Abstract
A multiplex reverse transcription-PCR method was optimized to monitor the duration of excretion of Sabin poliovirus strains in stools of vaccinees following administration of the first dose of the trivalent oral vaccine. The assay detected approximately 1 50% tissue culture infective dose of each poliovirus serotype spiked into cell culture media. Although PCR inhibitors were frequently encountered in the stool specimens, a 1:20 dilution of the extracted RNA was sufficient to obtain a positive PCR result. Analysis of 195 stool specimens collected from 26 vaccinees showed that poliovirus types 1, 2, and 3 were identified more frequently by PCR than by tissue culture isolation. The percentages of specimens positive by PCR for poliovirus types 1, 2, and 3 were 67.2, 82.6, and 53.8, respectively. In contrast, the culture method identified types 1, 2, and 3 virus in 55.4, 64.1, and 27.7% of the samples, respectively. Poliovirus type 2 excretion was detected by PCR in practically all of the oral poliovirus vaccine recipients for 4 to 8 weeks following vaccination. In contrast, excretion of type 1 and 3 viruses was more variable, with a range of 1 to 8 weeks. Shedding of type 3 virus ceased in ∼70% of vaccinees within a week after immunization. In addition to an enhanced sensitivity for the detection of poliovirus, this PCR method permits the direct characterization of virus in stool specimens without further passage in culture, which may select for genetic variants that may not accurately reflect the virus composition in the original specimen.
The live trivalent oral poliovirus (PV) vaccine (OPV) derived from the attenuated Sabin PV strains has been proven to be safe and efficacious in preventing the transmission of wild-type PV, resulting in the elimination of practically all cases of poliomyelitis in the United States (34). Despite the overwhelming success of the live PV vaccine in reducing morbidity and mortality, an average of eight cases of vaccine-associated paralytic poliomyelitis (VAPP) were reported each year between 1980 and 1994 in the United States (28). Type 2 and 3 PVs have been implicated in 90% of VAPP cases. It is well documented that the Sabin OPV strains undergo genetic changes upon replication in the gut of the vaccine recipient (for a review, see reference 13). In some instances, the genetic change is a reversion in the nucleotide sequence to that found in the wild-type progenitor. One of the best-characterized attenuating mutations in the Sabin strains is located in the 5′ noncoding region (5′ NCR) (for a review, see reference 20). The link between reversion at this locus in virus shed from vaccinees and VAPP is not clear. Reversion of the attenuated nucleotide in the 5′ NCR has been identified in type 1 (14, 27), type 2 (19), and type 3 (12) PV strains (PV1, PV2, and PV3, respectively) isolated from patients with VAPP. In contrast, vaccine-like viruses exhibiting the reverted 5′ NCR nucleotide are routinely recovered from stools from healthy vaccinees who do not develop paralytic disease (3, 7, 10, 35).
As the sole manufacturer of OPV in the United States, investigators at Wyeth-Lederle Vaccines and Pediatrics are interested in the duration of shedding and genetic characterization of vaccine-related viruses shed in the stools of infants that have been fed OPV. Currently, the identification and characterization of PV in stool specimens collected from OPV vaccinees rely on virus isolation in susceptible tissue culture cells (23). A second culture step in which virus is neutralized in the presence of serotype-specific antiserum pools is usually required to identify the serotypes of the viral isolates. A drawback of the culture method is that amplification in culture may select for a virus whose genetic composition is not representative of that of the virus in the original stool specimen. Another disadvantage of the culture method is that it can take anywhere from 1 to 3 weeks to obtain a result. We wanted to use a rapid reverse transcription (RT)-PCR method to identify PV serotypes directly in stool samples collected from OPV recipients enrolled in an ongoing vaccine study. Most of the RT-PCR assays in use at the time that our study began used conserved 5′ NCR primers that are generic for all members of the enterovirus genus or that do not differentiate PVs (1, 4, 15, 30, 41). PCR assays that distinguish PV from non-PV enteroviruses (2, 11, 26) and that differentiate Sabin and wild-type PV strains (32) have been reported, but most of these methods rely on a tissue culture step.
A multiplex RT-PCR method that allows the simultaneous identification of Sabin PV1, PV2, and PV3 vaccine strains in a single reaction has been reported (39). Contemporary wild-type PV strains are not detected. The PCR primers map to the region of the PV genome encoding the amino terminus of the VP1 capsid protein just upstream of the major antigenic site. Nucleotide sequence heterogeneity in this region among the three Sabin PV serotypes allows discrimination. The PCR assay was reported to be highly sensitive for the detection of purified Sabin viral RNA, but when this assay was applied to the detection of PV directly in clinical specimens, 21% of the culture-positive samples produced a negative PCR result. This outcome was attributed in part to the components of some stool samples that inhibit PCR amplification. We chose to optimize this PCR assay for the direct detection of vaccine-related PV in stool specimens and to implement it to monitor the duration of PV excretion in first-dose OPV recipients who were participants in an ongoing vaccine study. In this report, we describe assay modifications that significantly improved the detection of PV in stool samples known to contain PCR inhibitors. A comparative analysis of stool samples collected from first-dose OPV recipients over 8 weeks revealed that PCR was more sensitive than culture for the detection of PV1, PV2, and PV3. The enhanced sensitivity of detection of PV in stool specimens, coupled with a rapid time to the retrieval of assay results, makes this RT-PCR method an attractive alternative to culture for the identification of Sabin-related PV vaccine strains in stool specimens.
MATERIALS AND METHODS
Study population and specimen collection.
Subjects consisted of infants from an ongoing PV vaccine study. Twenty-two infants received their first dose of OPV (Orimune; Lederle Laboratories, Pearl River, N.Y.) at age 2 months and seven infants received OPV at age 5 months. The OPV contained 106.0 to 107.0, 105.1 to 106.1, and 105.8 to 106.8 50% tissue culture infective doses (TCID50s) of Sabin PV1, PV2, and PV3 per dose, respectively. Two stool specimens, one for culture and the other for PCR analysis, were collected in fecalyzer units (Evsco Pharmaceuticals, Buena, N.J.). Stool specimens were obtained from the study subjects before (day 0) and on days 1, 2, 3, 4, 7, 14, 21, 28, 42, and 56 following OPV administration. The specimens were kept frozen until processing. Approximately 260 stool specimens from 29 vaccinees were available for testing, but not all of the specimens met the criteria for inclusion in the analyses whose results are presented in Tables 3 and 4.
TABLE 3.
Proportion of PV-positive stool specimens from OPV vaccinees detected by RT-PCR and culture
| Virus | No. of specimens positive/no. of specimens tested (%)
|
|
|---|---|---|
| RT-PCR | Culture | |
| PV1 | 131/195 (67.2) | 108/195 (55.4) |
| PV2 | 161/195 (82.6) | 125/195 (64.1) |
| PV3 | 105/195 (53.8) | 54/195 (27.7) |
| Negative | 20/195 (10.3) | 50/195 (25.6) |
TABLE 4.
Proportion of OPV vaccinees shedding PV by RT-PCR and culture at the indicated day postvaccinationa
| Virus | No. of specimens positive for shedding/total no. of specimens tested (%)
|
|||
|---|---|---|---|---|
| RT-PCR
|
Culture
|
|||
| dpvb 7 | dpv 28 | dpv 7 | dpv 28 | |
| PV1 | 13/20 (65) | 11/16 (68.8) | 12/17 (70.6) | 8/14 (57.1) |
| PV2 | 18/20 (90) | 14/16 (87.5) | 17/17 (100) | 4/14 (28.6) |
| PV3 | 7/20 (35) | 6/16 (37.5) | 4/17 (23.5) | 4/14 (28.6) |
The proportions of OPV vaccinees shedding PV are the same as the percentage PV-positive stool specimens.
dpv, day postvaccination.
Viruses.
The Sabin type 1, 2, and 3 strains used for preparation of positive controls for the RT-PCR assay and the spiked medium samples used for determination of PCR and culture assay sensitivities were obtained from monovalent pools of PV used in the manufacture of the Orimune vaccine. Viral RNA for PCR analysis was extracted from this material as described below for the stool samples.
Stool sample preparation for culture.
A 10% (wt/wt) suspension of the stool specimen was prepared in tissue culture medium and was clarified by centrifugation at 3,000 × g at 4°C for 10 min. The supernatant was filtered through 0.2-μm-pore-size Millex GV filters (Millipore Inc., Bedford, Mass.). Stool samples weighing less than 0.1 g were resuspended in 2 ml of medium and were processed as described above.
Culture isolation and identification of PV in stool samples.
PV was isolated and typed in Vero cells by a single culture amplification step which differs from the conventional culture method that relies on two amplification steps: the first to obtain a virus isolate and the second to determine the serotype of the isolate. The type of PV isolated was identified by the method of Lim and Benyesh-Melnick (18) and Melnick (22). Polyclonal horse antisera specific to either Sabin PV1, Sabin PV2, or Sabin PV3 were mixed in the following combinations: anti-PV1 plus anti-PV2, anti-PV1 plus anti-PV3 anti-PV2 plus anti-PV3, and anti-PV1 plus anti-PV2 plus anti-PV3. Prior to mixing, the individual sera were diluted so that they contained 20 times the amount of antibody required to neutralize 100 TCID50s of virus. At this concentration, the antisera did not exhibit cross-neutralizing activity. Fifty microliters of each of the four antiserum pools was mixed with a 50-μl aliquot of fivefold dilutions ranging from 10−0.5 to 10−5.0 of the filtered stool suspension, and the mixtures were plated in triplicate in a 96-well tissue culture plate. The virus and serum mixtures were incubated at 37°C for 2 h. Vero cells (104/well) were added to the wells, and the plates were incubated at 33.5°C. The plates were observed daily for a cytopathic effect. When a cytopathic effect was observed in the presence of the pool of anti-PV2 plus anti-PV3, the virus was identified as PV1. Similarly, PV2 was identified from wells containing a pool of anti-PV1 plus anti-PV3, and PV3 was identified from wells containing a pool of anti-PV1 plus anti-PV2. Non-PV, if present, could be identified from wells containing the pool of anti-PV1, anti-PV2, and anti-PV3. Sufficient antisera were available to neutralize all the virus of a specific type present in the sample, as evidenced by the lack of virus breakthrough in any of the samples incubated in the presence of antisera to all three PV serotypes. Results were usually apparent on day 3 or 4. Final readings were made on days 7 to 10. The TCID50s per gram of stool were calculated by the Reed-Muench method (17).
Preparation of stool and virus-spiked samples for RT-PCR.
A 10% (wt/wt) suspension of the stool specimen was prepared in tissue culture medium. Stool samples weighing less than 0.2 g were suspended in a minimum volume of 2 ml. The specimen was clarified by centrifugation at 3,200 × g at 4°C for 15 min. An aliquot of the supernatant was subjected to a high-speed centrifugation at 16,000 × g at 4°C for 15 min to remove residual debris. A total of 400 μl of the supernatant was treated with 100 μl of 5× lysis buffer (2.5% sodium dodecyl sulfate, 100 mM Tris [pH 7.5], 25 mM EDTA, 100 mM NaCl, 0.5 mg of proteinase K per ml) at 56°C for 30 min. The sample was extracted with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1; pH 5.2). Sodium acetate (pH 5.2) was added to 0.25 M, and the RNA was precipitated with 2 volumes of ethanol and collected by centrifugation. The RNA pellet was rinsed with 70% ethanol, air dried, and resuspended in 50 μl of sterile, RNase-free water. A 1:20 dilution of the RNA was prepared, and 10 μl of the undiluted sample and 10 μl of the diluted sample were amplified by RT-PCR. Virus-spiked samples containing known amounts of Sabin type 1, 2, and 3 viruses were prepared in tissue culture medium to determine the sensitivity of the RT-PCR assay for PV detection. These samples were not subjected to the clarification steps used for stool specimens, but the remaining processing steps were identical. Each time that a series of stool specimens from an OPV recipient was processed, two virus-negative tissue culture medium controls were interspersed within the series and were processed in parallel to monitor for contamination due to previously amplified PCR products or sample cross contamination.
Direct identification of PV in stool samples by RT-PCR. (i) RT-PCR primers and probes.
The primers for RT-PCR and the probes for the detection of Sabin virus PCR products were modified from those described previously (39, 40). The oligonucleotide sequences and their genome nucleotide positions are presented in Table 1. There are three primer pairs consisting of a forward (F) and a reverse (R) primer that are specific for each Sabin virus serotype and a corresponding antisense detection probe (P). PCR primers amplified 97-, 78-, and 53-bp products for Sabin type 1, 2, and 3 viruses, respectively. High-pressure liquid chromatography-purified oligonucleotides were obtained from Bio-Synthesis, Inc. (Lewisville, Tex.) and were reconstituted in RNase-free water.
TABLE 1.
Primers and probes used to determine Sabin PV serotype identity by RT-PCRa
| Designationb | Map positionc | Sequence (5′ to 3′) | Specificity |
|---|---|---|---|
| S1F | 2485–2503 | AGGTCAGATGCTTGAAAGC | Sabin PV1 |
| S1R | 2564–2581 | TCCACTGGCTTCAGTGTT | |
| S1-P | 2515–2545 | CGTTGCCGCCCCCACCGTTTCACGGACTGTG | |
| S2F | 2507–2526 | CCGTTGAAGGGATTACTAAA | Sabin PV2 |
| S2R-1B | 2567–2584 | GACCGCTCGGCTTGTGTC | |
| S2-P | 2529–2559 | GCTATTGGTGGAAGTCGGGGGAACCAATGCA | |
| S3F | 2508–2524 | AGGGCGCCCTAACTTTG | Sabin PV3 |
| S3R-1B | 2542–2560 | AGTATCAGGTAAGCTATCC | |
| S3-PT | 2522–2545 | ATCCTGTTGCTTCGGGAGTGACAA |
(ii) RT and PCR amplification.
RT and PCR were performed in a single step with a buffer compatible with both enzymatic reactions as described previously, (39, 40), with modifications. A total of 10 μl of sample RNA that was denatured at 95°C for 3 min and snap-cooled on ice was amplified in a 100-μl multiplex reaction mixture consisting of 50 mM Tris (pH 8.8 at 25°C), 70 mM KCl, 5 mM MgCl2, 10 mM dithiothreitol, 0.2 mM each dGTP, dATP, dCTP, and dTTP, 20 pmol of each Sabin serotype primer (S1F-S1R, S2F–S2R-1B, S3F–S3R-1B), 10 U of human placental RNase inhibitor (Boehringer Mannheim, Indianapolis, Ind.), 5 U of avian myeloblastosis virus reverse transcriptase (Promega, Madison, Wis.), and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer, Branchburg, N.J.).
RT was performed at 42°C for 30 min, followed by heat inactivation of the reverse transcriptase enzyme at 95°C for 3 min. PCR was performed in a Perkin-Elmer DNA thermal cycler 480 for 35 cycles by using a touchdown protocol (9): denaturation at 94°C for 45 s, annealing for 45 s at 62°C and stepping down in 1°C increments to 58°C, and extension at 72°C for 1 min (an extra 9 min at 72°C was used after the final cycle). Two cycles were run at each annealing temperature from 62 to 59°C, and the remaining 27 cycles were run at an annealing temperature of 58°C. A typical thermal cycler run was completed in 3.5 h.
(iii) Detection of RT-PCR products by solution hybridization with radiolabeled probes.
A total of 20 pmol of each of the antisense oligonucleotide probes S1-P, S2-P, and S3-PT specific for Sabin PV1, Sabin PV2, and Sabin PV3, respectively, was 5′ end labeled with [γ-32P]ATP (specific activity, 3,000 Ci/mmol; Amersham, Arlington Heights, Ill.) and polynucleotide kinase (Boehringer Mannheim) according to the enzyme manufacturer’s instructions. Unincorporated radioactivity was removed by centrifugation of the labeling reaction mixture through a Bio-spin 6 column (Bio-Rad Laboratories, Hercules, Calif.) according to the manufacturer’s instructions. Probes were diluted in 66 mM NaCl–44 mM EDTA (pH 8), and 10 μl containing 105 cpm per serotype probe was added to 10 μl of undiluted PCR product for solution hybridization as described previously (39), with modifications. Samples were denatured at 95°C for 5 min and annealed at 57°C for 30 min. The hybrids were electrophoresed on a nondenaturing 15% polyacrylamide gel (acrylamide to bis-acrylamide at 29:1; 17 by 15 by 0.08 cm) run with TBE (89 mM Tris-borate, 2 mM EDTA [pH 8.3]). Autoradiography of the wet gel was performed with Hyperfilm MP (Amersham) and intensifying screens at −70°C for 0.5 to 2 h.
RESULTS
Optimization of stool sample preparation to remove inhibitors of RT-PCR.
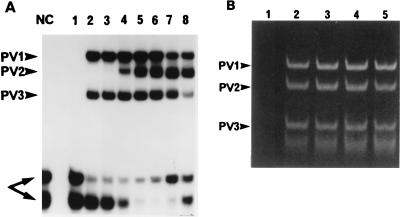
Although the first two available stool series collected from OPV recipients did not inhibit PCR amplification or hybridization detection, the stool samples from the next few OPV recipients that we analyzed showed significant inhibition. This was not surprising since numerous reports have described inhibitors in stool specimens which interfere with PCR assays for the detection of group A rotavirus (38), Norwalk virus (8, 16), and Salmonella strains (37). Each series of stool specimens was evaluated for inhibition by spiking undiluted and diluted extracts from the day 0 prevaccination sample with a Sabin PV strain RNA control and amplifying the sample along with an unspiked extract and control by RT-PCR. If no PV PCR products were detected in the undiluted spiked sample, the stool specimen was considered to be inhibitory. A number of reagents that are reported to remove stool inhibitors were tested, including Chelex-100 (Bio-Rad) metal chelating resin, the cationic detergent cetyltrimethylammonium bromide (16, 31), and Trizol LS (Life Technologies, Grand Island, N.Y.), which is a solution of phenol-guanidine isothiocyanate, but none of these proved to be adequately effective. Dilution of the RNA extracted from the stool specimens was also investigated. A twofold dilution series (1:5 to 1:80) of an inhibitory prevaccination virus-negative extract collected from a stool specimen from vaccinee M was spiked with 104 TCID50 equivalents of Sabin type 1, 2, and 3 PV RNA and amplified by RT-PCR. The results of solution hybridization detection of the PCR products with probe annealing at 62°C are presented in Fig. 1A. The undiluted extract (lane 1) did not generate any Sabin PV hybridization signals. Sabin PV1 and PV3 signals were recovered with a 1:5 dilution (lane 2), but detection of the Sabin PV2 signal required at least a 1:20 dilution of the extract (lane 4). Dilution beyond 1:20 (lanes 5 and 6) resulted in an enhanced Sabin PV2 signal.
FIG. 1.
Recovery of Sabin virus RT-PCR signals by dilution of an inhibitory extract of stool from vaccinee M. PV PCR products, indicated by arrowheads, were detected by OH assay with a cocktail of 32P-labeled Sabin serotype-specific probes (A) and for a subset of samples from panel A by ethidium bromide staining of products electrophoresed in a 15% polyacrylamide gel (B). (A and B) Lanes: 1, Undiluted stool; 2, 1:5 dilution; 3, 1:10 dilution; 4, 1:20 dilution; 5, 1:40 dilution. (A) Lanes: NC, no RNA template negative control; 6, stool diluted 1:80; 7, no stool extract; 8, positive control extract from OPV vaccinee W. Arrows identify unhybridized probes at the bottom of panel A.
The RT-PCR products that were analyzed by the oligomer hybridization (OH) assay (Fig. 1A, lanes 1 to 5) were electrophoresed in a polyacrylamide gel and stained with ethidium bromide (Fig. 1B) to determine if the Sabin PV2 signal inhibition was at the level of RT-PCR amplification or hybridization. The undiluted extract (Fig. 1B, lane 1) did not support amplification of the viral RNA of any Sabin serotype. Surprisingly, all diluted extracts tested (lanes 2 to 5) showed comparable amounts of the 78-bp Sabin PV2 PCR product. These data indicated that the extract of the stool specimen obtained from vaccinee M before vaccination contained inhibitors of RT-PCR amplification as well as hybridization detection. It should be noted that inhibitory stool samples from vaccinee M collected after vaccination with OPV as well as those collected from some of the other OPV recipients included in this study showed similar levels of production of Sabin PV2 PCR product, but for these samples there was inefficient product detection by hybridization. In contrast, some stool samples (from vaccinee W) did not contain inhibitory substances and generated strong PV2 signals in the OH assay (Fig. 1A, lane 8) under the assay conditions.
Undiluted extracts from postvaccination series of stool specimens that were collected from additional subjects and that were inhibitory to the RT-PCR were diluted 1:20 and tested by the RT-PCR assay. The 1:20 dilution was chosen because it was the dilution that resulted in the recovery of the PV2 signal in the OH assay (Fig. 1A, lane 4). For all samples, the 1:20 dilution led to the detection of additional PV PCR-positive stool samples that would have been missed if only the undiluted stool extracts were analyzed. Taking into consideration the fact that assay sensitivity may be compromised if diluted material is tested, the PCR method was modified to include tests of both undiluted and 1:20-diluted stool extracts to maximize the sensitivity of virus detection.
OH assay optimization to improve PV2 detection from inhibitory stool specimens.
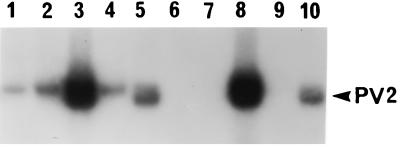
Inefficient detection of PV2 from inhibitory stool specimens suggested that the conditions used for hybridization, such as the temperature of probe annealing and the molarity of the salt in the probe diluent, may not be optimal for these samples. Since stool sample composition is variable, one can envision that specimen-associated salts, metals, or proteins that are carried over into the hybridization reaction mixture may create suboptimal conditions. The hybridization protocol (39) specified a probe annealing temperature of 62°C. The results obtained with a 5°C drop in the annealing temperature of the Sabin 2 probe (S2-P) to 57°C were compared to those obtained at 62°C with a series of RT-PCR products derived from inhibitory and noninhibitory stool specimens as well as Sabin virus-spiked medium controls. Lowering of the annealing temperature had a dramatic effect on the detection of PV2 products in the OH assay, as shown in Fig. 2. Inhibitory stool samples that were not detected at 62°C (lanes 6, 7, and 9) were detected at 57°C (lanes 1, 2, and 4). A 52°C annealing temperature was also tested, but the hybridization background increased with no significant enhancement in the sensitivity of PV detection over that observed at 57°C. The use of 57°C as the annealing temperature for the Sabin serotype-specific probes resulted in enhanced detection of PV2 without compromising the detection of PV1 and PV3. The PV serotype specificities of the probes were confirmed at the optimized 57°C annealing temperature (data not shown). It should be noted that computer-predicted melting temperatures for the S1-P, S2-P, and S3-PT probes were 66, 61, and 52°C, respectively.
FIG. 2.
Recovery of PV2 hybridization signals from inhibitory stool extracts by decreasing the S2-P probe annealing temperature in the OH assay from 62 to 57°C. Inhibitory undiluted extracts of stool specimens from OPV vaccinees, along with extracts that did not interfere with RT-PCR, were amplified, and the PCR products were detected by the OH assay with the PV2-specific probe S2-P at 57°C (lanes 1 to 5) and 62°C (lanes 6 to 10). A 30-min autoradiograph is shown. The PV2 PCR product is indicated by the arrowhead. Lanes 1 and 6, inhibitory extract of stool from vaccinee M; lanes 2 and 7, inhibitory extract of stool from vaccinee 34; lanes 3 and 8, noninhibitory extract of stool from vaccinee W; lanes 4 and 9, inhibitory extract of stool prepared from prevaccination stool (day 0) from vaccinee M spiked with Sabin PV1, PV2, and PV3; lanes 5 and 10, extract prepared from Sabin PV2 spiked in culture medium (no stool).
Sensitivity of RT-PCR versus that of culture for PV detection in virus-spiked samples.
The sensitivity of RT-PCR versus that of culture for the detection of PV was evaluated (Table 2) Serial fivefold dilutions of Sabin PV1, PV2, and PV3 were spiked in tissue culture medium (20 to 12,500 TCID50s/ml per serotype), and aliquots were analyzed by culture and PCR as described in Materials and Methods. Undiluted samples were tested by both methods to maximize the potential for virus detection. Note that the culture method evaluated approximately a twofold greater volume of each spiked sample than was amplified by RT-PCR. As indicated in Table 2, the dilution containing the least amount of Sabin PV (20 TCID50s/ml) was positive by RT-PCR but was negative by culture. Taking into account the difference in the proportion of sample tested by the methods, in the absence of stool extract, RT-PCR was 5- to 10-fold more sensitive than culture for PV detection. This sensitivity reflects a best-case situation that probably would not be duplicated with stool specimens due to the presence of substances that inhibit RT-PCR or that are toxic to tissue culture.
TABLE 2.
Sensitivity of RT-PCR versus that of culture for PV detection
| Titer (TCID50s/ml)a | Culture
|
RT-PCR
|
||
|---|---|---|---|---|
| TCID50 | PV detection | TCID50 | PV detection | |
| 12,500 | 1,875 | + | 1,000 | + |
| 2,500 | 375 | + | 200 | + |
| 500 | 75 | + | 40 | + |
| 100 | 15 | + | 8 | + |
| 20 | 3 | − | 1.6b | + |
Titer represents amount of each serotype per milliliter of sample.
A value of <1 TCID50 did not generate a PCR signal.
Sensitivity of RT-PCR versus that of culture for PV detection in stool specimens from OPV recipients.
A total of 195 stool samples collected from 26 vaccinees that yielded data by both the RT-PCR and the culture methods were included to assess PV detection sensitivity. The proportions of PV1-, PV2-, and PV3-positive specimens and virus-negative specimens by the two methods are presented in Table 3. Note that a single stool sample may be positive for multiple PV serotypes. RT-PCR detected more PV-positive specimens than culture, and this was the case for each PV serotype. The RT-PCR method identified 12% more PV1-positive samples and 18% more PV2-positive samples than culture. Most striking, twice as many specimens were positive for PV3 by RT-PCR than by culture. In addition, a higher proportion of specimens that were negative for PV of any serotype was identified by culture (25.6%) than by RT-PCR (10.3%). All three PV serotypes in a single specimen were identified in 42.6% of the specimens by RT-PCR but only 24.1% of the specimens by culture. RT-PCR was more efficient in identifying multiple PV serotypes.
A total of 35, 41, and 58 stool samples were culture negative and PCR positive for PV1, PV2, and PV3, respectively. Results for a single stool specimen could be discrepant for one or more PV serotypes. This large number of discrepant results was not surprising since RT-PCR was more sensitive than culture for the detection of PV in stool specimens (Table 3). It is highly improbable that these discrepant results represent false-positive PCR results because (i) numerous negative controls were routinely clean and (ii) the distribution was not random in that ∼80% of the samples with discrepant results either were collected on day 1, when the vaccine virus most likely did not have sufficient time to transit the gut, or were the last specimen(s) collected in the series and were the specimens most temporally removed from the day of immunization, when virus titers were expected to be waning. The few samples from within the middle of a series with discrepant results were in almost all cases flanked by samples that were both culture and PCR positive, indicating that the virus titers in the samples with discrepant results may have been too low for detection by culture. The preponderance of samples with discrepant results for PV3 detection most likely reflects a lower replication efficiency of this vaccine component in the gastrointestinal tract due to interference by PV1 and PV2, resulting in reduced virus titers that culture may not have been sensitive enough to pick up. To minimize the potential for false-positive results by PCR due to contamination, stool sample preparation, PCR amplification, and OH detection were performed in separate rooms with dedicated pipettes with filter-plugged tips and single-use reagent aliquots. Tissue culture medium-negative controls were interspersed with stool samples from vaccinees and were processed in parallel. Multiple negative controls minus RNA template were included in the PCR. On the rare occasion that any of the negative controls was positive, the analysis was invalidated and repeated.
A total of 12, 5, and 8 stool samples were culture positive and PCR negative for PV1, PV2, and PV3, respectively. Repeat testing of the majority of the samples with discrepant culture-positive and PCR-negative results in tests with a single Sabin PV serotype primer pair for the maximization of detection resulted in a negative PCR result. It should be noted that specimens with discrepant results (culture positive and PCR negative) were often found to inhibit the RT-PCR.
RT-PCR versus culture analysis of PV shedding in first-dose OPV recipients.
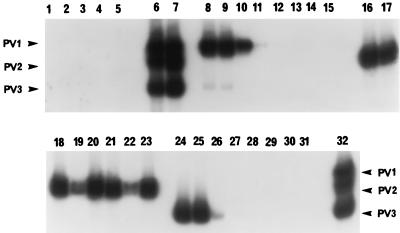
The duration of virus shedding was assessed by RT-PCR for 20 OPV recipients and by culture for 18 vaccinees (samples from 2 subjects were toxic to culture) who received their first dose of vaccine at the age of 2 months. The results for two subjects were not included in the analysis because their prevaccination stool sample (day 0) was positive for PV of one or more serotypes. In addition, data collected from seven vaccinees who received their first vaccine dose at 5 months of age were excluded. The results of RT-PCR analysis of the stool specimen series from 2-month-old vaccinee Z are presented in Fig. 3 to illustrate a representative virus shedding profile. The undiluted stool extracts of this series were not inhibitory. PV1 was shed out to day 7, and a faint signal for PV1 was detected again at day 28 postvaccination. PV2 was shed out to day 28, and PV3 was detected only up to day 3 postvaccination.
FIG. 3.
Duration of PV shedding from an OPV recipient (vaccinee Z). Stool samples were collected before OPV administration (day 0) and on various days (days 1, 2, 3, 4, 7, 14, 21, and 28) within 1 month after OPV administration. Undiluted stool extracts were amplified, and PCR products were detected by OH assay with either a mixture of all three Sabin serotype-specific 32P-labeled probes (lanes 1 to 7 and 32) or individual probes S1-P (lanes 8 to 15), S2-P (lanes 16 to 23), and S3-PT (lanes 24 to 31). A 2-h autoradiograph is shown. The PV PCR products are indicated by arrowheads. Lanes: 1 to 3, PCR-negative controls minus RNA template; 4, extraction-negative control; 5, sample from day 0, 6, PCR-positive control; 7, PCR-positive control spiked into an extract of a stool obtained on day 0; 8, 16, and 24, sample from day 1; 9, 17, and 25, samples from day 2; 10, 18, and 26, sample from day 3; 11, 19, and 27, sample from day 4; 12, 20, 28, sample from day 7; 13, 21, and 29, sample from day 14; 14, 22, and 30, sample from day 21; lanes 15, 23, and 31, sample from day 28; 32, extraction-positive control.
The shedding of PV by the OPV recipients as assessed by RT-PCR and culture is summarized in Table 4. The percentages of vaccinees excreting PV1, PV2, and PV3 at day 7 and PV1 and PV3 at day 28 as determined by RT-PCR and culture were comparable. In contrast, the percentage of vaccinees shedding PV2 at day 28 as determined by culture was significantly reduced compared to that determined by RT-PCR. As assessed by the PCR method, PV2 was still being excreted at day 28 postvaccination by almost all of the vaccinees, and 80% (four of five) of the vaccinees continued to shed virus at day 56 (data not shown), which was about the time when the infants were scheduled to receive their second dose of vaccine. Detection of PV1 excretion by RT-PCR was more variable, with some subjects ceasing virus shedding within 1 week of vaccination and others excreting virus anywhere from 1 to 8 weeks following immunization. Shedding of PV3 was similar to that of PV1, but the majority of vaccinees ceased shedding of virus within 1 week postvaccination. The proportions of vaccinees whose last PV-positive stool specimen as detected by RT-PCR was collected at day 28 or beyond were 68.8% (11 of 16), 93.8% (15 of 16), and 43.8% (7 of 16) for PV1, PV2, and PV3, respectively. The mean durations of PV excretion on the basis of when the last PV-positive stool sample was detected by RT-PCR were 24.4, 32.7, and 17.9 days for PV1, PV2, and PV3, respectively. In contrast, the mean durations of excretion as detected by culture were 19.3, 20.5, and 13.5 days for PV1, PV2, and PV3, respectively. In comparison with RT-PCR, the culture method was deficient in detecting PV2 in many samples collected at 3 weeks or more following vaccination. The PV titers per gram of stool derived from culture analysis of the specimens ranged from 2.57 to 7.0 log10 TCID50s over the entire sample collection period. There were no differences in the range of titers of the PV1, PV2, and PV3 serotypes shed in the stool specimens.
DISCUSSION
In this report, we describe the optimization and implementation of a multiplex RT-PCR method for the detection and serotype identification of Sabin vaccine viruses directly from stool specimens collected from first-dose OPV recipients. The assay had to be modified from the published format (39, 40) to improve the sensitivity of detection of virus in clinical stool specimens which may contain inhibitory substances. We found that use of a 1:20 dilution of the stool extract consistently allowed recovery of virus-positive hybridization signals from inhibitory samples. Optimization of the hybridization conditions revealed that a reduction in the probe annealing temperature, from 62°C as specified by the original assay format to 57°C, enhanced the detection of PV2 in inhibitory stool specimens.
The sensitivity of the optimized RT-PCR assay for PV detection, determined with a virus-spiked medium titration series, was approximately 1 TCID50 for each serotype, and this sensitivity was 5- to 10-fold greater than that of isolation by culture. Evaluation of series of stool specimens from OPV recipients by RT-PCR and culture resulted in a higher frequency of detection of PV1, PV2, and PV3 by RT-PCR and a lower frequency of virus-negative samples by RT-PCR. It should be noted that isolation by culture required an initial 1:5 dilution of the filtered, clarified stool suspension to avoid the culture toxicity associated with some specimens, and use of this dilution could have contributed to the reduced sensitivity of the culture method. Additionally, use of a sample volume greater than 150 μl for evaluation by culture as well as a more sensitive cell line for PV isolation, such as primary monkey kidney cells, may have boosted the sensitivity of culture. The culture method appeared to be biased for the detection of PV1 in stool specimens. This may be due to increased shedding of virus of this serotype as a result of the titer of the PV1 component in the Orimune dose, which is greater than the titers of the PV2 and PV3 components.
The frequency of PV-positive stool samples by culture (Table 4) was compared to that reported in other clinical studies examining virus shedding in stool specimens from first-dose recipients of the Orimune vaccine. Cohen-Abbo et al. (6) reported the recovery of PV1, PV2, and PV3 in 30, 45, and 20% of stool specimens, respectively, collected 1 month postvaccination. Our Vero cell culture method yielded a much higher frequency of PV1 isolation, but the frequency of PV2 isolation was reduced. The different cell lines used for PV propagation, the different virus growth conditions, and the different antisera used for virus typing may account for the observed discrepancies. A study in the United Kingdom by Ramsay et al. (29) showed that 49, 48, and 12% of stool specimens collected weekly for up to 4 weeks postvaccination were positive for PV1, PV2, and PV3, respectively, whereas the rates of positivity were 58.3, 66.7, and 30%, respectively, when the stool specimens obtained at days 7, 14, 21, and 28 postvaccination in this study were examined. However, these results are not comparable because in the study performed in the United Kingdom, the OPV (manufactured by Wellcome) that was administered has a formulation different from that of the Orimune vaccine and has lower potencies of the three Sabin strains per dose compared to the potencies in the Orimune vaccine used in the United States. These factors could account for the reduced level of virus shedding in the stool.
The virus shedding profiles of the vaccinees compiled from the RT-PCR and culture data were concordant. Discrepancies, which for the most part were a virus-negative culture result versus a positive result by PCR, could be attributed to the enhanced sensitivity of PCR for the detection of all three PV types in stool specimens. Culture was particularly inefficient for the isolation of PV3 from samples collected at early times postvaccination (<1 week). The PV3 component of OPV is not as effective as the PV1 and PV2 components at inducing seroconversion in vaccinees after administration of the first vaccine dose (6) due to replication interference by PV1 and PV2, resulting in reduced levels of shedding of PV3 in stool specimens. The level of reduction of shedding may be such that it is below the threshold of the sensitivity of detection by culture. Culture was not efficient in detecting PV2 from samples collected at later times postvaccination (≥3 weeks). PV2 replicates best in the gastrointestinal tract and thus elicits a strong mucosal antibody response. The inability of culture to detect PV2 at late times postvaccination may be the result of the presence in the gut of large amounts of neutralized virus (complexed with antibody) that would not be detected by an infectivity assay but that could be detected by PCR.
The RT-PCR assay was optimized to enhance PV detection in stool samples and was implemented to track virus excretion in OPV vaccinees enrolled in an ongoing vaccine study. The RT-PCR assay can yield results in 2 days, whereas culture results are not available for 1 to 3 weeks. Assay time can be further reduced to a single day without a significant loss of sensitivity by omitting the hybridization step and visualizing the amplified PCR products in ethidium bromide-stained gels. However, hybridization provides the benefit of detection of PV in samples containing low virus titers, it can confirm assay specificity, and it can detect low-level PCR contamination, if it is present. Another advantage of RT-PCR is that it can be performed in laboratories without tissue culture capabilities with minimal expense.
In a move to eliminate the rare cases of polio that result from OPV, the Advisory Committee on Immunization Practices of the federal Centers for Disease Control and Prevention has recommended a change in the polio vaccination schedule from the current practice of administering OPV only at 2, 4, and 6 months of age to a sequential schedule of injection of enhanced-potency inactivated polio vaccine (IPV) at 2 and 4 months followed by the administration of two doses of OPV at 12 to 18 months and 4 to 6 years of age. It is believed that the immunity acquired from the first two doses of inactivated vaccine, which is unlikely to cause paralytic polio, should be sufficient to protect the very small number of children who contract disease from the oral vaccine. The RT-PCR assay could be used to monitor the impact of the change in the vaccination schedule on virus shedding. In addition, RT-PCR could be implemented to evaluate how new OPV formulations, with changes in the ratio and/or titer of Sabin strains or OPV produced by an altered manufacturing procedure (change in cell substrate), affect PV excretion in the stools of vaccinees. Mallet et al. (21) recently reported on the use of a type-specific nested PCR assay to assess the shedding of Sabin strains in stools from vaccinees who were administered OPV prepared in either primary monkey kidney or Vero cells to appraise the bioequivalence of these two vaccines.
The RT-PCR assay can be modified further to evaluate the frequency of 5′ NCR revertants directly in the stools of children immunized with a mixed IPV-OPV schedule. A study involving the Orimune vaccine reported an increased frequency of occurrence of PV revertants and an increased duration of virus excretion in the stools of vaccinees who were immunized with IPV followed by OPV compared to those in the stools of control vaccinees who were immunized only with OPV (3, 25). The concern is the possibility of an increased risk of VAPP in vaccinees and unimmunized contacts by transmission of viruses with an enhanced neurovirulence potential. To assess revertant frequencies, it is preferable to assay the virus composition in stools by a direct method without culture amplification that could affect the proportion of detectable revertants, leading to a biased result. Analysis of mutants by PCR and restriction enzyme cleavage has been described to quantify reverted virus in monovalent Sabin vaccine (5), but this technique is not applicable for use with stool samples from individuals immunized with a trivalent vaccine. It may be possible to capture a specific virus type or RNA from clinical samples via serotype-specific antibodies or oligonucleotide probes by a magnetic bead approach and use the selected material for analysis of mutants by PCR and restriction enzyme cleavage or a similar site-specific PCR analysis. It should be noted that PCR, unlike culture, cannot distinguish live infectious virus and inactivated virus (neutralized with antibody). Therefore, the clinical relevance of the revertant population as determined by PCR is a point of concern. In order to correlate revertants shed in stools with virus transmission and associated disease, it is important to demonstrate that the virus being detected consists of live replicating particles.
The exquisite sensitivity and specificity of the RT-PCR method for the detection of PV in stool specimens described here coupled with a rapid assay time underscore its utility for the assessment of the impact of the recent change in the U.S. polio vaccination schedule on virus excretion and reversion.
REFERENCES
- 1.Abebe A, Johansson B, Abens J, Strannegard O. Detection of enteroviruses in faeces by polymerase chain reaction. Scand J Infect Dis. 1992;24:265–273. doi: 10.3109/00365549209061331. [DOI] [PubMed] [Google Scholar]
- 2.Abraham R, Chonmaitree T, McCombs J, Prabhakar B, Lo Verde P T, Ogra P L. Rapid detection of poliovirus by reverse transcription and polymerase chain amplification: application for differentiation between poliovirus and nonpoliovirus enterovirus. J Clin Microbiol. 1993;31:395–399. doi: 10.1128/jcm.31.2.395-399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abraham R, Minor P, Dunn G, Modlin J F, Ogra P L. Shedding of virulent poliovirus revertants during immunization with oral poliovirus vaccine after prior immunization with inactivated polio vaccine. J Infect Dis. 1993;168:1105–1109. doi: 10.1093/infdis/168.5.1105. [DOI] [PubMed] [Google Scholar]
- 4.Chapman N M, Tracy S, Gauntt C J, Fortmueller U. Molecular detection and identification of enteroviruses using enzymatic amplification and nucleic acid hybridization. J Clin Microbiol. 1990;28:843–850. doi: 10.1128/jcm.28.5.843-850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chumakov K M, Powers L B, Noonan K E, Roninson I B, Levenbook I S. Correlation between amount of virus with altered nucleotide sequence and the monkey test for acceptability of oral poliovirus vaccine. Proc Natl Acad Sci USA. 1991;88:199–203. doi: 10.1073/pnas.88.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen-Abbo A, Culley B S, Reed G W, Sannella E C, Mace R L, Robertson S E, Wright P F. Seroresponse to trivalent oral poliovirus vaccine as a function of dosage interval. Pediatr Infect Dis J. 1995;14:100–106. doi: 10.1097/00006454-199502000-00004. [DOI] [PubMed] [Google Scholar]
- 7.Contreras G, Dimock K, Furesz J, Gardell C, Hazlett D, Karpinski K, McCorkle G, Wu L. Genetic characterization of Sabin types 1 and 3 poliovaccine virus following serial passage in the human intestinal tract. Biologicals. 1992;20:15–26. doi: 10.1016/s1045-1056(05)80003-x. [DOI] [PubMed] [Google Scholar]
- 8.De Leon R, Matsui S M, Baric R S, Herrmann J E, Blacklow N R, Greenberg H B, Sobsey M D. Detection of Norwalk virus in stool specimens by reverse transcriptase-polymerase chain reaction and nonradioactive oligoprobes. J Clin Microbiol. 1992;30:3151–3157. doi: 10.1128/jcm.30.12.3151-3157.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Don R H, Cox P T, Wainwright B J, Baker K, Mattick J S. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 1991;19:4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn G, Begg N T, Cammack N, Minor P D. Virus excretion and mutation by infants following primary vaccination with live oral poliovaccine from two sources. J Med Virol. 1990;32:92–95. doi: 10.1002/jmv.1890320205. [DOI] [PubMed] [Google Scholar]
- 11.Egger D, Pasamontes L, Ostermayer M, Bienz K. Reverse transcription multiplex PCR for differentiation between polio- and enteroviruses from clinical and environmental samples. J Clin Microbiol. 1995;33:1442–1447. doi: 10.1128/jcm.33.6.1442-1447.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evans D M A, Dunn G, Minor P D, Schild G C, Cann A J, Stanway G, Almond J W, Currey K, Maizel J V., Jr Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovaccine genome. Nature (London) 1985;314:548–550. doi: 10.1038/314548a0. [DOI] [PubMed] [Google Scholar]
- 13.Friedrich F. Genomic modifications in Sabin vaccine strains isolated from vaccination-associated cases, healthy contacts and healthy vaccinees. Acta Virol. 1996;40:157–170. [PubMed] [Google Scholar]
- 14.Georgescu M-M, Delpeyroux F, Tardy-Panit M, Balanant J, Combiescu M, Combiescu A A, Guillot S, Crainic R. High diversity of poliovirus strains isolated from the central nervous system from patients with vaccine-associated paralytic poliomyelitis. J Virol. 1994;68:8089–8101. doi: 10.1128/jvi.68.12.8089-8101.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyypiä T, Auvinen P, Maaronen M. Polymerase chain reaction for human picornaviruses. J Gen Virol. 1989;70:3261–3268. doi: 10.1099/0022-1317-70-12-3261. [DOI] [PubMed] [Google Scholar]
- 16.Jiang X, Wang J, Graham D Y, Estes M K. Detection of Norwalk virus in stool by polymerase chain reaction. J Clin Microbiol. 1992;30:2529–2534. doi: 10.1128/jcm.30.10.2529-2534.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lennette E H. General principles underlying laboratory diagnosis of viral and rickettsial infections. In: Lennette E H, Schmidt N J, editors. Diagnostic procedures for viral and rickettsial infections. 4th ed. Washington, D.C: American Public Health Association; 1969. pp. 1–65. [Google Scholar]
- 18.Lim K A, Benyesh-Melnick M. Typing of viruses by combinations of antiserum pools: application to typing of enteroviruses. J Immunol. 1960;84:309–317. [PubMed] [Google Scholar]
- 19.Macadam A J, Pollard S R, Ferguson G, Skuce R, Wood D, Almond J W, Minor P D. Genetic basis of attenuation of the Sabin type 2 vaccine strain of poliovirus in primates. Virology. 1993;192:18–26. doi: 10.1006/viro.1993.1003. [DOI] [PubMed] [Google Scholar]
- 20.Macadam A J, Stone D M, Almond J W, Minor P D. The 5′ noncoding region and virulence of poliovirus vaccine strains. Trends Microbiol. 1994;2:449–454. doi: 10.1016/0966-842x(94)90803-6. [DOI] [PubMed] [Google Scholar]
- 21.Mallet L, Pelloquin F, Brigaud M, Caudrelier P, Bandet R, Xueref C, Fuchs F, Gibelin N, Goldman C, Moulin J C, de Fraipont F, Montagnon B, Peyron L, Aymard M. Comparative study of poliovirus excretion after vaccination of infants with two oral polio vaccines prepared on Vero cells or on primary monkey kidney cells. J Med Virol. 1997;52:50–60. [PubMed] [Google Scholar]
- 22.Melnick J L. Enteroviruses. In: Lennette E H, editor. Laboratory diagnosis of viral infections. New York, N.Y: Marcel Dekker, Inc.; 1985. pp. 241–256. [Google Scholar]
- 23.Melnick J L. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 655–712. [Google Scholar]
- 24.Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, Genba Y, Nakano Y, Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci USA. 1982;79:5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogra P L, Faden H S, Abraham R, Duffy L C, Sun M, Minor P D. Effect of prior immunity on the shedding of virulent revertant virus in feces after oral immunization with live attenuated poliovirus vaccines. J Infect Dis. 1991;164:191–194. doi: 10.1093/infdis/164.1.191. [DOI] [PubMed] [Google Scholar]
- 26.Olive D M, Al-Mufti S, Al-Mulla W, Khan M A, Pasca A, Stanway G, Al-Nakib W. Detection and differentiation of picornaviruses in clinical samples following genomic amplification. J Gen Virol. 1990;71:2141–2147. doi: 10.1099/0022-1317-71-9-2141. [DOI] [PubMed] [Google Scholar]
- 27.Otelea D, Guillot S, Furione M, Combiescu A A, Balanant J, Candrea A, Crainic R. Genomic modifications in naturally occurring neurovirulent revertants of Sabin 1 polioviruses. Dev Biol Stand. 1993;78:33–38. [PubMed] [Google Scholar]
- 28.Prevots D R, Sutter R W, Strebel P M, Wharton M, Hadler S C. Poliomyelitis prevention in the United States: introduction of a sequential vaccination schedule of inactivated poliovirus vaccine followed by oral poliovirus vaccine. Morbid Mortal Weekly Rep. 1997;46:1–25. [PubMed] [Google Scholar]
- 29.Ramsay M E, Begg N T, Gandhi J, Brown D. Antibody response and viral excretion after live polio vaccine or a combined schedule of live and inactivated polio vaccines. Pediatr Infect Dis J. 1994;13:1117–1121. doi: 10.1097/00006454-199412000-00009. [DOI] [PubMed] [Google Scholar]
- 30.Rotbart H A. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol. 1990;28:438–442. doi: 10.1128/jcm.28.3.438-442.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos N, Gouvea V. Improved method for purification of viral RNA from fecal specimens for rotavirus detection. J Virol Methods. 1994;46:11–21. doi: 10.1016/0166-0934(94)90012-4. [DOI] [PubMed] [Google Scholar]
- 32.Schweiger B, Schreier E, Böthig B, López-Pila J M. Differentiation of vaccine and wild-type polioviruses using polymerase chain reaction and restriction enzyme analysis. Arch Virol. 1994;134:39–50. doi: 10.1007/BF01379105. [DOI] [PubMed] [Google Scholar]
- 33.Stanway G, Hughes P J, Mountford R C, Reeve P, Minor P D, Schild G C, Almond J W. Comparison of the complete nucleotide sequences of the genomes of the neurovirulent poliovirus P3/Leon/37 and its attenuated Sabin vaccine derivative P3/Leon 12a1b. Proc Natl Acad Sci USA. 1984;81:1539–1543. doi: 10.1073/pnas.81.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strebel P M, Sutter R W, Cochi S L, Biellik R J, Brink E W, Kew O M, Pallansch M A, Orenstein W A, Hinman A R. Epidemiology of poliomyelitis in the United States one decade after the last reported case of indigenous wild virus-associated disease. Clin Infect Dis. 1992;14:568–579. doi: 10.1093/clinids/14.2.568. [DOI] [PubMed] [Google Scholar]
- 35.Tatem J M, Weeks-Levy C, Mento S J, DiMichele S J, Georgiu A, Waterfield W F, Sheip B, Costalas C, Davies T, Ritchey M B, Cano F R. Oral poliovirus vaccine in the United States: molecular characterization of Sabin type 3 after replication in the gut of vaccinees. J Med Virol. 1991;35:101–109. doi: 10.1002/jmv.1890350206. [DOI] [PubMed] [Google Scholar]
- 36.Toyoda H, Kohara M, Kataoka Y, Suganuma T, Omata T, Imura N, Nomoto A. Complete nucleotide sequences of all three poliovirus serotype genomes: implication for genetic relationship, gene function and antigenic determinants. J Mol Biol. 1984;174:561–585. doi: 10.1016/0022-2836(84)90084-6. [DOI] [PubMed] [Google Scholar]
- 37.Widjojoatmodjo M N, Fluit A C, Torensma R, Verdonk G P H T, Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3198. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilde J, Eiden J, Yolken R. Removal of inhibitory substances from human fecal specimens for detection of group A rotaviruses by reverse transcriptase and polymerase chain reaction. J Clin Microbiol. 1990;28:1300–1307. doi: 10.1128/jcm.28.6.1300-1307.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang C-F, De L, Holloway B P, Pallansch M A, Kew O M. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 1991;20:159–179. doi: 10.1016/0168-1702(91)90107-7. [DOI] [PubMed] [Google Scholar]
- 40.Yang C-F, De L, Yang S-J, Gómez J R, Cruz J R, Holloway B P, Pallansch M A, Kew O M. Genotype-specific in vitro amplification of sequences of the wild type 3 polioviruses from Mexico and Guatemala. Virus Res. 1992;24:277–296. doi: 10.1016/0168-1702(92)90124-r. [DOI] [PubMed] [Google Scholar]
- 41.Zoll G J, Melchers W J G, Kopecka H, Jambroes G, van der Poel H J A, Galama J M D. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J Clin Microbiol. 1992;30:160–165. doi: 10.1128/jcm.30.1.160-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]