Abstract
Clostridiodes difficile (C. difficile) was ranked an “urgent threat” by the Centers for Disease Control and Prevention (CDC) in 2019. C. difficile infection (CDI) is the most common healthcare-associated infection (HAI) in the United States of America as well as the leading cause of antibiotic-associated gastrointestinal disease. C. difficile is a gram-positive, rod-shaped, spore-forming, anaerobic bacterium that causes infection of the epithelial lining of the gut. CDI occurs most commonly after disruption of the human gut microflora following the prolonged use of broad-spectrum antibiotics. However, the recurrent nature of this disease has led to the hypothesis that biofilm formation may play a role in its pathogenesis. Biofilms are sessile communities of bacteria protected from extracellular stresses by a matrix of self-produced proteins, polysaccharides, and extracellular DNA. Biofilm regulation in C. difficile is still incompletely understood, and its role in disease recurrence has yet to be fully elucidated. However, many factors have been found to influence biofilm formation in C. difficile, including motility, adhesion, and hydrophobicity of the bacterial cells. Small changes in one of these systems can greatly influence biofilm formation. Therefore, the biofilm regulatory system would need to coordinate all these systems to create optimal biofilm-forming physiology under appropriate environmental conditions. The coordination of these systems is complex and multifactorial, and any analysis must take into consideration the influences of the stress response, quorum sensing (QS), and gene regulation by second messenger molecule cyclic diguanosine monophosphate (c-di-GMP). However, the differences in biofilm-forming ability between C. difficile strains such as 630 and the “hypervirulent” strain, R20291, make it difficult to assign a “one size fits all” mechanism to biofilm regulation in C. difficile. This review seeks to consolidate published data regarding the regulation of C. difficile biofilms in order to identify gaps in knowledge and propose directions for future study.
Author summary
Clostridioides difficile (C. difficile) is an opportunistic bacterial pathogen that causes infection of the human gut epithelium following disruption of the normal gut microflora, usually by broad-spectrum antibiotics. C. difficile infection (CDI) is recurrent in 20% to 30% of cases and can lead to significant health-related complications such as pseudomembranous colitis and, in severe cases, death. The impact and cost of this pathogen on healthcare systems are significant, and some aspects of the pathogen’s lifestyle in the host are, as yet, unknown. It is hypothesised that C. difficile exists in the gut as a biofilm due to the infection’s severity and recurrent nature. The biofilm mode of bacterial growth can protect the cells from external factors such as antibiotic treatment, physiological processes, and the immune system. However, biofilm regulation in C. difficile is not yet fully characterised, and in this review, we consolidate published primary research on C. difficile biofilm regulation to gain a comprehensive overview of the factors involved and how they may interact to enable biofilm development within a host.
Introduction
Clostridioides difficile (C. difficile) is a gram-positive, rod-shaped, obligatory anaerobic bacterium with a low genome GC content (630 GC = 29.06%) [1]. This spore-forming anaerobe is found asymptomatically in 1% to 3% of healthy adult humans [2]. Additionally, it is also found in many other animals including diverse mammals, reptiles, and birds, all with the potential for zoonotic transfer to humans (for further consideration of this aspect, readers are referred to the following recent reviews [3–5]). Broad-spectrum antibiotic treatment can disrupt the normal microbiota in the gut causing dysbiosis of the gut microbiome [6,7]. This presents an opportunity for the rapid growth of C. difficile, leading to C. difficile infection (CDI) [2].
C. difficile has been ranked an “urgent threat” by the Centers for Disease Control and Prevention (CDC) in 2019 as it is the most common cause of healthcare-associated infection (HAI) in the United States of America [8,9] as well as the leading cause of antibiotic-associated gastrointestinal disease [9,10]. However, CDI has become increasingly prevalent as a community associated infection in younger, healthier patients [11,12]. Recent data on the impact of C. difficile in Europe and, specifically, the United Kingdom, are limited and have been described as urgently needed [13]. An epidemiological study on HAIs conducted between 2016 to 2017 found almost 45% of healthcare-associated gastrointestinal infections in Europe were caused by C. difficile [14].
The recurrent nature of CDI, affecting 20% to 30% of cases, adds to the already significant health service costs associated with treatment of this infection [15–17]. The real global impact of CDI is unclear; however, considering the US reported half a million CDI cases and 29,000 deaths in 2012, with an average annual cost between US$5.4 and US$6.3 billion [18], it is estimated to be significantly higher than this (for a comprehensive review, please see [19]).
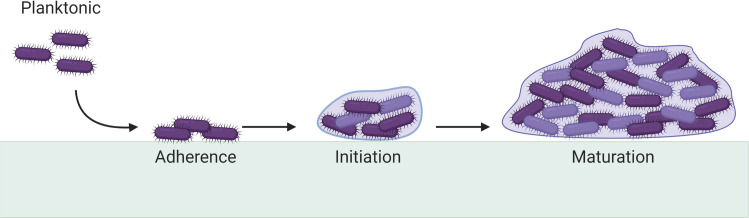
It has been hypothesised that C. difficile’s virulence and recurrence are due to its ability to form biofilms in the gut [20]. Biofilms (Fig 1) are sessile communities of microbes living within self-produced, hydrated extracellular matrices generally composed of proteins, polysaccharides, and nucleic acids [21–24]. The specific structural and compositional properties of a biofilm depend on the bacterial species, which produces the biofilm, and the environmental conditions it is exposed to [25,26]. Infections where bacteria form biofilms are characterised as persistent, chronic, and extremely difficult to treat due to the biofilms’ inherent increase in antibiotic resistance [27,28].
Fig 1. Stages of biofilm formation.
Free-floating planktonic cells adhere to a surface and begin microcolony formation with production of a hydrated extracellular matrix of proteins, polysaccharides, and nucleic acids. A mature biofilm may contain subpopulations of cells existing in various states where dispersion of the biofilm may occur due to physical disruption or cell signalling. Adapted from [29]. Created with BioRender.com.
C. difficile has been successfully grown in multispecies biofilms in vivo with species representative of the human gut microbiome [30]. The in vitro human gut model found C. difficile spores could be conserved within the multispecies biofilms to cause recurrence [30]. In vivo studies using animal models have also shown that C. difficile is capable of forming multicellular structures such as microbial aggregates in the gut [31–34].
Further indication that C. difficile biofilms are important in pathogenesis of disease is the observation that bile salts such as deoxycholate (DOC), in the presence of fermentable sugars found in the gut, can significantly increase biofilm formation as a mechanism of the stress response [35]. Several factors influence bacterial ability to form biofilms, including adherence and motility [36–39]. Development of mutant strains of C. difficile and their use as models to assess the resulting effects of the mutations on biofilm formation have led to some understanding of how biofilms are regulated in C. difficile [20,40–44]. The aim of this review, therefore, is to consolidate and critically analyse published information on the factors known to regulate biofilm formation. We intend to provide the scientific community with a comprehensive understanding of what coordinates the necessary cellular features within C. difficile to create optimal conditions for biofilm formation.
Cells in a sessile state
Cells within biofilms are described as being in a nonmotile, sessile state where they are attached to a surface. The switch between motile and sessile lifestyles in a wide range of bacteria—including C. difficile—is controlled by second messenger molecule, cyclic diguanosine monophosphate (c-di-GMP) [39]. c-di-GMP controls gene expression by acting on riboswitches located upstream of the controlled genes [39,45–47]. Riboswitch Cdi-1-3 is activated by c-di-GMP to down-regulate the FlgB operon containing 29 flagellar genes in the switch from motile to sessile lifestyle [45].
However, flagellar expression is also controlled by phase variation in all C. difficile strains except 630, which is effectively locked in a “flagella on” state [48]. Low levels of intracellular c-di-GMP allow for transcription of the downstream inversion site named Cdi4 in strain R20291, which can change between an “on” or “off” position [48]. DNA inversion by site-specific recombination of Cdi4 located upstream of the flgB operon into the “flagella off” position down-regulates flagellar expression by posttranscriptionally destabilising or degrading the mRNA potentially by a trans-acting element, thereby inhibiting flagellar expression on the cell surface [48].
Flagella are primarily responsible for the swimming motility of C. difficile in planktonic cultures [49–51]. Investigations of gene expression in established C. difficile biofilms compared to planktonic cultures agree that flagellar genes are decreased in expression in biofilms [38,52]. Hypervirulent C. difficile strain R20291, originally isolated from an outbreak in Stoke Mandeville Hospital, UK, formed biofilms with visibly reduced expression of flagella when grown for 7 days on glass beads [38,53]. This was reflected by a significant reduction in expression of the major flagellar filament gene, fliC, by 2.54-fold when compared to planktonic counterparts, indicating that flagella are down-regulated in an established biofilm [38]. Expression of several other flagellar biosynthesis genes were decreased in a 72-hour 630Δerm biofilm including flhA, flbD, flgE, and flgD, again showing that down-regulation of flagella components are required for biofilm formation [52]. It is unsurprising that flagella would be down-regulated when entering a sessile state, such as biofilm formation, as flagellar assembly and production is an energetically expensive process [54].
In contrast, Dapa and colleagues suggested that flagella play an important role in the late stages of biofilm formation as an R20291 fliC ClosTron disruption mutant exhibited decreased biofilm after 5 days compared to the parent strain [36]. However, Valiente and colleagues found that their R20291 fliC ClosTron mutant cells were completely lacking in flagella but had formed aggregates and mature 6-day biofilms similar to the parent R20291 strain [55]. The differences in these studies could be a result of a difference in the mutations, growth conditions, or indeed be related to the vigour of washing planktonic cells from biofilms.
Further investigations into specific glycan modifications of C. difficile flagella have also been undertaken via ClosTron mutagenesis of several glycosyltransferase genes, which are responsible for type B posttranscriptional modifications of flagella in ribotype 027 strains of C. difficile, including strain R20291 [55]. Type B modifications add glycan chains to flagella containing O-linked N-acetyl hexosamine, 2 rhamnoses with or without methylation, and a novel sulfonated peptidyl amidoglycan [55]. The addition of these glycan chains did not affect flagellar assembly in cells of R20291 but appeared to significantly increase biofilm formation and aggregation [55]. This was suggested to be due to an increase in cell hydrophobicity in the mutant C. difficile R20291 that lacked this particular flagellar posttranscriptional modification [55]. Only 2 of the 5 mutants created exhibited reduced motility, yet all 5 mutants showed a similar increase in biofilm formation. Valiente and colleagues thus concluded that motility does not heavily influence biofilm formation, but hydrophobicity, arising from the presence of glycan chains on the flagella, could enhance biofilm formation through increased aggregation [55].
Cell surface components and adherence in C. difficile biofilms
Poquet and colleagues analysed expression of genes associated with the biosynthetic pathways of certain cell surface components in biofilm and planktonic 630Δerm using microarray analysis of extracted RNA. They concluded that both the cell envelope and cell wall are modelled differently in biofilms compared to planktonic cells due to an up-regulation of phospholipid metabolism, Acyl Carrier proteins and fatty acid synthesis in the biofilm model [52]. This is similar to biofilm formation processes in Bacillus subtilis, in which fatty acid synthesis is required to build a hydrophobic environment within the biofilm in order to hold the complex structure together [56].
Membrane-bound glycopolymers such as PSII (Polysaccharide II) teichoic acids are conserved polysaccharide antigens, expressed at the cell surface of C. difficile [52,57]. The lcpB gene is involved in depositing PSII teichoic acids at the cell surface, and its expression was increased by 24.93-fold in 72-hour 630Δerm biofilm cells compared to planktonic cells [52]. This could be significant as a ClosTron mutation in the lcpB gene in strain 630Δerm significantly increased biofilm formation at 24 hours (p < 0.05), a phenotypic effect that was partially restored by complementation of the lcpB gene [44]. The lcpB mutant exhibited elongation and thickening of the cells along with curvature, abnormalities in septa, and slower, suboptimal growth phenomena, which have been seen in other C. difficile mutants exhibiting increased biofilm formation [41,44,58]. This could indicate that, although the lcpB gene itself does not regulate biofilm formation, the changes to the cell surface caused by the mutation could influence biofilm formation via knock-on effects on hydrophobicity of the cell surface and resultant cellular aggregation, similar to those observed in flagellar protein (FliC) mutants [55].
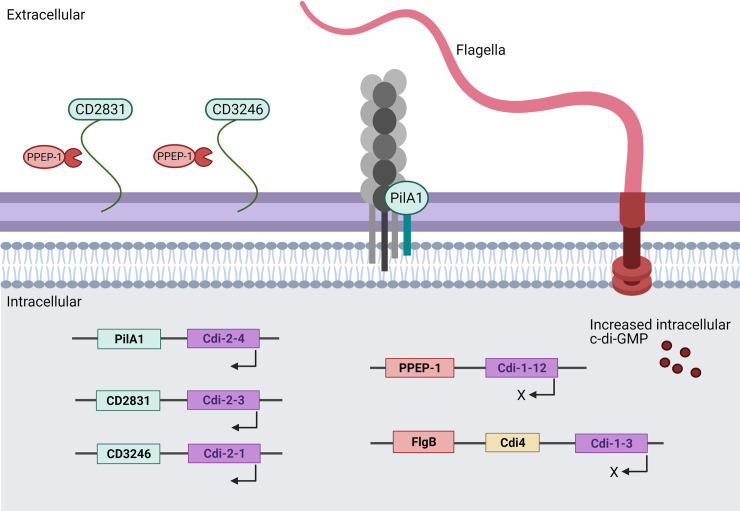
Other cell surface structures such as type IV pili are also known to influence biofilm formation in C. difficile, specifically with regard to their role in initial surface adherence [21,22,38,59,60]. Like the flagellar operon, the pilA1 major pilin gene (CD3513) is positively regulated by c-di-GMP acting on the upstream riboswitch, Cdi-2-4 (Fig 2) [45,60]. The pilA1 gene was up-regulated in 1-week-old biofilms along with pilK, pilU, and pilV when compared to planktonic growth in C. difficile R20291 [38]. A pilA1 disruption mutant of R20291 was created that had no visible pili and that produced thinner biofilms—measured by confocal microscopy—with critically reduced biomass at a 24-hour time point [38]. However, by day 7, there was no significant difference in biofilm biomass or thickness between the pilA1 mutant and the R20291 parent strain [38]. This led to the conclusion that pili are required for initial biofilm formation, most likely in the adhesion step [59], but not in the late stages of biofilm formation [38]. A similar effect was also seen in P. aeruginosa where, although pili-driven motility was not essential, it positively contributed to biofilm formation [61]. Further investigation of gene expression in the pilA1 mutant at the point of adherence could help elucidate which putative adhesins are up-regulated to compensate for the lack of pili and which led to wild-type (WT) levels of biofilm formation in C. difficile R20291 at 7 days.
Fig 2. Regulation of cell surface components under high intracellular levels of c-di-GMP acting on specific riboswitches (purple).
Down-regulated components in the presence of increased intracellular c-di-GMP are shown in red to include flagella and PPEP-1, which is responsible for cleavage of CD2831 and CD3246. Up-regulated components are shown in green, namely CD2831, CD3246, and PilA1, causing an increase in surface PilA1 structures. Created with BioRender.com. c-di-GMP, cyclic diguanosine monophosphate.
A separate study investigating gene expression in 630Δerm biofilms at 72 hours grown in a microfermentor found that the genes for type IV pili biogenesis, including pliA, were up-regulated compared to planktonic cells (Table 1), in a similar pattern to that observed in 7-day-old R20291 biofilms [38,52]. However, a separate study has shown that although type IV pili affect biofilm formation in both 630Δerm and R20291, they are of more critical importance in the latter as there was a higher promoter activity of the pilA1 gene in R20291 [60]. This has led to the suggestion that the significantly increased expression of pilA1 in R20291 biofilms when compared to planktonic culture, and which was not observed in the 630Δerm model, is responsible for the increased biofilm-forming capability of R20291, which has also been reported in previous studies [36,41,60]. However, reports of 630 and 630Δerm biofilms with significantly greater biofilm biomass than R20291 have also been published, which could highlight the need for standardised methods for biomass measurement [20,62].
Table 1. Cell surface associated component genes’ expression in 630Δerm 72-hour biofilm cells, compared to planktonically grown cells.
| Gene ID | Gene name | Gene function/product | Fold change |
|---|---|---|---|
| Fatty acid biogenesis | |||
| CD1183 | acpP | ACP | 9.92 |
| CD1062 | acpP | ACP | 9.45 |
| Cell wall | |||
| CD2766 | lcpB | Ligase anchoring polysaccharide to the cell wall | 24.93 |
| CD2762 | uppS | Putative undecaprynyl phosphate synthetase | −1.23 |
| Exported proteins | |||
| CD2831 | Putative adhesin | 6.77 | |
| CD2830 | zmp1 | Extracellular zinc metalloprotease | −1.16 |
| CD0873 | Adhesin and sugar binding lipoprotein (ABC transport) | 65.34 | |
| Type IV pili biogenesis | |||
| CD3513 | pilA1 | Putative pilin protein | 6.11 |
| CD2305 | pilW | Putative pilin protein | 11.31 |
| CD3504 | Putative type IV prepilin peptidase, A24A family | 6.45 | |
| CD3506 | Conserved hypothetical protein | 6.19 | |
| CD3507 | Putative type IV pilin | 10.78 | |
| CD3508 | Putative type IV pilin | 12.91 | |
| CD3509 | Putative type IV pilus assembly protein | 10.13 | |
| CD3510 | Putative membrane protein | 9.78 | |
| CD3511 | Putative type IV pilus secretion protein | 9.25 | |
| Flagellum biogenesis | |||
| CD0234 | csrA | Carbon storage regulator homolog CsrA | −1.35 |
| CD0254 | flgD | Basal-body rod modification protein FlgD | −1.24 |
| CD0255 | flgE | Flagellar hook protein FlbE (distal rod protein) | −1.29 |
| CD0255A | flbD | Flagellar protein FlbD | −1.28 |
| CD0263 | flhA | Flagellar biosynthesis protein FlhA | −1.19 |
630Δerm biofilm was grown in a continuous flow microfermentor for 72 hours with gene expression measured by microarray. Negative integers represent decreased expression. (Data from [52] transformed to show absolute fold changes).
ACP, acyl carrier protein.
Additional proteins with an adhesin function in C. difficile including cwp66 and CD0802 were both down-regulated in 7-day-old R20291 biofilms propagated on glass coverslips compared to planktonic cells [38]. Expression of the cell wall–binding cysteine protease, cwp84, wasn’t significantly different between the biofilm and planktonic grown cells of strain R20291 [38]; however, when the gene was disrupted by a ClosTron insertion in this strain, biofilm formation was dramatically decreased at 1, 3, and 5 days (p < 0.05) [36]. Alternatively, another study in 630Δerm measured a significant increase in biofilm formation by a cwp84 mutant (p < 0.05) [37]. This may indicate that cwp84, like pilA1, is regulated differently in 630Δerm and R20291 [60] and therefore could play a more significant role in R20291 biofilm formation. A comparative controlled study would be required to investigate this.
The Cwp84 protease is also responsible for cleavage of SlpA into the 2 S-layer protein subunits that form the outer S-layer cover on vegetative C. difficile cells [63–65]. Richards and colleagues investigated the influence of the S-layer on biofilm formation by creating deletion mutations in the SLCT-11 S-layer cassette of C. difficile strain Ox247 (ribotype 005) [40]. Mutations in orf2, orf3, orf4, orf7, orf16, and orf19 of the SLCT-11 caused an average decrease in cell length of approximately 15%, and the ability to sporulate was eliminated in orf2 and orf19 mutants [40]. While the orf2 mutant was more weakly adherent to Caco-2 cells, it was the only mutant that formed significantly more biofilm in 24-well microtiter plates at 24- and 72-hour time points compared to the parent strain (p < 0.05) as quantified using the crystal violet assay [40]. It was concluded that the effects on biofilm formation of mutating cwp84 in C. difficile were not due to the effect this has on SlpA [36,37,40].
Several cell surface associated proteins in C. difficile are regulated by c-di-GMP including CD2795 (cell wall binding protein, Cwp11), CD2796 (cell surface protein, Cwp10), and CD2797 (calcium-binding adhesion protein) [45]. CD2795, encoding Cwp11, has been identified as a protein released in the “secretome” during biofilm formation, while cwp10 expression was found to be increased by 9.13-fold in 630Δerm biofilms when compared with planktonic cells [37,52].
Cell wall adhesin CD2831 is characterised as a microbial surface component recognising adhesive matrix molecule (MSCRAMM) as it has been found to bind to immobilised collagen on ELISA plates [39]. CD2831 and CD3246 are both cell surface proteins positively regulated by c-di-GMP through riboswitches Cdi-2-3 and Cdi-2-1, respectively [45,62]. Both are cleaved from the cell surface by CD2830, a zinc-dependent metalloprotease (Zmpl) named PPEP-1 (pro-pro endopeptidase), which is negatively regulated by c-di-GMP acting on riboswitch Cdi-1-12 [39,62,66,67]. Cdi-2-1 up-regulates CD3246, a cell surface protein that produced a significant increase in biofilm formation when expressed ectopically using an inducible plasmid [45].
CD2830 was found to be the most down-regulated gene—by 62.61-fold—when c-di-GMP levels were artificially increased using an inducible plasmid, whereas CD2831 was the most up-regulated gene (by 42.51-fold) [45]. This correlates well with observations of gene expression in 630Δerm biofilm compared to planktonic cells (Table 1), where CD2831 expression increased by 6.77-fold and CD2830 expression decreased by 1.16-fold [52].
Overexpression of CD2831 using an inducible promoter on a plasmid increased 630Δerm biofilm proportionately [39]. Similarly, a ClosTron mutant of CD2831 in strain 630 showed a significant reduction in early biofilm at 24 hours (p < 0.05) but did not exhibit significant effects on attachment or late biofilm formation [62]. This mutation could be complimented using an inducible plasmid containing the CD2831 gene to restore biofilm to the original phenotype [62]. Furthermore, increased expression of CD2831 or CD3246 using an inducible plasmid in a PPEP-1 mutant of strain 630 caused a significant increase in early biofilm formation ((p < 0.01) or (p < 0.05), respectively) [62].
Other cell surface structures, namely CD3392, CD0183, and CD3145, which are not regulated by c-di-GMP, also produced significant reductions in biofilm formation (p < 0.01, p < 0.001, and p < 0.05, respectively) when genetically disrupted by ClosTron mutagenesis [62]. CD3392 is a cell wall protein containing an SPKTG motif anchored to the cell wall via sortase, which was present in the biofilm matrix of strain 630Δerm [62]. Whereas CD0183 and CD3145 (CbpA) are anchored to the cell wall independently of sortase but contain SPSTG and SPQTG motifs, respectively [62,68]. A role for CD0183 and CbpA in biofilm formation has yet to be determined [47,62,69]; however, CbpA is a novel cell surface exposed adhesin with a high affinity for binding to collagens I and V [62,69], and CD0183 with suspected autolytic properties has been shown to be positively controlled by small RNA RCd2, which itself is induced when cells enter exponential growth phase [47].
Taken together, the above observations by a range of independent groups suggest that CD2831 and CbpA could play a role in pathogenesis by binding to native collagen-producing cells found on the host extracellular matrix after toxin activity has exposed the fibronectin and collagen [39,69,70]. CD2831 may also aid virulence by inhibiting the formation of the C1 complement complex by binding to the C1q component of the host complement pathway [39], thereby limiting the host’s immune response. This observation is important for understanding of the pathogenesis of C. difficile infection and disease as it suggests that CD2831 and CpbA could be responsible for C. difficile adherence to the gut epithelium via host collagen exposed through the action of C. difficile toxins, while at the same time possibly limiting the immune response.
Toxin production in C. difficile biofilms
C. difficile tcdA and tcdB genes encode toxin A and toxin B proteins, respectively, and are located within the pathogenicity locus (PaLoc) of the genome [71]. The other genes required for toxin synthesis are also encoded within PaLoc; tcdR and tcdC are both involved in regulation and tcdE, which has a role in toxin secretion [72,73]. Toxin production by C. difficile is positively controlled by up-regulation of tcdR expression by effect of sigma factor sigD (originally annotated and named by Aubry and colleagues as fliA) [74,75]. However, sigD is negatively controlled by c-di-GMP levels and is also responsible for regulation of the fliC gene [74]. Consequently, sigD has been identified as a key virulence regulator as it can lead to decreased expression of both flagella and toxin production under elevated c-di-GMP levels [74].
An RNA-seq comparison of gene expression in C. difficile R20291 biofilm and planktonic cultures revealed a statistically significant increase in tcdB expression by 2.83-fold (q < 0.05) in biofilms compared to planktonic culture, but without a corresponding increase in tcdA expression [38]. However, when comparing toxin expression in biofilm compared to colony growth, both tcdA and tcdB exhibited significant decreases in expression in the biofilm culture (by 3.25-fold and 2.89-fold, respectively) [38]. Differential expression of tcdA and tcdB was also found in C. difficile 630Δerm biofilms grown in a continuous-flow microfermentor for 72 hours [52]. However, in this case, tcdA expression had decreased by only 1.03-fold, and tcdB expression was not significantly different [52]. The reported differences between these studies, with respect to the bacterial strains used, culture conditions, and time of RNA extraction make direct comparisons difficult. However, it raises the possibility that, under comparable conditions, the 2 strains may well behave similarly with regard to toxin gene expression in biofilms [38,42,76].
In C. difficile 630 cells cultured in human faecal water, expression of tcdA was decreased by 4-fold (p < 0.001) [77]. It was suggested that this was due to a decrease in metabolism of carbohydrates into butyrate [77]. Nonetheless, while toxin expression decreased in the presence of FW, the expression of sporulation genes significantly increased—by up to 300-fold in some cases [77]. This indicates that FW components induce sporulation, potentially aiding the transmission of C. difficile, but that FW components alone do not necessarily play a role in disease onset by inducing toxin production [77].
Sporulation in C. difficile
C. difficile produces metabolically dormant structures called spores that are resistant to oxygen and heat stress as well as some disinfecting agents [78,79]. Spores, as the transmissible agent, are therefore important for the spread of C. difficile as they allow the pathogen to survive outside of the host [78–80]. The Spo0A master regulator in B. subtilis is responsible for driving 3 possible pathways: sporulation, production of an extracellular matrix, or production of toxins, which sacrifice some cells in order to gain nutrients, a process referred to as cannibalism [20,36,81–83]. Low levels of phosphorylated Spo0A causes matrix production in B. subtilis, whereas increasing phosphorylation results in spore production [81]. Disruption of spo0A in C. difficile strain R20291 resulted in complete inhibition of sporulation and significantly smaller biofilms than the parent strain at 3 and 6 days (p < 0.05) when grown in microtiter plates as measured by crystal violet assay [20]. These smaller spo0A mutant biofilms were more easily detached and dispersed when gently agitated in tissue culture flasks, compared to the parent strain biofilm [20]. This reduction in biofilm in the C. difficile R20291 Spo0A mutant led the authors to conclude that the role of spo0A in C. difficile is analogous to that of the Spo0A regulator in B. subtilis [20]. However, the effects on biofilm formation and dispersal could also be an indication that the spo0A regulator mutation influences other processes such as adherence, an aspect not examined by Dawson and colleagues in their 2012 paper [20].
A more recent study by Dawson and colleagues examined sporulation frequency within biofilms for 5 C. difficile strains including 630 and R20291 and found similarities in biofilm composition between strains [62]. A positive correlation between sporulation frequency and biofilm biomass was found along with a positive correlation with eDNA in the matrix [62]. It was suggested that the increased eDNA and sporulation correlating with increased biofilm biomass could arise from induction of sporulation within the biofilms resulting in cell lysis to release eDNA and intracellular proteins into the matrix [62]. The importance of eDNA as a key component in C. difficile biofilms was proven by successful inhibition and disruption of C. difficile biofilms using DNase enzymes [35,62]. However, identification of other proteins in the matrix, namely Cwp19 and phage protein (phiCD24-1), which both contribute to cell lysis and so would also elevate eDNA, indicates a complexity to the biofilm formation process and can explain why disruption of single genes cannot produce an all-or-nothing biofilm phenotype where many processes are involved [62]. Additionally, the authors also found that 630 and R20291 had significantly greater numbers of spores in their biofilms, compared to planktonic supernatants, which were suggested to aid recurrence of infection through dispersal of the biofilm and reseeding spores elsewhere in the gut [62].
A study performed by Tijerina-Rodríguez and colleagues to determine the differences in recurrent-CDI (R-CDI) cases and nonrecurrent-CDI (NR-CDI) clinical samples found that there was no significant difference in distribution of C. difficile ribotypes between R-CDI and NR-CDI samples; however, 81% of 102 samples examined contained RT-027 strains of C. difficile [84]. Significantly higher rates of sporulation were found in R-CDI isolates compared to NR-CDI isolates grown in 7-day mono-biofilms (p = 0.015), which corresponded with R-CDI biofilms having higher expression of both spo0A and sigH, the alternative sigma factor for controlling sporulation [84]. This also fits with the theory of Dawson and colleagues discussed above that biofilms with increased sporulation frequency can reseed and lead to recurrent infections [62]. Crucially, however, there was no significant difference in biofilm formation between the 2 groups (R-CDI and NR-CDI) at 7 days as measured by the crystal violet assay [84]. This suggests that increased spo0A expression does not influence biofilm formation significantly and that sporulation may play a larger role in recurrence of disease than biofilm formation [84].
The role of the stress response in C. difficile biofilms
Bacteria respond to stressors, such as adverse and fluctuating conditions, by altering gene expression to adapt to new environments [85]. Biofilm formation by bacteria is recognised as a stress response, as the production of an extracellular matrix shields the bacterial cells from external pressures such as oxygen stress and antibiotics [20,86–88]. Several factors within the biofilm can contribute to recalcitrance to antibiotics such as polarity of the matrix inhibiting drug diffusion and reduced metabolic activity within the cells preventing antibiotic uptake into the cells [27].
Subinhibitory antibiotic concentrations are known to cause a stress response in many bacteria including Escherichia coli, Vibrio cholerae, and Staphylococcus aureus, where they also lead to increased expression of virulence factors [89]. C. difficile strains that showed susceptibility to antibiotics such as metronidazole produced significantly more biofilm in the presence of subinhibitory concentrations of this antibiotic [86,90]. The regulatory processes and mechanism underpinning this observation were not confirmed in these studies; however, subinhibitory concentrations of antibiotics have been found to trigger the stress response in other bacteria [89]. Consequently, treatment of CDI with lower concentrations of ineffective antibiotics could worsen the disease state or lead to recurrence.
A pattern of stress responses inducing morphological changes is seen in many bacterial species [89]. For example, a disruption mutation in the prkC gene in 630Δerm resulted in the mutant producing significantly increased biofilm under bile salt–induced stress conditions (p < 0.05) [57]. The prkC gene encodes a membrane-associated serine/threonine (Ser/Thr) kinase enzyme with 2 penicillin-binding and Ser/Thr kinase-associated (PASTA) domains, but its function is, as yet, uncharacterised in C. difficile [57]. The prkC mutant strain produced significantly increased biofilm after 24 hours, but only in the presence of compounds that stressed the cell envelope, such as the bile salt DOC, or the antibiotic polymyxin B [57]. Other effects caused by the mutation were a >100% elongation of the cells, in which abnormal septa were identified by confocal and transmission electron microscopy, along with replication defects, reduced motility, and increased aggregation [57].
The DnaK protein is also an important component of the C. difficile stress response and has also been found in the biofilm matrix [58,62]. It is a molecular chaperone that aids protein folding and is up-regulated under heat stress conditions [91]. A ClosTron insertion mutation in the dnaK gene of 630Δerm disrupted DnaK production and generated a less thermotolerant strain that exhibited suboptimal growth at 37°C and reduced motility due most likely to the lack of flagella on the cell surface as revealed by TEM [58]. However, disruption of the dnaK gene also resulted in a significant increase in biofilm formation, as well as approximately 50% elongation of the cells into filamentous rods [58].
Similar effects were seen when other elements of the C. difficile stress response were investigated. The SOS response is a regulatory network within the stress response of bacterial cells that reacts to cellular damage and, in particular, DNA damage [41,85,89]. The SOS response is regulated by proteins LexA and RecA [89]. The global transcriptional repressor, LexA, temporarily halts cellular division in C. difficile and activates processes for cellular DNA repair [41,92]. Disruption of the lexA gene in C. difficile R20291 resulted in elongated, filamentous cells, as well as increased biofilm production compared to the parent strain [41]. These observations are similar to those made with the dnaK mutant in 630Δerm, thus emphasising the effects of stress on biofilm formation [58].
While physiological stress is clearly implicated in biofilm formation, other signalling processes initiated by favourable growth conditions that result in an increase in cell number and density are also present in bacteria [93,94]. This method of communication is referred to as quorum sensing (QS).
The role of quorum sensing in C. difficile biofilms
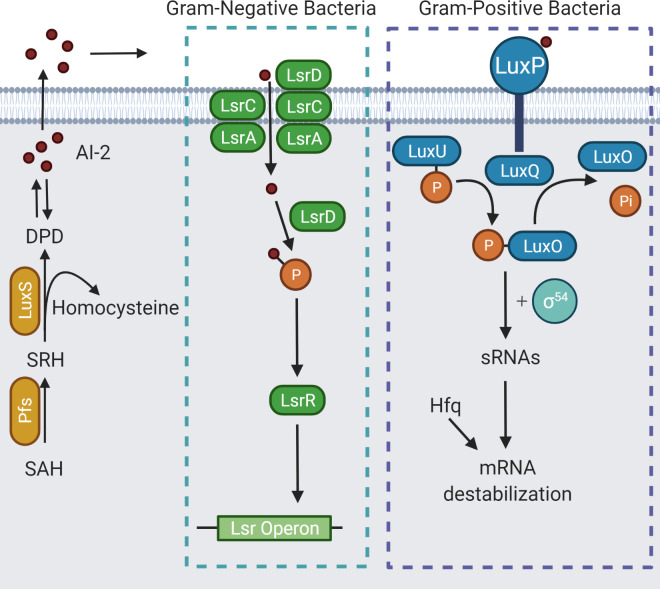
QS is a system that regulates gene expression in bacterial cells by using signalling molecules called autoinducers that build up as a direct result of increasing cell population density [93,94]. The LuxS QS system exists in both gram-positive and gram-negative bacteria and uses signalling molecules collectively known as autoinducer-2 (AI-2) [95,96]. AI-2 molecules act on cell surface receptors not yet identified in C. difificile to generate an intracellular signalling cascade that results in gene regulation (Fig 3) [42,95,96].
Fig 3. Overview of the LuxS QS system in both gram-negative and gram-positive bacteria.
Both systems use the same signalling molecules, collectively named AI-2. Created with BioRender.com adapted from [96]. AI-2, autoinducer-2; QS, quorum sensing.
A luxS ClosTron mutant in the hypervirulent C. difficile strain R20291 was defective in production of AI-2 molecules, as measured by induction of bioluminescence in the bioreporter strain Vibrio harveyi BB170, which responds only to AI-2 signals [42,97]. As a result of this mutation in luxS, the mutant R20291 was also deficient in biofilm formation [42]. Chemical complementation using the compound 4,5-dihydroxy-2,3-pentanedione (DPD), which spontaneously and reversibly forms AI-2, showed that a concentration of 100 nM DPD could restore biofilm formation to WT levels, thereby confirming that both AI-2 and LuxS are involved in biofilm formation in C. difficile [42,95]. However, the receptor for AI-2 in C. difficile has yet to be identified [42]. RNA-seq analysis of gene expression in the R20291 luxS mutant biofilm cells revealed 3 genes with increased expression: CDR20291_2554 (crr), a PTS system glucose-specific transporter subunit IIA; CDR20291_2927, a cellobiose phosphate-degrading protein; and CDR20291_2930 (treA), a trehalose-6-phosphate hydrolase [42]. A total of 18 genes exhibited decreased expression compared to the WT; these genes corresponded, in the main, to 2 putative prophage regions [42]. The prophage regions, therefore activated by AI-2, have been linked to induction of cell lysis that produces eDNA, which, as mentioned above, is necessary for biofilm formation in C. difficile [36,42,62,98].
However, a separate study that investigated gene expression in biofilms, planktonic cells, and colony-grown C. difficile R20291 reported no significant difference in expression of the luxS gene between the growth models [38]. It was suggested that this could be due to the RNA being extracted from a 7-day-old biofilm in the later stages of growth, where it might be expected that there would be differences in gene expression compared to the early stages of biofilm formation [38]. AI-2 production by planktonic C. difficile has been shown to increase to a maximum at 8 hours, correlating to mid to late log phase of growth, declining thereafter [76]. Lee and Song did not, however, investigate biofilm formation alongside this observation. Biofilm initiation in C. difficile has been shown to occur between 6 and 12 hours, which could feasibly correlate with the peak in AI-2 seen at 8 hours [38,42,76,90].
In hypervirulent C. difficile strains such as those belonging to RT-017 or RT-027, the whole agr operon, agrACDB (agr2), encodes another QS system [53,99,100]. This is similar to the agr QS locus, agrACDB in S. aureus, which is a global regulator of virulence gene expression [99]. However, the partial locus, agrDB (agr1), has been found in all C. difficile genomes analysed to date [53,100]. The agr2 QS system uses the small secreted cyclic autoinducing peptide (AIP) encoded by agrD, which is processed and exported by the transmembrane protein encoded by agrB [99]. A study investigating the difference between clinical isolates of C. difficile from R-CDI cases and NR-CDI cases found no significant difference in biofilm formation between the 2 groups but found a significant increase in sporulation in R-CDI isolate biofilms (p = 0.015) [84]. There was also no significant difference in luxS expression; however, agrD1 expression was significantly increased by 14.02 relative to 16S rRNA expression (p = 0.001) in R-CDI isolates when compared to NR-CDI isolates [84].
The agr QS system has also been associated with toxin regulation as allelic exchange deletion mutants of agrB1D1 in the agr1 partial locus of 630 and R20291 were deficient in both toxin A and B production [100]; an agrA insertional inactivation (ClosTron) mutation in R20291 also resulted in significantly reduced expression of tcdA compared to the parent R20291 strain as well as reduced flagellar operon expression [99]. On the other hand, an R20291 luxS disruption mutant deficient in AI-2 production exhibited no change in toxin RNA expression compared to the WT, indicating that the LuxS QS system has perhaps minimal influence on toxin production in C. difficile [42]. This is reinforced by the differences in expression of tcdA and tcdB when luxS expression remains unchanged in R20291 biofilms grown for 7 days on glass beads [38].
The insertion mutation in the agrA gene of C. difficile strain R20291 also resulted in decreased expression of 3 genes encoding diguanylate cyclase (DGC) and phosphodiesterase (PDE) enzymes responsible for c-di-GMP production [99]. This implies that the agr QS system is also involved with in regulation of production of c-di-GMP [99].
The regulation of c-di-GMP in C. difficile
Soutourina and colleagues hypothesised that biofilm formation and motility in C. difficile is controlled by c-di-GMP acting on riboswitches, a type of small noncoding RNA molecule which, in C. difficile and other bacteria, act directly or indirectly on mRNA to control protein production [39,45–47]. Upon binding of c-di-GMP to a riboswitch, transcription of the associated mRNA molecule can be decreased by premature termination or positively controlled via a complex alternative splicing mechanism [47]. Small increases of 2- to 6-fold in intracellular concentrations of c-di-GMP by a nisin-inducible plasmid in strain 630 compared to empty vector control have been shown to be sufficient to cause a change in gene expression in C. difficile as exemplified by a significant reduction in swarming motility [101]. However, it has also been reported that cellular response to c-di-GMP levels within the cell can vary between C. difficile strains, for example, R20291 and 630, as R20291 has some of the lowest number of metabolic genes controlled by c-di-GMP [101,102]. Intracellular c-di-GMP levels are controlled by the actions of DGC and PDE enzymes [45].
A nisin-inducible plasmid overexpressing dccA, encoding a DGC enzyme responsible for the conversion of 2 guanosine triphosphate (GTP) molecules into c-di-GMP, has been used to increase intracellular c-di-GMP levels [45,102]. Artificial fluctuations in intracellular c-di-GMP concentration have been shown to influence swimming motility and flagellar gene expression, as well as aggregation and type IV pili in 630Δerm (Table 2); therefore, it is unsurprising that increasing intracellular c-di-GMP concentrations significantly increased biofilm formation at 24 hours as measured by crystal violet staining [8,45,59,60,101]. A plasmid containing dccA induced by anhydrotetracycline (ATc) was used to increase c-di-GMP levels in strain 630 [62]. A significant increase in early biofilm formation at 24 hours was found (p < 0.001 with 25 ng/ml ATc), but by 72 hours, the mature biofilms with increased c-di-GMP showed no significant increase in biofilm formation compare to the WT [62]. Therefore, it was concluded that c-di-GMP plays a greater role in early biofilm formation rather than the later stages [62]. However, other effects of artificially increasing intracellular c-di-GMP have also been reported such as defective cell separation, resulting in chains of C. difficile cells with abnormal septa, similar to the cell elongation mentioned above in dnaK, lexA, prkC, and lcpB mutant strains, which all exhibited increased biofilm formation [41,44,57,58,66].
Table 2. Changes in c-di-GMP riboswitch-controlled gene expression resulting from increased intracellular c-di-GMP in Clostridioides difficile strain 630Δerm.
| Class | Riboswitch | Downstream gene | Encoded protein | Fold change |
|---|---|---|---|---|
| Class I | Cdi-1-1 | CD1990 | SH3 domain containing protein | 31.09 |
| Cdi-1-2 | CD2797 | Calcium binding adhesion protein | −7.14 | |
| Cdi-1-3 | CD0245 | FlgB operon encoding 29 flagellar genes | −15.06 | |
| Cdi-1-8 | CD19903 | Hypothetical protein | −7.78 | |
| Cdi-1-9 | CD2309 | Hypothetical protein | −4.66 | |
| Cdi-1-11 | CD33682 | Hypothetical protein | −8.90 | |
| Cdi-1-12 | CD2830 | Zmpl/PPEP-1 | −62.61 | |
| Class II | Cdi-2-1 | CD3246 | Cell surface protein | 4.29 |
| Cdi-2-2 | CD3267 | Two-component response regulator | 18.78 | |
| Cdi-2-3 | CD2831 | Putative adhesin predicted to bind collagen | 42.51 | |
| Cdi-2-4 | CD3513 | Type IV pili (PilA1) | 11.74 |
Fold changes in gene expression were measured by microarray analysis of planktonically grown 630Δerm expressing plasmid encoded pDccA compared to a vector control. Plasmid induced expression of dccA by 1 μg/ml nisin was used to increase intracellular c-di-GMP levels. (Adapted from [45]).
c-di-GMP, cyclic diguanosine monophosphate; PPEP-1, pro-pro endopeptidase-1; Zmpl, zinc-dependent metalloprotease.
It is unknown if the extent of the artificial increase in intracellular c-di-GMP by the inducible plasmid is likely under physiological conditions, so what is seen could be an overestimation of the effects, whereas the observations of gene expression within a biofilm model, in which c-di-GMP was not artificially influenced, could represent a more realistic assessment of the effects of c-di-GMP [45,52]. Some studies have reported undetectable levels of c-di-GMP in strain 630 using liquid chromatography–tandem mass spectrometry (LC–MS/MS) with a limit of detection of 0.5 ng/mg dry weight [66]. Nonetheless, Purcell and colleagues recorded levels of c-di-GMP in planktonic C. difficile 630 cells to be 15 to 50 nM, which is low in comparison to other bacteria [101,103].
The effect of nutrient availability on intracellular c-di-GMP concentrations could explain why C. difficile biofilm formation is enhanced in the presence of glucose in the media [8,90]. Nutrient availability modulates CodY (CD1275), a transcriptional repressor that is activated by GTP [8]. However, GTP is also required to make c-di-GMP in a process that is regulated by PdcA, a PDE enzyme that degrades c-di-GMP [8]. Interestingly, the PdcA enzyme is also regulated by CodY but is dependent on GTP concentrations [8]. Therefore, at low GTP levels, PdcA transcription is active to produce PdcA enzyme molecules, which break down c-di-GMP into GTP. However, with high GTP levels, CodY is activated and acts to down-regulate PdcA transcription to prevent PdcA enzyme-mediated degradation of c-di-GMP in a negative feedback loop. CodY has also been found to bind to the tcdR toxin gene promoter in the presence of GTP, also linking increased toxin expression to elevated nutrient availability [104]. Consequently, increasing nutrient availability increases c-di-GMP levels, which can, in turn, act on relevant riboswitches to promote biofilm formation in C. difficile [8,45].
Regulation of dccA, a DGC that synthesises c-di-GMP and CD1412, a putative PDE that degrades c-di-GMP, has also been linked to the CD2214-2215 two-gene operon [45,47,52]. In 630, CD2214 encodes a protein homologous to the biofilm and sporulation repressor, sinR, from B. subtilis [52,105]. When disrupted by ClosTron mutagenesis in strain 630Δerm, the CD2214-2215 two-gene operon was found to regulate many pathways including sporulation, the stress response, toxin synthesis, and membrane transport as well as metabolism and motility potentially through the effects of c-di-GMP [52,106]. Therefore, it has been suggested that the CD2214-CD2215 two-gene operon could be responsible for the regulation of some 15% of genes that are differentially expressed in biofilm formation, including genes involved in cellular metabolism and transition between growth stages [52].
Conclusions
Biofilm formation and bacterial over growth in the gut is a serious threat because of increasing antibiotic resistance, making treatments less effective [107]. The complexity of C. difficile biofilm regulation and the differences between strains clearly indicate that there will almost certainly be no “silver bullet” solution to this growing and challenging problem. This review is timely in that it has identified stark differences in regulatory systems between the most intensely investigated C. difficile strains, namely 630 and R20291, making it difficult to assign a “one size fits all” paradigm to biofilm regulation in this important organism. Although R20291 has a lower number of metabolic genes controlled by c-di-GMP than 630 [101,102], there appears to be a regulatory link between c-di-GMP and the full agr1 QS system that is found in R20291, which is not found in 630 [99]. In addition, opposite effects on biofilm formation in disruption mutants of cwp84 in both 630Δerm and R20291 have been observed [36,37], and there is what appears to be a more vital role for type IV pili in R20291 biofilm formation when compared to 630 [60]. Variation in flagellar machinery and associated genes has also been observed between R20291 and 630 strains with respect to arrangement and posttranscriptional modifications as well as phase variation capabilities [55]; however, we can conclude that flagellar genes, and functional flagella, are down-regulated in biofilms [38,52].
The lack of studies to confirm observations made by individual research groups makes it difficult to conclude an absolute pathway for biofilm regulation. Perhaps, importance should be placed on focusing efforts to elucidate the biofilm formation pathway in few select C. difficile strains before expanding research to increasing numbers of strains with highly variable results. The use of gene expression studies should, in future, allow us to learn more about biofilm regulation in different C. difficile strains, with the potential for development of improved treatment options for this opportunistic pathogen.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–786. Available from: https://www.nature.com/articles/ng1830. doi: 10.1038/ng1830 [DOI] [PubMed] [Google Scholar]
- 2.Vedantam G, Clark A, Chu M, McQuade R, Mallozzi M, Viswanathan VK, et al. Clostridium difficile infection: Toxins and non-toxin virulence factors, and their contributions to disease establishment and host response. Gut Microbes. 2012:121–34. doi: 10.4161/gmic.19399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gould LH, Limbago B. Clostridium difficile in food and domestic animals: A new foodborne pathogen. Clin Infect Dis. 2010:577–582. doi: 10.1086/655692 [DOI] [PubMed] [Google Scholar]
- 4.Songer JG, Anderson MA. Clostridium difficile: An important pathogen of food animals. Anaerobe. 2006:1–4. doi: 10.1016/j.anaerobe.2005.09.001 [DOI] [PubMed] [Google Scholar]
- 5.Hensgens MPM, Keessen EC, Squire MM, Riley TV, Koene MGJ, De Boer E, et al. Clostridium difficile infection in the community: A zoonotic disease? Clin Microbiol Infect. Blackwell Publishing Ltd. 2012:635–45. doi: 10.1111/j.1469-0691.2012.03853.x [DOI] [PubMed] [Google Scholar]
- 6.Buffie CG, Jarchum I, Equinda M, Lipuma L, Gobourne A, Viale A, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108:4554–61. doi: 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell EB, McKee RW, Courson DS, Garrett EM, McBride SM, Cheney RE, et al. A nutrient-regulated cyclic diguanylate phosphodiesterase controls Clostridium difficile biofilm and toxin production during stationary phase. Infect Immun. 2017;85. doi: 10.1128/IAI.00347-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.CDC. Antibiotic resistance threats in the United States. US Dep Heal Hum Serv CDC. 2019. 10.15620/cdc:82532 [DOI]
- 10.Lessa FC, Mu Y, Bamberg WM, Beldavs ZG, Dumyati GK, Dunn JR, et al. Burden of Clostridium difficile infection in the United States. N Engl J Med. 2015;372:825–34. doi: 10.1056/NEJMoa1408913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Roo AC, Regenbogen SE, De Roo AC, Regenbogen SE. Clostridium difficile Infection: An Epidemiology Update. Clin Colon Rectal Surg. 2020;33:49–57. doi: 10.1055/s-0040-1701229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guh AY, Mu Y, Winston LG, Johnston H, Olson D, Farley MM, et al. Trends in U.S. Burden of Clostridioides difficile Infection and Outcomes. N Engl J Med. 2020;382:1320–30. doi: 10.1056/NEJMoa1910215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kampouri E, Croxatto A, Prod’hom G, Guery B. Clostridioides difficile Infection. Still a Long Way to Go. J Clin Med. 2021;10:389. doi: 10.3390/jcm10030389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suetens C, Latour K, Kärki T, Ricchizzi E, Kinross P, Moro ML, et al. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: Results from two european point prevalence surveys, 2016 to 2017. Eur Secur. 2018;23:1800516. doi: 10.2807/1560-7917.ES.2018.23.46.1800516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tresman R, Goldenberg SD. Healthcare resource use and attributable cost of Clostridium difficile infection: A micro-costing analysis comparing first and recurrent episodes. J Antimicrob Chemother. 2018;73:2851–5. doi: 10.1093/jac/dky250 [DOI] [PubMed] [Google Scholar]
- 16.Cornely OA, Miller MA, Louie TJ, Crook DW, Gorbach SL. Treatment of first recurrence of clostridium difficile infection: Fidaxomicin versus vancomycin. Clin Infect Dis. 2012;55. doi: 10.1093/cid/cis462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guery B, Menichetti F, Anttila VJ, Adomakoh N, Aguado JM, Bisnauthsing K, et al. Extended-pulsed fidaxomicin versus vancomycin for Clostridium difficile infection in patients 60 years and older (EXTEND): a randomised, controlled, open-label, phase 3b/4 trial. Lancet Infect Dis. 2018;18:296–307. doi: 10.1016/S1473-3099(17)30751-X [DOI] [PubMed] [Google Scholar]
- 18.Zhang S, Palazuelos-Munoz S, Balsells EM, Nair H, Chit A, Kyaw MH, et al. Cost of hospital management of Clostridium difficile infection in United States-a meta-analysis and modelling study. BMC Infect Dis. 2016;16. doi: 10.1186/s12879-016-1786-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balsells E, Shi T, Leese C, Lyell I, Burrows J, Wiuff C, et al. Global burden of Clostridium difficile infections: A systematic review and meta-analysis. J Glob Health. 2019;9. doi: 10.7189/jogh.09.010407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dawson LF, Valiente E, Faulds-Pain A, Donahue EH, Wren BW, Popoff MR. Characterisation of Clostridium difficile Biofilm Formation, a Role for Spo0A. PLoS ONE. 2012;7. doi: 10.1371/journal.pone.0050527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001:135–8. doi: 10.1016/s0140-6736(01)05321-1 [DOI] [PubMed] [Google Scholar]
- 22.Davey ME, O’Toole GA. Microbial Biofilms: from Ecology to Molecular Genetics. Microbiol Mol Biol Rev. 2000;64:847–67. doi: 10.1128/MMBR.64.4.847-867.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487 [DOI] [PubMed] [Google Scholar]
- 24.Kavanaugh JS, Flack CE, Lister J, Ricker EB, Ibberson CB, Jenul C, et al. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio. 2019;10. doi: 10.1128/mBio.01137-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shrout JD, Chopp DL, Just CL, Hentzer M, Givskov M, Parsek MR. et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–77. doi: 10.1111/j.1365-2958.2006.05421.x [DOI] [PubMed] [Google Scholar]
- 26.Rossi E, Paroni M, Landini P. Biofilm and motility in response to environmental and host-related signals in Gram negative opportunistic pathogens. J Appl Microbiol. Blackwell Publishing Ltd. 2018:1587–602. doi: 10.1111/jam.14089 [DOI] [PubMed] [Google Scholar]
- 27.Lebeaux D, Ghigo J-M, Beloin C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol Mol Biol Rev. 2014;78:510–43. doi: 10.1128/MMBR.00013-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bjarnsholt T. The role of bacterial biofilms in chronic infections. APMIS Suppl. 2013:1–51. doi: 10.1111/apm.12099 [DOI] [PubMed] [Google Scholar]
- 29.Rasamiravaka T, Labtani Q, Duez P, El Jaziri M. The formation of biofilms by pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. Biomed Res Int. 2015. doi: 10.1155/2015/759348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Normington C, Moura IB, Bryant JA, Ewin DJ, Clark EV, Kettle MJ, et al. Biofilms harbour Clostridioides difficile, serving as a reservoir for recurrent infection. NPJ Biofilms Microbiomes. 2021;7. doi: 10.1038/s41522-021-00184-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soavelomandroso AP, Gaudin F, Hoys S, Nicolas V, Vedantam G, Janoir C, et al. Biofilm structures in a mono-associated mouse model of Clostridium difficile infection. Front Microbiol. 2017;8. doi: 10.3389/fmicb.2017.02086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spencer J, Leuzzi R, Buckley A, Irvine J, Candlish D, Scarselli M, et al. Vaccination against Clostridium difficile using toxin fragments: Observations and analysis in animal models. Gut Microbes. 2014;5:225–32. doi: 10.4161/gmic.27712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semenyuk EG, Poroyko VA, Johnston PF, Jones SE, Knight KL, Gerding DN, et al. Analysis of bacterial communities during Clostridium difficile infection in the mouse. Infect Immun. 2015;83:4383–91. doi: 10.1128/IAI.00145-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Buckley AM, Spencer J, Candlish D, Irvine JJ, Douce GR. Infection of hamsters with the UK clostridium difficile ribotype 027 outbreak strain R20291. J Med Microbiol. 2011;60:1174–80. doi: 10.1099/jmm.0.028514-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubois T, Tremblay YDN, Hamiot A, Martin-Verstraete I, Deschamps J, Monot M, et al. A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. NPJ Biofilms Microbiomes. 2019;5. doi: 10.1038/s41522-019-0087-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol. 2013;195:545–55. doi: 10.1128/JB.01980-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantaléon V, Soavelomandroso AP, Bouttier S, Briandet R, Roxas B, Chu M, et al. The Clostridium difficile protease Cwp84 Modulates both biofilm formation and cell- surface properties. PLoS ONE. 2015;10. doi: 10.1371/journal.pone.0124971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maldarelli GA, Piepenbrink KH, Scott AJ, Freiberg JA, Song Y, Achermann Y, et al. Type IV pili promote early biofilm formation by Clostridium difficile. Pathog Dis. 2016;74. doi: 10.1093/femspd/ftw061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arato V, Gasperini G, Giusti F, Ferlenghi I, Scarselli M, Leuzzi R, et al. Dual role of the colonization factor CD2831 in Clostridium difficile pathogenesis. Sci Rep. 2019;9. doi: 10.1038/s41598-019-42000-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards E, Bouché L, Panico M, Arbeloa A, Vinogradov E, Morris H, et al. The S-layer protein of a Clostridium difficile SLCT-11 strain displays a complex glycan required for normal cell growth and morphology. J Biol Chem. 2018;293:18123–37. doi: 10.1074/jbc.RA118.004530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walter BM, Cartman ST, Minton NP, Butala M, Rupnik M. The SOS response master regulator LexA is associated with sporulation, motility and biofilm formation in clostridium difficile. PLoS ONE. 2015;10. doi: 10.1371/journal.pone.0144763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slater RT, Frost LR, Jossi SE, Millard AD, Unnikrishnan M. Clostridioides difficile LuxS mediates inter-bacterial interactions within biofilms. Sci Rep. 2019;9. doi: 10.1038/s41598-019-46143-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gu H, Qi H, Chen S, Shi K, Wang H, Wang J, et al. Carbon storage regulator CsrA plays important roles in multiple virulence-associated processes of Clostridium difficile. Microb Pathog. 2018;121:303–9. doi: 10.1016/j.micpath.2018.05.052 [DOI] [PubMed] [Google Scholar]
- 44.Chu M, Mallozzi MJG, Roxas BP, Bertolo L, Monteiro MA, Agellon A, et al. A Clostridium difficile Cell Wall Glycopolymer Locus Influences Bacterial Shape, Polysaccharide Production and Virulence. PLoS Pathog. 2016;12. doi: 10.1371/journal.ppat.1005946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKee RW, Harvest CK, Tamayo R. Cyclic Diguanylate Regulates Virulence Factor Genes via Multiple Riboswitches in Clostridium difficile. mSphere. 2018;3. doi: 10.1128/mSphere.00423-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ternan NG. Small regulatory RNA molecules in bacteria. OA Microbiol 2013;1:1–8. Available from: https://pure.ulster.ac.uk/en/publications/small-regulatory-rna-molecules-in-bacteria-3. [Google Scholar]
- 47.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, et al. Genome-Wide Identification of Regulatory RNAs in the Human Pathogen Clostridium difficile. PLoS Genet. 2013;9. doi: 10.1371/journal.pgen.1003493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anjuwon-Foster BR, Tamayo R. A genetic switch controls the production of flagella and toxins in Clostridium difficile. PLoS Genet. 2017;13. doi: 10.1371/journal.pgen.1006701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baban ST, Kuehne SA, Barketi-Klai A, Cartman ST, Kelly ML, Hardie KR, et al. The Role of Flagella in Clostridium difficile Pathogenesis: Comparison between a Non-Epidemic and an Epidemic Strain. PLoS ONE. 2013;8. doi: 10.1371/journal.pone.0073026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Twine SM, Reid CW, Aubry A, McMullin DR, Fulton KM, Austin J, et al. Motility and flagellar glycosylation in Clostridium difficile. J Bacteriol. 2009;191:7050–62. doi: 10.1128/JB.00861-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dingle TC, Mulvey GL, Armstrong GD. Mutagenic analysis of the clostridium difficile flagellar proteins, flic and flid, and their contribution to virulence in hamsters. Infect Immun. 2011;79:4061–7. doi: 10.1128/IAI.05305-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poquet I, Saujet L, Canette A, Monot M, Mihajlovic J, Ghigo JM, et al. Clostridium difficile Biofilm: Remodeling metabolism and cell surface to build a sparse and heterogeneously aggregated architecture. Front Microbiol. 2018;9. doi: 10.3389/fmicb.2018.02084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, et al. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol. 2009;10. doi: 10.1186/gb-2009-10-9-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ternan NG, Jain S, Srivastava M, McMullan G. Comparative transcriptional analysis of clinically relevant heat stress response in Clostridium difficile strain 630. PLoS ONE. 2012;7. doi: 10.1371/journal.pone.0042410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Valiente E, Bouché L, Hitchen P, Faulds-Pain A, Songane M, Dawson LF, et al. Role of glycosyltransferases modifying type B flagellin of emerging hypervirulent Clostridium difficile lineages and their impact on motility and biofilm formation. J Biol Chem. 2016;291:25450–61. doi: 10.1074/jbc.M116.749523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedrido ME, de Oña P, Ramirez W, Leñini C, Goñi A, Grau R. et al. Spo0A links de novo fatty acid synthesis to sporulation and biofilm development in Bacillus subtilis. Mol Microbiol. 2013;87:348–67. doi: 10.1111/mmi.12102 [DOI] [PubMed] [Google Scholar]
- 57.Cuenot E, Garcia-Garcia T, Douche T, Gorgette O, Courtin P, Denis-Quanquin S, et al. The Ser/Thr Kinase PrkC participates in cell wall homeostasis and antimicrobial resistance in clostridium difficile. Infect Immun. 2019;87. doi: 10.1128/IAI.00005-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jain S, Smyth D, O’Hagan BMG, Heap JT, McMullan G, Minton NP, et al. Inactivation of the dnaK gene in Clostridium difficile 630 Δerm yields a temperature-sensitive phenotype and increases biofilm-forming ability. Sci Rep. 2017;7. doi: 10.1038/s41598-017-17583-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKee RW, Aleksanyan N, Garrett EM, Tamayo R. Type IV Pili Promote Clostridium difficile Adherence and persistence in a mouse model infection. Infect Immun. 2018:1–13. doi: 10.1128/IAI [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purcell EB, McKee RW, Bordeleau E, Burrus V, Tamayo R. Regulation of Type IV Pili contributes to surface behaviors of historical and epidemic strains of Clostridium difficile. J Bacteriol. 2016;198:565–77. doi: 10.1128/JB.00816-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Deligianni E, Pattison S, Berrar D, Ternan NG, Haylock RW, Moore JE, et al. Pseudomonas aeruginosa cystic fibrosis isolates of similar RAPD genotype exhibit diversity in biofilm forming ability in vitro. BMC Microbiol. 2010;10. doi: 10.1186/1471-2180-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dawson LF, Peltier J, Hall CL, Harrison MA, Derakhshan M, Shaw HA, et al. Extracellular DNA, cell surface proteins and c-di-GMP promote biofilm formation in Clostridioides difficile. Sci Rep. 2021;11. doi: 10.1038/s41598-020-78437-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fagan RP, Fairweather NF. Biogenesis and functions of bacterial S-layers. Nat Rev Microbiol. 2014:211–22. doi: 10.1038/nrmicro3213 [DOI] [PubMed] [Google Scholar]
- 64.Kirk JA, Banerji O, Fagan RP. Characteristics of the Clostridium difficile cell envelope and its importance in therapeutics. Microb Biotechnol. John Wiley and Sons Ltd; 2017:76–90. doi: 10.1111/1751-7915.12372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chapetón Montes D, Candela T, Collignon A, Janoir C. Localization of the Clostridium difficile cysteine protease Cwp84 and insights into its maturation process. J Bacteriol. 2011;193:5314–21. doi: 10.1128/JB.00326-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peltier J, Shaw HA, Couchman EC, Dawson LF, Yu L, Choudhary JS, et al. Cyclic diGMP Regulates Production of Sortase Substrates of Clostridium difficile and Their Surface Exposure through ZmpI Protease-mediated Cleavage. J Biol Chem. 2015;290:24453–69. doi: 10.1074/jbc.M115.665091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hensbergen PJ, Klychnikov OI, Bakker D, Dragan I, Kelly ML, Minton NP, et al. Clostridium difficile secreted Pro-Pro endopeptidase PPEP-1 (ZMP1/CD2830) modulates adhesion through cleavage of the collagen binding protein CD2831. FEBS Lett. 2015;589:3952–8. doi: 10.1016/j.febslet.2015.10.027 [DOI] [PubMed] [Google Scholar]
- 68.Peltier J, Shaw HA, Wren BW, Fairweather NF. Disparate subcellular location of putative sortase substrates in Clostridium difficile. Sci Rep. 2017;7. doi: 10.1038/s41598-017-08322-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tulli L, Marchi S, Petracca R, Shaw HA, Fairweather NF, Scarselli M, et al. Cbpa: A Novel Surface Exposed Adhesin Of Clostridium Difficile Targeting Human Collagen. Cell Microbiol. 2013;15:1674–87. doi: 10.1111/cmi.12139 [DOI] [PubMed] [Google Scholar]
- 70.Pothoulakis C. Effects of Clostridium difficile Toxins on Epithelial Cell Barrier. Ann N Y Acad Sci. 2000;915:347–56. doi: 10.1111/j.1749-6632.2000.tb05263.x [DOI] [PubMed] [Google Scholar]
- 71.Cohen SH, Tang YJ, Silva J. Analysis of the pathogenicity locus in Clostridium difficile strains. J Infect Dis. 2000;181:659–63. doi: 10.1086/315248 [DOI] [PubMed] [Google Scholar]
- 72.Govind R, Dupuy B. Secretion of Clostridium difficile toxins A and B requires the holin-like protein tcdE. PLoS Pathog. 2012;8. doi: 10.1371/journal.ppat.1002727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matamouros S, England P, Dupuy B. Clostridium difficile toxin expression is inhibited by the novel regulator TcdC. Mol Microbiol. 2007;64:1274–88. doi: 10.1111/j.1365-2958.2007.05739.x [DOI] [PubMed] [Google Scholar]
- 74.McKee RW, Mangalea MR, Purcell EB, Borchardt EK, Tamayo R. The second messenger cyclic Di-GMP regulates Clostridium difficile toxin production by controlling expression of sigD. J Bacteriol. 2013;195:5174–85. doi: 10.1128/JB.00501-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aubry A, Hussack G, Chen W, KuoLee R, Twine SM, Fulton KM, et al. Modulation of toxin production by the flagellar regulon in Clostridium difficile. Infect Immun. 2012;80:3521–32. doi: 10.1128/IAI.00224-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee ASY, Song KP. LuxS/autoinducer-2 quorum sensing molecule regulates transcriptional virulence gene expression in Clostridium difficile. Biochem Biophys Res Commun. 2005;335:659–66. doi: 10.1016/j.bbrc.2005.07.131 [DOI] [PubMed] [Google Scholar]
- 77.Ternan NG, Moore ND, Smyth D, McDougall GJ, Allwood JW, Verrall S, et al. Increased sporulation underpins adaptation of Clostridium difficile strain 630 to a biologically–relevant faecal environment, with implications for pathogenicity. Sci Rep. 2018;8. doi: 10.1038/s41598-018-35050-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dawson LF, Valiente E, Donahue EH, Birchenough G, Wren BW. Hypervirulent clostridium difficile pcr-ribotypes exhibit resistance to widely used disinfectants. PLoS ONE. 2011;6. doi: 10.1371/journal.pone.0025754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, et al. Global Analysis of the Sporulation Pathway of Clostridium difficile. PLoS Genet. 2013;9:1003660. doi: 10.1371/journal.pgen.1003660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, et al. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun. 2012;80:2704–11. doi: 10.1128/IAI.00147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio. 2010;1. doi: 10.1128/mBio.00035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.López D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in bacillus subtilis. Mol Microbiol. 2009;74:609–18. doi: 10.1111/j.1365-2958.2009.06882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Claverys JP, Håvarstein LS. Cannibalism and fratricide: Mechanisms and raisons d’être. Nat Rev Microbiol. 2007:219–29. doi: 10.1038/nrmicro1613 [DOI] [PubMed] [Google Scholar]
- 84.Tijerina-Rodríguez L, Villarreal-Treviño L, Baines SD, Morfín-Otero R, Camacho-Ortíz A, Flores-Treviño S, et al. High sporulation and overexpression of virulence factors in biofilms and reduced susceptibility to vancomycin and linezolid in recurrent Clostridium [Clostridioides] difficile infection isolates. PLoS ONE. 2019;14. doi: 10.1371/journal.pone.0220671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Erill I, Campoy S, Barbé J. Aeons of distress: An evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev. 2007;31:637–56. doi: 10.1111/j.1574-6976.2007.00082.x [DOI] [PubMed] [Google Scholar]
- 86.Vuotto C, Moura I, Barbanti F, Donelli G, Spigaglia P. Subinhibitory concentrations of metronidazole increase biofilm formation in Clostridium difficile strains. Pathog Dis. 2016;74. doi: 10.1093/femspd/ftv114 [DOI] [PubMed] [Google Scholar]
- 87.Hoffman LR, D’Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–5. doi: 10.1038/nature03912 [DOI] [PubMed] [Google Scholar]
- 88.Wu S, Li X, Gunawardana M, Maguire K, Guerrero-Given D, Schaudinn C, et al. Beta- lactam antibiotics stimulate biofilm formation in non-typeable Haemophilus influenzae by up-regulating carbohydrate metabolism. PLoS ONE. 2014;9. doi: 10.1371/journal.pone.0099204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kelley WL. Lex marks the spot: The virulent side of SOS and a closer look at the LexA regulon. Mol Microbiol. 2006:1228–38. doi: 10.1111/j.1365-2958.2006.05444.x [DOI] [PubMed] [Google Scholar]
- 90.Alotaibi B. The role of Clostridium difficile surface structures in virulence and gut colonization. Ulster University. 2017. [Google Scholar]
- 91.Jain S, Graham C, Graham RLJ, McMullan G, Ternan NG. Quantitative proteomic analysis of the heat stress response in Clostridium difficile strain 630. J Proteome Res. 2011;10:3880–90. doi: 10.1021/pr200327t [DOI] [PubMed] [Google Scholar]
- 92.Johnston JL, Sloan J, Fyfe JAM, Davies JK, Rood JI. The recA gene from Clostridium perfringens is induced by methyl methanesulphonate and contains an upstream Cheo box. Microbiology. 1997;143:885–90. doi: 10.1099/00221287-143-3-885 [DOI] [PubMed] [Google Scholar]
- 93.Fuqua WC, Winans SC, Greenberg EP. Quorum Sensing in Bacteria: the LuxR-LuxI Family of Cell Density-Responsive Transcriptional Regulators. J Bacteriol. 1994. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC205046/. doi: 10.1128/jb.176.2.269-275.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Proctor CR, McCarron PA, Ternan NG. Furanone quorum-sensing inhibitors with potential as novel therapeutics against Pseudomonas aeruginosa. J Med Microbiol. Microbiology Society. 2020:195–206. doi: 10.1099/jmm.0.001144 [DOI] [PubMed] [Google Scholar]
- 95.Vendeville A, Winzer K, Heurlier K, Tang K, Hardie K. Making “sense” of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nature. 2005;3:383–396. Available from: https://www.nature.com/articles/nrmicro1146. doi: 10.1038/nrmicro1146 [DOI] [PubMed] [Google Scholar]
- 96.Wang Y, Liu B, Grenier D, Yi L. Regulatory mechanisms of the LuxS/AI-2 system and bacterial resistance. Antimicrob Agents Chemother. 2019;63. doi: 10.1128/AAC.01186-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bassler BL, Wright M, Silverman MR. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol Microbiol. 1994;13:273–86. doi: 10.1111/j.1365-2958.1994.tb00422.x [DOI] [PubMed] [Google Scholar]
- 98.Semenyuk EG, Laning ML, Foley J, Johnston PF, Knight KL, Gerding DN, et al. Spore formation and toxin production in Clostridium difficile biofilms. PLoS ONE. 2014;9. doi: 10.1371/journal.pone.0087757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martin MJ, Clare S, Goulding D, Faulds-Pain A, Barquist L, Browne HP, et al. The agr locus regulates virulence and colonization genes in clostridium difficile 027. J Bacteriol. 2013;195:3672–81. doi: 10.1128/JB.00473-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Darkoh C, Odo C, Dupont HL. Accessory gene regulator-1 locus is essential for virulence and pathogenesis of Clostridium difficile. mBio. 2016;7. doi: 10.1128/mBio.01237-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. Cyclic diguanylate inversely regulates motility and aggregation in clostridium difficile. J Bacteriol. 2012;194:3307–16. doi: 10.1128/JB.00100-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bordeleau E, Fortier LC, Malouin F, Burrus V. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 2011;7. doi: 10.1371/journal.pgen.1002039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Simm R, Morr M, Remminghorst U, Andersson M, Römling U. Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal Biochem. 2009;386:53–8. doi: 10.1016/j.ab.2008.12.013 [DOI] [PubMed] [Google Scholar]
- 104.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol. 2007;66:206–19. doi: 10.1111/j.1365-2958.2007.05906.x [DOI] [PubMed] [Google Scholar]
- 105.Cairns LS, Hobley L, Stanley-Wall NR. Biofilm formation by Bacillus subtilis: New insights into regulatory strategies and assembly mechanisms. Mol Microbiol. Blackwell Publishing Ltd; 2014:587–598. doi: 10.1111/mmi.12697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Girinathan BP, Ou J, Dupuy B, Govind R. Pleiotropic roles of Clostridium difficile sin locus. PLoS Pathog. 2018;14. doi: 10.1371/journal.ppat.1006940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vuotto C, Donelli G, Buckley A, Chilton C. Clostridium difficile biofilm. Advances in Experimental Medicine and Biology. Springer New York LLC. 2018:97–115. doi: 10.1007/978-3-319-72799-8_7 [DOI] [PubMed] [Google Scholar]