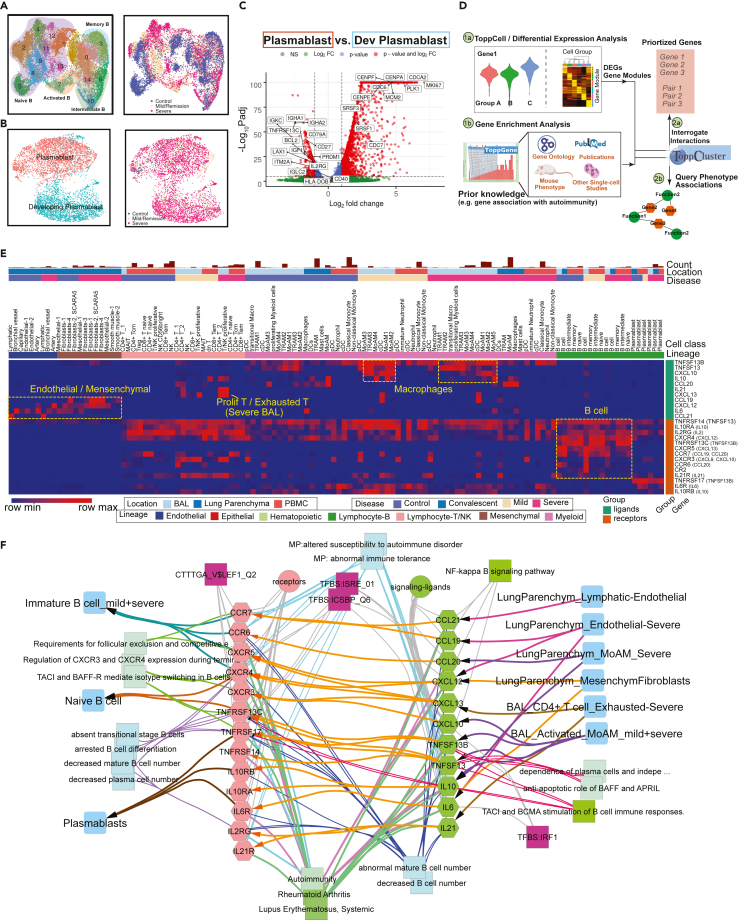
Figure 5.
Implicating a multi-lineage cell network capable of driving extrafollicular B cell maturation and the emergence of humoral autoimmunity in COVID-19 patients
(A) Uniform Manifold Approximation and Projections (UMAPs) of sub-clusters (Left) and COVID-19 conditions (Right) of B cells after integration of peripheral blood mononuclear cells (PBMC) and bronchoalveolar lavage (BAL) datasets.
(B) UMAPs of subtypes (Left) and COVID-19 conditions (Right) of plasmablasts after integration of PBMC and BAL datasets.
(C) Volcano plot depicts differentially expressed genes between plasmablasts and developing plasmablasts. Student t-tests were applied and p values were adjusted by the Benjamini-Hochberg procedure. Thresholds of adjusted p values and log fold changes in the volcano plot are 1.0∗10-6 and 1.0, respectively.
(D) Workflow of discovering and prioritizing candidate genes related to a disease-specific phenotype with limited understanding.
(E) The heatmap shows the normalized expression levels of candidate ligands and receptors for COVID-19 autoimmunity in multiple compartments in healthy donors and COVID-19 patients. Binding ligands of receptor genes were shown in parentheses on the right. Hot spots of expression are highlighted.
(F) Network analysis of autoimmunity-associated gene expression by COVID-19 cell types. Prior knowledge associated gene associations include GWAS, OMIM, mouse knockout phenotype, and additional recent manuscripts were selected from ToppGene enrichment results of differentially expressed ligands and receptors and shown on the network. Orange arrows present the interaction directions from ligands (green) to receptors (pink) on B cells. Annotations for these genes, including single-cell co-expression (blue), mouse phenotype (light blue), transcription factor binding site (purple) and signaling pathways (green) are shown. See also Figures S14, S15 and Table S5.