Abstrtact
Motivation
Viruses are ubiquitous in the living world, and their ability to infect more than one host defines their host range. However, information about which virus infects which host, and about which host is infected by which virus, is not readily available.
Results
We developed a web-based tool called the Viral Host Range database to record, analyze and disseminate experimental host range data for viruses infecting archaea, bacteria and eukaryotes.
Availability and implementation
The ViralHostRangeDB application is available from https://viralhostrangedb.pasteur.cloud. Its source code is freely available from the Gitlab instance of Institut Pasteur (https://gitlab.pasteur.fr/hub/viralhostrangedb).
1 Introduction
Viral genomic data are expanding, and their in silico analysis poses many challenges, including how to predict the likely host of a given virus (de Jonge et al., 2020; Dzunkova et al., 2019; Kieft et al., 2020; Li et al., 2020; Santiago-Rodriguez and Hollister, 2019). The gold standard for host identification remains the experimental evidence, which can take a long time and considerable effort to obtain. Four years passed between the prediction of Bacteroidetes as the putative host for crAssphage (the most abundant human gut bacteriophage) and the first experimental evidence that the strain Bacteroidetes intestinalis APC919/174 serves as a host for ϕcrAss001 (Dutilh et al., 2014; Shkoporov et al., 2018).
The GenBank (Sayers et al., 2019) database might be expected to provide information about the host of a virus, but these records mostly identify the host only to genus or species level, which is insufficient. For instance, the host indicated for bacteriophage T4 is the bacterium Escherichia coli, with no identification of a strain, which is as imprecise as indicating that human cells are the host for HIV-1. For a non-expert, such information suggests that any E. coli strain can be infected by bacteriophage T4, or that any human cell can be infected by HIV-1. Another public resource that could be used is the International Committee on Taxonomy of Viruses (ICTV) (Lefkowitz et al., 2018). However, host is not indicated in the data available from the ICTV website (talk.ictvonline.org). Finally, it is possible to search in microbial collections (ATCC; www.atcc.org, DSMZ; www.dsmz.de) the host associated with a deposited virus, but, unfortunately, these resources contain data for only limited numbers of published virus-host pairs.
Over and above the identification of a single host for virus propagation, virus–host range is another characteristic that is not readily available from public data sources. For viruses infecting multicellular organisms, including humans, in particular, the determination of host range is limited by the ability to grow cell lines. By contrast, for unicellular organisms, the number of hosts to be tested is very large, but unfortunately data are rarely published under an exploitable format. Interestingly, bacteriophage host range data are as old as the first article naming these viruses, published in 1917 by d’Herelle, in which bacteriophages infecting a Shiga strain were reported to be unable to infect Flexner or Hiss strains (d'Herelle, 1917).
For decades, viral host range tests were routinely performed for the typing of bacteria (Sabat et al., 2013; Sechter et al., 2000). Nowadays, host ranges are being determined for an increasing number of bacteriophages to identify candidates for phage therapy. This treatment for bacterial infections was originally proposed in 1917, and is used regularly in some countries (Georgia, Poland) (d'Herelle, 1917; Kutateladze, 2015). Its use is now expanding worldwide to treat infections caused by antibiotic-resistant pathogens (Corbellino et al., 2019; Dedrick et al., 2019; Jennes et al., 2017; Schooley et al., 2017). Consequently, semi-automated systems for high-throughput host range tests have been developed (http://www.aphage.com/the-science/). However, only the small number of positive outputs from these tests are finally used, with the bulk of the information obtained discarded and, thus, unavailable.
Another major challenge is the integration of host range data into a single searchable and analysis tool. Viral host range data are, by definition, a variable, which should be regenerated dynamically following the acquisition of new data.
2 Materials and methods
2.1 Data availability
The ViralHostRangeDB application is available from https://viralhostrangedb.pasteur.cloud. Its source code is freely available from the Gitlab instance of Institut Pasteur (https://gitlab.pasteur.fr/hub/viralhostrangedb), under the terms of the MIT license, together with detailed documentation (https://hub.pages.pasteur.fr/viralhostrangedb/) including instructions for use, deployment and administration purposes. A demonstration server can be run directly from a docker image (https://hub.docker.com/r/viralhostrangedb/demo), providing a way of testing all features of the application, including the privileges and (in)visibility of private data sources.
2.2 Architecture
The architecture of the ViralHostRangeDB web application is based on the Django Web Framework and the PostgreSQL database. Data are displayed, on the server side, in the Django REST framework. This environment provides efficient and safe data storage as well as tight control access. The application, its database and routine processes (backup, email notifications, virus/host identifier analysis, etc.), are hosted on a Kubernetes cluster (https://kubernetes.io/), providing high availability, scalability and fail-over. The global software quality of the application is ensured through unit test scenarios covering 99% of the code base.
2.3 Importing data
Any authenticated user can contribute datasets via the top menu. Datasets can be uploaded as Excel files as detailed in the online documentation (https://hub.pages.pasteur.fr/viralhostrangedb/compatible_file.html). Excel data files are imported with the Pandas and xlrd Python packages (McKinney, 2017). During the mapping of the responses of a file onto the global scheme, the thresholds suggested to users are calculated with the NumPy (Oliphant, 2006) and Scikit-learn (Pedregosa et al., 2011) packages. The NCBI identifiers describing the host and virus strains are validated with Entrez web services (Sayers et al., 2020) which are queried with the BioPython (Cock et al., 2009) package.
2.4 Privacy
The access to uploaded datasets can be finely controlled, by restricting it to the uploader only, sharing it with a specific set of other users, or making it public. It is also possible to set permissions for the edition of a dataset for each user. Private data sources can be accessed only by explicitly authorized users, regardless of whether the user is a curator or a privileged administrator. To secure edition operations on the datasets, all modifications are logged and stored in histories, to allow rollback.
2.5 Search tool
The web interface allows the interrogation of datasets. A ‘search module’, accessible either through a quick search box or through a specific advanced search page, can be used to discover datasets through full text and specific filters (e.g. host or virus names, contributor, publication, etc.). The exploration module, accessible from the top menu or from the search results, provides the main functionality of the application: the ability to compare the responses of any number of hosts to any number of viruses, across all the datasets accessible.
3 Results
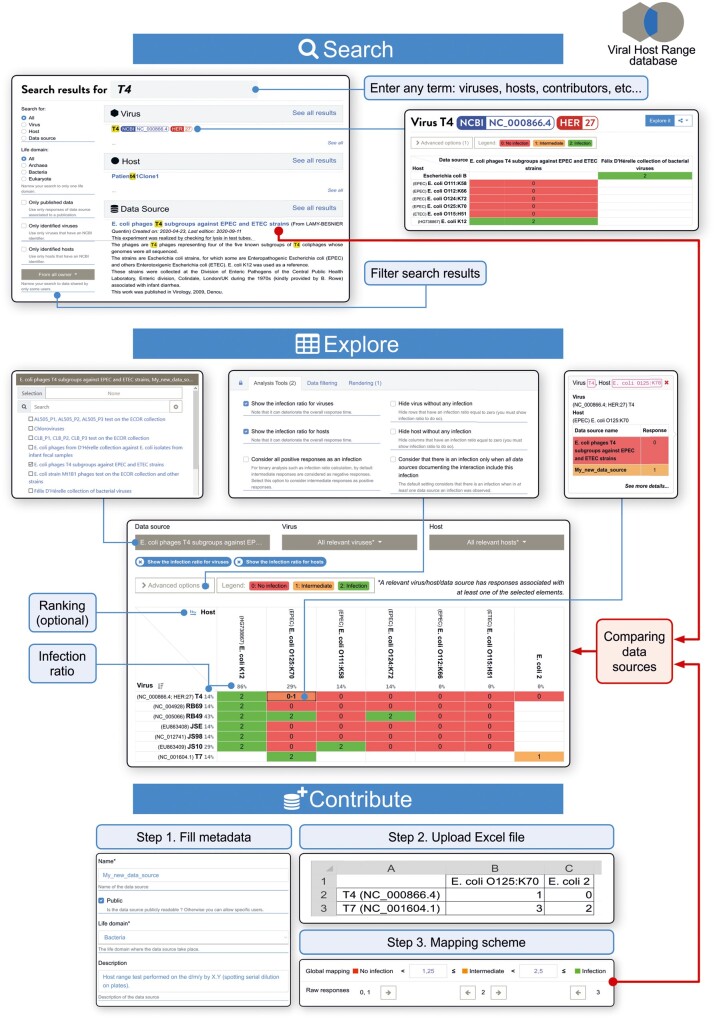
We circumvented the challenges associated with virus–host range analysis, by designing the Viral Host Range database (VHRdb, https://viralhostrangedb.pasteur.cloud/), which compiles experimental host range data provided by contributors. This open web-based resource can be used to explore and analyze publicly accessible data with a powerful search engine that scans data and metadata (virus or host names, contributor name, location, GenBank accession number, etc.). Not only can users find a virus, but they can also immediately identify the set of hosts on which it has been tested, across all the available data. Filters, analysis and display settings can facilitate rapid visualization of the most relevant information, such as the highest host range score or the most susceptible host (Fig. 1). Importantly, when discrepancies between datasets are detected, they are highlighted and direct access is provided to the source data, for further investigations.
Fig. 1.
Diagram presenting the main functionalities of the Viral Host Range database. The top panel (Search) introduces the search tool and links to subsequent information. The bottom panel (Contribute) presents the main steps that contributors must achieve to record new data. Shown in the middle panel (Explore) is an example of results obtained from dataset comparison, using the datasets selected from the searched results displayed in the top panel and the newly contributed data displayed in the bottom panel (red arrows). Main tools and options to select, rank and display data are also indicated
We designed a user-guided process for uploading data compatible with the VHRdb mapping tool, to facilitate comparisons of datasets. This mapping tool is the cornerstone of VHRdb, translating the contributor’s original (numerical) data into a unified ranking system. The mapping tool was designed to allow each contributor to classify the results of virus–host interaction tests into a maximum of three responses: ‘0’, for ‘no infection’; ‘2’ for ‘infection’ and ‘1’ for ‘intermediate’, corresponding to any interaction that is different from ‘0’ and ‘2’. Then, contributors can readily compare their results with publicly available datasets (curated by administrators to ensure that the database remains homogeneous). If kept private, data are neither accessible to, nor curated by administrators. Analysis across a restricted number of datasets is also possible, to focus on specificities associated with one or several viruses or hosts.
Another issue affecting the accurate appreciation of a virus–host range is the lack of precise characterizations of tested hosts. In particular, most of clinical isolates used to determine the host range of bacteriophages for phage therapy applications are not sequenced. In addition, viruses themselves evolve over time and adapt their host range to the available hosts (Rothenburg and Brennan, 2020). The VHRdb therefore handles GenBank accession numbers for both viruses and hosts, as a solution to provide unique identifiers.
In addition to the identification of suitable hosts for viruses and the cross-analysis of experimental tests, we anticipate that the VHRdb will become a resource for the development of machine learning approaches, which require large amounts of data, to improve the prediction of the host of a virus, or even the receptor that it uses (Leite et al., 2018; Young et al., 2020). It could also be used more directly by clinicians, who will increasingly have access to the genome sequences of pathogens. If the strain infecting a patient is closely related to a tested strain present in the VHRdb, candidate bacteriophages are immediately identified, shortening the time required to develop an appropriate treatment. The VHRdb will also provide opportunities to address fundamental questions in virology, from ecological dynamics to the molecular mechanisms underlying virus–host interactions.
The VHRdb is a unique, publicly accessible resource for the community of microbial virologists, for the rapid identification, characterization and dissemination of data for virus–host interactions of broad interest to the educational, scientific and medical communities and to private sector entities developing applications.
At the time of publication, the VHRdb holds 15 753 interactions obtained from 739 viruses infecting 1 664 archaeal, bacterial or protist hosts, including the entire Felix d’Herelle collection of bacteriophages.
Acknowledgements
We warmly thank Roger Carlson, David Dunigan, Mart Krupovic, Sylvain Moineau, Marie-Agnès Petit, Catherine Schouler and Denise Tremblay, and all current and former members of the laboratory of L. Debarbieux for contributing data to the VHRdb. We thank the IT Department of Institut Pasteur, including Thomas Menard, in particular, for providing access to the Kubernetes cluster and initial training. We thank Jean-François Charles for assistance in designing the figure. Q.L.-B. is funded by the École Doctorale FIRE—Programme Bettencourt.
Funding
This work has been supported by grant ANR-19-AMRB-0002 to L.D.
Conflict of Interest: none declared.
Contributor Information
Quentin Lamy-Besnier, Bacteriophage, Bacterium, Host Laboratory, Department of Microbiology, Institut Pasteur, Paris F-75015, France; Université de Paris, Paris, France.
Bryan Brancotte, Hub de Bioinformatique et Biostatistique, Département Biologie Computationnelle, Institut Pasteur, Paris F-75015, France.
Hervé Ménager, Hub de Bioinformatique et Biostatistique, Département Biologie Computationnelle, Institut Pasteur, Paris F-75015, France.
Laurent Debarbieux, Bacteriophage, Bacterium, Host Laboratory, Department of Microbiology, Institut Pasteur, Paris F-75015, France.
References
- Cock P.J. et al. (2009) Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics, 25, 1422–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbellino M. et al. (2019) Eradication of a multi-drug resistant, carbapenemase-producing Klebsiella pneumoniae isolate following oral and intra-rectal therapy with a custom-made, lytic bacteriophage preparation. Clin. Infect. Dis., 70, 1998–2001. [DOI] [PubMed] [Google Scholar]
- d'Herelle F. (1917) Sur un microbe invisible antagoniste des bacilles dysentériques. C. R. Acad. Sci. Paris, 165, 373–375. [Google Scholar]
- Dedrick R.M. et al. (2019) Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat Med., 25, 730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge P.A. et al. (2020) Adsorption sequencing as a rapid method to link environmental bacteriophages to hosts. iScience, 23, 101439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilh B.E. et al. (2014) A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun., 5, 4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzunkova M. et al. (2019) Defining the human gut host-phage network through single-cell viral tagging. Nat. Microbiol., 4, 2192–2203. [DOI] [PubMed] [Google Scholar]
- Jennes S. et al. (2017) Use of bacteriophages in the treatment of colistin-only-sensitive Pseudomonas aeruginosa septicaemia in a patient with acute kidney injury-a case report. Crit. Care, 21, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft K. et al. (2020) VIBRANT: automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome, 8, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutateladze M. (2015) Experience of the Eliava Institute in bacteriophage therapy. Virol. Sin., 30, 80–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz E.J. et al. (2018) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res., 46, D708–D717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite D.M.C. et al. (2018) Computational prediction of inter-species relationships through omics data analysis and machine learning. BMC Bioinform., 19, 420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. et al. (2020) A deep learning-based method for identification of bacteriophage-host interaction. IEEE/ACM Trans. Comput. Biol. Bioinform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W. (2017) Python for Data Analysis: Data Wrangling with Pandas, NumPy, and IPython. O'Reilly Media, Newton, MA. [Google Scholar]
- Oliphant T.E. (2006) A Guide to NumPy. Trelgol Publishing. [Google Scholar]
- Pedregosa F. et al. (2011) Scikit-learn: machine learning in Python. J. Mach. Learn. Res., 12, 2825–2830. [Google Scholar]
- Rothenburg S., Brennan G. (2020) Species-specific host-virus interactions: implications for viral host range and virulence. Trends Microbiol., 28, 46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabat A.J. et al. (2013) Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill., 18, 20380. [DOI] [PubMed] [Google Scholar]
- Santiago-Rodriguez T.M., Hollister E.B. (2019) Human virome and disease: high-throughput sequencing for virus discovery, identification of phage-bacteria dysbiosis and development of therapeutic approaches with emphasis on the human gut. Viruses, 11, 656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W. et al. (2019) GenBank. Nucleic Acids Res., 47, D94–D99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayers E.W. et al. (2020) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 48, D9–D16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R.T. et al. (2017) Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob. Agents Chemother., 61, e00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechter I. et al. (2000) Twenty-three years of Klebsiella phage typing: a review of phage typing of 12 clusters of nosocomial infections, and a comparison of phage typing with K serotyping. Clin. Microbiol. Infect., 6, 233–238. [DOI] [PubMed] [Google Scholar]
- Shkoporov A.N. et al. (2018) PhiCrAss001 represents the most abundant bacteriophage family in the human gut and infects Bacteroides intestinalis. Nat. Commun., 9, 4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young F. et al. (2020) Predicting host taxonomic information from viral genomes: a comparison of feature representations. PLoS Comput. Biol., 16, e1007894. [DOI] [PMC free article] [PubMed] [Google Scholar]