Abstract
Numerous assays which use conserved DNA or rRNA sequences as targets for amplification have been described for the diagnosis of tuberculosis. However, these techniques have not been applied successfully to the monitoring of therapeutic efficacy owing to the persistence of amplifiable nucleic acid beyond the point at which smears and cultures become negative. Semiquantitative analysis of rRNA has been used to reduce the time required for antimicrobial susceptibility testing of Mycobacterium tuberculosis, although growth for up to 5 days in the presence of some drugs is still required to discriminate resistant strains. The purpose of the present study was to determine whether quantitative analysis of M. tuberculosis mRNA could be used to assess bacterial viability and to illustrate the application of this technique to rapid determination of drug susceptibility. Levels of mRNA encoding the 85B protein (α-antigen), IS6110 DNA, and 16S rRNA were compared in parallel cultures of M. tuberculosis that were treated with either no drug, 0.2 μg of isoniazid per ml, or 1 μg of rifampin per ml. Exposure of sensitive strains to isoniazid or rifampin for 24 h reduced the levels of 85B mRNA to <4 and <0.01%, respectively, of those present in control cultures without drug. In contrast, the levels of IS6110 DNA and 16S rRNA did not diminish over the same period. Strains which were resistant to either isoniazid or rifampin demonstrated no reduction in 85B mRNA in the presence of the drug to which they were nonresponsive. Quantitative analysis of 85B mRNA offers a potentially useful tool for the rapid determination of M. tuberculosis drug susceptibility and for the monitoring of therapeutic efficacy.
The resurgence of tuberculosis in the United States over the past decade and its continued dominance as a cause of morbidity and mortality (36) have focused attention on the need for more rapid and reliable means of diagnosis. Numerous assays for the detection of DNA and rRNA sequences which are specific for the Mycobacterium tuberculosis complex have now been described (11, 22, 23, 37, 42, 44). Although beneficial to the initial diagnosis of infection, such assays have proven to be an inappropriate substitute for conventional microbiological methods of monitoring the response of patients to therapy. Mitchison (30) has stated that the most important criterion for successful treatment of tuberculosis, besides the prevention of drug resistance, is sputum conversion and sterilization of active lesions. Typically, smears and cultures become negative within 2 to 3 months of the start of effective chemotherapy. However, we have demonstrated the persistence of M. tuberculosis DNA in sputum >12 months after the start of treatment and >6 months after culture conversion for some patients (20). Similarly, Moore et al. (33) observed a poor correlation between smear and culture results and the presence of M. tuberculosis 16S rRNA in sputum from patients who were treated with a standard four-drug regimen. As a result of these and other studies, molecular diagnostic assays that use DNA and rRNA targets have not been approved by the U.S. Food and Drug Administration for use with specimens from patients receiving antituberculosis therapy.
We have previously described a reverse transcriptase (RT) PCR (RT-PCR) for the mRNA encoding the M. tuberculosis complex 85B protein (α-antigen) (9), one of the most abundantly expressed proteins in both broth cultures and human mononuclear phagocytes (18, 26, 45). The purpose of the present study was to validate the use of this mRNA species as a marker for bacterial viability and to demonstrate the ability of a quantitative RT-PCR to discriminate between drug-sensitive and drug-resistant strains of M. tuberculosis in vitro. Such a distinction has grown increasingly important with the emergence of multidrug-resistant organisms and the high mortality rates associated with infections with such organisms (13, 14, 35). Prokaryotic mRNA, in contrast to DNA and rRNA, is rapidly degraded, with a typical half-life of 3 min (2, 3, 43). As a result, an mRNA-based amplification assay is likely to detect only viable organisms and be a good indicator of susceptibility to antimicrobial drugs and/or chemotherapeutic efficacy. Our original qualitative RT-PCR assay for 85B mRNA has been converted to a quantitative format with the Applied BioSystems Prism 7700 Sequence Detection System (19), which offers greater speed and reproducibility than is possible with conventional quantitative competitive PCR assays (8). We describe a comparison of the results of culture in the presence of either isoniazid (INH) or rifampin (RIF) and those of quantitative assays for M. tuberculosis 85B mRNA and IS6110 DNA and a semiquantitative RT-PCR assay for mycobacterial 16S rRNA.
MATERIALS AND METHODS
Strains.
The following strains of M. tuberculosis H37Rv were obtained from the American Type Culture Collection (ATCC): ATCC 27294 (sensitive to both INH and RIF), ATCC 35838 (RIF resistant; MIC, 250 μg/ml), and ATCC 33823 (INH and streptomycin resistant; MICs, 25 and 500 μg/ml, respectively).
Culture conditions.
All cultures were incubated at 37°C in an atmosphere of 5% CO2. Liquid cultures were grown in Dubos broth (Difco Laboratories, Detroit, Mich.) containing 10% Dubos medium albumin, 0.5% glycerol, and 0.1% Tween 80. Colony counts for viable bacilli were performed by preparing serial 10-fold dilutions of cells in fresh broth without antimycobacterial drugs and plating on Dubos agar (Difco) containing 10% Dubos oleic albumin complex, 0.5% glycerol, and 0.1% Tween 80.
Each of the three strains of M. tuberculosis was grown to the mid-logarithmic phase in Dubos broth and was adjusted to a McFarland standard of 2 to 3, equivalent to approximately 106 CFU/ml. Standardized suspensions were diluted to 104 to 105 CFU/ml in 75 ml of fresh medium and were incubated for a further 4 days in 225-cm2 tissue culture flasks with gentle agitation (40 rpm). Cultures were then split into three equal volumes to which was added either no drug (control), 0.2 μg of INH per ml, or 1.0 μg of RIF per ml (final concentrations). Each volume of culture was subdivided into 3-ml aliquots in screw-cap plastic tubes (16 by 150 mm) which were incubated without agitation for up to a further 72 h. Viable counts were performed after 0, 24, and 72 h by plating on solid medium, and at each time point, 1-ml aliquots were frozen at −70°C for subsequent extraction of DNA and RNA. Three samples were collected at time zero, and two more were collected at each time point thereafter.
Nucleic acid extraction.
To ensure consistent efficiency of nucleic acid recovery within each strain of M. tuberculosis, samples collected under the three different growth conditions (control without drug, plus INH, or plus RIF) were processed simultaneously. Duplicate samples were extracted at each time point. Extraction controls comprising standardized aliquots of M. tuberculosis ATCC 27294 were also included in each run. Bacteria were lysed with a FastPrep FP120 cell disrupter and Blue FastRNA Tubes (both from Bio 101, Inc., La Jolla, Calif.). RNA was extracted by a modified guanidine-acid phenol lysis procedure as described previously (9). DNA was recovered from the same sample by backextraction of the interface layers with a basic salt solution followed by ethanol precipitation (8).
Quantitative PCR for IS6110 DNA.
IS6110 DNA was quantified with an ABI Prism 7700 Sequence Detection System (Applied BioSystems Division, Perkin-Elmer, Foster City, Calif.) as described by DesJardin et al. (8). In brief, a 163-bp region of IS6110 was amplified with primers IS6 (5′-GGCTGTGGGTAGCAGACC) and IS7 (5′-CGGGTCCAGATGGCTTGC), which correspond to nucleotides 807 to 824 and 969 to 952, respectively, of the insertion sequence (29). An oligonucleotide detector probe with the sequence 5′-TGTCGACCTGGGCAGGGTTCG was labeled at its 5′ and 3′ termini with 5-carboxyfluorescein (FAM) and N,N,N′,N′-tetramethyl-6-carboxyrhodamine (TAMRA), respectively. The probe hybridizes to the intervening region between the two PCR primers and is degraded during the course of amplification by the 5′-3′ exonuclease activity of the Taq DNA polymerase (21). This results in release of the reporter dye (FAM) from the proximity of the quencher dye (TAMRA) and an increase in fluorescence proportional to the amount of specific PCR product generated (27). Duplicate amplification reactions were performed on each DNA sample by using standard buffer conditions and the following cycling parameters: 50°C for 2 min and 95°C for 5 min, followed by 40 cycles of 94°C for 30 s and 68°C for 1 min. The quantity of DNA in each reaction was determined during the exponential phase of amplification from the cycle threshold (CT), which is defined as the fractional cycle number that reflects a positive PCR result, and with reference to a standard curve generated by amplification of known amounts of IS6110 DNA. The cycle threshold was set at 10 times the standard deviation of the mean baseline emission between cycles 3 and 15 of amplification. To compensate for background fluorescence, the signal generated by the reporter dye was normalized by using the TAMRA emission spectrum (19). The majority of samples became positive after between 17 and 35 cycles of amplification.
Quantitative RT-PCR for 85B mRNA.
The 85B mRNA was quantified following the synthesis of cDNA by using avian myeloblastosis RT (Boehringer-Mannheim, Indianapolis, Ind.). Briefly, 5 μl of a total volume of 100 μl of extracted RNA was reverse transcribed in a 20-μl reaction volume (9). The reaction mixtures were incubated in a GeneAmp PCR System 9600 thermal cycler (Perkin-Elmer) at 42°C for 1 h, and the reactions were terminated by heating at 95°C for 5 min. Parallel reactions were performed without the addition of AMV RT in order to monitor for DNA contamination of the RNA extract. Five microliters of the reverse transcription mixture was subsequently amplified by PCR.
The efficiency of reverse transcription was determined with in vitro transcripts of a modified M. tuberculosis 85B gene. The ∼600-bp EcoRI-SacII fragment of the M. tuberculosis H37Rv 85B gene (10) was amplified by PCR, cloned into the plasmid vector pBluescript KS+ (Stratagene, La Jolla, Calif.), and modified by insertion at the NarI site of an 80-bp fragment of the Mycobacterium avium plasmid pLR7 (1). In vitro transcripts were generated from the pBluescript T3 RNA polymerase promoter with a MAXIscript T3 Kit (Ambion, Austin, Tex.) according to the manufacturer’s instructions. Transcripts were determined to be of the correct size by denaturing gel electrophoresis and were quantified by spectrophotometry. Comparison with in vitro transcripts of unmodified 85B mRNA without the 80-bp insert demonstrated equivalent reverse transcription and amplification efficiencies for both targets. Dilutions of control transcripts ranging from 102 to 104 molecules/μl in 10 ng of yeast carrier RNA (Ambion) per μl were included in each RT-PCR assay. The efficiency of reverse transcription was defined as the ratio of the number of input control mRNA targets to the observed number of DNA molecules detected in each reaction, as determined by reading from a standard curve for amplified 85B DNA. Typical efficiencies ranged from 10 to 30%. However, since all RT-PCRs for a given strain were performed simultaneously, variations in reverse transcription efficiency did not influence the overall pattern of the results.
Following reverse transcription, amplification and detection of 85B cDNA were performed with an ABI Prism 7700 Sequence Detection System with the PCR primers described by DesJardin et al. (9) and a dually labeled oligonucleotide detector probe with the sequence 5′-(FAM)-TCGAGTGACCCGGCATGGGAGCGT-3′-(TAMRA). PCR was performed with 50-μl reaction mixtures containing 10 mM Tris-HCl (pH 8.3); 50 mM KCl; 3 mM MgCl2; 0.2 μM PCR primers; 100 nM detector probe; 200 μM dATP, dCTP, and dGTP and 400 μM dUTP (all deoxynucleoside triphosphates were from Pharmacia, Piscataway, N.J.); 5 μg of bovine serum albumin (Pharmacia); 500 ng of yeast RNA (Ambion), 1 U of uracil-DNA-glycosylase (New England Biolabs, Beverly, Mass.); and 1 U of Taq DNA polymerase (BioLase; ISC BioExpress, Kaysville, Utah). Amplification was carried out by using the following cycling parameters: 50°C for 2 min and 95°C for 5 min, followed by 40 cycles of 94°C for 30 s and 68°C for 1 min. As with the IS6110 DNA assay, the TAMRA emission spectrum was used to standardize for background fluorescence. The threshold value of fluorescence for a positive PCR result was set at 10 times the standard deviation of the mean baseline emission between cycles 3 and 15 of amplification. The quantity of DNA in each reaction mixture was determined from the CT value with reference to a standard curve generated by amplification of known amounts of target DNA from Mycobacterium bovis ATCC 19210. Standards containing between 5 and 78,125 copies of 85B DNA per reaction mixture were included each time that an assay was run. Graphs of the starting DNA concentration against the CT value were consistently linear over this range of input targets, and CT values for samples and standards were all obtained at between 18 and 35 cycles of amplification.
PCR amplification of each reverse transcription reaction was performed twice by using separate standard curves, and the mean value obtained from the replicate determinations was used in subsequent calculations. The number of molecules of 85B mRNA per milliliter of culture was calculated by dividing the observed number of DNA molecules per reaction mixture by the efficiency of reverse transcription as determined with the control RNA transcripts (see above) and by correcting for the dilution factors involved in reverse transcription and PCR amplification. In each case, to correct for DNA contamination of the extracted RNA, the amount of 85B DNA detected in the absence of RT was subtracted from the value obtained when the enzyme was included in the reaction mixture. No samples assayed in the absence of RT yielded values of >10% of those in the presence of the enzyme, indicating that there was no significant contamination of RNA with DNA.
Semiquantitative analysis of 16S rRNA.
RT-PCR for mycobacterial 16S rRNA was performed as described previously (9) with 5 μl of a 1:10,000 dilution of the RNA extracted from each sample. In each case, control reactions to monitor DNA contamination were performed with 5 μl of a 1:10 dilution of the extracted RNA. PCR amplification was carried out with 32P-labeled oligonucleotide primers, and the products were visualized by autoradiography following electrophoresis through 8% polyacrylamide gels. As controls for the efficiency of amplification, PCRs for the 16S rRNA gene were performed with dilutions of genomic DNA from M. bovis ATCC 19210.
RESULTS
Rationale.
The purpose of the present study was to determine whether quantitative assays for DNA, rRNA, and mRNA could provide a useful alternative to conventional culture for assessing the viability of M. tuberculosis following exposure to two frontline antimycobacterial agents. Parallel cultures of three isogenic strains of M. tuberculosis were exposed to either no drug, 0.2 μg of INH per ml, or 1 μg of RIF per ml for up to 72 h. Viable counts were performed after 0, 24, and 72 h and aliquots were withdrawn at each time point for nucleic acid extraction. The DNA and RNA levels in each sample were quantified by PCR assay.
IS6110 DNA and 85B mRNA.
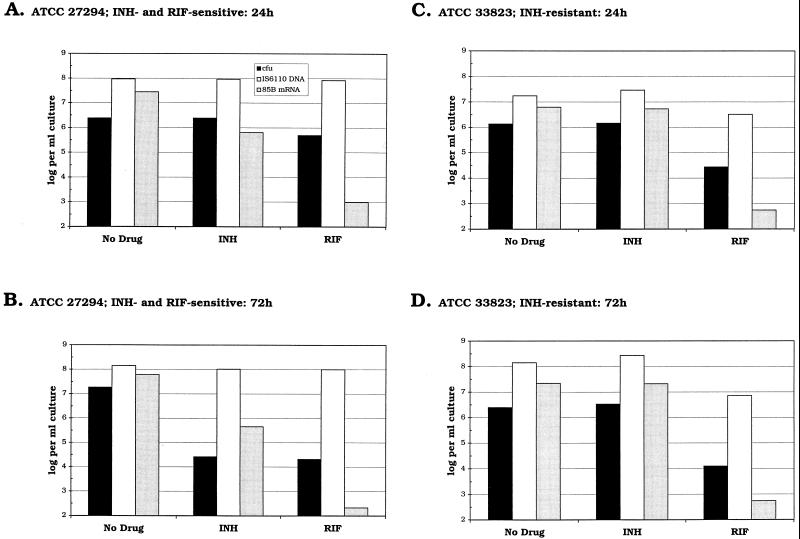
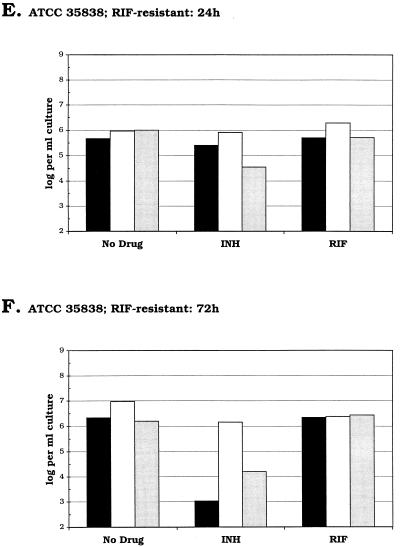
Figures 1A and B show the results of conventional culture and quantitative analysis of IS6110 DNA and 85B mRNA for M. tuberculosis ATCC 27294, which is sensitive to both INH and RIF. Relative to the control culture without drug, no appreciable decrease in viable counts or IS6110 DNA levels was observed after 24 h of exposure to either antimicrobial agent. In contrast, the levels of 85B mRNA present in the INH- and RIF-treated cultures were 2 and 0.003%, respectively, of those detected in the control (Fig. 2A). After 72 h, viable counts for the INH- and RIF-treated cultures were similar, although the amount of 85B mRNA present was >1,000-fold higher in the presence of INH than in the presence of RIF. In neither of the drug-treated cultures were IS6110 DNA levels observed to decline over the course of the experiment.
FIG. 1.
Charts showing log10 numbers of CFU (■), numbers of copies of IS6110 DNA (□), and numbers of copies of 85B mRNA ( ) per milliliter of culture for each of the three strains of M. tuberculosis after 24 h (A, C, and E) or 72 h (B, D, and F) of exposure to either no drug, 0.2 μg of INH per ml, or 1 μg of RIF per ml. The nucleic acid levels presented here are the mean values obtained from extraction of duplicate samples.
) per milliliter of culture for each of the three strains of M. tuberculosis after 24 h (A, C, and E) or 72 h (B, D, and F) of exposure to either no drug, 0.2 μg of INH per ml, or 1 μg of RIF per ml. The nucleic acid levels presented here are the mean values obtained from extraction of duplicate samples.
FIG. 2.
Charts showing the quantity of 85B mRNA detected at each time point as a percentage of that present in parallel control cultures in the absence of either INH or RIF. Drugs were added to cultures of M. tuberculosis at time zero. Cultures were exposed to either 0.2 μg of INH per ml (•) or 1 μg of RIF per ml (▴). (A) INH- and RIF-sensitive strain ATCC 27294; (B) INH-resistant strain ATCC 33823; (C) RIF-resistant strain ATCC 35838.
A similar pattern of mRNA expression was observed on exposure of INH-resistant strain ATCC 33823 to RIF (Fig. 1C and D and Fig. 2B). After 24 h the level of mRNA in the RIF-treated culture was <0.01% of that present in the control culture. In contrast, similar levels of 85B mRNA were detected in both the drug-free control culture and in the culture exposed to INH. Colony counts and IS6110 DNA levels obtained after 24 h in the presence of RIF were 2 and 19%, respectively, of those obtained with the control culture.
With RIF-resistant strain ATCC 35838, equivalent amounts of 85B mRNA were detected in the control and RIF-treated cultures (Fig. 1E and F and Fig. 2C). In contrast, exposure of this strain to INH for 24 h reduced the level of detectable 85B mRNA by >95%, which was similar to the reduction observed with the other INH-sensitive strain, ATCC 27294. Meanwhile, after 24 h viable counts were reduced by <50% relative to those for the control culture and IS6110 DNA levels were reduced by <13%.
16S rRNA.
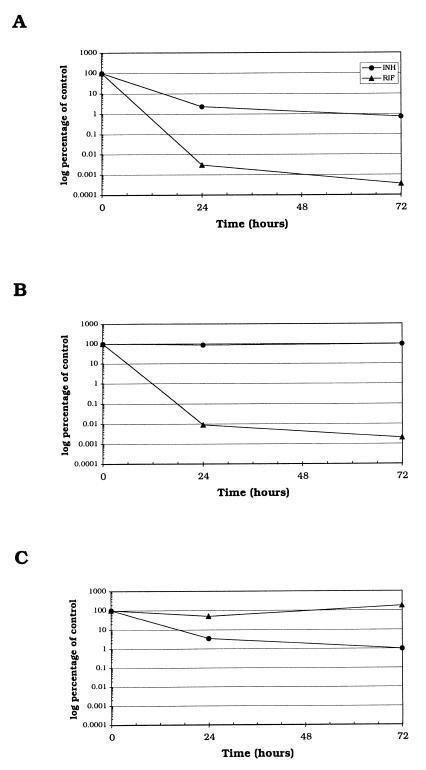
Semiquantitative analysis of 16S rRNA levels was performed by RT-PCR amplification of 1:10,000 dilutions of the RNA isolated at each time point. It was not possible to discern any decrease in rRNA levels at dilutions lower than 1:10,000, regardless of the length of exposure to either INH or RIF. This was due to saturation of the PCR mixture with the large number of 16S rRNA targets present in each cell (∼4,000), such that all specimens yielded bands of similar intensities.
We were able to detect a slight reduction in the 16S rRNA level after 24 h of exposure of the drug-sensitive strain ATCC 27294 to RIF (Fig. 3). The extent of this reduction was less dramatic than that observed for 85B mRNA, and substantial amounts of 16S rRNA remained after exposure of cultures to RIF for 72 h. Over the time course of these experiments, both ATCC 27294 and ATCC 33823 exhibited similar 16S rRNA profiles in the presence of RIF.
FIG. 3.
Autoradiographs showing semiquantitative RT-PCR analysis of 16S rRNA levels in three strains of M. tuberculosis exposed to either no drug, 0.2 μg of INH per ml, or 1 μg of RIF per ml. (A) Drug-sensitive strain ATCC 27294; (B) INH-resistant strain ATCC 33823; (C) RIF-resistant strain ATCC 35838. Lanes: 1 and 2, time zero, prior to the addition of either INH or RIF; 3, 24 h, no drug; 4, 24 h, INH; 5, 24 h, RIF; 6, 72 h, no drug; 7, 72 h, INH; 8, 72 h, RIF; 9, RT buffer to which target was not added; 10, 5 × 104 genome equivalents of M. bovis ATCC 19210 DNA.
Exposure of the INH-sensitive strains ATCC 27294 and ATCC 35838 to INH for 24 h had no observable effect on the levels of 16S rRNA. However, after 72 h the levels of 16S rRNA in the INH-treated cultures were diminished relative to those in the control cultures.
DISCUSSION
Numerous molecular assays which target conserved DNA or rRNA sequences have been developed for the diagnosis of tuberculosis (11, 22, 23, 37, 42, 44). Although such techniques offer considerable time-saving advantages over conventional microbiology and have the potential for enhanced sensitivity, they have not been applied successfully in the role of the monitoring of therapeutic efficacy. Previous studies have demonstrated a poor correlation between the results of conventional microbiological follow-up among patients receiving standard multidrug antituberculosis therapy and the presence of M. tuberculosis DNA and rRNA in clinical specimens (15, 20, 33). This is presumably due to a combination of the shedding of intact dormant or dead bacilli from the focus of infection and an inherent resistance of these nucleic acid targets to degradation in long-lived macrophages. The purpose of the present study was to evaluate the use of mRNA, which is purported to have a short half-life in actively growing cells (2, 43), as a marker of bacterial viability. The mRNA target that we chose to analyze was that encoding the M. tuberculosis 85B antigen. This protein is known to be highly expressed in both broth (18, 45) and macrophage (18, 26) M. tuberculosis culture systems, and it was reasonable to predict that viable bacilli would possess a corresponding abundance of the encoding mRNA.
We compared the levels of 85B mRNA, IS6110 DNA, and 16S rRNA in cultures of M. tuberculosis that were treated with either INH or RIF. Under the conditions that we used, exposure of sensitive strains of M. tuberculosis to 1 μg of RIF per ml for 24 h reduced the levels of 85B mRNA to <0.01% of those present in the absence of the drug. In contrast, the levels of IS6110 DNA and 16S rRNA did not diminish over the same period. These results are consistent with direct inhibition of transcription by binding of RIF to the RNA polymerase β subunit (4, 34) and with the thesis that the mRNA encoding the 85B protein is relatively short-lived. Preliminary data from our laboratory indicate that 85B mRNA has a half-life of ∼5 min (unpublished data).
INH-sensitive strains that were exposed to 0.2 μg of INH per ml for 24 h exhibited levels of 85B mRNA that were 2 to 4% of those detected in control cultures and >100-fold higher than those found in cultures treated with RIF. Although we examined only a single concentration of each of these drugs, the more gradual effect of INH on mRNA expression is thought to reflect its indirect influence on transcription through the inhibition of cell wall metabolism and the fact that INH is itself not bactericidal but must first be converted to an active form which in turn blocks mycolic acid synthesis (4, 34). Even though INH may preclude cell division, it is not unreasonable to expect transcription of 85B mRNA to continue for some time following exposure to the drug. On the basis of this evidence, a decay in 85B mRNA levels in the presence of INH appears to be a predictor of bactericidal activity and drug susceptibility.
Although strains of M. tuberculosis that were susceptible to INH and RIF exhibited marked reductions in 85B mRNA expression within 24 h of exposure to these drugs, this was followed by a much slower rate of decline over the following 48 h. Mitchison (30, 31) has proposed that within an infected host there exist different populations of cells, with each population metabolizing at different rates and having corresponding unique susceptibilities to various antimycobacterial agents. The evidence obtained here suggests that, even within a homogeneous culture system such as the one used in the present study, there may be subpopulations of cells which metabolize at a reduced rate and thereby avoid the rapid bactericidal effects of INH and RIF. In separate studies, we have observed a similar biphasic pattern of decline in M. tuberculosis 85B mRNA levels in sputum from patients receiving a standard four-drug regimen (7). To date, all the patients who have been monitored have responded to treatment as determined both by conventional culture and by molecular analysis. As a consequence, it is unclear at this time how drug resistance would be reflected in the mRNA profile. It is possible that susceptibility to RIF could mask resistance to one or more of the other antituberculosis drugs. However, any reversal in the downward trend in mRNA levels would be an indication of treatment failure and the likelihood of emerging drug resistance or noncompliance. Even in such cases, it is likely that mRNA analysis would still provide a more rapid assessment of the efficacy of a treatment regimen than is currently possible by conventional means.
In the present study, the two RIF-sensitive strains of M. tuberculosis exhibited very low levels of mRNA after 24 h of exposure to the drug, yet viable counts remained relatively high. This must indicate an ability of the organism to recover from the inhibitory effects of RIF once the drug is removed by dilution in fresh broth and the bacteria are plated on solid medium. Continued exposure to RIF nevertheless resulted in a decrease in the number of organisms capable of growth in vitro. Prolonged incubation with RIF therefore appears to be necessary to bring about a loss of viability, yet we were able to predict this at a much earlier stage from analysis of mRNA expression. Importantly, the RIF-resistant strain of M. tuberculosis did not demonstrate a reduction in either viable counts or 85B mRNA levels in the presence of the drug.
Recently, Western blot analysis was reported to show that exposure of M. tuberculosis cultures to INH induced production of two of the three proteins of the antigen 85 complex (16). We observed no evidence of such induction in our culture system, although this could be due to differences in the growth conditions used in the two studies, the choice of time points for sampling, or the fact that it is the 85A and 85C proteins rather than the 85B protein which are induced by INH. INH is thought to be rapidly transported into the bacterial cell, and examination of samples closer to the point of addition of the drug would permit evaluation of the drug’s immediate effects on gene expression before its bactericidal activity is manifest.
In conjunction with improvements in disease diagnosis brought about by the introduction of molecular assays, there has been a renewed emphasis on the need for susceptibility testing of mycobacterial isolates. This has been led by a marked increase in the proportion of drug-resistant isolates and, in particular, those resistant to more than one drug (12, 36, 38). Rapid reporting of susceptibilities facilitates timely modification of the treatment regimen and is a key element in limiting the spread of drug-resistant tuberculosis (38, 40). In particular, rapid detection of resistance to RIF is important because it is regarded as a marker for multidrug-resistant tuberculosis (39) and is associated with poor clinical outcome (17).
The conventional proportional method of determining susceptibility to antituberculosis drugs by plating on solid medium requires at least 3 weeks of incubation before results are known. Even with the BACTEC liquid culture system, susceptibility tests may require up to 14 days. Several gene amplification assays for the detection of specific genetic mutations which are responsible for resistance to RIF, INH, ethambutol, fluoroquinolones, and ethionamide are now available. However, these assays have limited practical value owing to incomplete understanding of all the mutations associated with the development of resistance. Molecular methods which provide a direct measurement of bacterial metabolism can potentially circumvent this problem. Previous studies have examined the application of quantitative assays for 16S rRNA in the determination of mycobacterial drug susceptibility (25, 28, 32, 41). As in the present investigation, the stability of the rRNA target sequence typically necessitated incubation in the presence of the antimycobacterial agent for between 3 and 5 days to obtain reliable discrimination of drug-sensitive and drug-resistant isolates. Cangelosi et al. (5) recently described a hybridization assay for M. tuberculosis pre-16S rRNA which permitted detection of susceptibility to RIF within 24 h of initial exposure to the drug. This system was, however, unable to detect any depletion of pre-16S rRNA in the presence of either INH or ethambutol. Use of a qualitative RT-PCR assay for M. tuberculosis 85B mRNA for the determination of susceptibility to INH has been reported previously (24). However, the inability to compare levels of mRNA between cultures necessitated prolonged incubation in order to demonstrate a loss of signal in the presence of the drug. Furthermore, the assay lacked the appropriate controls to account for possible DNA contamination of RNA extracts. In contrast, using a quantitative RT-PCR assay we were able to discriminate susceptible and resistant strains within 24 h of exposure to either INH or RIF, and as alluded to above, it is likely that this could be accomplished with much shorter periods of incubation. Although we did not correlate our results with those of conventional proportional methods of susceptibility testing, the apparent rapid turnover of 85B mRNA offers the potential of an improved system of susceptibility testing.
Currently, the major drawback to mRNA-based assays of bacterial viability are the difficulties associated with preparing and handling a highly labile RNA target and, in particular, in obtaining RNA that is free of contaminating DNA. The procedure for the isolation of mycobacterial RNA that has been developed by DesJardin et al. (9) works well for both cultured organisms and sputum, yet it is not suitable for use in a clinical setting owing to its reliance on repeated extraction with organic solvents. However, with improvements in methodology, analysis of mRNA expression may compete with conventional microbiology as the “gold standard” for susceptibility testing and chemotherapeutic monitoring.
ACKNOWLEDGMENTS
This work was supported by a Research Agreement between the University of Arkansas and Becton Dickinson & Company and the Tuberculosis Research, Prevention & Control Unit (NIH contract N01-AI45244).
We thank Ying Chen, Shirley Haun, Maria Winters, and Robert Pruss for excellent technical assistance and Joseph H. Bates for helpful discussions.
Lucy DesJardin and Tobin Hellyer contributed equally to this study.
REFERENCES
- 1.Beggs M L, Crawford J T, Eisenach K D. Isolation and sequencing of the replication region of Mycobacterium avium plasmid pLR7. J Bacteriol. 1995;177:4836–4840. doi: 10.1128/jb.177.17.4836-4840.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belasco J G, Nilsson G, von Gabain A, Cohen S N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986;46:245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- 3.Ben-Hamida F, Schlessinger D. Synthesis and breakdown of ribonucleic acid in Escherichia coli starving for nitrogen. Biochim Biophys Acta. 1966;119:183–191. doi: 10.1016/0005-2787(66)90049-9. [DOI] [PubMed] [Google Scholar]
- 4.Blanchard J S. Molecular mechanisms of drug resistance in Mycobacterium tuberculosis. Annu Rev Biochem. 1996;65:215–239. doi: 10.1146/annurev.bi.65.070196.001243. [DOI] [PubMed] [Google Scholar]
- 5.Cangelosi G A, Brabant W H, Britschgi T B, Wallis C K. Detection of rifampin- and ciprofloxacin-resistant Mycobacterium tuberculosis by using species-specific assays for precursor rRNA. Antimicrob Agents Chemother. 1996;40:1790–1795. doi: 10.1128/aac.40.8.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coleman G, Brown S. The effect of rifampicin on the stability of the messenger ribonucleic acid of Bacillus amyloliquefaciens as determined by DNA:RNA hybridization. J Gen Microbiol. 1976;92:200–206. doi: 10.1099/00221287-92-1-200. [DOI] [PubMed] [Google Scholar]
- 7.DesJardin L E, Chen Y, Perkins M D, Teixiera L, Cave M D, Eisenach K D. Microbial markers as surrogates for response to chemotherapy of tuberculosis. Am J Respir Crit Care Med. 1996;155:A255. doi: 10.1164/ajrccm.160.1.9811006. [DOI] [PubMed] [Google Scholar]
- 8.DesJardin L E, Chen Y, Teixeira L, Perkins M D, Cave M D, Eisenach K D. Quantification of IS6110 DNA in sputum during the treatment of tuberculosis. J Clin Microbiol. 1998;36:1964–1968. doi: 10.1128/jcm.36.7.1964-1968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DesJardin L E, Perkins M D, Teixeira L, Cave M D, Eisenach K D. Alkaline decontamination of sputum specimens adversely affects stability of mycobacterial mRNA. J Clin Microbiol. 1996;34:2435–2439. doi: 10.1128/jcm.34.10.2435-2439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Wit L, Palou M, Content J. Nucleotide sequence of the 85B-protein gene of Mycobacterium bovis BCG and Mycobacterium tuberculosis. DNA Seq. 1994;4:267–270. [PubMed] [Google Scholar]
- 11.Eisenach K D, Sifford M D, Cave M D, Bates J H, Crawford J T. Detection of Mycobacterium tuberculosis in sputum samples using a polymerase chain reaction. Am Rev Respir Dis. 1991;144:1160–1163. doi: 10.1164/ajrccm/144.5.1160. [DOI] [PubMed] [Google Scholar]
- 12.Ellner J J, Hinman A R, Dooley S W, Fischl M A, Sepkowitz K A, Goldberger M J, Shinnick T M, Iseman M D, Jacobs W R. Tuberculosis symposium: emerging problems and promise. J Infect Dis. 1993;168:537–551. doi: 10.1093/infdis/168.3.537. [DOI] [PubMed] [Google Scholar]
- 13.Fischl M A, Daikos G L, Uttamchandani R B, Poblete R B, Moreno J N, Reyes R R, Boota A M, Thompson L M, Cleary T J, Oldham S A, Saldana M J, Lai S. Clinical presentation and outcome of patients with HIV infection and tuberculosis caused by multiple-drug-resistant bacilli. Ann Intern Med. 1992;117:184–190. doi: 10.7326/0003-4819-117-3-184. [DOI] [PubMed] [Google Scholar]
- 14.Frieden T R, Fine Sherman L, Lay Maw K, Fujiwara P I, Crawford J T, Nivin B, Sharp V, Hewlett D, Brudney K, Alland D, Kreiswirth B N. A multi-institutional outbreak of highly drug-resistant tuberculosis: epidemiology and clinical outcomes. JAMA. 1996;276:1229–1235. [PubMed] [Google Scholar]
- 15.Gamboa F, Manterola J M, Vinado B, Matas L, Gimenez M, Lonca J, Manzano J R, Rodrigo C, Cardona P J, Padilla E, Dominguez J, Ausina V. Direct detection of Mycobacterium tuberculosis complex in nonrespiratory specimens by Gen-Probe Amplified Mycobacterium tuberculosis Direct Test. J Clin Microbiol. 1997;35:307–310. doi: 10.1128/jcm.35.1.307-310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garbe, T. R., N. S. Hibler, and V. Deretic. Isoniazid induces expression of the antigen 85 complex in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 40:1754–1756. [DOI] [PMC free article] [PubMed]
- 17.Goble M, Iseman M D, Madsen L A, Waite D, Ackerson L, Horsburgh C R. Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N Engl J Med. 1993;328:527–532. doi: 10.1056/NEJM199302253280802. [DOI] [PubMed] [Google Scholar]
- 18.Harth G, Lee B-Y, Wang J, Clemens D L, Horwitz M A. Novel insights into the genetics, biochemistry, and immunocytochemistry of the 30-kilodalton major extracellular protein of Mycobacterium tuberculosis. Infect Immun. 1996;64:3038–3047. doi: 10.1128/iai.64.8.3038-3047.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 20.Hellyer T J, Fletcher T W, Bates J H, Stead W W, Templeton G L, Cave M D, Eisenach K D. Strand displacement amplification and the polymerase chain reaction for monitoring response to treatment in patients with pulmonary tuberculosis. J Infect Dis. 1996;173:934–941. doi: 10.1093/infdis/173.4.934. [DOI] [PubMed] [Google Scholar]
- 21.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iovannisci D M, Winn-Deen E S. Ligation amplification and fluorescence detection of Mycobacterium tuberculosis DNA. Mol Cell Probes. 1993;7:35–43. doi: 10.1006/mcpr.1993.1005. [DOI] [PubMed] [Google Scholar]
- 23.Jonas V, Alden M J, Curry J I, Kamisango K, Knott C A, Lankford R, Wolfe J M, Moore D F. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by amplification of rRNA. J Clin Microbiol. 1993;31:2410–2416. doi: 10.1128/jcm.31.9.2410-2416.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jou N-T, Yoshimori R B, Mason G R, Louie J S, Liebling M R. Single-tube, nested, reverse transcriptase PCR for detection of viable Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:1161–1165. doi: 10.1128/jcm.35.5.1161-1165.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawa D E, Pennell D R, Kubista L N, Schell R F. Development of a rapid method for determining the susceptibility of Mycobacterium tuberculosis to isoniazid using the Gen-Probe DNA hybridization system. Antimicrob Agents Chemother. 1989;33:1000–1005. doi: 10.1128/aac.33.7.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee B-Y, Horwitz M A. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Invest. 1995;96:245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak K J, Flood S J A, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 28.Martin-Casabona N, Xairo Mimo D, Gonzalez T, Rossello J, Arcalis L. Rapid method for testing susceptibility of Mycobacterium tuberculosis by using DNA probes. J Clin Microbiol. 1997;35:2521–2525. doi: 10.1128/jcm.35.10.2521-2525.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McAdam R A, Hermans P W M, van Soolingen D, Zainuddin Z F, Catty D, van Embden J D A, Dale J W. Characterization of a Mycobacterium tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol. 1990;4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 30.Mitchison D A. Basic mechanisms of chemotherapy. Chest. 1979;76S:771S–781S. doi: 10.1378/chest.76.6_supplement.771. [DOI] [PubMed] [Google Scholar]
- 31.Mitchison D A. Understanding the chemotherapy of tuberculosis—current problems. J Antimicrob Chemother. 1992;29:477–493. doi: 10.1093/jac/29.5.477. [DOI] [PubMed] [Google Scholar]
- 32.Miyamoto J, Koga H, Kohno S, Tashiro T, Hara K. New drug susceptibility test for Mycobacterium tuberculosis using the hybridization protection assay. J Clin Microbiol. 1996;34:1323–1326. doi: 10.1128/jcm.34.5.1323-1326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore D F, Curry J I, Knott C A, Jonas V. Amplification of rRNA for assessment of treatment response of pulmonary tuberculosis patients during antimicrobial therapy. J Clin Microbiol. 1996;34:1745–1749. doi: 10.1128/jcm.34.7.1745-1749.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pablos-Mendez A, Sterling T R, Frieden T R. The relationship between delayed or incomplete treatment and all-cause mortality in patients with tuberculosis. JAMA. 1996;276:1223–1228. doi: 10.1001/jama.1996.03540150025026. [DOI] [PubMed] [Google Scholar]
- 36.Raviglione M C, Snider D E, Kochi A. Global epidemiology of tuberculosis: morbidity and mortality of a worldwide epidemic. JAMA. 1995;273:220–226. [PubMed] [Google Scholar]
- 37.Shah J S, Liu J, Buxton D, Hendricks A, Robinson L, Radcliffe G, King W, Lane D, Olive D M, Klinger J D. Q-Beta replicase-amplified assay for detection of Mycobacterium tuberculosis directly from clinical specimens. J Clin Microbiol. 1995;33:1435–1441. doi: 10.1128/jcm.33.6.1435-1441.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shinnick T M, Good R C. Diagnostic mycobacteriology laboratory practices. Clin Infect Dis. 1995;21:291–299. doi: 10.1093/clinids/21.2.291. [DOI] [PubMed] [Google Scholar]
- 39.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 40.Tenover F C, Crawford J T, Huebner R E, Geiter L J, Horsburgh C R, Good R C. The resurgence of tuberculosis: is your laboratory ready? J Clin Microbiol. 1993;31:767–770. doi: 10.1128/jcm.31.4.767-770.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van der Vliet G M E, Schepers P, Schukkink R A F, van Gemen B, Klatser P R. Assessment of mycobacterial viability by RNA amplification. Antimicrob Agents Chemother. 1994;38:1959–1965. doi: 10.1128/aac.38.9.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Vliet G M E, Schukkink R A F, van Gemen B, Schepers P, Klatser P R. Nucleic acid sequence-based amplification (NASBA) for the identification of mycobacteria. J Gen Microbiol. 1993;139:2423–2429. doi: 10.1099/00221287-139-10-2423. [DOI] [PubMed] [Google Scholar]
- 43.Von Gabain A, Belasco J G, Schottel J L, Chang A C Y, Cohen S N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Walker G T, Fraiser M S, Schram J L, Little M C, Nadeau J G, Malinowski D P. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiker H G, Harboe M. The antigen 85 complex: a major secretion product of Mycobacterium tuberculosis. Microbiol Rev. 1992;56:648–661. doi: 10.1128/mr.56.4.648-661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]