Key Points
PEP-619WW mice have superior control of immunogenic tumors.
PEP-619WW mice had increased tumor-infiltrating lymphocytes.
Tumor-infiltrating lymphocytes have a more activated phenotype in PEP-619WW mice.
Abstract
The 1858C>T allele of the tyrosine phosphatase PTPN22 (causing amino acid substitution R620W in encoded protein lymphoid tyrosine phosphatase) is present in 5–10% of the North American population and is strongly associated with numerous autoimmune diseases. Although much research has been done to define how this allele potentiates autoimmunity, the influence PTPN22 and its proautoimmune allele have in tumor immunity is poorly defined. To interrogate the role this allele may have in the antitumor immune response, we used CRISPR/Cas9 to generate mice in which the ortholog of lymphoid tyrosine phosphatase, PEST domain–enriched protein (PEP), is mutated at position 619 to produce the relevant proautoimmune mutation (R619W). Results of this study show that mice homozygous for this alteration (PEP-619WW) resist tumor growth as compared with wild-type mice. Consistent with these results, tumors from PEP-619WW mice have more CD45 infiltrates containing more activated CD8 T cells and CD4 T cells. In addition, there are more conventional dendritic cell type 1 (cDC1) cells and fewer myeloid-derived suppressor cells in tumors from PEP-619WW mice. Interestingly, the tumor-infiltrating PEP-619WW cDC1 cells have decreased PD-L1 expression compared with cDC1 cells from PEP-wild-type mice. Taken together, our data show that the proautoimmune allele of Ptpn22 drives a strong antitumor response in innate and adaptive immune cells resulting in superior control of tumors.
Introduction
Allelic variation in genes associated with regulating immune responses potentially impact an individual’s response to foreign and self-antigens. A minor allele of the protein tyrosine phosphatase nonreceptor type 22 (PTPN22) has been extensively studied for its contribution to autoimmunity (1–3). Genome-wide association studies identified a tryptophan-encoding common allelic variant, at amino acid 620 (R620W), that is associated with an increased risk for multiple autoimmune diseases, such as type 1 diabetes, rheumatoid arthritis, and systemic lupus erythematosus (3–12). This allele is present in 5–15% of the North American and European populations and is considered the highest non-HLA autoimmune risk allele variant (2, 3). Therefore, understanding how PTPN22 and its proautoimmune allele regulate variation in immune cell functions is of great importance.
In humans, PTPN22 encodes the protein lymphoid tyrosine phosphatase (Lyp), the expression of which is confined to bone marrow–derived cells and thus expressed in all immune cells (4). To better understand the role of this gene, researchers have often used Ptpn22 knockout mice, which are deficient in the expression of the Lyp ortholog, PEST domain–enriched protein (PEP; PEP-null) (13). These studies have shed light on the role of Lyp/PEP in multiple immune cell types. In lymphocytes, Lyp/PEP tempers TCR and BCR signaling through dephosphorylation of Src kinases (13, 14). Binding partners enabling such activity include TNFR-associated factor 3 (TRAF3) and CSK (15). In myeloid cells, the interaction of Lyp/PEP with TRAF3 promotes TLR activation and type I IFN production (16). Through these studies, researchers have identified mechanisms that may contribute to the pathogenesis of multiple autoimmune disorders. In addition, we and others found that mice lacking Ptpn22 (PEP-null) exhibit less of an exhausted T cell phenotype during chronic viral infection (17, 18), suggesting Ptpn22 contributes to the generation of T cell exhaustion. Furthermore, Brownlie et al. (19, 20) have demonstrated that adoptively transferred OT-1 CD8 T cells lacking Ptpn22 have increased function against tumor-associated Ags. These studies (including our own) were performed using PEP-null mice rather than mice expressing the equivalent of the human proautoimmune allele; it remains to be determined whether the human allele would affect disease pathogenesis in a nonautoimmune context.
Although some studies have found that lack of Ptpn22 is similar in phenotype to the R620W mutation (R619W in mice), there are also a number of differences. This may be because of the fact that the substitution of arginine to tryptophan within the C-terminal region of the protein does not directly affect phosphatase activity of this enzyme (21). Rather, the presence of tryptophan disrupts the ability of the enzyme to bind with other proteins, such as CSK and TRAF, which help direct the phosphatase to appropriate substrates (22, 23). As an example of differing consequences, whereas PEP-null mice exhibit an increased frequency and potency of T regulatory cells (Tregs) as compared with wild-type (WT) (24), this same Treg phenotype is not observed in naive mice bearing the Ptpn22 proautoimmune allele (1). However, it has been reported that under lymphopenic conditions, both PEP-null and proautoimmune allele-expressing CD4 T cells have a proliferative advantage over WT CD4 T cells and have a higher propensity to differentiate into Tregs (25). Despite its importance to human health, the Ptpn22 alternative allele has not been studied in the context of the antitumor response and the accompanying T cell activation and exhaustion.
In this study, we use C57BL/6 mice mutated using CRISPR/Cas9 to express the murine ortholog of the Ptpn22 proautoimmune allele (PEP-619WW) and well-established models of syngeneic murine tumors to define the effect this allele has on tumor growth and the antitumor response.
Materials and Methods
Mice
Both males and females ranging from 6 to 12 wk of age were used in this study. Animals were housed in general housing conditions at Scripps Research-La Jolla. All animal studies were reviewed and approved by Scripps Research Institutional Animal Care and Use Committee (protocol no. 06-0291). C57BL/6 WT mice were originally purchased from Jackson Labs and then bred and maintained in Scripps Animal Facility. Ptpn22 proautoimmune allele-bearing mice (PEP-619WW) were generated using CRISPR/Cas9 technology on a C57BL/6 background using methods previously reported (5). In short, four nucleotides were replaced on exon 14 of Ptpn22 to insert the BspEI restriction site and cause an arginine (R)-to-tryptophan (W) amino acid substitution at amino acid position 619. Genotypes were confirmed through PCR using the following primers that flank the mutated region of Ptpn22: Forward-5′-AGCTGATGAAAATGTCCTATTGTGA-3′ and Reverse-5′-GTCCCACTGCATTCTGGTGA-3′. After amplification, PCR products are digested overnight at 37°C with the restriction enzyme, BspEI, which is unique to mutated mice. Digested PCR products are run on an agarose gel to visualize digested bands (Supplemental Fig. 4).
Rag1−/− C57BL/6 mice were a gift from Dr. David Nemazee (Scripps Research-La Jolla) and then were maintained in our colony. These mice were crossed with our PEP-619WW C57BL/6 mice to create Rag1+/− PEP-WT/619W heterozygotes. These mice were backcrossed to Rag1−/− PEP-WT to generate Rag1−/− PEP-WT/619W heterozygous mice. These mice were bred to each other to eventually generate mice that lacked lymphocytes and were homozygous for the proautoimmune allele of Ptpn22, which we denote as Rag1−/− PEP-619WW mice (breeding scheme is diagrammed in Supplemental Fig. 1). These mice were genotyped by lack of circulating lymphocytes and with the PCR genotyping protocol described earlier.
Tumor culture and growth
All tumor cell lines were cultured in DMEM supplemented with 10% heat-inactivated FCS, 1% glutamine and gentamicin, and 50 mM β-mercaptoethanol at 37°C, 5% CO2. In addition, B16-OVA cells were cultured with geneticin (G418) for at least two passes before implanting in mice. Cell lines are passed once they are at 80% confluency. Cells are not cultured for longer than 3 mo.
One day before tumor implant, the left flank of mice is shaved. Although tumor cells are in growth phase, based on confluency of plate (less than 80% confluent), cells are harvested, counted, and resuspended in sterile HBSS for implantation into mice. Tumor cells are implanted s.c. in the shaven area of mice at the following amounts (all in a total volume of 100 μL): 2 × 105 B16-OVA, 1 × 106 MC38, 2 × 105 Lewis lung carcinoma (LLC) cells, or 2 × 105 B16F10 cells. Mice were monitored, and tumor growth was measured at indicated time points. Tumor volume (mm3) was calculated with the following equation: (Length × width2)/2. Once the tumor reached 30 mm in any direction or became ulcerated, the mouse was removed from the study. Tumors were defined as immunogenic or poorly immunogenic based on previous work in the field (26, 27).
Flow cytometry and Abs
Tumors were excised and placed into HBSS with 2% FBS. Samples were minced and incubated with STEMCELL Spleen dissociation media (STEMCELL Technologies, Vancouver, BC, Canada) according to the manufacturer’s instructions. After incubation, the minced sample was smashed and filtered through a 40-μM filter to create a single-cell suspension. Single-cell suspension was counted and resuspended to the desired concentration (dependent on experiment) in HBSS with 2% FBS. Single-cell suspensions were used for staining and flow cytometric analysis. Cells were stained in serum-free HBSS.
All flow cytometry was completed on a spectral cytometer, the Cytek Aurora with a four-laser or five-laser system (405, 488, 640, 561, and 355 nm [five lasers only]). Single-color stain OneComp eBeads (Thermo Fisher USA) were used for unmixing. Unmixed files were analyzed using FlowJo Software (BD Biosciences, San Diego, CA). Abs used in various combinations (depending on experiment) are as follows: Ghost Viability Dye (v510, 1:1000 dilution; Tonbo Biosciences, San Diego, CA), CD45 (BV570/BV605, 1:200, clone 30-F11; Biolegend), CD3e (PE-Cy5/AF532, 1:200, clone 145-2C11; Tonbo Biosciences/Thermo Fisher), CD4 (PerCP/BV605, 1:200, clone RM4-5; Tonbo Biosciences/Biolegend), CD8a (allophycocyanin-Cy7/allophycocyanin-H7/allophycocyanin, 1:200, clone 53-6.7; BD Biosciences), CD11c (PE-Cy5.5, 1:100, clone N418; Thermo Fisher), CD11b (PerCP-Cy5.5, 1:200, clone M1/70; Biolegend), F4/80 (Pacific Orange, 1:100, clone BM8; Thermo Fisher), PDCA-1 (Pacific Blue, 1:200, clone 129C1; Biolegend), CD80 (BV421, 1:200, clone 16-10A1), CD86 (BV605, 1:200, clone GL1; Biolegend), PD-1 (PE-Cy7, 1:200, clone J43.1; Tonbo Biosciences), PD-L1 (PE/BV711/PE-Cy7, 1:100, clone 10F.9G2; Tonbo Biosciences/Biolegend), CD44 (AF700, 1:200, clone IM7; Biolegend), CD62L (FITC, 1:100, MEL-14; Tonbo Biosciences), Ly6C (BV785, 1:200, clone HK1.4; Biolegend), Ly6G (PE-eFlour610, 1:200, clone IA8; Invitrogen), CD206 (AF647, 1:100, clone CO68C2; Biolegend), CD209b (allophycocyanin, 1:200, clone 22D1; Tonbo Biosciences), NK1.1 (FITC, 1:100, clone PK136; Biolegend), CD19 (BV711, 1:400, clone 6D5; Biolegend), B220 (allophycocyanin-Cy5.5, 1:200, clone RA3-6B2; Invitrogen), MHC class II (MHC II) I-Ab (FITC, 1:200, clone AF6-120.1; Biolegend), CXCR5 (BV605, 1:100, clone L138D7; Biolegend), Tbet (allophycocyanin/AF647, 1:200; Biolegend), and Foxp3 (PE, 1:100, clone FJK-16s; Invitrogen). Cells were stained in HBSS, at 4°C, in the dark. If intracellular staining for transcription factors was required, Tonbo Biosciences FoxP3 Fix/Perm kit was used per manufacturer’s instructions (Tonbo Biosciences).
Density of immune cells in tumors was determined by the following equation: Total count CD45+ cells in tumor/tumor volume postresection (mm3).
NanoString spatial GeoMX digital spatial profiling
At 14 d postimplant of B16-OVA tumors, tumors from PEP-WT and PEP-619WW mice were harvested, fixed in 10% neutral-buffered formalin, and paraffin embedded. The three mean tumor sizes from each genotype were cut, put onto slides, and sent to NanoString (Seattle, WA) for GeoMX digital spatial profiling (DSP). Tissue sections were stained for morphology markers DNA-SYTO13, Pmel/S100b-Alexa 532, and CD45-Alexa 594 and a set of 53 photocleavable probes. Using these morphology markers, we selected a total of 12 regions of interest (ROIs) per genotype based on CD45-rich regions spaced throughout each tissue section. Each ROI was exposed to UV illumination, and the eluent was collected and transferred into individual wells of a microtiter plate. Once the 12 ROIs were processed, indexing oligos were hybridized to NanoString optical barcodes for digital counting on the nCouter. Digital counts from barcodes corresponding to protein probes were then normalized to housekeeping counts, area, and CD45 probe. Differences in protein expression (measured by signal-to-noise ratio) between genotypes were determined by t test with Welch’s correction.
The list of target proteins was as follows: Aryl hormone receptor, androgen receptor, B7-H3, Batf3, CD11b, CD11c, CD227/IL17RA, CD14, CD163, CD19, CD27, CD28, CD31, CD34, CD3e, CD4, CD40, CD40L, CD44, CD45, CD86, CD8a, CTLA-4, Epcam, estrogen receptor, F4/80, Fibronectin, Foxp3, GAPDH, GFP, GITR, Granzyme B, Her2, Histone H3, ICOS, IFNgR, Ki67, Lag3, Ly6G/C, MHC II, OX40L, PanCK, PD-1, PD-L1, Pmel17, RbIgG, RbIgG2a, RbIgG2b, S100b, S6, SMA, Tim-3, and VISTA.
Statistics
Statistical tests were completed with GraphPad Prism Software. Specific tests and p values, which are determined as significant, are stated in the figure legends.
Results
Effects of the Ptpn22 proautoimmune allele on tumor growth kinetics
To determine whether the human relevant proautoimmune allele of Ptpn22 (PEP-619WW) influenced tumor control, we compared the growth of an immunogenic tumor cell line, B16-OVA, between PEP-WT and PEP-619WW mice. B16-OVA tumors were significantly smaller in PEP-619WW mice than in PEP-WT mice at 1 month postimplant (Fig. 1A–C). The role of the immune system in combating tumor growth is well established (28). Using Rag1−/− mice, which lack T and B lymphocytes, crossed to our PEP-619WW mice (Rag1−/− PEP-619WW mice; Supplemental Fig. 1A), we tested the necessity of adaptive immunity to control tumors in our mice bearing the Ptpn22 proautoimmune allele. Rag1−/− PEP-619WW mice showed no detectable difference in B16-OVA tumor volume compared with Rag1−/− PEP-WT mice (Supplemental Fig. 1B–D), suggesting the adaptive response is required for the 619WW mutation to control B16-OVA tumors.
FIGURE 1.
Ptpn22 proautoimmune allele-bearing mice have less antigenic tumor burden. PEP-WT (A, D, G, and J) or PEP-619WW (B, E, H, and K) mice were implanted (s.c.) with 2 × 105 B16-OVA cells (n = 8 each genotype) (A–C), 1 × 106 MC38 cells (n = 23 PEP-WT; n = 24 PEP-619WW) (D and E), 5 × 105 LLC cells (n = 14 PEP-WT; n = 13 PEP-619WW) (G–I), or 2 × 105 B16F10 (n = 15 PEP-WT; n = 14 PEP-619WW) (J–L). Tumor width and length were measured, and volume was calculated. Individual mice for each tumor type shown in (A), (B), (D), (E), (G), (H), (J), and (K). Mean with SEM shown in (C), (F), (I), and (L). Significance was determined using a t test with Welch’s correction at each time point to compare the two genotypes; *p < 0.05. Data shown are pooled from independent experiments (three for B16-OVA, three for MC38, two for LLC, three for B16-F10).
In addition, we tested growth of MC38, another immunogenic tumor line, and found growth was also significantly delayed in PEP-619WW mice (Fig. 1D–F). In contrast, there was no detectable difference in growth of LLC cells between PEP-WT and PEP-619WW mice (Fig. 1G–I). Also, when implanted with B16F10 cells, which are relatively poorly immunogenic, tumor growth was highly variable in PEP-619WW mice, whereas growth in PEP-WT mice was more consistent (Fig. 1J, 1K). About one-third of the PEP-619WW mice had delayed growth, and one mouse did not develop a palpable tumor until almost a month postimplant (Fig. 1K). Despite these few mice that seemed to have superior control of B16F10 tumors, as a group there is no significant difference between B16F10 tumor growth in PEP-WT and PEP-619WW mice (Fig. 1L). Taken together, these results suggest the expression of PEP-619WW can inhibit the growth of immunogenic tumors, yet is less effective against poorly immunogenic tumor lines.
Ptpn22 proautoimmune allele influences tumor-infiltrating immune cells
With PEP-619WW mice having increased tumor control of B16-OVA and MC38 tumors, we set out to define any changes within the immune microenvironment that may be contributing to this delayed tumor growth. We isolated tumor-infiltrating lymphocytes at 14 d after B16-OVA tumor implant. At this time point, tumors are still small, but there are sizable numbers of tumor-infiltrating lymphocytes, which are responsible for immune control of the tumor. In addition, this time point is when the size of tumors is comparable in PEP-WT and PEP-619WW mice and would provide insight into immune changes occurring just before divergence in growth kinetics. Tumors from PEP-619WW mice were found to have a high frequency of tumor-infiltrating CD45+ cells (Fig. 2A, 2B) at day 14. We also examined the density of CD45+ cells at a later time point, day 30, when B16-OVA tumors are different sizes. The density of CD45+ tumors was highly variable in both genotypes. There was a trend of increased CD45+ density in B16-OVA tumors from PEP-619WW at both time points (Fig. 1C). In preparing for an in-depth histochemical analysis of immune-rich regions in the tumor, tumor sections from PEP-WT and PEP-619WW mice were stained with anti-mouse CD45. This staining confirmed the flow cytometry data, allowing visualization of CD45+ infiltrate in the tumors from PEP-619WW mice (Fig. 2E). Tumors from PEP-619WW mice have more CD45+ cells throughout the tumor. In addition, the CD45 staining appeared to be more uniform in tumors from PEP-619WW mice, whereas CD45+ staining in tumors from PEP-WT mice was only in distinctive regions in the tissue (Fig. 2E). CD45+ tumor infiltration is more robust in mice bearing the Ptpn22 proautoimmune allele.
FIGURE 2.
Tumors from PEP-619WW mice have more immune cell infiltrate. B16-OVA tumor-infiltrating CD45+ cells were identified by flow cytometry. (A) Representative flow cytometry plots (lymphocytes > single-cell forward light scatter > single-cell side light scatter > live > CD45+) and (B) quantification of CD45+ tumor infiltrate, determined by flow cytometry. Density of CD45 cells in tumor (count total CD45/volume postresection tumor) at (C) 14 and (D) 30 d posttumor implant. (E) Representative microscopic images from NanoString GeoMX DSP platform indicating regions of interest within these tumor sections showing CD45 stain (red). Scale bar across top, 2 mm. Zoomed-in images of representative regions of tumors from PEP-WT (i and ii). Scale bar in (i), 1 mm; scale bar in (ii), 2 mm. PEP-619WW (iii and iv). Scale bar in (iii), 1 mm; scale bar in (iv), 1 mm. Flow cytometry data shown are from representative experiment. Flow cytometry experiments were repeated at least three times.
Tumors from PEP-619WW mice have more activated T cell populations
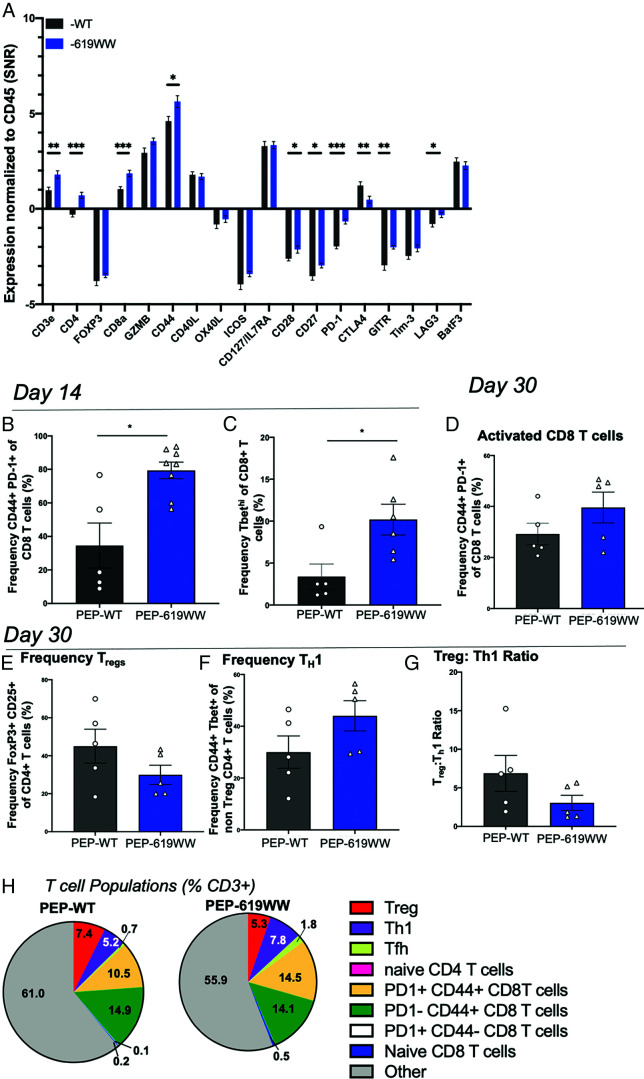
Considering the correlation between increased immune infiltration in tumors from PEP-619WW mice and reduced tumor growth, it was important to define the infiltrating immune cell types within the tumor-infiltrating milieu. We more closely examined T cell activation and myeloid cell activation. Using the NanoString Spatial Genomics GeoMX DSP platform, we were able to conduct an in-depth profile of immune-related proteins within the immune-dense regions of the tumor (day 14 postimplant), which were identified with anti-mouse CD45-Alexa 532 stain overlaying with DNA-SYTO13. We selected 12 ROIs in geometrically distinct areas selected over three tumor sections for each genotype, where each section was from a different tumor (Supplemental Fig. 2A, 2B). To define the differences within the T cell compartment, we first focused on the relative expression of T cell–related proteins normalized to CD45 expression within each ROI, which is measured by the signal-to-noise ratio. This revealed a significant increase in the expressed levels of CD3e, CD4, CD8a, CD44, CD28, CD27, PD-1, GITR, and Lag-3 in the immune-rich regions of tumors from PEP-619WW mice (Fig. 3A). In addition, there is a decrease of CTLA-4 abundance within the immune-rich regions of tumors from PEP-619WW mice (Fig. 3A).
FIGURE 3.
Enhanced T cell activation in tumors from PEP-619WW mice. (A) Relative abundance of indicated proteins (normalized to CD45+) within CD45+-rich regions of 14 d postimplant B16-OVA tumors; data are from NanoString GeoMX DSP platform. Flow cytometry analysis of whole tumor at 14 d postimplant. (B) Frequency CD44+ PD1+ Cd8 T cells (lymphocyte > single cell × 2 > live > CD45+ > CD3+ CD19− > CD8a+ CD4− >CD44+ PD-1+) and (C) Tbethi CD8 T cell (lymphocyte > single cell × 2 > live > CD45+ > CD3+ CD19− > CD8a+ CD4− > Tbethi). (D) Frequency of activated CD8 T cells at 30 d (same gating strategy as in B). Frequency of (E) Tregs (lymphocyte > single cell × 2 > live > CD45+ > CD3+ CD19− > CD8a− CD4+ > Foxp3+ CD25+), (F) Th1 cells (lymphocyte > single cell × 2 > live > CD45+ > CD3+ CD19− > CD8a− CD4+ > Foxp3− > CD44+ Tbet+), (G) ratio of Treg to Th1 (%Treg/%Th1), and (H) breakdown of multiple T cell phenotypes 30 d postimplant. T follicular helper (Tfh) cells identified as non-Tregs > CXCR5+ PD-1+. t-test with Welch’s correction, *p < 0.05, **p < 0.01, ***p < 0.001. Data shown are from a representative experiment. Experiments were repeated three times. SNR, single-to-noise ratio.
Although this data set allowed us to investigate immune-rich regions of the tumors, it did not look at the tumor as a whole or the level of expression of T cell–related markers on specific types of T cells. As such, we next turned to flow cytometry to globally examine T cell activation at a single-cell level. Within PEP-619WW mice, the CD8 T cell compartment exhibited a more activated phenotype as measured by CD44 and Tbet expression at 14 d postimplant (Fig. 3B, 3C). The increased CD8 T cell activation present at 14 d in tumors from PEP-619WW mice persisted at 1 mo postimplant. At this later time point, tumors from PEP-619WW retained an increased frequency of activated CD8 T cells (CD44+ PD-1+ CD8 T cells) (Fig. 3D, 3H). At 14 d after tumor implant, there was no difference detected in the amount of intratumoral Tregs or Th1 cells. In addition, there was no difference detected in the expression levels of PD-1 or Lag3 on intratumoral CD4 T cells at 14 d after tumor implant between the genotypes (Supplemental Fig. 3F–I). However, at 30 d after implant, PEP-619WW had altered CD4 T cell phenotypes present within the tumor as compared with tumors from PEP-WT mice. Tumors from PEP-619WW mice had a decreased frequency of Tregs (Foxp3+ CD25+ CD4 T cells) and increased frequency of Th1 cells (non-Tregs CD44+ Tbet+ CD4 T cells) (Fig. 3E, 3F, 3H). This rendered a significantly lower Treg/Th1 ratio in tumors from PEP-619WW mice (Fig. 3G). Importantly, there were no differences in the number of Tregs, naive CD4s, activated CD4s, and activated CD8 T cells in the spleens of naive animals of either genotype, a phenotype that was previously reported as a characteristic of PEP-null mice (Supplemental Fig. 3A–D) (24). There was also an increased proportion of T follicular helper cells in tumors from PEP-619WW mice compared with PEP-WT mice at both 14 and 30 d postimplant (Fig. 3H and Supplemental Fig. 3E).
PEP-619WW mice have increased myeloid cell activation in the tumor microenvironment
Although T cells are necessary for proper tumor control, the dendritic cell (DC) and myeloid compartment is also critical in supporting a cytotoxic T cell response (29, 30). In addition, the presence of suppressive myeloid cells can counteract a strong T cell–infiltrating response through expression of T cell inhibitory ligands (like PD-L1) and suppressive cytokine secretion (29). Again, using the NanoString Spatial genomics GeoMX/DSP platform, we focused on immune-dense regions within the tumors from PEP-WT and PEP-619WW mice to broadly examine myeloid- and tumor-associated markers. Tumors from PEP-619WW mice had a significant increase in the abundance of cellular identification markers CD19 and CD11c. In addition, tumors from PEP-619WW mice had increased amounts of T cell–activating receptors CD40 and MHC II within the immune-rich regions (Fig. 4A). Also, these same areas had a lesser abundance of myeloid-derived suppressor cell (MDSC) marker CD163, tumor-associated receptors B7-H3, and androgen receptor, which have been previously correlated with poor tumor clearance (Fig. 4A) (31–33).
FIGURE 4.
Enhanced myeloid cell activation in tumors from PEP-619WW mice. (A) Relative protein abundance (normalized to CD45) of indicated protein target from CD45-rich regions of interest in B16-OVA tumors from PEP-WT and PEP-619WW mice, 14 d postimplant. Data are from the NanoString GeoMX DSP platform. Tumors were harvested at 14 d postimplant and put in a single-cell suspension for flow cytometry analysis. (B) Frequency of tumor CD103+ cDCs of CD45+ cells (lymphocytes > single cells × 2 > live > CD45+ > CD19− CD3− > F4/80− > CD11c+ MHC II+ > CD103+ > CD8a+ CD11b−). (C) Density of CD103+ cDCs in tumors at 14 d postimplant. (D) Representative histogram (gray filled = unstained; black line = PEP-WT; blue line = PEP-619WW) and (E) quantification of PD-L1 surface expression on tumor-infiltrating CD103+ cDCs at 14 d postimplant. (F) Frequency of CD103+ cDCs of CD45+ tumor-infiltrating cells (lymphocytes > single cells × 2 > live > CD45+ > CD19− CD3− > F4/80− > CD11c+ MHC II+ > CD103+) 30 d postimplant. (G) Expression of PD-L1 on intratumoral CD103+ cDCs (gray filled = unstained; black line = PEP-WT; blue line = PEP-619WW); histogram is from concatenated CD103+ cDC population from one representative experiment and (H) quantified PD-L1 expression (geometric mean fluorescence intensity [gMFI]) on CD11b+ cDCs (lymphocytes > single cells × 2 > live > CD45+ > CD19− CD3− > F4/80− > CD11c+ MHC II+ > CD8a− CD11b+). Significance was determined using a t test with Welch’s correction, *p < 0.05, **p < 0.01, ***p < 0.001. Data shown are from a representative experiment. Experiments were repeated at least three times.
Next, we turned to flow cytometry to examine the immunostimulatory versus immunoinhibitory phenotype of the myeloid cells within the tumor microenvironment from PEP-WT and PEP-619WW mice at a single-cell level. We focused on DCs within the tumor microenvironment because these are critical for proper T cell activation and immune-mediated tumor clearance (30). Tumors from PEP-619WW mice had a higher proportion and density of conventional DC (cDC) type 1 (cDC1) cells (F4/80− CD11c+ MHC II+ CD103+) (Fig. 4B, 4C) at day 14 postimplant. In addition, these cDC1 cells had significantly lower levels of PD-L1 expression (Fig. 4D, 4E). At day 30 postimplant, tumors harvested from PEP-619WW mice also had more cDC1 cells (F4/80− CD11c+ CD103+) (Fig. 4F). At this later time point, there was similar expression of PD-L1 on cDC1 cells (Fig. 4G). Infiltrating cDC2 cells (F4/80− CD11c+ MHCII+ CD8a− CD11b+) had consistently high levels of PD-L1 expression (Fig. 4H).
Discussion
There is a strong genetic link associated with autoimmunity that drives overt immune activation (2, 34). Given this relationship, we wanted to know whether expression of a proautoimmune allele could drive a positive disease outcome in situations in which overt immune activation is favorable, such as cancer. Previously, using mice deficient in Ptpn22 expression, we and others have demonstrated that the protein tyrosine phosphatase Ptpn22 drives T cell exhaustion during viral infection (17, 18). Also, Brownlie et al. (19, 20) showed CD8 T cells lacking Ptpn22 were superior as adoptive cell transfer therapy of tumors compared with WT CD8 T cells. Recently, another group showed Ptpn22 knockout mice had enhanced rejection of MC38 and Hepa1-6.x1 during immunotherapy with checkpoint blockade (35). This same study briefly looked at the spontaneous rejection of Hepa1-6.x1 cells in mice bearing the Ptpn22 proautoimmune allele (PEP-619WW mice). There was a modest increase in spontaneous rejection compared with WT animals (35). However, the study did not look at other tumor lines or elaborate on the underlying immunological differences between the WT mice and the mice bearing the proautoimmune allelic variant of Ptpn22.
To test any influence the PTPN22 proautoimmune allelic variant has on tumor growth and the antitumor immune response, we used CRISPR/Cas9 to generate C57BL/6 mice that endogenously express the murine ortholog of the human relevant Ptpn22 variant (PEP-619WW). We tested the capacity of our PEP-619WW mice to control both immunogenic (B16-OVA and MC38) and poorly immunogenic (B16F10 and LLC) tumors (Fig. 1). After implantation of B16-OVA cells, PEP-619WW mice exhibited significantly better tumor control without any therapeutic intervention (Fig. 1A–C). PEP-619WW mice also had superior control of another immunogenic tumor, MC38 (Fig. 1D–F). However, when PEP-619WW mice were implanted with poorly immunogenic tumor line B16F10 melanoma cells, this significant increase in tumor control was mitigated (Fig. 1J–L). Furthermore, like their WT counterparts, PEP-619WW mice were not able to control the poorly immunogenic LLC tumor (Fig. 1G–I). These data suggest that the enhanced tumor control seen in PEP-619WW mice was driven by the presence of novel tumor-expressed Ags. Future studies will investigate the role of Ag specificity in enhancing tumor control in the PEP-619WW mice.
It is known that T cell activation is critical for the cytotoxic destruction of malignancies and is associated with improved survival of melanoma (36, 37). Indeed, using Rag1−/− mice, which lack lymphocytes, expressing PEP-619WW in the innate compartment can no longer control B16-OVA tumors (Supplemental Fig. 1B–D). In our immune-competent mice, tumors from PEP-619WW had more immune cell infiltration at both early (14 d postimplant) and later (30 d postimplant) time points (Fig. 2). When we focused on the immune-rich regions of the tumors (at day 14) using the NanoString GeoMX DSP platform, there was increased relative expression of CD3 and CD8a, suggesting more CD8 T cell infiltration in tumors from PEP-619WW mice (Fig. 3A). Furthermore, there was more CD44 and PD-1 expression, which are upregulated upon T cell activation, in tumors from PEP-619WW mice (Fig. 3A). Lastly, there was less CTLA-4 and more CD28 relative expression in tumors from PEP-619WW compared with PEP-WT (Fig. 3A). CTLA-4 binds to CD80, restricting further activation through the interaction of CD80 and CD28 (38, 39). Lower levels of CTLA-4 and increased levels of CD28 within the tumors from PEP-619WW suggest that these T cells are receiving more activating than suppressive signals. Using the GeoMX platform, we were able to focus on immune-rich regions in the tumors. This was important because tumors from the PEP-619WW mice had more CD45 infiltration overall. As such, we wanted to determine whether the immune epicenters of the tumors differed between the PEP-WT and PEP-619WW mice. Although this did allow us to address the global differences within those immune rich-regions, this platform does not allow the user to focus on a single-cell level or the tumor as a whole, which was a significant limitation. When markers are distinct for a single-cell subset (such as Foxp3), this does not matter as much. However, when a marker is shared among many cell types, such as CD44, the results are less specific and you must follow up with subsequent methods to address any differences of shared markers at a single-cell level.
To assess T cell activation at a single-cell level, we used flow cytometry. Flow cytometric analysis confirmed that tumors from PEP-619WW mice had significantly more activated CD8 T cells infiltrating at 14 d postimplant (Fig. 3C, 3D). We also show that tumors from PEP-619WW have a persistent CD8 T cell activation present at 30 d posttumor implant (Fig. 3E). It is well characterized that the proautoimmune allele of Ptpn22 can enhance CD8 T cell activation (40); to our knowledge, this is the first such demonstration within the tumor microenvironment. Future research will assess the killing capacity and functionality of the tumor-infiltrating CD8 T cells from PEP-619WW mice and whether expressing this proautoimmune allele has any advantage for adoptive cell therapy, as seen for T cells lacking Ptpn22 (19, 20).
CD4 T cell subsets also play a key role in the antitumor response (41, 42). Previous studies have shown that Ptpn22 and its proautoimmune allele do influence CD4 T cell activation and function (40). Tregs are associated with poor tumor control, because these cells can temper cytotoxic T cell function (39). Mice lacking Ptpn22 have more Tregs and increased potency of Treg function compared with WT mice (24, 43). However, mice bearing the proautoimmune allele of Ptpn22 do not have an increased amount of Tregs under homeostatic conditions (1) (Supplemental Fig. 3). In this study, we show that at 1 month postimplant, PEP-619WW actually have fewer tumor-infiltrating Tregs compared with tumors from PEP-WT mice (Fig. 3F, 3G, 3H). In addition, tumors from PEP-619WW mice had more Th1 and T follicular helper cells, both of which are reported to correlate with positive outcomes in cancer, compared with tumors from PEP-WT mice (42) (Fig. 3G, 3H). Taken together, these data suggest that the Ptpn22 proautoimmune allele promotes a more activated tumor-infiltrating T cell population.
Another important consideration for proper tumor control is the presence of MDSCs. Ptpn22 can influence the polarization of macrophages (44, 45). Within immune-rich regions, we did observe that PEP-WT mice had more CD163 relative expression, suggesting a higher frequency of MDSCs present in the tumor (Fig. 4A). Although tumors from PEP-619WW mice had a high level of CD11b expression, they did not have high levels of CD163 expression, suggesting these were not MDSCs in the tumor microenvironment (Fig. 4A).
Within these immune-rich regions, we also observe increased expression of CD11c, suggesting more DCs, in tumors from the PEP-619WW mice (Fig. 4A). Tumors from PEP-619WW mice also exhibited greater expression levels of CD40 and MHC II, both of which serve as activators of the T cell response (Fig. 4A). In addition, there is less inhibitory checkpoint receptor VISTA expression in tumors from PEP-619WW mice (46) (Fig. 4A). Taken together, this suggests that the tumor-infiltrating DCs in PEP-619WW are better equipped for T cell activation.
To interrogate the DCs at a single-cell level, we again turned to flow cytometry. Confirming the GeoMX DSP data, there were more tumor-infiltrating DCs in PEP-619WW, specifically cDC1 cells (Fig. 4B, 4C, 4F). cDC1 cells are critical for priming a CD8 T cell response and mounting a robust and proper antitumor immune response, which leads to better tumor control (30). More cDC1 cells have been associated with better tumor control and a more activated CD8 T cell phenotype. Not only were the cDC1 cells more abundant in the PEP-619WW tumor-bearing mice, but these cells also had significantly less inhibitory receptor PD-L1 surface expression (Fig. 4D, 4E). To our knowledge, we are the first to put forward a link between this proautoimmune allele and PD-L1 expression. The difference in PD-L1 expression between PEP-619WW and PEP-WT cells was specific to the cDC1 compartment at earlier time points (day 14), and cDC2 cells (CD11b+ CD8a− CD11c+ MHC II+) did not exhibit a difference in PD-L1 expression within the tumor (Fig. 4H), nor did CD103+ cDCs at 30 d postimplant. Clinically, the blockade of PD-L1 and its cognate receptor PD-1 has had success in promoting tumor clearance (47, 48). In the B16-OVA system, tumor control is greater when anti-PD-L1 is paired with an Ag-loaded DC immunization and OVA-specific T cells adoptive transfer therapy (49). Future studies will define the molecular mechanism of the PEP-619WW molecule that is affecting PD-L1 expression and further define whether this difference in PD-L1 expression between the two genotypes is required for tumor control.
This study explores the role of the tyrosine phosphatase PTPN22 proautoimmune allele on tumor control. Broadly, we have determined that proautoimmune alleles can influence the antitumor response by promoting a more proinflammatory environment and overcoming the anti-inflammatory signals present during tumor growth. Interestingly, the presence of this proautoimmune allele affects both the T cell and non-T cell compartment to promote better control of tumor growth. In humans, the PTPN22 proautoimmune allele, rs2476601, is associated with a lower incidence of melanoma and other malignant neoplasms of the skin (C43–C44) (p = 1.8196e−8; Gene Atlas UK biobank population). Currently, we do not know whether the enhancement in the T cell response, myeloid cell activation, or both is driving the difference in tumor control between our two strains of mice. Transfer of PEP-619WW T cells into a lymphopenic environment (such as into a Rag1−/− mouse or lethally irradiated animal) preferentially differentiate into Tregs at a higher proportion compared with PEP-WT T cells (25). Therefore, isolating lymphocytes or myeloid cells as the sole Ptpn22 differing cell through adoptive transfer techniques into immune deficient hosts does not properly recapitulate the phenotypes observed in our immune-competent mice. Future studies will need to examine which PEP-619WW-bearing immune cells are necessary and/or sufficient to control tumors, as well as understand whether these results could translate into improved therapies for patients. Results of this study provide a platform to further investigate the concerted relationships between immune cells to achieve a desired phenotype, as well as determine whether and how other proautoimmune alleles affect the antitumor immune response.
Supplementary Material
Acknowledgments
We thank Scripps Research Flow Cytometry Core, Histology and Microscopy Core, and Vivarium staff for their expertise and assistance in this work. We also thank C. Alvarez for help with cutting tissue sections, as well as careful reading and editing of this manuscript, and L. Zhang, NanoString, for patience and assistance in going through the data from the NanoString GeoMX DSP platform. Since the creation of PEP-619WW mice, K. Mowen has passed away and is greatly missed.
R.C.O. conceptualized, designed, and performed experiments; analyzed results; and wrote and edited the manuscript. K. Marquardt provided reagents, bred and genotyped mice, and performed experiments. K. Mowen was the principal investigator whose laboratory generated the PEP-619WW mice on the C57BL/6 background. L.A.S. conceptualized and designed experiments, reviewed data, and wrote and edited the manuscript.
This work was supported by Department of Health and Human Services/National Institute of Allergy and Infectious Diseases/National Institutes of Health Grants U01 AI130842 (to L.A.S.) and T32 AI007354 27 (fellowship to support R.C.O.).
Linda A. Sherman is a Distinguished Fellow of AAI.
The online version of this article contains supplemental material.
- cDC
- conventional dendritic cell
- cDC1
- conventional DC type 1
- DC
- dendritic cell
- DSP
- digital spatial profiling
- LLC
- Lewis lung carcinoma
- Lyp
- lymphoid tyrosine phosphatase
- MDSC
- myeloid-derived suppressor cell
- MHC II
- MHC class II
- PEP
- PEST domain–enriched protein
- PTPN22
- protein tyrosine phosphatase nonreceptor type 22
- ROI
- region of interest
- TRAF
- TNFR-associated factor
- Treg
- T regulatory cell
- WT
- wild-type
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Dai X., James R. G., Habib T., Singh S., Jackson S., Khim S., Moon R. T., Liggitt D., Wolf-Yadlin A., Buckner J. H., Rawlings D. J.. 2013. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J. Clin. Invest. 123: 2024–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burn G. L., Svensson L., Sanchez-Blanco C., Saini M., Cope A. P.. 2011. Why is PTPN22 a good candidate susceptibility gene for autoimmune disease? FEBS Lett. 585: 3689–3698. [DOI] [PubMed] [Google Scholar]
- 3.Bottini N., Vang T., Cucca F., Mustelin T.. 2006. Role of PTPN22 in type 1 diabetes and other autoimmune diseases. Semin. Immunol. 18: 207–213. [DOI] [PubMed] [Google Scholar]
- 4.Cohen S., Dadi H., Shaoul E., Sharfe N., Roifman C. M.. 1999. Cloning and characterization of a lymphoid-specific, inducible human protein tyrosine phosphatase, Lyp. Blood 93: 2013–2024. [PubMed] [Google Scholar]
- 5.Lin X., Pelletier S., Gingras S., Rigaud S., Maine C. J., Marquardt K., Dai Y. D., Sauer K., Rodriguez A. R., Martin G., et al. 2016. CRISPR-Cas9-Mediated Modification of the NOD Mouse Genome With Ptpn22R619W Mutation Increases Autoimmune Diabetes. Diabetes 65: 2134–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez L. M., Anaya J. M., Gonzalez C. I., Pineda-Tamayo R., Otero W., Arango A., Martín J.. 2005. PTPN22 C1858T polymorphism in Colombian patients with autoimmune diseases. Genes Immun. 6: 628–631. [DOI] [PubMed] [Google Scholar]
- 7.Di Y., Zhong S., Wu L., Li Y., Sun N.. 2015. The Association between PTPN22 Genetic Polymorphism and Juvenile Idiopathic Arthritis (JIA) Susceptibility: An Updated Meta-Analysis. Iran. J. Public Health 44: 1169–1175. [PMC free article] [PubMed] [Google Scholar]
- 8.Chiaroni-Clarke R. C., Li Y. R., Munro J. E., Chavez R. A., Scurrah K. J., Pezic A., Akikusa J. D., Allen R. C., Piper S. E., Becker M. L., et al. 2015. The association of PTPN22 rs2476601 with juvenile idiopathic arthritis is specific to females. Genes Immun. 16: 495–498. [DOI] [PubMed] [Google Scholar]
- 9.Cao Y., Liu K., Tian Z., Hogan S. L., Yang J., Poulton C. J., Falk R. J., Li W.. 2015. PTPN22 R620W polymorphism and ANCA disease risk in white populations: a metaanalysis. J. Rheumatol. 42: 292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X. H., Ma A. G., Han X. X., Chen L., Liang H., Aishan-Litifu, Abudumijit-Ablez, Xue F.. 2017. Protein tyrosine phosphatase nonreceptor type 22 (PTPN22) gene single nucleotide polymorphisms and its interaction with T2DM on pulmonary tuberculosis in Chinese Uygur population. Oncotarget 8: 65601–65608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang H., Wang Z., Rani P. L., Fu X., Yu W., Bao F., Yu G., Li J., Li L., Sun L., et al. 2017. Identification of PTPN22, ST6GAL1 and JAZF1 as psoriasis risk genes demonstrates shared pathogenesis between psoriasis and diabetes. Exp. Dermatol. 26: 1112–1117. [DOI] [PubMed] [Google Scholar]
- 12.Wang W., Meng X., Liu Y., Ma X., Zhang Q., Li C., Li C., Ren L.. 2017. Association Between Protein Tyrosine Phosphatase Non-Receptor Type 22 (PTPN22) Polymorphisms and Risk of Ankylosing Spondylitis: A Meta-analysis. Med. Sci. Monit. 23: 2619–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hasegawa K., Martin F., Huang G., Tumas D., Diehl L., Chan A. C.. 2004. PEST domain-enriched tyrosine phosphatase (PEP) regulation of effector/memory T cells. Science 303: 685–689. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J., Zahir N., Jiang Q., Miliotis H., Heyraud S., Meng X., Dong B., Xie G., Qiu F., Hao Z., et al. 2011. The autoimmune disease-associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat. Genet. 43: 902–907. [DOI] [PubMed] [Google Scholar]
- 15.Wallis A. M., Wallace E. C., Hostager B. S., Yi Z., Houtman J. C. D., Bishop G. A.. 2017. TRAF3 enhances TCR signaling by regulating the inhibitors Csk and PTPN22. Sci. Rep. 7: 2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y., Shaked I., Stanford S. M., Zhou W., Curtsinger J. M., Mikulski Z., Shaheen Z. R., Cheng G., Sawatzke K., Campbell A. M., et al. 2013. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity 39: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maine C. J., Teijaro J. R., Marquardt K., Sherman L. A.. 2016. PTPN22 contributes to exhaustion of T lymphocytes during chronic viral infection. Proc. Natl. Acad. Sci. USA 113: E7231–E7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jofra T., Galvani G., Kuka M., Di Fonte R., Mfarrej B. G., Iannacone M., Salek-Ardakani S., Battaglia M., Fousteri G.. 2017. Extrinsic Protein Tyrosine Phosphatase Non-Receptor 22 Signals Contribute to CD8 T Cell Exhaustion and Promote Persistence of Chronic Lymphocytic Choriomeningitis Virus Infection. Front. Immunol. 8: 811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brownlie R. J., Garcia C., Ravasz M., Zehn D., Salmond R. J., Zamoyska R.. 2017. Resistance to TGFβ suppression and improved anti-tumor responses in CD8+ T cells lacking PTPN22. Nat. Commun. 8: 1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brownlie R. J., Wright D., Zamoyska R., Salmond R. J.. 2019. Deletion of PTPN22 improves effector and memory CD8+ T cell responses to tumors. JCI Insight 4: e127847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vang T., Congia M., Macis M. D., Musumeci L., Orrú V., Zavattari P., Nika K., Tautz L., Taskén K., Cucca F., et al. 2005. Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat. Genet. 37: 1317–1319. [DOI] [PubMed] [Google Scholar]
- 22.Gregorieff A., Cloutier J.-F., Veillette A.. 1998. Sequence requirements for association of protein-tyrosine phosphatase PEP with the Src homology 3 domain of inhibitory tyrosine protein kinase p50(csk). J. Biol. Chem. 273: 13217–13222. [DOI] [PubMed] [Google Scholar]
- 23.Cloutier J. F., Veillette A.. 1996. Association of inhibitory tyrosine protein kinase p50csk with protein tyrosine phosphatase PEP in T cells and other hemopoietic cells. EMBO J. 15: 4909–4918. [PMC free article] [PubMed] [Google Scholar]
- 24.Maine C. J., Marquardt K., Cheung J., Sherman L. A.. 2014. PTPN22 controls the germinal center by influencing the numbers and activity of T follicular helper cells. J. Immunol. 192: 1415–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knipper J. A., Wright D., Cope A. P., Malissen B., Zamoyska R.. 2020. PTPN22 Acts in a Cell Intrinsic Manner to Restrict the Proliferation and Differentiation of T Cells Following Antibody Lymphodepletion. Front. Immunol. 11: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lechner M. G., Karimi S. S., Barry-Holson K., Angell T. E., Murphy K. A., Church C. H., Ohlfest J. R., Hu P., Epstein A. L.. 2013. Immunogenicity of murine solid tumor models as a defining feature of in vivo behavior and response to immunotherapy. J. Immunother. 36: 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong W., Myers J. S., Wang F., Wang K., Lucas J., Rosfjord E., Lucas J., Hooper A. T., Yang S., Lemon L. A., et al. 2020. Comparison of the molecular and cellular phenotypes of common mouse syngeneic models with human tumors. BMC Genomics 21: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth M. J., Dunn G. P., Schreiber R. D.. 2006. Cancer immunosurveillance and immunoediting: the roles of immunity in suppressing tumor development and shaping tumor immunogenicity. In Advances in Immunology, Academic Press, San Diego, CA, p. 1–50. [DOI] [PubMed] [Google Scholar]
- 29.Haas L., Obenauf A. C.. 2019. Allies or enemies—the multifaceted role of myeloid cells in the tumor microenvironment. Front. Immunol. 10: 2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Böttcher J. P., Reis e Sousa C., Sousa C.. 2018. The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer 4: 784–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang S., Wei W., Zhao Q.. 2020. B7-H3, a checkpoint molecule, as a target for cancer immunotherapy. Int. J. Biol. Sci. 16: 1767–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickey T. E., Selth L. A., Chia K. M., Laven-Law G., Milioli H. H., Roden D., Jindal S., Hui M., Finlay-Schultz J., Ebrahimie E., et al. 2021. The androgen receptor is a tumor suppressor in estrogen receptor-positive breast cancer. Nat. Med. 27: 310–320. [DOI] [PubMed] [Google Scholar]
- 33.Fujita K., Nonomura N.. 2019. Role of androgen receptor in prostate cancer: a review. World J. Mens Health 37: 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramos P. S., Shedlock A. M., Langefeld C. D.. 2015. Genetics of autoimmune diseases: insights from population genetics. J. Hum. Genet. 60: 657–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cubas R., Khan Z., Gong Q., Moskalenko M., Xiong H., Ou Q., Pai C., Rodriguez R., Cheung J., Chan A. C.. 2020. Autoimmunity linked protein phosphatase PTPN22 as a target for cancer immunotherapy. J. Immunother. Cancer 8: e001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Leun A. M., Thommen D. S., Schumacher T. N.. 2020. CD8+ T cell states in human cancer: insights from single-cell analysis. Nat. Rev. Cancer 20: 218–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clemente C. G., Mihm M. C. Jr., Bufalino R., Zurrida S., Collini P., Cascinelli N.. 1996. Prognostic value of tumor infiltrating lymphocytes in the vertical growth phase of primary cutaneous melanoma. Cancer 77: 1303–1310. [DOI] [PubMed] [Google Scholar]
- 38.Krummel M. F., Allison J. P.. 1995. CD28 and CTLA-4 have opposing effects on the response of T cells to stimulation. J. Exp. Med. 182: 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker L. S. K.2013. Treg and CTLA-4: two intertwining pathways to immune tolerance. J. Autoimmun. 45: 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bottini N., Peterson E. J.. 2014. Tyrosine phosphatase PTPN22: multifunctional regulator of immune signaling, development, and disease. Annu. Rev. Immunol. 32: 83–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bos R., Sherman L. A.. 2010. CD4+ T-cell help in the tumor milieu is required for recruitment and cytolytic function of CD8+ T lymphocytes. Cancer Res. 70: 8368–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim H.-J., Cantor H.. 2014. CD4 T-cell subsets and tumor immunity: the helpful and the not-so-helpful. Cancer Immunol. Res. 2: 91–98. [DOI] [PubMed] [Google Scholar]
- 43.Brownlie R. J., Miosge L. A., Vassilakos D., Svensson L. M., Cope A., Zamoyska R.. 2012. Lack of the phosphatase PTPN22 increases adhesion of murine regulatory T cells to improve their immunosuppressive function. Sci. Signal. 5: ra87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M., Beauchemin H., Popovic N., Peterson A., d’Hennezel E., Piccirillo C. A., Sun C., Polychronakos C.. 2017. The common, autoimmunity-predisposing 620Arg > Trp variant of PTPN22 modulates macrophage function and morphology. J. Autoimmun. 79: 74–83. [DOI] [PubMed] [Google Scholar]
- 45.Chang H. H., Miaw S. C., Tseng W., Sun Y. W., Liu C. C., Tsao H. W., Ho I. C.. 2013. PTPN22 modulates macrophage polarization and susceptibility to dextran sulfate sodium-induced colitis. J. Immunol. 191: 2134–2143. [DOI] [PubMed] [Google Scholar]
- 46.Huang X., Zhang X., Li E., Zhang G., Wang X., Tang T., Bai X., Liang T.. 2020. VISTA: an immune regulatory protein checking tumor and immune cells in cancer immunotherapy. J. Hematol. Oncol. 13: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong H., Strome S. E., Salomao D. R., Tamura H., Hirano F., Flies D. B., Roche P. C., Lu J., Zhu G., Tamada K., et al. 2002. Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat. Med. 8: 793–800. [DOI] [PubMed] [Google Scholar]
- 48.Turajlic S., Gore M., Larkin J.. 2018. First report of overall survival for ipilimumab plus nivolumab from the phase III Checkmate 067 study in advanced melanoma. Ann. Oncol. 29: 542–543. [DOI] [PubMed] [Google Scholar]
- 49.Pilon-Thomas S., Mackay A., Vohra N., Mulé J. J.. 2010. Blockade of programmed death ligand 1 enhances the therapeutic efficacy of combination immunotherapy against melanoma. J. Immunol. 184: 3442–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.