SUMMARY
Exosomes are nanoparticles secreted by all cell types and are a large component of the broader class of nanoparticles termed extracellular vesicles (EVs). Once secreted, exosomes gain access to the interstitial space and ultimately the circulation, where they exert local paracrine or distal systemic effects. Because of this, exosomes are important components of an intercellular and intraorgan communication system capable of carrying biologic signals from one cell type or tissue to another. The exosomal cargo consists of proteins, lipids, miRNAs, and other RNA species, and many of the biologic effects of exosomes have been attributed to miRNAs. Exosomal miRNAs have also been used as disease biomarkers. The field of exosome biology and metabolism is rapidly expanding, with new discoveries and reports appearing on a regular basis, and it is possible that potential therapeutic approaches for the use of exosomes or miRNAs in metabolic diseases will be initiated in the near future.
INTRODUCTION
The study of exosomes is a rapidly evolving field that intersects almost all areas of physiology and pathophysiology. Exosomes are heterogeneous and represent a component of a broader class of extracellular vesicles (EVs) released by almost all cell types. Secreted exosomes enter the interstitial space and ultimately gain access to the circulation to serve as critical components in a diverse array of intercellular crosstalk and communication systems. Exosomes are bilayer vesicular nanoparticles within the class of EVs and range in size from 30 to 150 nm and are released from cells through a specific, multi-stepped exocytotic process (Doyle and Wang, 2019; Kalluri and LeBleu, 2020; Li et al., 2019c). Microvesicles (MVs), sometimes referred to as ectosomes, are larger EVs that bud off from the plasma membrane with a size of 100 nm–1 μm, and apoptotic bodies are an even larger component of EVs (50–5,000 nm in diameter) that are released by dying apoptotic cells (Doyle and Wang, 2019; Kalluri and LeBleu, 2020; Carnino et al., 2020). In this review, we will focus specifically on exosomes, their biogenesis, cargo, and biologic potential.
An initial concept in the exosome field was that these nanoparticles serve as a waste disposal system allowing cells to package and secrete unwanted cellular components into the extracellular environment. While this may still be partly correct, more recent studies have focused on the importance of exosome effectors of various physiologic and disease-related events and as biomarkers of disease. Indeed, the diverse signaling properties of exosomes have now been well recognized in numerous publications spanning a variety of fields. Here, we will review the current understanding of exosomes in the field of metabolism as disease biomarkers and effectors of metabolic regulation. Since exosomes are secreted from cells in a regulated manner, enter the circulation, and are taken up by distal cells where they exert biologic effects, exosome biology has many similarities to a classical endocrine system. Indeed, emerging evidence indicates that the loading of specific cargos into exosomes, as well as their secretion, can be regulated, drawing further analogies to a typical endocrine system. Enriched populations of exosomes can be isolated from cell culture media and various biofluids using a number of techniques based on size, density, and membrane protein constituents (Dang et al., 2020), as will be discussed later. There is no single standard, well-accepted technique for exosome isolation, but the methodologies are rapidly advancing (Théry et al., 2018). Despite the technical limitations of exosome isolation methodologies, the heterogeneity of these particles, and the various means of characterization, many important discoveries have already emerged from the exosome field. To a considerable extent, these efforts have been energized by the important potential of circulating exosomes as physiologic regulators and potential therapeutic vehicles and as disease biomarkers.
As an integral part of normal cellular function, almost all cell types release a variety of nanovesicles into the extracellular environment. These vesicles, commonly termed EVs, contain a number of cargo components, such as proteins, lipids, and a variety of RNA species, including mRNAs, long noncoding RNAs, and microRNAs (miRNAs) (O’Brien et al., 2020). Some of these proteins and lipids are expressed on the surface, while others are embedded in the lipid bilayer enclosing these vesicles with many other components contained within the vesicular lumen. Among these EVs, exosomes are of particular interest because their biogenesis and exocytotic secretory pathway comprises specific intracellular mechanisms that can be regulated to determine exosome composition, function, and perhaps targeting. Environmental and cellular cues such as stress, inflammation, or cell-cycle events can change the specific cargos loaded into exosomes and can also regulate the overall process of exosome release (Gurunathan et al., 2021).
Using the circulation as a transit system, exosomes arrive at distal cellular sites where they can bind to cell surfaces and undergo endocytosis through specific mechanisms. Recent evidence suggests that the mechanisms of exosome uptake can be regulated and that classes of exosomes may contain specific targeting molecules leading to some degree of specificity toward recipient tissues (Murphy et al., 2019). Exosomes can also undergo transcytosis, allowing them to cross the blood-brain barrier, gaining access to the CNS (Ramirez et al., 2018). The International Society of Extracellular Vesicles (ISEV) recommends the phrase EVs as a general term describing all nanoparticles released by cells with a lipid bilayer (Théry et al., 2018). Exosomes are an important component of EVs, and various techniques exist to isolate enriched populations of exosomes. However, exosome isolation techniques are still evolving, and current methodologies yield preparations that are highly enriched in exosomes but still contain smaller amounts of other EV subtypes, such as MVs. Using a reliable method for exosome preparation with adequate characterization of the resulting vesicles, it is reasonable to use the term exosomes in describing these preparations if the appropriate caveats are acknowledged. With respect to characterization, the ISEV recommends the measurement of at least three protein markers that are characteristic of exosomes, including at least one transmembrane protein, such as the tetraspanins (CD63, CD81, and CD82), integrins, or others. In addition, one cytosolic protein is required (e.g., TSG101, ALIX, and syntenin) and one negative protein marker (e.g., albumin and ribosomal proteins) (Théry et al., 2018). In addition to these criteria, cell-type-specific exosomal proteins have been identified in leukocytes (CD37 and CD53), endothelial cells (ECs) (PECAM1), mesenchymal stem cells (MSCs) (CD90), and others (Théry et al., 2018). Throughout the rest of this review, the term exosomes will be used when appropriate.
EXOSOME BIOGENESIS
Exosomes are derived from endosomal structures that originate through endocytosis of invaginated endosomes from the plasma membrane (Jella et al., 2018; Figure 1). These early sorting endosomes eventually mature into late sorting endosomes, which can then generate multivesicular endosomal structures. From these latter particles, multivesicular bodies (MVBs) develop by the inward invagination of the endosomal membrane, forming numerous blebs within these MVBs, often termed intraluminal vesicles (ILVs) (Jella et al., 2018). These ILV-containing MVBs can fuse with lysosomes comprising a degradative pathway. Alternatively, they can undergo a specific exocytotic process whereby they ultimately fuse with the plasma membrane, releasing the invaginated exosomes into the extracellular space (Zhang et al., 2019). Multiple proteins and lipids have been described that participate in the process of exosome biogenesis. For example, Rab proteins regulate vesicular traffic and exosome formation (Blanc and Vidal, 2018). In addition, other proteins assemble into four components of the endosomal sorting complex required for transport (ESCRT-0, -I, -II, and -III) along with accessory proteins such as ALIX and others (Colombo et al., 2013; Vietri et al., 2020). These complexes are involved in MVB and ILV biogenesis. The ESCRT-0 complex recognizes and sequesters ubiquitinated proteins in the endosomal membrane, whereas the ESCRT-I and -II complexes are responsible for deformation of membranes into buds with sequestered cargo. ESCRT-III components subsequently drive vesicle scission (Colombo et al., 2013). Tetraspanins are transmembrane proteins that induce membrane-curved structures enabling vesicle formation (Andreu and Yáñez-Mó, 2014). Finally, various lipid-modifying enzymes such as sphingomyelinase generate ceramides that promote vesicle formation (Wortzel et al., 2019). However, much work remains to be done to elucidate specific roles of these intracellular components in the exosomal biogenesis/exocytosis process. A complicating factor is that many of the proteins involved in these processes also serve other intracellular functions, so that gain- or loss-of-function studies may simultaneously perturb exosomal biogenesis/exocytosis and other intracellular events. The processes underlying exosome biogenesis remain to be fully defined and the reader is referred to several excellent reviews on this topic (Bebelman et al., 2018; Zhang et al., 2019).
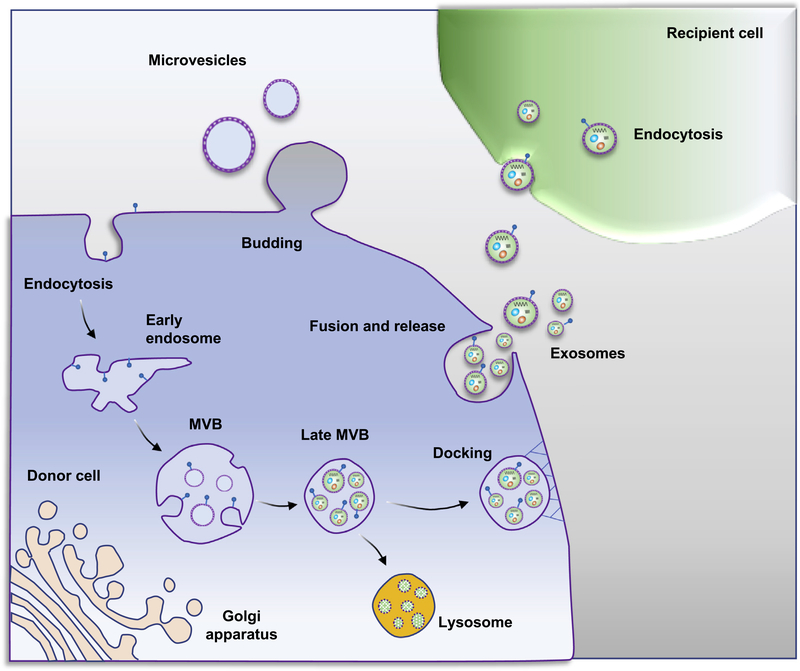
Figure 1. Exosome and microvesicle biogenesis pathways.
Microvesicles bud directly from the plasma membrane, and exosomes are generated by inward budding of the multivesicular body (MVB) lipid bilayer membrane. MVB fusion with the plasma membrane is a tightly regulated multistep process that includes MVB trafficking along microtubules and docking at the plasma membrane for further exosome release. Alternatively, MVBs can fuse with lysosomes as part of the degradative process. Initially, exosomes bind to the cell surface of recipient cells through protein-protein or receptor-ligand interactions, which can initiate signaling cascades that activate endocytotic pathways. The exosome cargo confers the biologic effects of exosomes on recipient cells.
EXOSOME UPTAKE BY RECIPIENT CELLS
After exosomes arrive at recipient cells, internalization occurs through a two-step process. Initially, exosomes bind to the cell surface through protein-protein or receptor-ligand interactions, which can initiate signaling cascades that activate endocytotic pathways (Horibe et al., 2018).
Uptake through membrane-bound receptors
The role of exosome surface proteins has been demonstrated by treating exosomes with proteinase K showing reduced uptake by recipient cells (Inder et al., 2014; Smyth et al., 2014). Many proteins have been identified as membrane-bound receptors enabling the binding of exosomes to cells. This can include integrin-tetraspanin complexes (Hazawa et al., 2014; Jankovičová et al., 2020), and/or binding of the exosomal ephrin receptor (Eph) to membranal ephrin expressed on recipient cells (Irie et al., 2009; Gong et al., 2016; Sato et al., 2019). Another example is the macrophage receptor with collagenous structure (MARCO), which can mediate exosome internalization by macrophages (Kanno et al., 2020). In addition, exosome membrane proteins also modulate circulating kinetics. This was shown by finding that protease treatment of exosomes prior to IV administration resulted in delayed clearance. Since exosome surface proteins are influenced by the protein repertoire of the parental cell, exosome membrane composition can vary based on the cellular physiological state. Exosomes are enriched in glycoproteins, and since receptor-mediated exosome uptake can be glycan dependent (Williams et al., 2019), disruption of native glycosylation alters uptake in recipient cell types (Williams et al., 2019). Whether glycosylation is directly involved in cell recognition and/or active entry into cells is unclear.
Endocytosis
Endocytosis is the most frequently reported mechanism for exosome uptake by either clathrin-dependent or -independent pathways (Mulcahy et al., 2014). Clathrin-mediated endocytosis is a well-understood process for the uptake of extracellular materials, and inhibition of this pathway reduces the uptake of exosomes (Murphy et al., 2019; Mulcahy et al., 2014). Clathrin-independent endocytosis primarily involves caveolin 1, RhoA, and ARF6 (Sandvig et al., 2008). Caveolins can mediate the formation of specific microdomains within the plasma membrane, also known as lipid rafts, which are entry points for exosomes (Murphy et al., 2019).
Macropinocytosis, phagocytosis, and fusion
Macropinocytosis involves the uptake of larger quantities of extracellular fluid into a macropinosome in a process that depends on actin polymerization (Lin et al., 2020). During macropinocytosis, plasma membrane protrusions driven by actin filaments form an invagination that nonspecifically endocytoses extracellular fluid and small particles (Lin et al., 2020). Numerous examples of exosome uptake through this pathway have been reported (Fitzner et al., 2011; Svensson et al., 2013; Tian et al., 2014).
Macrophages, dendritic cells, and other cell types also take up exosomes via phagocytosis (Mulcahy et al., 2014). Phagocytosis may represent a selective process for exosome clearance by immune cells (Feng et al., 2010; McKelvey et al., 2015).
Fusion is another potential mechanism for exosome internalization. During cell surface membrane fusion, the two distinct lipid bilayer membranes come into close proximity and form a hemifusion stalk. This stalk expands, and a hemifusion diaphragm bilayer appears, followed by fusion pore opening, facilitating the mixing of two hydrophobic cores (Mulcahy et al., 2014). The potential role of general processes versus targeted mechanisms directing specific exosome subtypes for uptake into selected recipient cells is an important and emerging topic and a key subject for future research.
EVs CONTAIN A DIVERSE ARRAY OF CARGOES
The encapsulated cargoes of EVs are protected from degradation and remain relatively stable in different biofluids. These cargoes include proteins, lipids, metabolites, amino acids, various RNA species, and DNA (Veziroglu and Mias, 2020; Mohan et al., 2020; Yáñez-Mó et al., 2015).
Exosome cargoes
The exosome cargo confers the biologic effects of exosomes on neighboring or distal cells. The lipid composition of exosomes includes sphingolipids, cholesterol, phosphatidylserine, saturated fatty acids, and ceramides (Trajkovic et al., 2008; Skotland et al., 2020), all of which can be found in plasma membranes. A direct role for ceramide during the budding of ILVs into the lumen of MVBs was shown using neutral sphingomyelinase inhibitors that reduced overall exosome secretion (Menck et al., 2017). The proteome of exosomes includes membrane trafficking-related proteins, such as the tetraspanins (CD63, CD81, CD82, and CD9), that are recruited to exosomes by ALIX- and ESCRT-III-dependent pathways (Larios et al., 2020). Exosomes can also be enriched in heat-shock proteins (Hsp60, Hsp70, and Hsp90), integrins, and MHC class II proteins (Clayton et al., 2005). Exosomes do not simply represent the protein composition of the parental cells. Certain proteins are enriched in exosomes by selective protein cargo sorting controlled by post-translational modifications (Carnino et al., 2020). One of these is ubiquitination of cargo proteins required for their binding to ESCRT complexes (Carnino et al., 2020).
Among the different exosomal RNA species, miRNAs and mRNAs are the most well studied (Carnino et al., 2020). Valadi et al. have demonstrated that some exosomal mRNAs are intact and can undergo translation to functional proteins in recipient cells (Valadi et al., 2007). The process by which RNA species are sorted into the exosome can be selective, and these mechanisms have been recently reviewed (Wei et al., 2021; O’Brien et al., 2020).
Exosomal DNA (exoDNA) can include both nuclear and mitochondria DNA in the form of single-stranded or double-stranded DNA. ExoDNA can be located on the surface or inside the vesicle. The mechanisms of DNA packaging into exosomes have remained unclear (Wortzel et al., 2019).
MV cargoes
MVs also contain the aforementioned cargoes, and several mechanisms regulate the recruitment of cargo proteins into the MV lumen, including the ESCRT (Yáñez-Mó et al., 2015). In addition, cytoplasmic proteins bind to the plasma membrane at the MV luminal creation sites. This binding is based on post-translational modifications such as myristoylation and palmitoylation, and these proteins concentrate at the plasma membrane domains of MV budding (Meldolesi, 2018). Since MVs cover a significant and heterogeneous size range (50 nM–1 μM) (Konoshenko et al., 2018), it would be intriguing to assess differences between MV vesicle size and cargo contents.
Different RNA species exist in MVs, including mRNA, non-coding RNA, and others (Turchinovich et al., 2019). Both miRNAs (including guide strands and passenger strands) and pre-miRNAs can be packaged into MVs to deliver information (Shu et al., 2019). The exact mechanism for packaging miRNAs into MVs is not yet clear, but differences in miRNA content between MVs and their parental cells suggest selective loading of miRNAs into MVs (Groot and Lee, 2020). DNA cargo within MVs includes single-stranded DNA, double-stranded genomic DNA, and mitochondrial DNA. The DNA varies in length, largely dependent on the size of the MVs with which it is associated (Elzanowska et al., 2021; Bruno et al., 2020). Intact MV DNA can be >2 Mbp and is associated with histones (H2B), suggesting the presence of intact chromosomal DNA (Vagner et al., 2018).
BIOLOGY OF miRNA GENERATION, PROCESSING, LOADING, AND SECRETION
miRNAs are ~22-nt-long non-coding RNAs that are critical post-transcriptional regulators of gene expression in metazoans and plants and have an essential role in the post-transcriptional regulation of gene expression (Filipowicz et al., 2008). miRNAs regulate a large array of cellular functions, and changes in their expression are associated with several human pathophysiological states. Many miRNAs are expressed in a tissue-specific or developmental-stage-specific manner, contributing to cell-type-specific profiles of mRNA and protein expression (Ivey and Srivastava, 2015; Kloosterman and Plasterk, 2006).
miRNA processing
Pri-miRNAs are hairpin-shaped precursor miRNA molecules either transcribed by RNA polymerase II from independent genes or introns of protein-coding genes (Figure 2). The pri-miRNAs act as substrates for two enzymes, Drosha and Dicer, members of the RNase III family. Drosha cleavage results in an ~70-nt pre-miRNA exported to the cytoplasm, where Dicer processes it to an ~22-bp miRNA/miRNA duplex (Michlewski and Cáceres, 2019; Han et al., 2004). One strand of this duplex, representing the mature miRNA, is incorporated into the miRNA-induced silencing complex (miRISC). Within the miRISC, miRNAs pair to their target mRNAs. This process involves a 6- to 8-nt miRNA seed sequence that binds to complementary nucleotides within the 3′ UTR of the target mRNA, leading to translational repression or deadenylation and degradation (O’Brien et al., 2020; Catalanotto et al., 2016). Argonaute (AGO) proteins, which directly interact with miRNAs, and glycine-tryptophan protein of 182 kDa (GW182) are essential factors in the assembly and function of miRISCs and represent key steps in the miRNA repression pathway. In the process of miRNA maturation, both Drosha and Dicer are assisted by several cofactors or accessory proteins, such as SMAD nuclear interacting protein (SNIP1) and adenosine deaminases (ADARs) (Eulalio et al., 2009; Krol et al., 2010). The main components of miRISC include miRNAs, mRNA targets of miRNAs, GW182, and AGO2. The AGO2 protein binds to U or A at the 5′ end of miRNAs and plays an essential role in mediating the formation of mRNA:miRNA pairs, followed by translational repression or degradation of the mRNA target (O’Brien et al., 2020).
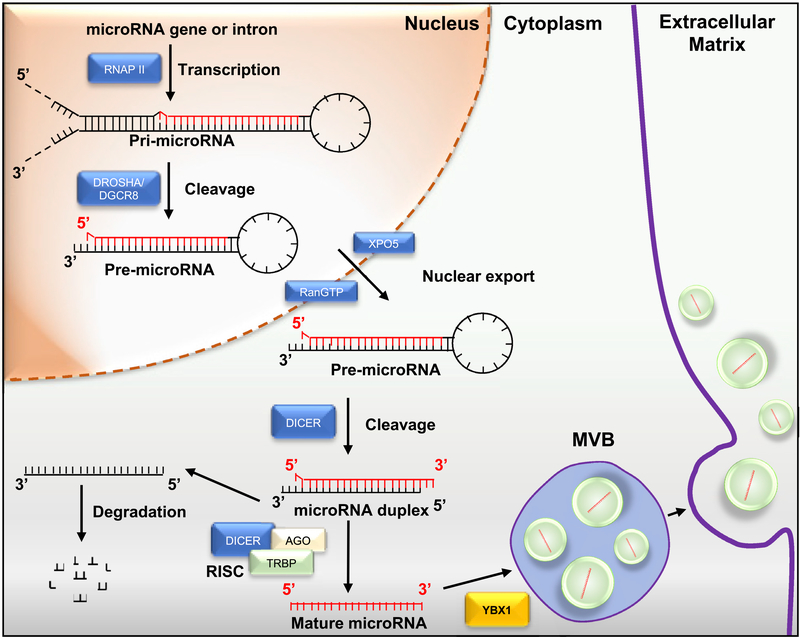
Figure 2. miRNA biogenesis.
Pri-miRNAs are hairpin-shaped precursor miRNA molecules transcribed by either RNA polymerase II (RNAPII) from independent genes or introns of protein-coding genes. The pri-miRNAs are cleaved by Drosha/DGCR8 into pre-miRNA and exported to the cytoplasm from the nuclei mediated by Exportin 5 (XPO5) binding. Dicer cleavage generates an miRNA duplex intermediate. Trans-activation-responsive RNA-binding protein (TRBP) and Argonaute (AGO) protein assemble into the RNA-induced silencing complex (RISC). One miRNA strand is transferred to AGO protein, resulting in the formation of RISC. AGO2 binds to one of the miRNA strands to form the mature miRNA, which will be selectively incorporated into exosomes. After miRNA maturation, the RNA-binding protein YBX1 also sorts miRNAs into exosomes.
miRNA loading
Both the process of MVB/exosome biogenesis and miRNA-sequence-specific determinants may modulate miRNA sorting into exosomes. As discussed earlier, ESCRT plays an important role in MVB biogenesis and exocytosis, but knockdown of ESCRT proteins does not affect miRNA sorting to exosomes (Kosaka et al., 2010). Interestingly, knockdown of Alix, an ESCRT-III accessory protein, did not affect the number of released EVs but induced a decrease in secreted miRNAs (Iavello et al., 2016). Within exosomes, the miRNAs contain common sequences, or EXO-motifs, that facilitate binding to RNA-binding proteins (RBPs), such as heterogeneous nuclear ribonucleoproteins (hnRNPA2B1) and SYNCRIP (Villarroya-Beltri et al., 2013; Santangelo et al., 2016). Some studies demonstrate that short sequence motifs overrepresented in miRNAs, such as GGAG and UGCA found in miR-198 and miR-601, control sorting into exosomes, and directed mutagenesis of these motifs modulates miRNA cargoes (Villarroya-Beltri et al., 2013).
Recent studies have also shown the connections between AGO2 and exosomal miRNA sorting, and AGO2 has been identified in exosomal protein analyses by mass spectrometry (MS) or western blotting (Zhang et al., 2015a; Goldie et al., 2014; Melo et al., 2014). It is known that knockout of AGO2 decreases the types or abundance of preferentially exported miRNAs in HEK293T-derived exosomes (Guduric-Fuchs et al., 2012). Other evidence also shows the relationship between miRISC and exosomal miRNA sorting. Thus, YBX1 (Y-box protein I), but not AGO2, binds to miR-223 and miR-144 and can control packaging into vesicles (Ung et al., 2014; Shurtleff et al., 2016). YBX1 also co-localizes with members of the RISC, including GW182, which can be found in exosomes (Goodier et al., 2007; Gallois-Montbrun et al., 2007). Thus, the main components of miRISC are co-localized with MVBs (Gibbings et al., 2009; Wang et al., 2019). In summary, specific sequences present in miRNAs may guide their incorporation into exosomes, whereas other proteins may control the sorting of exosomal miRNAs in a sequence-independent fashion.
Exosomal miRNA secretion
The proportion of miRNAs is higher in exosomes than in their parent cells, and several studies show that miRNAs are not randomly incorporated into exosomes (Guduric-Fuchs et al., 2012; Li et al., 2012; Zhao et al., 2019). Thus, a key functional point is that analysis of miRNA expression levels in various cell lines and their secreted exosomes shows that the miRNA content of cells compared to their exosomes is quite different. For example, Guduric-Fuchs et al. have shown that miR-150, miR-142–3p, and miR-451, among others, preferentially enter exosomes. In this context, it should be noted that the cellular content of miRNAs represents a mixture of endogenous miRNAs, as well as the miRNAs that enter the cell via exosomes derived from other cell types (Guduric-Fuchs et al., 2012). In this way, the miRNA cargo of a secreted exosome that enters the circulation from one cell type can significantly affect the miRNA profile of a recipient cell. It would be of interest to determine whether miRISC loading and mRNA translational repression are the same or different for endogenously produced miRNAs versus those that enter the cells through uptake of exosomes.
MECHANISMS OF miRNA ACTION AND TARGET IDENTIFICATION
As mentioned earlier, miRNAs inhibit the translation of target mRNAs within the RISC, where they bind to RPBs such as Argonaute (Ciafrè and Galardi, 2013). Through seed sequence binding to the mRNA 3′ UTR region, target mRNAs are loaded into the RISC, where translational arrest can occur (Gu et al., 2009). While the seed sequence interactions with mRNA 3′ UTR regions apply generally to miRNA/target mRNA pairs, exceptions to these rules have been demonstrated (Didiano and Hobert, 2008; Bartel, 2009; Moore et al., 2015). In some cases, miRNA sequences outside of the seed sequence or mRNA nucleotides not within the mRNA 3′ UTR are involved in the binding interaction.
miRNA sequencing studies demonstrate that exosomes harvested from blood or conditioned media (CM) of specific cell types contain hundreds of different miRNAs (Bhome et al., 2018; Van den Brande et al., 2018). However, the number of miRNA transcripts per exosome particle can be quite small given the very large number of exosomal particles in blood or CM (Bhome et al., 2018). If a particular exosomal preparation exerts a metabolic effect, it can be a daunting task to identify the individual miRNA or group of miRNAs responsible for the measured biologic effect. However, strategies for identifying the miRNA(s) of interest have been used. First, one needs to thoroughly sequence all the miRNAs within the exosomal preparation. While hundreds may be detected, most of these miRNAs are expressed at very low levels and are unlikely to exert biologic effects when taken up by recipient cells. Thus, the more abundant miRNAs are most likely to contain the miRNA(s) of interest (Yáñez-Mó et al., 2015). Since miRNAs generally downregulate their mRNA targets, it is reasonable to assume that the miRNA(s) of interest that lead to a given biologic effect will exhibit differential increased expression in exosomes derived from the cell type of interest (e.g., cells from obese animals compared to lean controls) (Veziroglu and Mias, 2020; Sigismund et al., 2012). This strategy allows one to focus on the more highly expressed exosomal miRNAs that are also differentially expressed in the condition of interest. This greatly reduces the total number of candidate miRNAs, and if the biologic effect of interest can be reduced to a cell-based screening assay, then all the candidate miRNAs can be screened for activity. This provides a methodology to identify the key miRNA, or group of miRNAs, responsible for the biologic effect observed.
If one identifies an exosomal miRNA that produces the desired biologic effect, then the next step is to define the mechanism. miRNAs exert their biologic effects by inhibiting the translation of their target mRNAs, but identifying the target mRNA for a specific miRNA can be difficult. However, strategies have been proposed that can often lead to tangible results. Using various bioinformatic tools such as TargetScan and PicTar, one can identify all the mRNA targets that could theoretically interact with a given miRNA seed sequence. This typically yields a relatively large number (dozens or more) of theoretical mRNA targets for a given miRNA. One can then use transcriptome assessments such as RNA sequencing (RNA-seq) to identify the mRNAs suppressed by a given miRNA in the target tissue. Since not all theoretical mRNA targets will be expressed in the target tissue of interest, and only a subset will be downregulated by applying the miRNA, this approach can reduce the number of potential mRNA targets to be considered. With luck, only a relatively small number of potential mRNA targets will fulfil these criteria. These can then be studied on an individual basis to identify the biologically relevant mRNA target of a given miRNA.
Due to the proximity of RBPs, miRNAs, and target mRNAs within the RISC, another general approach involves immunoprecipitation methods, which can be used to successfully identify miRNA/mRNA target pairs (Thomson et al., 2011). As a general approach, whole-cell lysates can be crosslinked with various reagents to stabilize RBP/miRNA/mRNA interactions. The RBPs (e.g., Argonaute 1) can then be immunoprecipitated, followed by reversal of the crosslinks and sequencing of the precipitated mRNAs and miRNAs (Lin and Miles, 2019; Wessels et al., 2019). With this approach, one can match the miRNAs to target mRNAs by assessing miRNA (usually seed sequence) complementarity to potential mRNA targets (Helwak et al., 2013; Nussbacher and Yeo, 2018; Van Nostrand et al., 2016).
NON-miRNA EXOSOME CARGO
Although less well studied than exosomal miRNAs, different types of proteins such as enzymes, hormones, cytokines, ligands, and others can be components of exosome cargo released from metabolic tissues and can be functional in recipient cells. For example, hypoxic adipocytes release exosomes containing enzymes related to de novo lipogenesis (Sano et al., 2014). Also, hepatocyte-derived exosomes can contain active arginase-1 that modulates the metabolism of circulating arginine, regulating vascular function (Royo et al., 2017).
The secretion of adiponectin from adipocytes can also include exosome-mediated pathways. Eicosapentanoic acid (EPA) and docosahexaenoic acid (DHA) treatment increase adiponectin in the exosome fraction (DeClercq et al., 2015), and exosomes isolated from mouse serum were associated with adiponectin. However, the proportion of total circulating adiponectin in exosomes is relatively small, suggesting that adiponectin within exosomes may not have robust biologic effects. Another hormone found in adipocyte exosomes is resistin. Rong et al. showed that melatonin decreases exosomal trafficking of resistin from adipocytes to hepatocytes, mitigating ER stress-induced hepatic steatosis (Rong et al., 2019).
Exosomal lipid cargo
Exosomal lipids can also be functional in the progression of metabolic disease. For example, sphingosine 1-phosphate (S1P) contained in endothelial-derived exosomes can enhance migration of hepatic stellate cells (HSCs) (Wang et al., 2015a). Interestingly, circulating exosomes containing C16:0 ceramide- and S1P-enriched lipid species are progressively increased in the plasma of obese patients with simple steatosis and non-alcoholic steatohepatitis (NASH) patients with early fibrosis (Kakazu et al., 2016). In addition, adipocytes can release exosomesized, lipid-filled vesicles that become a source of lipids in local macrophages (Flaherty et al., 2019).
ADIPOSE TISSUE EXOSOME BIOLOGY
Increased adipose tissue mass is a hallmark of obesity and is present in the great majority of individuals with type 2 diabetes mellitus (T2DM) (Kusminski et al., 2016; Ghaben and Scherer, 2019). In obesity, both in the presence and absence of diabetes, chronic tissue inflammation, particularly in adipose tissue and liver, is an important contributor to insulin resistance (Hotamisligil and Erbay, 2008; Lackey and Olefsky, 2016; Lee and Olefsky, 2021; Saltiel, 2021; Ferrante, 2013). Obesity-induced chronic inflammation is not confined to adipose and liver since there are reports of proinflammatory pathway activation in the CNS, the gastrointestinal tract, muscle, and pancreatic islets, consistent with an intra- and interorgan network of crosstalk in these complex heterogeneous disorders (Lee and Olefsky, 2021). A sizable literature exists exploring various cytokines, adipokines, lipid species, and other factors released from adipose tissue that may contribute to systemic metabolic dysfunction (Lago et al., 2007; Hernandez et al., 2021; Vegiopoulos et al., 2017).
Since exosomes can be released from adipose tissue and work locally in a paracrine manner or enter the circulation to have systemic effects, they have been increasingly studied as effectors of intercellular signaling. As discussed earlier, exosomes contain a variety of cargo components, and many of the biologic effects of these particles have been attributed to their miRNA content (Figure 3). While evaluation of adipose tissue vesicles in the blood is still limited by the absence of adipocyte-derived specific markers, highly expressed adipocyte proteins such as aP2/FABP4, perilipin-1, adiponectin, or PPAR have been identified in circulating EVs (Connolly et al., 2018). Nonetheless, their use as specific adipocyte markers can be confounded because their expression varies according to adipocyte differentiation state and/or hypertrophy, and many of these proteins are also expressed in adipose tissue macrophages (ATMs). Using a fat-specific knockout of the miRNA-processing enzyme Dicer, Thomou et al. showed that adipocytes were major contributors to circulating exosomal miRNAs (Thomou et al., 2017) (Figure 4A). Conversely, tracing plasma EVs in mice expressing a fluorescent protein (TdTomato) specifically in adipocytes, Flaherty et al. normalized the abundance of the fluorescent protein level of plasma exosomes to the tetraspanin CD63 and suggested that adipocyte-derived exosomes represented a minority of the circulating exosomes (Flaherty et al., 2019). Further work will be needed to evaluate the exact contribution of adipocyte vesicles to the circulating pool of EVs (Amosse et al., 2018).
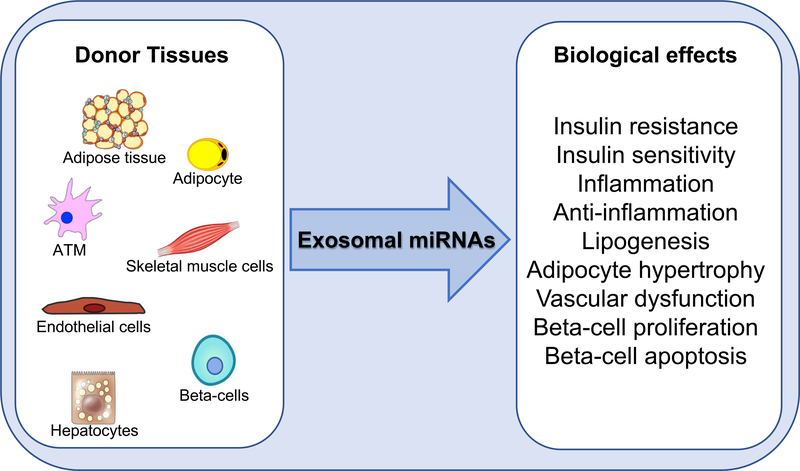
Figure 3. Exosomal miRNA biological effects.
Exosomal miRNAs secreted by different metabolic tissues have effects on insulin signaling, inflammation, vascular function, adipocyte biology, and beta cell physiology.
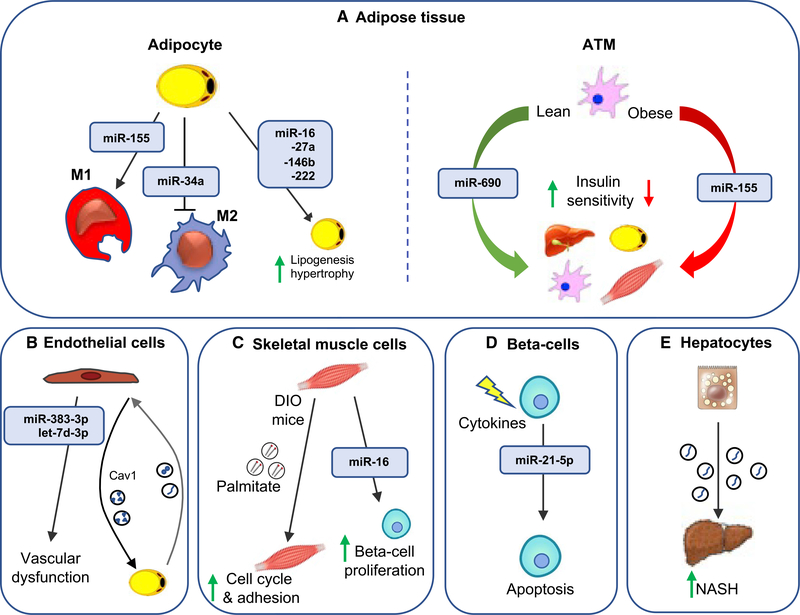
Figure 4. Exosomal miRNA effects on recipient cells.
Exosomal miRNAs mediate communication between donor cells and recipient cells, playing an important role in metabolic signaling.
(A) Adipocyte exosomal miRNAs can modulate macrophage polarization and lipogenesis and hypertrophy in adipocytes. Additionally, adipose tissue macrophage (ATM) exosomes can regulate insulin sensitivity through the delivery of miR-690 to the liver, adipocytes, ATMs, and skeletal muscle.
(B) Inflammatory factors induce the release of endothelial cell (EC) exosomes that contain miR-383–3p and let-7d-3p. ECs transfer cav1-containing exosomes to adipocytes, which reciprocate by releasing exosomes taken up by ECs.
(C) Skeletal muscle-derived exosomes induce cell cycle and adhesion in skeletal muscle as well as proliferation of isolated mouse beta cells.
(D) Cytokine-treated beta cell lines and human islets lead to enhanced secretion of exosomal miR-21–5p. Exosomes from cytokine-treated beta cells induce apoptosis in the recipient cells.
(E) Lipotoxic hepatocytes secrete exosomes that activate hepatic stellate cells, promoting the fibrotic NASH phenotype.
In studies using whole blood as a source of circulating exosomes, the overall findings show that exosomes from obese subjects produce glucose intolerance when administered to lean mice and miRs 122, 192, 27a-3p, and 27b-3p have been implicated in these effects (Castaño et al., 2018; Párrizas et al., 2015; Jones et al., 2017) (Figure 4A). In some cases, mimics of these miRNAs reproduced the effects on glucose tolerance. Castaño et al. also showed that circulating exosomes from obese mice promoted glucose intolerance and insulin resistance when administered to lean mice (Castaño et al., 2018).
WHOLE ADIPOSE TISSUE EXOSOMES
There are a number of studies showing that exosomes derived from both human and mouse obese adipose tissue can inhibit insulin sensitivity (Kita et al., 2019; Dang et al., 2019; Pan et al., 2019; Zhang et al., 2015b). Exosomes prepared from obese human adipose tissue exhibit differential expression of a variety of miRNAs that influence insulin signaling. For example, exosomes harvested from obese adipose tissue explants can directly promote insulin resistance (Kranendonk et al., 2014), and in one of these studies, miR-141–3p appeared to play a causative role (Dang et al., 2019). Other studies using exosomes derived from adipose tissue explants from obese mice or humans have also shown biologic effects to impair insulin sensitivity (Zhang et al., 2015b). Similar effects of exosomes from obese adipose tissue were reported by Kranendonk et al. (2014). Pan et al. found that miR-34a was upregulated in visceral fat of obese subjects and observed that knockout of miR-34a in mice protected them from obesity-induced glucose intolerance and insulin resistance (Pan et al., 2019). They further reported that miR-34a expression led to decreased ATM content in obese adipose tissue with an increase in the percentage of M2-like and a decrease in M1-like ATMs, indicating a potential mechanism for the insulin-sensitizing effects of this exosomal miRNA. Deng et al. also showed that exosome-like particles from obese mouse adipose tissue can be taken up by circulating monocytes and activate the subsequent proinflammatory differentiation program once they differentiate into macrophages (Deng et al., 2009; Flaherty et al., 2019) (Figure 4A). Using human adipose tissue explants, Ferrante et al. found that exosomes obtained from obese versus lean subjects contain a number of miRNAs that were differentially expressed (Ferrante et al., 2015). They found that a subset of these miRNAs modulated cellular signaling pathways relevant to insulin action. However, it is important to note that adipose tissue is composed of a variety of different cell types (adipocytes, macrophages, immune cell types, and ECs), and therefore, when studying adipose tissue explant-derived exosomes, the specific cell type producing the biologically relevant exosomes cannot be determined.
ADIPOCYTE EXOSOMES
Using adipocytes obtained from obese animals or high glucose-treated 3T3L1 adipocytes, the harvested adipocyte exosomes directly induce monocyte differentiation into inflammatory macrophages (Eguchi et al., 2015). In a similar vein, Pan et al. found that exosomes derived from adipocytes can inhibit the M2-like macrophage polarization state, and this effect was attributed to miR-34a (Pan et al., 2019) (Figure 4A). Thomou et al. used their adipocyte-specific dicer knockout mice to assess the biologic effects of adipocyte-derived exosomes (Thomou et al., 2017). They found that exosomes released from brown adipocytes express high levels of miR-99b, which can enter the circulation and travel to the liver, where they inhibit FGF21 expression. It is possible that corresponding changes in blood levels of FGF21 have metabolic consequences in obesity and/or T2DM. Adipose-derived exosomal miRNAs also have paracrine functions as exosomes released from large adipocytes contain miR-16, miR-27a, miR-146b, and miR-222, which can be transferred to small adipocytes to stimulate lipogenesis and adipocyte hypertrophy (Müller et al., 2011) (Figure 4A).
Adipocyte exosomes can also contain non-miRNA cargoes that exert distant biologic effects. For example, Crewe et al. show that exosomes produced by adipocytes from obese mice contain mitochondrial particles and mitochondria proteins that exhibit oxidative damage but still display respiratory function. These exosomes can be released from adipocytes and taken up by cardiomyocytes in the heart where they trigger production of reactive oxygen species (ROS). This exerts protective effects in the heart against oxidative stress and ischemia/reperfusion injury. In this manner, adipocyte-derived exosomes in obesity can actually exert cardioprotective effects in certain circumstances (Crewe et al., 2021). Exosomes themselves are heterogeneous in terms of size, cargo content, and cell origin, and with current techniques, it is difficult to make a “pure” population of exosomes. While highly enriched exosome preparations can be produced, it is possible that some of the reports in the exosome/metabolism field include studies of preparations of exosomes heavily mixed with other EV subtypes.
ADIPOSE TISSUE MACROPHAGE EXOSOMES
Obesity is characterized by a substantial accumulation of ATMs. These ATMs have been denoted as either classically activated proinflammatory M1 or alternatively activated anti-inflammatory M2 cells. However, the terms M1 or M2 do not adequately apply to in vivo macrophage populations such as ATMs. These labels relate to bone marrow-derived macrophages (BMDMs) that have been polarized in vitro by treatment with either LPS to generate M1 BMDMs or IL-13/IL-4 to create M2 BMDMs. Macrophage populations in vivo may have similarities to the M1 or M2 state, but they are not the same (Li et al., 2019a; Xiong et al., 2019; Jaitin et al., 2019; Seidman et al., 2020; Zhao et al., 2017). Detailed bulk RNA-seq or single-cell RNA-seq studies have demonstrated that there are discrete clusters of macrophage subpopulations within these general categories (Li et al., 2019a; Xiong et al., 2019; Jaitin et al., 2019; Seidman et al., 2020; Zhao et al., 2017).Recent papers often use the phrases M1-like or M2-like to indicate this concept when describing in vivo macrophage populations. In obesity, the increased number of ATMs involves an accumulation of proinflammatory M1-like cells (Boutens and Stienstra, 2016). These macrophages produce proinflammatory cytokines, ROS, and nitric oxide (NO), which all play a role in eliminating exogenous pathogens (Lee and Olefsky, 2021).
Many studies have shown that these M1-like ATMs are major contributors to the insulin-resistant state (Russo and Lumeng, 2018; Hill et al., 2014; Hotamisligil and Erbay, 2008; Lackey and Olefsky, 2016; Lee and Olefsky, 2021; Saltiel, 2021; Ferrante, 2013) (Figure 4A). This has prompted studies of ATM-derived exosomes. Thus, Fuchs et al. showed that plasma- and adipose tissue-derived exosomes from obese subjects caused decreased insulin signaling in myotubes and hepatocytes (Fuchs et al., 2021). Furthermore, exosomes harvested from ATMs derived from obese mice directly cause insulin resistance in vitro when incubated with adipocytes, primary hepatocytes, or L6 myocytes (Ying et al., 2017). These “obese” ATM exosomes were also given intravenously to chow-fed lean mice. Interestingly, these “obese” exosome-treated lean recipient mice developed glucose intolerance, hyperinsulinemia, and insulin resistance comparable to that observed in obese mice (Ying et al., 2017). Since body weight and food intake were unchanged by in vivo exosome treatment, these findings indicate that “obese” ATM exosomes can directly cause insulin resistance. Another study by Liu et al. reported that miR-29a was a key pathogenic RNA within obese ATM exosomes causing insulin resistance (Liu et al., 2019). In contrast, ATM exosomes derived from lean mice led to the opposite phenotype (Ying et al., 2017). Thus, treatment of insulin target cells (adipocytes, myocytes, or primary hepatocytes) with “lean” ATM exosomes directly caused enhanced insulin signaling with an improvement in overall cellular insulin sensitivity. In vivo treatment of insulin-resistant, hyperglycemic obese mice with “lean” ATM exosomes resulted in improved glucose tolerance with enhanced insulin sensitivity. Overall, one could view these “lean” ATM exosomes as an effective therapeutic modality, improving metabolic dysfunction in obesity. These studies also showed that either the “obese” or “lean” ATM exosomes exert their metabolic effects entirely through their miRNA cargo since depletion of miRNAs from these exosomes by knockout of proteins critical for processing or loading of miRNAs into exosomes, such as Drosha or YBX1, abrogated all the exosome-mediated effects. In other words, miRNA-free “lean” or “obese” exosomes did not exert metabolic effects either in vitro or in vivo.
ATMs in lean mice generally have an anti-inflammatory phenotype and are often termed alternatively activated, or M2-like. More in-depth studies of “lean” ATM exosomes are challenging due to the paucity of ATMs in the lean state. However, bone marrow-derived progenitor cells can be cultured in vitro with IL-13 and IL-4, which drives them toward a more classical M2 polarization state. Although ATMs from lean mice and M2 BMDMs are comparable, they are not identical, and certain phenotypic and transcriptomic differences exist (Li et al., 2019a). Nevertheless, a recent report shows that exosomes harvested from M2 BMDMs cause improved insulin signaling in adipocytes, myocytes, and primary hepatocytes in vitro, and when administered intravenously to obese mice, they improve glucose tolerance and enhance insulin sensitivity (Ying et al., 2021). miRNA sequencing of exosome preparations yields a relatively complete picture of all the miRNAs. Using exosomal miRNA sequencing, a bioinformatics approach, and cell-based screening assays, Ying et al. showed that miR-690 was the major miRNA component in these M2 BMDM exosomes leading to the beneficial metabolic effects (Figure 4B). Indeed, treating insulin target cells in vitro or obese mice in vivo with a synthetic mimic of miR-690 recapitulated all the favorable metabolic effects of the M2 BMDM or “lean” ATM exosomes. A synthetic antagomir of miR-690 blocked these effects, indicating the specificity of the findings. Interestingly, both “lean” ATM exosomes and M2 BMDM exosomes expressed comparably high levels of miR-690. These M2 BMDM exosomes or the miR-690 mimic also caused a robust anti-inflammatory effect in obese mice leading to a decrease in M1-like and an increase in M2-like ATMs. Similar findings were noted in vitro in which miR-690 promoted an anti-inflammatory polarization phenotype in BMDMs. These studies provide an example of exosomal miRNA biology leading to the identification of a specific miRNA, which might have future therapeutic potential as an insulin sensitizer if translated into humans.
Conceptually similar results were obtained by Zhang et al., who found that exosomes generated from adipocytes isolated from high-fat diet mice promoted macrophage differentiation to the pro-inflammatory M1-like state (Zhang et al., 2015b). These investigators attributed these effects to exosomal miR-155 (Figure 4A). Not surprisingly, once these macrophages were polarized to the proinflammatory state, CM from these cells directly caused decreased insulin sensitivity in skeletal muscle cell lines. Subsequent studies showed that macrophages activated via LPS treatment produce exosomes that inhibit insulin-stimulated glucose transport in adipocytes (De Silva et al., 2018).
Other studies in human and mouse macrophages have supported a role for M1-like macrophage-derived exosomes in promoting macrophage proinflammatory pathways. For example, in both human and mouse macrophage exosomes, miR-155 has been identified as a potent pro-inflammatory factor by repressing the expression of PPARγ, Socs1, or Socs6 (O’Connell et al., 2007; Ying et al., 2017; Wang et al., 2017; Ge et al., 2021). Chen et al. showed that LPS stimulation of the human macrophage THP-1 cell line leads to the release of exosomes that can activate the stellate cell fibrogenic program, and they implicated miR-103–3p in this process (Chen et al., 2020b). If this phenomenon also applies to hepatic macrophages during the development of hepatic inflammation and fibrosis, then this could represent a NASH-enhancing mechanism.
ENDOTHELIAL CELL EXOSOMES
EC-derived exosomes are released upon activation or apoptosis. EC exosomes constitute a large subclass of all circulating vesicles in peripheral blood (5%–15%), although a sizeable proportion of circulating plasma exosomes are also derived from platelets and erythrocytes (Combes et al., 1999; Dignat-George and Boulanger, 2011; Markiewicz et al., 2013; Arraud et al., 2014). EC exosomes can impair vascular function through pro-coagulative (Combes et al., 1999) and pro-inflammatory functions (Buesing et al., 2011), as well as by mitigating NO production from ECs (Brodsky et al., 2004; Densmore et al., 2006) (Figure 4B).
Proteomic analyses showed that secreted EC exosomes contain proteins that are also found in the originating ECs, and some can be transferred into recipient cells (Liu et al., 2013). Other inflammatory factors, including interleukin-1 (IL-1), interferon-γ (IFN-γ), and bacterial LPS, can induce the release of EC exosomes, which contain specific miRNAs that were present in significantly lower amounts compared to vesicles derived from unstimulated ECs (Yamamoto et al., 2015). Crewe et al. used a cav1 mouse model to show that neighboring ECs transfer cav1-containing exosomes to adipocytes in vivo, which reciprocate by releasing exosomes taken up by ECs (Crewe et al., 2018) (Figure 4B). Because EC exosomes can contain signaling molecules derived from the circulation, one can speculate that the effect of EC exosomes on adipocyte physiology is dependent on the type of signals in the blood at any given time. This mechanism would enable transfer of plasma constituents from ECs to adipocytes and may be regulated by feeding status and obesity, suggesting that exosomes participate in the tissue response to changes in the systemic nutrient state. If so, it follows that EC exosomes could customize the functional response of adipocytes to systemic changes in metabolism. The potentially wide-ranging implications of the EC-adipocyte communication axis for adipocyte function and whole-body metabolism are still unclear (Crewe et al., 2018).
SKELETAL MUSCLE EXOSOMES
Several studies have shown both paracrine and endocrine effects of skeletal muscle-derived exosomes in the maintenance of muscle homeostasis and communication with other tissues (Qin and Dallas, 2019; Li et al., 2019b). Muscle cells, such as myoblasts and myotubes, are a source of exosomes expressing Tsg101 and Alix protein markers, along with other proteins involved in signal transduction (Rome et al., 2019; Forterre et al., 2014; Guescini et al., 2010). As myoblasts differentiate into myotubes, the pattern of secreted proteins changes. Proteomic analysis showed that most of the exosomal proteins are derived from skeletal muscle cytosol. Of these proteins, only 10% have a canonical secretory sequence, and 43% were identified as non-classical secreted proteins lacking an N-terminal secretory sequence (Safdar et al., 2016). Exosomes may play a role in skeletal muscle differentiation and maturation as many proteins involved in myoblast to myotube formation are found in exosomes (Le Bihan et al., 2012).
Emerging evidence suggests that exercise enhances the biogenesis of MVBs and the resulting exosomes (Garner et al., 2020). Subsequently, exosome release following exercise can carry exerkines that might modulate intercellular communication (Safdar and Tarnopolsky, 2018). Several papers have suggested that these might modulate aging (Bertoldi et al., 2018), T2DM (Li et al., 2019b; Safdar et al., 2016), cardiovascular diseases (CVDs) (Bei et al., 2017; Zhou et al., 2019; Ma et al., 2018), immunity (Lancaster and Febbraio, 2005; Wu and Liu, 2018), and sarcopenia (Rong et al., 2020).
It is known that palmitate-induced skeletal muscle insulin resistance triggered the release of a new population of myotube-derived exosomes enriched in palmitate. (Aswad et al., 2014). Palmitate found in myotube-derived exosomes from diet-induced obese (DIO) mice was transferred to recipient myotubes and resulted in the upregulation of genes involved in cell cycle and cellular adhesion (Figure 4C). Among them, the pro-inflammatory cytokine IL-6 and the cell-cycle regulator cyclin D1 were strongly upregulated, whereas Glut4 was downregulated.
EXOSOMES AND BETA CELL FUNCTION
Exosomes have been implicated in regulating pancreatic beta cell function. For instance, skeletal muscle-derived exosomes from mice fed with a standard diet enriched with 20% palmitate induced the proliferation of the beta cell MIN6B1 cell line as well as in isolated mouse islets (Marchand et al., 2016). These skeletal muscle-derived exosomes were enriched in miR-16 that was transferred to the MIN6B1 cells and regulated Ptch1, a gene involved in pancreas development (Marchand et al., 2016) (Figure 4C).
Several studies have also shown potential beneficial effects of MSC-derived exosomes on beta cells. Intravenous injections of MSC-exosomes reduced fasting blood glucose levels, restored basal insulin levels (Sabry et al., 2020), and promoted beta cell regeneration, improving insulin secretion (Mahdipour et al., 2019) in diabetic rats. Furthermore, MSC-exosomes protected beta cells against hypoxia-induced apoptosis via enriched exosomal miR-21 (Chen et al., 2020a).
Beta cells also secrete exosomes in response to stimuli or stress and can affect neighboring cells. Guay et al. have shown that exosomes secreted from MIN6B1 and INS-1 can efficiently transfer miRNA to MIN6B1 recipient cells (Guay et al., 2015). Furthermore, exosomes from cytokine-treated MIN6B1 cells induce apoptosis in the recipient MIN6B1 cells mediated by AGO2 (Guay et al., 2015). Lakhter et al. showed that cytokine-treated beta-cell lines and human islets led to enhanced secretion of exosomal miR-21–5p (Lakhter et al., 2018) (Figure 4D). While beta-cells clearly secrete exosomes that can work locally in a paracrine manner on neighboring beta-cells, whether beta-cell exosomes can enter the circulation to produce systemic effects is problematic since their contribution to the total circulating pool of exosomes would be negligible.
EXOSOMES IN NAFLD AND NASH
Non-alcoholic fatty liver disease (NAFLD) is an extremely common feature of obese and T2DM individuals, and ~30% of these subjects ultimately develop NASH. NASH is associated with increased risk for CVD and is a precursor condition to cirrhosis and hepatocellular cancer. Within the liver, intercellular communication plays an important role in the progression of inflammation and fibrosis, which are major hallmarks of NASH. In the NASH liver, hepatocytes exhibit steatosis and lipotoxicity, Kupffer cells (KCs) and recruited hepatic macrophages (RHMs) promote liver inflammation, and HSCs are the primary drivers of fibrosis. All liver cells, including hepatocytes, HSCs, hepatic macrophages, and liver sinusoidal ECs, are capable of exosome release (Sung et al., 2018), suggesting that paracrine-mediated intercellular crosstalk between liver cell types could be an important contributing mechanism to hepatic disease.
In healthy conditions, hepatocytes produce exosomes containing sphingosine kinase 2, contributing to cell survival, growth, and proliferation (Nojima et al., 2016). In addition, hepatocyte-derived exosomes can mediate other biologic effects through their miRNA cargo. For example, in the early stages of obesity, hepatocyte exosomes are enriched in miR-3075, which can produce robust insulin-sensitizing effects both in vitro and in vivo when given to obese mice. A major target for miR-3075 is FA2H, which participates in the production of ceramides. Since FA2H is suppressed by miR-3075, decreased ceramide levels may provide at least part of the mechanism for the insulin-sensitizing effects of this miRNA. Interestingly, in chronic obesity, hepatocyte exosomes no longer carry appreciable amounts of miR-3075 and do not have insulin-sensitizing effects. Indeed, hepatocyte-derived exosomes from chronic obese mice promote insulin resistance by stimulating proinflammatory activation and polarization of macrophages. In this way, the initial response of hepatocytes is to produce insulin-sensitizing exosomes, and this might be viewed as a compensatory phenomenon to mitigate the full development of insulin resistance in obesity. As time goes on and chronic obesity ensues, this compensatory response is lost and hepatocyte exosomes promote insulin resistance through their proinflammatory actions (Yudong Ji et al., 2021).
Hepatocytes can also release exosomes that modulate the transcriptional program of neighboring hepatocytes and non-parenchymal cells toward inflammation and fibrosis (Hernández et al., 2020). One of the key events in NAFLD progression is the accumulation of potential harmful lipids in hepatocytes, leading to hepatocyte lipotoxicity (Alkhouri et al., 2009). Liver lipotoxicity increases hepatocyte exosome release, which might contribute to inflammation and fibrosis (Figure 4E). For example, lipotoxicity-induced hepatocyte exosomes can stimulate the expression and secretion of pro-inflammatory cytokines, contributing to local inflammation (Hirsova et al., 2016). Another study showed that lipid-induced hepatocyte exosomes are released in a caspase-3-dependent manner and promote EC activation (Povero et al., 2013). These exosomes also stimulated HSC activation via delivery of miR-128–3p that suppresses PPAR-γ expression (Povero et al., 2015). In addition, exosomes carrying miR-1 were released from steatotic hepatocytes and mediated pro-inflammatory effects in ECs via downregulation of KLF4 and activation of the NF-κB pathway (Jiang et al., 2020). Lipotoxicity-induced hepatocyte exosomes are enriched in chemokine (C-X-C motif) ligand 10 (CXCL10), which is a macrophage chemoattractant (Ibrahim et al., 2016). This results in recruitment and M1-like polarization of hepatic macrophages (Tomita et al., 2016), as well as monocyte adhesion (Dai et al., 2020). Moreover, hepatocyte exosomes can contribute to hepatic recruitment of monocyte-derived macrophages by promoting monocyte adhesion via integrin β1-dependent mechanisms (Guo et al., 2019). Thus, hepatocyte exosomes can activate KCs and promote the recruitment and activation of RHMs during NASH development (Figure 4E).
Data from different diet-induced animal models of NASH have shown that blood exosome concentration increases with disease progression in a time-dependent manner (Kakazu et al., 2016; Povero et al., 2013). These exosomes can amplify inflammation through multiple mechanisms such as macrophage activation and monocyte chemotaxis (Hernández et al., 2020). The release of hepatocyte-derived exosomes was reduced by inactivating the DR5 signaling pathway or inhibiting Rho-associated protein kinase 1 (ROCK1), suggesting that ROCK1 inhibition in NASH mice could lead to a reduction in exosome release with less liver inflammation and fibrosis (Hirsova et al., 2016).
More recently, human liver stem cell (HLSC)-derived exosomes have been used to treat mice with diet-induced steatohepatitis (Bruno et al., 2020). Treatment significantly downregulated hepatic profibrotic and pro-inflammatory gene expression and ameliorated the histological abnormalities in mice with NASH. Proteomic analysis of sinusoidal-derived exosomes identified various anti-inflammatory proteins, which may have contributed to the observed beneficial effects (Bruno et al., 2020).
Several reports have shown that lipotoxic hepatocytes can release exosomes that stimulate activation and proliferation of HSCs, with increased expression of a variety of genes in the fibrotic pathway. Exosomal miRNAs 28–3P and 192 have been implicated in these effects. Interestingly, exosomes derived from lipotoxic hepatocytes can express Vanin-1 on the surface, and treatment with Vanin-1 neutralizing Abs prevents exosome-mediated HSC activation. HSCs themselves are another important source of exosomes that contribute to liver injury in NASH. Activation or trans-differentiation of HSCs results in insoluble collagen deposition and distortion of the normal macro- and micro-anatomical structures of the liver (Friedman, 2008). During liver injury, exosomes from activated HSCs containing the pro-fibrogenic connective tissue growth factor (CTGF) can activate other HSCs (Charrier et al., 2014). Additionally, intercellular communication between quiescent and activated HSCs via exosomes may also modulate fibrosis since activated HSCs can release exosomes that activate quiescent HSCs. In this regard, a role for miR-214 in regulating the expression of alpha-smooth muscle actin and collagen in activated HSCs has been demonstrated (Chen et al., 2015).
In summary, intercellular signaling among hepatocytes, KCs, RHMs, and HSCs plays a key role in the progression from NAFLD to NASH, and local communication through exosomes may be integral to this process. Despite the abundant evidence of intercellular communication within the liver, it should be pointed out that there is also evidence that signals extrinsic to the liver have an important impact on NAFLD and NASH. This could include cytokines and adipokines from obese adipose tissue, signals from a leaky gastrointestinal tract such as LPS, and exosomes originating outside the liver, all of which can travel to the liver through the circulation, promoting NAFLD and NASH.
EXOSOMES AS BIOMARKERS FOR METABOLIC DISEASE
With respect to biomarker studies, exosomal cargo represents a way to sample the metabolic state within cells at a given point in time. With this line of reasoning, circulating exosomal cargo can be thought of as a liquid biopsy of the metabolic state of the originating cell type. In the best case, a biomarker would have specificity for the etiology or complications of a particular disease. With respect to obesity, while it is not necessary to utilize a circulating biomarker to diagnose the obese state, it would be of high importance to identify biomarkers that might predict the development of obesity, weight loss success, or recidivism. To the extent that biomarker studies focus on circulating exosomes, an important caveat must be kept in mind. The circulating exosomal pool is the composite of all exosomes released from a large variety of different cell types. Therefore, depending on the disease in question, exosomal signatures from the relevant disease-related cell types may be blunted or confounded by the presence of circulating exosomal cargo derived from multiple other cells or tissues. In addition, when focusing on exosomes as biomarkers, another important consideration is that many of the earlier studies did not use current methods to produce well-characterized enriched exosome preparations, so some of the reported miRNA signatures may reside to an unknown extent in non-exosomal EV components.
Previous studies have shown that changes in circulating miRNA profiles are one of the physiological responses to the development of metabolic diseases (Hsieh et al., 2015; Nunez Lopez et al., 2016; Ortega et al., 2013; Pescador et al., 2013; Villard et al., 2015; Wang et al., 2015b; Willeit et al., 2017; Wu et al., 2015). Thus, the potential of circulating miRNAs as signatures for metabolic diseases has been studied in both humans and animal models. In evaluating biomarker studies of this type, it is important to distinguish whether the miRNAs of interest are within exosomes, bound to blood proteins, or circulating free in the circulation. Among a group of differentially expressed miRNAs, Ortega et al. found that miR-140–5p abundance was greater in the blood of morbidly obese patients (BMI ≥ 40 Kg/m2) than obese men (40 Kg/m2 > BMI ≥ 30 Kg/m2) (Ortega et al., 2013). After surgery-induced weight loss, circulating miR-140–5p was markedly reduced, suggesting that this miRNA could be a biomarker for the change of fat mass. In addition, circulating miRNAs could be predictive signatures suggesting which obese patients are at greatest risk for T2DM development. In the population-based Malmӧ Diet and Cancer Study Cardiovascular Cohort, Gallo et al. reported that circulating miR-483–5p level was associated with the incidence of T2DM and CVD (Gallo et al., 2018). In addition, numerous studies have reported changes in circulating miRNA profiles associated with obesity or other metabolic diseases (Nunez Lopez et al., 2016; Wang et al., 2015b; Willeit et al., 2017; Wu et al., 2015). However, very few miRNA candidates were concordant across these studies, and different patterns were observed. These inconsistent results may relate to technical differences, and some studies have assessed all circulating miRNAs, while others have assessed EV miRNAs, with only a few examining exosomal miRNAs, and the miRNA cargo among these different components is different. In addition, the methods of miRNA measurements are not uniform, with some reports using chromatin immunoprecipitation (ChIP)-based technologies and others using direct sequencing to assess the miRNA expression signatures. Interestingly, an increase in the absolute level of circulating EVs in obesity has been a consistent observation in several studies.
Ghai et al. reported a set of miRNAs that are significantly decreased in plasma exosomes of T2DM patients after metformin treatment (Ghai et al., 2019). More importantly, among these metformin-repressed exosome miRNAs, most were expressed at greater levels in treated T2DM patients than in healthy humans. In contrast, these metformin-mediated changes in miRNA abundance were barely detected in whole plasma or exosome-depleted plasma. Another early study by Povero et al. found that in choline-deficient diet-induced liver steatosis, most of the circulating miR-122 was contained within exosomes, while in lean control mice, miR-122 was mostly bound to Argonaute 2 in the circulation independent of exosomes (Povero et al., 2014). In addition, Endzeliņš et al. suggest that miRNAs within plasma exosomes could be a more predictable source of miRNAs than whole plasma (Endzeliņš et al., 2017). Exosomes can be secreted by most cell types, and it has been a challenge to distinguish the cell-specific origin of exosomes and their miRNA cargos in biofluids. Emerging evidence indicates that some exosomes harbor cell-specific surface proteins that allow precipitation from biofluids. For example, Prattichizzo et al. found that both platelet and activated EC-derived exosomes carry CD31, allowing them to enrich CD31+ exosomes from human plasma using a CD31 antibody-conjugated magnetic bead-based precipitation method (Prattichizzo et al., 2021). They found that a group of specific miRNAs within CD31+ plasma exosomes from T2DM patients are tightly associated with the incidence of adverse cardiovascular events. Although many studies have profiled miRNAs within exosomes isolated from CM after ex vivo culture of primary cells, these exosome miRNA profiles may differ from those expressed under physiological conditions in vivo. Thus, methods to more precisely measure cell-specific circulating exosome miRNAs will greatly advance this area of the biomarker field. Taken together, these findings support the concept that circulating miRNAs could be sensitive signatures for metabolic diseases, but future studies will be required to validate this concept.
Exosome protein cargo composition is also associated with certain cellular functions, suggesting that exosome protein signatures could be potential biomarkers. Thus, Freeman et al. have reported that human plasma exosomes harbored various proteins associated with insulin signaling and that lower levels of phospho-S6RP, phospho-GSK3β, and phospho-AKT within plasma exosomes were associated with greater HOMA-B levels (Freeman et al., 2018). In addition, greater HOMA-IR was significantly associated with less phospho-S6RP within plasma exosomes. Also, plasma exosomes isolated from T2DM patients contained less leptin receptor and phospho-IR than in healthy human plasma exosomes. These findings suggest that circulating exosome protein cargo may provide markers of beta cell dysfunction and/or insulin resistance. By comparing the protein profiles within plasma exosomes derived from healthy, pre-cirrhotic, and cirrhotic NASH humans, Povero et al. found a set of hepatocyte-derived exosome proteins that have potential as biomarkers for the diagnosis of NASH (Povero et al., 2020). However, the number of reports focusing on exosome proteins as biomarkers is quite limited, and future work will be necessary to explore this issue.
TRANSLATIONAL AND CLINICAL ASPECTS
Exosomes as therapeutics or drug delivery vehicles
Given the nature of exosomes as nanovesicles, which can transfer cargos across different biological barriers, exosomes have been explored as delivery vectors for various molecules, including proteins and different RNA species. Interestingly, some studies have found that the uptake of exosomes can be cell specific, which is an important feature for drug delivery (Hoshino et al., 2015, 2020). However, there is a paucity of clinical papers on this subject in metabolic diseases, although studies are now gearing up. In other fields, more significant progress has been made, and a couple of these studies will be briefly summarized as examples of what might emerge in the future in the field of metabolism. For example, Hoshino et al. suggested that tumor cell-derived exosomes harbor specific integrins, which direct exosome homing to specific organs (Hoshino et al., 2015). Indeed, an early study showed that exosomes expressing αv integrin-specific peptides efficiently deliver exosomes to αv integrin+ tumors in prostate, breast, cervical, and pancreatic cancer models (Sugahara et al., 2009). In addition, Alvarez-Erviti et al. engineered mouse dendritic cells to produce neuron-specific RVG (rabies viral glycoprotein) peptide expressing exosomes that can specifically deliver small interfering RNA (siRNA) cargos into neurons, microglia, and oligodendrocytes after intravenous injection (Alvarez-Erviti et al., 2011). Another study by Yang et al. also used RVG-expressing exosomes to deliver miR-124 into the infarction site in a mouse stroke model (Yang et al., 2017). Similar engineering approaches have been used in other studies. For example, Kooijmans et al. found that fusion of glycosylphosphatidylinositol anchor peptides enhanced the binding efficiency of exosomes harboring anti-EGFR nanobodies into EGFP-positive tumor cells (Kooijmans et al., 2016). Ongoing clinical studies are being conducted to validate these findings from animal models.
An increasing number of studies have shown the effects of exosomes in metabolic diseases. Exosomes from MSCs have been tested as therapeutics in several clinical studies because of their regenerative capacity, immune tolerance, and immunoregulatory functions. The first report of MSC-derived exosomes in humans showed a significant attenuation of acute graft-versus-host disease symptoms within the first week of MSC exosome treatment, which remained stable for 4 months (Kordelas et al., 2014). In another study, Nassar et al. found that treatment of patients with chronic kidney disease (CKD) with human umbilical cord blood-derived MSC exosomes attenuated inflammatory responses and the progression of CKD (Nassar et al., 2016). In addition, a clinical trial has been initiated to evaluate the effects of human umbilical cord blood-derived MSC exosomes on beta cell mass in type 1 diabetes patients (ClinicalTrials.gov identifier NCT02138331). However, results from this trial have not yet been reported. Another clinical trial is ongoing to evaluate the effects of ginger- or aloe-derived exosomes on insulin resistance and chronic inflammation in patients with polycystic ovarian syndrome (NCT03493984). With respect to the concept of providing human exosomes from a donor to a recipient, unwanted immunologic reactions are unlikely since exosome-containing whole-blood and plasma transfusions have been used for decades.
CONCLUSIONS AND FUTURE DIRECTIONS
Exosomes are nanoparticles secreted by all cell types that gain access to the interstitial space and ultimately the circulation. In this manner, exosomes represent a widespread signaling system for intercellular and interorgan crosstalk. Exosome secretion and cargo loading are regulated by nutritional inputs, cell stress, and a wide variety of physiologic or pathophysiologic states. Given these regulatory events and the ability of secreted exosomes from a given cell to act on recipient cells at distal sites, the exosomal system displays many features of classical endocrine signaling. The exosomal miRNA cargo can affect a variety of biologic pathways, and new studies appear on a regular basis identifying specific miRNAs, or groups of miRNAs, that regulate metabolic events. Undoubtedly new and improved methods will emerge to generate more highly purified exosome preparations, which will be useful to address outstanding questions concerning the regulation of exosome secretion, targeting of exosomes to specific destinations, and detailed mechanisms of exosome uptake by recipient cells. The use of circulating exosomes as a liquid biopsy method to identify disease biomarkers has already yielded positive results, and future studies are likely to identify improved biomarker signatures, which will prove clinically useful. Exosomes have substantial therapeutic potential, and clinical studies outside the field of metabolism are already underway. In the field of metabolism, future efforts could involve the administration of natural exosomes, artificial exosomes containing specific miRNA cargos, as well as other types of payloads loaded into exosomes. The field of exosome biology is moving at a rapid pace, and one can envision answers to some of the important questions in the near future.
ACKNOWLEDGMENTS
This study was funded by the U.S. National Institute of Diabetes and Digestive and Kidney Diseases (P30 DK063491, R00DK115998, and R01DK125560 to W.Y. and P30 DK063491 and DK101395 to J.M.O.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alkhouri N, Dixon LJ, and Feldstein AE (2009). Lipotoxicity in nonalcoholic fatty liver disease: not all lipids are created equal. Expert Rev. Gastroenterol. Hepatol 3, 445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, and Wood MJ (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol 29, 341–345. [DOI] [PubMed] [Google Scholar]
- Amosse J, Durcin M, Malloci M, Vergori L, Fleury A, Gagnadoux F, Dubois S, Simard G, Boursier J, Hue O, et al. (2018). Phenotyping of circulating extracellular vesicles (EVs) in obesity identifies large EVs as functional conveyors of macrophage migration inhibitory factor. Mol. Metab 18, 134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreu Z, and Yáñez-Mó M (2014). Tetraspanins in extracellular vesicle formation and function. Front. Immunol 5, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraud N, Linares R, Tan S, Gounou C, Pasquet JM, Mornet S, and Brisson AR (2014). Extracellular vesicles from blood plasma: determination of their morphology, size, phenotype and concentration. J. Thromb. Haemost 12, 614–627. [DOI] [PubMed] [Google Scholar]
- Aswad H, Forterre A, Wiklander OP, Vial G, Danty-Berger E, Jalabert A, Lamaziè re A, Meugnier E, Pesenti S, Ott C, et al. (2014). Exosomes participate in the alteration of muscle homeostasis during lipid-induced insulin resistance in mice. Diabetologia 57, 2155–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebelman MP, Smit MJ, Pegtel DM, and Baglio SR (2018). Biogenesis and function of extracellular vesicles in cancer. Pharmacol. Ther 188, 1–11. [DOI] [PubMed] [Google Scholar]
- Bei Y, Chen T, Banciu DD, Cretoiu D, and Xiao J (2017). Circulating exosomes in cardiovascular diseases. Adv. Exp. Med. Biol 998, 255–269. [DOI] [PubMed] [Google Scholar]
- Bertoldi K, Cechinel LR, Schallenberger B, Corssac GB, Davies S, Guerreiro ICK, Belló-Klein A, Araujo ASR, and Siqueira IR (2018). Circulating extracellular vesicles in the aging process: impact of aerobic exercise. Mol. Cell. Biochem 440, 115–125. [DOI] [PubMed] [Google Scholar]
- Bhome R, Del Vecchio F, Lee GH, Bullock MD, Primrose JN, Sayan AE, and Mirnezami AH (2018). Exosomal microRNAs (exomiRs): small molecules with a big role in cancer. Cancer Lett. 420, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc L, and Vidal M (2018). New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 9, 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutens L, and Stienstra R (2016). Adipose tissue macrophages: going off track during obesity. Diabetologia 59, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky SV, Zhang F, Nasjletti A, and Goligorsky MS (2004). Endothelium-derived microparticles impair endothelial function in vitro. Am. J. Physiol. Heart Circ. Physiol 286, H1910–H1915. [DOI] [PubMed] [Google Scholar]
- Bruno S, Pasquino C, Herrera Sanchez MB, Tapparo M, Figliolini F, Grange C, Chiabotto G, Cedrino M, Deregibus MC, Tetta C, and Camussi G (2020). HLSC-derived extracellular vesicles attenuate liver fibrosis and inflammation in a murine model of non-alcoholic steatohepatitis. Mol. Ther 28, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesing KL, Densmore JC, Kaul S, Pritchard KA Jr., Jarzembowski JA, Gourlay DM, and Oldham KT (2011). Endothelial microparticles induce inflammation in acute lung injury. J. Surg. Res 166, 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnino JM, Ni K, and Jin Y (2020). Post-translational modification regulates formation and cargo-loading of extracellular vesicles. Front. Immunol 11, 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaño C, Kalko S, Novials A, and Párrizas M (2018). Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 115, 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto C, Cogoni C, and Zardo G (2016). MicroRNA in control of gene expression: an overview of nuclear functions. Int. J. Mol. Sci 17, 1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrier A, Chen R, Chen L, Kemper S, Hattori T, Takigawa M, and Brigstock DR (2014). Exosomes mediate intercellular transfer of pro-fibrogenic connective tissue growth factor (CCN2) between hepatic stellate cells, the principal fibrotic cells in the liver. Surgery 156, 548–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chen R, Kemper S, Charrier A, and Brigstock DR (2015). Suppression of fibrogenic signaling in hepatic stellate cells by Twist1-dependent microRNA-214 expression: role of exosomes in horizontal transfer of Twist1. Am. J. Physiol. Gastrointest. Liver Physiol 309, G491–G499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Chen J, Cheng Y, Fu Y, Zhao H, Tang M, Zhao H, Lin N, Shi X, Lei Y, et al. (2020a). Mesenchymal stem cell-derived exosomes protect beta cells against hypoxia-induced apoptosis via miR-21 by alleviating ER stress and inhibiting p38 MAPK phosphorylation. Stem Cell Res. Ther 11, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Yao X, Yao H, Ji Q, Ding G, and Liu X (2020b). Exosomal miR-103–3p from LPS-activated THP-1 macrophage contributes to the activation of hepatic stellate cells. FASEB J. 34, 5178–5192. [DOI] [PubMed] [Google Scholar]
- Ciafrè SA, and Galardi S (2013). microRNAs and RNA-binding proteins: a complex network of interactions and reciprocal regulations in cancer. RNA Biol. 10, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A, Turkes A, Navabi H, Mason MD, and Tabi Z (2005). Induction of heat shock proteins in B-cell exosomes. J. Cell Sci 118, 3631–3638. [DOI] [PubMed] [Google Scholar]
- Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Théry C, and Raposo G (2013). Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci 126, 5553–5565. [DOI] [PubMed] [Google Scholar]
- Combes V, Simon AC, Grau GE, Arnoux D, Camoin L, Sabatier F, Mutin M, Sanmarco M, Sampol J, and Dignat-George F (1999). In vitro generation of endothelial microparticles and possible prothrombotic activity in patients with lupus anticoagulant. J. Clin. Invest 104, 93–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly KD, Wadey RM, Mathew D, Johnson E, Rees DA, and James PE (2018). Evidence for adipocyte-derived extracellular vesicles in the human circulation. Endocrinology 159, 3259–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crewe C, Joffin N, Rutkowski JM, Kim M, Zhang F, Towler DA, Gordillo R, and Scherer PE (2018). An endothelial-to-adipocyte extracellular vesicle axis governed by metabolic state. Cell 175, 695–708.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crewe C, Funcke JB, Li S, Joffin N, Gliniak CM, Ghaben AL, An YA, Sadek HA, Gordillo R, Akgul Y, et al. (2021). Extracellular vesicle-based interorgan transport of mitochondria from energetically stressed adipocytes. Cell Metab. 33, ■■■-■■■. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai S, Liu F, Qin Z, Zhang J, Chen J, Ding WX, Feng D, Ji Y, and Qin X (2020). Kupffer cells promote T-cell hepatitis by producing CXCL10 and limiting liver sinusoidal endothelial cell permeability. Theranostics 10, 7163–7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang SY, Leng Y, Wang ZX, Xiao X, Zhang X, Wen T, Gong HZ, Hong A, and Ma Y (2019). Exosomal transfer of obesity adipose tissue for decreased miR-141–3p mediate insulin resistance of hepatocytes. Int. J. Biol. Sci 15, 351–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang XTT, Kavishka JM, Zhang DX, Pirisinu M, and Le MTN (2020). Extracellular vesicles as an efficient and versatile system for drug delivery. Cells 9, 2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva N, Samblas M, Martínez JA, and Milagro FI (2018). Effects of exosomes from LPS-activated macrophages on adipocyte gene expression, differentiation, and insulin-dependent glucose uptake. J. Physiol. Biochem 74, 559–568. [DOI] [PubMed] [Google Scholar]
- DeClercq V, d’Eon B, and McLeod RS (2015). Fatty acids increase adiponectin secretion through both classical and exosome pathways. Biochim. Biophys. Acta 1851, 1123–1133. [DOI] [PubMed] [Google Scholar]
- Deng ZB, Poliakov A, Hardy RW, Clements R, Liu C, Liu Y, Wang J, Xiang X, Zhang S, Zhuang X, et al. (2009). Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 58, 2498–2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densmore JC, Signorino PR, Ou J, Hatoum OA, Rowe JJ, Shi Y, Kaul S, Jones DW, Sabina RE, Pritchard KA Jr., et al. (2006). Endothelium-derived microparticles induce endothelial dysfunction and acute lung injury. Shock 26, 464–471. [DOI] [PubMed] [Google Scholar]
- Didiano D, and Hobert O (2008). Molecular architecture of a miRNA-regulated 3′ UTR. RNA 14, 1297–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignat-George F, and Boulanger CM (2011). The many faces of endothelial microparticles. Arterioscler. Thromb. Vasc. Biol 31, 27–33. [DOI] [PubMed] [Google Scholar]
- Doyle LM, and Wang MZ (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8, 727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi A, Mulya A, Lazic M, Radhakrishnan D, Berk MP, Povero D, Gornicka A, and Feldstein AE (2015). Microparticles release by adipocytes act as “find-me” signals to promote macrophage migration. PLoS ONE 10, e0123110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzanowska J, Semira C, and Costa-Silva B (2021). DNA in extracellular vesicles: biological and clinical aspects. Mol. Oncol 15, 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endzeliņš E, Berger A, Melne V, Bajo-Santos C, Soboļevska K, Ābols A, Rodriguez M, Šantare D, Rudņickiha A, Lietuvietis V, et al. (2017). Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 17, 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Tritschler F, and Izaurralde E (2009). The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA 15, 1433–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, and Sui SF (2010). Cellular internalization of exosomes occurs through phagocytosis. Traffic 11, 675–687. [DOI] [PubMed] [Google Scholar]
- Ferrante AW Jr. (2013). The immune cells in adipose tissue. Diabetes Obes. Metab 15 (Suppl 3), 34–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante SC, Nadler EP, Pillai DK, Hubal MJ, Wang Z, Wang JM, Gordish-Dressman H, Koeck E, Sevilla S, Wiles AA, and Freishtat RJ (2015). Adipocyte-derived exosomal miRNAs: a novel mechanism for obesity-related disease. Pediatr. Res 77, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, and Sonenberg N (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet 9, 102–114. [DOI] [PubMed] [Google Scholar]
- Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, and Simons M (2011). Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J. Cell Sci 124, 447–458. [DOI] [PubMed] [Google Scholar]
- Flaherty SE 3rd, Grijalva A, Xu X, Ables E, Nomani A, and Ferrante AW Jr. (2019). A lipase-independent pathway of lipid release and immune modulation by adipocytes. Science 363, 989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forterre A, Jalabert A, Berger E, Baudet M, Chikh K, Errazuriz E, De Larichaudy J, Chanon S, Weiss-Gayet M, Hesse AM, et al. (2014). Proteomic analysis of C2C12 myoblast and myotube exosome-like vesicles: a new paradigm for myoblast-myotube cross talk? PLoS ONE 9, e84153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman DW, Noren Hooten N, Eitan E, Green J, Mode NA, Bodogai M, Zhang Y, Lehrmann E, Zonderman AB, Biragyn A, et al. (2018). Altered extracellular vesicle concentration, cargo, and function in diabetes. Diabetes 67, 2377–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman SL (2008). Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol. Rev 88, 125–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Samovski D, Smith GI, Cifarelli V, Farabi SS, Yoshino J, Pietka T, Chang SW, Ghosh S, Myckatyn TM, and Klein S (2021). Associations among adipose tissue immunology, inflammation, and exosomes and insulin sensitivity in people with obesity and nonalcoholic fatty liver disease. Gastroenterology. 10.1053/j.gastro.2021.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo W, Esguerra JLS, Eliasson L, and Melander O (2018). miR-483–5p associates with obesity and insulin resistance and independently associates with new onset diabetes mellitus and cardiovascular disease. PLoS ONE 13, e0206974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, and Malim MH (2007). Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol 81, 2165–2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner RT, Solfest JS, Nie Y, Kuang S, Stout J, and Gavin TP (2020). Multivesicular body and exosome pathway responses to acute exercise. Exp. Physiol 105, 511–521. [DOI] [PubMed] [Google Scholar]
- Ge X, Tang P, Rong Y, Jiang D, Lu X, Ji C, Wang J, Huang C, Duan A, Liu Y, et al. (2021). Exosomal miR-155 from M1-polarized macrophages promotes EndoMT and impairs mitochondrial function via activating NF-κB signaling pathway in vascular endothelial cells after traumatic spinal cord injury. Redox Biol. 41, 101932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaben AL, and Scherer PE (2019). Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol 20, 242–258. [DOI] [PubMed] [Google Scholar]
- Ghai V, Kim TK, Etheridge A, Nielsen T, Hansen T, Pedersen O, Galas D, and Wang K (2019). Extracellular vesicle encapsulated microRNAs in patients with type 2 diabetes are affected by metformin treatment. J. Clin. Med 8, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, and Voinnet O (2009). Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol 11, 1143–1149. [DOI] [PubMed] [Google Scholar]
- Goldie BJ, Dun MD, Lin M, Smith ND, Verrills NM, Dayas CV, and Cairns MJ (2014). Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 42, 9195–9208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Körner R, Gaitanos L, and Klein R (2016). Exosomes mediate cell contact-independent ephrin-Eph signaling during axon guidance. J. Cell Biol 214, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier JL, Zhang L, Vetter MR, and Kazazian HH Jr. (2007). LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol 27, 6469–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot M, and Lee H (2020). Sorting mechanisms for microRNAs into extracellular vesicles and their associated diseases. Cells 9, 1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S, Jin L, Zhang F, Sarnow P, and Kay MA (2009). Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol 16, 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guay C, Menoud V, Rome S, and Regazzi R (2015). Horizontal transfer of exosomal microRNAs transduce apoptotic signals between pancreatic beta-cells. Cell Commun. Signal 13, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, and Simpson DA (2012). Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics 13, 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guescini M, Guidolin D, Vallorani L, Casadei L, Gioacchini AM, Tibollo P, Battistelli M, Falcieri E, Battistin L, Agnati LF, and Stocchi V (2010). C2C12 myoblasts release micro-vesicles containing mtDNA and proteins involved in signal transduction. Exp. Cell Res 316, 1977–1984. [DOI] [PubMed] [Google Scholar]
- Guo Q, Furuta K, Lucien F, Gutierrez Sanchez LH, Hirsova P, Krishnan A, Kabashima A, Pavelko KD, Madden B, Alhuwaish H, et al. (2019). Integrin β1-enriched extracellular vesicles mediate monocyte adhesion and promote liver inflammation in murine NASH. J. Hepatol 71, 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S, Kang MH, and Kim JH (2021). A comprehensive review on factors influences biogenesis, functions, therapeutic and clinical implications of exosomes. Int. J. Nanomedicine 16, 1281–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, and Kim VN (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 18, 3016–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazawa M, Tomiyama K, Saotome-Nakamura A, Obara C, Yasuda T, Gotoh T, Tanaka I, Yakumaru H, Ishihara H, and Tajima K (2014). Radiation increases the cellular uptake of exosomes through CD29/CD81 complex formation. Biochem. Biophys. Res. Commun 446, 1165–1171. [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, and Tollervey D (2013). Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández A, Arab JP, Reyes D, Lapitz A, Moshage H, Bañales JM, and Arrese M (2020). Extracellular vesicles in NAFLD/ALD: from pathobiology to therapy. Cells 9, 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez JD, Li T, Rau CM, LeSuer WE, Wang P, Coletta DK, Madura JA 2nd, Jacobsen EA, and De Filippis E (2021). ω−3PUFA supplementation ameliorates adipose tissue inflammation and insulin-stimulated glucose disposal in subjects with obesity: a potential role for apolipoprotein E. Int. J. Obes 45, 1331–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AA, Reid Bolus W, and Hasty AH (2014). A decade of progress in adipose tissue macrophage biology. Immunol. Rev 262, 134–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsova P, Ibrahim SH, Krishnan A, Verma VK, Bronk SF, Werneburg NW, Charlton MR, Shah VH, Malhi H, and Gores GJ (2016). Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology 150, 956–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horibe S, Tanahashi T, Kawauchi S, Murakami Y, and Rikitake Y (2018). Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer 18, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, et al. (2020). Extracellular vesicle and particle biomarkers define multiple human cancers. Cell 182, 1044–1061.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil GS, and Erbay E (2008). Nutrient sensing and inflammation in metabolic diseases. Nat. Rev. Immunol 8, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh CH, Rau CS, Wu SC, Yang JC, Wu YC, Lu TH, Tzeng SL, Wu CJ, and Lin CW (2015). Weight-reduction through a low-fat diet causes differential expression of circulating microRNAs in obese C57BL/6 mice. BMC Genomics 16, 699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iavello A, Frech VS, Gai C, Deregibus MC, Quesenberry PJ, and Camussi G (2016). Role of Alix in miRNA packaging during extracellular vesicle biogenesis. Int. J. Mol. Med 37, 958–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim SH, Hirsova P, Tomita K, Bronk SF, Werneburg NW, Harrison SA, Goodfellow VS, Malhi H, and Gores GJ (2016). Mixed lineage kinase 3 mediates release of C-X-C motif ligand 10-bearing chemotactic extracellular vesicles from lipotoxic hepatocytes. Hepatology 63, 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inder KL, Ruelcke JE, Petelin L, Moon H, Choi E, Rae J, Blumenthal A, Hutmacher D, Saunders NA, Stow JL, et al. (2014). Cavin-1/PTRF alters prostate cancer cell-derived extracellular vesicle content and internalization to attenuate extracellular vesicle-mediated osteoclastogenesis and osteoblast proliferation. J. Extracell. Vesicles 3. 10.3402/jev.v3.23784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N, Takada Y, Watanabe Y, Matsuzaki Y, Naruse C, Asano M, Iwakura Y, Suda T, and Matsuo K (2009). Bidirectional signaling through ephrinA2-EphA2 enhances osteoclastogenesis and suppresses osteoblastogenesis. J. Biol. Chem 284, 14637–14644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, and Srivastava D (2015). microRNAs as developmental regulators. Cold Spring Harb. Perspect. Biol 7, a008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, Lundgren P, Bleriot C, Liu Z, Deczkowska A, et al. (2019). Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178, 686–698.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovičová J, Sečová P, Michalková K, and Antalíková J (2020). Tetraspanins, more than markers of extracellular vesicles in reproduction. Int. J. Mol. Sci 21, 7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jella KK, Nasti TH, Li Z, Malla SR, Buchwald ZS, and Khan MK (2018). Exosomes, their biogenesis and role in inter-cellular communication, tumor microenvironment and cancer immunotherapy. Vaccines (Basel) 6, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Chen Q, Wang W, Ling Y, Yan Y, and Xia P (2020). Hepatocyte-derived extracellular vesicles promote endothelial inflammation and atherogenesis via microRNA-1. J. Hepatol 72, 156–166. [DOI] [PubMed] [Google Scholar]
- Jones A, Danielson KM, Benton MC, Ziegler O, Shah R, Stubbs RS, Das S, and Macartney-Coxson D (2017). miRNA signatures of insulin resistance in obesity. Obesity (Silver Spring) 25, 1734–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakazu E, Mauer AS, Yin M, and Malhi H (2016). Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1α-dependent manner. J. Lipid Res 57, 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, and LeBleu VS (2020). The biology, function, and biomedical applications of exosomes. Science 367, eaau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S, Hirano S, Sakamoto T, Furuyama A, Takase H, Kato H, Fukuta M, and Aoki Y (2020). Scavenger receptor MARCO contributes to cellular internalization of exosomes by dynamin-dependent endocytosis and macropinocytosis. Sci. Rep 10, 21795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita S, Maeda N, and Shimomura I (2019). Interorgan communication by exosomes, adipose tissue, and adiponectin in metabolic syndrome. J. Clin. Invest 129, 4041–4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, and Plasterk RH (2006). The diverse functions of microRNAs in animal development and disease. Dev. Cell 11, 441–450. [DOI] [PubMed] [Google Scholar]
- Konoshenko MY, Lekchnov EA, Vlassov AV, and Laktionov PP (2018). Isolation of extracellular vesicles: general methodologies and latest trends. BioMed Res. Int 2018, 8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooijmans SA, Aleza CG, Roffler SR, van Solinge WW, Vader P, and Schiffelers RM (2016). Display of GPI-anchored anti-EGFR nanobodies on extracellular vesicles promotes tumour cell targeting. J. Extracell. Vesicles 5, 31053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordelas L, Rebmann V, Ludwig AK, Radtke S, Ruesing J, Doeppner TR, Epple M, Horn PA, Beelen DW, and Giebel B (2014). MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 28, 970–973. [DOI] [PubMed] [Google Scholar]
- Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, and Ochiya T (2010). Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem 285, 17442–17452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranendonk ME, Visseren FL, van Herwaarden JA, Nolte-’t Hoen EN, de Jager W, Wauben MH, and Kalkhoven E (2014). Effect of extracellular vesicles of human adipose tissue on insulin signaling in liver and muscle cells. Obesity (Silver Spring) 22, 2216–2223. [DOI] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D, et al. (2010). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631. [DOI] [PubMed] [Google Scholar]
- Kusminski CM, Bickel PE, and Scherer PE (2016). Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat. Rev. Drug Discov 15, 639–660. [DOI] [PubMed] [Google Scholar]
- Lackey DE, and Olefsky JM (2016). Regulation of metabolism by the innate immune system. Nat. Rev. Endocrinol. 12, 15–28. [DOI] [PubMed] [Google Scholar]
- Lago F, Dieguez C, Gómez-Reino J, and Gualillo O (2007). Adipokines as emerging mediators of immune response and inflammation. Nat. Clin. Pract. Rheumatol 3, 716–724. [DOI] [PubMed] [Google Scholar]
- Lakhter AJ, Pratt RE, Moore RE, Doucette KK, Maier BF, DiMeglio LA, and Sims EK (2018). Beta cell extracellular vesicle miR-21–5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 61, 1124–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster GI, and Febbraio MA (2005). Exosome-dependent trafficking of HSP70: a novel secretory pathway for cellular stress proteins. J. Biol. Chem 280, 23349–23355. [DOI] [PubMed] [Google Scholar]
- Larios J, Mercier V, Roux A, and Gruenberg J (2020). ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol 219, e201904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan MC, Bigot A, Jensen SS, Dennis JL, Rogowska-Wrzesinska A, Lainé J, Gache V, Furling D, Jensen ON, Voit T, et al. (2012). In-depth analysis of the secretome identifies three major independent secretory pathways in differentiating human myoblasts. J. Proteomics 77, 344–356. [DOI] [PubMed] [Google Scholar]
- Lee YS, and Olefsky J (2021). Chronic tissue inflammation and metabolic disease. Genes Dev. 35, 307–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhu D, Huang L, Zhang J, Bian Z, Chen X, Liu Y, Zhang CY, and Zen K (2012). Argonaute 2 complexes selectively protect the circulating microRNAs in cell-secreted microvesicles. PLoS ONE 7, e46957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Menoret A, Farragher C, Ouyang Z, Bonin C, Holvoet P, Vella AT, and Zhou B (2019a). Single cell transcriptomics based-MacSpectrum reveals novel macrophage activation signatures in diseases. JCI Insight 5, e126453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Liu H, Ma C, Chen Y, Wang J, and Yang Y (2019b). Exosomes are the novel players involved in the beneficial effects of exercise on type 2 diabetes. J. Cell. Physiol 10.1002/jcp.28319. [DOI] [PubMed] [Google Scholar]
- Li X, Corbett AL, Taatizadeh E, Tasnim N, Little JP, Garnis C, Daugaard M, Guns E, Hoorfar M, and Li ITS (2019c). Challenges and opportunities in exosome research-perspectives from biology, engineering, and cancer therapy. APL Bioeng. 3, 011503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, and Miles WO (2019). Beyond CLIP: advances and opportunities to measure RBP-RNA and RNA-RNA interactions. Nucleic Acids Res. 47, 5490–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin XP, Mintern JD, and Gleeson PA (2020). Macropinocytosis in different cell types: similarities and differences. Membranes (Basel) 10, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Huang W, Zhang R, Wu J, Li L, and Tang Y (2013). Proteomic analysis of TNF-α-activated endothelial cells and endothelial microparticles. Mol. Med. Rep 7, 318–326. [DOI] [PubMed] [Google Scholar]
- Liu T, Sun YC, Cheng P, and Shao HG (2019). Adipose tissue macrophage-derived exosomal miR-29a regulates obesity-associated insulin resistance. Biochem. Biophys. Res. Commun 515, 352–358. [DOI] [PubMed] [Google Scholar]
- Ma T, Chen Y, Chen Y, Meng Q, Sun J, Shao L, Yu Y, Huang H, Hu Y, Yang Z, et al. (2018). MicroRNA-132, delivered by mesenchymal stem cell-derived exosomes, promote angiogenesis in myocardial infarction. Stem Cells Int. 2018, 3290372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdipour E, Salmasi Z, and Sabeti N (2019). Potential of stem cell-derived exosomes to regenerate b islets through Pdx-1 dependent mechanism in a rat model of type 1 diabetes. J. Cell. Physiol 234, 20310–20321. [DOI] [PubMed] [Google Scholar]
- Marchand L, Jalabert A, Meugnier E, Van den Hende K, Fabien N, Nicolino M, Madec AM, Thivolet C, and Rome S (2016). miRNA-375 a sensor of glucotoxicity is altered in the serum of children with newly diagnosed type 1 diabetes. J. Diabetes Res 2016, 1869082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markiewicz M, Richard E, Marks N, and Ludwicka-Bradley A (2013). Impact of endothelial microparticles on coagulation, inflammation, and angiogenesis in age-related vascular diseases. J. Aging Res 2013, 734509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKelvey KJ, Powell KL, Ashton AW, Morris JM, and McCracken SA (2015). Exosomes: mechanisms of uptake. J. Circ. Biomark 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi J (2018). Exosomes and ectosomes in intercellular communication. Curr. Biol 28, R435–R444. [DOI] [PubMed] [Google Scholar]
- Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, et al. (2014). Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menck K, Sönmezer C, Worst TS, Schulz M, Dihazi GH, Streit F, Erdmann G, Kling S, Boutros M, Binder C, and Gross JC (2017). Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J. Extracell. Vesicles 6, 1378056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G, and Cáceres JF (2019). Post-transcriptional control of miRNA biogenesis. RNA 25, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan A, Agarwal S, Clauss M, Britt NS, and Dhillon NK (2020). Extracellular vesicles: novel communicators in lung diseases. Respir. Res 21, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Scheel TK, Luna JM, Park CY, Fak JJ, Nishiuchi E, Rice CM, and Darnell RB (2015). miRNA-target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun 6, 8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy LA, Pink RC, and Carter DR (2014). Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 3. 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller G, Schneider M, Biemer-Daub G, and Wied S (2011). Microvesicles released from rat adipocytes and harboring glycosylphosphatidylinositol-anchored proteins transfer RNA stimulating lipid synthesis. Cell. Signal 23, 1207–1223. [DOI] [PubMed] [Google Scholar]
- Murphy DE, de Jong OG, Brouwer M, Wood MJ, Lavieu G, Schiffelers RM, and Vader P (2019). Extracellular vesicle-based therapeutics: natural versus engineered targeting and trafficking. Exp. Mol. Med 51, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar W, El-Ansary M, Sabry D, Mostafa MA, Fayad T, Kotb E, Temraz M, Saad AN, Essa W, and Adel H (2016). Umbilical cord mesenchymal stem cells derived extracellular vesicles can safely ameliorate the progression of chronic kidney diseases. Biomater. Res 20, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima H, Freeman CM, Schuster RM, Japtok L, Kleuser B, Edwards MJ, Gulbins E, and Lentsch AB (2016). Hepatocyte exosomes mediate liver repair and regeneration via sphingosine-1-phosphate. J. Hepatol 64, 60–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez Lopez YO, Garufi G, and Seyhan AA (2016). Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst 13, 106–121. [DOI] [PubMed] [Google Scholar]
- Nussbacher JK, and Yeo GW (2018). Systematic discovery of RNA binding proteins that regulate microRNA levels. Mol. Cell 69, 1005–1016.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K, Breyne K, Ughetto S, Laurent LC, and Breakefield XO (2020). RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol 21, 585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell RM, Taganov KD, Boldin MP, Cheng G, and Baltimore D (2007). MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 104, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega FJ, Mercader JM, Catalán V, Moreno-Navarrete JM, Pueyo N, Sabater M, Gómez-Ambrosi J, Anglada R, Fernández-Formoso JA, Ricart W, et al. (2013). Targeting the circulating microRNA signature of obesity. Clin. Chem 59, 781–792. [DOI] [PubMed] [Google Scholar]
- Pan Y, Hui X, Hoo RLC, Ye D, Chan CYC, Feng T, Wang Y, Lam KSL, and Xu A (2019). Adipocyte-secreted exosomal microRNA-34a inhibits M2 macrophage polarization to promote obesity-induced adipose inflammation. J. Clin. Invest 129, 834–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Párrizas M, Brugnara L, Esteban Y, González-Franquesa A, Canivell S, Murillo S, Gordillo-Bastidas E, Cussó R, Cadefau JA, García-Roves PM, et al. (2015). Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J. Clin. Endocrinol. Metab 100, E407–E415. [DOI] [PubMed] [Google Scholar]
- Pescador N, Pérez-Barba M, Ibarra JM, Corbatón A, Martínez-Larrad MT, and Serrano-Ríos M (2013). Serum circulating microRNA profiling for identification of potential type 2 diabetes and obesity biomarkers. PLoS ONE 8, e77251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero D, Eguchi A, Niesman IR, Andronikou N, de Mollerat du Jeu X, Mulya A, Berk M, Lazic M, Thapaliya S, Parola M, et al. (2013). Lipid-induced toxicity stimulates hepatocytes to release angiogenic microparticles that require Vanin-1 for uptake by endothelial cells. Sci. Signal 6, ra88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero D, Eguchi A, Li H, Johnson CD, Papouchado BG, Wree A, Messer K, and Feldstein AE (2014). Circulating extracellular vesicles with specific proteome and liver microRNAs are potential biomarkers for liver injury in experimental fatty liver disease. PLoS ONE 9, e113651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero D, Panera N, Eguchi A, Johnson CD, Papouchado BG, de Araujo Horcel L, Pinatel EM, Alisi A, Nobili V, and Feldstein AE (2015). Lipid-induced hepatocyte-derived extracellular vesicles regulate hepatic stellate cell via microRNAs targeting PPAR-γ. Cell. Mol. Gastroenterol. Hepatol 1, 646–663.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povero D, Yamashita H, Ren W, Subramanian MG, Myers RP, Eguchi A, Simonetto DA, Goodman ZD, Harrison SA, Sanyal AJ, et al. (2020). Characterization and proteome of circulating extracellular vesicles as potential biomarkers for NASH. Hepatol. Commun 4, 1263–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prattichizzo F, Matacchione G, Giuliani A, Sabbatinelli J, Olivieri F, de Candia P, De Nigris V, and Ceriello A (2021). Extracellular vesicle-shuttled miRNAs: a critical appraisal of their potential as nano-diagnostics and nanotherapeutics in type 2 diabetes mellitus and its cardiovascular complications. Theranostics 11, 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, and Dallas SL (2019). Exosomes and extracellular RNA in muscle and bone aging and crosstalk. Curr. Osteoporos. Rep 17, 548–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez SH, Andrews AM, Paul D, and Pachter JS (2018). Extracellular vesicles: mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 15, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome S, Forterre A, Mizgier ML, and Bouzakri K (2019). Skeletal muscle-released extracellular vesicles: state of the art. Front. Physiol 10, 929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong B, Feng R, Liu C, Wu Q, and Sun C (2019). Reduced delivery of epididymal adipocyte-derived exosomal resistin is essential for melatonin ameliorating hepatic steatosis in mice. J. Pineal Res 66, e12561. [DOI] [PubMed] [Google Scholar]
- Rong S, Wang L, Peng Z, Liao Y, Li D, Yang X, Nuessler AK, Liu L, Bao W, and Yang W (2020). The mechanisms and treatments for sarcopenia: could exosomes be a perspective research strategy in the future? J. Cachexia Sarcopenia Muscle 11, 348–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo F, Moreno L, Mleczko J, Palomo L, Gonzalez E, Cabrera D, Cogolludo A, Vizcaino FP, van-Liempd S, and Falcon-Perez JM (2017). Hepatocyte-secreted extracellular vesicles modify blood metabolome and endothelial function by an arginase-dependent mechanism. Sci. Rep 7, 42798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo L, and Lumeng CN (2018). Properties and functions of adipose tissue macrophages in obesity. Immunology 155, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabry D, Marzouk S, Zakaria R, Ibrahim HA, and Samir M (2020). The effect of exosomes derived from mesenchymal stem cells in the treatment of induced type 1 diabetes mellitus in rats. Biotechnol. Lett 42, 1597–1610. [DOI] [PubMed] [Google Scholar]
- Safdar A, and Tarnopolsky MA (2018). Exosomes as mediators of the systemic adaptations to endurance exercise. Cold Spring Harb. Perspect. Med 8, a029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safdar A, Saleem A, and Tarnopolsky MA (2016). The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat. Rev. Endocrinol 12, 504–517. [DOI] [PubMed] [Google Scholar]
- Saltiel AR (2021). Insulin signaling in health and disease. J. Clin. Invest 131, e142241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandvig K, Torgersen ML, Raa HA, and van Deurs B (2008). Clathr-in-independent endocytosis: from nonexisting to an extreme degree of complexity. Histochem. Cell Biol 129, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano S, Izumi Y, Yamaguchi T, Yamazaki T, Tanaka M, Shiota M, Osada-Oka M, Nakamura Y, Wei M, Wanibuchi H, et al. (2014). Lipid synthesis is promoted by hypoxic adipocyte-derived exosomes in 3T3-L1 cells. Biochem. Biophys. Res. Commun 445, 327–333. [DOI] [PubMed] [Google Scholar]
- Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, and Tripodi M (2016). The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 17, 799–808. [DOI] [PubMed] [Google Scholar]
- Sato S, Vasaikar S, Eskaros A, Kim Y, Lewis JS, Zhang B, Zijlstra A, and Weaver AM (2019). EPHB2 carried on small extracellular vesicles induces tumor angiogenesis via activation of ephrin reverse signaling. JCI Insight 4, e132447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman JS, Troutman TD, Sakai M, Gola A, Spann NJ, Bennett H, Bruni CM, Ouyang Z, Li RZ, Sun X, et al. (2020). Niche-specific reprogramming of epigenetic landscapes drives myeloid cell diversity in nonalcoholic steatohepatitis. Immunity 52, 1057–1074.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Z, Tan J, Miao Y, and Zhang Q (2019). The role of microvesicles containing microRNAs in vascular endothelial dysfunction. J. Cell. Mol. Med 23, 7933–7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R (2016). Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife 5, e19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Confalonieri S, Ciliberto A, Polo S, Scita G, and Di Fiore PP (2012). Endocytosis and signaling: cell logistics shape the eukaryotic cell plan. Physiol. Rev 92, 273–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skotland T, Sagini K, Sandvig K, and Llorente A (2020). An emerging focus on lipids in extracellular vesicles. Adv. Drug Deliv. Rev 159, 308–321. [DOI] [PubMed] [Google Scholar]
- Smyth TJ, Redzic JS, Graner MW, and Anchordoquy TJ (2014). Examination of the specificity of tumor cell derived exosomes with tumor cells in vitro. Biochim. Biophys. Acta 1838, 2954–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, and Ruoslahti E (2009). Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 16, 510–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Kim J, and Jung Y (2018). Liver-derived exosomes and their implications in liver pathobiology. Int. J. Mol. Sci 19, 3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Mörgelin M, and Belting M (2013). Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem 288, 17713–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, Antoniou A, Arab T, Archer F, Atkin-Smith GK, et al. (2018). Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomou T, Mori MA, Dreyfuss JM, Konishi M, Sakaguchi M, Wolfrum C, Rao TN, Winnay JN, Garcia-Martin R, Grinspoon SK, et al. (2017). Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 542, 450–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson DW, Bracken CP, and Goodall GJ (2011). Experimental strategies for microRNA target identification. Nucleic Acids Res. 39, 6845–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, and Xiao ZD (2014). Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem 289, 22258–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Freeman BL, Bronk SF, LeBrasseur NK, White TA, Hirsova P, and Ibrahim SH (2016). CXCL10-mediates macrophage, but not other innate immune cells-associated inflammation in murine nonalcoholic steatohepatitis. Sci. Rep 6, 28786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brügger B, and Simons M (2008). Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 319, 1244–1247. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, Drapkina O, and Tonevitsky A (2019). Transcriptome of extracellular vesicles: state-of-the-art. Front. Immunol 10, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ung TH, Madsen HJ, Hellwinkel JE, Lencioni AM, and Graner MW (2014). Exosome proteomics reveals transcriptional regulator proteins with potential to mediate downstream pathways. Cancer Sci. 105, 1384–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner T, Spinelli C, Minciacchi VR, Balaj L, Zandian M, Conley A, Zijlstra A, Freeman MR, Demichelis F, De S, et al. (2018). Large extracellular vesicles carry most of the tumour DNA circulating in prostate cancer patient plasma. J. Extracell. Vesicles 7, 1505403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, and Lötvall JO (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol 9, 654–659. [DOI] [PubMed] [Google Scholar]
- Van den Brande S, Gijbels M, Wynant N, Santos D, Mingels L, Gansemans Y, Van Nieuwerburgh F, and Vanden Broeck J (2018). The presence of extracellular microRNAs in the media of cultured Drosophila cells. Sci. Rep 8, 17312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. (2016). Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat. Methods 13, 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegiopoulos A, Rohm M, and Herzig S (2017). Adipose tissue: between the extremes. EMBO J. 36, 1999–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veziroglu EM, and Mias GI (2020). Characterizing extracellular vesicles and their diverse RNA contents. Front. Genet 11, 700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M, Radulovic M, and Stenmark H (2020). The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol 21, 25–42. [DOI] [PubMed] [Google Scholar]
- Villard A, Marchand L, Thivolet C, and Rome S (2015). Diagnostic value of cell-free circulating microRNAs for obesity and type 2 diabetes: a meta-analysis. J. Mol. Biomark. Diagn 6, 251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, and Sánchez-Madrid F (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Ding Q, Yaqoob U, de Assuncao TM, Verma VK, Hirsova P, Cao S, Mukhopadhyay D, Huebert RC, and Shah VH (2015a). Exosome adherence and internalization by hepatic stellate cells triggers sphingosine 1-phosphate-dependent migration. J. Biol. Chem 290, 30684–30696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Hong J, Cao Y, Shi J, Gu W, Ning G, Zhang Y, and Wang W (2015b). Elevated circulating microRNA-122 is associated with obesity and insulin resistance in young adults. Eur. J. Endocrinol 172, 291–300. [DOI] [PubMed] [Google Scholar]
- Wang C, Zhang C, Liu L, A X, Chen B, Li Y, and Du J (2017). Macrophage-derived mir-155-containing exosomes suppress fibroblast proliferation and promote fibroblast inflammation during cardiac injury. Mol. Ther 25, 192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yu F, Ding H, Wang Y, Li P, and Wang K (2019). Emerging function and clinical values of exosomal microRNAs in cancer. Mol. Ther. Nucleic Acids 16, 791–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Chen Q, Lin L, Sha C, Li T, Liu Y, Yin X, Xu Y, Chen L, Gao W, et al. (2021). Regulation of exosome production and cargo sorting. Int. J. Biol. Sci 17, 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels HH, Lebedeva S, Hirsekorn A, Wurmus R, Akalin A, Mukherjee N, and Ohler U (2019). Global identification of functional microRNA-mRNA interactions in Drosophila. Nat. Commun 10, 1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, Barwari T, Whitehead M, Ramírez CM, Goedeke L, et al. (2017). Circulating microRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes 66, 347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Pazos R, Royo F, González E, Roura-Ferrer M, Martinez A, Gamiz J, Reichardt NC, and Falcón-Pérez JM (2019). Assessing the role of surface glycans of extracellular vesicles on cellular uptake. Sci. Rep 9, 11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wortzel I, Dror S, Kenific CM, and Lyden D (2019). Exosome-mediated metastasis: communication from a distance. Dev. Cell 49, 347–360. [DOI] [PubMed] [Google Scholar]
- Wu CX, and Liu ZF (2018). Proteomic profiling of sweat exosome suggests its involvement in skin immunity. J. Invest. Dermatol 138, 89–97. [DOI] [PubMed] [Google Scholar]
- Wu L, Dai X, Zhan J, Zhang Y, Zhang H, Zhang H, Zeng S, and Xi W (2015). Profiling peripheral microRNAs in obesity and type 2 diabetes mellitus. APMIS 123, 580–585. [DOI] [PubMed] [Google Scholar]
- Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao XY, Ji Y, Li C, Guo L, et al. (2019). Landscape of intercellular crosstalk in healthy and NASH liver revealed by single-cell secretome gene analysis. Mol. Cell 75, 644–660.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Niida S, Azuma E, Yanagibashi T, Muramatsu M, Huang TT, Sagara H, Higaki S, Ikutani M, Nagai Y, et al. (2015). Inflammation-induced endothelial cell-derived extracellular vesicles modulate the cellular status of pericytes. Sci. Rep 5, 8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Mó M, Siljander PR, Andreu Z, Zavec AB, Borràs FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, et al. (2015). Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang X, Chen X, Wang L, and Yang G (2017). Exosome mediated delivery of miR-124 promotes neurogenesis after ischemia. Mol. Ther. Nucleic Acids 7, 278–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Riopel M, Bandyopadhyay G, Dong Y, Birmingham A, Seo JB, Ofrecio JM, Wollam J, Hernandez-Carretero A, Fu W, et al. (2017). Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell 171, 372–384.e12. [DOI] [PubMed] [Google Scholar]
- Ying W, Gao H, Dos Reis FCG, Bandyopadhyay G, Ofrecio JM, Luo Z, Ji Y, Jin Z, Ly C, and Olefsky JM (2021). MiR-690, an exosomalderived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab. 33, 781–790.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudong Ji ZL, Gao H, Gomes Dos Reis FC, Bandyopadhyay G, Jin Z, Manda KA, Isaac R, Yang M, Fu W, Ying W, et al. (2021). Hepatocyte-derived exosomes from early onset obese mice promote insulin sensitivity through miR-3075. Nat. Metab 10.1038/542255-021-00444-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Li L, Li M, Guo C, Yao J, and Mi S (2015a). Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13, 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shi L, Mei H, Zhang J, Zhu Y, Han X, and Zhu D (2015b). Inflamed macrophage microvesicles induce insulin resistance in human adipocytes. Nutr. Metab. (Lond.) 12, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Liu H, and Tang WH (2019). Exosomes: biogenesis, biologic function and clinical potential. Cell Biosci. 9, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao YL, Tian PX, Han F, Zheng J, Xia XX, Xue WJ, Ding XM, and Ding CG (2017). Comparison of the characteristics of macrophages derived from murine spleen, peritoneal cavity, and bone marrow. J. Zhejiang Univ. Sci. B 18, 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Sun X, and Li L (2019). Biogenesis and function of extracellular miRNAs. ExRNA 1, 38. [Google Scholar]
- Zhou H, Wang B, Yang Y, Jia Q, Qi Z, Zhang A, Lv S, and Zhang J (2019). Exosomes in ischemic heart disease: novel carriers for bioinformation. Biomed. Pharmacother 120, 109451. [DOI] [PubMed] [Google Scholar]