Common wheat supplies vast amounts of dietary carbohydrate and protein for over 60% of world population. With continued economic development, demand for high‐yielding varieties with premium grain quality is increasing. Grain quality is a multigenic trait that is simultaneously affected by many factors, and it is more complicated in hexaploid wheat. CRISPR/Cas9‐based genome editing provides great opportunities to create more allelic variations in a much faster and precise manner (Li et al., 2020a, 2020b; Sanchez‐Leon et al., 2018; Zhang et al., 2020a, 2020b). In this study, four grain quality‐related genes viz. pinb, waxy, ppo and psy, which involved in wheat grain hardness, starch quality and dough colour, respectively, were targeted for genome editing. Specific sgRNAs were selected, and corresponding vectors were constructed for Agrobacterium‐mediated transformation in wheat cultivar Fielder (Figure 1a). CRISPR‐targeted mutations were identified by Hi‐TOM sequencing (Figure 1b; Liu et al., 2019). The editing efficiency (EE) including editing rates (mutant plants/total plants) and editing ratio (editing reads/total reads) increased significantly across generations (Figure 1c). The complex genome composition of wheat makes it difficult to edit all alleles simultaneously. Despite of this, most of the mutants had 100% EE in T2 and T3 generation. Mutant plants with the same mutant type in A, B and D subgenomes, such as pinb‐47, waxy‐2, ppo‐7 and psy‐13 were chosen for analysis (Figure 1c). The expression of all the four target genes decreased significantly in the mutants compared with wildtype (WT), and it was almost undetectable during the whole grain development stages (Figure 1d).
Figure 1.
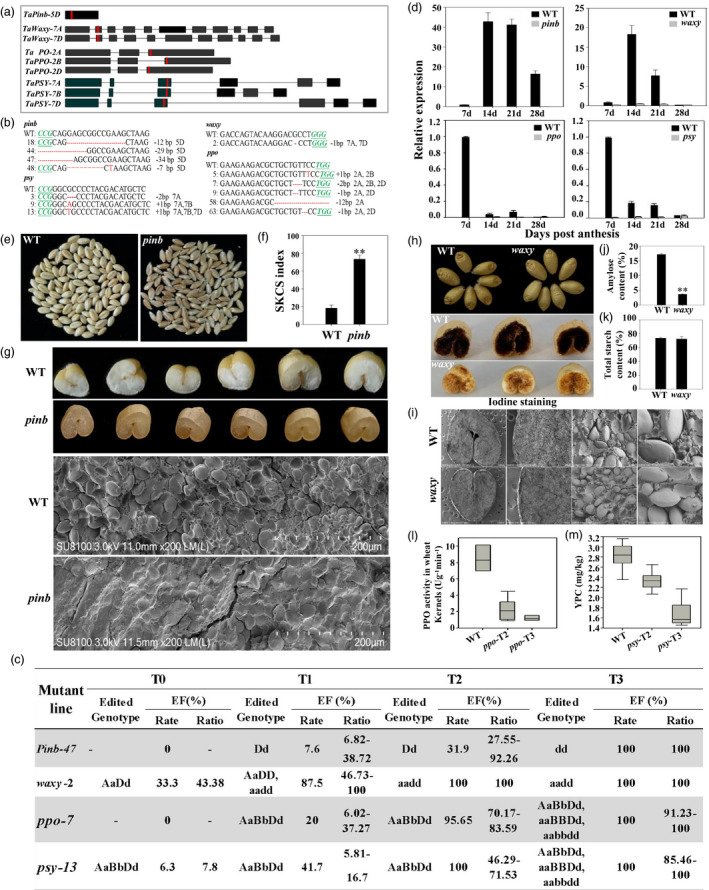
CRISPR/Cas9‐mediated genome editing for generating pinb, waxy, ppo and psy mutants in common wheat. (a) Gene structure and sgRNA selection in target genes. Introns are shown as lines, and exons are shown as black boxes. Target sites are indicated in red. (b) CRISPR/Cas9‐induced mutagenesis of target genes. PAM sites are underlined and indicated in italics in green. Mutation sites are indicated in red. (c) Detailed analysis of CRISPR‐induced mutation of the four target genes across T0, T1, T2 and T3 generation. EE, Editing Efficiency; editing rate = mutant plants/total plants×100%; editing ratio = editing reads/total reads × 100%. (d) Expression level of the four target genes in developing grains of WT and mutants. Quantitative reverse‐transcription PCR (qRT‐PCR) was used for analysis. The Actin gene was used for normalization. (e) Grain phenotypes of CRISPR‐pinb mutants and WT. (f) The SKCS index of pinb and WT. (g) SEM of endosperms in pinb mutants and WT. (h) Grain phenotypes of CRISPR‐waxy mutants and WT. Top panel, comparison of grain appearance between WT and mutants. Bottom panel, iodine‐staining of endosperm in cross‐sections of seeds. (i) SEM of endosperms in waxy mutants and WT. (j) Amylose content in waxy mutants and WT. (k) Total starch content in waxy mutants and WT. (l) PPO activity in kernels of ppo mutants and WT. (m) YPC in grains from psy mutants and WT. Data are presented as means ± standard error from three biological replicates. *P < 0.05, **P < 0.01.
Grain hardness is the most important and defining quality in wheat. Pinb is a single‐copy gene located on chromosome 5DS, whose absence or alteration by mutation could result in hard texture (Giroux and Morris, 1998). The sgRNA was designed in the coding sequence of pinb gene (Figure 1a; Zhang et al., 2019a). Mutations were not detected in T0 plants, but first appeared in T1 lines. The EE increased and the highest editing ratio achieved 92.26% in T2 generation. T3 homozygous plants with 34‐bp deletion that led to frameshift mutations were investigated (Figure 1c). The cross‐section of mutant seeds displayed eminent translucent phenotype while that of WT showed different appearance of white colour (Figure 1e, 1g). Grain hardness index based on the single kernel characterization system (SKCS, Giroux and Morris, 1998) was significantly elevated in CRISPR‐pinb mutants. The SKCS index for pinb seeds was 73.4 (from 67 to 80 ) in average, while that for WT was only 15–24 with an average of 18.2 (Figure 1f). Thus, we successfully converted soft wheat into hard wheat. Observation of cross‐sectional grain structure with scanning electron microscope (SEM) showed different adhesion levels between starch granules and protein matrix. Starch granules were loose and separated with protein matrix in WT, while deeply trapped and tightly integrated with protein matrix in mutants (Figure 1g). Multiple pinb alleles had been previously detected from different wheat cultivars across different geographic regions, but most of them were single nucleotide insertion, deletion and substitution with SKCS index around 60 (Chen et al., 2013). SKCS index was significantly elevated in CRISPR‐induced mutants, which may be attributed to the large deletion of 34‐bp in the coding region. The new allele reported herein with a much higher SKCS index hence broadens genetic resource for wheat grain hardness improvement.
Grain starch components have important influences on wheat flour quality. Waxy is a key enzyme in amylose synthesis in wheat endosperm, which is encoded by Wx‐A1, Wx‐B1 and Wx‐D1 located on 7A, 4A and 7D chromosomes, respectively (Yamamori et al., 1994). In order to obtain glutinous wheat, we constructed a binary vector to target all functional waxy homoeoalleles. We designed an sgRNA targeting the second exon of waxy gene. Sequence amplification and analysis confirmed that Wx‐B1 was deficient in Fielder. A 1‐bp deletion in both A and D subgenomes which caused premature translation termination was found in waxy‐2. The edited genotype was inherited to next generation without occurrence of new mutation. T2 plants with homozygous mutations were identified. The CRISPR‐edited grains had appearance of being a bit whiter and opaquer compared with WT (Figure 1h). SEM showed no significant differences in starch structure between waxy and WT (Figure 1i). Endosperm in cross‐sections of WT turned dark blue when stained with iodine, while waxy turned red‐brown which was a strong evidence for glutinous wheat (Figure 1h). As expected, waxy showed a significant reduction in amylose content (3.6%) compared with WT (17.2%) (Figure 1j), while the total starch contents of mutants (72.5%) and WT (73.6%) were similar (Figure 1k). Thus, we have successfully mutated waxy gene to generate glutinous wheat with lower amylose content.
Polyphenol oxidase (PPO) activity and yellow pigment content (YPC) are the two important parameters determining the colour of wheat products (Li et al., 2015). PPO catalyses phenols oxidation into dark‐coloured products, a feature often undesirable for wheat end‐use products. Therefore, developing wheat cultivars with low PPO activity has always been an important goal in wheat breeding. One sgRNA was designed to target the conserved sites of the third exon of all three homoeologs (Figure 1a). No mutation was detected in ppo‐7 of T0. Mutation of 2‐bp deletion was obtained in T1 and passed to T3 generation, which were selected for further analysis. The PPO activity in wheat kernel was 9.0 U g−1 min−1 in WT. T2 plants with editing ratio 70.17–83.59% showed an average of 2.4 U g−1 min−1, and T3 mutants with 100% editing ratio were 1.24 U g−1 min−1, which was significantly lower than WT (Figure 1l). It indicated negative relationship between edited ratios and PPO activity.
Phytoene synthase (PSY) catalyses a vital step in carotenoid biosynthesis, generally recognized as the most important regulatory enzyme in the pathway. PSY homoeologs (TaPSY‐7A, 7B and 7D) were edited by the same sgRNA (Figure 1a). Mutants with a ‘T’ insertion at all three homoeologs were chosen. The edited reads showed negative relationship with YPC content (Figure 1m). YPC quantification of T2 plants with editing ratio 46.29–71.53% was 2.33 mg/kg, and T3 homozygous mutants achieved 1.68 mg/kg in average, which was significantly decreased than WT (2.81 mg/kg) (Figure 1m). PSY editing decreased downstream metabolites in carotenoid biosynthesis pathway.
In conclusion, new allelic variations of the target genes (pinb, waxy, ppo and psy) were created in Fielder through Agrobacterium‐delivered CRISPR/Cas9 system. Editing ratio of the mutants had positive relationship with the phenotype. When editing ratio achieved 100% in the frameshift homozygous mutants, gene expression was almost undetectable, and phenotypes different from WT were obviously observed. Furthermore, many of the mutants had segregated out the CRISPR/Cas9 transgene. Thus, we had successfully obtained new wheat germplasms with improved grain quality in hardness, starch composition and dough colour. We envision these new wheat germplasms with improved grain quality can be used as donor parents to improve grain quality of elite wheat cultivars through backcross breeding.
Conflicts of interest
The authors declare no conflicts of interest.
Author Contributions
S.Z., Y.Q. and G.L. designed the experiment, analysed data and wrote the manuscript. R.Z and J.L. performed Hi‐TOM sequencing. J.G. and Y.L. performed wheat transformation. W.L. and G.S. managed the wheat plants and collected samples.
Acknowledgements
We appreciated Qixin Sun’s group of China Agricultural University for providing pBUE411 vector. This work was funded by Agricultural Variety Improvement Project of Shandong Province (2019LZGC015), National Natural Science Foundation of China (32072006), Natural Science Foundation of Shandong Province (ZR2020MC097) and Major Scientific and Technological Innovation Projects of Shandong Key R & D Plan (2020CXGC010805). The work was also supported by the startup funds from University of Maryland, College Park.
Zhang, S. , Zhang, R. , Gao, J. , Song, G. , Li, J. , Li, W. , Qi, Y. , Li, Y. and Li, G. (2021) CRISPR/Cas9‐mediated genome editing for wheat grain quality improvement. Plant Biotechnol. J. 10.1111/pbi.13647
Contributor Information
Yulian Li, Email: liyulian01@163.com.
Genying Li, Email: lgy111@126.com.
References
- Chen, F. , Li, H. and Cui, D. (2013) Discovery, distribution and diversity of puroindoline‐D1 genes in bread wheat from five countries (Triticum aestivum L.). BMC Plant Biol. 13, 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux, M.J. and Morris, C.F. (1998) Wheat grain hardness results from highly conserved mutations in the friabilin components puroindoline a and b. Proc. Nat. Acad. Sci. USA, 95, 6262–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Jiao, G. , Sun, Y. , Chen, J. , Zhong, Y. , Yan, L. , Jiang, D. et al. (2020b) Modification of starch composition, structure and properties through editing of TaSBEIIa in both winter and spring wheat varieties by CRISPR/Cas9. Plant Biotechnol. J. 19, 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Zhang, S. , Zhang, R. , Gao, J. , Qi, Y. , Song, G. , Li, W. et al. (2020a) Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol. J. 19, 427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Wu, Y. , Wang, T. , Li, L. , Lu, L. et al. (2015) Polyphenol oxidase acitivty and yellowpigment content in Aegilops tauschii, Triticum turgidum, Triticum aestivum, synthetic hexaploid wheat and its parents. J. Cereal Sci. 65, 192–201. [Google Scholar]
- Liu, Q. , Wang, C. , Jiao, X. , Zhang, H. , Song, L. , Li, Y. , Gao, C. et al. (2019) Hi‐TOM: a platform for high‐throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Leon, S. , Gil‐Humanes, J. , Ozuna, C.V. , Gimenez, M.J. , Sousa, C. , Voytas, D.F. and Barro, F. (2018) Low‐gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 16, 902–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamori, M. , Nakamura, T. , Endo, T.R. and Nagamine, T. (1994) Waxy protein deficiency and chromosomal location of coding genes in common wheat. Theor. Appl. Genet. 89, 179–184. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Zhang, R. , Gao, J. , Gu, T. , Song, G. , Li, W. , Li, D. et al. (2019a) High efficient and heritable targeted mutagenesis in wheat via the Agrobacterium tumerfaciens‐mediated CRISPR/Cas9 system. Int. J. Mol. Sci. 20, 4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Hua, L. , Gupta, A. , Tricoli, D. , Edwards, K. , Yang, B. and Li, W. (2019b) Development of an Agrobacterium‐delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 17, 1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]


