Abstract
The experience of individuals with Coronavirus Disease 2019 (COVID‐19) ranges from asymptomatic to life threatening multi‐organ dysfunction. Specific HLA alleles may affect the predisposition to severe COVID‐19 because of their role in presenting viral peptides to launch the adaptive immune response to severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). In this population‐based case–control study in the midwestern United States, we performed high‐resolution HLA typing of 234 cases hospitalized for COVID‐19 in the St. Louis metropolitan area and compared their HLA allele frequencies with those of 22,000 matched controls from the National Marrow Donor Program (NMDP). We identified two predisposing alleles, HLA‐DRB1*08:02 in the Hispanic group (OR = 9.0, 95% confidence interval: 2.2–37.9; adjusted p = 0.03) and HLA‐A*30:02 in younger African Americans with ages below the median (OR = 2.2, 1.4–3.6; adjusted p = 0.01), and several candidate alleles with potential associations with COVID‐19 in African American, White, and Hispanic groups. We also detected risk‐associated amino acid residues in the peptide binding grooves of some of these alleles, suggesting the presence of functional associations. These findings support the notion that specific HLA alleles may be protective or predisposing factors to COVID‐19. Future consortium analysis of pooled cases and controls is warranted to validate and extend these findings, and correlation with viral peptide binding studies will provide additional evidence for the functional association between HLA alleles and COVID‐19.
Keywords: COVID‐19, disease association, HLA, SARS‐CoV‐2
Abbreviations
- COVID‐19
Coronavirus Disease 2019
- NGS
Next‐generation sequencing
- NMDP
National Marrow Donor Program
- ONT
Oxford Nanopore Technologies
- OR
Odds Ratio
- PCR
Polymerase chain reaction
- RT‐PCR
Reverse transcriptase‐polymerase chain reaction
- SARS‐CoV‐2
Severe acute respiratory syndrome coronavirus 2
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has swept across the globe causing the Coronavirus Disease 2019 (COVID‐19) pandemic1 and unprecedented disease burden.2 In the United States alone, the total cases and total deaths reached 25.7 million and 435,151, respectively, as of January 31, 2021 (www.cdc.gov). In addition to known COVID‐19 cases, 86% of all infections are predicted to be undocumented and likely to be mild.3 Patients with moderate to severe COVID‐19 likely represent only a fraction of the total number of infected individuals.4
SARS‐CoV‐2 appears to elicit highly heterogeneous innate and adaptive immune responses, leading to drastically different outcomes. Although older age and certain comorbidities are known to contribute to increased mortality,5, 6, 7 younger and seemingly healthy patients are not completely protected from severe COVID‐19.8 The broad demographics and range of severity among COVID‐19 patients have been reported by multiple epidemiologic studies from different regions in the United States.7, 8, 9 Importantly, some self‐reported symptoms of COVID‐19 show increased correlation among monozygotic twins,10 suggesting that the predisposition to symptomatic COVID‐19 may be heritable. Therefore, it is imperative to elucidate the immunogenetic underpinning of the diverse host responses to determine who is at risk for COVID‐19 and why.
HLA molecules play an essential role in the defense against viral infections. HLA present peptides derived from viral proteins to T cell receptors to initiate adaptive immunity mediated by pathogen‐specific T and B cells.11 Class I HLA molecules, encoded by HLA‐A, ‐B, and ‐C genes, are ubiquitously expressed and present peptides to CD8+ T cells; class II HLA molecules, heterodimers encoded by HLA‐DRA/DRB1/3/4/5, ‐DQA1/DQB1, ‐DPA1/DPB1, are primarily expressed on antigen‐presentation cells and present peptides to CD4+ T cells. To accommodate peptides derived from a broad spectrum of pathogens, diverse HLA molecules are encoded by thousands of different alleles in the human population.12 However, as each individual has at most two alleles per locus, an individual's peptide repertoire is more limited than that of the population. Some HLA molecules may be disadvantageous, compared with others, in presenting peptides derived from SARS‐CoV‐2, as suggested by in silico modelings.13 However, there is no published data on HLA allele frequencies in COVID‐19 patients of different population categories in the US.
Several HLA alleles have been associated with the susceptibility to and different outcomes of SARS caused by SARS‐CoV in 2003.14, 15, 16, 17, 18 The association of HLA alleles and COVID‐19 has been examined in several populations, primarily in China19 and Italy.20, 21, 22 This study aims to identify HLA alleles associated with moderate to severe COVID‐19 among several patient populations in the St. Louis metropolitan area, as compared with matched population controls. We hypothesize that HLA alleles predisposing to symptomatic infection by SARS‐CoV‐2 are enriched in patients hospitalized for COVID‐19. We conducted a population‐based case–control study, focusing on classical HLA genes typed by next‐generation sequencing, and performed analyses at the allele and protein sequence levels.
2. MATERIALS AND METHODS
2.1. Study population
The study was approved by the Human Research Protection Office of Washington University in St. Louis (IRB ID #: 202004002) and the Institutional Review Board of Mercy Hospital (IRB ID #:1599032–2). Cases consisted of adult inpatients between the ages of 18 and 90 years, who were hospitalized for COVID‐19 at Barnes‐Jewish Hospital or Mercy Hospital, two of the largest medical centers serving the St. Louis metropolitan area. All cases had a remnant, EDTA‐anticoagulated blood specimen available in the clinical laboratories. SARS‐CoV‐2 infections were confirmed by real‐time reverse transcriptase‐polymerase chain reaction (RT‐PCR) testing of nasopharyngeal swabs. A total of 234 cases were enrolled and HLA typed during the study period from March 26 to July 7, 2020.
A total of 22,000 population controls were randomly selected from the National Marrow Donor Program (NMDP) volunteer adult donor registry recruited since 2005. Controls were matched for one of four self‐identified “race/ethnic” population categories on the NMDP registry donor recruitment form (African American, Asian Pacific Islander, Whites, and Hispanics), gender, age quartiles, and the first digit of zip codes. A total of 10,000 controls were retrieved for the Whites. Because of the limited availability of minority donors in the NMDP, 4000 controls were retrieved for each of the other groups. Because of the registry recruitment policy of NMDP, the maximum control age (60 years) was younger than the oldest cases. Considering this caveat and the increased comorbidities of elderly patients, ad hoc analysis was performed for younger African Americans (n = 76) and Whites (n = 27) with ages below the medians of 64 and 68 years, respectively, and their matched population controls (4000 for the African Americans and 10,000 for the Whites); the sample sizes of Asian Pacific Islanders and Hispanics were too small to be dichotomized. Population controls from NMDP were HLA typed at high‐resolution typing primarily by next‐generation sequencing, and also by sequence‐specific oligonucleotide or Sanger sequence‐based typing.23
2.2. Clinical data collection
The following clinical data were collected for all cases by retrospective chart review and entered into a REDCap database24: demographics (age, gender, self‐reported population category, zip code, and BMI), comorbidities (diabetes mellitus, chronic lung diseases, and cardiovascular diseases), duration of hospitalization, ICU admission, mechanical ventilation, and time of last encounter or death.
2.3. HLA typing of cases
Genomic DNA was extracted from remnant peripheral blood specimens using the EZ1 DNA Blood 350 μl Kit (Qiagen, Hilden, Germany). A total of 192 samples were typed by the AllType assay (One Lambda, West Hills, CA) on the Ion Chef/S5 Ion Torrent platform.25 A total of 42 samples were amplifed using the NGS LR kit (One Lambda, West Hills, CA) and sequenced following the SQK‐LSK109 protocol on the R10.3 MinION flow cells (FLO‐MIN111, Oxford Nanopore Technologies).26 Genotypes of HLA‐A, ‐B, ‐C, ‐DPA1, ‐DPB1, ‐DQA1, ‐DQB1, ‐DRB1, ‐DRB3/4/5 genes were assigned based on key‐exon sequences (G groups) and limited to the 2‐field resolution.
2.4. Statistical analysis
The demographics of cases and controls were reported with standard descriptive statistics, including counts, proportions, and medians and ranges, as appropriate. All association analyses were performed for each population separately. For the allele association analysis at the HLA‐A, ‐B, ‐C, ‐DRB1, and ‐DQB1 loci, frequencies of alleles in cases were compared with those in controls by Fisher's exact test using the pyHLA package (version 1.1.1).27 The default allelic genetic model was used to compare one allele against other alleles grouped together, and the default minimal allele frequency of 0.05 was applied. Associations with unadjusted p value <0.05 were reported as candidate alleles of interest. Multiple comparisons of alleles with frequencies of 0.05 or higher were adjusted in the above analyses by controlling the false discovery rate at 5% using the Benjamini–Hochberg procedure,28 and an adjusted p value <0.05 was considered statistically significant. The amino acid association analysis was also performed using the pyHLA package using default options. Amino acid associations with unadjusted p value below 0.05 were further examined if they were carried by protecting or predisposing alleles. The locations of these alleles were visualized within available crystal structures using PyMOL (Molecular Graphics System, Version 2.4.1, Schrödinger, LLC.) to determine their relevance to peptide presentation.
3. RESULTS
3.1. Demographics and characteristics of the study population
Cases consisted of 167 African‐Americans, 56 Whites, 7 Asian Pacific Islanders, and 4 Hispanics. The baseline demographics for cases and controls are shown for each population in Table 1. While the geographic location, gender ratio, and median age were well matched between the cases and controls, the cases were skewed toward older ages because of the maximum age of 60 years in the NMDP controls (Table 1).
TABLE 1.
Demographics of cases and controls
| Total | Gender (%) | Age | |||||
|---|---|---|---|---|---|---|---|
| Male | Female | Median | Low | High | |||
| African Americans | Cases | 167 | 55.1 | 44.9 | 64 | 15 | 89 |
| Controls | 4000 | 53.0 | 47.0 | 53.5 | 18 | 60 | |
| Asian Pacific Islanders | Cases | 7 | 71.4 | 28.6 | 55 | 50 | 88 |
| Controls | 4000 | 75.0 | 25.0 | 52.5 | 18 | 60 | |
| Whites | Cases | 56 | 67.9 | 32.1 | 68 | 23 | 87 |
| Controls | 10,000 | 65.0 | 35.0 | 55.5 | 18 | 60 | |
| Hispanics | Cases | 4 | 75.0 | 25.0 | 48 | 32 | 57 |
| Controls | 4000 | 75.0 | 25.0 | 53.5 | 18 | 60 | |
All COVID‐19 cases were confirmed by positive RT‐PCR testing and hospitalized for treatment. A total of 121 patients (51.7%) were admitted to intensive care units (ICU), and 75 patients (32.1%) received mechanical ventilation. The overall mortality rate was 21.8% among the cases. A broad range of co‐morbidities were documented with chronic cardiac disease (73.5%) and diabetes (41.9%) being the most common. The clinical characteristics of cases were listed for each population in Table 2.
TABLE 2.
Clinical characteristics of all cases
| African Americans | Asian Pacific Islanders | Whites | Hispanics | |
|---|---|---|---|---|
| Case counts | 167 | 7 | 56 | 4 |
| BMI, mean (SD) | 30.5 (9.3)a | 25 (3.2) | 29.6 (7.9) | 27.6 (7.8) |
| Co‐morbidities, n (%) | ||||
| Chronic lung disease | 28 (16.8) | 0 (0) | 16 (28.6) | 0 (0) |
| HTN/cardiac disease | 126 (75.4) | 3 (42.9) | 42 (75) | 1 (25) |
| Diabetes | 73 (43.7) | 3 (42.9) | 21 (37.5) | 1 (25) |
| End‐stage renal disease | 29 (17.3) | 0 (0) | 12 (21.4) | 1 (25) |
| Autoimmune disease | 8 (4.8) | 0 (0) | 5 (8.9) | 0 (0) |
| Cancer | 12 (7.2) | 0 (0) | 2 (3.6) | 0 (0) |
| Trauma | 4 (2.4) | 0 (0) | 2 (3.6) | 0 (0) |
| Surgery | 7 (4.2) | 0 (0) | 4 (7.1) | 2 (50) |
| Sepsis | 42 (25.1) | 3 (42.9) | 23 (41) | 3 (75) |
| None | 13 (7.8) | 3 (42.9) | 2 (3.6) | 1 (25) |
| ICU admission, n (%) | 87 (52.1) | 4 (57.1) | 27 (48.2) | 3 (75) |
| Mechanical ventilation, n (%) | 52 (31.1) | 3 (42.9) | 17 (30.4) | 3 (75) |
| Death, n (%) | 32 (19.2) | 3 (42.9) | 15 (26.8) | 1 (25) |
Abbreviations: BMI, body‐mass index; HTN, hypertension; ICU, intensive care unit; SD, standard deviation.
One missing value.
3.2. HLA association at the allele level
Because of limited sample size and statistical power, we examined alleles with overall frequencies above 5% in the primary analysis. We identified one protective allele in African Americans, and five predisposing alleles in Whites and Hispanics. Table 3 shows the counts and frequencies of these alleles in cases and controls, overall frequencies, unadjusted and adjusted p values, odds ratios (OR), and the 95% confidence intervals of ORs. Only HLA‐DRB1*08:02 in Hispanics remained statistically significant after adjusting for multiple comparisons (OR = 9.0, adjusted p = 0.03). HLA‐DRB1*08:02 was detected in three of the four heterozygous cases with an allele frequency of 37.5%, while its frequency in the matched population control was 6.2% (Table 3). Among other groups of the cases, the frequencies of HLA‐DRB1*08:02 were 0%, 0.9%, and 7.1% in the African Americans, Whites, and Asian Pacific Islanders, respectively; the allele frequencies in the corresponding population control groups are 0.3%, 0.1%, and 0.7%. Results for all alleles analyzed in the four populations were provided in Table S1.
TABLE 3.
HLA alleles associated with COVID‐19 (Minimal allele frequency = 5%)
| Group | Allele | A_case | B_case | A_ctrl | B_ctrl | F_case | F_ctrl | Freq | p_unadj | OR | L95 | U95 | p_adj |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African Americans | HLA‐B*42:01 | 10 | 324 | 472 | 7528 | 0.0299 | 0.0590 | 0.0578 | 0.02 | 0.5 | 0.3 | 0.9 | 0.14 |
| Whites | HLA‐B*35:01 | 13 | 99 | 1153 | 18,847 | 0.1161 | 0.0576 | 0.0580 | 0.01 | 2.1 | 1.2 | 3.8 | 0.10 |
| HLA‐A*11:01 | 13 | 99 | 1272 | 18,728 | 0.1161 | 0.0636 | 0.0639 | 0.03 | 1.9 | 1.1 | 3.5 | 0.16 | |
| Hispanics | HLA‐DRB1*08:02 | 3 | 5 | 498 | 7502 | 0.3750 | 0.0622 | 0.0626 | 0.01 | 9.0 | 2.2 | 37.9 | 0.03 |
| HLA‐C*04:01 | 4 | 4 | 1253 | 6747 | 0.5000 | 0.1566 | 0.1570 | 0.02 | 5.4 | 1.3 | 21.6 | 0.07 | |
| HLA‐DQB1*04:02 | 3 | 5 | 771 | 7229 | 0.3750 | 0.0964 | 0.0967 | 0.03 | 5.6 | 1.3 | 23.6 | 0.14 | |
| Younger African Americans (Ad hoc analysis) | HLA‐A*30:02 | 21 | 131 | 534 | 7466 | 0.1382 | 0.0668 | 0.0681 | 0.0017 | 2.2413 | 1.4022 | 3.5825 | 0.0134 |
| Younger Whites (Ad hoc analysis) | HLA‐A*11:01 | 7 | 47 | 1171 | 18,829 | 0.1296 | 0.0585 | 0.0587 | 0.0377 | 2.3948 | 1.0801 | 5.3098 | 0.1886 |
Abbreviations: adj, adjusted for multiple testing of alleles with frequency above 5%; A_case, Count of this allele in cases; A_ctrl, Count of this allele in controls; B_case, Count of other alleles in cases; B_ctrl, Count of other alleles in controls; F, frequency; Freq, overall frequency; L95, lower bound 95% confidence interval; OR, odds ratio; unadj, unadjusted; U95, upper bound 95% confidence interval.
In the ad hoc analysis of younger African Americans with ages below the median against their matched population controls, HLA‐A*30:02 was associated with an increased risk of COVID‐19 (OR = 2.2, unadjusted p = 0.0017, adjusted p = 0.01). Among other groups of the cases, HLA‐A*30:02 was not detected in the Whites, Asian Pacific Islanders, or Hispanics; the allele frequencies in the corresponding population control groups are 1%, 0.1%, and 2.2%. The frequencies of HLA‐A*30:02 were 13.8% and 6.7% in the patients and population controls, respectively (Table 3). Among younger Whites, one predisposing allele, HLA‐A*11:01, was detected (OR = 2.4, unadjusted p = 0.04); however, it was no longer statistically significant after adjusting for multiple comparisons (Table 3). Results for all alleles analyzed in the younger African Americans and Whites were provided in Table S2.
3.3. HLA association at the amino acid residue level
In African Americans, we identified two potentially protective amino acid residues in the peptide‐binding groove of HLA‐B, a serine at position 24 and a threonine at position 163 (Table 4 and Figure 1A), which are carried by the protective candidate allele HLA‐B*42:01 (Table 3). No amino acid residues were associated with COVID‐19 in the Whites that were carried by the two potential predisposing alleles identified in the allele association analysis.
TABLE 4.
Amino acid residues carried by candidate alleles and associated with COVID‐19
| Group | Gene | Residue | A_case | B_case | A_ctrl | B_ctrl | F_case | F_ctrl | Freq | p | OR | ACR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| African Americans | HLA‐B | Ser24a | 81 | 86 | 2380 | 1620 | 0.4850 | 0.5950 | 0.5906 | 0.006 | 0.6 | B*07:02,B*07:05,B*07:09,B*07:104,B*08:01,B*08:12,B*08:23,B*14:01,B*14:02,B*14:03,B*14:05,B*14:09,B*14:22,B*15:03,B*15:13,B*15:18,B*15:37,B*15:47,B*15:52,B*18:01,B*37:01,B*38:01,B*39:01,B*39:06,B*39:09,B*39:10,B*39:11,B*39:13,B*39:60,B*40:12,B*42:01,B*42:02,B*48:01,B*81:01,B*82:01,B*82:02 |
| HLA‐B | Thr163a | 44 | 123 | 1472 | 2528 | 0.2635 | 0.3680 | 0.3638 | 0.007 | 0.6 | B*08:01,B*08:12,B*08:23,B*14:01,B*14:02,B*14:03,B*14:05,B*14:09,B*14:22,B*18:01,B*18:04,B*37:01,B*38:01,B*38:02,B*39:01,B*39:02,B*39:06,B*39:09,B*39:10,B*39:11,B*39:13,B*39:15,B*39:60,B*41:01,B*41:02,B*41:03,B*42:01,B*42:02,B*55:01,B*55:02 | |
| Hispanics | HLA‐C | Ser11a | 4 | 0 | 1547 | 2453 | 1.0000 | 0.3868 | 0.3874 | 0.022 | 14.3 | C*01:02,C*01:10,C*04:01,C*04:04,C*04:120,C*04:54,C*14:02,C*14:03,C*14:06 |
| HLA‐DQB1 | Glu70a | 3 | 1 | 714 | 3286 | 0.7500 | 0.1785 | 0.1791 | 0.020 | 10.7 | DQB1*03:25,DQB1*04:02 | |
| HLA‐DQB1 | Asp71 | 3 | 1 | 714 | 3286 | 0.7500 | 0.1785 | 0.1791 | 0.020 | 10.7 | DQB1*03:25,DQB1*04:02 | |
| HLA‐DQB1 | Phe9a | 4 | 0 | 1257 | 2743 | 1.0000 | 0.3143 | 0.3149 | 0.010 | 19.6 | DQB1*04:02,DQB1*06:02 | |
| HLA‐DQB1 | Leu56 | 3 | 1 | 714 | 3286 | 0.7500 | 0.1785 | 0.1791 | 0.020 | 10.7 | DQB1*03:25,DQB1*04:02 | |
| HLA‐DRB1 | Leu74a | 3 | 1 | 660 | 3340 | 0.7500 | 0.1650 | 0.1656 | 0.016 | 11.8 | DRB1*08:01,DRB1*08:02,DRB1*08:03,DRB1*08:04,DRB1*08:06,DRB1*08:07,DRB1*08:10,DRB1*08:30 | |
| HLA‐DRB1 | Ser189 | 3 | 1 | 566 | 3434 | 0.7500 | 0.1415 | 0.1421 | 0.010 | 14.2 | DRB1*08:02,DRB1*08:03,DRB1*08:04,DRB1*08:06,DRB1*08:07,DRB1*08:10 | |
| HLA‐DRB1 | Gly13a | 3 | 1 | 771 | 3229 | 0.7500 | 0.1928 | 0.1933 | 0.025 | 9.8 | DRB1*08:01,DRB1*08:02,DRB1*08:03,DRB1*08:04,DRB1*08:06,DRB1*08:07,DRB1*08:10,DRB1*08:30,DRB1*12:01,DRB1*12:02,DRB1*14:04 | |
| HLA‐DRB1 | Tyr16 | 3 | 1 | 771 | 3229 | 0.7500 | 0.1928 | 0.1933 | 0.025 | 9.8 | DRB1*08:01,DRB1*08:02,DRB1*08:03,DRB1*08:04,DRB1*08:06,DRB1*08:07,DRB1*08:10,DRB1*08:30,DRB1*12:01,DRB1*12:02,DRB1*14:04 | |
| Younger African Americans (Ad hoc analysis) | HLA‐A | Asn77a | 51 | 25 | 2204 | 1796 | 0.6711 | 0.5510 | 0.5532 | 0.04703 | 1.65 | A*01:01,A*01:02,A*01:03,A*01:09,A*23:01,A*23:54,A*23:56,A*24:02,A*24:03,A*24:07,A*24:21,A*26:01,A*26:08,A*26:12,A*29:01,A*29:02,A*29:40,A*30:02,A*30:04,A*36:01,A*36:03,A*43:01,A*80:01 |
| HLA‐A | Arg152a | 18 | 58 | 552 | 3448 | 0.2368 | 0.1380 | 0.1398 | 0.01896 | 1.97 | A*30:02,A*80:01 |
Abbreviations: A, residue present; ACR, alleles where the residue is present; B, residue absent; ctrl, controls; F, frequency; Freq, overall frequency; OR, odds ratio calculated with Haldane's correction of Woolf's method; unadj, unadjusted.
FIGURE 1.
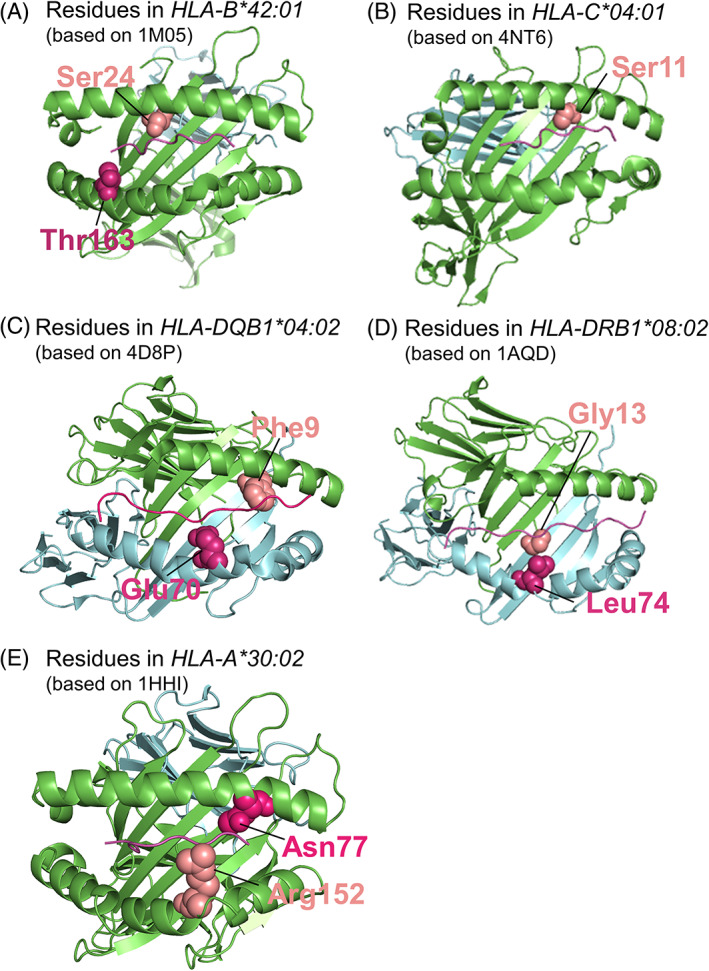
Location of amino acid residues associated with COVID‐19. Ribbon models were created using PyMOL and representative structures from the Protein Data Bank (PDB). The PDB ID is listed for each model. Amino acid residues of interest are highlighted as spheres in salmon or magenta. Class I α chain and β2‐microglobulin are colored green and blue, respectively, in A, B, and E. Class II α chain and β chain are colored green and blue, respectively, in C and D
In Hispanics, we found nine potentially predisposing residues carried by HLA‐C*04:01, ‐DQB1*04:02, and ‐DRB1*08:02 (Table 4); five of these residues were located in the peptide binding grooves of respective molecules (Figure 1B–D).
Finally, in the ad hoc analysis of younger African Americans, we identified two predisposing residues located in the peptide binding groove of HLA‐A*30:02 (Table 4; Figure 1E).
4. DISCUSSION
In this study, we identified HLA‐DRB1*08:02 and HLA‐A*30:02 as potential risk factors for symptomatic SARS‐CoV‐2 infection in the Hispanics and younger African Americans, respectively, relative to their matched population controls. We also report several potentially protective and predisposing candidate alleles found in the African Americans, Whites, and Hispanics as well as several amino acid residues with potential implications in altered peptide presentation during the immune response to SARS‐CoV‐2. The study followed a prespecified protocol for case enrollment and data analysis, and ad hoc analysis was performed for younger African Americans and Whites. High‐quality genotyping data of cases and controls enabled the analysis at allele and amino acid levels. Despite the modest sample size, this is the first report on the HLA and COVID‐19 associations in cases of diverse populations in the United States. The preliminary findings in the Hispanics and younger African Americans are novel. It is of paramount importance to examine the immunogenetics of COVID‐19 in these minority populations with doubled to tripled rates of hospitalization and mortality compared with white and non‐hispanic populations.29 Our findings support the notion that specific HLA alleles may contribute to the protection from or predisposition to severe COVID‐19. Although the experimental evidence for a functional association remains lacking, the discovery of several associated amino acids in the peptide‐binding grooves of both class I and II molecules is consistent with the role of HLA‐restricted peptide presentation in the susceptibility to symptomatic SARS‐CoV‐2 infection.
Our findings add to the growing literature on the interaction between HLA and COVID‐19 from studies that vary in their approaches and study designs. Several epidemiology studies investigated the correlation between HLA genotype frequencies and regional prevalence or fatality rates of COVID‐19. The frequencies of HLA‐A*11:01 in 21 countries correlated negatively with the fatality rates of COVID‐19 in corresponding countries,30 while HLA‐A*02:01 was reported to be associated with increased risk for COVID‐19.31 In Italy, higher regional frequencies of HLA‐B*44 and ‐C*01 independently correlated with a faster local growth rate of SARS‐CoV‐2 infections; at the haplotype level, Pisanti et al reported the positive correlation between regional frequencies of the HLA‐A*01:01 g‐B*08:01 g‐C*07:01 g‐DRB1*03:01 g haplotype and the local prevalence and mortality of COVID‐19.32 Although these epidemiological studies indicate protective or permissive roles of HLA related factors in SARS‐CoV‐2 infection, their findings have not been replicated across studies.
Case–control studies offered another opportunity to uncover the immunogenetic underpinning of COVID‐19. While a recent genome‐wide association study (GWAS) performed in Italian and Spanish populations did not identify significant signals for COVID‐19 in the HLA region,22 several case–control studies from China and Italy reported a few significant associations. Wang and colleagues reported significantly increased counts of HLA‐B*15:27 (n = 8; 4.9% of cases) in 82 mild to severe COVID‐19 cases in Zhejiang province, China, as compared with 3548 controls from a local marrow donor registry. HLA‐C*07:29 also reached statistical significance in this study but only occurred once (0.6% of cases), which may not be reliable. Novelli et al observed increased frequencies of HLA‐B*27:07, ‐DRB1*15:01, and ‐DQB1*06:02 in 99 Italian COVID‐19 patients as compared with 1017 Italian individuals previously typed in the authors' laboratory.21
Another case–control study by Amoroso et al reported that HLA‐DRB1*08 was associated with almost doubled risk for COVID‐19 in solid‐organ transplant recipients and waitlisted candidates in Italy; HLA‐DRB1*08 positive COVID‐19 patients also had significantly increased mortality.20 These results, despite being from a population of transplant recipients and candidates, were consistent with our suggestive finding of increased HLA‐DRB1*08:02 among a small number of Hispanic COVID‐19 patients. While HLA‐DRB1*08:02 is more frequently found in Hispanics in North and South Americas, HLA‐DRB1*08:01 is the dominant HLA‐DRB1*08 allele found in Europe and is probably carried by most of the HLA‐DRB1*08‐positive cases in the Amoroso study (Figure S1).33 Of note, both HLA‐DRB1*08:01 and 08:02 share the two risk‐associated amino acid residues, 13Gly and 74Leu, which we hypothesize might be responsible for their poor binding affinity with SARS‐CoV‐2 peptides. Although our finding was limited to three observations among four Hispanics, these cases were not known to be related and did not share a common haplotype.
HLA‐A*30:02 has been associated with an increased risk for type 1 diabetes,34 while its role in viral infection control has not been widely known. In a preprint article, HLA‐A*30:02 was found to be enriched among COVID‐19 patients compared with controls without COVID‐19, although the association was not statistical significant after adjusting for multiple comparisons.35 The study appeared to be underpowered with 100 COVID‐19 patients and 26 controls, and the descent or ethnicity of the study population was not reported. Our finding of HLA‐A*30:02 as a risk factor for COVID‐19 among younger African Americans is consistent this earlier report and needs to be confirmed by further studies.
To establish a functional association between HLA and susceptibility to severe COVID‐19, future research will need to demonstrate the unproductive presentation of viral peptides by specific HLA molecules. However, because of the large size of the SARS‐CoV‐2 peptide repertoire and the diversity of HLA in the human population, in silico modeling has been frequently used to narrow down risk‐associated alleles and to identify peptide targets for vaccine development.13, 36, 37, 38, 39 As the modeling strategies and tools differ among studies,36 various predisposing alleles have been reported as expected.13, 31, 40, 41 Some studies also correlated lower class I peptide binding capacity with disease severity among COVID‐19 patients,42 or predicted altered binding of variant viral peptides and specific HLA alleles.43 In one of the most comprehensive analysis of HLA binding affinities of viral peptides, a list of strongest and weakest binders of SARS‐CoV‐2 peptides were predicted.40 One predisposing candidate allele found in our study, HLA‐C*04:01, was among the weakest binders of all viral peptides in this study.40 Finally, the HLA‐DRB1*08 allele group was predicted to be unable to bind SARS‐CoV‐2 peptides at high affinities by Amoroso et al20 and our own modeling (data not shown), which supports the finding of HLA‐DRB1*08:02 as a potential predisposing allele in our study.
Our study has important limitations. Although the sample size is comparable to most single‐center case–control studies in the literature, ideally thousands of cases may be pooled in a consortium setting for each population to maximize the power for detailed mapping of HLA‐disease associations. Therefore, protective and risk alleles with low to moderate effect sizes might have been missed in our study. We also used gender and geography‐matched population controls from a donor registry to demonstrate an enrichment of risk alleles among hospitalized COVID‐19 cases. Age matching was also performed to an extent that was limited in that stem cell donors older than age 60 are not recruited, however little evidence exists to support that HLA frequencies vary substantially by age within self‐reported ethnic categories in the general population. The comorbidities of the population controls were not available, so we could not control for comorbidities in this study, thus any potential interactions with HLA would remain undetected. The disease status of the controls was unknown, thus when compared with similar‐sized studies where known disease‐free controls are utilized, this study design has a higher likelihood that true associations would remain undetected. However, our approach of using stem cell donors has a benefit that a larger number of controls are available. An alternative study design would enroll COVID‐19 patients with no or mild symptoms as controls from the same location as the cases, which could allow the identification of HLA alleles associated with moderate to severe COVID‐19. However, this strategy may limit the sample size of controls available for the study. Additionally, the disease severity of COVID‐19 may be dynamic, requiring longitudinal follow up for symptoms.
In summary, we conducted a population‐based case–control study involving multiple populations in the midwest of United States and identified HLA‐DRB1*08:02, HLA‐A*30:02, and several other candidate alleles with increased or decreased frequencies among hospitalized COVID‐19 patients compared with matched population controls. As the suggestive finding in Hispanics was based on a small number of cases, caution is needed in their interpretation. We also determined the amino acid residues in these alleles that may be involved in peptide presentation during the immune response to SARS‐CoV‐2. Future consortium analysis of pooled cases and controls is warranted to validate and extend these findings, and correlation with peptide binding studies will provide additional evidence supporting the functional association between HLA alleles and severe COVID‐19.
FUNDING INFORMATION
Research fund from Washington University School of Medicine Department of Pathology and Immunology (C.L.) and NIH NIAID award number R41 AI142919‐01 (C.L.).
CONFLICT OF INTEREST
The authors have declared no conflicting interests.
AUTHOR CONTRIBUTIONS
Designed the study: Emily Schindler, Christopher W. Farnsworth, Loren Gragert, and Chang Liu. Collected specimens: Emily Schindler, Karl Hock, Christopher W. Farnsworth, and Chang Liu. Collected clinical data: Emily Schindler and Chang Liu. Performed HLA typing: Brian F. Duffy and Chang Liu. Performed data analysis and wrote the manuscript: Marian Dribus, Loren Gragert, and Chang Liu. All authors read and approved the final version of the manuscript.
Supporting information
Figure S1. Frequencies and distributions of common alleles in the DRB1*08 group.
Table S1. Alleles examined in the primary analysis.
Table S2. Alleles examined in the ad hoc analysis.
ACKNOWLEDGMENTS
We thank Megan Arb of Center for Clinical Studies of Washington University School of Medicine for helping with the data collection. We thank National Marrow Donor Program/Be The Match for providing control data and Martin Maiers for kindly reviewing the manuscript. We thank One Lambda Inc for providing the AllType and NGS LR kits and sequencing reagents for the Ion Chef/Ion Torrent S5 platform for this study.
Schindler E, Dribus M, Duffy BF, et al. HLA genetic polymorphism in patients with Coronavirus Disease 2019 in Midwestern United States. HLA. 2021;98(4):370‐379. 10.1111/tan.14387
Emily Schindler and Marian Dribus contributed to the work equally.
Funding information National Institutes of Health, Grant/Award Number: R41 AI142919‐01; Washington University School of Medicine Department of Pathology and Immunology
Contributor Information
Loren Gragert, Email: lgragert@tulane.edu.
Chang Liu, Email: cliu32@wustl.edu.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1.Zhou P, Yang X‐L, Wang X‐G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J, Chen X, Deng X, et al. Disease burden and clinical severity of the first pandemic wave of COVID‐19 in Wuhan, China. Nat Commun. 2020;11(1):5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS‐CoV‐2). Science. 2020;368(6490):489‐493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Z, McGoogan JM. Characteristics of and important lessons from the Coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 7,2314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323(13):1239‐1242. [DOI] [PubMed] [Google Scholar]
- 5.Gandhi RT, Lynch JB, Del Rio C. Mild or moderate Covid‐19. N Engl J Med. 2020;383:1757‐1766. 10.1056/NEJMcp2009249 [DOI] [PubMed] [Google Scholar]
- 6.Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID‐19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020;46(5):846‐848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC COVID‐19 Response Team . Preliminary estimates of the prevalence of selected underlying health conditions among patients with Coronavirus Disease 2019 ‐ United States, February 12–March 28, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(13):382‐386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold JAW, Wong KK, Szablewski CM, et al. Characteristics and clinical outcomes of adult patients hospitalized with COVID‐19 ‐ Georgia, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69(18):545‐550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garg S, Kim L, Whitaker M, et al. Hospitalization rates and characteristics of patients hospitalized with laboratory‐confirmed Coronavirus disease 2019 ‐ COVID‐NET, 14 states, March 1–30, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(15):458‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams FMK, Freidin MB, Mangino M, et al. Self‐reported symptoms of covid‐19 including symptoms most predictive of SARS‐CoV‐2 infection, are heritable. bioRxiv. Published online April 24, 2020. doi: 10.1101/2020.04.22.20072124 [DOI] [PubMed]
- 11.Murphy K, Travers P, Walport M. Janeway's Immunobiology. 7th ed.New York, NY: Garland Science; 2012. [Google Scholar]
- 12.Robinson J, Barker DJ, Georgiou X, Cooper MA, Flicek P, Marsh SGE. IPD‐IMGT/HLA Database. Nucleic Acids Res. 2020;48(D1):D948‐D955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen A, David JK, Maden SK, et al. Human leukocyte antigen susceptibility map for severe acute respiratory syndrome Coronavirus 2. J Virol. 2020;94(13):e00510‐20. 10.1128/JVI.00510-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin M, Tseng H‐K, Trejaut JA, et al. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med Genet. 2003;4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng MHL, Lau K‐M, Li L, et al. Association of human‐leukocyte‐antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J Infect Dis. 2004;190(3):515‐518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S‐F, Chen K‐H, Chen M, et al. Human‐leukocyte antigen class I Cw 1502 and class II DR 0301 genotypes are associated with resistance to severe acute respiratory syndrome (SARS) infection. Viral Immunol. 2011;24(5):421‐426. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y‐MA, Liang S‐Y, Shih Y‐P, et al. Epidemiological and genetic correlates of severe acute respiratory syndrome coronavirus infection in the hospital with the highest nosocomial infection rate in Taiwan in 2003. J Clin Microbiol. 2006;44(2):359‐365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keicho N, Itoyama S, Kashiwase K, et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum Immunol. 2009;70(7):527‐531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang W, Zhang W, Zhang J, He J, Zhu F. Distribution of HLA allele frequencies in 82 Chinese individuals with coronavirus disease‐2019 (COVID‐19). HLA. 2020;96(2):194‐196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amoroso A, Magistroni P, Vespasiano F, et al. HLA and AB0 polymorphisms may influence SARS‐CoV‐2 infection and COVID‐19 severity. Transplantation. 2021;105(1):193‐200. [DOI] [PubMed] [Google Scholar]
- 21.Novelli A, Andreani M, Biancolella M, et al. HLA allele frequencies and susceptibility to COVID‐19 in a group of 99 Italian patients. HLA. 2020;96(5):610‐614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Severe Covid‐19 GWAS Group , Ellinghaus D, Degenhardt F, et al. Genomewide association study of severe Covid‐19 with respiratory failure. N Engl J Med. 2020;383(16):1522‐1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012;80(1):1‐11. [DOI] [PubMed] [Google Scholar]
- 24.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)–a metadata‐driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C, Duffy BF, Weimer ET, et al. Performance of a multiplexed amplicon‐based next‐generation sequencing assay for HLA typing. PLoS One. 2020;15(4):e0232050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu C, Yang X, Duffy BF, et al. High‐resolution HLA typing by long reads from the R10.3 Oxford nanopore flow cells. Hum Immunol. 2021;82(4):288‐295. 10.1016/j.humimm.2021.02.005 [DOI] [PubMed] [Google Scholar]
- 27.Fan Y, Song Y‐Q. PyHLA: tests for the association between HLA alleles and diseases. BMC Bioinformatics. 2017;18(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289‐300. [Google Scholar]
- 29.CDC . COVID‐19 Hospitalization and Death by Race/Ethnicity. Accessed February 13, 2021. https://www.cdc.gov/coronavirus/2019-ncov/covid-data/investigations-discovery/hospitalization-death-by-race-ethnicity.html
- 30.Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. SARS‐CoV‐2 genomic variations associated with mortality rate of COVID‐19. J Hum Genet. 2020;65(12):1075‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomita Y, Ikeda T, Sato R, Sakagami T. Association between HLA gene polymorphisms and mortality of COVID‐19: an in silico analysis. Immun Inflamm Dis. 2020;8(4):684‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pisanti S, Deelen J, Gallina AM, et al. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid‐19. J Transl Med. 2020;18(1):352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solberg OD, Mack SJ, Lancaster AK, et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta‐analytic review of 497 population studies. Hum Immunol. 2008;69(7):443‐464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noble JA, Valdes AM, Bugawan TL, Apple RJ, Thomson G, Erlich HA. The HLA class I A locus affects susceptibility to type 1 diabetes. Hum Immunol. 2002;63(8):657‐664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren RL, Birol I. Retrospective in silico HLA predictions from COVID‐19 patients reveal alleles associated with disease prognosis. medRxiv. Published online November 2, 2020. doi: 10.1101/2020.10.27.20220863 [DOI]
- 36.Prachar M, Justesen S, Steen‐Jensen DB, et al. Identification and validation of 174 COVID‐19 vaccine candidate epitopes reveals low performance of common epitope prediction tools. Sci Rep. 2020;10(1):20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed SF, Quadeer AA, McKay MR. Preliminary identification of potential vaccine targets for the COVID‐19 Coronavirus (SARS‐CoV‐2) based on SARS‐CoV immunological studies. Viruses. 2020;12(3):254. 10.3390/v12030254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee CH, Koohy H. In silico identification of vaccine targets for 2019‐nCoV. F1000Res. 2020;9:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS‐CoV‐2. Cell Host Microbe. 2020;27(4):671‐680.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barquera R, Collen E, Di D, et al. Binding affinities of 438 HLA proteins to complete proteomes of seven pandemic viruses and distributions of strongest and weakest HLA peptide binders in populations worldwide. HLA. 2020;96(3):277‐298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pretti MAM, Galvani RG, Vieira GF, Bonomo A, Bonamino MH, Boroni M. Class I HLA allele predicted restricted antigenic coverages for spike and nucleocapsid proteins are associated with deaths related to COVID‐19. Front Immunol. 2020;11:3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Iturrieta‐Zuazo I, Rita CG, García‐Soidán A, et al. Possible role of HLA class‐I genotype in SARS‐CoV‐2 infection and progression: a pilot study in a cohort of Covid‐19 Spanish patients. Clin Immunol. 2020;219:108572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Sousa E, Ligeiro D, Lérias JR, et al. Mortality in COVID‐19 disease patients: correlating the association of major histocompatibility complex (MHC) with severe acute respiratory syndrome 2 (SARS‐CoV‐2) variants. Int J Infect Dis. 2020;98:454‐459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Frequencies and distributions of common alleles in the DRB1*08 group.
Table S1. Alleles examined in the primary analysis.
Table S2. Alleles examined in the ad hoc analysis.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


