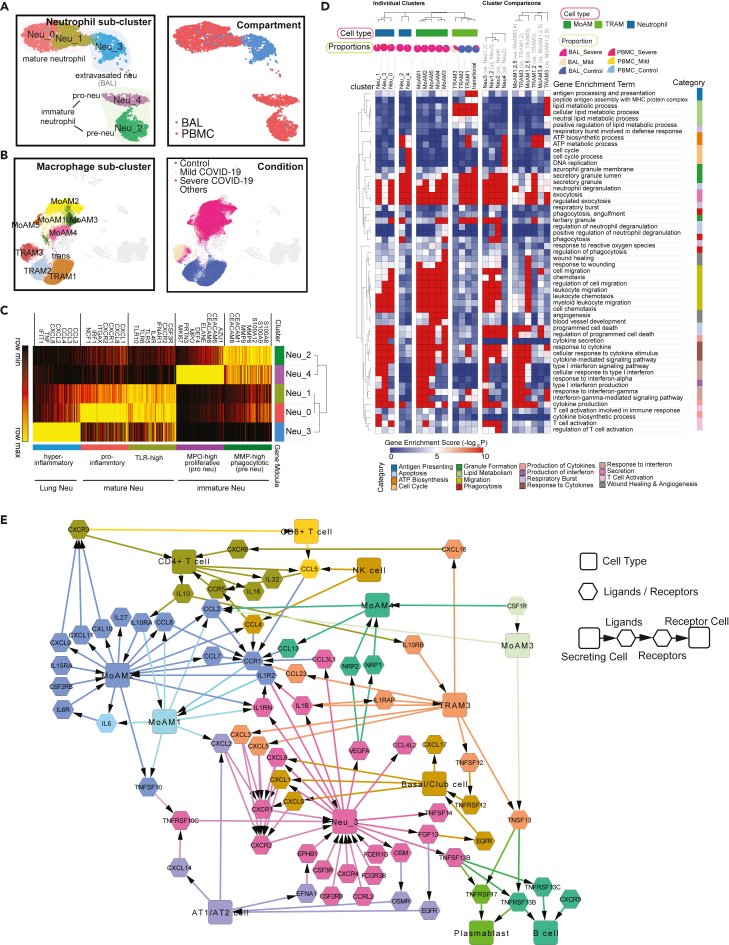
Figure 3.
Functional analysis of compartment-specific immature and subtype-differentiated neutrophils and monocytic macrophages in COVID-19 patients
(A) Five sub-clusters and three cell groups were identified after the integration of neutrophils in peripheral blood mononuclear cells (PBMC) and bronchoalveolar lavage (BAL) (Left). The distribution of compartments is shown on the right.
(B) Sub-clusters (Left) and COVID-19 conditions (Right) of monocyte-derived macrophages and tissue-resident macrophages were identified after integration of BAL datasets.
(C) Heatmap of gene modules from ToppCell with top 200 upregulated genes for each neutrophil sub-cluster. Important neutrophil-associated genes and inferred roles of sub-clusters were shown on two sides.
(D) Heatmap of associations between subclusters of neutrophils and macrophages and myeloid-cell-associated pathways (Gene Ontology). Gene modules with 200 upregulated genes for sub-clusters were used for enrichment in ToppCluster. Additionally, enrichment of top 200 differentially expressed genes (DEGs) for comparisons in Figures S5D and S6B were appended on the right. Gene enrichment scores, defined as −log10(adjusted p value), were calculated as the strength of associations. Pie charts showed the proportions of COVID-19 conditions in each cluster.
(E) Gene interaction network in the BAL of severe patients. Highly expressed ligands and receptors of each cell type in the BAL of severe patients were selected based on Figure S8. Among them, genes with unique and distinct expression patterns in each cell type were chosen, for example, CCL17 for cDCs and CXCR1 for neutrophils. Interaction was inferred using both CellChat database and embedded cell interaction database in ToppCell. The molecular interaction between cell types is represented as a flow of arrows in such a way: secreting cells → ligands → receptors → receptor cells, where rectangles represent various cell types and hexagons represent ligands and receptors. The color for a gene is consistent with the cell type with the highest expression level. See also Figures S5–S11 and Table S3.