSB repair pathway, such as the HR pathway or the NHEJ pathway. Identification of additional proteins in the FA nuclear complex may elucidate its biochemical function.
ACKNOWLEDGMENTS
We thank C. Mathew for the BD32 (FA-A) cell line and H. Joenje for EUFA143 and EUFA316 cells.
This research was supported by NIH grants R01-HL5725 and PO1-CA39542. I.G.H. is supported by a fellowship from the Cancer Research Institute. A.D.D. is a Scholar of the Leukemia Society of America.
REFERENCES
- 1.Auerbach A D, Buchwald M, Joenje H. Fanconi anemia. In: Vogelstein B, Kinzler K W, editors. The genetic basis of human cancer. New York, N.Y: McGraw Hill; 1997. [Google Scholar]
- 2.Baumann P, West S C. Role of the human RAD51 protein in homologous recombination and double-stranded break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 3.Buchwald M. Complementation groups: one or more per gene. Nat Genet. 1995;11:228–230. doi: 10.1038/ng1195-228. [DOI] [PubMed] [Google Scholar]
- 4.Chu G. Double strand break repair. J Biol Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 5.Cumming R C, Liu J M, Youssoufian H, Buchwald M. Suppression of apoptosis in hematopoietic factor-dependent progenitor cell lines by expression of the FAC gene. Blood. 1996;88:4558–4567. [PubMed] [Google Scholar]
- 6.D’Andrea A D, Grompe M. Molecular biology of Fanconi anemia: implications for diagnosis and therapy. Blood. 1997;90:1725–1736. [PubMed] [Google Scholar]
- 7.de Winter J P, Waisfisz Q, Rooimans M A, van Berkel C G M, Bosnoyan-Collins L, Alon N, Carreau M, Bender O, Demuth I, Schindler D, Pronk J C, Arwert F, Hoehn H, Digweed M, Buchwald M, Joenje H. The Fanconi anaemia group G gene is identical with human XRCC9. Nat Genet. 1998;20:281–283. doi: 10.1038/3093. [DOI] [PubMed] [Google Scholar]
- 8.Escarceller M, Buchwald M, Singleton B K, Jeggo P A, Jackson S P, Moustacchi E, Papadopoulo D. Fanconi anemia C gene product plays a role in the fidelity of blunt DNA end-joining. J Mol Biol. 1998;279:375–385. doi: 10.1006/jmbi.1998.1784. [DOI] [PubMed] [Google Scholar]
- 9.The Fanconi Anemia/Breast Cancer Consortium. Positional cloning of the Fanconi anaemia group A gene. Nat Genet. 1996;14:324–328. doi: 10.1038/ng1196-324. [DOI] [PubMed] [Google Scholar]
- 9a.Garcia-Higuera, I. Unpublished data.
- 10.Hoatlin M E, Christianson T A, Keeble W W, Hammond A T, Zhi Y, Heinrich M C, Tower P A, Bagby G C., Jr The Fanconi anemia group C gene product is located in both the nucleus and cytoplasm of human cells. Blood. 1998;91:1418–1425. [PubMed] [Google Scholar]
- 11.Jachymczyk W J, von Borstel R C, Mowat M R, Hastings P J. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol Gen Genet. 1981;182:196–205. doi: 10.1007/BF00269658. [DOI] [PubMed] [Google Scholar]
- 12.Joenje H, Arwert F, Eriksson A W, de Koning H, Oostra A B. Oxygen-dependence of chromosomal aberrations in Fanconi’s anaemia. Nature. 1981;290:142. doi: 10.1038/290142a0. [DOI] [PubMed] [Google Scholar]
- 13.Joenje H, Oostra A B, Wijker M, di Summa F M, van Berkel C G M, Rooimans M A, Ebell W, van Weel M, Pronk J C, Buchwald M, Arwert F. Evidence for at least eight Fanconi anemia genes. Am J Hum Genet. 1997;61:940–944. doi: 10.1086/514881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joenje H, Ten F L, Oostra A, Berkel C V, Rooimans M, Schroeder S, Kurth T, Wegner R, Gille J, Buchwald M, Arwert F. Classification of Fanconi anemia patients by complementation analysis: evidence for a fifth genetic subtype. Blood. 1995;86:2156. [PubMed] [Google Scholar]
- 15.Kaiser T N, Lojewski A, Dougherty C, Juergens L, Sahar E, Latt S A. Flow cytometric characterization of the response of Fanconi’s anemia cells to mitomycin C treatment. Cytometry. 1982;2:291–297. doi: 10.1002/cyto.990020505. [DOI] [PubMed] [Google Scholar]
- 16.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 17.Kruyt F A E, Hoshino T, Liu J M, Joseph P, Jaiswal A K, Youssoufian H. Abnormal microsomal detoxification implicated in Fanconi anemia group C by interaction of the FAC protein with NADPH cytochrome P450 reductase. Blood. 1998;92:3050–3056. [PubMed] [Google Scholar]
- 18.Kruyt F A E, Youssoufian H. The Fanconi anemia proteins FAA and FAC function in different cellular compartments to protect against cross-linking agent cytotoxicity. Blood. 1998;92:2229–2236. [PubMed] [Google Scholar]
- 19.Kupfer G, Näf D, Garcia-Higuera I, Wasik J, Cheng A, Yamashita T, Tipping A, Morgan N, Mathew C G, D’Andrea A D. A patient-derived mutant form of the Fanconi anemia protein, FANCA, is defective in nuclear accumulation. Exp Hematol. 1999;27:587–593. doi: 10.1016/s0301-472x(99)00022-3. [DOI] [PubMed] [Google Scholar]
- 20.Kupfer G, Yamashita T, Naf D, Suliman A, Asano S, D’Andrea A D. The Fanconi anemia protein, FAC, binds to the cyclin-dependent kinase, cdc2. Blood. 1997;90:1047–1054. [PubMed] [Google Scholar]
- 20a.Kupfer, G. M., and A. D’Andrea. Unpublished data.
- 21.Kupfer G M, Naf D, Suliman A, Pulsipher M, D’Andrea A D. The Fanconi anemia proteins, FAA and FAC, interact to form a nuclear complex. Nat Genet. 1997;17:487–490. doi: 10.1038/ng1297-487. [DOI] [PubMed] [Google Scholar]
- 22.Levran O, Erlich T, Magdalena N, Gregory J J, Batish S D, Verlander P C, Auerbach A D. Sequence variation in the Fanconi anemia gene FAA. Proc Natl Acad Sci USA. 1997;94:13051–13056. doi: 10.1073/pnas.94.24.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Buchwald M, Walsh C E, Young N S. Fanconi anemia and novel strategies for therapy. Blood. 1994;84:3995–4007. [PubMed] [Google Scholar]
- 24.Liu N, Lamerdin J E, Tebbs R S, Schild D, Tucker J D, Shen M R, Brookman K W, Siciliano M J, Walter C A, Fan W, Narayana L S, Zhou Z-Q, Adamson A W, Sorensen K J, Chen D J, Jones N J, Thompson L H. XRCC2 and XRCC3, new human Rad51-family members, promote chromosome stability and protect against DNA cross-links and other damages. Mol Cell. 1998;1:783–793. doi: 10.1016/s1097-2765(00)80078-7. [DOI] [PubMed] [Google Scholar]
- 25.Liu N, Lamerdin J E, Tucker J D, Zhou Z-Q, Walter C A, Albala J S, Busch D B, Thompson L H. The human XRCC9 gene corrects chromosomal instability and mutagen sensitivities in CHO UV40 cells. Proc Natl Acad Sci USA. 1997;94:9232–9237. doi: 10.1073/pnas.94.17.9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lo Ten Foe J R, Rooimans M A, Bosnoyan-Collins L, et al. Expression cloning of a cDNA for the major Fanconi anemia gene, FAA. Nat Genet. 1996;14:320–323. doi: 10.1038/ng1196-320. [DOI] [PubMed] [Google Scholar]
- 27.Magana-Schwencke N, Henriques J A, Chanet R, Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc Natl Acad Sci USA. 1982;79:1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marathi U K, Howell S R, Ashmun R A, Brent T P. The Fanconi anemia complementation group C protein corrects DNA interstrand cross-link-specific apoptosis in HSC536N cells. Blood. 1996;88:2298–2305. [PubMed] [Google Scholar]
- 29.Näf D, Kupfer G M, Suliman A, Lambert K, D’Andrea A D. Functional activity of the Fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol Cell Biol. 1998;18:5952–5960. doi: 10.1128/mcb.18.10.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ory D, Neugeboren B, Mulligan R. A stable human-derived packaging cell line for production of high-titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–11406. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papavassiliou A G, Treeier M, Chavrier C, Bohmann D. Targeted degradation of c-Fos, but not v-Fos, by a phosphorylation-dependent signal on c-Jun. Science. 1992;258:1941–1944. doi: 10.1126/science.1470918. [DOI] [PubMed] [Google Scholar]
- 32.Pulsipher M, Kupfer G M, Naf D, Suliman A, Lee J-S, Jakobs P, Grompe M, Joenje H, Sieff C, Guinan E, Mulligan R, D’Andrea A D. Subtyping analysis of Fanconi anemia by immunoblotting and retroviral gene transfer. Mol Med. 1998;4:468–479. [PMC free article] [PubMed] [Google Scholar]
- 33.Rathbun R, Faulkner G, Ostroski M, Christianson T, Hughes G, Jones G, Cahn R, Maziarz R, Royle G, Keeble W, Heinrich M, Grompe M, Tower P, Bagby G. Inactivation of the Fanconi anemia group C gene augments interferon-gamma induced apoptotic responses in hematopoietic cells. Blood. 1997;90:974. [PubMed] [Google Scholar]
- 34.Ridet A, Guillouf C, Duchaud E, Cundari E, Fiore M, Moustacchi E, Rosselli F. Deregulated apoptosis is a hallmark of the Fanconi anemia syndrome. Cancer Res. 1997;57:1722–1730. [PubMed] [Google Scholar]
- 35.Schaeffer L, Moncollin V, Roy R, Staub A, Mezzina M, Sarasin A, Weeda G, Hoeijmakers J H, Egly J M. The ERCC2/DNA repair protein is associated with the class II BTF2/TFIIH transcription factor. EMBO J. 1994;13:2388–2392. doi: 10.1002/j.1460-2075.1994.tb06522.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith J, Andrau J C, Kallenbach S, Laquerbe A, Doyen N, Papadopoulo D. Abnormal rearrangements associated with V(D)J recombination in Fanconi anemia. J Mol Biol. 1998;281:815–825. doi: 10.1006/jmbi.1998.1971. [DOI] [PubMed] [Google Scholar]
- 37.Strathdee C A, Duncan A M V, Buchwald M. Evidence for at least four Fanconi anemia genes including FACC on chromosome 9. Nat Genet. 1992;1:196–198. doi: 10.1038/ng0692-196. [DOI] [PubMed] [Google Scholar]
- 38.Strathdee C A, Gavish H, Shannon W R, Buchwald M. Cloning of cDNAs for Fanconi’s anaemia by functional complementation. Nature. 1992;356:763–767. doi: 10.1038/356763a0. [DOI] [PubMed] [Google Scholar]
- 39.van Vuuren A J, Appeldoorn E, Odijk H, Yasui A, Jaspers N G J, Bootsma D, Hoeijmakers J H J. Evidence for a repair enzyme complex involving ERCC1 and complementing activities of ERCC4, ERCC11 and xeroderma pigmentosum group F. EMBO J. 1993;12:3693–3701. doi: 10.1002/j.1460-2075.1993.tb06044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver D T. What to do at an end: DNA double-strand-break repair. Trends Genet. 1995;11:388–392. doi: 10.1016/s0168-9525(00)89121-0. [DOI] [PubMed] [Google Scholar]
- 41.Whitney M A, Royle G, Low M J, Kelly M A, Axthelm M K, Reifsteck C, Olson S, Braun R E, Heinrich M C, Rathbun R K, Bagby G C, Grompe M. Germ cell defects and hematopoietic hypersensitivity to gamma-interferon in mice with a targeted disruption of the Fanconi anemia C gene. Blood. 1996;88:49–58. [PubMed] [Google Scholar]
- 42.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 43.Yamashita T, Barber D L, Zhu Y, Wu N, D’Andrea A D. The Fanconi anemia polypeptide FACC is localized to the cytoplasm. Proc Natl Acad Sci USA. 1994;91:6712–6716. doi: 10.1073/pnas.91.14.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita T, Kupfer G M, Naf D, Suliman A, Joenje H, Asano S, D’Andrea A D. The Fanconi anemia pathway requires FAA phosphorylation and FAA/FAC nuclear accumulation. Proc Natl Acad Sci USA. 1998;95:13085–13090. doi: 10.1073/pnas.95.22.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Youssoufian H. Localization of Fanconi anemia C protein to the cytoplasm of mammalian cells. Proc Natl Acad Sci USA. 1994;91:7975–7979. doi: 10.1073/pnas.91.17.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]