Summary
De novo allopolyploidization in Brassica provides a very successful model for reconstructing polyploid genomes using progenitor species and relatives to broaden crop gene pools and understand genome evolution after polyploidy, interspecific hybridization and exotic introgression. B. napus (AACC), the major cultivated rapeseed species and the third largest oilseed crop in the world, is a young Brassica species with a limited genetic base resulting from its short history of domestication, cultivation, and intensive selection during breeding for target economic traits. However, the gene pool of B. napus has been significantly enriched in recent decades that has been benefit from worldwide effects by the successful introduction of abundant subgenomic variation and novel genomic variation via intraspecific, interspecific and intergeneric crosses. An important question in this respect is how to utilize such variation to breed crops adapted to the changing global climate. Here, we review the genetic diversity, genome structure, and population‐level differentiation of the B. napus gene pool in relation to known exotic introgressions from various species of the Brassicaceae, especially those elucidated by recent genome‐sequencing projects. We also summarize progress in gene cloning, trait‐marker associations, gene editing, molecular marker‐assisted selection and genome‐wide prediction, and describe the challenges and opportunities of these techniques as molecular platforms to exploit novel genomic variation and their value in the rapeseed gene pool. Future progress will accelerate the creation and manipulation of genetic diversity with genomic‐based improvement, as well as provide novel insights into the neo‐domestication of polyploid crops with novel genetic diversity from reconstructed genomes.
Keywords: polyploid crop, Brassica, gene pool, exotic introgressions, genomic changes, genomic‐based improvement
Introduction
Rapeseed is an important oilseed crop cultivated worldwide, with a yield of approximately 75 million tons and 37.58 million hm2 acreage in recent years. Rapeseed accounts for 13% of worldwide oil production and is third behind oil palm (35%) and soybean (28%) (USDA). The widely cultivated double‐low rapeseed (low erucic acid and low glucosinolate content in seeds), also known as canola (Brassica napus), not only provides a healthy and nutritionally balanced edible oil for humans, but is also one of the most important sources of protein in animal feed. Rapeseed is also a soil‐improving crop that can improve the soil nutrient status (Agegnehu et al., 2014; Chen et al., 2018a), accumulate beneficial bacteria (Zhang et al., 2017a), and suppress disease (Fang et al., 2016a) when incorporated into many crop rotation systems, and it is an important source of bioenergy, including biodiesel and industrial oil. Rapeseed breeding and industrial development are inseparable from the innovative utilization of germplasm resources. Here, we summarize the research progress to date, challenges, countermeasures and future perspectives on the evaluation, innovation and utilization of B. napus germplasm resources, as well as the progress of genomic‐based tools such as molecular marker‐assisted selection (MAS), whole genome‐wide predictions and genome editing.
The origin, evolution, and genetic diversity of Brassica napus
Brassica napus (2n = 38, AACC) is an allotetraploid that was formed by interspecific crosses between the diploid ancestors of B. rapa (2n = 20, AA) and B. oleracea (2n = 18, CC) in the last 10 000 years (An et al., 2019; Bancroft et al., 2011; Chalhoub et al., 2014; Kimber and McGregor, 1995; Prakash and Hinata, 1980; Sun et al., 2017; UN, 1935). There are records to indicate that B. napus may have multiple independent origins, including the generally speculated origin in Europe (Chalhoub et al., 2014; Prakash et al., 2011) and the possible origin in Asia (Liu, 2000). Unfortunately, it is challenging to determine its precise origins without wild B. napus. Studies have shown that the A genome of B. napus may have evolved from a European turnip ancestor (B. rapa ssp. rapa) (Lu et al., 2019; Yang et al., 2016) and that the C genome may have been contributed by the wild Brassica species B. montana (2n = 18) (Song et al., 1997) or by the common ancestor of Kohlrabi, cauliflower, broccoli, and Chinese kale (Lu et al., 2019). The European origin has been supported by recent population evolutionary analysis based on genomic and transcriptome sequences (An et al., 2019; Lu et al., 2019).
As a young allotetraploid species with less than 500 years of cultivation history (Allender and King, 2010), B. napus was initially cultivated as vegetables including B. napus subsp. rapifera (swede and rutabaga) and B. napus subsp. pabularia (fodder rape and Siberian kale) for human consumption and animal fodder, and was then gradually domesticated as an oilseed crop (B. napus subsp. oleifera) after the industrial revolution (Allender and King, 2010; An et al., 2019; Applequist and Ohlson, 1972; Damania et al., 1997). The ‘double‐low’ (low glucosinolate and erucic acid content in seeds) breeding performed since the 1950s (Krzymanski, 1978; Stefansson and Kondra, 1975) and local genetic improvements for early maturity and heterosis, such as those made in China (Fu et al., 1989; Liu, 2000; Song et al., 2020; Zou et al., 2019) (Figure 1), have rapidly established rapeseed as a major global oil crop and greatly improved its industrial value and status.
Figure 1.
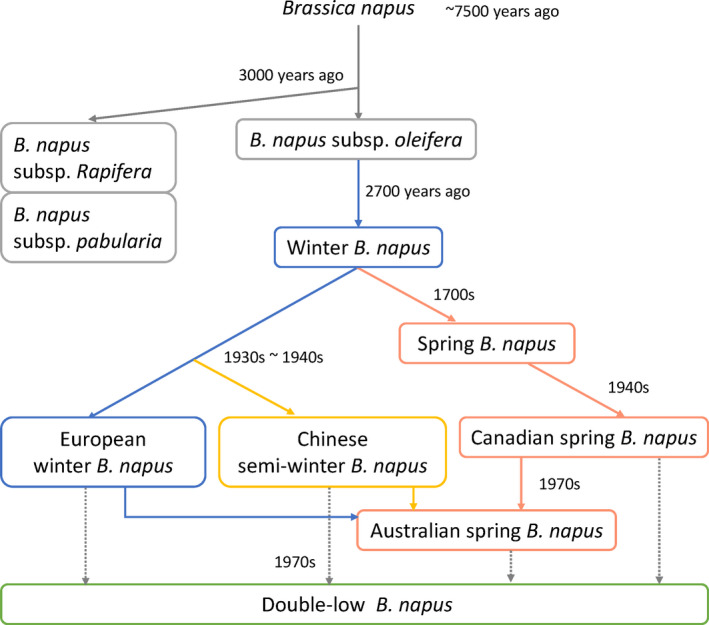
Cultivation and domestication process and genetic groups of Brassica napus. The arrow from European winter B. napus to Australian spring B. napus represents the introductions from Europe to Australia, including both winter type and spring type B. napus. The arrow from Chinese semi‐winter B. napus to Australian spring B. napus also included the introductions from Japan to Australia.
The oilseed types of B. napus can be divided into spring types (no vernalization requirement; grown mainly in North America, Scandinavia and Eastern Europe), semi‐winter types (low to moderate vernalization requirement; grown in China and Europe) and winter types (strict vernalization requirement; grown mostly in Europe) (Liu, 2000). Winter‐type B. napus is the original type that was first cultivated in Europe in the late Middle Ages (Fussell, 1955). Then, spring‐type B. napus was differentiated in approximately 1700 (Bonnema, 2012) and spread to England in the late 18th century, followed by Canada in the 1940s and Australia in the 1970s (Cowling, 2007). The Chinese semi‐winter type of B. napus originated in Poland and made its way to China through Japan (Wu et al., 2018).
There are tens of thousands of diverse germplasm resources of rapeseed and its related species stored in the major international germplasm banks in China, the United States, the United Kingdom, Korea, India, Germany, Canada, Australia, Russia, Japan and the Netherlands (Knee et al., 2011; Li et al., 2020a). However, we should take into account that B. napus is the youngest Brassica species with limited genetic diversity (Cowling, 2007; Fu and Gugel, 2010) due to the limited history of cultivation and domestication (Prakash et al., 2011), traditional breeding methods, and long‐term double‐low breeding in the 1950s and 1980s (Friedt and Snowdon, 2010). Therefore, to break through this breeding block and further expand the genetic diversity of B. napus, innovative germplasm improvement based on hybridization, genomic analysis, (pre) breeding and genetic manipulation is imperative.
Broadening the genetic diversity of the Brassica napus gene pool
Approaches such as hybridization, physical and chemical mutagenesis, and genetic engineering have often been used to introduce novel variation and to broaden the genetic basis of rapeseed; the former, which includes intraspecific and interspecific hybridization, has been the most extensive and achievable way to date, and is still ongoing in various breeding programs worldwide.
Germplasm innovation and utilization based on intraspecific hybridization
Crosses are frequently made between B. napus lines from groups with different subspecies, ecotypes or complementary traits to produce new germplasm or to increase heterosis (Kebede et al., 2010; Qian et al., 2009; Udall et al., 2004). For example, some of the inbred lines derived from rutabaga (B. napus var. napobrassica) × spring‐type B. napus crosses gave higher seed yield and had greater oil content than their spring‐type B. napus parent (Shiranifar et al., 2020). Besides, through hybridization between winter‐type B. napus and spring‐type B. napus, clubroot (Rahman et al., 2011) and blackleg resistance (Light et al., 2011) from winter‐type B. napus have been introduced into spring‐type B. napus, and a determinate inflorescence strain that is beneficial for reducing plant height has been obtained (Li et al., 2018b). In addition, hybrids derived from spring‐type, semi‐winter type, and winter‐type B. napus showed strong heterosis (Table 1), and the spring × winter‐type hybrids have been successfully used in commercial breeding after solving flowering problems (Dang and Chen, 2015).
Table 1.
Exotic introgressions and genomic changes of Brassica napus
| Crops | Hybridization method | Trait introduction | Reference |
|---|---|---|---|
| Intraspecific hybridization | |||
| Spring‐type × semi‐winter type | Pollination | Heterosis | Qian et al. (2007); Yao et al. (2013) |
| Spring‐type × winter‐type | Pollination | Determinate inflorescence | Li et al. (2018b) |
| Clubroot resistance | Fredua‐Agyeman and Rahman (2016); Rahman et al. (2011) | ||
| Black leg resistance | Light et al. (2011) | ||
| Heterosis | Dang and Chen (2015); Quijada et al. (2006) | ||
| Winter‐type × semi‐winter type | Pollination | Heterosis | Qian et al. (2009) |
| rutabaga (B. napus var. napobrassica) × spring‐type | Pollination | Higher seed yield and oil content | Shiranifar et al. (2020) |
| Introgressions of the genetic components of a single species within Brassica | |||
| B. napus × B. rapa | Pollination | Early maturity | Liu (2000) |
| Clubroot resistance | Hirani et al. (2016); Liu et al. (2018); Zhan et al. (2015) | ||
| Heterosis | Zhang et al. (2015) | ||
| B. napus × B. oleracea | Pollination | Expanded genetic diversity | Iftikhar et al. (2018) |
| Sclerotinia resistance | Mei et al. (2020) | ||
| Self‐incompatibility | Ripley and Beversdorf (2003) | ||
| Heterosis | Li et al. (2014) | ||
| B. oleracea ×B. napus | Embryo rescue | Heterosis | Kaminski et al. (2020) |
| B. juncea × B. napus | Pollination | Pod shattering resistance | Prakash and Chopra (1988) |
| Yellow seeds | Liu et al. (2010) | ||
| Multilocular silique | Chen et al. (2018b) | ||
| Blackleg resistance | Dixelius (1999); Rashid et al. (2018); Schelfhout et al. (2006) | ||
| Early maturity | Zaman et al. (2019b) | ||
| B. napus × B. carinata | Repeated pollination | Determinate inflorescence | Tu et al. (2020) |
| Pollination | Blackleg resistance | Fredua‐Agyeman et al. (2014); Navabi et al. (2010) | |
| Pod shattering resistance | Dhaliwal et al. (2017) | ||
| B. fruticulosa × B. napus | Embryo culture | Chen et al. (2011) | |
| Synthetic B. napus | |||
| B. rapa × B. oleracea and B. oleracea × B. rapa | Ovary culture | Song et al. (1993) | |
| Pollination and protoplast fusion | Becker et al. (1995) | ||
| Ovule culture | Lu et al. (2001) | ||
| Ovary culture, embryo culture | Yellow seeds | Wen et al. (2008) | |
| B. rapa × B. oleracea | Ovary culture, embryo culture | Large seeds | Fu et al. (2012b) |
| Embryo rescue | Sclerotinia resistance | Ding et al. (2019) | |
| B. oleracea × B. rapa | Gaeta et al. (2007); Lukens et al. (2006); Xiong et al. (2011) | ||
| Yellow seeds | Rahman (2001) | ||
| Embryo rescue | Heterosis | Girke et al. (2012); Samans et al. (2017) | |
| B. carinata × B. rapa | Pollination | Heterosis, high linoleic acid and high linolenic acid contents, nitrogen efficiency | Chen et al. (2010); Qian et al. (2005); Qian et al. (2003); Wang et al. (2015); Xiao et al. (2010); Zou et al. (2018); Zou et al. (2010) |
| B. juncea × B. carinata | Pollination | Heterosis | Chatterjee et al. (2016) |
| Exotic introgressions from Brassicaceae beyond interspecific hybridizations with B. napus | |||
| B. napus × O. violaceus | Protoplast fusion | Red flowers, higher oleic and linoleic acid contents | Ding et al. (2013); Fu et al. (2018); Gautam et al. (2016); Ge et al. (2009); Hua and Li (2006); Ma and Li (2007) |
| B. napus × radish cabbage | Pollination | Clubroot resistance | Zhan et al. (2017) |
| B. napus × R. sativus | Protoplast fusion | Fertility restored | Sakai et al. (1996) |
| Flower culture method | Vigor | Metz et al. (1995) | |
| Embryo rescue | Clubroot resistance | Diederichsen et al. (2015) | |
| B. napus × I. indigotica | Protoplast fusion | Resistance to influenza virus, cytoplasmic male sterile | Du et al. (2009); Kang et al. (2014); Tu et al. (2020) |
| B. napus × S. alba | Protoplast fusion | Yellow seeds | Wang et al. (2014) |
| Electrofusion | Sclerotinia resistance, yellow seeds | Li et al. (2009) | |
| B. napus × L. fendleri | Pollination and embryo culture | Increased linoleic acid and linolenic acid content | Du et al. (2008) |
| B. napus × S. arvensis | Protoplast fusion | Cytoplasmic male sterile | Cheng et al. (2008) |
| B. napus × C. bursa‐pastoris | Pollination | Early maturity, double‐low quality, Sclerotinia resistance | Chen et al. (2009); Chen et al. (2007); Zhang et al. (2013) |
Germplasm innovation and exotic introgressions based on interspecific hybridization
Interspecific hybridization is important in promoting speciation, evolution, and the production and transfer of rich genomic variation, and has often been used for crop genetic improvement and enrichment of the available germplasm pool (Abbott, 1992; Mallet, 2005; Rieseberg and Carney, 1998). Compared with B. napus, other Brassica species show rich genetic diversity (Warwick, 2011) and obvious genetic differences and subgenomic variation relative to B. napus (Chalhoub et al., 2014; Parkin et al., 2014; Zou et al., 2016a). These species also have many desirable traits and high genetic diversity, as shown in Table S1 and Figure 2 (Quezada‐Martinez et al., 2021; Warwick, 2011). These traits have been or could be further introduced into the B. napus gene pool to provide genetic diversity, and germplasm types suited to changing environments and industry requirements. It is reported that the root exudates of the wild or related species would be a benefit for the improvement of the agricultural output and reduction of environmental impacts (Preece and Penuelas, 2020). We would also pay attention on the advantageous root exudates of these wild or related species and introducing it to B. napus.
Figure 2.

Diverse germplasm from Brassica and its related species. A, B, and C are images of B. rapa, where B shows a winter‐type oilseed B. rapa variety bred by Gansu Agricultural University; D, E, and F are images of B. nigra; G (Li et al., 2014), H (Fu et al., 2012b), I–L are images of B. oleracea; M–Q are images of B. juncea, where M shows a trilocular variety J163‐4 (Xu et al., 2017), N shows a variety with yellow seeds, O shows a variety with purple leaves, and P shows a leafy branching line (Muntha et al., 2019); R–W are images of B. carinata lines, where R and S show plants in seeding stages with green and purple leaves, respectively, T shows a plants at flowering time, U shows the determinate inflorescence of B. carinata, V shows a milky white flower, and W shows yellow seeds (Guo et al., 2012); X and Y shows the seeding stage and flowering stage of B. fruticulosa (Chen et al., 2011). a shows flowering plants of Orychophragmus violaceus; b and c show flower buds and flowering plant of Isatis indigotica (Du et al., 2009); d shows flowering plant of synthetic Brassicoraphanus (RRCC) (Zhan et al., 2017); e and f show the seedling stage and flowering stage of Lesquerella fendleri; and g shows a flowering plant of Raphanus sativus.
Germplasm introgressions from species that share a genome with Brassica napus
The presence of “shared” Brassica genomes with high levels of sequence similarity and organization between multiple species (Chalhoub et al., 2014; Liu et al., 2014; Parkin et al., 2014), has offered a unique opportunity for germplasm improvement (FitzJohn et al., 2007; Katche et al., 2019). The diploid progenitor B. rapa can spontaneously hybridize and backcross with B. napus (Table 1) (Hansen et al., 2001) and has most commonly been used in the breeding of Chinese semi‐winter‐type B. napus (Liu, 2000; Zou et al., 2019) as well as Canadian spring‐type B. napus (Attri and Rahman, 2018). Modern genome sequencing analysis has also clearly identified abundant imported components from B. rapa in Chinese B. napus varieties; the latter shows unique footprints of differentiation that have arisen during the process of artificial selection, breeding and environmental adaptation, resulting in a semi‐winter‐type subpopulation with obvious genomic differences from its European ancestors (Sun et al., 2017; Wu et al., 2018; Zou et al., 2019).
Although the diploid progenitor B. oleracea shows low cross‐compatibility with B. napus (Myers, 2006; Quazi, 1988), with frequent sterility in the resulting interspecific hybrids (Li et al., 2014), several useful traits including self‐incompatibility (Ripley and Beversdorf, 2003), Sclerotinia resistance (Ding et al., 2013; Mei et al., 2020), expanded genetic diversity (Bennett et al., 2008; Iftikhar et al., 2018) and improved heterosis potential (Li et al., 2014) have been successfully transferred into B. napus.
With the deterioration of the global climate, the allotetraploid rapeseed B. juncea (2n = 36, AABB) has become a possible alternative to B. napus in the low moisture regions (Paritosh et al., 2014) of Canada and Australia, and is often used for the improvement of B. napus in terms of blackleg disease resistance (Figure 3E) (Dixelius, 1999; Rashid et al., 2018), early maturity (Zaman, 1989), yellow seededness (Liu et al., 2010), pod shatter resistance (Prakash and Chopra, 1988) and multilocular silique traits that have potential for high yield (Chen et al., 2018b). Chinese hybrid varieties Huangaizao, Xiangzayou 631 and Xiangzayou 518, with early maturity, yellow seeds, and a high oil content, has been bred using this approach (Yang et al., 2021).
Figure 3.
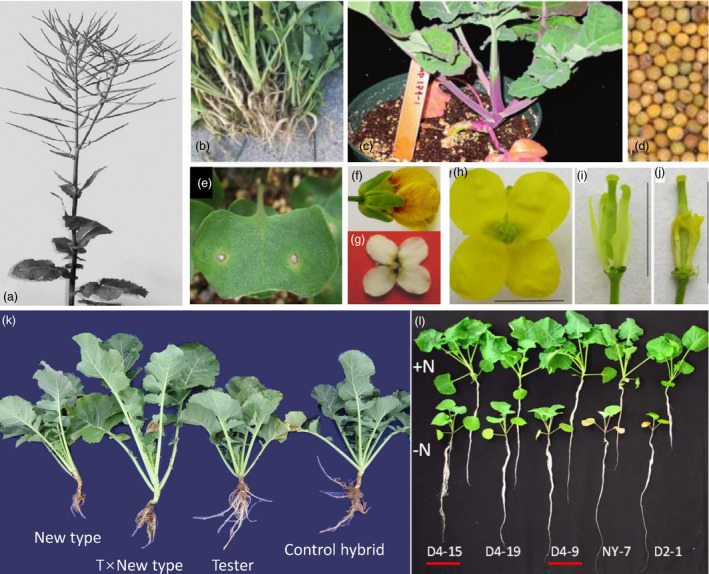
Rich favourable trait variation of Brassica napus lines with introgressions from other species. A shows a plant at the seeding stage with a determinate inflorescence (Tu et al., 2020); B shows clubroot‐resistant seedlings in the field (Zhan et al., 2015); C shows a plant with purple stems (Navabi et al., 2011); D shows yellow seeds (Wen et al., 2008); E shows a plant with blackleg resistance (Rashid et al., 2018); F shows a red flower (Fu et al., 2018); G shows a white flower (Fu et al., 2012b); H, I, and J show the flowers of a male sterile line, where I and J are flowers with sepals and petals removed (Kang et al., 2014); K shows the subgenomic heterosis of new‐type B. napus; and L shows the growth contrast of N‐efficient new‐type B. napus lines D4‐15 and D4‐9 under high‐N and low‐N conditions at the seedling stage (Wang, 2014).
Brassica carinata (Ethiopian mustard, 2n = 34, BBCC) is a very important oilseed crop that has formed slightly earlier than B. napus but later than B. juncea (Song et al., 2021a) and been cultivated in Ethiopia and neighbouring countries for centuries (Alemayehu and Becker, 2002), and it is a promising and competitive feedstock for bio‐based fuel industries (Seepaul et al., 2019). Desirable traits, such as determinate inflorescences, which are suitable for mechanized production (Figure 3A) (Tu et al., 2020), blackleg resistance (Fredua‐Agyeman et al., 2014; Navabi et al., 2010; Navabi et al., 2011), and pod shatter resistance (Dhaliwal et al., 2017), have been introduced from B. carinata to B. napus.
Synthetic Brassica napus as a subpopulation with a novel genomic composition
In addition to introducing certain superior genes or alleles, a creative approach for germplasm innovation has involved the synthesis of B. napus through hybridization between species with AA and CC genome. As an example, synthetic B. napus (ArArCoCo) was created by crossing diploid B. oleracea (CoCo) and B. rapa (ArAr) (superscript represents the first letter of the species name) (Table 1): some lines showed yellow seeds (Figure 3D) (Rahman, 2001; Wen et al., 2008), large seeds and white flowers (Figure 3G) (Fu et al., 2012b), and improved Sclerotinia resistance (Ding et al., 2019). The second kind of synthetic B. napus (ArArCcCc) was created by hybridization between artificially synthesized hexaploidy (ArArBcBcCcCc, created by B. carinata and B. rapa) (Jiang et al., 2007; Tian et al., 2010) and B. napus, followed by more than 10 rounds of self‐pollination, MAS, cytological observation, and trait selection (Hu et al., 2019; Li et al., 2006; Xiao et al., 2010). Lines with high linoleic and linolenic acid content, nitrogen use efficiency (Figure 3L) (Wang et al., 2015), and disease resistance have been selected from this gene pool, and some lines have been used in the breeding of Chinese hybrid varieties Youyan 50 (Zhang et al., 2014) and Huayouza72. In another approach, synthetic B. napus (AjAjCcCc) was created by interspecific hybridization between B. juncea (AjAjBjBj) and B. carinata (BcBcCcCc) followed by chromosome doubling and several generations of self‐pollination and trait selection (Chatterjee et al., 2016). These synthetic B. napus have shown abundant genetic and phenotypic variation, significant genetic differentiation when compared to traditional B. napus, and strong subgenomic heterosis potential as a subpopulation of B. napus (Figure 3K) (Abel et al., 2005; Chen et al., 2010; Girke et al., 2012; Hu et al., 2020; Seyis et al., 2006), and they comprise a rich source of genetic variation for the improvement of established B. napus types.
Exotic introgressions based on intergeneric and intertribal hybridization
Beyond Brassica, rich genetic resources in the Brassicaceae (Warwick, 2011) are valuable for introducing favourable traits to B. napus as well as for exploring new crops. For example, the ornamental plant Orychophragmus violaceus contains a low erucic acid content and a high number of branches, pods and seeds (Figure 2a) (Luo et al., 1994) and can potentially produce special industrial fatty acids in seed oil (Li et al., 2018c). Radish (Raphanus sativus, 2n = 18, RR) has clubroot resistance (Zhan et al., 2017) and cytoplasmic male sterility traits (Ogura, 1968). The medicinal plant Isatis indigotica (2n = 14, Figure 2b,c) shows resistance to diseases caused by bacteria and viruses (Kang et al., 2020). White mustard (Sinapis alba, 2n = 24) has yellow seeds, resistance to biotic and abiotic stresses, pod shatter resistance, and beneficial ingredients for humans (Kumari et al., 2020). Some Brassicaceae species with insect resistance have also been identified, as reviewed in Herve (2018).
The development of tissue culture and protoplast fusion technologies promotes intergeneric and even intertribal hybridization in rapeseed by introducing various favourable traits and creating new germplasm (FitzJohn et al., 2007; Katche et al., 2019) (Table 1). B. napus lines with additional chromosome fragments of O. violaceus that showed red flowers (Figure 3F) and higher oleic and linoleic acid contents were created by intergeneric hybridization between O. violaceus and B. napus (Table 1). Radish cabbage (RRCC) (Figure 2d), raparadish (AARR), allopolyploid alien addition lines and substitution lines with chromosomes from R. sativus (Chen and Wu, 2008; Hagimori et al., 1992; Karpechenko, 1928; Lange et al., 1989; Metz et al., 1995) have been created as a bridge to transfer cytoplasmic male sterility (Bannerrot et al., 1974), fertility‐restorers (Sakai et al., 1996), beet cyst nematode resistance (Peterka et al., 2004), and clubroot resistance (Diederichsen et al., 2015; Zhan et al., 2017) traits from R. sativus to B. napus. A complete set of B. napus monosomic alien addition lines was obtained with each line carrying a chromosome of I. indigotica and showed obvious virus resistance (Du et al., 2009; Kang et al., 2014). Through crossing with I. indigotica, S. alba, and S. arvensis, B. napus lines with cytoplasmic male sterility traits were created (Figure 3H–J) (Cheng et al., 2008; Kang et al., 2014; Wang et al., 2014). In addition, after crossing Capsella bursa‐pastoris (2n = 32) (Figure 2e,f) with double‐high B. napus, new double‐low B. napus lines with Sclerotinia resistance and early maturity were selected (Chen et al., 2007; Zhang et al., 2013).
Reconstructed B. napus genome by exotic germplasm introgressions
With the development of genetics and genomics technologies (van Dijk et al., 2018; Luo et al., 2020; Minoche et al., 2015; Sedlazeck et al., 2018), especially the de novo assembly of the genomes and pangenome of B. napus and its related species (Table S2), an increasing number of studies have shown that introgressions of exotic germplasm not only broaden the genetic base of B. napus but can also lead to major genomic changes. These changes vary with the relationship between species, the degree of exotic introgression, and the selection stress on the hybrid progeny (Table 1). Therefore, in‐depth studies on genomic structural changes caused by ancient events and new introgressions of related species, as well as their influence on traits and heterosis, will provide a theoretical framework for understanding and manipulating the reconstructed rapeseed genome.
Genomic variation in B. napus and related species
Brassica napus is an allotetraploid with a relatively complex genome (1130 Mb) (Chalhoub et al., 2014). Since the first genome assembly of B. napus was released, high‐quality genomes of five cultivars have been revealed based on state‐of‐the‐art sequencing technologies, with improvements in the size of the assembly from 634.19 Mb to 1008 Mb and an average N50 scaffold size from 777.3 Kb to 57.88 Mb, respectively (Table S2). In addition, a 1.8 Gb pangenome of eight varieties containing approximately 150 000 genes was constructed (Song et al., 2020). These genomic analyses showed that the A and C genomes of B. napus are highly homologous, and homeologous exchange events (HEs) are ongoing processes that have greatly promoted the generation of novel, large‐scale structural variation in B. napus (Chalhoub et al., 2014). Significant differentiation, such as asymmetric subgenomic evolution, different subgenomic recombination frequencies, and small‐ to mid‐scale chromosomal structural variations between different ecological types, has also been detected in B. napus (An et al., 2019; Chawla et al., 2021; Kianian and Quiros, 1992; Samans et al., 2017; Song et al., 2020; Zou et al., 2019). Moreover, this genetic variation has been linked to quantitative trait loci for very different traits including chlorophyll content (Qian et al., 2016), flowering time (Vollrath et al., 2021; Wu et al., 2018), germination (Nguyen et al., 2016) and disease resistance (Dolatabadian et al., 2020; Gabur et al., 2020).
For the rest of the Brassica species, high‐quality reference genomes have been released (Table S2). Because of the differences in interspecific hybridization events, speciation and polyploidization, geographic isolation, domestication, and cultivation, comparative genomics analysis based on genomes have revealed abundant subgenomic variation between the A, B, and C genomes present in different species, that is, Ar/Aj/An, Bn/Bj/Bc, and Co/Cn/Cc (Chalhoub et al., 2014; Liu et al., 2014; Perumal et al., 2020; Song et al., 2021a; Wang et al., 2011; Yang et al., 2016). Genome assembly has greatly promoted the functional genomics and breeding of crops, but the existing genome resources are far from comprehensive. In future, focusing on pangenomic variation at the genome structural level using multiple reference genomes to detect presence–absence variation may shed further light on individual genomic differences between lines, genetic groups and ecotypes as a basis for breeding.
Genome changes caused by interspecific hybridization
Both genomic introgressions of related species and resynthesis of Brassica species have led to rich subgenomic variation within Brassica, and new genetic variation has additionally been created by interspecific hybridization‐induced “genomic shock”. Numerous chromosomal structural variations, including inversions and translocations, the disappearance of parental alleles, the generation of new alleles, and the imprinting of transposon activity, have been detected in B. napus lines with B. rapa introgressions (Zou et al., 2011). These genetic variations arose from the introduction of existing subgenomic variation, as well as from new variation induced by interspecific hybridization; in addition, considerable variation associated with heterosis was revealed by QTL analysis (Fu et al., 2012a; Zou et al., 2011).
Synthetic B. napus (ArArCoCo) is known to be meiotically unstable with frequent chromosomal exchanges between the A and C subgenomes, which can result in genomic rearrangements and structural variants (Song et al., 1995; Szadkowski et al., 2010; Xiong et al., 2011). Other changes may involve gene expression and epigenetic modifications (Chen and Pikaard, 1997; Gaeta et al., 2007; Lukens et al., 2006; Ran et al., 2016; Xu et al., 2009), transposable element activation and small RNA changes (Albertin et al., 2006; Fu et al., 2016; Hurgobin et al., 2018; Palacios et al., 2019), changes in tRNA (Wei et al., 2014), and the rapid mutation of repetitive sequences (Gao et al., 2014). Synthetic B. napus (ArArCcCc), whose genome was mostly replaced by B. rapa and B. carinata, showed a stable genome after generations of self‐pollination and recurrent selection but still contained numerous structural variation that was significantly different from that of traditional B. napus (Hu et al., 2019; Zou et al., 2018). The characteristics of these different structural variations and their influence on genome stability, trait improvement, and heterosis are worth further study.
Genomic changes caused by distant hybridization
For the genetic changes caused by intergeneric hybridization, male parental chromosome elimination is very common in offspring (Chaudhary et al., 2013; Houben et al., 2011; Tayeng et al., 2012). In hybridizations between Brassica species and other genera, progeny usually retain the chromosomes from Brassica female parents and eliminate partial or total chromosomes from male parents, as reviewed in Li (2020). In addition, the elimination of C genome chromosomes has also been observed in the hybridization between B. napus and I. indigotica (Tu et al., 2010), Lesquerella fendleri (Du et al., 2008), O. violaceus (Cheng et al., 2002; Hua and Li, 2006), and Crambe abyssinica (Zhu et al., 2016). Compared with intra‐ and interspecific hybridization, genomic changes in intergeneric hybridization are more difficult to capture (Li, 2020), and aneuploid progeny may have increased genomic instability, including chromosome missegregation, mitotic recombination, mutations, and increased DNA damage (Gautam et al., 2016). For example, two new kinds of B. napus (2n = 38, AACC) were extracted from the intergeneric allohexaploid AACCOO (2n = 66) (Gautam et al., 2016) and allotetraploid ACPP (2n = 38) (Chen et al., 2009) after loss of the entire O genome from O. violaceus and the Ca genome from C. bursa‐pastoris. This new B. napus showed the same chromosome composition as its B. napus parents, but genomic changes, including alien introgression, loss and gain of DNA segments, transposon mobilization, and extensive DNA methylation alteration, occurred after polyploidization and aneuploidization (Gautam et al., 2016; Zhang et al., 2013).
Challenges and approaches for further exploring and expanding the rapeseed gene pool
Notably, the gene pool of B. napus was significantly enriched with genetic and genomic diversity through various crosses, which introduced valuable traits, and provided innovative approaches for heterosis breeding. However, we should also note that exploring this diversity for breeding utilization has been hindered by technical limitations. First, the offspring from crosses, especially from distant crosses, often have genome instability, unusual chromosome and allele segregation, and linkage drag of deleterious traits, which requires generations of backcrossing, targeted trait selection (Becker et al., 1995; Seyis et al., 2003), and recurrent selection (Hu et al., 2019) to remove. Second, due to the limited studies of the donor species, most of the improvements in these germplasms are based on inefficient and inaccurate blind selection without the assistance of genomic information. Finally, the acquisition of large‐scale interspecific hybridization is still difficult even with the help of modern techniques, and innovative and high‐throughput methods to create genetic variation are needed. With recent biotechnological developments, there has been rapid progress in dissecting the rapeseed genome diversity, population differentiation, and gene functions, and in manipulating genes and genomes with marker‐assisted selection, transgenics, and genome editing. It is time to further improve the breeding potential of this germplasm, especially germplasm with exotic introgression, with the assistance of genome‐based approaches (Figure 4).
Figure 4.
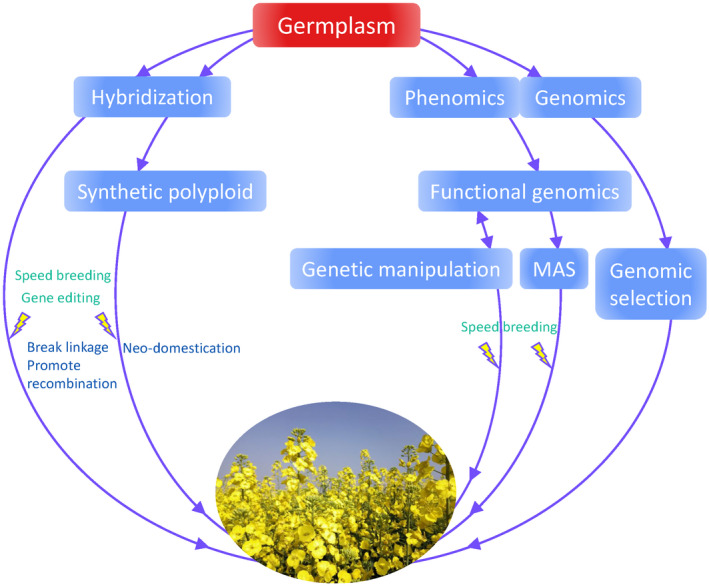
Approaches for further exploring the rapeseed gene pool. MAS represents marker‐assisted selection.
High throughput genotyping and phenotyping
The rapid development of genome sequencing technology has greatly promoted genomic analysis and provides a foundational tool for crop improvement by facilitating rapid selection in modern breeding (Bevan et al., 2017). First, abundant genomic information is available for Brassica and Brassicaceae germplasm, including high‐quality reference genomes (Table S2), organelle genomes (Mohd Saad et al., 2021), pangenomes (Golicz et al., 2016; Hurgobin et al., 2018; Song et al., 2020), gene annotations, transcriptomes, and high‐throughput genotypes for thousands of cultivars and breeding lines that have been identified by SNP chips and sequencing approaches (An et al., 2019; Cheng et al., 2016; Lu et al., 2019; Schmutzer et al., 2015; Wu et al., 2018; Zhang et al., 2017c). These data were stored in public platforms for sharing and remaining including the Brassica Database (http://39.100.233.196), the Brassica napus Genome Browser (https://wwwdev.genoscope.cns.fr/brassicanapus/) (Chalhoub et al., 2014), BnPedigome (http://ibi.zju.edu.cn/bnpedigome/index.php) (Zou et al., 2019), BnaSNPDB (http://rapeed.zju.edu.cn:3838/bnasnpdb) (Yan et al., 2020), and BnPIR (http://cbi.hzau.edu.cn/bnapus/) (Song et al., 2021b). High‐throughput genotyping by whole‐genome sequencing, target segment sequencing and SNP‐chip array is ongoing for Brassica germplasm, and these data resources and genotyping techniques are becoming increasingly inexpensive and convenient for exploring Brassica pangenomic variation, and especially for identifying and introducing favourable genes and alleles into the B. napus gene pool.
Genotyping and phenotyping are important in the analysis of the genetics architecture of traits and the improvement of crops. However, the laborious and time‐consuming traditional manual scoring of phenotypes, which is much slower than high‐throughput genotyping, has become the rate‐limiting step (Fahlgren et al., 2015). In recent years, fast, affordable, accurate and non‐destructive high‐throughput phenotyping platforms based on various imaging techniques, remote‐sensing tools, cloud storage, and machine learning have received widespread attention (Araus and Kefauver, 2018; Shakoor et al., 2017; Yang et al., 2020) and have been used in many crops in the field or lab. For rapeseed, researchers have begun to investigate the use of high‐throughput phenotyping to dynamically screen plant growth architecture within rapeseed intervarietal substitution lines and associated populations, using image‐based traits that reflect shoot growth and predict the final yield (Knoch et al., 2020; Li et al., 2020b). In addition, an unmanned aerial vehicle‐based hyperspectral imaging technique has been used to estimate rapeseed seedpods maturity (Singh et al., 2021), flower number (Wan et al., 2018), plant vegetation fraction and flower fraction (Fang et al., 2016b). However, this technique is being developed and needs to be improved.
Genes/QTLs, MAS, GS and their applications in (pre) breeding
The vast amounts of genotyping and phenotyping information now available have greatly facilitated functional genomics research with thousands of QTLs and candidate genes identified based on high‐density genetic maps, genome‐wide association analysis, and QTL‐seq (Harper et al., 2012; Liu et al., 2020a; Luo et al., 2017; Ogura and Busch, 2015; Raman et al., 2016; Wei et al., 2016). 57 QTL and 787 marker–trait associations for 47 image‐based traits were identified based on high‐throughput phenotyping (Knoch et al., 2020; Li et al., 2020b). In addition, some important genes were cloned, and their functions were verified, including the genes reviewed in (Hu et al., 2017), the abiotic stress‐responsive genes reviewed in (Chikkaputtaiah et al., 2017), and genes related to flowering time, male sterility, plant architecture, seed weight, pod length, oil and fatty acid contents, pod shatter resistance, blackleg resistance, leaf colour, and nutrition absorption (Table 2).
Table 2.
Important trait related genes cloned in Brassica napus in recent years
| Trait | Trait related genes | Reference |
|---|---|---|
| Flowering time | BnFLC.A2 | Chen et al. (2018c) |
| BnFLC.A10 | Hou et al. (2012); Yin et al. (2020) | |
| BnaSDG8 | Jiang et al. (2018) | |
| Male sterility | BnaC9.Tic40 | Xia et al. (2016) |
| MS5 | Xin et al. (2016); Xin et al. (2020) | |
| Plant architecture | BnaTFL1 | Sriboon et al. (2020) |
| BnD14 | Stanic et al. (2021) | |
| BnaRGA | Yang et al. (2017) | |
| BnaIAA7 | Cheng et al. (2021) | |
| Seed weight and pod length | BnaA9.CYP78A9 | Shi et al. (2019) |
| BnaEOD3 | Khan et al. (2020) | |
| BnaUPL3.C03 | Miller et al. (2019) | |
| Oil content | BnSFAR4, BnSFAR5 | Karunarathna et al. (2020) |
| Bnaorf188 | Liu et al. (2019) | |
| Oil and fatty acid | BnaA.FAD2 | Okuzaki et al. (2018) |
| Phytic acid | BnITPK | Sashidhar et al. (2020) |
| Pod shattering resistance | BnJAG | Zaman et al. (2019a) |
| BnaA.ALCa, BnaC.ALC.a | Braatz et al. (2017) | |
| BnIND | Zhai et al. (2019) | |
| Leaf colour | BnaC07.HO1 | Zhu et al. (2017) |
| Boron absorption | BnaA3.NIP5;1 | Hua et al. (2016) |
| BnaA9.WRKY47 | Feng et al. (2020) | |
| BnaC4.BOR1;1c | Zhang et al. (2017b) | |
| Blackleg resistance | LepR3 | Larkan et al. (2013) |
| Rlm9 | Larkan et al. (2020) |
Functional genomics has resulted in many trait‐related markers being developed and has been applied in genetic improvement based on marker‐assisted selection, whereby single‐ or multigenes traits can be tracked using linked markers. This method has been successfully applied in almost all crop breeding programs for gene pyramiding and gene transgression (Dubcovsky, 2004; Hickey et al., 2019; Maharajan et al., 2018). In rapeseed, MAS technology has typically been used for selecting lines with recessive genic male sterility (Huang et al., 2012; Wang et al., 2007), self‐compatibility (Tochigi et al., 2011), clubroot resistance (Zhan et al., 2015), sclerotinia stem rot resistance (Ding et al., 2021), and high oleic acid content (Spasibionek et al., 2020; Zhao et al., 2019). Combined with the speed‐breeding technique that shortens the breeding life cycle through controlled light and temperature (Hickey et al., 2019), the MAS method will accelerate the improvement of rapeseed. However, we should note that many traits are qualitative and complex, and many functional genes, especially genes with small effects, are far from being fully studied, and this complexity could restrict the utilization of MAS. Except for these important agronomic and quality traits, identifying genes controlling cross‐compatibility and recombination perhaps will accelerate the germplasm resource innovation based on hybridization.
In addition to MAS, the genome‐wide selection (GS, or genome‐wide prediction), which predicts the breeding value of the testing population according to its genotype and the prediction model developed based on the genotype and phenotype of the training population, has become a popular tool in plant breeding since the late 2000s (Bernardo, 2016). Genome‐wide selection has also been studied in many crops and applied in maize and soybean (Bernardo, 2016). Prediction using different models, marker sizes, traits, heterosis, and genetic effects, has been investigated with DH populations and hybrid populations, and the results support the high potential of the genomic prediction in rapeseed (Table 3). In addition, the high prediction ability for hybrids whose parents contain exotic introgression of related species was also presented (Table 3, Hu et al., 2020). Genome‐wide selection was influenced by population size, the genetic relationship between the training set and prediction set, marker number, and the environment, which suggests that we can construct and improve the prediction models with suitable training populations, universal and affordable phenotypic and genotypic information, in‐depth and multidimensional omics, meteorology, and advanced algorithms.
Table 3.
Studies of genome‐wide selection in Brassica napus
| Populations | Traits | Marker | Model | Prediction ability | Reference |
|---|---|---|---|---|---|
| 391 winter type DH lines | SY, PH, FT, PC, OC, GLU | 253 SNP | RR‐BLUP | 0.41–0.84 (GLU to PH) | Wurschum et al. (2014) |
| BayesB | 0.34–0.81 (SY to PH) | ||||
| TN DH population | FT | 1248 SNP | RR‐BLUP, RKHS, Bayesian LASSO, BayesA, Bayes B, Random Forest, SVM (linear kernel), SVM (Gaussian kernel) | 0.638, 0.639, 0.639, 0.645, 0.644, 0.611, 0.593, 0.651 | Li et al. (2015) |
| 477 parents and 950 hybrids | SY, OY, OC, GLU, FT, SE, LS | 24 403 SNP | RR‐BLUP | 0.45, 0.75, 0.81, 0.61, 0.56, 0.29, 0.39 | Jan et al. (2016) |
| TN DH population | OC, PC, EAC, LEN, SAC, GLU | 60K DNA array | RR‐BLUP, BayesCπ, EG‐BLUP, GBLUP, MAS | 0.76, 0.66, 0.89, 0.76, 0.81, 0.79 | Zou et al. (2016b) |
| TN DH population and 318 hybrids | SY | 60K DNA array | GBLUP (A) | 0.49 (A), | Liu et al. (2017) |
| GBLUP (A + D) | 0.65 (A + D), | ||||
| GBLUP (A + D + E) | 0.72 (A + D + E) | ||||
| 225 parents and 448 hybrids | SY, TSW, SE, FT, OC, PC, GLU | 60K DNA array | GCA RR‐BLUP | 0.35–0.82 (SY to GLU) | Werner et al. (2018) |
| GCA + SCA RR‐BULP | |||||
| RR‐BULP + de novo GWAS | |||||
| GCA BayesB | |||||
| GCA + SCA BayesB | |||||
| GCA BBR + SCA BayesB | |||||
| 67 parents and 363 hybrids | SY, FT, SN, TSW, GLU, EAC, OC, OLE, LEI, LEN | 43 106 (SNPT) + 5496 (SNPS) | GBLUP (SNPT + A + D) | 0.73, 0.97, 0.77, 0.65, 0.97, 0.99, 0.70, 0.99, 0.91, 0.83 | Hu et al. (2020) |
| GBLUP (SNPS + A + D) | |||||
| GBLUP (SNPT+S + A + D) | |||||
| GBLUP (SNPT + SNPS + A + D) | |||||
| GBLUP (SNPT+S + A + D + E) | |||||
| A DH population with 148 lines | SY, FT, MAT, FD, TSW, OC, PC, GLU, SAT | 368 SNP | GBLUP | 0.14, 0.54, 0.58, 0.53, 0.66, 0.42, 0.55, 0.56, 0.47 | Koscielny et al. (2020) |
| 377 parents and 750 hybrids | SY, OY, SE, PC, FT, OC, GLU, LA, Biovolume, PH, MPH‐LA, MPH‐PH, MPH‐Biovolume | 13 201 SNP + 19 479 transcripts + 154 primary metabolites | GBLUP, RKHS | 0.32, 0.53, 0.27, 0.53, 0.64, 0.70, 0.61, 0.61, 0.59, 0.46, 0.42, 0.37 | Knoch et al. (2021) |
| 218 plants | Sclerotinia stem rot resistance | 24 634 SNP | LMM (A), LMM (A + AA), Bayes A, Bayes B, Bayes C, LASSO, BRR | 0.74, 0.76, 0.56, 0.69, 0.68, 0.63, 0.70 | Derbyshire et al. (2021) |
SY, seed yield; PH, plant height; LA, leaf area; FT, flowering time; FD, flowering duration; MAT, number of days to maturity; PC, protein content; OC, Oil content; GLU, glucosinolate content; OY, oil yield; SE, seedling emergence; LS, lodging resistance; EAC, erucic acid content; SAT, saturated fatty acid content; LEN, linolenic acid content; SAC, stearic acid content; SN, seed number per pod; TSW, thousand seeds weight; OLE, oleic acid content; LEI, linoleic acid content. BLUP, best linear unbiased prediction; RR‑BLUP, ridge regression BLUP; BBR, Bayesian Ridge Regression; GBLUP, genomic best linear unbiased prediction; EG‐BLUP, extended GBLUP; LMM, linear mixed models; RKHS, reproducing kernel Hilbert space regression based on Gaussian kernels; MAS, marker‐assisted selection; MPH, middle parent heterosis; GCA, general combining ability; SCA, specific combining ability; A, additive effects; D, dominance effects; E, epistatic interaction effects; SNPT, SNP markers identified with traditional B. napus reference genome; SNPS, species specific introgression SNP markers.
Despite the vast amounts of genetic, phenotypic and functional genomics information, these methods have been underutilized because they are not well integrated but are scattered across different programs and populations. Recently, researchers reviewed previous studies in rice, constructed a comprehensive map of rice quantitative trait nucleotides with inferred effects, and developed a genome navigation system for use in breeding (Wei et al., 2021). In addition, accumulated data from B. napus or even Brassica overall should also be unified and incorporated to develop an integrated platform with automated, unified, and incorporated genotyping and phenotyping. Importantly, high‐speed universal applications across wide genetic backgrounds will be convenient and time‐saving in taking full advantage of the genetic information in MAS and GS, providing opportunities for genome‐guided breeding.
Genetic manipulation
Despite introducing valuable traits through hybridization and MAS during breeding, genetic manipulation techniques, including transgenic production and gene editing that can precisely introduce or modify desired traits have gradually become key technologies in functional genomics and modern crop breeding. Transgenic technology which can specifically introduce a gene that is nonexistent in the target species has already been applied in B. napus breeding (reviewed by Ton et al., 2020 and Mohd Saad et al., 2021), resulting in varieties and lines with desirable traits such as herbicide resistance (Smyth and Phillips, 2001), hybrid breeding systems (male sterility or restoration) (Mariani et al., 1992), increased levels of the anticancer compound glucoraphanin (Liu et al., 2012), novel components such as docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) (Walsh et al., 2016), insect resistance (reviewed by (Herve, 2018)), and abiotic stress‐responsive tolerance (reviewed by Chikkaputtaiah et al., 2017).
Recently, gene editing has become the most popular and promising technique for functional genomics, and it promises to revolutionize crop improvement by creating new non‐transgenic varieties in a fast, efficient, and technically simple way without the use of transgenes (Schenke and Cai, 2020; Shelake et al., 2019; Songstad et al., 2017; Subedi et al., 2020). Gene editing makes it possible to generate targeted deletions, insertions, gene knockouts, and point mutations, to modulate gene expression by targeting transcription factors or epigenetic machinery to DNA, or to target and modify RNA (Broeders et al., 2020). This method has been successfully used in dozens of species to create desirable traits in major crops (Ezure and Miura, 2018; Malzahn et al., 2017; Manghwar et al., 2020; Manghwar et al., 2019; Metje‐Sprink et al., 2020; Zaman et al., 2019b), and dozens of gene‐edited crops have been approved by a few countries (Editorial, 2021). During the process of studying gene function in B. napus, gene‐edited lines with a higher seed yield, early flowering, improved architecture, orange flowers, pod shatter resistance, multilocular silique, yellow seeds, a higher oil content, an improved fatty acid composition in the seed, environmental stress tolerance, herbicide resistance, and disease resistance have been created (Table 4). The application of these techniques will accelerate functional genomics and result in the development of crops with desired traits that can contribute to increased yield potential under a changing global environment (Lyzenga et al., 2021).
Table 4.
The germplasm resources created by gene editing in Brassica napus
| Gene | Phenotype | Gene function | Reference |
|---|---|---|---|
| BnaEOD3 genes | Shorter siliques, smaller seeds, more seeds per silique, higher seed yield | EOD3 (ENHANCER3 OF DA1) plays a key role in controlling the seed size and silique length in tomato and Arabidopsis thaliana | Khan et al. (2020) |
| BnaSDG8.A and BnaSDG8.C | Early flowering | SDG8 (SET DOMAIN GROUP 8) is a pleiotropic gene involved in several plant biological processes, including flowering time and plant size | Jiang et al. (2018) |
| BnaTFL1 genes | Early flowering, altered plant architecture | TFL1 (TERMINAL FLOWER 1) is a flowering inhibitor and controls the identity of shoot meristem during the plant life span | Sriboon et al. (2020) |
| BnD14 | Improved architecture and seed yield | Strigolactone receptor | Stanic et al. (2021) |
| BnSPL3 genes | Developmental delay | SPL3 (SQUAMOSA PROMOTER BINDING PROTEIN‐LIKE 3) is key floral activator which acts upstream of LEY, FUL and AP1 in Arabidopsis | Li et al. (2018a) |
| BnaRGA genes | Decreased plant height | RGA (REPRESSOR OF GA1‐3) acts as a master repressor in gibberellic signalling | Yang et al. (2017) |
| BnaRGA and BnaIAA7 genes | Decreased plant height | Rapid turnover of IAA proteins is essential for normal auxin response | Cheng et al. (2021) |
| BnaA03.MAX1 and BnaC03.MAX1 | Semi‐dwarf, more branches, more siliques, increased yield | MAX1 (MORE AXILLARY GROWTH 1) encodes a cytochrome P450 monooxygenase (CYP711A1), which is a carlactone oxidase that catalyses the SL biosynthesis | Zheng et al. (2020) |
| BnaA09.ZEP and BnaC09.ZEP | Orange flowers | The nuclear‐encoded plastid enzyme zeaxanthin epoxidase (ZEP) plays a critical role in carotenoid biosynthesis | Liu et al. (2020b) |
| BnaMLPK | Self‐incompatibility | M‐locus protein kinase (MLPK) is thought to interact with the activated SRK, and control self‐incompatibility | Chen et al. (2019) |
| BnS6‐SMI2 | Self‐incompatibility | SCR‐methylation‐inducing region 2 (Smi2): the Smi2 of the dominant S locus generates small interfering RNAs (siRNAs), which suppresses the expression of the recessive S locus SCR by siRNA‐mediated DNA methylation in B. rapa | Dou et al. (2021) |
| MS5 | Genic male sterility | MS5 mediates early meiotic progression | Xin et al. (2020) |
| BnAP2 | Typical sepal carpeloid | A‐functional genes AP2 is required for sepal and petal development | Zhang et al. (2018a) |
| BnA10.LMI1 | Lobed leaves | A LATE MERISTEM IDENTITY1 (LMI1)‐like gene (BnA10.LMI1) encoding an HD‐Zip I transcription factor is the causal gene underlying the BnLLA10 locus, and BnLLA10, is responsible for the lobed‐leaf shape in rapeseed | Hu et al. (2018) |
| BnJAG genes | Pod shatter resistance | The Arabidopsis JAGGED (JAG) gene is a key factor implicated in the regulatory web of dehiscence fruit | Zaman et al. (2019a) |
| BnaA.ALC.a and BnaC.ALC.a | Pod shatter resistance | The Arabidopsis myc/bHLH gene ALCATRAZ (ALC) enables cell separation in fruit dehiscence | Braatz et al. (2017) |
| BnIND genes | Pod shatter resistance | IND (INDEHISCENT) is important for the formation of both the lignified and separation layers of the valve margin | Zhai et al. (2019) |
| BnCLV genes | Multilocular silique | The CLAVATA (CLV) pathways act in a feedback loop to regulate many aspects of stem cell function, including cell fate, proliferation, and growth in Arabidopsis | Yang et al. (2018) |
| BnaTT2 genes | Yellow seeds, increased oil content, higher linoleic acid, and linolenic acid | TT (Transparent Testa) genes are involved in the flavonoid biosynthetic pathway. TT2 regulates proanthocyanidin biosynthesis in seeds | Xie et al. (2020) |
| BnTT8 genes | Same as above | TT8 is a central component of the well‐conserved complex that controls flavonoid accumulation in various crops | Zhai et al. (2020) |
| BnSFAR4, BnSFAR5 | Higher oil content | SFAR (SEED FATTY ACID REDUCER) genes have a significant effect on seed oil content | Karunarathna et al. (2020) |
| BnaA.FAD2 genes | Increased oleic acid in seed | FAD2 (FATTY ACID DESATURASE 2) catalyses the desaturation of oleic acid (C18:1) to linoleic acid (C18:2) | Huang et al. (2020); Okuzaki et al. (2018) |
| BnITPK genes | Reduced phytic acid in seeds | Enzyme ITPK (inositol tetrakisphosphate kinase) catalyses the penultimate step for the synthesis of PA in plants | Sashidhar et al. (2020) |
| BnLPAT2, BnLPAT5 genes | Enlarged oil bodies and increased accumulation of starch in mature seeds | Lysophosphatidic acid acyltransferase (LPAT), a key enzyme in the Kennedy pathway, catalyses fatty acid chains into 3‐phosphoglycerate and promotes further production of oil in the form of triacylglycerol | Zhang et al. (2019) |
| BnaRGA genes | Drought tolerance | RGA (REPRESSOR Of GA7‐3) is a nuclear protein that negatively regulates the gibberellin signal transduction pathway; | Wu et al. (2020b) |
| BnALS genes | Herbicide resistance | Acetolactate synthase (ALS), a key enzyme for the biosynthesis of branchedchain amino acids, is the target site of several important herbicides | Cheng et al. (2021); Wu et al. (2020a) |
| BnWRKY11 and BnWRKY70 | Sclerotinia resistance | Many WRKY transcription factors associates with disease resistance in Arabidopsis | Sun et al. (2018) |
| CRT1a | Verticillium longisporum resistance | Loss of function of CRT1a (calreticulin) strongly reduces plant susceptibility to V. longisporum in both A. thaliana and B. napus | Probsting et al. (2020) |
| BnaA9.WRKY47 | Increased adaptation to low boron stress | WRKY‐mediated gene expression is involved in various stress responses, such as pathogen defence, cold resistance, salt tolerance and nutritional stresses, and in developmental and metabolic processes | Feng et al. (2020) |
In addition, gene editing has been used to construct genome‐wide mutant libraries in rice (Lu et al., 2017; Meng et al., 2017), soybean (Bai et al., 2020), and maize (Liu et al., 2020c); these libraries are of great value for functional genomics and crop improvement and could be rapidly developed for other crops (Zhang et al., 2018b). Despite the use of small‐scale variations, gene editing is beginning to be used to promote recombination at specific genomic regions in yeast and tomato, which is promising for generating recombinant individuals and breaking genetic linkage (Lyzenga et al., 2021). Gene editing has also been used for de novo domestication of wild tomato, groundcherry, and allotetraploid rice with several edited domestication traits (Lemmon et al., 2018; Li et al., 2018d; Yu et al., 2021; Zsögön et al., 2018); this method maintains the genetic diversity and valuable traits of the wild plant while increasing yield productivity, and avoids the lengthy crossings and selections of naturally occurring genetic mutations required for traditional domestication (Zhu and Zhu, 2021). Despite the great challenges of decoding the genetic/epigenetic basis of beneficial agronomic traits and integrating functional genomics discoveries with genome editing designs, these attempts highlight the potential of rapidly creating novel crops with a domesticated plant ideotype from wild plants and orphan crops using advanced biotechnologies (Hickey et al., 2019; Xie and Liu, 2021; Zsögön et al., 2017). For Brassica species, through the use of gene editing, we may be able to (i) create lines with desirable traits or mutation libraries; (ii) accelerate the introgression of elite traits by interspecific hybridization and MAS processes with increased recombination rates or targeted recombination; and (iii) facilitate the neo‐domestication of synthetic Brassica polyploids and wild species by rapidly editing genes related to domestication, genome stability, and recombination.
Outlook
Allotetraploid B. napus is the youngest Brassica species (Song et al., 2021a); it has a complex genome that is accompanied by chromosomal structural variation and characteristics that have driven and will continue to drive its evolution. In addition, the large‐scale interspecific hybridization has accumulated unique resources with rich genetic diversity and genomic structural variants (Song et al., 1995; Udall et al., 2005). However, this germplasm was created based on traditional breeding techniques that are often imprecise and long‐term processes (Subedi et al., 2020), and their value in trait improvement and heterosis utilization will be improved and fully exploited with the assistance of omics analysis, genome‐wide selection, speed breeding, and gene editing technology. First, it is very important to comprehensively dissect the complex genomic structures and pangenome diversity across the Brassica species, which could help build a genome‐wide atlas for genome‐based improvement. In the future, further reductions in long‐reads sequencing costs and increases in population sizes that can be used to detect pangenomic variations are needed for this approach. Second, functional genomics and genome‐editing approaches for important genes and pathways should be further advanced with reference to model plant species. Caution needs to be taken when using this approach due to gene subfunctionalization and genome structural variations in this young polyploid species; the functions, structures, copy numbers, and physical positions of the genes could change due to polyploidization, genomic structural variations caused by interspecific and intraspecific crosses, and domestication, adaptation and selection. Third, practical whole genome‐wide predictions along with marker‐assisted selection on target traits should be explored and performed at the population level to improve breeding values with high efficiency and low costs. High‐throughput phenomics and genomics, population genetics, and rapidly developed functional genomics will yield comprehensive and precise (pre) breeding. Last, and very importantly, along with the accumulating genome sequences, genotypes, phenotypes, populations, germplasms, and other genomic and genetic resources, an automatic and integrated platform could be developed and would significantly improve theoretical and breeding research in rapeseed. This platform will require international efforts regarding sharing, integrating, and exchanging data. These finding would revolutionize rapeseed genomic improvement, contribute to the sustainable development of the rapeseed industry, and provide new insights into the genome evolution and breeding design of B. napus and even other novel Brassica allopolyploids.
Conflict of interest
The authors declare that they have no competing interests.
Author contributions
JZ conceptualized the manuscript; DH, JJ, and JZ drafted the manuscript; DH prepared Figures, Tables and Supplementary Tables; JJ prepared Figure 1, Figure 3, and Table 1; JZ, RJS, ASM, JS, and JM contributed to critical revisions of the manuscript. All authors approved the final version for submission.
Supporting information
Table S1 Favourable traits reported in different Brassica species.
Table S2 Summary of the publicly available genome assemblies of Brassica and related species.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31861133016, 31970564) and the Deutsche Forschungsgemeinschaft (DFG; grants MA6473/2‐1, MA6473/3‐1 and SN14/22‐1). ASM is also partially funded by DFG under Germany’s Excellence Strategy – EXC 2070‐390732324. DDH is also partially funded by the National Natural Science Foundation of China (32000397). We acknowledged Dr. Jochen C. Reif from Department of Breeding Research, Leibniz Institute of Plant Genetics and Crop Plant Research (IPK) for his constructive suggestion for this review. We acknowledged the reviewers and editors for their constructive and helpful revision suggestions.
Hu, D. , Jing, J. , Snowdon, R. J. , Mason, A. S. , Shen, J. , Meng, J. and Zou, J. (2021) Exploring the gene pool of Brassica napus by genomics‐based approaches. Plant Biotechnol. J., 10.1111/pbi.13636
References
- Abbott, R.J. (1992) Plant invasions, interspecific hybridization and the evolution of new plant taxa. Trends Ecol. Evol. 7, 401–405. [DOI] [PubMed] [Google Scholar]
- Abel, S. , Möllers, C. and Becker, H.C. (2005) Development of synthetic Brassica napus lines for the analysis of “fixed heterosis” in allopolyploid plants. Euphytica, 146, 157–163. [Google Scholar]
- Agegnehu, G. , Lakew, B. and Nelson, P.N. (2014) Cropping sequence and nitrogen fertilizer effects on the productivity and quality of malting barley and soil fertility in the Ethiopian highlands. Arch. Agron. Soil Sci. 60, 1261–1275. [Google Scholar]
- Albertin, W. , Balliau, T. , Brabant, P. , Chevre, A.M. , Eber, F. , Malosse, C. and Thiellement, H. (2006) Numerous and rapid nonstochastic modifications of gene products in newly synthesized Brassica napus allotetraploids. Genetics, 173, 1101–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemayehu, N. and Becker, H. (2002) Genotypic diversity and patterns of variation in a germplasm material of Ethiopian mustard (Brassica carinata A. Braun). Genet. Resour. Crop Ev. 49, 573–582. [Google Scholar]
- Allender, C.J. and King, G.J. (2010) Origins of the amphiploid species Brassica napus L. investigated by chloroplast and nuclear molecular markers. BMC Plant Biol. 10, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An, H. , Qi, X.S. , Gaynor, M.L. , Hao, Y. , Gebken, S.C. , Mabry, M.E. , McAlvay, A.C. et al. (2019) Transcriptome and organellar sequencing highlights the complex origin and diversification of allotetraploid Brassica napus . Nat. Commun. 10, 2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applequist, L.A. and Ohlson, R. (1972) Rapeseed. Missouri, USA: Elsevier. [Google Scholar]
- Araus, J.L. and Kefauver, S.C. (2018) Breeding to adapt agriculture to climate change: affordable phenotyping solutions. Curr. Opin. Plant Biol. 45, 237–247. [DOI] [PubMed] [Google Scholar]
- Attri, R. and Rahman, H. (2018) Introgression of allelic diversity from genetically distinct variants of Brassica rapa into Brassica napus canola and inheritance of the B. rapa alleles. Crop Pasture Sci. 69, 94–106. [Google Scholar]
- Bai, M.Y. , Yuan, J.H. , Kuang, H.Q. , Gong, P.P. , Li, S.N. , Zhang, Z.H. , Liu, B. et al. (2020) Generation of a multiplex mutagenesis population via pooled CRISPR‐Cas9 in soya bean. Plant Biotechnol. J. 18, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancroft, I. , Morgan, C. , Fraser, F. , Higgins, J. , Wells, R. , Clissold, L. , Baker, D. et al. (2011) Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 29, 762–766. [DOI] [PubMed] [Google Scholar]
- Bannerrot, H. , Boulidard, L. , Couderon, Y. and Temple, J. (1974) Transfer of cytoplasmic male sterility from Raphanus sativus to Brassica oleracea . In Proc Eucarpia Meet Cruciferae ( Wills, A. B. & North, C. , eds), pp. 52–54. Dundee:Scottish Hortic Res Inst, Invergavrie. [Google Scholar]
- Becker, H.C. , Engqvist, G.M. and Karisson, B. (1995) Comparison of rapeseed cultivars and resynthesized lines based on allozyme and RFLP markers. Theor. Appl. Genet. 91, 63–67. [DOI] [PubMed] [Google Scholar]
- Bennett, R.A. , Thiagarajah, M.R. , King, J.R. and Rahman, M.H. (2008) Interspecific cross of Brassica oleracea var. alboglabra and B. napus: effects of growth condition and silique age on the efficiency of hybrid production, and inheritance of erucic acid in the self‐pollinated backcross generation. Euphytica, 164, 593–601. [Google Scholar]
- Bernardo, R. (2016) Bandwagons I, too, have known. Theor. Appl. Genet. 129, 2323–2332. [DOI] [PubMed] [Google Scholar]
- Bevan, M.W. , Uauy, C. , Wulff, B.B.H. , Zhou, J. , Krasileva, K. and Clark, M.D. (2017) Genomic innovation for crop improvement. Nature, 543, 346–354. [DOI] [PubMed] [Google Scholar]
- Bonnema, G. (2012) Diversity and taxonomy of brassica oil crops. In Genetics, Genomics and Breeding of Oilseed Brassicas ( Edwards, D. , Batley, J. , Parkin, I.A. and Kole, C. , eds), pp. 47–72. Boca Raton, FL: CRC Press. [Google Scholar]
- Braatz, J. , Harloff, H.J. , Mascher, M. , Stein, N. , Himmelbach, A. and Jung, C. (2017) CRISPR‐Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 174, 935–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeders, M. , Herrero‐Hernandez, P. , Ernst, M.P.T. , van der Ploeg, A.T. and Pijnappel, W. (2020) Sharpening the molecular scissors: advances in gene‐editing technology. iScience, 23, 100789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S.Y. , Parkin, I.A.P. , Tang, H.B. , Wang, X.Y. , Chiquet, J. et al. (2014) Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chatterjee, D. , Banga, S. , Gupta, M. , Bharti, S. , Salisbury, P.A. and Banga, S.S. (2016) Resynthesis of Brassica napus through hybridization between B. juncea and B. carinata . Theor. Appl. Genet. 129, 977–990. [DOI] [PubMed] [Google Scholar]
- Chaudhary, H.K. , Tayeng, T. , Kaila, V. and Rather, S.A. (2013) Use of asynchrony in flowering for easy and economical polyhaploid induction in wheat following Imperata cylindrica‐mediated chromosome elimination approach. Plant Breed. 132, 155–158. [Google Scholar]
- Chawla, H.S. , Lee, H. , Gabur, I. , Vollrath, P. , Tamilselvan‐Nattar‐Amutha, S. , Obermeier, C. , Schiessl, S.V. et al. (2021) Long‐read sequencing reveals widespread intragenic structural variants in a recent allopolyploid crop plant. Plant Biotechnol. J. 19, 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C.P. , Xiao, L. and Du, D.Z. (2018b) Advances in multilocular rapeseed. Chin. J. Oil Crop Sci. 40, 446–451. [In Chinese with English abstract]. [Google Scholar]
- Chen, F. , Yang, Y. , Li, B. , Liu, Z. , Khan, F. , Zhang, T. , Zhou, G. et al. (2019) Functional analysis of M‐Locus protein kinase revealed a novel regulatory mechanism of self‐incompatibility in Brassica napus L. Int. J. Mol. Sci. 20, 3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.F. , Ge, X.H. , Du, X.Z. , Zhao, Z.G. and Li, Z.Y. (2009) Genetic and histological characterization of a novel recessive genic male sterile line of Brassica napus derived from a cross with Capsella bursa‐pastoris . Euphytica, 167, 31–37. [Google Scholar]
- Chen, H.F. , Wang, H. and Li, Z.Y. (2007) Production and genetic analysis of partial hybrids in intertribal crosses between Brassica species (B. rapa, B. napus) and Capsella bursa‐pastoris . Plant Cell Rep. 26, 1791–1800. [DOI] [PubMed] [Google Scholar]
- Chen, H.G. and Wu, J.S. (2008) Characterization of fertile amphidiploid between Raphanus sativus and Brassica alboglabra and the crossability with Brassica species. Genet. Resour. Crop Ev. 55, 143–150. [Google Scholar]
- Chen, J.P. , Ge, X.H. , Yao, X.C. , Feng, Y.H. and Li, Z.Y. (2011) Synthesis and characterization of interspecific trigenomic hybrids and allohexaploids between three cultivated Brassica allotetraploids and wild species Brassica fruticulosa . Afr. J. Biotechnol. 10, 12171–12176. [Google Scholar]
- Chen, L. , Dong, F.M. , Cai, J. , Xin, Q. , Fang, C.C. , Liu, L. , Wan, L.L. et al. (2018c) A 2.833‐kb insertion in BnFLC.A2 and its homeologous exchange with BnFLC.C2 during breeding selection generated early‐flowering rapeseed. Mol. Plant, 11, 222–225. [DOI] [PubMed] [Google Scholar]
- Chen, S. , Liu, S.W. , Zheng, X. , Yin, M. , Chu, G. , Xu, C.M. , Yan, J.X. et al. (2018a) Effect of various crop rotations on rice yield and nitrogen use efficiency in paddy‐upland systems in southeastern China. Crop J. 6, 576–588. [Google Scholar]
- Chen, S. , Zou, J. , Cowling, W.A. and Meng, J. (2010) Allelic diversity in a novel gene pool of canola‐quality Brassica napus enriched with alleles from B. rapa and B. carinata . Crop Pasture Sci. 61, 483–492. [Google Scholar]
- Chen, Z.J. and Pikaard, C.S. (1997) Epigenetic silencing of RNA polymerase I transcription: a role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 11, 2124–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, B.F. , Seguin‐Swartz, G. and Somers, D.J. (2002) Cytogenetic and molecular characterization of intergeneric hybrids between Brassica napus and Orychophragmus violaceus . Genome, 45, 110–115. [DOI] [PubMed] [Google Scholar]
- Cheng, F. , Sun, R. , Hou, X. , Zheng, H. , Zhang, F. , Zhang, Y. , Liu, B. et al. (2016) Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea . Nat. Genet. 48, 1218–1224. [DOI] [PubMed] [Google Scholar]
- Cheng, H. , Hao, M. , Ding, B. , Mei, D. , Wang, W. , Wang, H. , Zhou, R. et al. (2021) Base editing with high efficiency in allotetraploid oilseed rape by A3A‐PBE system. Plant Biotechnol. J. 19, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J.H. , Li, Y.C. , Hu, Q. , Mei, D.S. , Li, Y.D. , Xu, Y.S. and Wang, W.M. (2008) Molecular identification and distinctness of NSa male sterile cytoplasm in Brassica napus . Acta Agron. Sin. 34, 1946–1952. [In Chinese with English abstract]. [Google Scholar]
- Chikkaputtaiah, C. , Debbarma, J. , Baruah, I. , Havlickova, L. , Boruah, H.P.D. and Curn, V. (2017) Molecular genetics and functional genomics of abiotic stress‐responsive genes in oilseed rape (Brassica napus L.): a review of recent advances and future. Plant Biotechnol. Rep. 11, 365–384. [Google Scholar]
- Cowling, W.A. (2007) Genetic diversity in Australian canola and implications for crop breeding for changing future environments. Field Crop Res. 104, 103–111. [Google Scholar]
- Damania, A.B. , Valkoun, J. , Willcox, G. and Qualset, C.O. (1997) The Origins of Agriculture and Crop Domestication: The Harlan Symposium. California, CA: ICARDA, IPGRI, FAO and UC/GRCP Publishers. [Google Scholar]
- Dang, B. and Chen, J. (2015) Genetic diversity increases heterosis‐The sprinter Brassica napus project. In 14th International Rapeseed Congress, pp. 43. Sascatoon, Saskatchewan, Canada 43. [Google Scholar]
- Derbyshire, M.C. , Khentry, Y. , Severn‐Ellis, A. , Mwape, V. , Saad, N.S.M. , Newman, T.E. , Taiwo, A. et al. (2021) Modeling first order additive x additive epistasis improves accuracy of genomic prediction for sclerotinia stem rot resistance in canola. Plant Genome, e20088. [DOI] [PubMed] [Google Scholar]
- Dhaliwal, I. , Mason, A.S. , Banga, S. , Bharti, S. , Kaur, B. , Gurung, A.M. , Salisbury, P.A. et al. (2017) Cytogenetic and molecular characterization of B‐genome introgression lines of Brassica napus L. Genes Genomes Genet. 7, 77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichsen, E. , Zou, J. , Meng, J. and Frauen, M. (2015) Clubroot control in oilseed rape using host resistance – potential and challenges. In 14th International Rapeseed Congress, pp. 331. Saskatoon, Saskatchewan, Canada 331. [Google Scholar]
- van Dijk, E.L. , Jaszczyszyn, Y. , Naquin, D. and Thermes, C. (2018) The third revolution in sequencing technology. Trends Genet. 34, 666–681. [DOI] [PubMed] [Google Scholar]
- Ding, L.N. , Li, T. , Guo, X.J. , Li, M. , Liu, X.Y. , Cao, J. and Tan, X.L. (2021) Sclerotinia stem rot resistance in rapeseed: recent progress and future prospects. J. Agric. Food Chem. 69, 2965–2978. [DOI] [PubMed] [Google Scholar]
- Ding, Y.J. , Mei, J.Q. , Li, Q.F. , Liu, Y. , Wan, H.F. , Wang, L. , Becker, H.C. et al. (2013) Improvement of Sclerotinia sclerotiorum resistance in Brassica napus by using B. oleracea . Genet. Resour. Crop Ev. 60, 1615–1619. [Google Scholar]
- Ding, Y.J. , Mei, J.Q. , Wu, Q.N. , Xiong, Z.Y. , Li, Y.H. , Shao, C.G. , Wang, L. et al. (2019) Synchronous improvement of subgenomes in allopolyploid: a case of Sclerotinia resistance improvement in Brassica napus . Mol. Breed. 39, 10. [Google Scholar]
- Dixelius, C. (1999) Inheritance of the resistance to Leptosphaeria maculans of Brassica nigra and B. juncea in near‐isogenic lines of B. napus . Plant Breed. 118, 151–156. [Google Scholar]
- Dolatabadian, A. , Bayer, P.E. , Tirnaz, S. , Hurgobin, B. , Edwards, D. and Batley, J. (2020) Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 18, 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, S.W. , Zhang, T. , Tu, J.X. , Shen, J.X. , Yi, B. , Wen, J. , Fu, T.D. et al. (2021) Generation of novel self‐incompatible Brassica napus by CRISPR/Cas9. Plant Biotechnol. J, 19, 875–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, X.Z. , Ge, X.H. , Zhao, Z.G. and Li, Z.Y. (2008) Chromosome elimination and fragment introgression and recombination producing intertribal partial hybrids from Brassica napus x Lesquerella fendleri crosses. Plant Cell Rep. 27, 261–271. [DOI] [PubMed] [Google Scholar]
- Du, X.Z. , Ge, X.H. , Yao, X.C. , Zhao, Z.G. and Li, Z.Y. (2009) Production and cytogenetic characterization of intertribal somatic hybrids between Brassica napus and Isatis indigotica and backcross progenies. Plant Cell Rep. 28, 1105–1113. [DOI] [PubMed] [Google Scholar]
- Dubcovsky, J. (2004) Marker‐assisted selection in public breeding programs: the wheat experience. Crop Sci. 44, 1895–1898. [Google Scholar]
- Editorial (2021) Next‐generation crop engineering. Nat. Plants, 7, 241. [DOI] [PubMed] [Google Scholar]
- Ezure, H. and Miura, K. (2018) Genome editing technologies for plant physiology. Plant Physiol. Biochem. 131, 1. [DOI] [PubMed] [Google Scholar]
- Fahlgren, N. , Feldman, M. , Gehan, M.A. , Wilson, M.S. , Shyu, C. , Bryant, D.W. , Hill, S.T. et al. (2015) A versatile phenotyping system and analytics platform reveals diverse temporal responses to water availability in Setaria . Mol. Plant, 8, 1520–1535. [DOI] [PubMed] [Google Scholar]
- Fang, S. , Tang, W. , Peng, Y. , Gong, Y. , Dai, C. , Chai, R. and Liu, K. (2016b) Remote estimation of vegetation fraction and flower fraction in oilseed rape with unmanned aerial vehicle data. Remote Sens. 8, 416. [Google Scholar]
- Fang, Y. , Zhang, L. , Jiao, Y. , Liao, J. , Luo, L. , Ji, S. , Li, J. et al. (2016a) Tobacco rotated with rapeseed for soil‐borne Phytophthora pathogen biocontrol: mediated by rapeseed root exudates. Front. Microbiol. 7, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, Y.N. , Cui, R. , Wang, S.L. , He, M.L. , Hua, Y.P. , Shi, L. , Ye, X.S. et al. (2020) Transcription factor BnaA9.WRKY47 contributes to the adaptation of Brassica napus to low boron stress by up‐regulating the boric acid channel gene BnaA3.NIP5;1 . Plant Biotechnol. J. 18, 1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzJohn, R.G. , Armstrong, T.T. , Newstrom‐Lloyd, L.E. , Wilton, A.D. and Cochrane, M. (2007) Hybridisation within Brassica and allied genera: evaluation of potential for transgene escape. Euphytica, 158, 209–230. [Google Scholar]
- Fredua‐Agyeman, R. , Coriton, O. , Huteau, V. , Parkin, I.A. , Chevre, A.M. and Rahman, H. (2014) Molecular cytogenetic identification of B genome chromosomes linked to blackleg disease resistance in Brassica napus × B. carinata interspecific hybrids. Theor. Appl. Genet. 127, 1305–1318. [DOI] [PubMed] [Google Scholar]
- Fredua‐Agyeman, R. and Rahman, H. (2016) Mapping of the clubroot disease resistance in spring Brassica napus canola introgressed from European winter canola cv. 'Mendel'. Euphytica, 211, 201–213. [Google Scholar]
- Friedt, W. and Snowdon, R. (2010) Oilseed rape. In Oil Crops. Handbook of Plant Breeding 4 ( Vollman, J. and Rajcan, I. , eds), pp. 91–126. Dordrecht, Heidelberg, London, New York: Springer. [Google Scholar]
- Fu, D.H. , Qian, W. , Zou, J. and Meng, J.L. (2012a) Genetic dissection of intersubgenomic heterosis in Brassica napus carrying genomic components of B. rapa . Euphytica, 184, 151–164. [Google Scholar]
- Fu, G. , Zhao, Z.G. and Du, D.Z. (2012b) Large seed resynthesized Brassica napus from hybrid of Brassica rapa and Brassica oleracea vir alboglabr . Chin. J. Oil Crop Sci. 34, 136–141. [In Chinese with English abstract]. [Google Scholar]
- Fu, T.D. , Yang, X.N. and Yang, G.S. (1989) Development and studies on Polima CMS in Brassica napus L. J. Huazhong Agric. Univ. 8, 201–207. [In Chinese]. [Google Scholar]
- Fu, W.Q. , Chen, D.Z. , Pan, Q. , Li, F.F. , Zhao, Z.G. , Ge, X.H. and Li, Z.Y. (2018) Production of red‐flowered oilseed rape via the ectopic expression of Orychophragmus violaceus OvPAP2. Plant Biotechnol. J. 16, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y.B. and Gugel, R.K. (2010) Genetic diversity of Canadian elite summer rape (Brassica napus L.) cultivars from the pre‐ to post‐canola quality era. Can. J. Plant Sci. 90, 23–33. [Google Scholar]
- Fu, Y. , Xiao, M.L. , Yu, H.S. , Mason, A.S. , Yin, J.M. , Li, J.N. , Zhang, D.Q. et al. (2016) Small RNA changes in synthetic Brassica napus . Planta, 244, 607–622. [DOI] [PubMed] [Google Scholar]
- Fussell, G.E. (1955) History of cole. Nature, 9, 48–51. [Google Scholar]
- Gabur, I. , Chawla, H.S. , Lopisso, D.T. , von Tiedemann, A. , Snowdon, R. and Obermeier, C. (2020) Gene presence‐absence variation associates with quantitative Verticillium longisporum disease resistance in Brassica napus . Sci. Rep. 10, 4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta, R.T. , Pires, J.C. , Iniguez‐Luy, F. , Leon, E. and Osborn, T.C. (2007) Genomic changes in resynthesized Brassica napus and their effect on gene expression and phenotype. Plant Cell, 19, 3403–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, C. , Yin, J. , Mason, A.S. , Tang, Z. , Ren, X. , Li, C. , An, Z. et al. (2014) Regularities in simple sequence repeat variations induced by a cross of resynthesized Brassica napus and natural Brassica napus . Plant Omics, 7, 35–46. [Google Scholar]
- Gautam, M. , Dang, Y.W. , Ge, X.H. , Shao, Y.J. and Li, Z.Y. (2016) Genetic and epigenetic changes in oilseed rape (Brassica napus L.) extracted from intergeneric allopolyploid and additions with Orychophragmus . Front. Plant Sci. 7, 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge, X.H. , Wang, J. and Li, Z.Y. (2009) Different genome‐specific chromosome stabilities in synthetic Brassica allohexaploids revealed by wide crosses with Orychophragmus . Ann. Bot. 104, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke, A. , Schierholt, A. and Becker, H.C. (2012) Extending the rapeseed gene pool with resynthesized Brassica napus II: heterosis. Theor. Appl. Genet. 124, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz, A.A. , Bayer, P.E. , Barker, G.C. , Edger, P.P. , Kim, H. , Martinez, P.A. , Chan, C.K.K. et al. (2016) The pangenome of an agronomically important crop plant Brassica oleracea . Nat. Commun. 7, 13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S.M. , Zou, J. , Li, R.Y. , Long, Y. , Chen, S. and Meng, J.L. (2012) A genetic linkage map of Brassica carinata constructed with a doubled haploid population. Theor. Appl. Genet. 125, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Hagimori, M. , Nagaoka, M. , Kato, N. and Yoshikawa, H. (1992) Production and characterization of somatic hybrids between the Japanese radish and cauliflower. Theor. Appl. Genet. 84, 819–824. [DOI] [PubMed] [Google Scholar]
- Hansen, L.B. , Siegismund, H.R. and Jorgensen, R.B. (2001) Introgression between oilseed rape (Brassica napus L.) and its weedy relative B. rapa L. in a natural population. Genet. Resour. Crop Ev. 48, 621–627. [Google Scholar]
- Harper, A.L. , Trick, M. , Higgins, J. , Fraser, F. , Clissold, L. , Wells, R. , Hattori, C. et al. (2012) Associative transcriptomics of traits in the polyploid crop species Brassica napus . Nat. Biotechnol. 30, 798–802. [DOI] [PubMed] [Google Scholar]
- Herve, M.R. (2018) Breeding for insect resistance in oilseed rape: challenges, current knowledge and perspectives. Plant Breed. 137, 27–34. [Google Scholar]
- Hickey, L.T. , Hafeez, A.N. , Robinson, H. , Jackson, S.A. , Leal‐Bertioli, S.C.M. , Tester, M. , Gao, C.X. et al. (2019) Breeding crops to feed 10 billion. Nat. Biotechnol. 37, 744–754. [DOI] [PubMed] [Google Scholar]
- Hirani, A.H. , Gao, F. , Liu, J. , Fu, G.H. , Wu, C.R. , Yuan, Y.X. , Li, W. et al. (2016) Transferring clubroot resistance from Chinese cabbage (Brassica rapa) to canola (B. napus). Can. J. Plant Pathol. 38, 82–90. [Google Scholar]
- Hou, J.N. , Long, Y. , Raman, H. , Zou, X.X. , Wang, J. , Dai, S.T. , Xiao, Q.Q. et al. (2012) A tourist‐like MITE insertion in the upstream region of the BnFLC.A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.). BMC Plant Biol. 12, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, A. , Sanei, M. and Pickering, R. (2011) Barley doubled‐haploid production by uniparental chromosome elimination. Plant Cell Tiss. Org. 104, 321–327. [Google Scholar]
- Hu, D.D. , Zhang, W.S. , Zhang, Y.K. , Chang, S.H. , Chen, L.L. , Chen, Y.Y. , Shi, Y.D. et al. (2019) Reconstituting the genome of a young allopolyploid crop, Brassica napus, with its related species. Plant Biotechnol. J. 17, 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, D.D. , Zhao, Y.S. , Shen, J.X. , He, X.X. , Zhang, Y.K. , Jiang, Y. , Snowdon, R. et al. (2020) Genome‐wide prediction for hybrids between parents with distinguished difference on exotic introgressions in Brassica napus . Crop J. 10.1016/j.cj.2020.11.002 [DOI] [Google Scholar]
- Hu, L.M. , Zhang, H. , Yang, Q.Y. , Meng, Q.W. , Han, S.Q. , Nwafor, C.C. , Khan, M.H.U. et al. (2018) Promoter variations in a homeobox gene, BnA10.LMI1, determine lobed leaves in rapeseed (Brassica napus L.). Theor. Appl. Genet. 131, 2699–2708. [DOI] [PubMed] [Google Scholar]
- Hu, Q. , Hua, W. , Yin, Y. , Zhang, X.K. , Liu, L.J. , Shi, J.Q. , Zhao, Y.G. et al. (2017) Rapeseed research and production in China. Crop J. 5, 127–135. [Google Scholar]
- Hua, Y.W. and Li, Z.Y. (2006) Genomic in situ hybridization analysis of Brassica napus x Orychophragmus violaceus hybrids and production of B. napus aneuploids. Plant Breed. 125, 144–149. [Google Scholar]
- Hua, Y. , Zhang, D. , Zhou, T. , He, M. , Ding, G. , Shi, L. and Xu, F. (2016) Transcriptomics‐assisted quantitative trait locus fine mapping for the rapid identification of a nodulin 26‐like intrinsic protein gene regulating boron efficiency in allotetraploid rapeseed. Plant Cell Environ. 39, 1601–1618. [DOI] [PubMed] [Google Scholar]
- Huang, H.B. , Cui, T.T. , Zhang, L.L. , Yang, Q.Y. , Yang, Y. , Xie, K.B. , Fan, C.C. et al. (2020) Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9‐mediated gene editing in Brassica napus . Theor. Appl. Genet. 133, 2401–2411. [DOI] [PubMed] [Google Scholar]
- Huang, Z. , Xiao, L. , Dun, X.L. , Xia, S.Q. , Yi, B. , Wen, J. , Shen, J.X. et al. (2012) Improvement of the recessive genic male sterile lines with a subgenomic background in Brassica napus by molecular marker‐assisted selection. Mol. Breed. 29, 181–187. [Google Scholar]
- Hurgobin, B. , Golicz, A.A. , Bayer, P.E. , Chan, C.K. , Tirnaz, S. , Dolatabadian, A. , Schiessl, S.V. et al. (2018) Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus . Plant Biotechnol. J. 16, 1265–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iftikhar, R. , Wang, X. and Rahman, H. (2018) Broadening the genetic base of Brassica napus canola by interspecific crosses with different variants of B. oleracea . Euphytica, 214, 133. [Google Scholar]
- Jan, H.U. , Abbadi, A. , Lucke, S. , Nichols, R.A. and Snowdon, R.J. (2016) Genomic prediction of testcross performance in canola (Brassica napus). PLoS One, 11, e0147769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, L. , Li, D.H. , Jin, L. , Ruan, Y. , Shen, W.H. and Liu, C.L. (2018) Histone lysine methyltransferases BnaSDG8.A and BnaSDG8.C are involved in the floral transition in Brassica napus . Plant J. 95, 672–685. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. , Tian, E. , Li, R. , Chen, L. and Meng, J. (2007) Genetic diversity of Brassica carinata with emphasis on the interspecific crossability with B. rapa . Plant Breed. 126, 487–491. [Google Scholar]
- Kaminski, P. , Marasek‐Ciolakowska, A. , Podwyszynska, M. , Starzycki, M. , Starzycka‐Korbas, E. and Nowak, K. (2020) Development and characteristics of interspecific hybrids between Brassica oleracea L. and B. napus L. Agronomy, 10, 1339. [Google Scholar]
- Kang, L. , Du, X.Z. , Zhou, Y.Y. , Zhu, B. , Ge, X.H. and Li, Z.Y. (2014) Development of a complete set of monosomic alien addition lines between Brassica napus and Isatis indigotica (Chinese woad). Plant Cell Rep. 33, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Kang, M.H. , Wu, H.L. , Yang, Q. , Huang, L. , Hu, Q.J. , Ma, T. , Li, Z.Y. et al. (2020) A chromosome‐scale genome assembly of Isatis indigotica, an important medicinal plant used in traditional Chinese medicine. Hortic. Res. 7, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpechenko, G.D. (1928) Polyploid hybrids of Raphanus sativus L. × Brassica oleracea L. Zeitschrift für Induktive Abstammungs‐ und Vererbungslehre, 48, 1–85. [Google Scholar]
- Karunarathna, N.L. , Wang, H.Y. , Harloff, H.J. , Jiang, L.X. and Jung, C. (2020) Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnol. J. 18, 2251–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katche, E. , Quezada‐Martinez, D. , Katche, E.I. , Vasquez‐Teuber, P. and Mason, A.S. (2019) Interspecific hybridization for Brassica crop improvement. Crop Breed. Genet. Genom. 1, e190007. [Google Scholar]
- Kebede, B. , Thiagarajah, M. , Zimmerli, C. and Rahman, M.H. (2010) Improvement of open‐pollinated spring rapeseed (Brassica napus L.) through introgression of genetic diversity from winter rapeseed. Crop Sci. 50, 1236–1243. [Google Scholar]
- Khan, M.H.U. , Hu, L.M. , Zhu, M.S. , Zhai, Y.G. , Khan, S.U. , Ahmar, S. , Amoo, O. et al. (2020) Targeted mutagenesis of EOD3 gene in Brassica napus L. regulates seed production. J. Cell Physiol. 236, 1996–2007. [DOI] [PubMed] [Google Scholar]
- Kianian, S.F. and Quiros, C.F. (1992) Trait inheritance, fertility, and genomic relationships of some n = 9 Brassica species. Genet. Resour. Crop Ev. 39, 165–175. [Google Scholar]
- Kimber, D.S. and McGregor, D.I. (1995) The species and their origin, cultivation and world production. In Brassica Oilseeds: Production and Utilization ( Kimber, D.S. and McGregor, D.I. , eds), pp. 1–9. Wallingford, CT: CABI Publishing. [Google Scholar]
- Knee, E.M. , Rivero, L. , Crist, D. , Grotewold, E. and Scholl, R. (2011) Germplasm and molecular resources. In Genetics and Genomics of the Brassicaceae ( Schmidt, R. and Bancroft, I. , eds), pp. 437–467. New York, NY: Springer. [Google Scholar]
- Knoch, D. , Abbadi, A. , Grandke, F. , Meyer, R.C. , Samans, B. , Werner, C.R. , Snowdon, R.J. et al. (2020) Strong temporal dynamics of QTL action on plant growth progression revealed through high‐throughput phenotyping in canola. Plant Biotechnol. J. 18, 68–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch, D. , Werner, C.R. , Meyer, R.C. , Riewe, D. , Abbadi, A. , Lucke, S. , Snowdon, R.J. et al. (2021) Multi‐omics‐based prediction of hybrid performance in canola. Theor. Appl. Genet. 134, 1147–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koscielny, C.B. , Gardner, S.W. , Technow, F. and Duncan, R.W. (2020) Linkage mapping and whole‐genome predictions in canola (Brassica napus) subjected to differing temperature treatments. Crop Pasture Sci. 71, 229–238. [Google Scholar]
- Krzymanski, J. (1978) Double low winter rape for Poland [Brassica napus]. In International Rapeseed Conference. June 12–16, 1978. Malmoe (Sweden).
- Kumari, P. , Singh, K.P. and Rai, P.K. (2020) Draft genome of multiple resistance donor plant Sinapis alba: an insight into SSRs, annotations and phylogenetics. PLoS One, 15, e0231002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, W. , Toxopeus, H. , Lubberts, J.H. , Dolstra, O. and Harrewijn, J.L. (1989) The development of raparadish (X Brassicoraphanus, 2n = 38), a new crop in agriculture. Euphytica, 40, 1–14. [Google Scholar]
- Larkan, N.J. , Lydiate, D.J. , Parkin, I.A.P. , Nelson, M.N. , Epp, D.J. , Cowling, W.A. , Rimmer, S.R. et al. (2013) The Brassica napus blackleg resistance gene LepR3 encodes a receptor‐like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 197, 595–605. [DOI] [PubMed] [Google Scholar]
- Larkan, N.J. , Ma, L.S. , Haddadi, P. , Buchwaldt, M. , Parkin, I.A.P. , Djavaheri, M. and Borhan, M.H. (2020) The Brassica napus wall‐associated kinase‐like (WAKL) gene Rlm9 provides race‐specific blackleg resistance. Plant J. 104, 892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, Z.H. , Reem, N.T. , Dalrymple, J. , Soyk, S. , Swartwood, K.E. , Rodriguez‐Leal, D. , Van Eck, J. et al. (2018) Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants, 4, 766–770. [DOI] [PubMed] [Google Scholar]
- Li, A.M. , Wei, C.X. , Jiang, J.J. , Zhang, Y.T. , Snowdon, R.J. and Wang, Y.P. (2009) Phenotypic variation in progenies from somatic hybrids between Brassica napus and Sinapis alba . Euphytica, 170, 289–296. [Google Scholar]
- Li, C. , Hao, M.Y. , Wang, W.X. , Wang, H. , Chen, F. , Chu, W. , Zhang, B.H. et al. (2018a) An efficient CRISPR/Cas9 platform for rapidly generating simultaneous mutagenesis of multiple gene homoeologs in allotetraploid oilseed rape. Front. Plant Sci. 9, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Feng, H. , Guo, C. , Yang, S. , Huang, W. , Xiong, X. , Liu, J. et al. (2020b) High‐throughput phenotyping accelerates the dissection of the dynamic genetic architecture of plant growth and yield improvement in rapeseed. Plant Biotechnol. J. 18, 2345–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, K.X. , Yao, Y.M. , Xiao, L. , Zhao, Z.G. , Guo, S.M. , Fu, Z. and Du, D.Z. (2018b) Fine mapping of the Brassica napus Bnsdt1 gene associated with determinate growth habit. Theor. Appl. Genet. 131, 193–208. [DOI] [PubMed] [Google Scholar]
- Li, L.X. , Chen, B.Y. , Yan, G.X. , Gao, G.Z. , Xu, K. , Xie, T. , Zhang, F.G. et al. (2020a) Proposed strategies and current progress of research and utilization of oilseed rape germplasm in China. J. Plant Genet. Resour. 21, 1–19. [In Chinese with English abstract]. [Google Scholar]
- Li, L. , Long, Y. , Zhang, L. , Dalton‐Morgan, J. , Batley, J. , Yu, L. , Meng, J. et al. (2015) Genome wide analysis of flowering time trait in multiple environments via high‐throughput genotyping technique in Brassica napus L. PLoS One, 10, e0119425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M.T. , Chen, X. and Meng, J.L. (2006) Intersubgenomic heterosis in rapeseed production with a partial new‐typed Brassica napus containing subgenome Ar firom B. rapa and Cc from Brassica carinata . Crop Sci. 46, 234–242. [Google Scholar]
- Li, Q.F. , Zhou, Q.H. , Mei, J.Q. , Zhang, Y.J. , Li, J.N. , Li, Z.Y. , Ge, X.H. et al. (2014) Improvement of Brassica napus via interspecific hybridization between B. napus and B. oleracea . Mol. Breed. 34, 1955–1963. [Google Scholar]
- Li, T. , Yang, X. , Yu, Y. , Si, X. , Zhai, X. , Zhang, H. , Dong, W. et al. (2018d) Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 36, 1160–1163. [DOI] [PubMed] [Google Scholar]
- Li, X.J. , Teitgen, A.M. , Shirani, A. , Ling, J. , Busta, L. , Cahoon, R.E. , Zhang, W. et al. (2018c) Discontinuous fatty acid elongation yields hydroxylated seed oil with improved function. Nat. Plants, 4, 711–720. [DOI] [PubMed] [Google Scholar]
- Li, Z.Y. (2020) Introgressive hybridization and germplasm innovation in Brassica Crops. J. Plant Genet. Resour. 21, 20–25. [In Chinese with English version]. [Google Scholar]
- Light, K.A. , Gororo, N.N. and Salisbury, P.A. (2011) Usefulness of winter canola (Brassica napus) race‐specific resistance genes against blackleg (causal agent Leptosphaeria maculans) in southern Australian growing conditions. Crop Pasture Sci. 62, 162–168. [Google Scholar]
- Liu, H.L. (2000) Genetics and Breeding of Rapeseed. Beijing: China Agricultural University Press. [Google Scholar]
- Liu, H.J. , Jian, L. , Xu, J. , Zhang, Q. , Zhang, M. , Jin, M. , Peng, Y. et al. (2020c) High‐throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell, 32, 1397–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Hao, W.J. , Liu, J. , Fan, S.H. , Zhao, W. , Deng, L.B. , Wang, X.F. et al. (2019) A novel chimeric mitochondrial gene confers cytoplasmic effects on seed oil content in polyploid rapeseed (Brassica napus). Mol. Plant, 12, 582–596. [DOI] [PubMed] [Google Scholar]
- Liu, P. , Zhao, Y. , Liu, G. , Wang, M. , Hu, D. , Hu, J. , Meng, J. et al. (2017) Hybrid performance of an immortalized F2 rapeseed population is driven by additive, dominance, and epistatic effects. Front. Plant Sci. 8, 815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Huang, H.B. , Yi, X.Q. , Zhang, Y.Y. , Yang, Q.Y. , Zhang, C.Y. , Fan, C.C. et al. (2020a) Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome‐wide association study. Plant Biotechnol. J. 18, 1472–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.Y. , Liu, Y.M. , Yang, X.H. , Tong, C.B. , Edwards, D. , Parkin, I.A.P. , Zhao, M.X. et al. (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5, 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.P. , Xu, A.X. , Liang, F.H. , Yao, X.Q. , Wang, Y. , Liu, X. , Zhang, Y. et al. (2018) Screening of clubroot‐resistant varieties and transfer of clubroot resistance genes to Brassica napus using distant hybridization. Breed. Sci. 68, 258–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Ye, S. , Yuan, G. , Ma, X. , Heng, S. , Yi, B. , Ma, C. et al. (2020b) Gene silencing of BnaA09.ZEP and BnaC09.ZEP confers orange color in Brassica napus flowers. Plant J. 104, 932–949. [DOI] [PubMed] [Google Scholar]
- Liu, Z.S. , Guan, C.Y. , Chen, S.Y. , Liu, S.Y. and Yan, L. (2010) Transfer of superior traits from Brassica juncea into Brassica napus . Agric. Sci. Technol. 11, 49–52. [Google Scholar]
- Liu, Z. , Hirani, A.H. , McVetty, P.B. , Daayf, F. , Quiros, C.F. and Li, G. (2012) Reducing progoitrin and enriching glucoraphanin in Brassica napus seeds through silencing of the GSL‐ALK gene family. Plant Mol. Biol. 79, 179–189. [DOI] [PubMed] [Google Scholar]
- Lu, C.M. , Zhang, B. , Kakihara, F. and Kato, M. (2001) Introgression of genes into cultivated Brassica napus through resynthesis of B. napus via ovule culture and the accompanying change in fatty acid composition. Plant Breed. 120, 405–410. [Google Scholar]
- Lu, K. , Wei, L.J. , Li, X.L. , Wang, Y.T. , Wu, J. , Liu, M. , Zhang, C. et al. (2019) Whole‐genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 10, 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y.M. , Ye, X. , Guo, R.M. , Huang, J. , Wang, W. , Tang, J.Y. , Tan, L.T. et al. (2017) Genome‐wide targeted mutagenesis in rice using the CRISPR/Cas9 system. Mol. Plant, 10, 1242–1245. [DOI] [PubMed] [Google Scholar]
- Lukens, L.N. , Pires, J.C. , Leon, E. , Vogelzang, R. , Oslach, L. and Osborn, T. (2006) Patterns of sequence loss and cytosine methylation within a population of newly resynthesized Brassica napus allopolyploids. Plant Physiol. 140, 336–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, C. , Fernie, A.R. and Yan, J.B. (2020) Single‐cell genomics and epigenomics: technologies and applications in plants. Trends Plant Sci. 25, 1030–1040. [DOI] [PubMed] [Google Scholar]
- Luo, P. , Lan, Z.Q. and Li, Z.Y. (1994) Orychophragmus violaceus, a potential edible‐oil crop. Plant Breed. 113, 83–85. [Google Scholar]
- Luo, Z.L. , Wang, M. , Long, Y. , Huang, Y.J. , Shi, L. , Zhang, C.Y. , Liu, X. et al. (2017) Incorporating pleiotropic quantitative trait loci in dissection of complex traits: seed yield in rapeseed as an example. Theor. Appl. Genet. 130, 1569–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga, W.J. , Pozniak, C.J. and Kagale, S. (2021) Advanced domestication: harnessing the precision of gene editing in crop breeding. Plant Biotechnol. J. 19, 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, N. and Li, Z.Y. (2007) Development of novel Brassica napus lines with canola quality and higher levels of oleic and linoleic acids derived from intergeneric hybrids between B. napus and Orychophragmus violaceus . Euphytica, 157, 231–238. [Google Scholar]
- Maharajan, T. , Ceasar, S.A. , Krishna, T.P.A. , Ramakrishnan, M. , Duraipandiyan, V. , Abdulla, A.D.N. and Ignacimuthu, S. (2018) Utilization of molecular markers for improving the phosphorus efficiency in crop plants. Plant Breed. 137, 10–26. [Google Scholar]
- Mallet, J. (2005) Hybridization as an invasion of the genome. Trends Ecol. Evol. 20, 229–237. [DOI] [PubMed] [Google Scholar]
- Malzahn, A. , Lowder, L. and Qi, Y.P. (2017) Plant genome editing with TALEN and CRISPR. Cell Biosci. 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar, H. , Li, B. , Ding, X. , Hussain, A. , Lindsey, K. , Zhang, X.L. and Jin, S.X. (2020) CRISPR/Cas systems in genome editing: methodologies and tools for sgRNA design, off‐target evaluation, and strategies to mitigate off‐target effects. Adv. Sci. 7, 1902312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghwar, H. , Lindsey, K. , Zhang, X.L. and Jin, S.X. (2019) CRISPR/Cas system: recent advances and future prospects for genome editing. Trends Plant Sci. 24, 1102–1125. [DOI] [PubMed] [Google Scholar]
- Mariani, C. , Gossele, V. , Debeuckeleer, M. , Deblock, M. , Goldberg, R.B. , Degreef, W. and Leemans, J. (1992) A chimeric ribonuclease‐inhibitor gene restores fertility to male sterile plants. Nature, 357, 384–387. [Google Scholar]
- Mei, J.Q. , Shao, C.G. , Yang, R.H. , Feng, Y.X. , Gao, Y. , Ding, Y.J. , Li, J.N. et al. (2020) Introgression and pyramiding of genetic loci from wild Brassica oleracea into B. napus for improving Sclerotinia resistance of rapeseed. Theor. Appl. Genet. 133, 1313–1319. [DOI] [PubMed] [Google Scholar]
- Meng, X.B. , Yu, H. , Zhang, Y.F. , Zhuang, F. , Song, X.G. , Gao, S.S. , Gao, C.X. et al. (2017) Construction of a genome‐wide mutant library in rice using CRISPR/Cas9. Mol. Plant, 10, 1238–1241. [DOI] [PubMed] [Google Scholar]
- Metje‐Sprink, J. , Sprink, T. and Hartung, F. (2020) Genome‐edited plants in the field. Curr. Opin. Biotech. 61, 1–6. [DOI] [PubMed] [Google Scholar]
- Metz, P.L.J. , Nap, J.P. and Stiekema, W.J. (1995) Hybridization of radish (Raphanus sativus L) and oilseed rape (Brassica napus L) through a flower‐culture method. Euphytica, 83, 159–168. [Google Scholar]
- Miller, C. , Wells, R. , McKenzie, N. , Trick, M. , Ball, J. , Fatihi, A. , Dubreucq, B. et al. (2019) Variation in expression of the HECT E3 Ligase UPL3 modulates LEC2 levels, seed size, and crop yields in Brassica napus . Plant Cell, 31, 2370–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoche, A.E. , Dohm, J.C. , Schneider, J. , Holtgrawe, D. , Viehover, P. , Montfort, M. , Sorensen, T.R. et al. (2015) Exploiting single‐molecule transcript sequencing for eukaryotic gene prediction. Genome Biol. 16, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Saad, N.S. , Severn‐Ellis, A.A. , Pradhan, A. , Edwards, D. and Batley, J. (2021) Genomics armed with diversity leads the way in Brassica improvement in a changing global environment. Front. Genet. 12, 600789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muntha, S.T. , Zhang, L.L. , Zhou, Y.F. , Zhao, X. , Hu, Z.Y. , Yang, J.H. and Zhang, M.F. (2019) Phytochrome A signal transduction 1 and CONSTANS‐LIKE 13 coordinately orchestrate shoot branching and flowering in leafy Brassica juncea . Plant Biotechnol. J. 17, 1333–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, J.R. (2006) Outcrossing Potential for Brassica Species and Implications for Vegetable Crucifer Seed Crops of Growing Oilseed Brassicas in the Willamette Valley. Oregon State University Extension Service. [Google Scholar]
- Navabi, Z.K. , Parkin, I.A. , Pires, J.C. , Xiong, Z. , Thiagarajah, M.R. , Good, A.G. and Rahman, M.H. (2010) Introgression of B‐genome chromosomes in a doubled haploid population of Brassica napus × B. carinata . Genome, 53, 619–629. [DOI] [PubMed] [Google Scholar]
- Navabi, Z.K. , Stead, K.E. , Pires, J.C. , Xiong, Z. , Sharpe, A.G. , Parkin, I.A. , Rahman, M.H. et al. (2011) Analysis of B‐genome chromosome introgression in interspecific hybrids of Brassica napus × B. carinata . Genetics, 187, 659–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen, T.C.T. , Obermeier, C. , Friedt, W. , Abrams, S.R. and Snowdon, R.J. (2016) Disruption of germination and seedling development in Brassica napus by mutations causing severe seed hormonal imbalance. Front. Plant Sci. 7, 1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura, H. (1968) Studies on the new male‐sterility in Japanese radish, with special reference to the utilization of this sterility towerds the practical raising of hybrid seeds. Memoirs Fac. Agric. Kagoshima Univ. 6, 39–78. [Google Scholar]
- Ogura, T. and Busch, W. (2015) From phenotypes to causal sequences: using genome wide association studies to dissect the sequence basis for variation of plant development. Curr. Opin. Plant Biol. 23, 98–108. [DOI] [PubMed] [Google Scholar]
- Okuzaki, A. , Ogawa, T. , Koizuka, C. , Kaneko, K. , Inaba, M. , Imamura, J. and Koizuka, N. (2018) CRISPR/Cas9‐mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus . Plant Physiol. Biochem. 131, 63–69. [DOI] [PubMed] [Google Scholar]
- Palacios, P.M. , Jacquemot, M.P. , Tapie, M. , Rousselet, A. , Diop, M. , Remoue, C. , Falque, M. et al. (2019) Assessing the response of small RNA populations to allopolyploidy using resynthesized Brassica napus allotetraploids. Mol. Biol. Evol. 36, 709–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paritosh, K. , Gupta, V. , Yadava, S.K. , Singh, P. , Pradhan, A.K. and Pental, D. (2014) RNA‐seq based SNPs for mapping in Brassica juncea (AABB): synteny analysis between the two constituent genomes A (from B. rapa) and B (from B. nigra) shows highly divergent gene block arrangement and unique block fragmentation patterns. BMC Genom. 15, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin, I.A.P. , Koh, C. , Tang, H.B. , Robinson, S.J. , Kagale, S. , Clarke, W.E. , Town, C.D. et al. (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea . Genome Biol. 15, R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumal, S. , Koh, C.S. , Jin, L. , Buchwaldt, M. , Higgins, E.E. , Zheng, C. , Sankoff, D. et al. (2020) A high‐contiguity Brassica nigra genome localizes active centromeres and defines the ancestral Brassica genome. Nat. Plants, 6, 929–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterka, H. , Budahn, H. , Schrader, O. , Ahne, R. and Schutze, W. (2004) Transfer of resistance against the beet cyst nematode from radish (Raphanus sativus) to rape (Brassica napus) by monosomic chromosome addition. Theor. Appl. Genet. 109, 30–41. [DOI] [PubMed] [Google Scholar]
- Prakash, S. and Chopra, V.L. (1988) Introgression of resistance to shattering in Brassica napus from Brassica juncea through non‐homologous recombination. Plant Breed. 101, 167–168. [Google Scholar]
- Prakash, S. and Hinata, K. (1980) Taxonomy, cytogenetics and origin of crop Brassica, a review. Opera Botanica, 55, 1–57. [Google Scholar]
- Prakash, S. , Wu, X.M. and Bhat, S.R. (2011) History, evolution, and domestication of Brassica crops. Plant Breed. Rev. 35, 19–84. [Google Scholar]
- Preece, C. and Penuelas, J. (2020) A return to the wild: root exudates and food security. Trends Plant Sci. 25, 14–21. [DOI] [PubMed] [Google Scholar]
- Probsting, M. , Schenke, D. , Hossain, R. , Hader, C. , Thurau, T. , Wighardt, L. , Schuster, A. et al. (2020) Loss of function of CRT1a (calreticulin) reduces plant susceptibility to Verticillium longisporum in both Arabidopsis thaliana and oilseed rape (Brassica napus). Plant Biotechnol. J. 18, 2328–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, L.W. , Qian, W. and Snowdon, R.J. (2016) Haplotype hitchhiking promotes trait coselection in Brassica napus . Plant Biotechnol. J. 14, 1578–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, W. , Chen, X. , Fu, D. , Zou, J. and Meng, J. (2005) Intersubgenomic heterosis in seed yield potential observed in a new type of Brassica napus introgressed with partial Brassica rapa genome. Theor. Appl. Genet. 110, 1187–1194. [DOI] [PubMed] [Google Scholar]
- Qian, W. , Li, Q. , Noack, J. , Sass, O. , Meng, J. , Frauen, M. and Jung, C. (2009) Heterotic patterns in rapeseed (Brassica napus L.): II. Crosses between European winter and Chinese semi‐winter lines. Plant Breed. 128, 466–470. [Google Scholar]
- Qian, W. , Liu, R. and Meng, J. (2003) Genetic effects on biomass yield in interspecific hybrids between Brassica napus and B. rapa . Euphytica, 134, 9–15. [Google Scholar]
- Qian, W. , Sass, O. , Meng, J. , Li, M. , Frauen, M. and Jung, C. (2007) Heterotic patterns in rapeseed (Brassica napus L.): I. Crosses between spring and Chinese semi‐winter lines. Theor. Appl. Genet. 115, 27–34. [DOI] [PubMed] [Google Scholar]
- Quazi, M.H. (1988) Interspecific hybrids between Brassica napus L. and B. oleracea L. developed by embryo culture. Theor. Appl. Genet. 75, 309–318. [Google Scholar]
- Quezada‐Martinez, D. , Addo Nyarko, C.P. , Schiessl, S.V. and Mason, A.S. (2021) Using wild relatives and related species to build climate resilience in Brassica crops. Theor. Appl. Genet. https://link.springer.com/content/pdf/10.1007/s00122-021-03793-3.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quijada, P.A. , Udall, J.A. , Lambert, B. and Osborn, T.C. (2006) Quantitative trait analysis of seed yield and other complex traits in hybrid spring rapeseed (Brassica napus L.): 1. Identification of genomic regions from winter germplasm. Theor. Appl. Genet. 113, 549–561. [DOI] [PubMed] [Google Scholar]
- Rahman, H. , Shakir, A. and Hasan, M.J. (2011) Breeding for clubroot resistant spring canola (Brassica napus L.) for the Canadian prairies: can the European winter canola cv. Mendel be used as a source of resistance? Can. J. Plant Sci. 91, 447–458. [Google Scholar]
- Rahman, M.H. (2001) Production of yellow‐seeded Brassica napus through interspecific crosses. Plant Breed. 120, 463–472. [Google Scholar]
- Raman, H. , Raman, R. , Coombes, N. , Song, J. , Prangnell, R. , Bandaranayake, C. , Tahira, R. et al. (2016) Genome‐wide association analyses reveal complex genetic architecture underlying natural variation for flowering time in canola. Plant Cell Environ. 39, 1228–1239. [DOI] [PubMed] [Google Scholar]
- Ran, L.P. , Fang, T.T. , Rong, H. , Jiang, J.J. , Fang, Y.J. and Wang, Y.P. (2016) Analysis of cytosine methylation in early generations of resynthesized Brassica napus . J. Integr. Agric. 15, 1228–1238. [Google Scholar]
- Rashid, M.H. , Hausner, G. and Fernando, W.G.D. (2018) Molecular and phenotypic identification of B‐genome introgression linked to Leptosphaeria maculans resistant gene Rlm6 in Brassica napus × B. juncea interspecific hybrids. Euphytica, 214, 205. [Google Scholar]
- Rieseberg, L.H. and Carney, S.E. (1998) Plant hybridization. New Phytol. 140, 599–624. [DOI] [PubMed] [Google Scholar]
- Ripley, V.L. and Beversdorf, W.D. (2003) Development of self‐incompatible Brassica napus: (I) introgression of S‐alleles from Brassica oleracea through interspecific hybridization. Plant Breed. 122, 1–5. [Google Scholar]
- Sakai, T. , Liu, H.J. , Iwabuchi, M. , KohnoMurase, J. and Imamura, J. (1996) Introduction of a gene from fertility restored radish (Raphanus sativus) into Brassica napus by fusion of X‐irradiated protoplasts from a radish restorer line and iodacetoamide‐treated protoplasts from a cytoplasmic male‐sterile cybrid of B. napus . Theor. Appl. Genet. 93, 373–379. [DOI] [PubMed] [Google Scholar]
- Samans, B. , Chalhoub, B. and Snowdon, R.J. (2017) Surviving a genome collision: genomic signatures of allopolyploidization in the recent crop species Brassica napus . Plant Genome, 10, 3. [DOI] [PubMed] [Google Scholar]
- Sashidhar, N. , Harloff, H.J. , Potgieter, L. and Jung, C.T. (2020) Gene editing of three BnITPK genes in tetraploid oilseed rape leads to significant reduction of phytic acid in seeds. Plant Biotechnol. J. 18, 2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schelfhout, C.J. , Snowdon, R. , Cowling, W.A. and Wroth, J.M. (2006) Tracing B‐genome chromatin in Brassica napus × B. juncea interspecific progeny. Genome, 49, 1490–1497. [DOI] [PubMed] [Google Scholar]
- Schenke, D. and Cai, D.G. (2020) Applications of CRISPR/Cas to improve crop disease resistance: beyond inactivation of susceptibility factors. iScience, 23, 101478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutzer, T. , Samans, B. , Dyrszka, E. , Ulpinnis, C. , Weise, S. , Stengel, D. , Colmsee, C. et al. (2015) Species‐wide genome sequence and nucleotide polymorphisms from the model allopolyploid plant Brassica napus . Sci. Data, 2, 150072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck, F.J. , Lee, H. , Darby, C.A. and Schatz, M.C. (2018) Piercing the dark matter: bioinformatics of long‐range sequencing and mapping. Nat. Rev. Genet. 19, 329–346. [DOI] [PubMed] [Google Scholar]
- Seepaul, R. , Marois, J. , Small, I.M. , George, S. and Wright, D.L. (2019) Carinata dry matter accumulation and nutrient uptake responses to nitrogen fertilization. Agron. J. 111, 2038–2046. [Google Scholar]
- Seyis, F. , Friedt, W. and Luhs, W. (2006) Yield of Brassica napus L. hybrids developed using resynthesized rapeseed material sown at different locations. Field Crop Res. 96, 176–180. [Google Scholar]
- Seyis, F. , Snowdon, R.J. , Luhs, W. and Friedt, W. (2003) Molecular characterization of novel resynthesized rapeseed (Brassica napus) lines and analysis of their genetic diversity in comparison with spring rapeseed cultivars. Plant Breed. 122, 473–478. [Google Scholar]
- Shakoor, N. , Lee, S. and Mockler, T.C. (2017) High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 38, 184–192. [DOI] [PubMed] [Google Scholar]
- Shelake, R.M. , Pramanik, D. and Kim, J.Y. (2019) Evolution of plant mutagenesis tools: a shifting paradigm from random to targeted genome editing. Plant Biotechnol. Rep. 13, 423–445. [Google Scholar]
- Shi, L.L. , Song, J.R. , Guo, C.C. , Wang, B. , Guan, Z.L. , Yang, P. , Chen, X. et al. (2019) A CACTA‐like transposable element in the upstream region of BnaA9.CYP78A9 acts as an enhancer to increase silique length and seed weight in rapeseed. Plant J. 98, 524–539. [DOI] [PubMed] [Google Scholar]
- Shiranifar, B. , Hobson, N. , Kebede, B. , Yang, R.C. and Rahman, H. (2020) Potential of rutabaga (Brassica napus var. napobrassica) gene pool for use in the breeding of B. napus canola. Crop Sci. 60, 157–171. [Google Scholar]
- Singh, K.D. , Duddu, H.S.N. , Vail, S. , Parkin, I. and Shirtliffe, S.J. (2021) UAV‐based hyperspectral imaging technique to estimate canola (Brassica napus L.) seedpods maturity. Can. J. Remote Sens. 47, 33–47. [Google Scholar]
- Smyth, S. and Phillips, P.W.B. (2001) Competitors co‐operating: establishing a supply chain to manage genetically modified canola. Int. Food Agribusiness Manage. Rev. 4, 51–66. [Google Scholar]
- Song, J.M. , Guan, Z.L. , Hu, J.L. , Guo, C.C. , Yang, Z.Q. , Wang, S. , Liu, D.X. et al. (2020) Eight high‐quality genomes reveal pan‐genome architecture and ecotype differentiation of Brassica napus . Nat. Plants, 6, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.M. , Liu, D.X. , Xie, W.Z. , Yang, Z. , Guo, L. , Liu, K. , Yang, Q.Y. et al. (2021b) BnPIR: Brassica napus pan‐genome information resource for 1,689 accessions. Plant Biotechnol. J. 19, 412–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K. , Lu, P. , Tang, K. and Osborn, T.C. (1995) Rapid genome change in synthetic polyploids of Brassica and its implications for polyploid evolution. Proc. Natl Acad. Sci. USA, 92, 7719–7723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, K.M. , Osborn, T.C. and Williams, P.H. (1997) Taxonomy based on nuclear RFLP analysis. In Recent Advance in Oilseed Brassicas ( Kalia, H.R. and Gupta, S.K. , eds), pp. 12–14. New Delhi: Kalyani Publishers. [Google Scholar]
- Song, K. , Tang, K. and Osborn, T.C. (1993) Development of synthetic Brassica amphidiploids by reciprocal hybridization and comparison to natural amphidiploids. Theor. Appl. Genet. 86, 811–821. [DOI] [PubMed] [Google Scholar]
- Song, X. , Wei, Y. , Xiao, D. , Gong, K. , Sun, P. , Ren, Y. , Yuan, J. et al. (2021a) Brassica carinata genome characterization clarifies U's triangle model of evolution and polyploidy in Brassica . Plant Physiol, 186, 388–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad, D.D. , Petolino, J.F. , Voytas, D.F. and Reichert, N.A. (2017) Genome editing of plants. Crit. Rev. Plant Sci. 36, 1–23. [Google Scholar]
- Spasibionek, S. , Mikolajczyk, K. , Cwiek‐Kupczynska, H. , Pietka, T. , Krotka, K. , Matuszczak, M. , Nowakowska, J. et al. (2020) Marker assisted selection of new high oleic and low linolenic winter oilseed rape (Brassica napusL.) inbred lines revealing good agricultural value. PLoS One, 15, e0233959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriboon, S. , Li, H.T. , Guo, C.C. , Senkhamwong, T. , Dai, C. and Liu, K.D. (2020) Knock‐out of TERMINAL FLOWER 1 genes altered flowering time and plant architecture in Brassica napus . BMC Genet. 21, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic, M. , Hickerson, N.M.N. , Arunraj, R. and Samuel, M.A. (2021) Gene‐editing of the strigolacotone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola). Plant Biotechnol. J. 19, 639–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson, B.R. and Kondra, Z.P. (1975) Tower summer rape. Can. J. Plant Sci. 55, 343–344. [Google Scholar]
- Subedi, U. , Ozga, J.A. , Chen, G.Q. , Foroud, N.A. and Singer, S.D. (2020) CRISPR/Cas‐mediated genome editing for the improvement of oilseed crop productivity. Crit. Rev. Plant Sci. 39, 195–221. [Google Scholar]
- Sun, F.M. , Fan, G.Y. , Hu, Q. , Zhou, Y.M. , Guan, M. , Tong, C.B. , Li, J.N. et al. (2017) The high‐quality genome of Brassica napus cultivar 'ZS11' reveals the introgression history in semi‐winter morphotype. Plant J. 92, 452–468. [DOI] [PubMed] [Google Scholar]
- Sun, Q.F. , Lin, L. , Liu, D.X. , Wu, D.W. , Fang, Y.J. , Wu, J. and Wang, Y.P. (2018) CRISPR/Cas9‐mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 19, 2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szadkowski, E. , Eber, F. , Huteau, V. , Lode, M. , Huneau, C. , Belcram, H. , Coriton, O. et al. (2010) The first meiosis of resynthesized Brassica napus, a genome blender. New Phytol. 186, 102–112. [DOI] [PubMed] [Google Scholar]
- Tayeng, T. , Chaudhary, H.K. and Kishore, N. (2012) Enhancing doubled haploid production efficiency in wheat (Triticum aestivum L. em. Thell) by in vivo colchicine manipulations in Imperata cylindrica‐mediated chromosome elimination approach. Plant Breed. 131, 574–578. [Google Scholar]
- Tian, E. , Jiang, Y. , Chen, L. , Zou, J. , Liu, F. and Meng, J. (2010) Synthesis of a Brassica trigenomic allohexaploid (B. carinata × B. rapa) de novo and its stability in subsequent generations. Theor. Appl. Genet. 121, 1431–1440. [DOI] [PubMed] [Google Scholar]
- Tochigi, T. , Udagawa, H. , Li, F. , Kitashiba, H. and Nishio, T. (2011) The self‐compatibility mechanism in Brassica napus L. is applicable to F1 hybrid breeding. Theor. Appl. Genet. 123, 475–482. [DOI] [PubMed] [Google Scholar]
- Ton, L.B. , Neik, T.X. and Batley, J. (2020) The use of genetic and gene technologies in shaping modern rapeseed cultivars (Brassica napus L.). Genes, 11, 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y.Q. , Sun, J. , Ge, X.H. and Li, Z.Y. (2010) Production and genetic analysis of partial hybrids from intertribal sexual crosses between Brassica napus and Isatis indigotica and progenies. Genome, 53, 146–156. [DOI] [PubMed] [Google Scholar]
- Tu, Y.Q. , Tang, J. , Zhang, Y. , Xin, J.J. , Tu, W.F. , Ji, H.L. and Dai, X.L. (2020) Innovation of new determinate inflorescence germplasm via hybridization between Brassica napus and Brassica carinata . J. Plant Genet. Resour. 21, 74–82. [In Chinese with English abstract]. [Google Scholar]
- Udall, J.A. , Quijada, P.A. , Polewicz, H. , Vogelzang, R. and Osborn, T.C. (2004) Phenotypic effects of introgressing Chinese winter and resynthesized Brassica napus L. germplasm into hybrid spring canola. Crop Sci. 44, 1990–1996. [Google Scholar]
- Udall, J.A. , Quijada, P.A. and Osborn, T.C. (2005) Detection of chromosomal rearrangements derived from homeologous recombination in four mapping populations of Brassica napus L. Genetics, 169, 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UN (1935) Genomic analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 7, 389–452. [Google Scholar]
- Vollrath, P. , Chawla, H.S. , Schiessl, S.V. , Gabur, I. , Lee, H.T. , Snowdon, R.J. and Obermeier, C. (2021) A novel deletion in FLOWERING LOCUS T modulates flowering time in winter oilseed rape. Theor. Appl. Genet. 134, 1217–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh, T.A. , Bevan, S.A. , Gachotte, D.J. , Larsen, C.M. , Moskal, W.A. , Merlo, P.A.O. , Sidorenko, L.V. et al. (2016) Canola engineered with a microalgal polyketide synthase‐like system produces oil enriched in docosahexaenoic acid. Nat. Biotechnol. 34, 881–887. [DOI] [PubMed] [Google Scholar]
- Wan, L. , Li, Y. , Cen, H. , Zhu, J. , Yin, W. , Wu, W. , Zhu, H. et al. (2018) Combining UAV‐based vegetation indices and image classification to estimate flower number in oilseed rape. Remote Sens. 10, 1484. [Google Scholar]
- Wang, G.L. (2014) Screening of high nitrogen efficient germplasms and its mechanism in new‐type Brassica napus . Huazhong Agric. Univ. [In Chinese]. [Google Scholar]
- Wang, G.L. , Ding, G.D. , Xu, F.S. , Cai, H.M. , Zou, J. and Ye, X.S. (2015) Genotype differences in photosynthetic characteristics and nitrogen efficiency of new‐type oilseed rape responding to low nitrogen stress. J. Agr. Sci. 153, 1030–1043. [Google Scholar]
- Wang, G.C. , He, J.P. , Hong, D.F. , Xie, Y.Z. , Xu, Z.H. , Liu, P.W. and Yang, G.S. (2007) Development of AFLP and SCAR markers linked to a recessive genic male sterile gene (Bnms3) in rapeseed for marker‐assisted selection. Korean J. Genet. 29, 481–487. [Google Scholar]
- Wang, J. , Gao, Y.N. , Kong, Y.Q. , Jiang, J.J. , Li, A.M. , Zhang, Y.T. and Wang, Y.P. (2014) Abortive process of a novel rapeseed cytoplasmic male sterility line derived from somatic hybrids between Brassica napus and Sinapis alba . J. Integr. Agric. 13, 741–748. [Google Scholar]
- Wang, X.W. , Wang, H.Z. , Wang, J. , Sun, R.F. , Wu, J. , Liu, S.Y. , Bai, Y.Q. et al. (2011) The genome of the mesopolyploid crop species Brassica rapa . Nat. Genet. 43, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Warwick, S.I. (2011) Brassicaceae in agriculture. In Genetics and Genomics of Brassicaceae ( Schmidt, R. and Bancroft, I. , eds), pp. 33–65. New York, NY: Springer. [Google Scholar]
- Wei, L.J. , An, Z.S. , Mason, A.S. , Xiao, M.L. , Guo, Y. , Yin, J.M. , Li, J.N. et al. (2014) Extensive tRNA gene changes in synthetic Brassica napus . J. Mol. Evol. 78, 38–49. [DOI] [PubMed] [Google Scholar]
- Wei, L.J. , Jian, H.J. , Lu, K. , Filardo, F. , Yin, N.W. , Liu, L.Z. , Qu, C.M. et al. (2016) Genome‐wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus . Plant Biotechnol. J. 14, 1368–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, X. , Qiu, J. , Yong, K. , Fan, J. , Zhang, Q. , Hua, H. , Liu, J. et al. (2021) A quantitative genomics map of rice provides genetic insights and guides breeding. Nat. Genet. 53, 243–253. [DOI] [PubMed] [Google Scholar]
- Wen, J. , Tu, J.X. , Li, Z.Y. , Fu, T.D. , Ma, C.Z. and Shen, J.X. (2008) Improving ovary and embryo culture techniques for efficient resynthesis of Brassica napus from reciprocal crosses between yellow‐seeded diploids B. rapa and B. oleracea . Euphytica, 162, 81–89. [Google Scholar]
- Werner, C.R. , Qian, L. , Voss‐Fels, K.P. , Abbadi, A. , Leckband, G. , Frisch, M. and Snowdon, R.J. (2018) Genome‐wide regression models considering general and specific combining ability predict hybrid performance in oilseed rape with similar accuracy regardless of trait architecture. Theor. Appl. Genet. 131, 299–317. [DOI] [PubMed] [Google Scholar]
- Wu, D. , Liang, Z. , Yan, T. , Xu, Y. , Xuan, L. , Tang, J. , Zhou, G. et al. (2018) Whole‐genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol. Plant, 12, 30–43. [DOI] [PubMed] [Google Scholar]
- Wu, J. , Chen, C. , Xian, G.Y. , Liu, D.X. , Lin, L. , Yin, S.L. , Sun, Q.F. et al. (2020a) Engineering herbicide‐resistant oilseed rape by CRISPR/Cas9‐mediated cytosine base‐editing. Plant Biotechnol. J. 18, 1857–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.J. , Yan, G.B. , Duan, Z.Q. , Wang, Z.J. , Kang, C.Y. , Guo, L. , Liu, K.D. et al. (2020b) Roles of the Brassica napus DELLA protein BnaA6.RGA, in modulating drought tolerance by interacting with the ABA signaling component BnaA10.ABF2. Front. Plant Sci. 11, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurschum, T. , Abel, S. and Zhao, Y.S. (2014) Potential of genomic selection in rapeseed (Brassica napus L.) breeding. Plant Breed. 133, 45–51. [Google Scholar]
- Xia, S.Q. , Wang, Z.X. , Zhang, H.Y. , Hu, K.N. , Zhang, Z.Q. , Qin, M.M. , Dun, X.L. et al. (2016) Altered transcription and neofunctionalization of duplicated genes rescue the harmful effects of a chimeric gene in Brassica napus . Plant Cell, 28, 2060–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y. , Chen, L. , Zou, J. , Tian, E. , Xia, W. and Meng, J. (2010) Development of a population for substantial new type Brassica napus diversified at both A/C genomes. Theor. Appl. Genet. 121, 1141–1150. [DOI] [PubMed] [Google Scholar]
- Xie, T. , Chen, X. , Guo, T.L. , Rong, H. , Chen, Z.Y. , Sun, Q.F. , Batley, J. et al. (2020) Targeted knockout of BnTT2 homologues for yellow‐seeded Brassica napus with reduced flavonoids and improved fatty acid composition. J. Agric. Food Chem. 68, 5676–5690. [DOI] [PubMed] [Google Scholar]
- Xie, X.R. and Liu, Y.G. (2021) De novo domestication towards new crops. Natl Sci. Rev. 8, nwab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Q. , Shen, Y. , Li, X. , Lu, W. , Wang, X. , Han, X. , Dong, F.M. et al. (2016) MS5 mediates early meiotic progression and its natural variants may have applications for hybrid production in Brassica napus . Plant Cell, 28, 1263–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, Q. , Wang, X. , Gao, Y. , Xu, D. , Xie, Z. , Dong, F. , Wan, L. et al. (2020) Molecular mechanisms underpinning the multiallelic inheritance of MS5 in Brassica napus . Plant J. 103, 1723–1734. [DOI] [PubMed] [Google Scholar]
- Xiong, Z. , Gaeta, R.T. and Pires, J.C. (2011) Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus . Proc. Natl Acad. Sci. USA, 108, 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, P. , Cao, S.Q. , Hu, K.N. , Wang, X.H. , Huang, W. , Wang, G. , Lv, Z.W. et al. (2017) Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci. Rep. 7, 3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y.H. , Zhong, L. , Wu, X.M. , Fang, X.P. and Wang, J.B. (2009) Rapid alterations of gene expression and cytosine methylation in newly synthesized Brassica napus allopolyploids. Planta, 229, 471–483. [DOI] [PubMed] [Google Scholar]
- Yan, T. , Wang, Q. , Maodzeka, A. , Wu, D. and Jiang, L. (2020) BnaSNPDB: an interactive web portal for the efficient retrieval and analysis of SNPs among 1,007 rapeseed accessions. Comput. Struct. Biotechnol. J. 18, 2766–2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, B. , Liu, Z.S. , Xiao, H.G. , Rao, Y. , Tang, R. , Zhang, C. and Wang, L.L. (2021) Advances on research and utilization of Brassica napus L using distant hybridization strategy. J. Plant Genet. Resour. 22, 593–602. [In Chinese with English abstract]. [Google Scholar]
- Yang, H. , Wu, J.J. , Tang, T. , Liu, K.D. and Dai, C. (2017) CRISPR/Cas9‐mediated genome editing efficiently creates specific mutations at multiple loci using one sgRNA in Brassica napus . Sci. Rep. 7, 7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J.H. , Liu, D.Y. , Wang, X.W. , Ji, C.M. , Cheng, F. , Liu, B.N. , Hu, Z.Y. et al. (2016) The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48, 1225–1232. [DOI] [PubMed] [Google Scholar]
- Yang, W. , Feng, H. , Zhang, X. , Zhang, J. , Doonan, J.H. , Batchelor, W.D. , Xiong, L. et al. (2020) Crop phenomics and high‐throughput phenotyping: past decades, current challenges, and future perspectives. Mol. Plant, 13, 187–214. [DOI] [PubMed] [Google Scholar]
- Yang, Y. , Zhu, K.Y. , Li, H.L. , Han, S.Q. , Meng, Q.W. , Khan, S.U. , Fan, C.C. et al. (2018) Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 16, 1322–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, Y.M. , Liu, H.D. , Xu, L. and Du, D.Z. (2013) Enhancing the heterosis of spring rapeseed varieties (Brassica napus L.) by using semi‐winter rapeseed varieties as parents. Acta Agron. Sin. 39, 118–125. [In Chinese with English abstract]. [Google Scholar]
- Yin, S. , Wan, M. , Guo, C.C. , Wang, B. , Li, H.T. , Li, G. , Tian, Y.Y. et al. (2020) Transposon insertions within alleles of BnaFLC.A10 and BnaFLC.A2 are associated with seasonal crop type in rapeseed. J. Exp. Bot. 71, 4729–4741. [DOI] [PubMed] [Google Scholar]
- Yu, H. , Lin, T. , Meng, X. , Du, H. , Zhang, J. , Liu, G. , Chen, M. et al. (2021) A route to de novo domestication of wild allotetraploid rice. Cell, 184, 1156–1170. [DOI] [PubMed] [Google Scholar]
- Zaman, M.W. (1989) Introgression in Brassica napus for adaptation to the growing conditions in Bangladesh. Theor. Appl. Genet. 77, 721–728. [DOI] [PubMed] [Google Scholar]
- Zaman, Q.U. , Chu, W. , Hao, M.Y. , Shi, Y.Q. , Sun, M.D. , Sang, S.F. , Mei, D.S. et al. (2019a) CRISPR/Cas9‐mediated multiplex genome editing of JAGGED gene in Brassica napus L. Biomolecules, 9, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman, Q.U. , Li, C. , Cheng, H.T. and Hu, Q. (2019b) Genome editing opens a new era of genetic improvement in polyploid crops. Crop J. 7, 141–150. [Google Scholar]
- Zhai, Y.G. , Cai, S.L. , Hu, L.M. , Yang, Y. , Amoo, O. , Fan, C.C. and Zhou, Y.M. (2019) CRISPR/Cas9‐mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 132, 2111–2123. [DOI] [PubMed] [Google Scholar]
- Zhai, Y.G. , Yu, K.D. , Cai, S.L. , Hu, L.M. , Amoo, O. , Xu, L. , Yang, Y. et al. (2020) Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J. 18, 1153–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan, Z.X. , Jiang, Y.F. , Zhu, Z.Y. , Zhang, C.S. , Yang, Q.Y. , Li, Q. , Hou, Z.K. et al. (2015) Development of close linked marker to PbBa8.1 conferring canola resistance to Plasmodiophora brassicae . Chin. J. Oil Crop Sci. 37, 766–771. [In Chinese with English abstract]. [Google Scholar]
- Zhan, Z.X. , Nwafor, C.C. , Hou, Z.K. , Gong, J.F. , Zhu, B. , Jiang, Y.F. , Zhou, Y.M. et al. (2017) Cytological and morphological analysis of hybrids between Brassicoraphanus, and Brassica napus for introgression of clubroot resistant trait into Brassica napus L. PLoS One, 12, e0177470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J.F. , Li, G.R. , Li, H.J. , Pu, X.B. , Jiang, J. , Chai, L. , Zheng, B.C. et al. (2015) Transcriptome analysis of interspecific hybrid between Brassica napus and B. rapa reveals heterosis for oil rape improvement. Int. J. Genomics, 2015, 230985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Nie, L.L. , Cheng, Q.Q. , Yin, Y.T. , Chen, K. , Qi, F.Y. , Zou, D.S. et al. (2019) Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR‐Cas9 system. Biotechnol. Biofuels, 12, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Chen, H.F. , He, M.L. , Zhao, Z.Q. , Cai, H.M. , Ding, G.D. , Shi, L. et al. (2017b) The boron transporter BnaC4.BOR1;1c is critical for inflorescence development and fertility under boron limitation in Brassica napus . Plant Cell Env. 40, 1819–1833. [DOI] [PubMed] [Google Scholar]
- Zhang, R.M. , Chen, D.L. and Li, Q.Y. (2014) Analysis of genetic distance between two parents and selection of parents resource of Youyan50. Seed, 24, 75–78. [In Chinese]. [Google Scholar]
- Zhang, W.S. , Hu, D.D. , Raman, R. , Guo, S.M. , Wei, Z.L. , Shen, X.Q. , Meng, J.L. et al. (2017c) Investigation of the genetic diversity and quantitative trait loci accounting for important agronomic and seed quality traits in Brassica carinata . Front. Plant Sci. 8, 615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.L. , Ge, X.H. , Shao, Y.J. , Sun, G.L. and Li, Z.Y. (2013) Genomic change, retrotransposon mobilization and extensive cytosine methylation alteration in Brassica napus introgressions from two intertribal hybridizations. PLoS One, 8, e56346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.X. , Zhang, R.J. , Gao, J.S. , Wang, X.C. , Fan, F.L. , Ma, X.T. , Yin, H.Q. et al. (2017a) Thirty‐one years of rice‐rice‐green manure rotations shape the rhizosphere microbial community and enrich beneficial bacteria. Soil Biol. Biochem. 104, 208–217. [Google Scholar]
- Zhang, Y. , Huang, S. , Wang, X. , Liu, J. , Guo, X. , Mu, J. , Tian, J. et al. (2018a) Defective APETALA2 genes lead to sepal modification in Brassica crops. Front. Plant Sci. 9, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y. , Massel, K. , Godwin, I.D. and Gao, C. (2018b) Applications and potential of genome editing in crop improvement. Genome Biol. 19, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Q. , Wu, J. , Cai, G.Q. , Yang, Q.Y. , Shahid, M. , Fan, C.C. , Zhang, C.Y. et al. (2019) A novel quantitative trait locus on chromosome A9 controlling oleic acid content in Brassica napus . Plant Biotechnol. J. 17, 2313–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, M. , Zhang, L. , Tang, M. , Liu, J. , Liu, H. , Yang, H. , Fan, S. et al. (2020) Knockout of two BnaMAX1 homologs by CRISPR/Cas9‐targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.). Plant Biotechnol. J. 18, 644–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, B. , Tu, Y.Q. , Zeng, P. , Ge, X.H. and Li, Z.Y. (2016) Extraction of the constituent subgenomes of the natural allopolyploid rapeseed (Brassica napus L.). Genetics, 204, 1015–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, L.X. , Yang, Z.H. , Zeng, X.H. , Gao, J. , Liu, J. , Yi, B. , Ma, C.Z. et al. (2017) Heme oxygenase 1 defects lead to reduced chlorophyll in Brassica napus . Plant Mol. Biol. 93, 579–592. [DOI] [PubMed] [Google Scholar]
- Zhu, X.G. and Zhu, J.K. (2021) Precision genome editing heralds rapid de vono domestication for new crops. Cell, 184, 1133–1134. [DOI] [PubMed] [Google Scholar]
- Zou, J. , Fu, D.H. , Gong, H.H. , Qian, W. , Xia, W. , Pires, J.C. , Li, R.Y. et al. (2011) De novo genetic variation associated with retrotransposon activation, genomic rearrangements and trait variation in a recombinant inbred line population of Brassica napus derived from interspecific hybridization with Brassica rapa . Plant J. 68, 212–224. [DOI] [PubMed] [Google Scholar]
- Zou, J. , Hu, D.D. , Liu, P.F. , Raman, H. , Liu, Z.S. , Liu, X.J. , Parkin, I.A.P. et al. (2016a) Co‐linearity and divergence of the A subgenome of Brassica juncea compared with other Brassica species carrying different A subgenomes. BMC Genomics, 17, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Hu, D. , Mason, A.S. , Shen, X. , Wang, X. , Wang, N. , Grandke, F. et al. (2018) Genetic changes in a novel breeding population of Brassica napus synthesized from hundreds of crosses between B. rapa and B. carinata . Plant Biotechnol. J. 16, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Mao, L. , Qiu, J. , Wang, M. , Jia, L. , Wu, D. , He, Z. et al. (2019) Genome‐wide selection footprints and deleterious variations in young Asian allotetraploid rapeseed. Plant Biotechnol. J. 17, 1998–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Zhao, Y. , Liu, P. , Shi, L. , Wang, X. , Wang, M. , Meng, J. et al. (2016b) Seed quality traits can be predicted with high accuracy in Brassica napus using genomic data. PLoS One, 11, e0166624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, J. , Zhu, J.L. , Huang, S.M. , Tian, E.T. , Xiao, Y. , Fu, D.H. , Tu, J.X. et al. (2010) Broadening the avenue of intersubgenomic heterosis in oilseed Brassica . Theor. Appl. Genet. 120, 283–290. [DOI] [PubMed] [Google Scholar]
- Zsögön, A. , Cermak, T. , Voytas, D. and Peres, L.E. (2017) Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: case study in tomato. Plant Sci. 256, 120–130. [DOI] [PubMed] [Google Scholar]
- Zsögön, A. , Cermak, T. , Naves, E.R. , Notini, M.M. , Edel, K.H. , Weinl, S. , Freschi, L. et al. (2018) De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 36, 1211–1216. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Favourable traits reported in different Brassica species.
Table S2 Summary of the publicly available genome assemblies of Brassica and related species.


