Abstract
Motivation
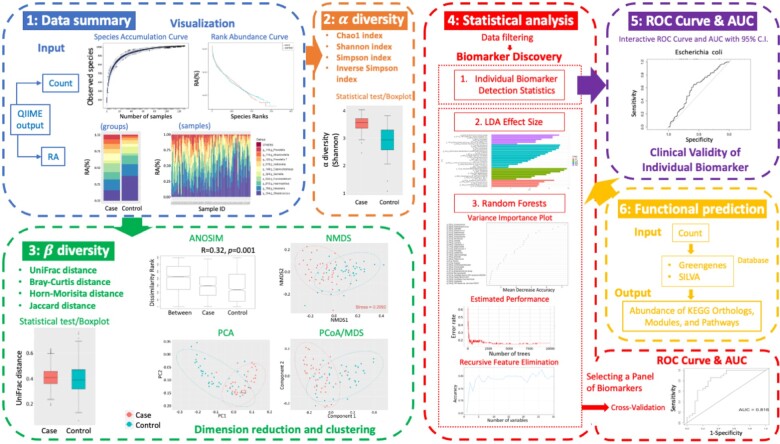
We proposed a wiSDOM (web-based inclusionary analysis Suite for Disease-Oriented Metagenomics) R Shiny application which comprises six functional modules: (i) initial visualization of sampling effort and distribution of dominant bacterial taxa among groups or individual samples at different taxonomic levels; (ii) statistical and visual analysis of α diversity; (iii) analysis of similarity (ANOSIM) of β diversity on UniFrac, Bray-Curtis, Horn-Morisita or Jaccard distance and visualizations; (iv) microbial biomarker discovery between two or more groups with various statistical and machine learning approaches; (v) assessment of the clinical validity of selected biomarkers by creating the interactive receiver operating characteristic (ROC) curves and calculating the area under the curve (AUC) for binary classifiers; and lastly (vi) functional prediction of metagenomes with PICRUSt or Tax4Fun.
Results
The performance of wiSDOM has been evaluated in several of our previous studies for exploring microbial biomarkers and their clinical validity as well as assessing the alterations in bacterial diversity and functionality. The wiSDOM can be customized and visualized as per users’ needs and specifications, allowing researchers without programming background to conduct comprehensive data mining and illustration using an intuitive browser-based interface.
Availability and implementation
The browser-based R Shiny interface can be accessible via (https://lun-ching.shinyapps.io/wisdom/) and freely available at (https://github.com/lunching/wiSDOM).
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Cumulative evidence from studies using the 16S rRNA amplicon sequencing technology in the past years has revealed that the compositional change in human microbiota is central to the development and progression of many human diseases (Cho and Blaser, 2012). Leading to a paradigm shift in clinical microbiology. Unlike shotgun metagenomics sequencing that is computationally laborious, 16S rRNA-based strategies, with fast turnaround, are cost-effective and applicable to high host DNA contamination (non-fecal samples). In these microbiome studies, raw sequence reads were initially processed and clustered into operational taxonomical units (OTUs) through a variety of well-developed tool suites, such as QIIME (Caporaso et al., 2010) and mothur (Schloss et al., 2009). Such OTU profiles could be highly dispersed and comprising many zeros, and in many analyses, the phylogenetic distance among these OTUs needs to be taken into account, posing enormous challenges in downstream data mining and presentation.
Current disease-oriented microbiome studies mostly aim to explore (i) the alterations in community compositions as measured by α diversity (within-sample diversity) or β diversity (between-sample diversity); (ii) discriminatory or differentially abundant microbes by classifying or comparative analyses; (iii) the clinical validity of potential microbial biomarkers and (iv) the functional changes in dysbiosis. Many statistical and machine learning approaches have been developed and publicly available for these investigations, however, these approaches aforementioned remain relatively sophisticated and require intensive research efforts due to being implemented by different programming languages such as R, Matlab, Python, etc. To address these burdens, we propose and implement a web-based inclusionary analysis Suite for Disease-Oriented Metagenomics, named wiSDOM, with browser-based R Shiny interface. Browser-based wiSDOM is freely available at the R Shiny server ‘shinyapps.io’ and can be downloaded and run locally at github repository (https://github.com/lunching/wiSDOM) for future update and maintenance. A user’s manual is provided in Supplementary Materials and wiSDOM github repository.
2 Program overview
Six functional modules are included to manage computational details in human microbiome research: (i) data summary; (ii) α diversity; (iii) β diversity; (iv) statistical analysis; (v) receiver operating characteristic (ROC) curve and area under the ROC curve (AUC) and (vi) functional prediction of metagenomes (Fig. 1). More information and function of each module are described in Supplementary Material. In addition, all R packages, their corresponding citations, and required input type of 16S rRNA data in each module are summarized in Supplementary Table S1.
Fig. 1.
Workflow and example output of wiSDOM
3 Discussions
Recent culture-free microbiome studies are largely exploratory, requiring intensive research efforts on data mining. Here, we present the web-based inclusionary analysis suite for disease-oriented metagenomics (wiSDOM), a free web platform with six function modules to manage advanced computational details in human microbiome research. wiSDOM is highly flexible for customization and can be broadly used without programming background, and the analyses in each module can be visualized as per users’ needs and specifications. Its performance has been evaluated in several of our previous studies for exploring microbial biomarkers and their clinical validity as well as assessing the alterations in bacterial diversity and functionality (Chiu et al., 2020; Su et al., 2020; Wu et al., 2020a,b,c). Due to limited space, a brief discussion regarding the comparisons of the wiSDOM with numerous freely available computational pipelines is provided in Supplementary Material.
To link dynamic bacterial consortia to human diseases, additional functions are needed to address several limitations of the present version. Although 16S rRNA amplicon sequencing appears to be the most commonly used strategy and has made important strides in clinical microbiology, deep shotgun metagenome sequencing usually offers more in-depth taxonomic characterization and functional insights into the human microbiomes than 16S rRNA-based methods. Currently, wiSDOM does not support taxonomy- and gene-centered analyses of shotgun data. Also unavailable is the integration of DADA2 (Callahan et al., 2016) to process sequencing reads into amplicon sequence variant (ASV) (Callahan et al., 2017) for subsequent data analysis and mining, although there remains a debate that ASVs should replace OTUs in marker gene analyses. Another issue is that wiSDOM does not provide correlation or association analysis as the connection between microbial communities and host factors has drawn more and more attention in microbiome studies (e.g. microbe-metabolite interaction). To fill these gaps, we intend to implement function modules to the wiSDOM in future releases.
Supplementary Material
Acknowledgements
The authors thank Mr. Rhian Resnick and Skyler Paulus for their aid with the use of high performance computing (HPC) service at Florida Atlantic University (FAU).
Funding
This work was supported by the National Institutes of Health [R01AG040211] and research grants from the Chang Gung Medical Foundation [CLRPG3J0012 and BMRPE97].
Conflict of Interest: none declared.
Contributor Information
Shih-Chi Su, Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung 204, Taiwan; Department of Dermatology, Drug Hypersensitivity Clinical and Research Center, Chang Gung Memorial Hospital, Linkou 333, Taiwan; Central Research Laboratory, XiaMen Chang Gung Hospital, Xiamen 361028, China.
James E. Galvin, Comprehensive Center for Brain Health, Department of Neurology, Miller School of Medicine, University of Miami, Miami, FL 33431, USA
Shun-Fa Yang, Institute of Medicine, Chung Shan Medical University, Taichung 402, Taiwan; Department of Medical Research, Chung Shan Medical University Hospital, Taichung 402, Taiwan.
Wen-Hung Chung, Whole-Genome Research Core Laboratory of Human Diseases, Chang Gung Memorial Hospital, Keelung 204, Taiwan; Department of Dermatology, Drug Hypersensitivity Clinical and Research Center, Chang Gung Memorial Hospital, Linkou 333, Taiwan.
Lun-Ching Chang, Department of Mathematical Sciences, Florida Atlantic University, Boca Raton, FL 33431, USA.
References
- Callahan B.J. et al. (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J, 11, 2639–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan B.J. et al. (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods, 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J.G. et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat. Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.Y. et al. (2020) Integration of metagenomics-metabolomics reveals specific signatures and functions of airway microbiota in mite-sensitized childhood asthma. Allergy, 75, 2846–2857. [DOI] [PubMed] [Google Scholar]
- Cho I., Blaser M.J. (2012) The human microbiome: at the interface of health and disease. Nat. Rev. Genet., 13, 260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D. et al. (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol., 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.C. et al. (2020) Oral microbial dysbiosis and its performance in predicting oral cancer. Carcinogenesis, doi: 10.1093/carcin/bgaa062. [DOI] [PubMed] [Google Scholar]
- Wu I.W. et al. (2020a) Integrative metagenomic and metabolomic analyses reveal severity-specific signatures of gut microbiota in chronic kidney disease. Theranostics, 10, 5398–5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu I.W. et al. (2020b) Compositional and functional adaptations of intestinal microbiota and related metabolites in CKD patients receiving dietary protein restriction. Nutrients, 12, 2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu I.W. et al. (2020c) Gut microbiota as diagnostic tools for mirroring disease progression and circulating nephrotoxin levels in chronic kidney disease: discovery and validation study. Int. J. Biol. Sci., 16, 420–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.