Abstract
Background—
Left ventricular noncompaction (LVNC) can occur in isolation or can co-occur with a cardiomyopathy phenotype or cardiovascular malformation. The yield of cardiomyopathy gene panel testing in infants, children, and adolescents with a diagnosis of LVNC is unknown. By characterizing a pediatric population with LVNC, we sought to determine the yield of cardiomyopathy gene panel testing, distinguish the yield of testing for LVNC with or without co-occurring cardiac findings, and define additional factors influencing genetic testing yield.
Methods and Results—
One hundred twenty-eight individuals diagnosed with LVNC at ≤21 years of age were identified, including 59% with idiopathic pathogenesis, 32% with familial disease, and 9% with a syndromic or metabolic diagnosis. Overall, 75 individuals had either cardiomyopathy gene panel (n=65) or known variant testing (n=10). The yield of cardiomyopathy gene panel testing was 9%. The severity of LVNC by imaging criteria was not associated with positive genetic testing, co-occurring cardiac features, pathogenesis, family history, or myocardial dysfunction. Individuals with isolated LVNC were significantly less likely to have a positive genetic testing result compared with those with LVNC and co-occurring cardiomyopathy (0% versus 12%, respectively; P<0.01).
Conclusions—
Genetic testing should be considered in individuals with cardiomyopathy co-occurring with LVNC. These data do not suggest an indication for cardiomyopathy gene panel testing in individuals with isolated LVNC in the absence of a family history of cardiomyopathy.
Keywords: cardiomyopathies, genetic testing, infant, pediatrics, phenotype
Left ventricular noncompaction (LVNC) is characterized by prominent myocardial trabeculations in a thick noncompacted layer adjacent to a thin compacted layer. LVNC has historically been thought to result from arrest of normal ventricular compaction of the myocardium that takes place during cardiogenesis.1 More recent data suggest additional pathogenic bases, including acquired forms of LVNC.2 LVNC has been reported in association with heart failure, arrhythmias, and embolic events and can present in isolation (iLVNC) or in combination with other types of cardiomyopathy (LVNC/cardiomyopathy) or cardiovascular malformation (LVNC/CVM).3–5 LVNC is recognized as a distinct form of cardiomyopathy by the American Heart Association but is not classified as such by the World Health Organization or the European Society of Cardiology.4 A growing body of evidence describing the extreme variability of the LVNC morphological spectrum has suggested that LVNC may be an anatomic variant of left ventricular (LV) structure rather than a disease in and of itself.2,6 In addition to the ongoing debate about what constitutes the LVNC phenotype, there remains uncertainty about a genetic contribution. Previous studies have identified variants in sarcomeric genes in 29% to 41% of primarily adult cohorts with LVNC.7–10 These studies, however, used various definitions for iLVNC, and many did not account for the co-occurrence of hypertrophic or dilated cardiomyopathy (DCM). Furthermore, these studies were in cohorts that were primarily adults, and the clinical genetic testing yield for children with LVNC remains uncertain. LVNC is also a known feature of several genetic syndromes, including Barth syndrome and chromosome 1p36 deletion syndrome, but the prevalence of familial, metabolic, and syndromic causes of LVNC in children and youth is largely unknown.11,12 The hypothesis of this study was that genetic testing yield would be lower among individuals with iLVNC compared with those with LVNC/cardiomyopathy. The objectives of this study were to characterize a pediatric population with LVNC, determine the yield of cardiomyopathy gene panel testing, distinguish the yield of testing by associated cardiac features (LVNC subtype), and define additional factors influencing genetic testing yield.
Methods
A retrospective medical record review of individuals evaluated at Cincinnati Children’s Hospital Medical Center between July 1, 2009, and December 31, 2012, was performed with approval from the Cincinnati Children’s Hospital Medical Center Institutional Review Board. Only those patients aged ≤21 years with a diagnosis of LVNC based on echocardiographic criteria, and confirmed by review of the cardiology clinic assessment, were included. Individuals with the following LVNC subtypes were included: (1) iLVNC; (2) co-occurring with hypertrophic cardiomyopathy (HCM), restrictive cardiomyopathy, or DCM; or (3) co-occurring with CVM. Individuals with cardiomyopathy secondary to chemotherapy, myocarditis, or environmental toxins were excluded. Study data were collected and managed using REDCap research electronic data capture tools hosted at Cincinnati Children’s Hospital Medical Center.13
Because there continues to be a lack of consensus about the diagnostic criteria of LVNC and concern about both the specificity and sensitivity of various approaches, we classified cases for analysis into 2 groups: stringent or standard imaging criteria. Both groups met published diagnostic criteria for LVNC at the time of the study. Stringent imaging criteria was defined as a 2 layered structure with a thin compact layer and a thick noncompact layer where the noncompact to compact ratio is >2:1 at end systole in the parasternal short axis, numerous deep trabeculations with blood are demonstrated in the recesses by color Doppler, and >3 trabeculations are visible in a single plane.14,15 Standard imaging criteria for LVNC was defined as any 2 of these 3 criteria. For all eligible subjects with a diagnosis of LVNC, the most recent internal echocardiographic images were reviewed by an echocardiographer. Older images were reviewed as needed for complete assessment. Figure 1 provides representative echocardiographic images for stringent and standard imaging criteria for LVNC.
Figure 1.

Imaging classifications in left ventricular noncompaction (LVNC). Echocardiographic images of an individual with a normal heart (A and D), an individual meeting standard imaging criteria for LVNC (B and E), and an individual meeting stringent imaging criteria for LVNC (C and F), as shown in the apical 4-chamber (A through C) and parasternal short-axis (D through F) views. The myocardium is excessively trabeculated with a thin compact layer in stringent cases, whereas the dimension of the noncompact layer and the degree of thinning of the compact layer varies in standard cases. In the example shown here, the standard case did not satisfy the ratio criteria (both the standard and stringent examples demonstrated blood flow between trabeculations by color Doppler, data not shown).
The cohort was analyzed as a whole and stratified into 3 groups for analysis. LVNC subtype groups included (1) iLVNC, (2) LVNC/cardiomyopathy, and (3) LVNC/CVM. Individuals with iLVNC met the echocardiographic criteria outlined above and did not have LV dilation or hypertrophy diagnostic of a cardiomyopathy phenotype, such as DCM or HCM. Individuals with iLVNC could have had unexplained systolic dysfunction. All individuals with CVM were included in the LVNC/CVM group, including those with a co-occurring cardiomyopathy. Individuals with CVM were grouped separately based on the most widely cited diagnostic criteria for LVNC, which requires the absence of coexisting cardiac anomalies.15 CVM was classified using a modified version of the National Birth Defects Prevention Study classification scheme.16,17
All patients underwent clinical cardiac and genetics evaluation. Evaluation included at least 1 visit with a clinical geneticist or genetic counselor. Demographic and clinical data, including presenting symptoms, medical history, cardiac testing (echocardiogram, ECG, Holter monitor, exercise test, and cardiac magnetic resonance imaging), family history, and referral indication, were collected. Individuals were considered to have myocardial dysfunction, specifically LV systolic dysfunction, if the ejection fraction z score was ≤−2 by echocardiography.
Genetic testing was ordered as clinically indicated and became increasingly comprehensive during the duration of the study. Gene panel testing was requested from 1 of 4 clinical testing laboratories and included HCM, DCM, and DCM/LVNC gene panels ranging in size from 11 to 38 genes. Table I in the Data Supplement provides further description of genes included in sequencing. We use the term cardiomyopathy gene testing to refer to variant, single gene, or gene panel testing for cardiomyopathy. Pathogenic or likely-pathogenic variants were considered positive genetic testing results. A variant of uncertain significance was not considered a positive test result. Likewise, a benign or presumed benign variant was considered negative. All gene variants were reviewed and interpretations confirmed or revised as indicated based on current clinical laboratory interpretation and American College of Medical Genetics and Genomics 2015 Standards and Guidelines.18 The yield of genetic testing was defined as the proportion of subjects who had positive genetic testing compared with the number of subjects who completed genetic testing.
Subjects were assigned a pathogenic category, including familial, syndromic, or idiopathic using categories set forth by the Pediatric Cardiomyopathy Registry.19 The familial classification applies to affected probands with documented cardiomyopathy (iLVNC, LVNC/cardiomyopathy, HCM, DCM, or restrictive cardiomyopathy) in a first-degree relative at the time of genetic testing. The syndromic group includes individuals with metabolic disease (inborn errors of metabolism or mitochondrial disease) or clinical criteria and dysmorphology for well-characterized genetic syndromes or patients with genetic testing identifying a syndromic cause. Individuals not meeting any of the above criteria were considered idiopathic. Family history data, including family history of sudden cardiac death, CVM, and cardiomyopathy, was also collected.
Statistical Analysis
Data analysis began by computing descriptive statistics for all relevant variables in the data set. Because of the exploratory nature of the study, all models specified here were tested at the unadjusted α=0.05 level. Data were analyzed using SAS v9.3.
We hypothesized that the LVNC imaging criteria of stringent or standard would be related to 5 specific cardiac disease characteristics. These characteristics were individually tested using contingency table analyses. Because of cell count variations, 3 characteristics (LVNC subphenotype, family history of cardiomyopathy, and myocardial dysfunction) were tested using traditional χ2 methods, and 2 characteristics (cardiomyopathy known variant testing results and pathogenesis) were tested using Fisher Exact Tests.
Our primary hypothesis, that gene variant status could be predicted from selected cardiac disease characteristics for children and youth with LVNC, was tested using Fisher Exact Tests. The outcome of interest was presence or absence of a positive cardiomyopathy gene panel test result, among those who had gene panel testing. The 6 characteristics tested included LVNC subtype, electrophysiology phenotype, pathogenesis, family history of sudden death, LVNC imaging features (stringent or standard), and myocardial dysfunction.
Results
Retrospective chart review identified 151 individuals with a diagnosis of LVNC at age ≤21 years. Echocardiographic images and reports confirming a diagnosis of LVNC were not available for review and did not meet LVNC criteria for 23 individuals, and these individuals were excluded from further analysis. In total, 128 individuals (70 men, 58 women) from 120 different families (8 sets of siblings) comprised the final cohort. Table 1 summarizes the clinical characteristics. The population was primarily white (72%) or black (26%) and non-Hispanic (98%). The cardiac diagnoses included 61 (48%) iLVNC, 42 (33%) LVNC/cardiomyopathy, and 25 (20%) LVNC/CVM. Within the LVNC/CVM group, 10 individuals also had a co-occurring cardiomyopathy. The vast majority of patients had an idiopathic (59%) or familial (32%) pathogenesis, whereas 9% had a syndromic or metabolic diagnosis. Ninety-four patients (73%) were diagnosed as having LVNC before age of 13 years. Individuals with iLVNC presented at older ages (77% presenting between ages 6 and 21) compared with individuals with LVNC/cardiomyopathy or LVNC/CVM (29% and 24%, respectively, presenting after the age of 6 years). Overall, 23% of the entire cohort and 30% of those with iLVNC presented for echocardiography secondary to a family history of sudden cardiac death, HCM, DCM, or LVNC. Clinical symptoms and examination findings prompted cardiac imaging in 54% of the individuals with iLVNC. Many individuals had >1 indication prompting referral for echocardiography. Symptoms included sudden cardiac arrest, syncope, chest pain, shortness of breath, and failure to thrive; physical examination findings included auscultation of a murmur and identification of an abnormal heart rhythm, including irregular heart beat and bradycardia. Ten individuals with iLVNC had cardiac imaging for other various indications, including follow-up of abnormal routine prenatal ultrasound, abnormal ECG screening before starting stimulant medication, systemic hypertension, genetic condition known to be associated with risk for cardiomyopathy, and prematurity.
Table 1.
Clinical Characteristics According to Cardiac Diagnosis
| Characteristic | All (n=128) | iLVNC (n=61) | LVNC/CM (n=42) | LVNC/CVM (n=25) |
|---|---|---|---|---|
| Sex, n (%) | ||||
| Men | 70 (55) | 35 (57) | 24 (57) | 11 (44) |
| Women | 58 (45) | 26 (43) | 18 (43) | 14 (56) |
| Age at presentation, n (%), y | ||||
| <1 | 36 (28) | 8 (13) | 23 (55) | 5 (20) |
| 1 to <6 | 27 (21) | 6 (10) | 7 (17) | 14 (56) |
| 6 to <13 | 31 (24) | 21 (35) | 6 (14) | 4 (16) |
| 13 to <18 | 30 (24) | 24 (39) | 4 (9) | 2 (8) |
| 18–21 | 4 (3) | 2 (3) | 2 (5) | 0 |
| Referral indication, n (%) | ||||
| Symptoms/examination findings | 75 (59) | 33 (54) | 28 (67) | 14 (56) |
| Family history of SCA/D or CM | 30 (23) | 18 (30) | 8 (19) | 4 (16) |
| Other | 23 (18) | 10 (16) | 6 (14) | 7 (28) |
| Second opinion | 41 (32) | 13 (21) | 21 (50) | 7 (28) |
| Patient status, n (%) | ||||
| Deceased | 2 (2) | 0 | 1 (2) | 1 (4) |
| Pathogenesis at evaluation, n (%) | ||||
| Idiopathic | 75 (59) | 36 (59) | 28 (67) | 11 (44) |
| Familial | 41 (32) | 21 (34) | 9 (21) | 11 (44) |
| Syndromic/metabolic | 12 (9) | 4 (7) | 5 (12) | 3 (12) |
| Family history, n (%) | ||||
| Cardiomyopathy | 33 (26) | 18 (30) | 8 (19) | 7 (28) |
| Sudden cardiac death | 19 (15) | 11 (18) | 6 (14) | 2 (8) |
| Cardiovascular malformation | 11 (9) | 5 (8) | 1 (2) | 5 (20) |
| LVNC designation, n (%) | ||||
| Stringent | 73 (57) | 35 (57) | 20 (48) | 18 (72) |
| Standard | 55 (43) | 26 (43) | 22 (52) | 7 (28) |
| Surgical repair of CVM | 7 (28) | |||
| Myocardial dysfunction, n (%) | 46 (36) | 10 (16) | 30 (71) | 6 (24) |
| Sudden cardiac arrest, n (%) | 3 (2) | 1 (2) | 1 (2) | 1 (4) |
| Cardiac transplant, n (%) | 1 (1) | 0 | 1 (2) | 0 |
| Syncope, n (%) | 14 (11) | 10 (16) | 3 (7) | 1 (4) |
| ICD present, n (%) | 7 (5) | 2 (3) | 5 (12) | 0 |
| Genetics evaluation | ||||
| Genetic counselor | 125 (98) | 59 (97) | 42 (100) | 24 (96) |
| Geneticist | 112 (88) | 48 (79) | 41 (98) | 23 (92) |
ICD indicates implantable cardioverter-defibrillator; iLVNC, isolated left ventricular noncompaction; LVNC/CM, left ventricular noncompaction with cardiomyopathy; LVNC/CVM, left ventricular noncompaction with cardiovascular malformation; and SCA/D, sudden cardiac arrest/death.
LVNC Diagnosis
Review of imaging identified that 57% of subjects met stringent imaging criteria for diagnosis of LVNC, whereas 43% fulfilled standard criteria. Genetic testing results (positive versus variant of uncertain significance/negative), cardiac subtype (iLVNC, LVNC/cardiomyopathy, and LVNC/CVM), pathogenesis (familial, idiopathic, syndromic, and metabolic), family history of cardiomyopathy, and myocardial dysfunction were compared between groups (Table 2). These analyses demonstrated that the clinical features analyzed do not vary with the stringency of applied imaging criteria.
Table 2.
Association of LVNC Imaging Features With Patient Clinical Characteristics
| Characteristic | Stringent Imaging Criteria (n=73) | Standard Imaging Criteria (n=55) | P Value |
|---|---|---|---|
| CM gene panel testing, n (%) | 40 (55) | 25 (45) | 0.48 |
| Positive | 9 (23) | 4 (16) | |
| Negative | 31 (77) | 21 (84) | |
| LVNC subphenotype, n (%) | 0.15 | ||
| iLVNC | 35 (48) | 26 (47) | |
| LVNC/CM | 20 (27) | 22 (40) | |
| LVNC/CVM | 18 (25) | 7 (13) | |
| Pathogenesis, n (%) | 0.65 | ||
| Familial | 20 (27) | 21 (38) | |
| Idiopathic | 45 (61) | 30 (54) | |
| Syndromic | 4 (6) | 2 (4) | |
| Metabolic | 4 (6) | 2 (4) | |
| Family history of CM,* n (%) | 0.45 | ||
| Present | 18 (25) | 15 (27) | |
| Absent | 54 (74) | 33 (60) | |
| Unknown | 1 (1) | 7 (13) | |
| Myocardial dysfunction, n (%) | 0.30 | ||
| Present | 29 (40) | 17 (31) | |
| Absent | 44 (60) | 38 (69) | |
iLVNC indicates isolated left ventricular noncompaction; LVNC/CM, left ventricular noncompaction with cardiomyopathy; and LVNC/CVM, left ventricular noncompaction with cardiovascular malformation.
Defined as a first- or second-degree relative with cardiomyopathy.
Genetic Testing and Results
Genetic testing for the cohort is summarized in Figure 2. Subjects were categorized as having syndromic, metabolic, familial, or idiopathic disease on their initial evaluation. An underlying genetic condition described previously in association with LVNC was identified in 9% of individuals. All 12 of these individuals had ≥1 genetic test, including but not limited to chromosome microarray, Noonan panel, and mitochondrial DNA sequencing (Figure 2). Based on this testing, 4 individuals were diagnosed with mitochondrial disease and 3 had a chromosome abnormality, including 1 individual with 1p36 deletion syndrome. Five individuals had a genetic syndrome, including Barth syndrome, malonic acidemia, and Noonan syndrome (Table II in the Data Supplement). Of the 116 subjects with an idiopathic or familial pathogenesis, 65 (56%) underwent cardiomyopathy gene panel testing, and an additional 10 (9%) had known familial variant testing. Overall, 17% (13/75) of individuals had a positive cardiomyopathy genetic testing result, and 33% (25/75) had a variant of uncertain significance. Likely-pathogenic and pathogenic variants were identified in genes reported previously in association with LVNC, including MYH7, MYBPC3, TPM1, and TNNT2. The specific genetic variants identified in the 13 patients with positive gene panel and known variant testing are summarized in Table 3. Of note, 3 individuals with iLVNC had positive known variant testing, all of whom had a family history of cardiomyopathy.
Figure 2.
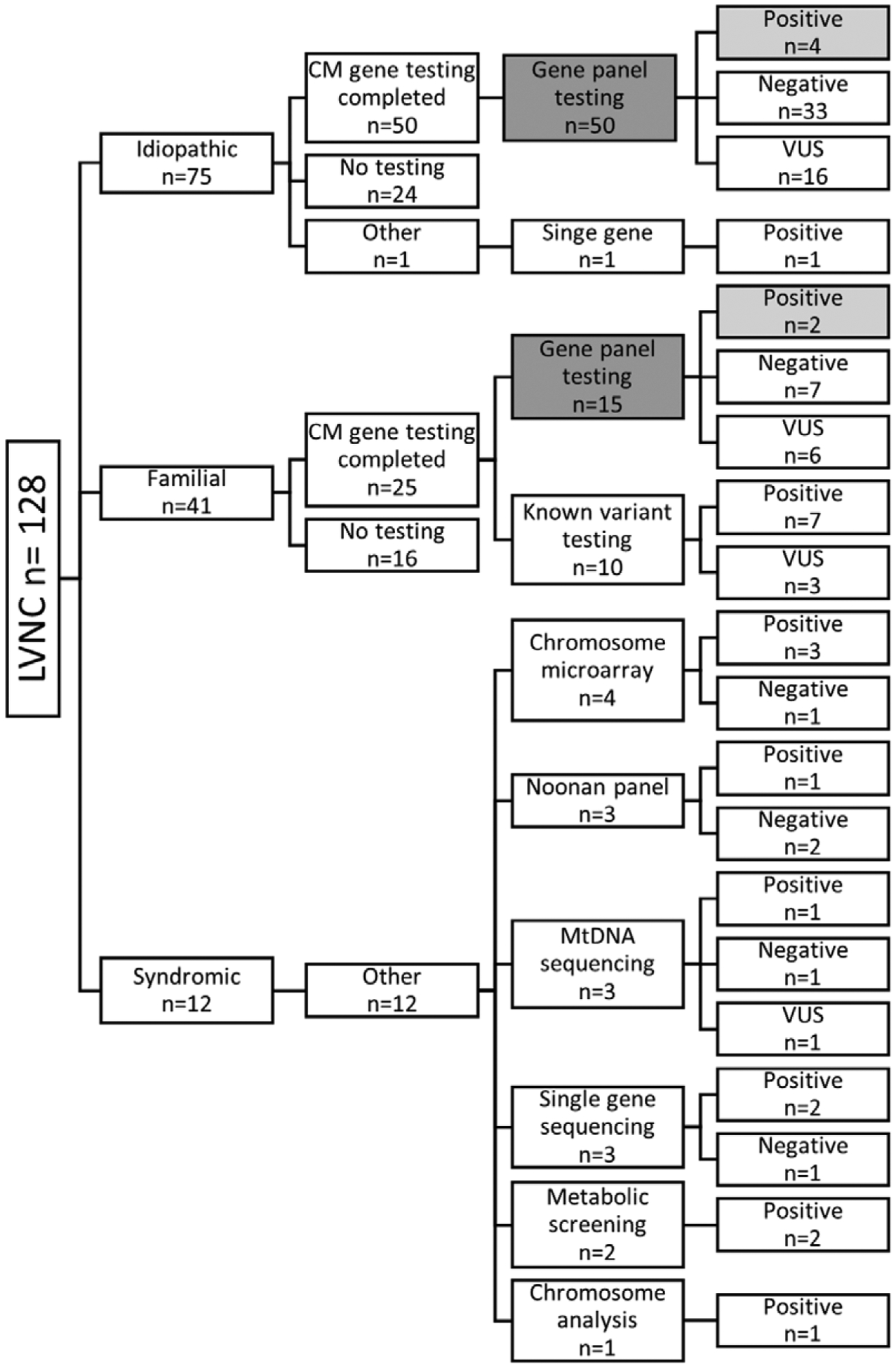
Genetic testing and results by pathogenesis. The data included in cardiomyopathy (CM) gene panel yield calculation is shown with the number of cardiomyopathy gene panels indicated by dark gray (n=65) and the number of positive results in light gray (n=6). Within the syndromic group, many of the 12 individuals had multiple genetic tests. LVNC indicates left ventricular noncompaction; and VUS, variant of uncertain significance.
Table 3.
Cardiomyopathy Gene Testing Results: Likely-Pathogenic and Pathogenic Variants
| Gene | Protein Variant | Genomic Variant | LVNC Subtype | CM Pathogenesis | Cardiac Phenotype | CM Gene Testing |
|---|---|---|---|---|---|---|
| MYH7 | Arg403Gln | 1208G>A | LVNC/CVM | Familial | HCM, septal | CM gene panel |
| MYH7 | Arg904Cys | 2710C>T | LVNC/CM | Idiopathic | DCM | CM gene panel |
| MYBPC3 | Trp702Valfs | 2373dupG | LVNC/CVM | Idiopathic | HCM, septal | CM gene panel |
| MYBPC3 | 3330+2T>G | LVNC/CM | Familial | HCM | CM gene panel | |
| MYBPC3 | 3330+5G>C | LVNC/CVM | Idiopathic | DCM/HCM, TAA | CM gene panel | |
| TPM1 | Glu272Gly | 815A>G | LVNC/CM | Idiopathic | DCM | CM gene panel |
| MYH7 | Lys184Gln | 550A>C | Isolated | Familial | LVNC | CM known variant |
| MYH7 | Lys184Gln | 550A>C | Isolated | Familial | LVNC | CM known variant |
| MYH7 | Arg294Gln | 746G>A | LVNC/CM | Familial | HCM | CM known variant |
| MYH7 | Arg403Gln | 1208G>A | LVNC/CM | Familial | HCM | CM known variant |
| MYH7 | Glu930Gln | 2788G>C | LVNC/CM | Familial | HCM | CM known variant |
| MYH7 | Glu930Gln | 2788G>C | LVNC/CM | Familial | HCM | CM known variant |
| TNNT2 | Lys210del | 629_631delAGA | Isolated | Familial | LVNC | CM known variant |
CM indicates cardiomyopathy; CVM, cardiovascular malformation; DCM, dilated cardiomyopathy; HCM, hypertrophic cardiomyopathy; LVNC, left ventricular noncompaction; and TAA, thoracic aortic aneurysm.
The yield of cardiomyopathy gene panel results is summarized in Table 4. The overall yield of gene panel testing was 9% with the highest rate among the LVNC/CVM group (30%), followed by the LVNC/cardiomyopathy group (12%). None of the individuals with iLVNC had a likely-pathogenic or pathogenic variant identified by gene panel testing. Of the 6 likely-pathogenic and pathogenic variants identified, 5 were absent from the Exome Aggregation Consortium, and the MYH7 variant, p.Arg904Cys (c.2710C>T), was identified in 1 individual resulting in a minor allele frequency of 8.238e-06.20 Of the 10 individuals with LVNC/CVM who had gene panel testing, 7 had a co-occurring cardiomyopathy. All 3 of the individuals who tested positive from this group had a co-occurring cardiomyopathy, specifically HCM.
Table 4.
Cardiomyopathy Gene Panel Yield*
| Testing and Results | All (n=116) | iLVNC (n=57) | LVNC/CM (n=37) | LVNC/CVM (n=22) |
|---|---|---|---|---|
| Cardiomyopathy gene panel, n (%) | 65 (56) | 30 (53) | 25 (60) | 10 (45) |
| Negative | 40 | 22 | 14 | 4 |
| VUS | 21 | 8 | 8† | 5† |
| Positive | 6 (9) | 0 (0) | 3 (12) | 3 (30) |
iLVNC indicates isolated left ventricular noncompaction; LVNC/CM, left ventricular noncompaction with cardiomyopathy; LVNC/CVM, left ventricular noncompaction with cardiovascular malformation; and VUS, variant of uncertain significance.
Excludes syndromic and metabolic cases.
Two individuals from the LVNC/CVM group with a VUS also had a positive result.
Because we grouped individuals with both CVM and cardiomyopathy in the LVNC/CVM group, we also calculated the yield of cardiomyopathy gene panel testing regardless of CVM status. The yield of testing with co-occurring cardiomyopathy (LVNC/cardiomyopathy [n=25] and LVNC/CVM with co-occurring cardiomyopathy [n=7]) was 19%.
Of the 41 individuals who did not have cardiomyopathy gene panel testing, 1 had primary arrhythmia testing and was found to have a RYR2 exon 3 deletion.21 The remaining 40 subjects did not have any cardiomyopathy gene testing. Genetic testing was not recommended for 23 (58%) individuals, including 13 (57%) with iLVNC, 3 (13%) with LVNC/cardiomyopathy, and 7 (30%) with LVNC/CVM; 13 (33%) had gene panel testing completed in a relative; 4 (10%) declined genetic testing or were lost to follow-up. Of the 13 who had gene panel testing completed in a relative, 12 relatives had negative genetic testing and 1 had variant of uncertain significance result.
Factors Influencing Cardiomyopathy Gene Panel Testing Yield
Cardiac disease characteristics’ influence on genetic testing yield was evaluated. The cardiovascular phenotype (iLVNC, LVNC/CVM, and LVNC/cardiomyopathy) was the only statistically significant predictor of positive gene panel results, and individuals with iLVNC were significantly less likely to have a positive genetic testing result compared with the LVNC/cardiomyopathy group (0% versus 12%, respectively; P<0.01). None of the other variables evaluated, including the presence of an electrophysiology phenotype (P=0.23), disease pathogenesis (P=0.32), family history of sudden cardiac death (P=0.57), stringent/standard imaging designation (P=0.64), or the presence of myocardial dysfunction (P=0.66), resulted in statistically significant influence on genetic testing yield.
Discussion
The Heart Rhythm Society and European Heart Rhythm Association guideline on genetic testing for the cardiomyopathies state that genetic testing for LVNC can be useful—a class IIa recommendation—however, specific guidelines for the occurrence of LVNC in isolation or with cardiomyopathy do not exist.22 Although data have suggested a genetic association for LVNC, most of the studies investigating genetic testing yield were in the adult population and did not clearly account for the co-occurrence of LVNC with cardiomyopathy or CVM.7,9 To our knowledge, the current study represents the largest cohort of pediatric aged individuals with LVNC for whom genetic testing and yield is reported.
In this study, none of the individuals with iLVNC had a positive cardiomyopathy gene panel result. This is in contrast to previous reports (41% and 29%, respectively).7,9 Hoedemakers et al reported on the yield of cardiomyopathy gene sequencing (16 sarcomeric and cytoskeletal genes) in 58 unrelated individuals, including 8 children, with iLVNC. Of the probands who completed genetic testing, 41% were found to have a variant interpreted to be pathogenic9. Although 24 individuals in this study had a diagnosis of heart failure, the pathogenic basis of the heart failure was not provided, suggesting that for some cases of LVNC, an associated cardiomyopathy may not have been specified. A second study published in 2011 identified a pathogenic variant in 1 of 8 sarcomeric genes (MYH7, ACTC1, TNNT2, TNNI3, MYL2, MYL3, TPM1, and MYBPC3) in 29% (18/63) of adult patients with a diagnosis of LVNC.7 The reasons for the discrepancy in yield between this study and these 2 prior studies may be the co-occurrence of LVNC with cardiomyopathy. Although Probst et al defined the cohort as isolated, the average ejection fraction among both mutation-positive and mutation-negative individuals was 38%, and the average LV diameter was 60 mm, which would be considered mild to moderately enlarged depending on sex. A difference in the genes included on various panels is a consideration; however, all of the genes in which variants were identified in this pediatric cohort were sequenced by Hoedemakers et al. Variant interpretation remains a challenge with not uncommon discrepancy between testing laboratories, and it is likely that some variants interpreted previously as pathogenic would no longer be considered clinically significant based on current criteria.18 The differences in yield between studies may also be because of inherent limitations in assigning a clinical diagnoses of LVNC and further support the need for improved diagnostic approaches, which incorporate clinical findings beyond imaging characterization of LV trabeculation.
Prior efforts to characterize the degree of LV trabeculation in healthy individuals have shown that varying degrees of LV trabeculation are common among control populations.23,24 All patients in our study who met imaging criteria for LVNC, however, were further subdivided based on fulfillment of stringent or standard imaging criteria in an attempt to refine the threshold for a clinical diagnosis of LVNC. The LVNC imaging findings were not a predictor of genetic testing yield, and the 2 groups did not differ across additional clinical variables. These findings suggest that the degree of LV trabeculation (once diagnostic criteria for LVNC are met) is not a predictor of genetic testing yield. These findings are in agreement with cardiac magnetic resonance studies in LVNC, which have shown that the degree of LV trabeculation lacks prognostic impact beyond more commonly known risk factors, such as LV dilation and systolic dysfunction.6
Up to 12% of individuals with LVNC are reported to have an additional, structural cardiac defect.3,25 There is some suggestion that individuals with CVM co-occurring with LVNC have poor postsurgical outcomes compared with individuals with CVM alone.26 The co-occurrence of LVNC with other cardiomyopathy phenotypes has also been reported. The Pediatric Cardiomyopathy Registry described 155 pediatric aged individuals with LVNC; 23% had iLVNC and 77% had LVNC/cardiomyopathy.27 In our single-site study of a pediatric cohort, 48% had iLVNC, 33% had LVNC/cardiomyopathy, and 20% had LVNC/CVM. Isolated LVNC was more prevalent in our cohort than reported previously. The distribution of subtypes was similar among the groups meeting stringent and standard imaging criteria for LVNC.
Our study limitations include those inherent to the challenges associated with the clinical diagnosis of LVNC and interpretation of gene variants. We tried to reduce potential for false-positives by echocardiographer systematic evaluation of images for LVNC criteria and confirmation of LVNC diagnosis. In addition, all individuals considered for inclusion were previously given a clinical diagnosis of LVNC by a pediatric cardiologist with heart failure expertise. Variant interpretation not uncommonly differs between clinical testing laboratories even when using current variant classification criteria.18,28 All genetic variants identified previously in this cohort were reviewed by a clinical testing laboratory and interpreted based on current published guidelines and available data.18 Despite these efforts, it is possible that some of the variants may be reclassified as additional data become available. In addition, all individuals with CVM were included in the LVNC/CVM group, including those with co-occurring cardiomyopathy. Thus, these data cannot adequately address the yield of cardiomyopathy gene panel testing in patients with LVNC and CVM without co-occurring cardiomyopathy. The genetic testing results in this study may reflect a lower limit of diagnostic yield because gene panels continue to expand (Table I in the Data Supplement). In addition, individuals did not have sequencing of genes encoding for proteins involved with other cardiac development processes. Mouse models have demonstrated the importance of the Notch signaling pathway in cardiac ventricular maturation, including trabeculation and compaction.29,30 MIB1 encodes a protein that functions as an E3 ubiquitin ligase and positively regulates Notch signaling. Two families with LVNC have been found to have variants in MIB1 that segregate with the phenotype in the family, suggesting a potential role of genes described previously in cardiovascular development and CVM with LVNC.31 Further investigation of genes important for cardiac development, cell cycle, and proliferation may provide insight into the genetics of LVNC.
Conclusion
In this pediatric cohort with LVNC, the majority of individuals had an idiopathic pathogenesis at the time of presentation, but 32% had familial disease and 9% had an underlying metabolic or syndromic genetic condition confirming the importance of a broad differential diagnosis, particularly in individuals diagnosed at young ages or with additional medical history suggestive of a unifying diagnosis. The only predictor of cardiomyopathy gene panel yield was the co-occurrence of LVNC with cardiomyopathy. None of the individuals with iLVNC had a positive gene panel result, whereas 12% of individuals with LVNC co-occurring with cardiomyopathy did. Two individuals with iLVNC had positive known variant testing, and both had a family history of cardiomyopathy. The absence of positive cardiomyopathy gene panel results in iLVNC suggests that LVNC may represent a benign anatomic variant in the absence of other cardiovascular disease findings. Our data suggest strong consideration of genetic testing for individuals with cardiomyopathy and co-occurring LVNC and for individuals with iLVNC and a family history of cardiomyopathy.
Supplementary Material
CLINICAL PERSPECTIVE.
Left ventricular noncompaction (LVNC) is characterized by prominent myocardial trabeculations in a thick noncompacted layer adjacent to a thin compacted layer. A growing body of evidence has suggested that LVNC may be an anatomic variant of left ventricular structure rather than a disease in and of itself. In addition to the ongoing debate about what constitutes the LVNC phenotype, there remains uncertainty about a genetic contribution with multiple pathogenic bases, including both genetic and acquired causes reported. These uncertainties leave healthcare providers with little to guide approach to clinical genetic testing for LVNC. We sought to characterize a pediatric population with LVNC, determine the yield of cardiomyopathy gene panel testing, and distinguish the yield of testing by LVNC subtype. LVNC subtype groups included (1) isolated LVNC, (2) LVNC with co-occurring cardiomyopathy, and (3) LVNC with co-occurring cardiovascular malformation. The yield of gene panel testing was 9% with the highest rate among the LVNC/cardiovascular malformation group (30%), followed by the LVNC/cardiomyopathy group (12%). Likely-pathogenic and pathogenic variants were identified in genes reported previously in association with LVNC (MYH7, MYBPC3, TPM1, and TNNT2). None of the individuals with LVNC isolation had a likely-pathogenic or pathogenic variant identified by gene panel testing. Individuals with LVNC isolation were significantly less likely to have a positive genetic testing result compared with the LVNC/cardiomyopathy group (0% versus 12%, respectively; P<0.01). Our data suggest strong consideration of genetic testing for individuals with cardiomyopathy and co-occurring LVNC and for individuals with LVNC and a family history of cardiomyopathy.
Acknowledgments
We thank the patients and their families.
Sources of Funding
Funding for this study was supported by the Heart Institute at Cincinnati Children’s Hospital Medical Center. This study was also supported, in part, by the Center for Clinical and Translational Science and Training Grant Award 1UL1TR001425-01 (REDCap).
Footnotes
The Data Supplement is available at http://circgenetics.ahajournals.org/lookup/suppl/doi:10.1161/CIRCGENETICS.117.001735/-/DC1.
Disclosures
None.
References
- 1.Gupta V, Poss KD. Clonally dominant cardiomyocytes direct heart morphogenesis. Nature. 2012;484:479–484. doi: 10.1038/nature11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arbustini E, Weidemann F, Hall JL. Left ventricular noncompaction: a distinct cardiomyopathy or a trait shared by different cardiac diseases? J Am Coll Cardiol. 2014;64:1840–1850. doi: 10.1016/j.jacc.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stähli BE, Gebhard C, Biaggi P, Klaassen S, Valsangiacomo Buechel E, Attenhofer Jost CH, et al. Left ventricular non-compaction: prevalence in congenital heart disease. Int J Cardiol. 2013;167:2477–2481. doi: 10.1016/j.ijcard.2012.05.095. [DOI] [PubMed] [Google Scholar]
- 4.Maron BJ, Towbin JA, Thiene G, Antzelevitch C, Corrado D, Arnett D, et al. ; American Heart Association; Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; Council on Epidemiology and Prevention. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 5.Biagini E, Ragni L, Ferlito M, Pasquale F, Lofiego C, Leone O, et al. Different types of cardiomyopathy associated with isolated ventricular noncompaction. Am J Cardiol. 2006;98:821–824. doi: 10.1016/j.amjcard.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 6.Andreini D, Pontone G, Bogaert J, Roghi A, Barison A, Schwitter J, et al. Long-term prognostic value of cardiac magnetic resonance in left ventricle noncompaction: a prospective multicenter study. J Am Coll Cardiol. 2016;68:2166–2181. doi: 10.1016/j.jacc.2016.08.053. [DOI] [PubMed] [Google Scholar]
- 7.Probst S, Oechslin E, Schuler P, Greutmann M, Boyé P, Knirsch W, et al. Sarcomere gene mutations in isolated left ventricular noncompaction cardiomyopathy do not predict clinical phenotype. Circ Cardiovasc Genet. 2011;4:367–374. doi: 10.1161/CIRCGENETICS.110.959270. [DOI] [PubMed] [Google Scholar]
- 8.Klaassen S, Probst S, Oechslin E, Gerull B, Krings G, Schuler P, et al. Mutations in sarcomere protein genes in left ventricular non-compaction. Circulation. 2008;117:2893–2901. doi: 10.1161/CIRCULATIONAHA.107.746164. [DOI] [PubMed] [Google Scholar]
- 9.Hoedemaekers YM, Caliskan K, Michels M, Frohn-Mulder I, van der Smagt JJ, Phefferkorn JE, et al. The importance of genetic counseling, DNA diagnostics, and cardiologic family screening in left ventricular non-compaction cardiomyopathy. Circ Cardiovasc Genet. 2010;3:232–239. doi: 10.1161/CIRCGENETICS.109.903898. [DOI] [PubMed] [Google Scholar]
- 10.Dellefave LM, Pytel P, Mewborn S, Mora B, Guris DL, Fedson S, et al. Sarcomere mutations in cardiomyopathy with left ventricular hyper-trabeculation. Circ Cardiovasc Genet. 2009;2:442–449. doi: 10.1161/CIRCGENETICS.109.861955. [DOI] [PubMed] [Google Scholar]
- 11.Shieh JT. Implications of genetic testing in noncompaction/hypertrabeculation. Am J Med Genet C Semin Med Genet. 2013;163C:206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kindel SJ, Miller EM, Gupta R, Cripe LH, Hinton RB, Spicer RL, et al. Pediatric cardiomyopathy: importance of genetic and metabolic evaluation. J Card Fail. 2012;18:396–403. doi: 10.1016/j.cardfail.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stöllberger C, Finsterer J. Left ventricular hypertrabeculation/non-compaction. J Am Soc Echocardiogr. 2004;17:91–100. doi: 10.1016/S0894-7317(03)00514-5. [DOI] [PubMed] [Google Scholar]
- 15.Jenni R, Oechslin E, Schneider J, Attenhofer Jost C, Kaufmann PA. Echocardiographic and pathoanatomical characteristics of isolated left ventricular non-compaction: a step towards classification as a distinct cardiomyopathy. Heart. 2001;86:666–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinton RB, McBride KL, Bleyl SB, Bowles NE, Border WL, Garg V, et al. Rationale for the cytogenomics of cardiovascular malformations consortium: a phenotype intensive registry based approach. J Cardiovasc Dev Dis. 2015;2:76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Botto LD, Lin AE, Riehle-Colarusso T, Malik S, Correa A; National Birth Defects Prevention Study. Seeking causes: classifying and evaluating congenital heart defects in etiologic studies. Birth Defects Res A Clin Mol Teratol. 2007;79:714–727. doi: 10.1002/bdra.20403. [DOI] [PubMed] [Google Scholar]
- 18.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ; ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grenier MA, Osganian SK, Cox GF, Towbin JA, Colan SD, Lurie PR, et al. Design and implementation of the North American Pediatric Cardiomyopathy Registry. Am Heart J. 2000;139(2 pt 3):S86–S95. [DOI] [PubMed] [Google Scholar]
- 20.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. ; Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell MJ, Czosek RJ, Hinton RB, Miller EM. Exon 3 deletion of ryanodine receptor causes left ventricular noncompaction, worsening catecholaminergic polymorphic ventricular tachycardia, and sudden cardiac arrest. Am J Med Genet A. 2015;167A:2197–2200. doi: 10.1002/ajmg.a.37140. [DOI] [PubMed] [Google Scholar]
- 22.Ackerman MJ, Priori SG, Willems S, Berul C, Brugada R, Calkins H, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA). Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 23.André F, Burger A, Loßnitzer D, Buss SJ, Abdel-Aty H, Gianntisis E, et al. Reference values for left and right ventricular trabeculation and noncompacted myocardium. Int J Cardiol. 2015;185:240–247. doi: 10.1016/j.ijcard.2015.03.065. [DOI] [PubMed] [Google Scholar]
- 24.Tizón-Marcos H, de la Paz Ricapito M, Pibarot P, Bertrand O, Bibeau K, Le Ven F, et al. Characteristics of trabeculated myocardium burden in young and apparently healthy adults. Am J Cardiol. 2014;114:1094–1099. doi: 10.1016/j.amjcard.2014.07.025. [DOI] [PubMed] [Google Scholar]
- 25.Agarwal A, Khandheria BK, Paterick TE, Treiber SC, Bush M, Tajik AJ. Left ventricular noncompaction in patients with bicuspid aortic valve. J Am Soc Echocardiogr. 2013;26:1306–1313. doi: 10.1016/j.echo.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Ramachandran P, Woo JG, Ryan TD, Bryant R, Heydarian HC, Jefferies JL, et al. The impact of concomitant left ventricular non-compaction with congenital heart disease on perioperative outcomes. Pediatr Cardiol. 2016;37:1307–1312. doi: 10.1007/s00246-016-1435-2. [DOI] [PubMed] [Google Scholar]
- 27.Jefferies JL, Wilkinson JD, Sleeper LA, Colan SD, Lu M, Pahl E, et al. ; Pediatric Cardiomyopathy Registry Investigators. Cardiomyopathy phenotypes and outcomes for children with left ventricular myocardial non-compaction: results from the Pediatric Cardiomyopathy Registry. J Card Fail. 2015;21:877–884. doi: 10.1016/j.cardfail.2015.06.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet Med. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Bücker S, Jungblut B, Böttger T, Cinnamon Y, Tchorz J, et al. Inhibition of Notch2 by Numb/Numblike controls myocardial compaction in the heart. Cardiovasc Res. 2012;96:276–285. doi: 10.1093/cvr/cvs250. [DOI] [PubMed] [Google Scholar]
- 30.Chen H, Zhang W, Sun X, Yoshimoto M, Chen Z, Zhu W, et al. Fkbp1a controls ventricular myocardium trabeculation and compaction by regulating endocardial Notch1 activity. Development. 2013;140:1946–1957. doi: 10.1242/dev.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luxán G, Casanova JC, Martínez-Poveda B, Prados B, D’Amato G, Mac-Grogan D, et al. Mutations in the NOTCH pathway regulator MIB1 cause left ventricular noncompaction cardiomyopathy. Nat Med. 2013;19:193–201. doi: 10.1038/nm.3046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


