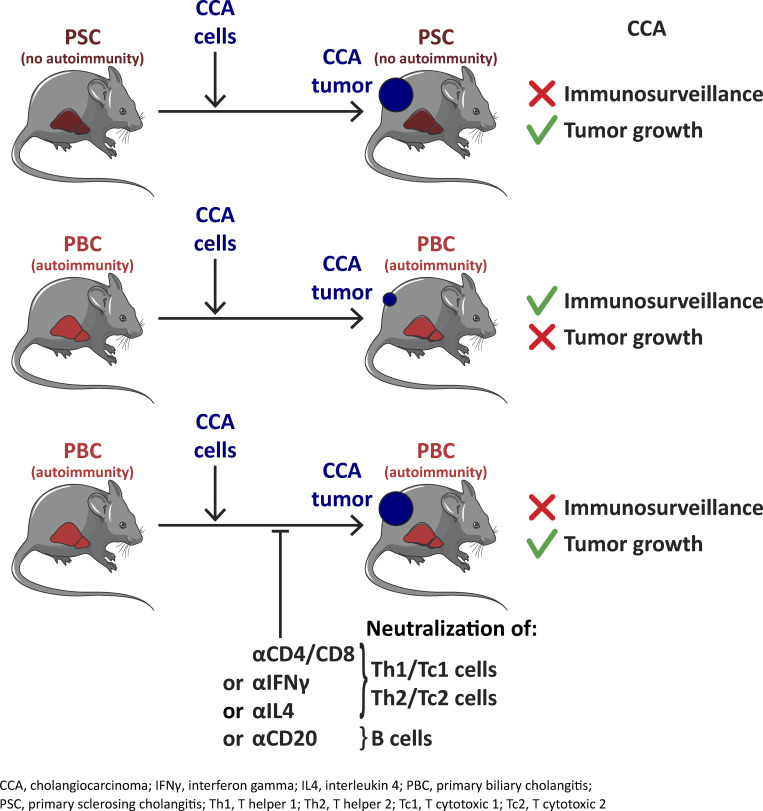
The present study shows in murine models that autoimmunity, which targets normal cholangiocytes upon primary biliary cholangitis, fuels the immunosurveillance of malignant cholangiocytes and protects from the emergence of cholangiocarcinoma.
Abstract
Cholangiocarcinoma (CCA) results from the malignant transformation of cholangiocytes. Primary sclerosing cholangitis (PSC) and primary biliary cholangitis (PBC) are chronic diseases in which cholangiocytes are primarily damaged. Although PSC is an inflammatory condition predisposing to CCA, CCA is almost never found in the autoimmune context of PBC. Here, we hypothesized that PBC might favor CCA immunosurveillance. In preclinical murine models of cholangitis challenged with syngeneic CCA, PBC (but not PSC) reduced the frequency of CCA development and delayed tumor growth kinetics. This PBC-related effect appeared specific to CCA as it was not observed against other cancers, including hepatocellular carcinoma. The protective effect of PBC was relying on type 1 and type 2 T cell responses and, to a lesser extent, on B cells. Single-cell TCR/RNA sequencing revealed the existence of TCR clonotypes shared between the liver and CCA tumor of a PBC host. Altogether, these results evidence a mechanistic overlapping between autoimmunity and cancer immunosurveillance in the biliary tract.
Graphical Abstract
Introduction
Chronic inflammation is a major contributor to cancer promotion and progression (Hanahan and Weinberg, 2011; Lin and Karin, 2007). On one hand, inflammation produces reactive oxygen and nitrogen species that can mutate DNA. On the other hand, the inflammatory environment abounds in multiple factors (i.e., cytokines, chemokines, growth factors, prostaglandins, and proteolytic enzymes) that support malignant cell proliferation, invasion, and metastasis; promote angiogenesis; and interfere with cancer immunosurveillance (Bruno et al., 2014; Taniguchi and Karin, 2018; Todoric et al., 2016). A plethora of clinical and epidemiological observations have validated the link between a prolonged inflammatory state and cancer incidence (Landskron et al., 2014; Multhoff et al., 2012; Todoric et al., 2016). Consolidating evidence came with the discovery that nonsteroidal anti-inflammatory drugs (e.g., aspirin) reduce the risk of developing several malignancies (Rothwell et al., 2018; Todoric et al., 2016). In sharp contrast, there is evidence that specific autoimmune reactions affecting nonessential cell types (e.g., melanocytes in vitiligo and thyrocytes in autoimmune thyroiditis) are coupled to improved immunosurveillance against tumors arising from such cells (i.e., melanoma and thyroid carcinoma, respectively; Failla et al., 2019; Kim et al., 2020; Moon et al., 2018; Myshunina et al., 2018; Nakamura et al., 2017; Paradisi et al., 2014; Teulings et al., 2013; Zitvogel et al., 2021). Hence, inflammatory and autoimmune processes may affect carcinogenesis in opposite directions.
Cholangiocarcinoma (CCA), which emerges from the bile duct–lining epithelial cells (i.e., cholangiocytes), is the second primary liver cancer after hepatocellular carcinoma (HCC). CCA is a highly lethal pathology with a 5-yr survival rate of 5–17% due to usually late diagnosis and the lack of efficient treatment options (Blechacz and Gores, 2008; Squadroni et al., 2017). CCA is largely resistant to chemotherapy. Surgical resection, which remains the sole curative option, is possible only in a minority (∼20%) of patients and is accompanied by frequent (>50%) local relapse (Banales et al., 2020). As is the case for other cancers, CCA and HCC evolve from unresolved inflammatory conditions. On one side, HCC mainly results from chronic viral hepatitis or alcohol-related cirrhosis or as a late-stage complication of nonalcoholic hepatosteatosis (El-Serag, 2012). On the other side, the main predisposing condition to CCA occurring in Western countries resides in primary sclerosing cholangitis (PSC).
PSC is a chronic cholangiopathy (i.e., a cholestatic liver disease in which cholangiocytes are primarily damaged). It is the second-most common chronic cholangiopathy, after primary biliary cholangitis (PBC; Banales et al., 2019). PSC increases not only the risk of CCA but also that of other neoplasms such as HCC, colon cancer, and gallbladder cancer (Razumilava et al., 2011). It is characterized by fibro-inflammatory strictures of the large bile ducts, which form the intra- and/or extrahepatic biliary tract. It occurs predominantly in men (female:male ratio of 1:2), and it is strongly associated with inflammatory bowel disease (Molodecky et al., 2011).
In contrast to PSC, PBC has proclivity toward women (female:male ratio of 10:1) and mainly affects small bile ducts (Myers et al., 2009; Sood et al., 2004). It harbors an autoimmune component responsible for a selective destruction of the bile ducts by inflammatory leukocytes, mainly T lymphocytes and plasma B cells. Serological markers of PBC include high levels of serum IgM and anti-mitochondrial autoantibodies. A loss of tolerance to the E2 subunit of the mitochondrial pyruvate dehydrogenase complex (PDC-E2) leads to cellular and humoral autoimmunity against this autoantigen (Gershwin and Mackay, 2008; Gulamhusein and Hirschfield, 2020; Wang et al., 2013).
Intriguingly, despite a higher prevalence of PBC than PSC (1.9–40.2 versus 0–16.2 per 100,000 inhabitants/year, respectively; Marchioni Beery et al., 2014), PBC has never been associated with CCA development (Parés and Rodés, 2003). Cases of CCA reported in PBC patients are extremely rare (Akisawa et al., 1998; Kulkarni and Beatty, 1977; Welzel et al., 2007) and have been reported mostly in the context of liver cirrhosis (which is a risk factor not only for CCA but also HCC) and without distinguishing between primary and secondary CCA (Welzel et al., 2007). Based on these epidemiological observations, we hypothesized that PBC-associated autoimmunity might be incompatible with CCA pathogenesis due to an improvement of immunosurveillance. In support of this assumption, type 1 T helper (Th1) and T cytotoxic (Tc1) effector cells are critical mediators of both autoimmunity and cancer immunosurveillance (Deng et al., 2019; Fridman et al., 2017; Knochelmann et al., 2018; Saxena et al., 2012; Zou and Restifo, 2010).
In the present study, we performed a side-by-side comparison of previously described murine models of autoimmune versus nonautoimmune cholangitis recapitulating the main features of PBC and PSC, respectively. Animals affected with these cholangiopathies were subsequently transplanted with syngeneic CCA cells in order to challenge our hypothesis. We observed that PBC-associated autoimmunity, but not PSC-related inflammation, prevented CCA outgrowth. Both CD4+ and CD8+ T cells were responsible for the PBC-associated immunosurveillance of CCA. In PBC mice, Th1/Tc1 and Th2/Tc2 cell subsets were notably enriched in the liver, detected in tumor-draining lymph nodes, and concentrated in CCA tissues compared with PSC mice or mice without cholangitis. T cells infiltrating PBC livers and CCA tissues shared multiple clonotypes that fell into the category of Tc1 cells. Altogether, our work provides evidence for a mechanistic overlapping between autoimmunity and cancer immunosurveillance.
Results
Establishment and comparison of the two preclinical models of cholangitis, PBC and PSC
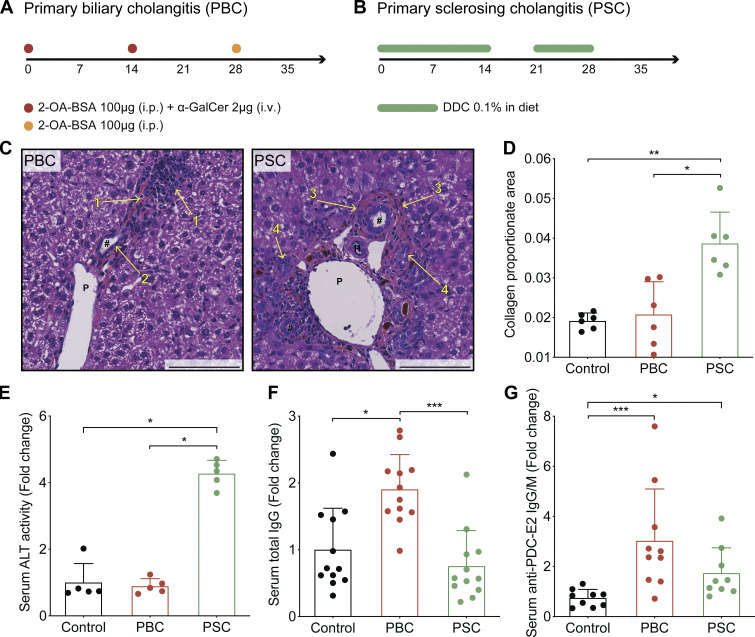
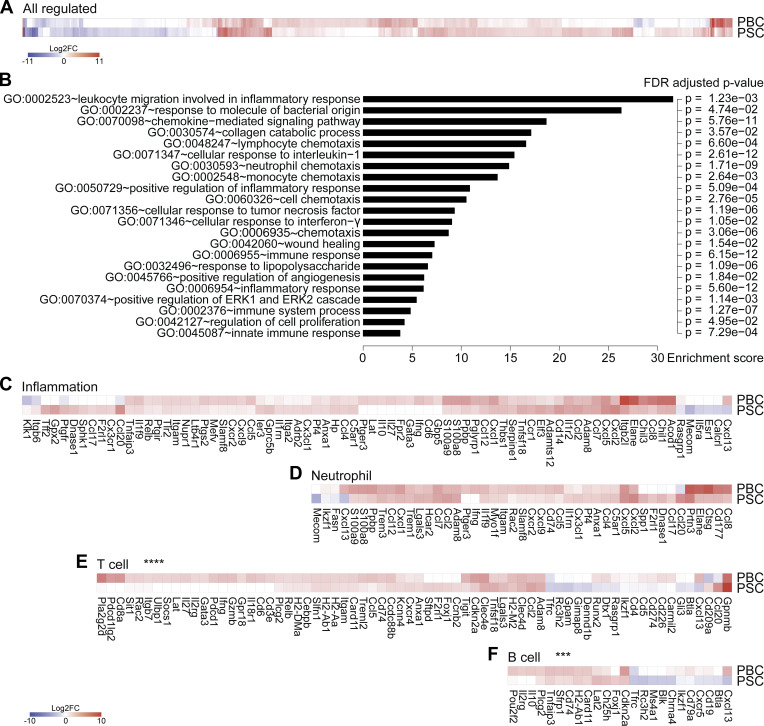
Nongenetic approaches were taken to induce two different types of cholangiopathies, PBC and PSC, in C57Bl/6 mice. Of note, existing genetic murine models of cholangitis (Bae et al., 2016; Concepcion and Medina, 2015; Fickert et al., 2014; Katsumi et al., 2015) were excluded as they rely on the constitutive deletion of immunological or metabolic factors that would impair cancer immunosurveillance. PBC was induced by i.p. challenge with 2-octynoic acid coupled to BSA (2OA-BSA) and i.v. injections of α-galactosylceramide (α-GalCer; a potent adjuvant that stimulates natural killer T [NKT] cells; Fig. 1 A), following established protocols known to break immune tolerance against the autoantigen PDC-E2; (Wakabayashi et al., 2008; Chang et al., 2014). PSC was induced by intermittent dietary challenge with 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC; Fig. 1 B), as previously described (Fickert et al., 2007; Ikenaga et al., 2017). DDC induces cholestasis by promoting the formation of intraductal porphyrin plugs. Both protocols yielded the expected histological stigmata of PBC and PSC (Fig. 1 C), with signs of fibrosis in PSC but not PBC (Fig. 1 D; Huang et al., 2013), as well as distinctive serological features such as an elevation in plasma transaminases in PSC but not PBC (Fig. 1 E) and increased serum levels of total IgG and anti–PDC-E2 IgG/M in PBC (Fig. 1, F and G). As expected, PBC but not PSC was associated with the induction of PDC-E2–reactive CD8+ and CD4+ T cells, as illustrated by the detection of splenocytes producing IFNγ in response to peptides derived from this autoantigen (data not shown). Transcriptomic analyses of the livers by RNA sequencing (RNA-seq) revealed mostly convergent alterations in the gene expression patterns when PBC and PSC were compared with normal, age- and sex-matched controls (Fig. 2 A and Table S1). Gene ontology (GO) term analyses of the modulated mRNAs revealed a significant enrichment of genes involved in inflammation, immune response, and chemotaxis of different leukocyte subsets (Fig. 2 B). The expression of genes listed under the GO terms “inflammation” and “neutrophil” was not significantly different (P > 0.5, Student t test) between PBC and PSC (Fig. 2, C and D). In sharp contrast, genes listed under the GO terms “T cell” and “B cell” were significantly (P < 0.001, Student t test) up-regulated in PBC compared with PSC livers (Fig. 2, E and F).
Figure 1.
Previously characterized mouse models of PBC and PSC cholangitis do recapitulate the main features of the corresponding human pathologies. (A) Experimental schedule of the induction of PBC in C57Bl/6 mice. Mice were injected at days 0 and 14 with 2OA-BSA (i.p.) plus α-GalCer (i.v.). At day 28, mice received one more dose of 2OA-BSA (i.p.). (B) Experimental schedule of the induction of PSC in C57Bl/6 mice. Mice were fed a diet containing 0.1% DCC for 2 wk. Then, after 1 wk of recovery on regular food, animals were fed again the DDC-supplemented diet during the fourth week. (C) Paraffin-embedded sections of PBC and PSC mouse livers at day 35 of cholangitis induction, stained with hematoxylin, eosin, and safran. #, bile duct; H, hepatic arteria; P, portal vein; 1, infiltrated leukocytes; 2, hyperplastic bile duct; 3, onion skin–like fibrosis; 4, ductular reaction. Porphyrin plugs (dark red) are observable in PSC liver section. Scale bars correspond to 100 µm. (D) Collagen proportionate area measured on Sirius red-stained sections of control, PBC, and PSC mouse livers harvested at day 35 of cholangitis induction. For each group, n = 6. (E) Relative ALT activity measured in the serum of control, PBC, and PSC mice at day 35 of cholangitis induction. For each group, n = 5. One representative experiment out of two is shown. (F) Relative level of total circulating IgG measured in control, PBC, and PSC mice at day 36. For each group, n = 12. One representative experiment out of two is shown. (G) Relative level of IgG and IgM specific to PDC-E2 measured in the serum of control, PBC, and PSC mice at day 35 of cholangitis induction. For control, PSC, and PBC groups, n = 9, 9, and 10 samples, respectively, pooled from two distinct experiments. (D–G) Graphs show individual and mean (±SD) values. P values were calculated by means of the Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Figure 2.
Livers from mice with PBC and PSC feature distinct adaptive immune cell signatures. (A–F) Following the experimental schedules illustrated in Fig. 1, A and B, hepatic gene expression was studied by transcriptomic analysis (RNA-seq) in healthy control (n = 4), PBC (n = 3), and PSC (n = 3) mice at day 35. (A) Fold change (≥log2|2|) of the 866 significantly modulated genes in PBC and PSC livers relative to healthy controls. Refer to Data S1 for a detailed heatmap and to Table S1 for individual and mean expression values of each gene. (B) GO term enrichment analysis of the genes up-regulated in both PBC and PSC livers (of note, genes down-regulated were not associated with a specific GO term enrichment). The bar chart displays the enrichment score of significantly enriched GO terms with their FDR-adjusted P value. (C–F) Focus on genes whose expression is modulated in PBC and PSC livers compared with healthy controls and listed under the GO terms “inflammation” (C), “neutrophil” (D), “T cell” (E), and “B cell” (F). P values of the paired Student t test of the comparisons between PBC and PSC profiles are indicated. ***, P < 0.001; ****, P < 0.0001. FC, fold-change; FDR, false discovery rate.
Altogether, these results support the contention that adaptive T and B lymphocyte–mediated immune responses play a quantitatively more important role in PBC than in PSC, in agreement with the literature that considers PBC as an autoimmune disease (Trivedi and Cullen, 2013; Tsuneyama et al., 2017) and PSC as a chronic inflammatory disease without a major autoimmune component (Dyson et al., 2018; Karlsen et al., 2017).
T cell–dependent control of CCA in PBC but not PSC hosts
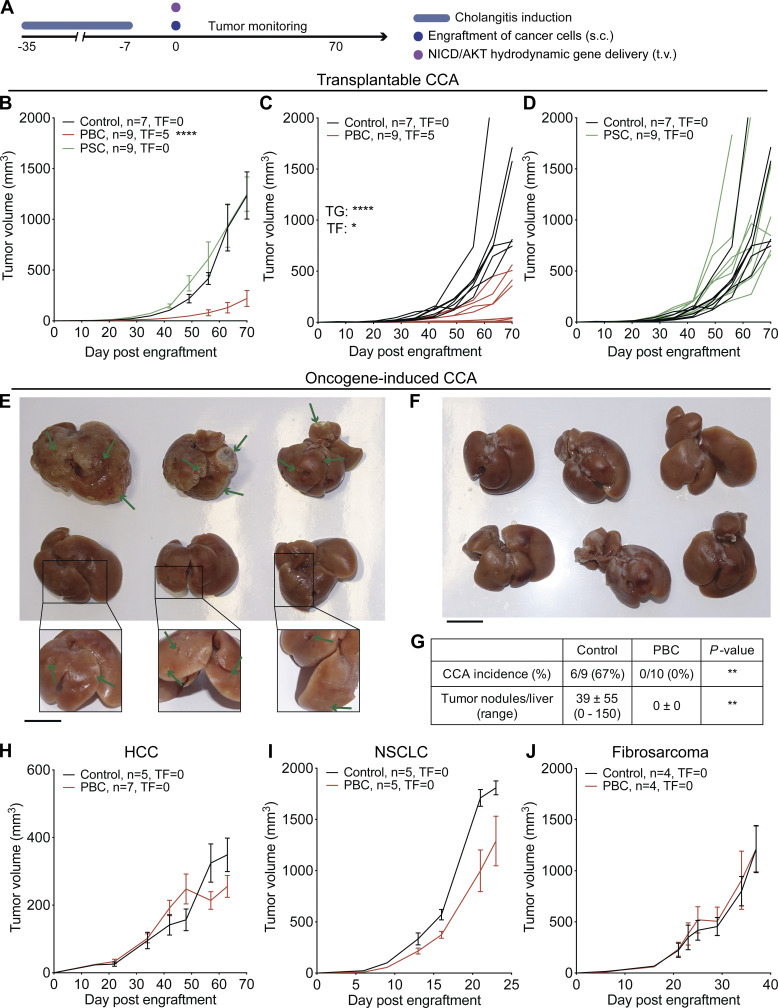
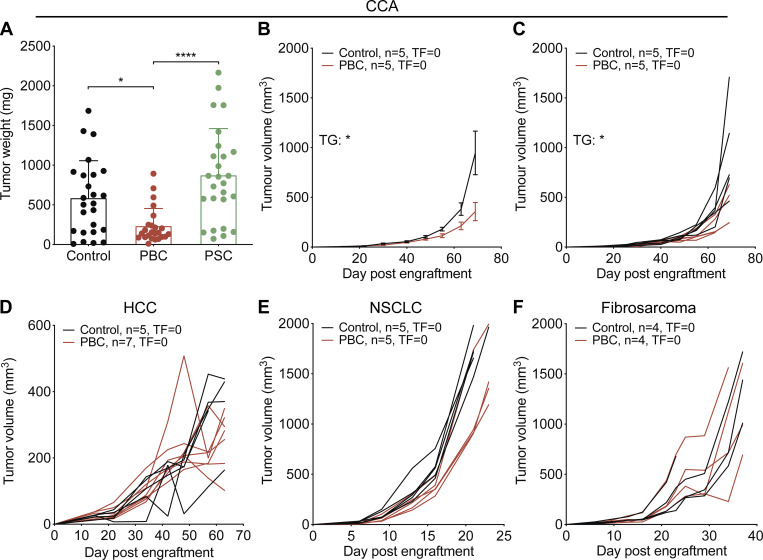
To investigate the possible impact of cholangitis on the immunosurveillance of CCA, we induced PBC and PSC in several cohorts of mice, as described above. 1 wk after the disease induction protocol had been completed, the animals were challenged by s.c. injection of various transplantable syngeneic cancer cell lines, all inoculated at normally tumorigenic doses (Fig. 3 A). Of note, a CCA cell line (SB1-JP4, which was selected from SB1 cells for accelerated tumor growth) grew indistinguishably in control mice and in mice with PSC, yet developed in a delayed fashion in mice with PBC (Fig. 3 B). This PBC-associated delay in CCA growth was highly significant (P < 0.0001, mixed-effects model) compared with control and PSC mice, with some tumors that even failed to develop in several (five out of nine) cases (Fig. 3, C and D; and Fig. S1 A). In addition, PBC caused a reduction in the s.c. growth of the parental SB1 CCA cell line (Fig. S1, B and C). More importantly, after the induction of PBC, the induction of orthotopic CCA by hydrodynamic injection of plasmids encoding the oncogenes NICD (Notch1 intracellular domain) and AKT (constitutively active myristoylated form) became inefficient. While six out of nine control mice developed macroscopically detectable CCA, all 10 PBC mice were resistant against cholangiocarcinogenesis (Fig. 3, E–G). In sharp contrast, s.c. models of HCC, nonsmall cell lung cancer (NSCLC), and fibrosarcoma indistinguishably progressed in control conditions and after PBC induction (Fig. 3, H–J; and Fig. S1, D–F).
Figure 3.
PBC but not PSC mediates specific control of CCA outgrowth. (A) Experimental schedule in C57Bl/6 mice. 1 wk after the end of cholangitis induction (see Fig. 1, A and B), syngeneic cancer cells were engrafted through s.c. injection and tumor growth (TG) was followed up (B–D and H–J). Alternatively, mice received plasmids encoding the oncogenes NICD and constitutively active AKT by hydrodynamic injection in the tail vein, and their liver was collected 5 wk later (E–G). (B–D) Tumor growth of transplantable SB1-JP4 CCA tumors in control, PBC, and PSC mice. For control group, n = 7, and for PBC and PSC groups, n = 9. One representative experiment out of two is shown. (E and F) Photographic images of control (E) and PBC (F) mouse livers after NICD/AKT plasmid injection. Green arrows indicate some tumor nodules. Scale bars correspond to 1 cm. For control group, n = 9, and for PBC group, n = 10. (G) Rate of CCA incidence and number of tumor nodules per liver in control and PBC mice that received the oncogenes NICD and AKT. P values were calculated using a Fisher’s exact test or a Mann–Whitney U test for comparing the rate of CCA incidence and the number of tumor nodules, respectively. (H–J) Growth of transplantable HCC (H), NSCLC (I), and fibrosarcoma (J) tumors in control and PBC mice. (H) For control group, n = 5, and for PBC group, n = 7. (I) For each group, n = 5. (J) For each group, n = 4. Graphs show mean (±SEM; B and H–J) or individual (C and D) tumor growth curves. Individual curves of HCC, NSCLC, and fibrosarcoma growth are displayed in Fig. S1, D–F. Regarding the comparison of the tumor growth curves, tumor growth P values were calculated by means of a linear mixed-effects model in comparison to control group. Regarding the comparison of the rate of tumor-free (TF) mice, pairwise tumor-free P values were calculated with Fisher’s exact test in comparison to control group. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. AKT, Protein kinase B; t.v., tail vein.
Figure S1.
PBC but not PSC prevents specifically the outgrowth of ectopic CCA. (A–F) Following the experimental schedules illustrated in Fig. 1, A and B, syngeneic cancer cells were engrafted through s.c. injection 1 wk after the end of cholangitis induction, and tumor growth followed. (A) Weight of CCA tumors implanted in control, PBC, and PSC mice at day 56/60 after SB1-JP4 cell line engraftment. Graph shows mean (±SD) values. P values were calculated by means of the Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons. For control, PBC, and PSC groups, n = 25, 24, and 26 tumors, respectively, pooled from three distinct experiments. (B–F) Growth of transplanted CCA SB1 (B and C), HCC (D), NSCLC (E), and fibrosarcoma (F) tumors in control and PBC mice. (B and C) For each group, n = 5. (D) For control and PBC groups, n = 5 and 7, respectively. (E) For each group, n = 5. (F) For each group, n = 4. Graphs show mean (±SEM) (B) or individual (C and D–F) tumor growth curves. For comparing the tumor growth (TG) curves or the rates of tumor-free (TF) mice, tumor growth and tumor-free P values were calculated by means of a linear mixed-effects model or Fisher’s exact test, respectively. *, P < 0.05; ****, P < 0.0001.
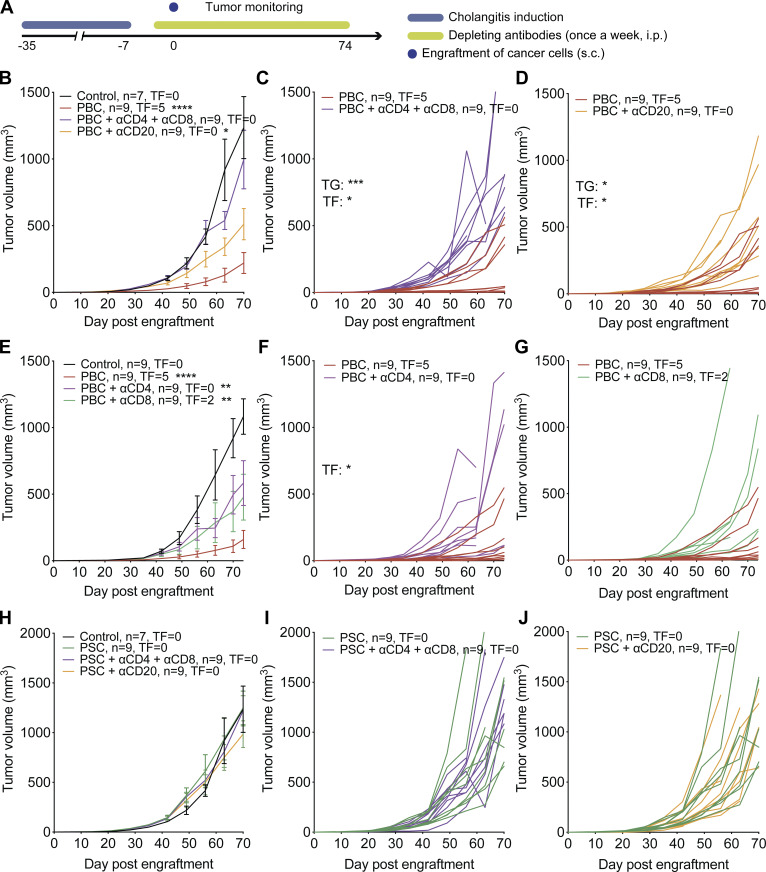
Next, we investigated whether adaptive immune responses mediated by T or B lymphocytes account for the CCA-specific tumor-suppressive effects of PBC. For this, we used monoclonal antibodies that deplete T cells (a cocktail of anti-CD4 and anti-CD8) or B cells (anti-CD20; Fig. 4 A). We found that T lymphocyte depletion abolished the control of CCA growth provided by PBC while B cell depletion had a partial effect (Fig. 4, B–D). Both CD4+ and CD8+ T cells contribute to improved CCA-specific immunosurveillance, because depletion of either T cell subset alone partially attenuated the CCA-protective effect of PBC (Fig. 4, E–G). In contrast, the depletion of T or B cells did not affect CCA progression in mice with PSC (Fig. 4, H–J).
Figure 4.
PBC-mediated protection against CCA relies on T lymphocytes. (A) Experimental schedule in C57Bl/6 mice. 1 wk after the end of cholangitis induction (see Fig. 1, A and B), mice were engrafted s.c. with syngeneic SB1-JP4 CCA cells (day 0). At days −2/−3 and +1, mice were injected i.p. with depleting antibodies targeting CD20, CD4, and/or CD8. These injections were repeated once a week. (B–J) Mean (±SEM; B, E, and H) or individual (C and D, F and G, I and J) growth curves of implanted SB1-JP4 CCA tumors in control, PBC, or PSC mice or PBC and PSC mice selectively depleted of CD4+ and/or CD8+ T lymphocytes or CD20+ B cells. (B–D) For control group, n = 7, and for PBC, or PBC + αCD4 + αCD8, or PBC + αCD20 groups, n = 9. One representative experiment out of two is shown. (E–G) For each group, n = 9. One representative experiment out of two is shown. (H–J) For control group, n = 7, and for PSC, or PSC + αCD4 + αCD8, or PSC + αCD20, n = 9. One representative experiment out of two is shown. Regarding the comparison of the tumor growth (TG) curves, tumor growth P values were calculated by means of a linear mixed-effects model in comparison to control group (B, E, and H), PBC group (C and D, F and G), or PSC group (I and J). Regarding the comparison of the rate of tumor-free (TF) mice, pairwise tumor-free P values were calculated with Fisher’s exact test on the incidence rate to control group (B, E, and H), PBC group (C and D, F and G), or PSC group (I and J). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Trend toward statistical significance was observed for the following comparison: tumor growth of PBC versus PBC+αCD4: P = 0.0544 (F).
Altogether, we conclude that PBC (but not PSC) specifically protects against CCA but not from other antigenically different malignancies through a T cell–dependent (and to a lesser extent B cell–dependent) mechanism.
Polarity of the T cell–mediated CCA immunosurveillance upon PBC
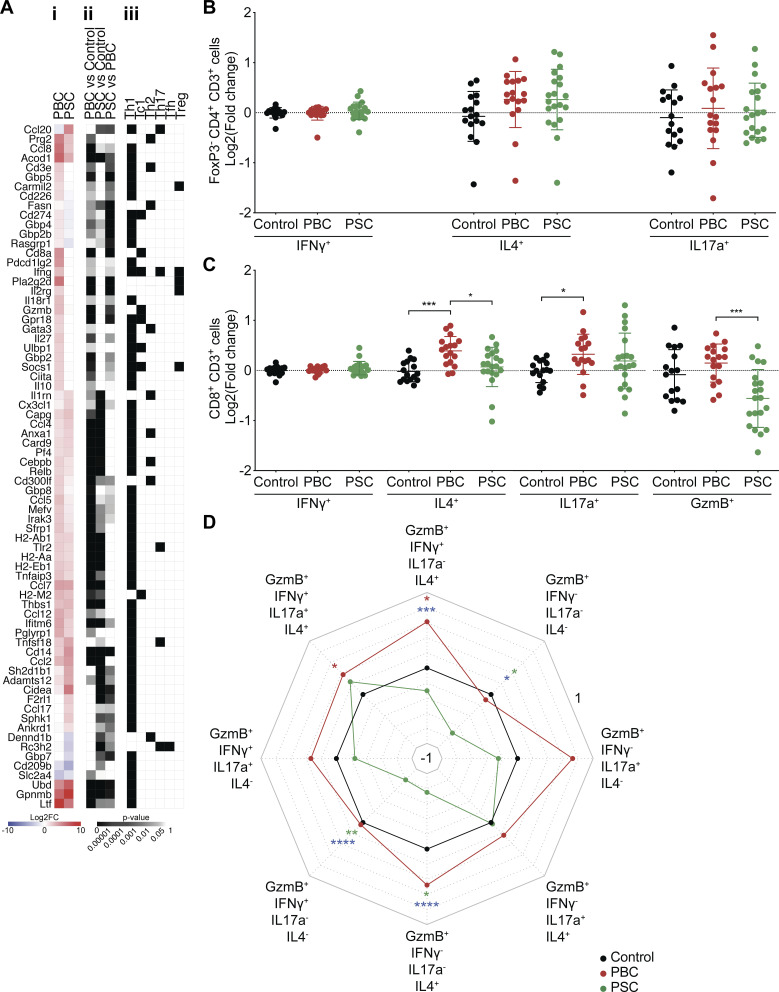
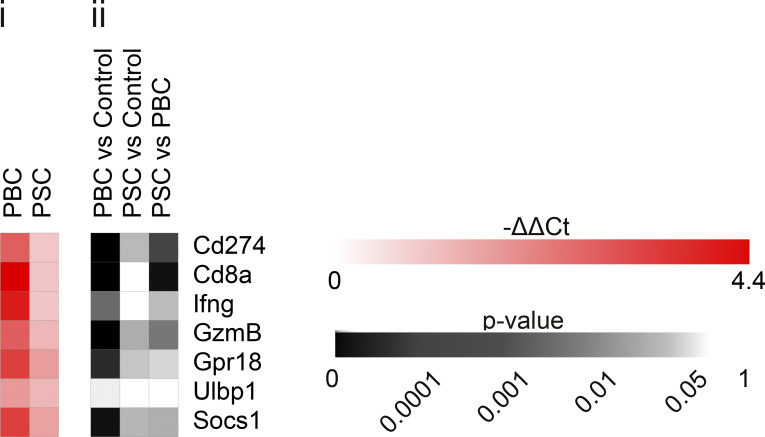
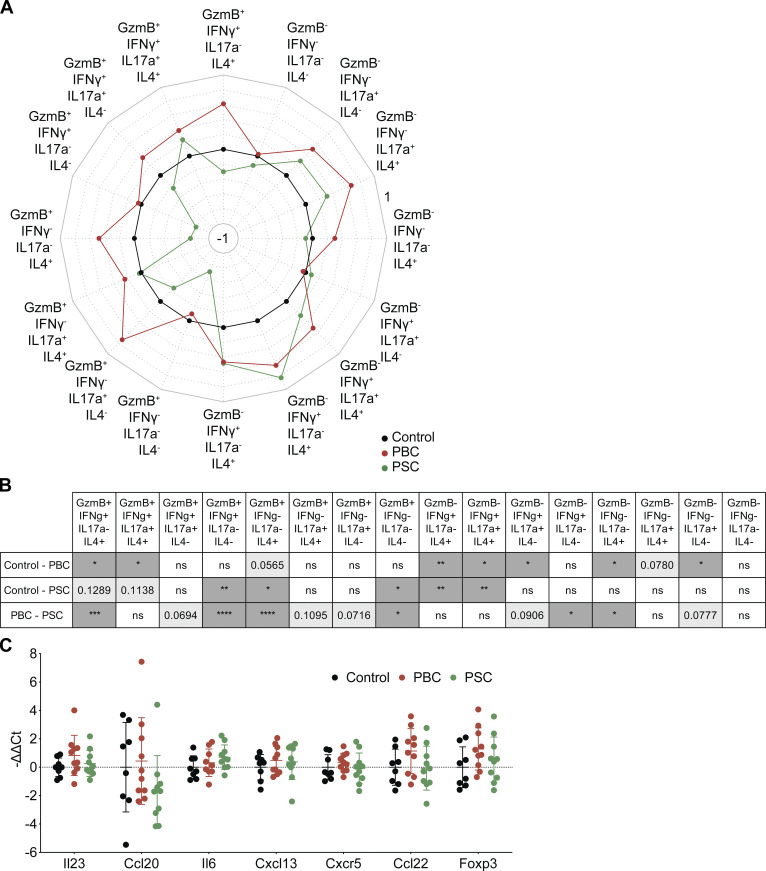
In tumor-naive hosts, deep analysis of the aforementioned liver RNA-seq dataset indicated that several genes specifically involved in Th1 and Tc1 responses were up-regulated in PBC livers with respect to controls and, in some cases, also with respect to PSC. Especially, this applies to Tc1-related genes encoding the markers CD8a, CD274 (best known as programmed death-ligand 1 [PD-L1]), UL16 binding protein 1 (ULBP1), IFNγ, G protein–coupled receptor 18 (GPR18), the transcription factor suppressor of cytokine signaling 1 (SOCS1), and the cytotoxic protease granzyme B (GzmB; Fig. 5 A). These PBC-induced Tc1-associated genes were validated by RT–quantitative PCR (RT-qPCR; Fig. S2).
Figure 5.
T cell polarization in hepatic and tumor-draining lymphatic tissues during cholangitis. (A) Following the experimental schedules illustrated in Fig. 1, A and B, gene expression was studied by transcriptomic analysis (RNA-seq) in liver samples of healthy control (n = 4), PBC (n = 3), and PSC (n = 3) mice at day 35. List of genes significantly modulated between the liver of PBC, PSC, and control mice and tagged with GO terms affiliated to T cell polarization. (A, i) The heatmap shows the fold-change (≥log2|2|) of gene expression in PBC and PSC livers relative to healthy controls. Genes involved in Tc1 response and up-regulated in PBC mice versus healthy and PSC animals are written in bold. (A, ii) The P value of each pairwise comparison is illustrated. (A, iii) Affiliation of each gene to a T cell lineage based on its related GO terms referring to Th1, Tc1, Th2, Th17, Tfh, and/or T reg cell polarization. (B–D) Following the experimental schedule illustrated in Fig. 3 A, the tumor-draining lymph node of control, PBC, and PSC mice was collected at day 56 after CCA engraftment. Cytokine production by T lymphocytes was analyzed by flow cytometry after a 24-h restimulation with anti-CD3 and anti–CD28-coated beads. (B and C) Graphs show individual (dots) and mean (±SD) log2(FC) values of single cytokine production in control, PBC, and PSC mice relative to control mice, in conventional CD4+ (B) or CD8+ (C) T cells. P values were calculated following a Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons. (D) Spider chart shows mean log2(FC) of the production of IFNγ, IL4, and/or IL17a by cytotoxic (GzmB+) lymphoid CD8+ T cells in control, PBC, and PSC mice relative to control mice. (B–D) For control, PBC, and PSC groups, n = 16, 17, and 20 samples, respectively, pooled from three distinct experiments. P values were calculated following a Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons. Red, green, and blue P values relate to PBC versus control, PSC versus control, and PBC versus PSC comparisons, respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Trend toward statistical significance was observed for the following comparisons: IL4+ cells in control versus PBC: P = 0.0510; control versus PSC: P = 0.1036 (B); GzmB+ cells in control versus PSC: P = 0.0782 (C); GzmB+ IFNγ+ IL17a+ IL4− cells in PBC versus PSC: P = 0.0694; GzmB+ IFNγ− IL17a− IL4+ cells in control versus PBC: P = 0.0565; and for GzmB+ IFNγ− IL17a+ IL4− cells PBC versus PSC: P = 0.0716 (D). Polyfunctionality of noncytotoxic (GzmB−) effector CD8+ T cells in the CCA-draining lymph node is included in Fig. S3 A and the corresponding P values in Fig. S3 B. FC, fold change.
Figure S2.
Relative expression of some genes related to Tc1 immune response in the liver of mice affected with PBC and PSC. Following the experimental schedules illustrated in Fig. 1, A and B, livers from healthy control (n = 4), PBC (n = 3), and PSC (n = 3) mice were collected at day 35, and hepatic gene expression was studied by RT-qPCR. (i) The heatmap shows the expression of genes involved in cytotoxic T cell response in PBC and PSC livers relative to healthy controls. (ii) The corresponding P value of each pairwise comparison is illustrated and was calculated by means of one-way ANOVA with Tukey’s pairwise multiple comparisons. Cd274, programmed cell death 1 ligand 1; Gpr18, G protein-coupled receptor 18; Socs1, suppressor of cytokine signaling 1; Ulbp1, UL16 binding protein 1.
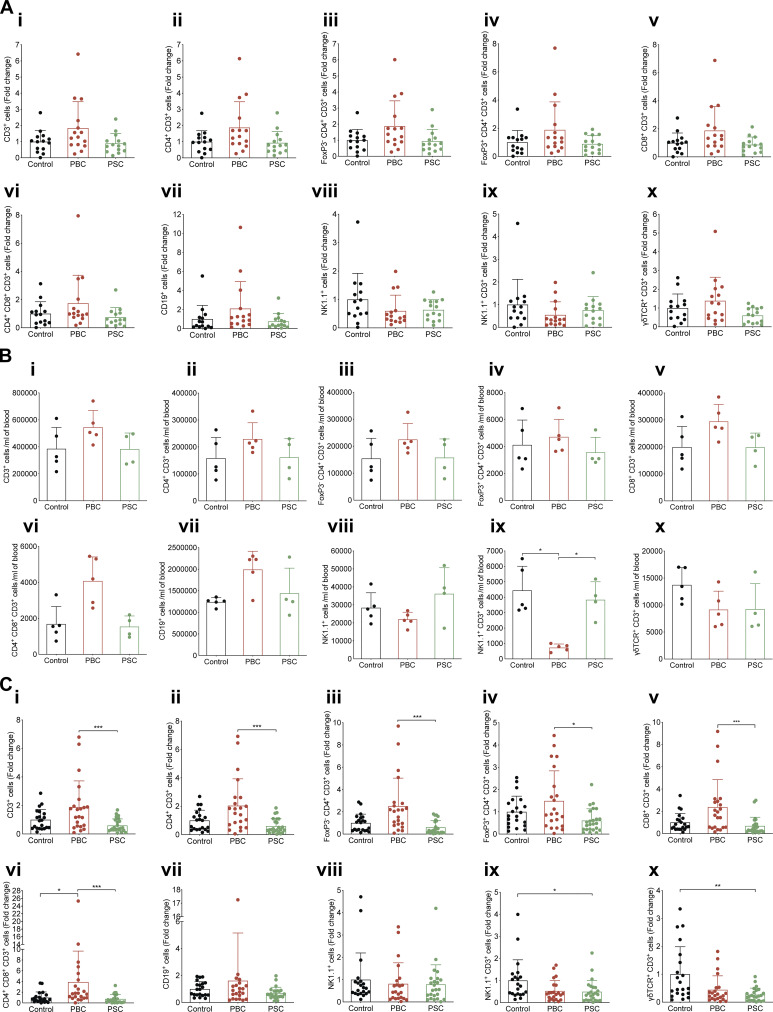
In CCA tumor–bearing hosts, lymphocyte populations and functions were investigated in the tumor and its periphery, namely draining lymph node and blood. In the lymph node draining the malignant lesion, the production of IL4 (by both conventional CD4+ T cells and CD8+ T cells) and that of IL17a (by CD8+ T cells) was elevated in mice with PBC (but not PSC; Fig. 5, B and C). Moreover, polyfunctional CD8+GzmB+ cells producing both IFNγ and IL4, together or not with IL17, were particularly abundant in CCA tumor–draining lymph nodes after PBC (Fig. 5 D; and Fig. S3, A and B), while no major changes in total T lymphocytes, conventional and regulatory CD4+ T cells, CD8+ T cells, total natural killer (NK), NKT, and γ/δ T lymphocytes were detected (Fig. S4 A).
Figure S3.
CD8+ T cell polyfunctionality within CCA tumor-draining lymph node during cholangitis. (A–C) Following the experimental schedule illustrated in Fig. 3 A, SB1-JP4 CCA tumors and their draining lymph nodes were collected from control, PBC, and PSC mice at day 56/60 after CCA engraftment. (A) Cytokine production by lymph node T lymphocytes was analyzed by flow cytometry after a 24-h restimulation with anti-CD3 and anti–CD28-coated beads. Spider chart shows mean log2(FC) of the CD8+ T cell production of IFNγ, IL4, IL17a, and/or GzmB in control, PBC, and PSC mice relative to control mice. For control, PBC, and PSC groups, n = 16, 17, and 20 samples, respectively, pooled from three distinct experiments. (B) Corresponding P values calculated by means of the Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (C) Expression of genes related to Th17/Tc17 (Il23, Ccl20, Il6), Tfh (Cxcl13, Cxcr5), and T reg cell (Ccl22, Foxp3) immune responses was measured by RT-qPCR within CCA tumors. For control, PBC, and PSC groups, n = 8, 10, and 10 samples, respectively, pooled from two distinct experiments. Graphs show individual and mean (±SD) relative expression values. P values were calculated by means of one-way ANOVA with Tukey’s pairwise multiple comparisons. Ccl20, C-C motif chemokine ligand 20; Ccl22, C-C motif chemokine ligand 22; Cxcl13, C-X-C motif chemokine ligand 13; Cxcr5, C-X-C motif chemokine receptor 5; FC, fold change; Foxp3, forkhead box P3.
Figure S4.
Lymphocytic cell subsets across several tissues during cholangitis. (A–C) Following the experimental schedule illustrated in Fig. 3 A, tumor-draining lymph nodes (A), blood (B), and tumors (C) from control, PBC, and PSC mice were collected at day 56 after SB1-JP4 CCA engraftment for flow cytometric analysis of the lymphocytic cell subset content. Relative counts (A and C) and absolute counts (per milliliter of blood; B) of total T lymphocytes (i); total (ii), conventional (iii), or regulatory (iv) CD4+ T cell, CD8+ T cell (v), and CD4+ CD8+ double-positive T cell (vi) subsets; total B cells (vii), NK (viii), NKT (ix), and γδ T (x) lymphocytes. Graphs show individual and mean (±SD) values. (A) For control, PBC, and PSC groups, n = 14, 15, and 14 samples, respectively, pooled from two distinct experiments. (B) For control, PBC, and PSC groups, n = 5, 5, and 4 samples, respectively. (C) For control, PBC, and PSC groups, n = 21, 22, and 24 samples, respectively, pooled from three distinct experiments. P values were determined by the Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Trend toward statistical significance was observed for the following comparisons: PBC versus PSC: P = 0.1187 (A, ii); PBC versus PSC: P = 0.1026 (A, iii); PBC versus PSC: P = 0.1271 (A, v); PBC versus PSC: P = 0.0968 (A, x); control versus PBC: P = 0.0573; PBC versus PSC: P = 0.0560 (B, vi); control versus PBC: P = 0.0852 (B, vii); control versus PSC: P = 0.0932 (C, i); control versus PBC: P = 0.0942 (C, iii); control versus PBC: P = 0.1031 (C, ix); control versus PBC: P = 0.0655 (C, x).
Meanwhile in the blood, the sole significant fluctuation associated with PBC was a trend toward an increase in circulating B and CD4+ CD8+ double-positive T cells and a decrease in NKT cells (Fig. S4 B).
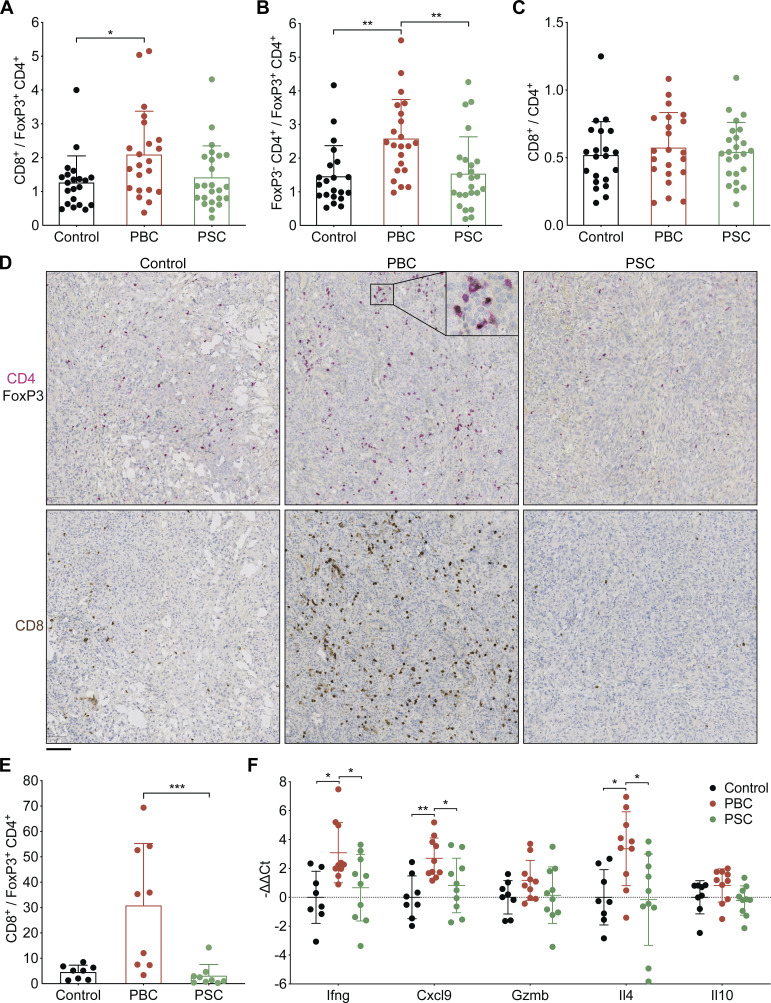
In the CCA microenvironment, cytofluorometric analyses of the T cell subpopulations infiltrating the malignant lesion (Fig. S4 C, i–vi) revealed an increased CD8+ T lymphocyte/regulatory FoxP3+CD4+ T cell (T reg cell) ratio (P = 0.0394; Fig. 6 A) and an enhanced ratio of conventional FoxP3−CD4+ T cells over T reg cells (P = 0.0022; Fig. 6 B) after PBC (but not PSC). The total CD8+/CD4+ T cell ratio remained unchanged (Fig. 6 C). Immunohistochemical analyses confirmed the elevation of the CD8+/CD4+FoxP3+ T reg cell ratio as an indicator of improved immunosurveillance following PBC (Fig. 6, D and E). No major variations in the total number of B, NK, and NKT cells were detected in either one of cholangitis-related diseases, while a decrease in γ/δ T lymphocytes was found in CCA tumors of both PBC and PSC (Fig. S4 C, vii–x).
Figure 6.
PBC affects T lymphocyte subsets infiltrating CCA tumors. (A–F) Following the experimental schedule illustrated in Fig. 3 A, SB1-JP4 CCA tumors were collected from control, PBC, and PSC mice at day 56/60 after CCA engraftment. T cell subsets were analyzed by flow cytometry (A–C) and immunohistochemistry (D and E), and the expression of T cell–associated effector and regulatory cytokines was measured by RT-qPCR (F). Ratios of either total CD8+ CD3+ T cells (A) or FoxP3− CD4+ CD3+ T cells (B) over FoxP3+ CD4+ CD3+ T reg cells, and of total CD8+ CD3+ T cells over total CD4+ CD3+ T cells (C) within CCA tumors. (A–C) For control, PBC, and PSC groups, n = 21, 22, and 24 samples, respectively, pooled from three distinct experiments. (D and E) PFA-fixed paraffin-embedded sections of implanted CCA tumors from control, PBC, and PSC mice were stained for CD4 (magenta), FoxP3 (black), or CD8 (brown) by immunohistochemistry (D), and the ratio of CCA-infiltrating CD8+ over FoxP3+ CD4+ cells was measured (E). (D) Scale bar corresponds to 100 µm. (E) For control, PBC, and PSC groups, n = 8, 9, and 9 samples, respectively, pooled from two experiments. (F) Relative expression of several genes encoding Th1/Tc1 (Ifng, Cxcl9, Gzmb) or Th2/Tc2 (Il4)-related effector or regulatory (Il10) molecules within CCA tumors. For control group, n = 8, and for PBC and PSC groups, n = 10 samples pooled from two experiments. Graphs show individual and mean (±SD) values. P values were calculated by means of the Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons (A–C and E) or one-way ANOVA with Tukey’s pairwise multiple comparisons (F). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Trend toward statistical significance was observed for the following comparisons: PBC versus PSC: P = 0.1218 (A); control versus PBC: P = 0.0646 (E); for Il10: PBC versus PSC: P = 0.1073 (F). PFA, paraformaldehyde.
Considering the limited amount of malignant tissue available in PBC animals due to anti-CCA immunosurveillance, we investigated tumor-infiltrating T cell subtypes by RT-qPCR. Similarly to the hepatic and lymphoid tissues, we observed a trend toward an increased expression of the type 1 cytokines IFNγ and chemokine C-X-C motif ligand 9 (CXCL9), as well as of the type 2 cytokine IL4, inside the CCA tissue of PBC hosts (Fig. 6 F), while multiple other cytokines associated with Th17, T follicular helper (Tfh), and T reg cells remained unaffected in CCA tumors of PBC mice compared with malignant tissues of PSC and control animals (Fig. S3 C).
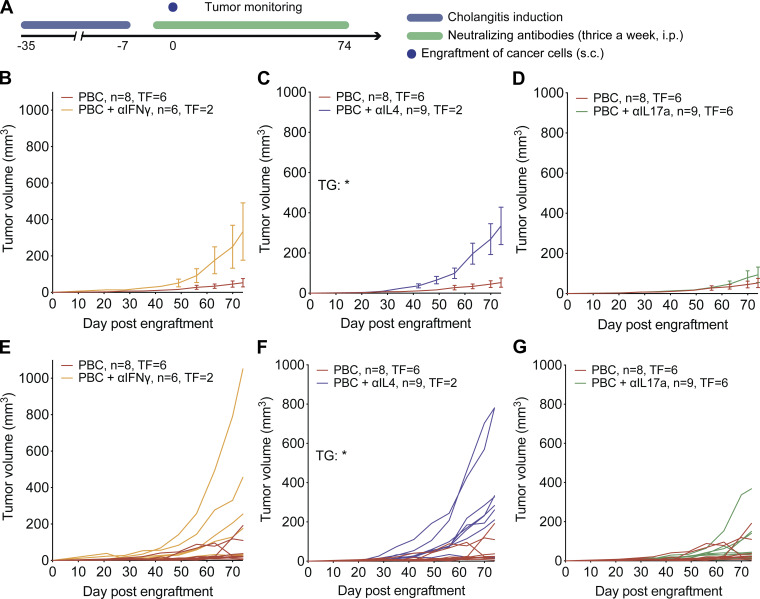
These results pointed to an active immune response within tumors evolving in the context of PBC. Intrigued by these observations, we decided to investigate the role of IL17a in PBC-induced anti-CCA immunosurveillance and compared IL17a with IFNγ, which is a well-established contributor to anticancer immune responses (Castro et al., 2018; Corthay et al., 2005; Dunn et al., 2006; Kalathil et al., 2019). Mice in which PBC had been induced received three weekly injections of either an isotype control antibody or antibodies that neutralize IL4, IL17a, or IFNγ as they were monitored for CCA growth (Fig. 7 A). Blockade of IFNγ showed a clear trend of a compromised PBC-elicited protection (P = 0.0513; Fig. 7, B and E), and a similar, significant (P = 0.0165) effect was observed for IL4 blockade (Fig. 7, C and F). In contrast, the neutralization of IL17a failed to affect PBC-induced immunosurveillance (Fig. 7, D and G). Altogether, these results suggest a major role for IFNγ and IL4 (but not for IL17a) in the PBC-associated autoimmunity that mediates CCA immunosurveillance.
Figure 7.
PBC-mediated protection against CCA requires IFNγ and IL4 but not IL17a. (A) Experimental schedule in C57Bl/6 mice. 1 wk after the end of cholangitis induction (see experimental schedule in Fig. 1 A), syngeneic SB1-JP4 CCA cells were engrafted into PBC mice (day 0). At day −1, +1, and +3 and then thrice a week until the end of the experiment, mice were injected with antibodies targeting effector cytokines. (B–G) Growth of SB1-JP4 CCA tumors in control or PBC mice or PBC mice injected with antibodies neutralizing either IFNγ (B and E), IL4 (C and F), or IL17a (D and G). For PBC, PBC + αIFNγ, PBC + αIL4, and PBC + αIL17 groups, n = 8, 6, 9, and 9 tumors, respectively, from one representative experiment out of two. Graphs show mean ± SEM (B–D) and individual (E–G) tumor growth (TG) curves. For comparing tumor growth curves, tumor growth P values were calculated by means of a linear mixed-effects model. For comparing the rates of tumor-free (TF) animals, pairwise tumor-free P values were calculated with Fisher’s exact test. *, P < 0.05. Trend toward statistical significance was observed for the following comparisons: TG of PBC versus PBC+αIFNγ: P = 0.0513 (B and E); TF of PBC versus PBC+αIL4: P = 0.0567 (C and F).
T cell clonotypes in PBC-elicited CCA immunosurveillance
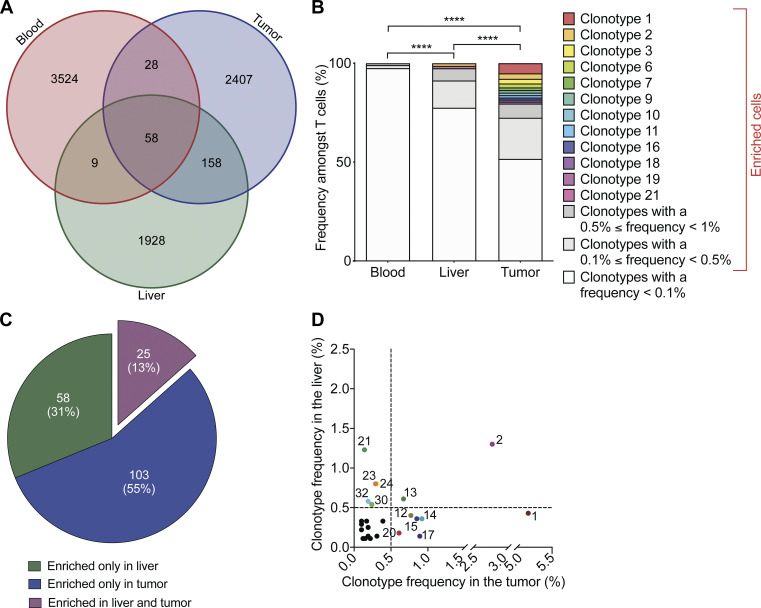
We wondered whether the specificity of T cells found in PBC livers and tumors developing after PBC might be shared to some extent, comparing them with circulating T cells from the blood. Single-cell TCR sequencing coupled to single-cell RNA sequencing (scTCR-seq/scRNA-seq) led to the identification of 8,112 distinct clonotypes (Fig. 8 A), with a hierarchy of clonotype enrichment that was significantly higher in PBC livers than in peripheral blood, as well as in CCA compared with blood and liver (Fig. 8 B). Indeed, 12 distinct clonotypes accounted for ∼20% of the tumor T cell infiltrate (Fig. 8 B). The overlap of clonotypes between liver and tumor was superior to the overlap between blood and liver or blood and tumor (Fig. 8 A). Among the 128 clonotypes enriched (i.e., accounting for >0.1% of total T cells infiltrating the tissue) in the tumor, 25 were also detected and enriched in the liver (Fig. 8 C and Table S2). Two clonotypes (#2 and #13) were particularly expanded, each representing over 0.5% of the total T cell population in both liver and tumor after PBC (Fig. 8 D).
Figure 8.
Some T cell clonotypes enriched in PBC liver also infiltrate CCA lesion. (A–D) Following the experimental schedule illustrated in Fig. 3 A, SB1-JP4 CCA tumor, liver, and blood of a PBC mouse were collected at day 70 after CCA engraftment. T cells of the malignant, hepatic, and blood tissues were sorted out and underwent single-cell transcriptomic analysis (scRNA-seq). Output data were used not only for T cell gene expression profiling but also for TCR sequencing, thus allowing characterization of clonotype. (A) Venn diagram of the T cell clonotype overlap between tumor, liver, and blood. (B) Proportion of T cell clonotypes among total T cells analyzed in each tissue. Clonotypes with a frequency >0.1% were considered enriched. Pairwise P values were calculated using Fisher’s exact test on the percentage of enriched T cell clonotypes. ****, P < 0.0001. (C) Number of enriched T cell clonotypes expanded only in tumor (n = 103), or liver (n = 58), or in both tissues (n = 25). (D) Proportion in the liver versus CCA tumor of each of the 25 T cell clonotypes enriched and shared between both tissues.
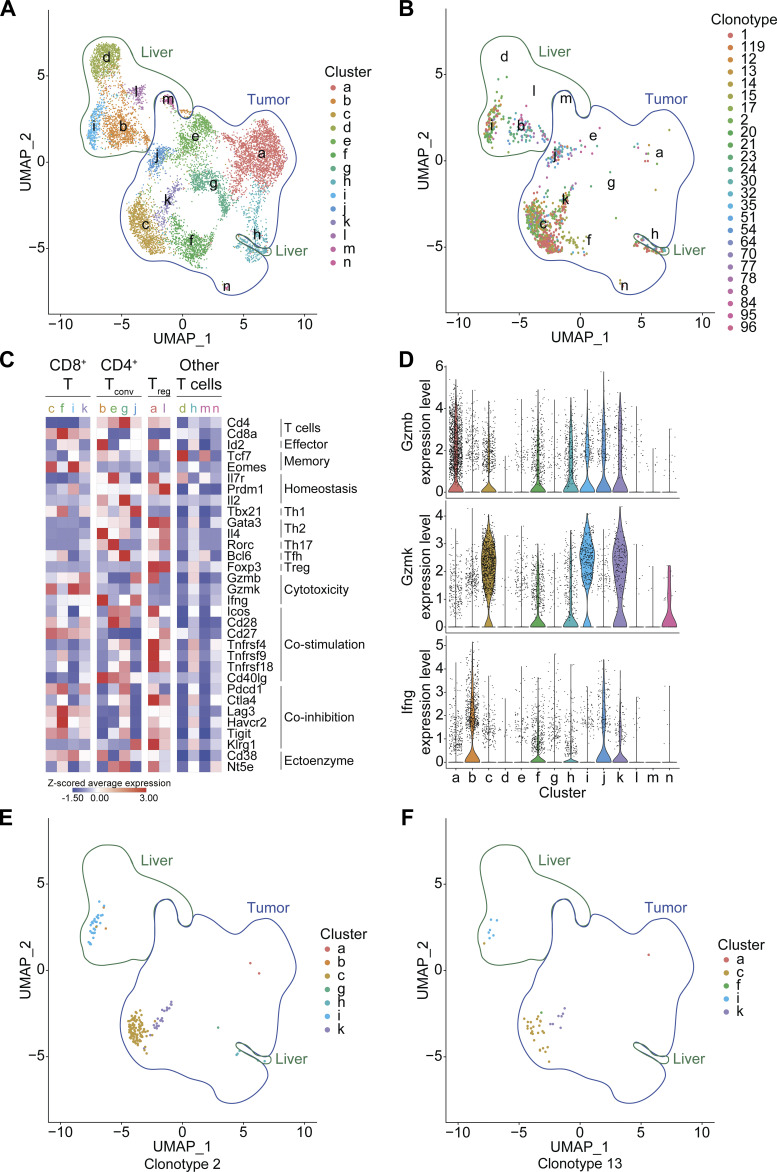
Bioinformatic analysis of the scRNA-seq data of total T cells isolated from the liver and tumor of a PBC mouse revealed a net distinction between the transcriptome of T lymphocytes located in the two organs (Fig. 9 A). Only one phenotypic cluster (h) was observed in both liver and tumor (Fig. 9 A). The 25 liver/tumor dual-enriched (>0.1%) clonotypes were spread across few clusters (mostly c, j, and k in the tumor; b and i in the liver; Fig. 9 B). Among these clusters, three corresponded to CD8+ T cells (c, i, and k) and two corresponded to conventional CD4+ T cells (b and j; Fig. 9 C). They exhibited a preponderant Tc1/Th1 polarization as illustrated by the expression of the enhancers Tbx21 and/or Eomes and of effector molecules, namely IFNγ for Th1 (b and j), or of the cytotoxic markers granzymes B and K for Tc1 (c, i, and k; Fig. 9, C and D; and Table S3). Interestingly, the two clusters containing most of the enriched CD4+ T cell clonotypes presented a multifunctional signature characterized by the coexpression of IFNγ along with either IL4 and IL2 (cluster b in the liver) or GzmB (cluster j in the tumor; Fig. 9, C and D; and Table S3). Clearly, the two most enriched clonotypes shared between CCA tumor and PBC liver (#2 in Fig. 9 E and #13 in Fig. 9 F) belonged to the category of CD8+ cytotoxic T lymphocytes (clusters c + k in tumor, and cluster i in liver; Fig. 9, E and F). Moreover, they showed signs of prolonged activation/exhaustion with the expression of Pdcd1 (encoding PD-1), Lag3, Harvc2/Tim3, and Tigit (Fig. 9 C), yet to a lesser extent than another cluster of CD8+ T cells (tumor cluster f) that contains only a few shared clonotypes (Fig. 9, B and F).
Figure 9.
Phenotype of the T cell clonotypes involved in PBC-elicited CCA immunosurveillance. (A–F) In silico analyses of scRNA-seq/scTCR-seq data revealed the phenotype of the 25 T cell clonotypes enriched in both liver and CCA tumor of a PBC mouse (Fig. 8 D). (A) Uniform manifold approximation and projection (UMAP) clustering projection of the total T lymphocytes sorted out from the tumoral and hepatic tissues of a PBC mouse. 14 phenotypic T cell clusters were detected (a–n). (B) UMAP clustering projection of the 25 T cell clonotypes enriched in both tumor and liver upon PBC. (C) Z-scored average expression of several genes associated with T cell function across the 14 clusters, consisting of four CD8+, four conventional (conv) CD4+, two regulatory (reg) CD4+, and four unassigned T cell clusters. (D) Violin plots displaying the expression level of Gzmb, Gzmk, and Ifng across the 14 clusters. (E and F) UMAP clustering projection of the T cell clonotypes #2 (E) and #13 (F) that were the most enriched in both CCA tumor and liver of the PBC mouse (Fig. 8 D).
In conclusion, multiple clonotypes enriched in PBC liver were also found in CCA tumors, where they exhibited a transcriptomic signature compatible with a function in anticancer immunosurveillance.
Discussion
Using previously characterized murine models of cholangitis, we demonstrated that PBC-associated autoimmunity but not PSC-related inflammation fuels the immunosurveillance of CCA and prevents its development in a Th1/Tc1 cell–dependent manner. The relevance of this finding is comforted by the remarkable, yet arguably imperfect, similarity between the cholestatic animal models and the human diseases. Indeed, the present work, supported by an extensive literature (Bae et al., 2016; Concepcion and Medina, 2015; Fickert et al., 2014; Katsumi et al., 2015; Mariotti et al., 2018; Wang et al., 2014), showed that the cholangitis, developing after the administration of 2OA-BSA/αGal-Cer or DDC to mice, recapitulates the main histopathological, serological, and immunological features of human PBC and PSC, respectively.
Mechanistically, peripheral depletion of T lymphocyte subsets demonstrated that the elimination of CCA upon PBC was relying on both CD4+ and CD8+ T cells. The PBC-induced immune response appeared specific to CCA, as it did not protect against other malignancies, including HCC. Transcriptomic analyses revealed an enrichment in mRNAs associated with Th1 and Tc1 lymphocyte subsets in PBC versus PSC livers. Flow cytometry–assisted immunophenotyping, together with gene expression assays, confirmed the presence of this T cell signature within CCA tumors and draining lymph nodes of mice with PBC but not PSC. Th1/Tc1–polarized subsets played a critical role in rejecting CCA tumors, as systemic neutralization of their prototypical effector cytokine IFNγ impaired PBC-mediated protection against CCA.
Given their recognized involvement in both autoimmunity and cancer immunosurveillance, Th1 CD4+ and Tc1 CD8+ T cells could be suspected to establish the mechanistic link between PBC and exacerbated immunosurveillance against CCA. On one hand, type 1 adaptive immunity has been shown to outweigh type 2 response during PBC (Yang et al., 2014). Elevation of the type 1 cytokines IFNγ and IL12, chemokines CXCL9 and CXCL10, and autoreactive CD8+ T lymphocytes are observed in the blood and liver of PBC patients and have been attributed a critical role in promoting and maintaining the autoimmune injury (Chuang et al., 2005; Kita et al., 2002a, 2002b; Shindo et al., 1996; Yamano et al., 2000; Yang et al., 2014). On the other hand, intratumoral infiltration of Th1 and cytotoxic CD8+ T cells correlates with favorable prognosis in multiple types of cancer (Fridman et al., 2013, 2017). CCA is no exception to this rule, and abundant tumor infiltration by CD8+ and CD4+ T lymphocytes coincides with improved outcome (Goeppert et al., 2013; Høgdall et al., 2018; Oshikiri et al., 2003; Paillet et al., 2020; Takagi et al., 2004). Additionally, a significant reduction of the T cell infiltrate was indicative of a switch from localized to metastatic CCA lesions (Tamma et al., 2019). Nevertheless, our results describe a rather unique situation where self-reactive Th1/Tc1 cells simultaneously mediate deleterious autoimmune and beneficial anticancer activities by concomitantly targeting normal and malignant cholangiocytes. Further investigations will aim at identifying the self-antigens responsible for this cross-reactivity, among which PDC-E2 is one candidate. They will substantiate the shared immune response and validate potential targets for the formulation of immunotherapeutic strategies against CCA (e.g., cancer vaccines; Pol et al., 2019; Pol et al., 2018; Pol et al., 2020a; Pol et al., 2020b; Pol et al., 2014).
Also, it will be important to examine the question as to whether autoimmunity may favor antitumor immunity outside the biliary tract. To our knowledge, a cancer-prophylactic role for autoimmunity has only been evoked in the case of vitiligo and melanoma. Indeed, a reduced incidence of melanoma has been reported in Caucasian patients with vitiligo, yet without further mechanistic insights (Kim et al., 2020; Paradisi et al., 2014; Teulings et al., 2013). In murine models of melanoma immunotherapy, leukoderma spread in treated animals that experienced complete remission and predicted a protection against a subsequent rechallenge with melanoma (Byrne et al., 2011; Lane et al., 2004; Pol et al., 2014). In the clinic, treatment-mediated induction of melanoma-specific immune responses coincides with the development of vitiligo, which is a positive prognostic marker (Failla et al., 2019; Nakamura et al., 2017). Furthermore, in preclinical models, it is possible to immunize mice with normal (nontransformed) ileal epithelial cells and to induce a protective immune response against colon cancer (Roberti et al., 2020).
In addition to the Th1/Tc1 cell subsets, future experimentations should conclude on a putative involvement of other adaptive immune compartments in CCA immunosurveillance triggered by PBC-related autoimmunity. In particular, our RNA-seq, gene expression, and flow cytometric analyses highlighted an enriched Th2/Tc2 signature (e.g., IL4, Gata3) in the liver, CCA tissue, and tumor-draining lymph node of PBC hosts compared with PSC and control animals. In this line, Th2-supportive double-positive CD4+ CD8+ T lymphocytes (Bohner et al., 2019) were increased in the blood of CCA-bearing PBC mice. Moreover, antibody-mediated neutralization of IL4 compromised the PBC-mediated reduction of CCA growth. As a matter of fact, an antitumoral activity of IL4 has previously been reported (Hemmerle and Neri, 2014; Tepper et al., 1989; Yu et al., 1993). Furthermore, IL4 expression in CD8+ T cells was more frequently associated with the cytotoxic protease GzmB under PBC. This could be a hint of the Tc2 involvement in the PBC-associated anti-CCA immune response. Of note, several studies highlighted that IL4-producing CD8+ T cells demonstrate a potent cytolytic activity, although less efficiently than Tc1 cells (Cerwenka et al., 1999; Kemp and Ronchese, 2001; St Paul and Ohashi, 2020; Vizler et al., 2000). Additionally, B cell depletion, yet incomplete, through administration of anti-CD20 antibodies significantly impaired PBC-mediated protection against CCA. Collectively, B lymphocytes and type 2 differentiated T lymphocytes may participate in CCA immunosurveillance in PBC hosts. This observation is original per se and of particular interest for the field of immuno-oncology. Indeed, whereas B cells play an undeniable role in PBC pathogenesis (Ma and Chen, 2019), they are recurrently detected within neoplastic microenvironments, but their pro- versus anti-tumoral function remains debated to date (Kaplon and Dieu-Nosjean, 2018; Shalapour and Karin, 2021; Yuen et al., 2016). Regarding CCA, one report has correlated a higher level of B cells infiltrated in the malignant bed with better prognosis (Goeppert et al., 2013). Therefore, B and Th2/Tc2 cell subsets might play a similar role as Th1/Tc1 cells in injuring the normal biliary tract and eliminating malignant cholangiocytes.
Additionally, we also observed an enrichment in the Th17/Tc17 cell lineages in the lymphoid tissues (but not in the CCA microenvironment) of PBC mice in comparison to PSC and control animals. This observation further supported the relevance of the murine model of PBC, as Th17 cell activity has been positively associated with progression toward advanced stages of the cholangitis (Qian et al., 2013; Yang et al., 2014). However, neutralization of the effector cytokine IL17a did not impede the protective immunity against ectopic CCA in mice affected with PBC. Thus, PBC-related Th17/Tc17 lymphocytes are not involved in CCA immunosurveillance, which is in line with the clinical observation that high intratumoral IL17a mRNA levels predict poor prognosis in patients with intrahepatic CCA (iCCA; Gu et al., 2012).
Molecular analyses using combined scTCR-seq and scRNA-seq led to the conclusion that PBC liver and CCA developing in the context of PBC contain multiple enriched clonotypes, often shared between both tissues. Clonotypes shared between peripheral blood T lymphocytes and liver or between blood and CCA were comparatively rare. Moreover, there was a clear tendency to oligoclonality in CCA (and to a lower extent in liver) compared with blood, allowing for a distinction between likely tumor-specific (as opposed to passenger) T cells. In the clinic, such T cell clonal expansion coincided with beneficial immune responses in oncological indications (Ikeda et al., 2017; Leruste et al., 2019). In our study, those clonotypes enriched in both PBC liver and CCA were not randomly distributed across the scRNA-seq–defined T cell clusters, but rather concentrated into defined subsets. In particular, the most enriched PBC liver/CCA tumor–shared clonotypes, which most likely mediate concomitant PBC-related autoimmunity and CCA immunosurveillance, fell into the category of Tc1 cells with prominent characteristics of cytotoxicity. More precisely, these CD8+ T cells expressed the proapoptotic proteases GzmB and/or GzmK, the master transcriptional regulators Eomes and/or Tbx21, the costimulatory molecule CD27, and some coinhibitory signals such as LAG-3, thus collectively evoking long-lived memory-like effector and/or exhausted phenotypes (Intlekofer et al., 2005; Kallies et al., 2020; Knox et al., 2014; Li et al., 2018; Martin and Badovinac, 2018; Winkler and Bengsch, 2020; Zheng et al., 2017). In the meantime, the vast majority of expanded tumoral CD4+ T cells clustered into the group of terminally differentiated cytotoxic CD4+ T cells, revealed by their expression of IFNγ, GzmB, and KLRG1 (Hirschhorn-Cymerman et al., 2012; Quezada et al., 2010). As opposed to the tumoral tissue, CD4+ T cell clonotypes enriched in the liver upon PBC appeared as type 1/2 hybrid cells previously described in controlling the inflammatory process (Peine et al., 2013). Although the exact nature of the MHC class I– and II–restricted autoantigens recognized by such clonotypes remains to be determined, the enrichment of T cells with identical TCRs in the PBC liver as well as in CCA strongly pleads for the existence of shared autoantigens relevant for PBC and CCA immunosurveillance. Obviously, these speculations are based on the use of rodent models. Hence, future investigations must focus on analyzing T cells infiltrating liver lesions in PBC, PSC, and CCA patients to unravel possible cross-reactivities between PBC and CCA in a clinically relevant setting.
Furthermore, some pathological features will require particular consideration. In particular, we should point out that PBC and PSC affect distinct regions of the biliary tree. Indeed, PBC results in progressive destruction of intrahepatic small bile ducts (especially interlobular ducts), whereas PSC mainly damages intrahepatic and extrahepatic large bile ducts (Banales et al., 2020). Meanwhile, iCCA shows spatial and morphological heterogeneity, thus raising speculations on plural cellular origins. Depending on the site of CCA emersion, mature cholangiocytes, biliary-like cells trans-differentiated from hepatocytes, or progenitor cells have been speculated as sources of malignant transformation due to their self-renewing potential and long life span (Moeini et al., 2021). On one side, small bile duct iCCAs grow as a tumor mass and may arise from hepatic progenitor cells or nonmucin-producing cuboidal cholangiocytes in Hering canals, ductules, and interlobular bile ducts. On the other side, large bile duct iCCA is preceded by precancerous lesions and characterized by sustained mucin production. It seems to arise from peribiliary glands or mucin-producing columnar cholangiocytes (Moeini et al., 2021). Thus, complementary studies will need to determine if PBC protects only against small bile duct iCCAs or if it can also prevent the emergence of large bile duct malignancies. Considering the rarity of CCA development upon PBC, the latter scenario can be envisioned. It would imply that the self-antigen(s) targeted by PBC/CCA cross-reactive T cells are presented on MHC molecules at the surface of malignant cholangiocytes regardless of their cellular origin. If so, vaccine strategies against such antigenic determinants or adoptive transfer of cognate T cells might have prophylactic benefit in patients at risk of developing CCA (e.g., PSC).
In conclusion, the present work revealed a connection between autoimmunity affecting the biliary tract and CCA immunosurveillance. It will be important to explore the similarities and differences between pathogenic autoimmunity (against normal cells) and beneficial autorecognition (of malignant cells) that are likely mediated by the same immune recognition mechanism. Does efficient antitumor immunity always come at the cost of cross-reactive autoimmunity against normal antigen? Would this explain why some cancers are relatively easy to cure, such as thyroid carcinoma (where immune elimination of normal thyrocytes can be easily palliated by hormonal substitution therapy) or melanoma (where autoimmunity only causes vitiligo), contrasting with others where definitive cure cannot be achieved, perhaps because the resulting autoimmune rejections (e.g., against pneumocytes in lung cancer or hepatocytes in liver cancer) cannot be tolerated by the organism? Future research will provide clues to these open questions.
Materials and methods
Mouse strains and accommodation
6–8-wk-old female C57Bl/6 mice were purchased from Envigo France. Animals were maintained in a temperature-controlled and specific pathogen–free environment, respecting cycles of 12 h of light and 12 h of darkness, and fed ad libitum with the SAFE A04 diet together with free access to water. Experiments were performed in compliance with European Union Directive 63/2010 and the protocols #8125–2016120818255662v1 and #25926-202006081459362 v2 approved by the Charles Darwin Ethics Committee for animal experimentation.
Cell lines and culture
The murine cell lines SB1 and SB1-JP4 (CCA), MCA205 (fibrosarcoma), TC-1 (nonsmall cell lung carcinoma), and Hep-55.1C (hepatoma; purchased from CLS – Cell Lines Service GmbH), all syngeneic in C57Bl/6 mice, were grown in DMEM supplemented with 10% (vol/vol) FBS and 100 IU/ml of penicillin G and 100 mg/ml streptomycin under standard culture conditions (37°C, 5% CO2, >90% humidity). The mouse CCA cell line SB1 was a kind gift from Pr. Gores’s laboratory (Mayo Clinic, Rochester, MN; Rizvi et al., 2018; Yamada et al., 2015).
Derivation of the SB1-JP4 cell line from a CCA tumor of SB1
In vivo derivation was performed as follows: 2 × 106 of SB1 cells were resuspended in 100 µl PBS and injected s.c. in the right flank of mice anesthetized with 2.5% isoflurane. 81 d after CCA cell engraftment, the largest tumor was harvested and dissociated under sterile condition for further in vitro cell culture. Cells were cultured in vitro for several days in DMEM supplemented with FBS and penicillin/streptomycin. After three passages, to get rid of the fibroblasts, the bulk cells were stained with the following fluorochrome-conjugated antibodies: anti-CD115 APC (clone AFS98; Thermo Fisher Scientific) and anti-CD45 APC-Cy7 (clone 30-F11; BD Biosciences). Double-negative cells were single-cell sorted by flow cytometry, then expanded. One clone (i.e., SB1-JP4) was selected for both its morphological similarity with the parental cell line and its ability to grow faster than the latter in vitro. The SB1-JP4 was used as the C57Bl/6 syngeneic CCA cell line for all in vivo experiments of the present study.
In vivo cholangitis mouse models
Model of autoimmune cholangitis mimicking PBC
PBC was induced in C57Bl/6 mice as described before (Chang et al., 2014; Wakabayashi et al., 2008). Mice were immunized with 100 µg of 2OA-BSA i.p. in a mix of PBS and CFA (Sigma-Aldrich) at day 0 and were reboosted at days 14 and 28 with 2OA-BSA and IFA (Sigma-Aldrich). Concomitantly with the two first injections of 2OA-BSA, mice were also injected i.v. with 2 µg of α-GalCer (Abcam) in PBS (Fig. 1 A).
Model of nonautoimmune cholangitis mimicking PSC
PSC was induced likewise as previously published in C57Bl/6 mice (Fickert et al., 2007; Ikenaga et al., 2017). They were fed the SAFE A04 diet containing 0.089%, in weight, of DDC (Sigma-Aldrich) for 2 wk (from day 0 to 14). After 1 wk of recovery with the regular diet, mice were fed again with the DDC-supplemented diet for an additional 1 wk (from day 21 to 28; Fig. 1 B).
Ex vivo serum analysis
Approximately 100 µl of blood was drawn from mice at day 35/36 after cholangitis induction started. After blood clotting and centrifugation, serum was collected for further analysis.
Alanine aminotransferase (ALT) activity
ALT enzymatic activity was measured by an ALT activity assay (Sigma-Aldrich) in 10 µl of mouse serum according to the supplier’s instructions.
Circulating IgG concentration
IgG concentration in serum was measured by a mouse IgG ELISA development kit (Mabtech), according to the supplier’s instructions, in MULTISORP Nunc plate (Dominique Dutscher) on 100 µl of serum diluted at 1:200,000.
Concentration of circulating IgG/M targeting PDC-E2
Anti–PDC-E2 concentration in serum was measured by a sandwich ELISA as follows: recombinant mouse PDC-E2 proteins (Abcam) were coated overnight at 4°C at a concentration of 1 µg/ml in a solution of NaCl 0.9% at pH 9.4 (Sigma-Aldrich) on a MaxiSorp Nunc Plate (Dominique Dutscher). 100 µl of mouse serum, diluted at 1:10 in incubation buffer (PBS with 0.05% Tween20 [Euromedex] and 0.1% BSA [Euromedex]) was then incubated for 2 h at room temperature. Finally, an alkaline phosphatase–conjugated goat anti-mouse IgG/M (Mabtech), diluted at 1:1,000 in incubation buffer, was added for 1 h at room temperature. The ELISA was revealed with 100 µl of the p-nitrophenyl-phosphate substrate solution (Mabtech), and the plate absorbance was read at 410 nm.
In vivo tumor growth experiment
Ectopic syngeneic tumor models
0.3 × 106 MCA205 cells, 0.3 × 106 of TC-1, 106 of Hep-55.1C, 3 × 106 SB1, or 3 × 106 SB1-JP4 cells were resuspended in 100 µl of PBS and injected s.c. in the right flank of mice under anesthesia with 2.5% isoflurane at 35 d after the beginning of cholangitis induction. Tumor growth was monitored via repeated measurements of the tumor size using a digital caliper. Tumor volume was calculated using the following formula: tumor size (mm3) = (length × width × height)/8 × 4/3 × pi. Mouse survival was monitored daily. Tumor size exceeding 2,000 mm3, tumor ulceration, weight loss superior to 20% compared with the beginning of the treatment, and poor body condition were considered as endpoints.
Oncogene-induced CCA model
iCCA was induced in mice through delivery to the liver of plasmids encoding the oncogenes AKT (constitutively active myristoylated AKT) and NICD, accompanied by the Sleeping Beauty transposase for oncogene integration into the cell host genome, as previously described (Fan et al., 2012). Mice received, via hydrodynamic injection in the tail vein, 2 ml of saline solution (0.9% NaCl) containing a mixture of the three plasmids (p): 20 µg of pNICD, 4 µg of pAKT, and 1 µg of pSleeping Beauty transposase. NICD and AKT were expressed under the EF1α promoter and the transposase under the CMV promoter. The plasmids were kindly provided by Dr. Diego Calvisi (University of Greifswald, Greifswald, Germany). Mouse livers were collected 5 wk after plasmid injection, before control mice became moribund.
In vivo lymphocyte depletion
CD4+ and/or CD8+ T cell depletion
T cell depletion was performed by i.p. injections of 100 µg of rat IgG2 antibodies targeting CD4 (clone GK1.5; BioXCell) and/or CD8 (clone 2.43; BioXCell). Injections were achieved 2–3 d before and 1 d after CCA cell engraftment, then repeated once a week throughout the whole course of the experiment to maintain T lymphocyte depletion.
B cell depletion
B cell depletion was performed by injecting 20 µg of rat IgG2 antibodies targeting CD20 (clone SA271G2; BioLegend). Injections were achieved 2–3 d before and 1 d after CCA cell engraftment, then repeated throughout the whole course of the experiment once a week to maintain B lymphocyte depletion. Isotype control (clone LTF-2; BioXCell) was delivered i.p. at the corresponding dose and time point. Peripheral T and B cell depletions were checked by flow cytometry on mouse blood after the fourth injection. Quantification of CD4+ and CD8+ T (CD3+CD4+ or CD3+CD8+) and B (CD19+B220+) lymphocytes showed a 98%, 97%, and 60% reduction of these cell subsets in comparison to healthy controls, respectively (Fig. S5 A).
Figure S5.
Flow cytometry analyses. (A) Flow cytometry plots of peripheral blood cells from control mice or PBC mice injected with anti–CD4/CD8 or anti-CD20 to deplete either CD4+ and CD8+ T cells or B cells, respectively. Following the experimental schedule illustrated in Fig. 4 A, PBC mice were repeatedly injected with the depleting antibodies. After the fourth injection of antibodies, blood was drawn for quantification of CD4+ and CD8+ T (CD3+ CD4+ or CD3+ CD8+) and B (CD19+ B220+) lymphocytes. (B–E) Gating strategies following the use of the four panels of fluorescent antibodies: “Cytokine production by lymphocytes” (B); “T lymphocytes” (C); “NK and B lymphocytes” (D); and “Lymphocytic subsets” (E). FSC-A, forward scatter-A; SSC-A, side scatter-A.
In vivo cytokine neutralization
IFNγ, IL4, or IL17a neutralization was performed by i.p. injections of 100 µg of rat IgG1 antibodies targeting IFNγ (clone R4-6A2; BioXCell), IL4 (clone 11B11; BioXcell), or IL17a (clone 17F3; BioXCell) at the following time points: 3 d and 1 d before CCA cell engraftment, then 1 d after, then repeated three times a week throughout the whole course of the experiment. Isotype control (clone HRPN; BioXCell) was delivered i.p. at the same dose and time points.
Ex vivo mouse tissue sampling and histological staining
Mouse caudate liver lobes were resected on day 35 after cholangitis induction start, or mouse tumors were collected at day 56 after CCA engraftment. Tissues were fixed in 4% paraformaldehyde (Euromedex). Samples were then dehydrated (successive baths of alcohol 70%, alcohol 100%, and isopropanol) and embedded in paraffin with LOGOS (dehydration/clearing processor; MM France). 4-μm-thick paraffin sections were performed with microtome and dried at 37°C until the next day. Hematoxylin, eosin, safran or Sirius red staining was performed on organs to study their morphology with a standard staining automate (Tech Inter).
Detection of cleaved CD8
The staining was performed using the Leïca Bond automated immunostainer instrument. Paraffin sections were processed for heat-induced antigen retrieval (ER2 corresponding EDTA buffer, pH 9) for 20 min at 100°C. Slides were incubated with the antibody (CD8 clone D4W2Z #98941, 1:400; CD11c clone D1V9Y #97585, 1:300) for 1 h at room temperature. Then, the slides were incubated with the Rabbit HRP PowerVision Kit (#PV6119; Leïca Biosystems). The signal was revealed with 3,3′-diaminobenzidine. Finally, the sections were counterstained by hematoxylin (Leïca Biosystems).
Detection of double markers CD4/FoxP3 simultaneously
The staining was performed using the Leïca Bond automated immunostainer instrument. Antigen retrieval was performed by incubating slides in ER2 buffer (pH 9.0) for 20 min at 100°C. Then, the antibodies were successively incubated for 1 h at room temperature and detected respectively by Bond Polymer Refine Red Detection (#DS9390; Leïca Biosystems) and by Bond Polymer Refine Detection kit (#DS9800; Leïca Biosystems). These slides were successively revealed by Red chromogen (Leïca Biosystems) and HIGHDEF Black HRP chromogen/substrate (#ADI-950-171-0030; Enzo Lifesciences). Finally, the sections were counterstained by hematoxylin (Leïca Biosystems).
Slide scanning
Each slide was scanned using the slide scanner Zeiss Axio Scan Z1 and visualized using the QuPath-0.2.3 software.
In silico histological image analysis
Image-based quantification of immune cells
Micrographs were analyzed using R software (https://www.r-project.org/) by means of EBImage (https://www.bioconductor.org/), RbioFormats (https://github.com/aoles/RbioFormats), and RniftyReg (CRAN) packages. After acquisition, a primary region of interest (ROI) was automatically defined by detecting tissue contours using the Otsu’s thresholding method on the grayscale-converted image. Each ROI was semi-automatically refined by removing necrotic and adipose regions, and its surface was computed. Cell nuclei were detected and numbered by using the red component of the hematoxylin, eosin, safran image; briefly, the image channel was converted into grayscale, normalized following a sigmoidal transformation, and thresholded to generate a nuclei mask, thereafter cleaned by morphological operations. Immune cells were detected and numbered using a similar approach, selecting the right image component according to the staining method. Finally, each population density was calculated as the cell count-to-tissue surface ratio.
Image-based quantification of collagen proportionate area
Micrographs (data not shown) were analyzed following similar procedure as for quantifying immune cells. Collagen ROI was defined as follows: each image was converted into grayscale, and a top hat filter was applied to enhance local contrast; the resulting image was thereafter normalized by a sigmoidal transformation, on which a threshold was finally applied. Collagen proportionate area was calculated by computing the ratio between collagen and total tissue surfaces.
Ex vivo tumor, blood, and lymph node processing
Blood sample processing
Approximately 300 µl of blood was drawn from mice and collected in a heparin-containing tube. Erythrocytes were removed using RBC lysis buffer (BioLegend), and the remaining cells were washed with PBS. Peripheral blood cells were resuspended at a concentration corresponding to 500 µl of the initial blood volume per milliliter of PBS.
Tumor processing
Tumors were resected, weighed, and transferred on ice in gentleMACS C tubes (Miltenyi Biotec) containing 1 ml of RPMI medium. Tumors were dissociated mechanically with scissors, then enzymatically using a mouse tumor dissociation kit (Miltenyi Biotec) and gentleMACS Octo Dissociator (Miltenyi Biotec) following the manufacturer’s instructions. Tumor homogenates were filtered through 70-µM MACS SmartStrainers (Miltenyi Biotec) and washed with PBS. Finally, bulk tumor cells were homogenized in PBS at a concentration corresponding to 250 mg of the initial tumor weight per milliliter.
Tumor-draining lymph node processing
The inguinal lymph node draining the s.c. CCA tumor was harvested and transferred on ice in a micro-tube containing RPMI. Lymph nodes were dissociated mechanically, filtered through 70-µM MACS SmartStrainers (Miltenyi Biotec), and washed with PBS. For each draining lymph node, bulk cells were homogenized in 400 µl of PBS.
Ex vivo unspecific restimulation of leukocytes
At day 56 after CCA cell injection, mouse tumor-draining lymph nodes were harvested and processed. In a 96-well cell culture plate, 11.5 × 104 leukocytes (equivalent to ∼8 × 104 T cells) were restimulated with 8 × 104 CD3/CD28–coated beads (Dynabeads Mouse T-Activator CD3/CD28; Thermo Fisher Scientific; T cells/beads ratio of 1:1) in 200 µl of serum-free CTL-Test PLUS Medium (ImmunoSpot) supplemented with 2 mM L-glutamine for 24 h. After 20 h, brefeldin A (BD GolgiPlug; BD Biosciences) was added to the medium at a 1:100 dilution for an additional 4 h. The total amount of cells was harvested after a 24-h incubation, stained for analysis of cytokine production with the panel “Cytokine production by lymphocytes” (see Flow cytometry section), and analyzed by flow cytometry as described below in the Flow cytometry section.
Ex vivo phenotyping of the lymphocytic cell subsets in the tumor, tumor-draining lymph node, and blood
At day 56 or 60 after CCA cell injection, blood samples, ectopic tumors, and tumor-draining lymph nodes (inguinal) were collected from mice and processed. 200 µl containing the equivalent of 50 mg of bulk tumor cells, or peripheral blood cells from 400 µl of blood, or leukocytes from half of a whole lymph node were stained for immunophenotyping with the panels “T lymphocytes,” or “NK and B lymphocytes,” or “Lymphocytic subsets” (see Flow cytometry section) and analyzed by flow cytometry as described in the Flow cytometry section. Absolute counts of leukocytes were normalized considering the following parameters: the weight of the harvested tumor or amount of blood drawn, the total volume of the processed tissue cell suspension, the proportion of the whole cell suspension stained, and the proportion of the stained cell suspension run through the flow cytometer.
Flow cytometry
Cell staining was performed as follows: cell viability was assessed with LIVE/DEAD Fixable Yellow dye (Thermo Fisher Scientific). Fc receptors were blocked with anti-mouse CD16/CD32 (BD Biosciences; clone 2.4G2, Mouse BD Fc Block) before surface staining with fluorescent antibodies (see below). Then, cells were fixed and permeabilized either with BD Cytofix/Cytoperm buffer (BD Biosciences) or with eBioscience Foxp3/Transcription Factor Staining Buffer (Thermo Fisher Scientific) when intranuclear staining of the transcription factor FoxP3 was required. Next, intracytoplasmic and intranuclear markers (i.e., FoxP3, GzmB, IFNγ, IL4, and IL17a) were stained with additional fluorescent antibodies. Four panels of fluorescent antibodies were applied for phenotyping immune cells. Panel 1 “Cytokine production by lymphocytes” consisted of anti–CD3-BV421 (145-2C11), anti–CD4-BUV496 (GK1.5), anti–CD8-PE (53–6.7), anti–CD25-BV786 (PC61), anti–CD45-BUV661 (30F11), anti–CD69-PE-Vio770 (REA937), anti–FoxP3-FITC (FJK-16s), anti–GzmB-PE-Dazzle594 (QA16A02), anti–IFNγ-APC (XMG1.2), anti–IL4-PerCP-Cy5.5 (11B11), anti–IL17a-APC-Cy7 (TCII-18H10), anti–NK1.1-BV605 (PK136), and anti–TCRγδ-BV711 (GL3). Panel 2 “T lymphocytes” consisted of anti–CD3-APC (17A2), anti–CD4-PerCP-Cy5.5 (RM4-5), anti–CD8-PE (53–6.7), anti–CD25-PE-Cy7 (PC61.5), anti–CD62L-APC-eFluor780 (MEL-14), anti–FoxP3 FITC (FJK-16s), and anti–PD-1-BV421 (RMP1-30). Panel 3 “NK and B lymphocytes” consisted of anti–B220-V450 (RA3-6B2), anti–CD3-FITC (17A2), anti–CD19-APC-H7 (ID3), anti–CD69-PE (H1-2F3), anti–NK1.1-PerCP-Cy5.5 (PK136), and anti–TCRγδ-APC (GL3). Panel 4 “Lymphocytic subsets” consisted of anti–B220-APC-Vio770 (REA755), anti–CD3-BV421 (145-2C11), anti–CD4-BUV496 (GK1.5), anti–CD8-PE (53–6.7), anti–CD19-PerCP-Cy5.5 (1D3), anti–CD25-BV786 (PC61), anti–CD27-PE-Dazzle594 (LG.3A10), anti–CD44-APC (IM7), anti–CD62L-PE-Vio770 (REA828), anti–CD45-BUV661 (30F11), anti–FoxP3-FITC (FJK-16s), anti–NK1.1-BV605 (PK136), and anti–TCRγδ-BV711 (GL3). Antibodies were purchased from BD Biosciences, Miltenyi Biotec, Thermo Fisher Scientific, and BioLegend. Finally, stained samples were run through a BD LSRFortessa X20 or BD LSR II flow cytometer (BD Biosciences). Data were acquired using BD FACSDiva 6.1.3 software (BD Biosciences) and analyzed using FlowJo 10.5.3 software (TreeStar, Inc.) according to the gating strategies displayed in Fig. S5, B–E.
Ex vivo RNA extraction
Mouse caudate liver lobes were harvested at day 35 after cholangitis induction start, and CCA mouse tumors were resected at day 56 after CCA cell injection. Both tissues were stored directly in RNAprotect Tissue Reagent solution (Qiagen) and stored at 4°C. Total RNA extraction and purification were performed using the RNeasy plus mini kit (Qiagen) and the Qiashredder homogenizer columns (Qiagen) according to the supplier’s instructions. Lysis and homogenization of the tissues were formerly achieved thanks to the Precellys 24 Tissue Homogenizer (Bertin Instruments) and hard tissue homogenizing CK28-R 2-ml tubes (Bertin Instruments) in the lysis buffer supplied in the RNeasy plus mini kit, at 5,000 rpm for 20 s (twice). RNA concentration in samples was then measured using a NanoDrop 1000 (Thermo Fisher Scientific).
In vitro RT-qPCR
Reverse transcription of RNA samples was performed using the SuperScript IV VILO Master Mix (Thermo Fisher Scientific) according to the manufacturer’s instructions. 1.8 mg of purified total RNA was converted into cDNA in 20 µl and diluted six times. qPCR was performed on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) using TaqMan Fast Advanced Master Mix and TaqMan Gene Expression Assays for the following murine genes according to the manufacturer’s instructions (Thermo Fisher Scientific): Ccl20 (Mn01268754_m1), Ccl22 (Mn00436439_m1), Cxcl9 (Mn00434946_m1), Cxcl13 (Mn00444534_m1), Cxcr5 (Mn00432086_m1), Cd8a (Mn01182107_g1), Cd274 (Mn00452054_m1), Foxp3 (Mn00475162_m1), Gzmb (Mn00442837_m1), Grp18 (Mn02620895_s1), Ifng (Mn01168134_m1), Il4 (Mn00445259_m1), Il6 (Mn00446190_m1), Il10 (Mn01288386_m1), Il17a (Mn00439619_m1), Il22 (Mn00444241_m1), Il23a (Mn00518984_m1), Socs1 (Mn00782550_s1), and Ubp1 (Mn00496724_m1). Expression results were normalized to Ppia signal (ΔCt) and to the expression mean of the control group (−ΔΔCt).
In vitro RNA-seq
Mouse caudate liver lobes were harvested on day 35 after cholangitis induction start, and total RNA was extracted. The libraries were prepared following the TruSeq mRNA protocol from Illumina, starting from 1 µg of high-quality total RNA. Paired end (2 × 75) sequencing was performed on an Illumina Nextseq 500 platform. Fluorometric Qubit RNA HS assay (Life Technologies) was applied to measure RNA concentration. RNA quality (RNA integrity number [RIN]) was assessed on the Agilent 2100 Bioanalyzer (Agilent Technologies). To build libraries, 1 µg of high-quality total RNA sample (RIN >8) was processed using TruSeq stranded mRNA kit (Illumina) according to the manufacturer’s instructions. Briefly, after purification of poly-A containing mRNA molecules, nucleic acids were fragmented and reverse-transcribed using random primers. Replacement of dTTP by dUTP during the second-strand synthesis allowed strand specificity to be achieved. Addition of a single A base to the cDNA was followed by ligation of adapters. Libraries were quantified by Qubit DNA HS Assay (Life Technologies), and library profiles were assessed using the DNA High Sensitivity LabChip kit on an Agilent Bioanalyzer. Libraries were sequenced on an Illumina Nextseq 500 instrument using 75–base-length read V2 chemistry in a paired-end mode. After sequencing, a primary analysis based on AOZAN software (ENS) was applied to demultiplex and control the quality of the raw data (based of FastQC modules/version 0.11.5; Perrin et al., 2017). Obtained fastq files were then aligned using Star algorithm (version 2.5.2b), and quality control of the alignment was realized with Picard tools (version 2.8.1). Reads were then counted using Feature count (version Rsubread 1.24.1), and the statistical analyses of the read counts were performed with the DESeq2 package version 1.14.1 to determine the proportion of differentially expressed genes between two samples.
In silico RNA-seq data analysis
RNA-seq analysis
RNA-seq data analysis was performed by GenoSplice technology (http://www.genosplice.com). Sequencing, data quality, reads repartition (e.g., for potential ribosomal contamination), and insert size estimation were performed using FastQC, Picard-Tools, Samtools, and rseqc. Reads were mapped using STARv2.4.0 (Dobin et al., 2013) on the mm10 Mouse genome assembly. Gene expression regulation study was performed as already described (Gacem et al., 2020; Romagnoli et al., 2020). Briefly, for each gene present in the Mouse FAST DB v2018_1 annotations, reads aligning on constitutive regions (which are not prone to alternative splicing) were counted. Based on these read counts, normalization and differential gene expression were performed using DESeq2 on R (v.3.2.5). Only genes expressed in at least one of the six compared experimental conditions were further analyzed. Genes were considered expressed if their fragments per kilobase of transcript per million fragments mapped (FPKM) value was >98% of the background FPKM value based on intergenic regions. Results were considered statistically significant for P values ≤0.05 and fold-changes ≥1.5. Significant GO terms and Kyoto Encyclopedia of Genes and Genomes pathways (Kanehisa et al., 2012) were retrieved using DAVID (Huang et al., 2009b).
Heatmap for the results visualization
Results were considered statistically significant for uncorrected P values ≤0.05 and fold-changes ≥2. Hierarchical Euclidean clustering and heatmaps were performed using Morpheus (https://software.broadinstitute.org/morpheus).
GO terms enrichment analysis
GO terms enrichment was analyzed using the DAVID Functional Annotation tool (https://david.ncifcrf.gov/home.jsp) on genes that were up-regulated in both conditions (PBC and PSC), or up-regulated in only one, or down-regulated for both cholangitis or only one (Huang et al., 2009a, 2009b).
Ex vivo T cell preparation for single-cell sequencing
Blood preparation
A soft RBC lysis protocol was used to preserve cell fitness and viability. Briefly, 500 µl PBS was added to blood, followed by 3.5 ml sterile water. After mixing, 400 µl PBS 10× was included before centrifugation. Cells were resuspended in PBS containing 0.5% BSA and 5 mM EDTA.
Liver preparation
Perfused livers were cut into small pieces and transferred to GentleMACS C tubes (Miltenyi Biotech) containing 3 ml of CO2 independent medium. After mechanical dissociation using GentleMACS Octo Dissociator (Miltenyi Biotech), the cell suspension was transferred into 50-ml tubes for extensive washing. Finally, the cell pellet was resuspended in PBS containing 0.5% BSA and 5 mM EDTA.
Tumor preparation
Tumors were cut into small pieces and placed into GentleMACS C tubes. Then, a cocktail of tissue-dissociating enzymes resuspended in 2.35 ml of RPMI medium was added (Mouse tumor dissociation kit; Miltenyi Biotech). GentleMACS C tubes containing the tissue were placed and incubated at 37°C onto a GentleMACS Octo Dissociator equipped with heaters (Miltenyi Biotech) for dissociation to occur. Next, cells were filtered through a 100-µm cell strainer and washed twice in PBS. Finally, the cell pellet was resuspended in PBS containing 0.5% BSA and 5 mM EDTA.
T cell sorting
CD4+ and CD8+ T cells from blood, liver, and tumor were sort-purified in a two-step procedure in which T cells were initially enriched in CD3+ cells using the MoJoSort mouse negative selection kit (BioLegend). Then, CD3+ cells were stained with a cocktail of antibodies targeting CD11c, Ly6C, Ly6G, CD19, F4/80, NK1.1 (all in FITC for Lineage negative exclusion), CD45, CD8, and CD4, together with DAPI, then sorted using BD FACSAria III (BD Biosciences) as DAPI−, Lineage−, CD45+, CD4+, or CD8+ cells. After sorting, cells were counted, and then CD4+ and CD8+ cells were mixed at 1:1 ratio. Final concentration was adjusted to 800 cells/µl (400 CD4+:400 CD8+ cells/µl).
In vitro scRNA-seq
scRNA-seq was performed on the 1:1 CD4+:CD8+ T cell mixture prepared from blood, tumor, or liver. A total of 10,000 cells per tissue were loaded onto the NextGEM 10× chip (10× Genomics). The RNA capture, barcoding, cDNA amplification, and library were performed according to the manufacturer’s recommendations (CG00207_Chronium NextGEM SingleCellV(D)J v1.1 user guide). 10× libraries were sequenced on NovaSeq (Illumina) SP (for TCR-seq) or S1 (for gene expression).
In silico scRNA-seq data analysis
Single-cell 5′ and V(D)J data processing
Single-cell 5′ and V(D)J data processing was performed by GenoSplice technology (http://www.genosplice.com). Assessment of the quality of the sequencing data was done using FastQC v0.11.2 on each single-cell 5′ and V(D)J sample. The analysis pipelines in Cell Ranger (10× Genomics, version 5.0.0) were used for single-cell sequencing data processing. V(D)J sequence assembly and paired clonotype calling were performed using cellranger vdj function, with reference refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0 for each sample and cellranger aggr function to integrate results. Read alignment and unique molecular identifier (UMI) quantifications were performed using cellranger count function with reference refdata-gex-mm10-2020-A.
Gene expression preprocessing
The two expression matrices containing the UMI counts were merged, and only the genes with UMI ≥ 1 in at least one cell were kept. The following filters were applied to generate a global matrix used in further analysis: cells with UMI ≥ 1,600, number of detected genes ≥700, and cells with UMI in mitochondrial genes ≤10%. For UMI normalization, Seurat 3.1.1 was used (Butler et al., 2018), and the global-scaling normalization method was applied with a scale factor of 10,000 and log-transformation of data. This was followed by a scaling linear transformation step.
Clustering and marker genes
Principal component analysis was performed on the scaled data, with a Jackstraw plot to choose how many principal components to retain as an input for Seurat clustering step. Clustering step was performed using default parameters, the Louvain algorithm as the clustering method, 11 principal components, and a resolution parameter defining the clusters’ granularity set to 0.6. Marker genes defining each cluster were found via differential expression testing, with a Wilcoxon rank sum test and a log fold-change threshold of 1.
Visualization of data
Single-cell 5′ and V(D)J were integrated in Seurat object. Functions vlnPlot, DimPlot, and DotPlot were used for data visualization.
Heatmap for gene expression visualization
Z-scored expression was calculated as follows: . Hierarchical Euclidean clustering and heatmaps have been performed using Morpheus (https://software.broadinstitute.org/morpheus).
In silico statistical analyses and data graphing
Linear mixed-effects models were applied for longitudinal comparison of tumor growth curves using TumGrowth web tool (https://kroemerlab.shinyapps.io/TumGrowth/; Enot et al., 2018). Other statistical analyses were performed using the software GraphPad Prism (v8) or R (v3.6.3). Tests performed are indicated in the figure legends and consisted of paired Student t test, Kruskal–Wallis H test with Dunn’s pairwise multiple comparisons, one-way or two-way ANOVA with Tukey’s pairwise multiple comparison, Mann-Whitney U test, or Fisher’s exact test. In the figures, statistically significant P values (P < 0.05) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001. Exact P values are displayed in the legend when 0.13 > P ≥ 0.05 to illustrate a trend toward significance. Outlier values were detected with the ROUT test. R or GraphPad Prism software was used for data graphing. Spider plots were generated by means of ggradar (https://github.com/ricardo-bion/ggradar) package.
Online supplemental material
Fig. S1 illustrates that PBC, but not PSC, prevents specifically the outgrowth of ectopic CCA. Fig. S2 shows the relative expression of some genes related to Tc1 immune response in the liver of mice affected with PBC and PSC. Fig. S3 displays CD8+ T cell polyfunctionality within CCA tumor-draining lymph node during cholangitis. Fig. S4 documents the lymphocytic cell subsets across several tissues during cholangitis. Fig. S5 depicts the gating strategies applied for flow cytometry phenotyping. Table S1 lists the individual and mean expression values of each gene differentially expressed in the liver of mice with PBC and PSC. Table S2 and Table S3 detail the characteristics of the enriched TCR clonotypes and T cell phenotypic clusters in the malignant and hepatic tissues of a PBC mouse. Data S1 is a detailed version of the heatmap displayed in Fig. 2 A and built from gene expression values listed in Table S1.
Supplementary Material
shows the overall profile of differentially expressed genes in the liver of mice affected with PBC and PSC.
shows enriched TCR clonotypes shared between the malignant and hepatic tissues of a PBC mouse and lists the 25 TCR clonotypes enriched in both tumor and liver of a PBC mouse.
shows characteristics of the T cell clusters in the malignant and hepatic tissues of a PBC mouse and expression level of immune markers related to T cell function across the 14 clusters of T lymphocytes infiltrated in the tumor and liver of a PBC mouse.
is a detailed version of the heatmap displayed in Fig. 2 A and built from gene expression values listed in Table S1.
Acknowledgments
We would like to thank Estelle Devêvre and Hélène Forher-Ting (Centre d'Histologie, d'Imagerie et de Cytométrie core facility, Cordeliers Research Center, Institut National de la Santé et de la Recherche Médicale [INSERM] U1138, Paris) and the staff of the histo-cytopathology core facility of the Gustave Roussy cancer campus (Villejuif, France), as well as the staff of the Genom'IC platform of the Cochin Institute (INSERM U1016, Paris) for their assistance in flow cytometry, histology, and transcriptomic analyses, respectively. We also thank Pr. Raoul Poupon for having supported this project since the very beginning.
J. Paillet is supported by the Association pour la lutte contre les maladies inflammatoires du foie et des voies biliaires. S. Gujar is supported by Dalhousie Medical Research Foundation, Canadian Cancer Society, and Canadian Institutes of Health Research. J.G. Pol is supported by the Association Française d'Hépatologie; Site de Recherche intégrée sur le Cancer Cancer Research and Personalized Medicine; Seerave Foundation; Multi-Organism Institute (ITMO) Aviesan Cancer (National Alliance for Life Sciences and Health); and Institut National du Cancer. G. Kroemer is supported by the Ligue contre le Cancer (équipe labellisée); Agence National de la Recherche (ANR) – Projets blancs; Agence National de la Recherche under the frame of E-Rare-2, the ERA-Net for Research on Rare Diseases; Association pour la recherche sur le cancer; Cancéropôle Ile-de-France; Chancellerie des universités de Paris (Legs Poix), Fondation pour la Recherche Médicale; a donation by Elior; European Research Area Network on Cardiovascular Diseases (MINOTAUR); Gustave Roussy Odyssea, the European Union Horizon 2020 Project Oncobiome; Fondation Carrefour; High-end Foreign Expert Program in China (GDW20171100085 and GDW20181100051), Institut National du Cancer; Inserm Program Heterogeneity of Tumors & Ecosystem; Institut Universitaire de France; LeDucq Foundation; the LabEx Immuno-Oncology; the Recherche Hospitalo-Universitaire Torino Lumière; the Seerave Foundation; the Site de Recherche intégrée sur le Cancer Stratified Oncology Cell DNA Repair and Tumor Immune Elimination; and Cancer Research and Personalized Medicine.
Author contributions: J. Paillet performed the experiments with support from C. Plantureux, S. Lévesque, J. Le Naour, G. Stoll, P. Caudana, J. Tosello Boari, N. Bloy, S. Lachkar, I. Martins, A. Checcoli, G. Autret, B. Lekbaby, J. Augustin, Y. Kim, and J.G. Pol. P. Opolon and A. Sauvat performed anatomopathological analyses. A. Delaune, N. Robil, P. de la Grange, J. Hamroune, and F. Letourneur contributed to transcriptomic data analyses. P.S.C. Leung, M.E. Gershwin, J.S. Zhu, and M.J. Kurth produced 2OA-BSA. S. Gujar, C. Coulouarn, L. Fouassier, L. Zitvogel, E. Piaggio, C. Housset, P. Soussan, M. Chiara Maiuri provided critical input to the project and edited the manuscript. G. Kroemer and J.G. Pol conceived and directed the project. G. Kroemer and J.G. Pol wrote the manuscript with input by J. Paillet.
Data availability
Raw transcriptome data reported in this paper are deposited and available from the Gene Expression Omnibus under accession no. GSE180289; GSE179993 for the RNA-Seq dataset (Fig. 2 and Fig. 5 A), and GSE180288 for the scTCR-seq/scRNA-seq dataset (Fig. 8 and Fig. 9). Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors.
References
- Akisawa, N., Maeda T., Tsuda K., Nishimori I., Morita M., Iwasaki S., Tomita A., Saibara T., Onishi S., Kiyoku Y., and Enzan H.. 1998. Primary biliary cirrhosis associated with cholangiocarcinoma. Dig. Dis. Sci. 43:2138–2142. 10.1023/A:1018831903371 [DOI] [PubMed] [Google Scholar]
- Bae, H.R., Leung P.S., Tsuneyama K., Valencia J.C., Hodge D.L., Kim S., Back T., Karwan M., Merchant A.S., Baba N., et al. 2016. Chronic expression of interferon-gamma leads to murine autoimmune cholangitis with a female predominance. Hepatology. 64:1189–1201. 10.1002/hep.28641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banales, J.M., Huebert R.C., Karlsen T., Strazzabosco M., LaRusso N.F., and Gores G.J.. 2019. Cholangiocyte pathobiology. Nat. Rev. Gastroenterol. Hepatol. 16:269–281. 10.1038/s41575-019-0125-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banales, J.M., Marin J.J.G., Lamarca A., Rodrigues P.M., Khan S.A., Roberts L.R., Cardinale V., Carpino G., Andersen J.B., Braconi C., et al. 2020. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 17:557–588. 10.1038/s41575-020-0310-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blechacz, B., and Gores G.J.. 2008. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 48:308–321. 10.1002/hep.22310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohner, P., Chevalier M.F., Cesson V., Rodrigues-Dias S.C., Dartiguenave F., Burruni R., Tawadros T., Valerio M., Lucca I., Nardelli-Haefliger D., et al. 2019. Double Positive CD4+CD8+ T Cells Are Enriched in Urological Cancers and Favor T Helper-2 Polarization. Front. Immunol. 10:622. 10.3389/fimmu.2019.00622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, A., Pagani A., Pulze L., Albini A., Dallaglio K., Noonan D.M., and Mortara L.. 2014. Orchestration of angiogenesis by immune cells. Front. Oncol. 4:131. 10.3389/fonc.2014.00131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A., Hoffman P., Smibert P., Papalexi E., and Satija R.. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36:411–420. 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, K.T., Côté A.L., Zhang P., Steinberg S.M., Guo Y., Allie R., Zhang W., Ernstoff M.S., Usherwood E.J., and Turk M.J.. 2011. Autoimmune melanocyte destruction is required for robust CD8+ memory T cell responses to mouse melanoma. J. Clin. Invest. 121:1797–1809. 10.1172/JCI44849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, F., Cardoso A.P., Gonçalves R.M., Serre K., and Oliveira M.J.. 2018. Interferon-Gamma at the Crossroads of Tumor Immune Surveillance or Evasion. Front. Immunol. 9:847. 10.3389/fimmu.2018.00847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerwenka, A., Morgan T.M., Harmsen A.G., and Dutton R.W.. 1999. Migration kinetics and final destination of type 1 and type 2 CD8 effector cells predict protection against pulmonary virus infection. J. Exp. Med. 189:423–434. 10.1084/jem.189.2.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, C.H., Chen Y.C., Yu Y.H., Tao M.H., Leung P.S., Ansari A.A., Gershwin M.E., and Chuang Y.H.. 2014. Innate immunity drives xenobiotic-induced murine autoimmune cholangitis. Clin. Exp. Immunol. 177:373–380. 10.1111/cei.12298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, Y.H., Lian Z.X., Cheng C.M., Lan R.Y., Yang G.X., Moritoki Y., Chiang B.L., Ansari A.A., Tsuneyama K., Coppel R.L., and Gershwin M.E.. 2005. Increased levels of chemokine receptor CXCR3 and chemokines IP-10 and MIG in patients with primary biliary cirrhosis and their first degree relatives. J. Autoimmun. 25:126–132. 10.1016/j.jaut.2005.08.009 [DOI] [PubMed] [Google Scholar]
- Concepcion, A.R., and Medina J.F.. 2015. Mouse models of primary biliary cirrhosis. Curr. Pharm. Des. 21:2401–2413. 10.2174/1381612821666150316121622 [DOI] [PubMed] [Google Scholar]
- Corthay, A., Skovseth D.K., Lundin K.U., Røsjø E., Omholt H., Hofgaard P.O., Haraldsen G., and Bogen B.. 2005. Primary antitumor immune response mediated by CD4+ T cells. Immunity. 22:371–383. 10.1016/j.immuni.2005.02.003 [DOI] [PubMed] [Google Scholar]
- Deng, Q., Luo Y., Chang C., Wu H., Ding Y., and Xiao R.. 2019. The Emerging Epigenetic Role of CD8+T Cells in Autoimmune Diseases: A Systematic Review. Front. Immunol. 10:856. 10.3389/fimmu.2019.00856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn, G.P., Koebel C.M., and Schreiber R.D.. 2006. Interferons, immunity and cancer immunoediting. Nat. Rev. Immunol. 6:836–848. 10.1038/nri1961 [DOI] [PubMed] [Google Scholar]
- Dyson, J.K., Beuers U., Jones D.E.J., Lohse A.W., and Hudson M.. 2018. Primary sclerosing cholangitis. Lancet. 391:2547–2559. 10.1016/S0140-6736(18)30300-3 [DOI] [PubMed] [Google Scholar]
- El-Serag, H.B. 2012. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 142:1264–1273.e1. 10.1053/j.gastro.2011.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enot, D.P., Vacchelli E., Jacquelot N., Zitvogel L., and Kroemer G.. 2018. TumGrowth: An open-access web tool for the statistical analysis of tumor growth curves. OncoImmunology. 7:e1462431. 10.1080/2162402X.2018.1462431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Failla, C.M., Carbone M.L., Fortes C., Pagnanelli G., and D’Atri S.. 2019. Melanoma and Vitiligo: In Good Company. Int. J. Mol. Sci. 20:5731. 10.3390/ijms20225731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, B., Malato Y., Calvisi D.F., Naqvi S., Razumilava N., Ribback S., Gores G.J., Dombrowski F., Evert M., Chen X., and Willenbring H.. 2012. Cholangiocarcinomas can originate from hepatocytes in mice. J. Clin. Invest. 122:2911–2915. 10.1172/JCI63212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickert, P., Stöger U., Fuchsbichler A., Moustafa T., Marschall H.U., Weiglein A.H., Tsybrovskyy O., Jaeschke H., Zatloukal K., Denk H., and Trauner M.. 2007. A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am. J. Pathol. 171:525–536. 10.2353/ajpath.2007.061133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fickert, P., Pollheimer M.J., Beuers U., Lackner C., Hirschfield G., Housset C., Keitel V., Schramm C., Marschall H.U., Karlsen T.H., et al. International PSC Study Group (IPSCSG) . 2014. Characterization of animal models for primary sclerosing cholangitis (PSC). J. Hepatol. 60:1290–1303. 10.1016/j.jhep.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, W.H., Dieu-Nosjean M.C., Pagès F., Cremer I., Damotte D., Sautès-Fridman C., and Galon J.. 2013. The immune microenvironment of human tumors: general significance and clinical impact. Cancer Microenviron. 6:117–122. 10.1007/s12307-012-0124-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridman, W.H., Zitvogel L., Sautès-Fridman C., and Kroemer G.. 2017. The immune contexture in cancer prognosis and treatment. Nat. Rev. Clin. Oncol. 14:717–734. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- Gacem, N., Kavo A., Zerad L., Richard L., Mathis S., Kapur R.P., Parisot M., Amiel J., Dufour S., de la Grange P., et al. 2020. ADAR1 mediated regulation of neural crest derived melanocytes and Schwann cell development. Nat. Commun. 11:198. 10.1038/s41467-019-14090-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershwin, M.E., and Mackay I.R.. 2008. The causes of primary biliary cirrhosis: Convenient and inconvenient truths. Hepatology. 47:737–745. 10.1002/hep.22042 [DOI] [PubMed] [Google Scholar]
- Goeppert, B., Frauenschuh L., Zucknick M., Stenzinger A., Andrulis M., Klauschen F., Joehrens K., Warth A., Renner M., Mehrabi A., et al. 2013. Prognostic impact of tumour-infiltrating immune cells on biliary tract cancer. Br. J. Cancer. 109:2665–2674. 10.1038/bjc.2013.610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, F.M., Gao Q., Shi G.M., Zhang X., Wang J., Jiang J.H., Wang X.Y., Shi Y.H., Ding Z.B., Fan J., and Zhou J.. 2012. Intratumoral IL-17+ cells and neutrophils show strong prognostic significance in intrahepatic cholangiocarcinoma. Ann. Surg. Oncol. 19:2506–2514. 10.1245/s10434-012-2268-8 [DOI] [PubMed] [Google Scholar]
- Gulamhusein, A.F., and Hirschfield G.M.. 2020. Primary biliary cholangitis: pathogenesis and therapeutic opportunities. Nat. Rev. Gastroenterol. Hepatol. 17:93–110. 10.1038/s41575-019-0226-7 [DOI] [PubMed] [Google Scholar]
- Hanahan, D., and Weinberg R.A.. 2011. Hallmarks of cancer: the next generation. Cell. 144:646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hemmerle, T., and Neri D.. 2014. The antibody-based targeted delivery of interleukin-4 and 12 to the tumor neovasculature eradicates tumors in three mouse models of cancer. Int. J. Cancer. 134:467–477. 10.1002/ijc.28359 [DOI] [PubMed] [Google Scholar]
- Hirschhorn-Cymerman, D., Budhu S., Kitano S., Liu C., Zhao F., Zhong H., Lesokhin A.M., Avogadri-Connors F., Yuan J., Li Y., et al. 2012. Induction of tumoricidal function in CD4+ T cells is associated with concomitant memory and terminally differentiated phenotype. J. Exp. Med. 209:2113–2126. 10.1084/jem.20120532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høgdall, D., Lewinska M., and Andersen J.B.. 2018. Desmoplastic Tumor Microenvironment and Immunotherapy in Cholangiocarcinoma. Trends Cancer. 4:239–255. 10.1016/j.trecan.2018.01.007 [DOI] [PubMed] [Google Scholar]
- Huang, W., Sherman B.T., and Lempicki R.A.. 2009a. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W., Sherman B.T., and Lempicki R.A.. 2009b. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang, Y., de Boer W.B., Adams L.A., MacQuillan G., Rossi E., Rigby P., Raftopoulos S.C., Bulsara M., and Jeffrey G.P.. 2013. Image analysis of liver collagen using sirius red is more accurate and correlates better with serum fibrosis markers than trichrome. Liver Int. 33:1249–1256. 10.1111/liv.12184 [DOI] [PubMed] [Google Scholar]
- Ikeda, Y., Kiyotani K., Yew P.Y., Sato S., Imai Y., Yamaguchi R., Miyano S., Fujiwara K., Hasegawa K., and Nakamura Y.. 2017. Clinical significance of T cell clonality and expression levels of immune-related genes in endometrial cancer. Oncol. Rep. 37:2603–2610. 10.3892/or.2017.5536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenaga, N., Peng Z.W., Vaid K.A., Liu S.B., Yoshida S., Sverdlov D.Y., Mikels-Vigdal A., Smith V., Schuppan D., and Popov Y.V.. 2017. Selective targeting of lysyl oxidase-like 2 (LOXL2) suppresses hepatic fibrosis progression and accelerates its reversal. Gut. 66:1697–1708. 10.1136/gutjnl-2016-312473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer, A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244. 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Kalathil, S.G., Hutson A., Barbi J., Iyer R., and Thanavala Y.. 2019. Augmentation of IFN-γ+ CD8+ T cell responses correlates with survival of HCC patients on sorafenib therapy. JCI Insight. 4:e130116. 10.1172/jci.insight.130116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallies, A., Zehn D., and Utzschneider D.T.. 2020. Precursor exhausted T cells: key to successful immunotherapy? Nat. Rev. Immunol. 20:128–136. 10.1038/s41577-019-0223-7 [DOI] [PubMed] [Google Scholar]
- Kanehisa, M., Goto S., Sato Y., Furumichi M., and Tanabe M.. 2012. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40(Database issue, D1):D109–D114. 10.1093/nar/gkr988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplon, H., and Dieu-Nosjean M.C.. 2018. [Which future for B lymphocytes infiltrating solid tumors: prognostic biomarker and/or therapeutic target?]. Med. Sci. (Paris). 34:72–78. 10.1051/medsci/20183401016 [DOI] [PubMed] [Google Scholar]
- Karlsen, T.H., Folseraas T., Thorburn D., and Vesterhus M.. 2017. Primary sclerosing cholangitis - a comprehensive review. J. Hepatol. 67:1298–1323. 10.1016/j.jhep.2017.07.022 [DOI] [PubMed] [Google Scholar]
- Katsumi, T., Tomita K., Leung P.S., Yang G.X., Gershwin M.E., and Ueno Y.. 2015. Animal models of primary biliary cirrhosis. Clin. Rev. Allergy Immunol. 48:142–153. 10.1007/s12016-015-8482-y [DOI] [PubMed] [Google Scholar]
- Kemp, R.A., and Ronchese F.. 2001. Tumor-specific Tc1, but not Tc2, cells deliver protective antitumor immunity. J. Immunol. 167:6497–6502. 10.4049/jimmunol.167.11.6497 [DOI] [PubMed] [Google Scholar]
- Kim, H.S., Kim H.J., Hong E.S., Kim K.B., Lee J.D., Kang T.U., and Ahn H.S.. 2020. The incidence and survival of melanoma and nonmelanoma skin cancer in patients with vitiligo: a nationwide population-based matched cohort study in Korea. Br. J. Dermatol. 182:907–915. 10.1111/bjd.18247 [DOI] [PubMed] [Google Scholar]
- Kita, H., Lian Z.X., Van de Water J., He X.S., Matsumura S., Kaplan M., Luketic V., Coppel R.L., Ansari A.A., and Gershwin M.E.. 2002a. Identification of HLA-A2-restricted CD8(+) cytotoxic T cell responses in primary biliary cirrhosis: T cell activation is augmented by immune complexes cross-presented by dendritic cells. J. Exp. Med. 195:113–123. 10.1084/jem.20010956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita, H., Matsumura S., He X.S., Ansari A.A., Lian Z.X., Van de Water J., Coppel R.L., Kaplan M.M., and Gershwin M.E.. 2002b. Quantitative and functional analysis of PDC-E2-specific autoreactive cytotoxic T lymphocytes in primary biliary cirrhosis. J. Clin. Invest. 109:1231–1240. 10.1172/JCI0214698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knochelmann, H.M., Dwyer C.J., Bailey S.R., Amaya S.M., Elston D.M., Mazza-McCrann J.M., and Paulos C.M.. 2018. When worlds collide: Th17 and Treg cells in cancer and autoimmunity. Cell. Mol. Immunol. 15:458–469. 10.1038/s41423-018-0004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox, J.J., Cosma G.L., Betts M.R., and McLane L.M.. 2014. Characterization of T-bet and eomes in peripheral human immune cells. Front. Immunol. 5:217. 10.3389/fimmu.2014.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, P.B., and Beatty E. Jr. 1977. Cholangiocarcinoma associated with biliary cirrhosis due to congenital biliary atresia. Am. J. Dis. Child. 131:442–444. 10.1001/archpedi.1977.02120170068013 [DOI] [PubMed] [Google Scholar]
- Landskron, G., De la Fuente M., Thuwajit P., Thuwajit C., and Hermoso M.A.. 2014. Chronic inflammation and cytokines in the tumor microenvironment. J. Immunol. Res. 2014:149185. 10.1155/2014/149185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane, C., Leitch J., Tan X., Hadjati J., Bramson J.L., and Wan Y.. 2004. Vaccination-induced autoimmune vitiligo is a consequence of secondary trauma to the skin. Cancer Res. 64:1509–1514. 10.1158/0008-5472.CAN-03-3227 [DOI] [PubMed] [Google Scholar]
- Leruste, A., Tosello J., Ramos R.N., Tauziède-Espariat A., Brohard S., Han Z.Y., Beccaria K., Andrianteranagna M., Caudana P., Nikolic J., et al. 2019. Clonally Expanded T Cells Reveal Immunogenicity of Rhabdoid Tumors. Cancer Cell. 36:597–612.e8. 10.1016/j.ccell.2019.10.008 [DOI] [PubMed] [Google Scholar]
- Li, J., He Y., Hao J., Ni L., and Dong C.. 2018. High Levels of Eomes Promote Exhaustion of Anti-tumor CD8+ T Cells. Front. Immunol. 9:2981. 10.3389/fimmu.2018.02981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, W.W., and Karin M.. 2007. A cytokine-mediated link between innate immunity, inflammation, and cancer. J. Clin. Invest. 117:1175–1183. 10.1172/JCI31537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W.T., and Chen D.K.. 2019. Immunological abnormalities in patients with primary biliary cholangitis. Clin. Sci. (Lond.). 133:741–760. 10.1042/CS20181123 [DOI] [PubMed] [Google Scholar]
- Marchioni Beery, R.M., Vaziri H., and Forouhar F.. 2014. Primary Biliary Cirrhosis and Primary Sclerosing Cholangitis: a Review Featuring a Women’s Health Perspective. J. Clin. Transl. Hepatol. 2:266–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti, V., Strazzabosco M., Fabris L., and Calvisi D.F.. 2018. Animal models of biliary injury and altered bile acid metabolism. Biochim. Biophys. Acta Mol. Basis Dis. 1864(4, 4 Pt B):1254–1261. 10.1016/j.bbadis.2017.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, M.D., and Badovinac V.P.. 2018. Defining Memory CD8 T Cell. Front. Immunol. 9:2692. 10.3389/fimmu.2018.02692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeini, A., Haber P.K., and Sia D.. 2021. Cell of origin in biliary tract cancers and clinical implications. JHEP Rep. 3:100226. 10.1016/j.jhepr.2021.100226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodecky, N.A., Kareemi H., Parab R., Barkema H.W., Quan H., Myers R.P., and Kaplan G.G.. 2011. Incidence of primary sclerosing cholangitis: a systematic review and meta-analysis. Hepatology. 53:1590–1599. 10.1002/hep.24247 [DOI] [PubMed] [Google Scholar]
- Moon, S., Chung H.S., Yu J.M., Yoo H.J., Park J.H., Kim D.S., and Park Y.J.. 2018. Associations between Hashimoto Thyroiditis and Clinical Outcomes of Papillary Thyroid Cancer: A Meta-Analysis of Observational Studies. Endocrinol. Metab. (Seoul). 33:473–484. 10.3803/EnM.2018.33.4.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multhoff, G., Molls M., and Radons J.. 2012. Chronic inflammation in cancer development. Front. Immunol. 2:98. 10.3389/fimmu.2011.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, R.P., Shaheen A.A., Fong A., Burak K.W., Wan A., Swain M.G., Hilsden R.J., Sutherland L., and Quan H.. 2009. Epidemiology and natural history of primary biliary cirrhosis in a Canadian health region: a population-based study. Hepatology. 50:1884–1892. 10.1002/hep.23210 [DOI] [PubMed] [Google Scholar]
- Myshunina, T.M., Guda B.D., Bolgov M.Y., Mikhailenko N.I., and Tronko N.D.. 2018. Differentiated thyroid carcinomas associated with chronic thyroiditis: biological and clinical properties. Exp. Oncol. 40:128–131. 10.31768/2312-8852.2018.40(2):128-131 [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Tanaka R., Asami Y., Teramoto Y., Imamura T., Sato S., Maruyama H., Fujisawa Y., Matsuya T., Fujimoto M., and Yamamoto A.. 2017. Correlation between vitiligo occurrence and clinical benefit in advanced melanoma patients treated with nivolumab: A multi-institutional retrospective study. J. Dermatol. 44:117–122. 10.1111/1346-8138.13520 [DOI] [PubMed] [Google Scholar]
- Oshikiri, T., Miyamoto M., Shichinohe T., Suzuoki M., Hiraoka K., Nakakubo Y., Shinohara T., Itoh T., Kondo S., and Katoh H.. 2003. Prognostic value of intratumoral CD8+ T lymphocyte in extrahepatic bile duct carcinoma as essential immune response. J. Surg. Oncol. 84:224–228. 10.1002/jso.10321 [DOI] [PubMed] [Google Scholar]
- Paillet, J., Kroemer G., and Pol J.G.. 2020. Immune contexture of cholangiocarcinoma. Curr. Opin. Gastroenterol. 36:70–76. [DOI] [PubMed] [Google Scholar]
- Paradisi, A., Tabolli S., Didona B., Sobrino L., Russo N., and Abeni D.. 2014. Markedly reduced incidence of melanoma and nonmelanoma skin cancer in a nonconcurrent cohort of 10,040 patients with vitiligo. J. Am. Acad. Dermatol. 71:1110–1116. 10.1016/j.jaad.2014.07.050 [DOI] [PubMed] [Google Scholar]
- Parés, A., and Rodés J.. 2003. Natural history of primary biliary cirrhosis. Clin. Liver Dis. 7:779–794. 10.1016/S1089-3261(03)00100-4 [DOI] [PubMed] [Google Scholar]
- Peine, M., Rausch S., Helmstetter C., Fröhlich A., Hegazy A.N., Kühl A.A., Grevelding C.G., Höfer T., Hartmann S., and Löhning M.. 2013. Stable T-bet(+)GATA-3(+) Th1/Th2 hybrid cells arise in vivo, can develop directly from naive precursors, and limit immunopathologic inflammation. PLoS Biol. 11:e1001633. 10.1371/journal.pbio.1001633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin, S., Firmo C., Lemoine S., Le Crom S., and Jourdren L.. 2017. Aozan: an automated post-sequencing data-processing pipeline. Bioinformatics. 33:2212–2213. 10.1093/bioinformatics/btx154 [DOI] [PubMed] [Google Scholar]
- Pol, J.G., Zhang L., Bridle B.W., Stephenson K.B., Rességuier J., Hanson S., Chen L., Kazdhan N., Bramson J.L., Stojdl D.F., et al. 2014. Maraba virus as a potent oncolytic vaccine vector. Mol. Ther. 22:420–429. 10.1038/mt.2013.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, J.G., Acuna S.A., Yadollahi B., Tang N., Stephenson K.B., Atherton M.J., Hanwell D., El-Warrak A., Goldstein A., Moloo B., et al. 2019. Preclinical evaluation of a MAGE-A3 vaccination utilizing the oncolytic Maraba virus currently in first-in-human trials. OncoImmunology. 8:e1512329. 10.1080/2162402X.2018.1512329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, J.G., Atherton M.J., Bridle B.W., Stephenson K.B., Le Boeuf F., Hummel J.L., Martin C.G., Pomoransky J., Breitbach C.J., Diallo J.S., et al. 2018. Development and applications of oncolytic Maraba virus vaccines. Oncolytic Virother. 7:117–128. 10.2147/OV.S154494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, J.G., Atherton M.J., Stephenson K.B., Bridle B.W., Workenhe S.T., Kazdhan N., McGray A.R., Wan Y., Kroemer G., and Lichty B.D.. 2020a. Enhanced immunotherapeutic profile of oncolytic virus-based cancer vaccination using cyclophosphamide preconditioning. J. Immunother. Cancer. 8:e000981. 10.1136/jitc-2020-000981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, J.G., Bridle B.W., and Lichty B.D.. 2020b. Detection of Tumor Antigen-Specific T-Cell Responses After Oncolytic Vaccination. Methods Mol. Biol. 2058:191–211. 10.1007/978-1-4939-9794-7_12 [DOI] [PubMed] [Google Scholar]
- Qian, C., Jiang T., Zhang W., Ren C., Wang Q., Qin Q., Chen J., Deng A., and Zhong R.. 2013. Increased IL-23 and IL-17 expression by peripheral blood cells of patients with primary biliary cirrhosis. Cytokine. 64:172–180. 10.1016/j.cyto.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Quezada, S.A., Simpson T.R., Peggs K.S., Merghoub T., Vider J., Fan X., Blasberg R., Yagita H., Muranski P., Antony P.A., et al. 2010. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J. Exp. Med. 207:637–650. 10.1084/jem.20091918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razumilava, N., Gores G.J., and Lindor K.D.. 2011. Cancer surveillance in patients with primary sclerosing cholangitis. Hepatology. 54:1842–1852. 10.1002/hep.24570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi, S., Fischbach S.R., Bronk S.F., Hirsova P., Krishnan A., Dhanasekaran R., Smadbeck J.B., Smoot R.L., Vasmatzis G., and Gores G.J.. 2018. YAP-associated chromosomal instability and cholangiocarcinoma in mice. Oncotarget. 9:5892–5905. 10.18632/oncotarget.23638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti, M.P., Yonekura S., Duong C.P.M., Picard M., Ferrere G., Tidjani Alou M., Rauber C., Iebba V., Lehmann C.H.K., Amon L., et al. 2020. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat. Med. 26:919–931. 10.1038/s41591-020-0882-8 [DOI] [PubMed] [Google Scholar]
- Romagnoli, M., Bresson L., Di-Cicco A., Pérez-Lanzón M., Legoix P., Baulande S., de la Grange P., De Arcangelis A., Georges-Labouesse E., Sonnenberg A., et al. 2020. Laminin-binding integrins are essential for the maintenance of functional mammary secretory epithelium in lactation. Development. 147:dev181552. 10.1242/dev.181552 [DOI] [PubMed] [Google Scholar]
- Rothwell, P.M., Cook N.R., Gaziano J.M., Price J.F., Belch J.F.F., Roncaglioni M.C., Morimoto T., and Mehta Z.. 2018. Effects of aspirin on risks of vascular events and cancer according to bodyweight and dose: analysis of individual patient data from randomised trials. Lancet. 392:387–399. 10.1016/S0140-6736(18)31133-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, A., Desbois S., Carrié N., Lawand M., Mars L.T., and Liblau R.S.. 2012. Tc17 CD8+ T cells potentiate Th1-mediated autoimmune diabetes in a mouse model. J. Immunol. 189:3140–3149. 10.4049/jimmunol.1103111 [DOI] [PubMed] [Google Scholar]
- Shalapour, S., and Karin M.. 2021. The neglected brothers come of age: B cells and cancer. Semin. Immunol.:101479. 10.1016/j.smim.2021.101479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo, M., Mullin G.E., Braun-Elwert L., Bergasa N.V., Jones E.A., and James S.P.. 1996. Cytokine mRNA expression in the liver of patients with primary biliary cirrhosis (PBC) and chronic hepatitis B (CHB). Clin. Exp. Immunol. 105:254–259. 10.1046/j.1365-2249.1996.d01-759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood, S., Gow P.J., Christie J.M., and Angus P.W.. 2004. Epidemiology of primary biliary cirrhosis in Victoria, Australia: high prevalence in migrant populations. Gastroenterology. 127:470–475. 10.1053/j.gastro.2004.04.064 [DOI] [PubMed] [Google Scholar]
- Squadroni, M., Tondulli L., Gatta G., Mosconi S., Beretta G., and Labianca R.. 2017. Cholangiocarcinoma. Crit. Rev. Oncol. Hematol. 116:11–31. 10.1016/j.critrevonc.2016.11.012 [DOI] [PubMed] [Google Scholar]
- St Paul, M., and Ohashi P.S.. 2020. The Roles of CD8+ T Cell Subsets in Antitumor Immunity. Trends Cell Biol. 30:695–704. 10.1016/j.tcb.2020.06.003 [DOI] [PubMed] [Google Scholar]
- Takagi, S., Miyagawa S., Ichikawa E., Soeda J., Miwa S., Miyagawa Y., Iijima S., Noike T., Kobayashi A., and Kawasaki S.. 2004. Dendritic cells, T-cell infiltration, and Grp94 expression in cholangiocellular carcinoma. Hum. Pathol. 35:881–886. 10.1016/j.humpath.2004.03.016 [DOI] [PubMed] [Google Scholar]
- Tamma, R., Annese T., Ruggieri S., Brunetti O., Longo V., Cascardi E., Mastropasqua M.G., Maiorano E., Silvestris N., and Ribatti D.. 2019. Inflammatory cells infiltrate and angiogenesis in locally advanced and metastatic cholangiocarcinoma. Eur. J. Clin. Invest. 49:e13087. 10.1111/eci.13087 [DOI] [PubMed] [Google Scholar]
- Taniguchi, K., and Karin M.. 2018. NF-κB, inflammation, immunity and cancer: coming of age. Nat. Rev. Immunol. 18:309–324. 10.1038/nri.2017.142 [DOI] [PubMed] [Google Scholar]
- Tepper, R.I., Pattengale P.K., and Leder P.. 1989. Murine interleukin-4 displays potent anti-tumor activity in vivo. Cell. 57:503–512. 10.1016/0092-8674(89)90925-2 [DOI] [PubMed] [Google Scholar]
- Teulings, H.E., Overkamp M., Ceylan E., Nieuweboer-Krobotova L., Bos J.D., Nijsten T., Wolkerstorfer A.W., Luiten R.M., and van der Veen J.P.. 2013. Decreased risk of melanoma and nonmelanoma skin cancer in patients with vitiligo: a survey among 1307 patients and their partners. Br. J. Dermatol. 168:162–171. 10.1111/bjd.12111 [DOI] [PubMed] [Google Scholar]
- Todoric, J., Antonucci L., and Karin M.. 2016. Targeting Inflammation in Cancer Prevention and Therapy. Cancer Prev. Res. (Phila.). 9:895–905. 10.1158/1940-6207.CAPR-16-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, P.J., and Cullen S.. 2013. Etiopathogenesis of primary biliary cirrhosis: an overview of recent developments. Hepatol. Int. 7:28–47. 10.1007/s12072-012-9362-7 [DOI] [PubMed] [Google Scholar]
- Tsuneyama, K., Baba H., Morimoto Y., Tsunematsu T., and Ogawa H.. 2017. Primary Biliary Cholangitis: Its Pathological Characteristics and Immunopathological Mechanisms. J. Med. Invest. 64:7–13. 10.2152/jmi.64.7 [DOI] [PubMed] [Google Scholar]
- Vizler, C., Bercovici N., Heurtier A., Pardigon N., Goude K., Bailly K., Combadière C., and Liblau R.S.. 2000. Relative diabetogenic properties of islet-specific Tc1 and Tc2 cells in immunocompetent hosts. J. Immunol. 165:6314–6321. 10.4049/jimmunol.165.11.6314 [DOI] [PubMed] [Google Scholar]
- Wakabayashi, K., Lian Z.X., Leung P.S., Moritoki Y., Tsuneyama K., Kurth M.J., Lam K.S., Yoshida K., Yang G.X., Hibi T., et al. 2008. Loss of tolerance in C57BL/6 mice to the autoantigen E2 subunit of pyruvate dehydrogenase by a xenobiotic with ensuing biliary ductular disease. Hepatology. 48:531–540. 10.1002/hep.22390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Budamagunta M.S., Voss J.C., Kurth M.J., Lam K.S., Lu L., Kenny T.P., Bowlus C., Kikuchi K., Coppel R.L., et al. 2013. Antimitochondrial antibody recognition and structural integrity of the inner lipoyl domain of the E2 subunit of pyruvate dehydrogenase complex. J. Immunol. 191:2126–2133. 10.4049/jimmunol.1301092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Yang G.X., Tsuneyama K., Gershwin M.E., Ridgway W.M., and Leung P.S.. 2014. Animal models of primary biliary cirrhosis. Semin. Liver Dis. 34:285–296. 10.1055/s-0034-1383728 [DOI] [PubMed] [Google Scholar]
- Welzel, T.M., Graubard B.I., El-Serag H.B., Shaib Y.H., Hsing A.W., Davila J.A., and McGlynn K.A.. 2007. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: a population-based case-control study. Clin. Gastroenterol. Hepatol. 5:1221–1228. 10.1016/j.cgh.2007.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, F., and Bengsch B.. 2020. Use of Mass Cytometry to Profile Human T Cell Exhaustion. Front. Immunol. 10:3039. 10.3389/fimmu.2019.03039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, D., Rizvi S., Razumilava N., Bronk S.F., Davila J.I., Champion M.D., Borad M.J., Bezerra J.A., Chen X., and Gores G.J.. 2015. IL-33 facilitates oncogene-induced cholangiocarcinoma in mice by an interleukin-6-sensitive mechanism. Hepatology. 61:1627–1642. 10.1002/hep.27687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano, T., Higashi T., Nouso K., Nakatsukasa H., Kariyama K., Yumoto E., Kobayashi Y., Yamamoto K., Iwagaki H., Yagi T., et al. 2000. Serum interferon-gamma-inducing factor/IL-18 levels in primary biliary cirrhosis. Clin. Exp. Immunol. 122:227–231. 10.1046/j.1365-2249.2000.01356.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.Y., Ma X., Tsuneyama K., Huang S., Takahashi T., Chalasani N.P., Bowlus C.L., Yang G.X., Leung P.S., Ansari A.A., et al. 2014. IL-12/Th1 and IL-23/Th17 biliary microenvironment in primary biliary cirrhosis: implications for therapy. Hepatology. 59:1944–1953. 10.1002/hep.26979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J.S., Wei M.X., Chiocca E.A., Martuza R.L., and Tepper R.I.. 1993. Treatment of glioma by engineered interleukin 4-secreting cells. Cancer Res. 53:3125–3128. [PubMed] [Google Scholar]
- Yuen, G.J., Demissie E., and Pillai S.. 2016. B lymphocytes and cancer: a love-hate relationship. Trends Cancer. 2:747–757. 10.1016/j.trecan.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. 2017. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 169:1342–1356.e16. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
- Zitvogel, L., Perreault C., Finn O.J., and Kroemer G.. 2021. Beneficial autoimmunity improves cancer prognosis. Nat. Rev. Clin. Oncol. 18:591–602. 10.1038/s41571-021-00508-x [DOI] [PubMed] [Google Scholar]
- Zou, W., and Restifo N.P.. 2010. T(H)17 cells in tumour immunity and immunotherapy. Nat. Rev. Immunol. 10:248–256. 10.1038/nri2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the overall profile of differentially expressed genes in the liver of mice affected with PBC and PSC.
shows enriched TCR clonotypes shared between the malignant and hepatic tissues of a PBC mouse and lists the 25 TCR clonotypes enriched in both tumor and liver of a PBC mouse.
shows characteristics of the T cell clusters in the malignant and hepatic tissues of a PBC mouse and expression level of immune markers related to T cell function across the 14 clusters of T lymphocytes infiltrated in the tumor and liver of a PBC mouse.
is a detailed version of the heatmap displayed in Fig. 2 A and built from gene expression values listed in Table S1.
Data Availability Statement
Raw transcriptome data reported in this paper are deposited and available from the Gene Expression Omnibus under accession no. GSE180289; GSE179993 for the RNA-Seq dataset (Fig. 2 and Fig. 5 A), and GSE180288 for the scTCR-seq/scRNA-seq dataset (Fig. 8 and Fig. 9). Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors.