Abstract
Silencing most gene expression from all but one X chromosome in female mammals provides a means to overcome X-linked gene expression imbalances with males. Central to establishing gene silencing on the inactivated X chromosome are the actions of the long noncoding RNA XIST that triggers the repackaging of the chosen X into facultative heterochromatin. While understanding the mechanisms through which XIST expression is regulated and mediates its affects has been a major focus of research since its discovery, less is known about the role XIST plays in maintaining chromatin at the human inactive X chromosome (Xi). Here we use genome engineering to delete the promoter of XIST to knockout expression from the Xi in non-cancerous diploid human somatic cells. Although some heterochromatin features exhibit limited change at the Xi, two of those assessed showed significant reductions including histone H2A monoubiquitylation at lysine 119 and histone H3 trimethylation at lysine 27, both of which are covalent histone modifications catalyzed by the Polycomb Repressive Complexes 1 and 2 respectively. Coupled with these reductions, we observed an occasional gain of euchromatin signatures on Xp, but despite these signs of chromatin instability, we did not observe appreciable changes in the reactivation of genes from the Xi. Collectively, these data are consistent with maintenance of dosage compensation at the Xi involving multiple redundant layers of gene silencing.
Keywords: X chromosome inactivation, XIST, Polycomb, Dosage compensation
Introduction
Early in the development of eutherian female mammals, one of the two X chromosomes is rendered transcriptionally silent, balancing X-linked gene expression with that of males (Lyon 1961). Once chosen, the same X is maintained as the inactive X chromosome (Xi) in all daughter cells derived from subsequent somatic cell divisions. Because the process occurs at a multi-cell stage and each cell makes an independent choice as to which X to silence, females are mosaic for X-linked mono-allelic expression. Central to this process of X chromosome inactivation (XCI) is one of the first characterized long noncoding RNAs called the X-inactive specific transcript (XIST). The XIST locus resides within the X inactivation center at Xq13 (Brown et al. 1991b), a physically defined region of the X (Brockdorff et al. 1991; Leppig et al. 1993) that is required in cis for XCI to occur. The gene is composed of a multi-exon, polyadenylated transcript (Brown et al. 1991a), and the orthologous gene can be found at the XIC in mouse (Borsani et al. 1991; Brockdorff et al. 1992). As the name indicates, XIST is expressed exclusively from the allele on the Xi and physically associates with the Xi territory in what is often referred to as an XIST cloud (Clemson et al. 1996). XIST induces the transition of the chosen Xi into facultative heterochromatin, which ultimately shuts down most expression from the chromosome (Carrel and Willard 2005), and under the light microscope the Xi can be observed in female nuclei as a densely staining mass, or Barr body, typically associated with the nuclear periphery or in close proximity to a nucleolus (Barr and Bertram 1949). Facultative heterochromatin of the Xi is characterized by numerous chromatin changes relative to its active X (Xa) counterpart. These include the acquisition of repressive histone modifications including di- and tri-methylation of lysine-9 (H3K9me3) (Boggs et al. 2002; Peters et al. 2002) and tri-methylation of lysine-27 (H3K27me3) (Plath et al. 2003; Silva et al. 2003) of histone H3, mono-ubiquitylation of histone H2A at lysine 119 (H2AK119Ub1) (de Napoles et al. 2004; Fang et al. 2004; Smith et al. 2004) and reduced levels of the euchromatic marker histone H3 di-methylated at lysine 4 (H3K4me2) (Boggs et al. 2002; Chadwick and Willard 2002). In addition to covalent modification of core histones, the histone variant macroH2A1 is enriched at the Xi (Costanzi and Pehrson 1998), as are a number of repressive chromatin proteins including structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) (Blewitt et al. 2008) and heterochromatin protein 1 (Chadwick and Willard 2003). Collectively, these and other acquired features serve to stably lock-in the silent state at the Xi.
Much of what we understand regarding the role of XIST in the early stages of XCI has come from studies in mouse (Loda and Heard 2019), although inducible human transgenes (Chow et al. 2007; Kelsey et al. 2015) and recent advances in human stem cell biology has provided insight into XCI mechanisms in humans (Patrat et al. 2020). While it is known that XIST is not required to maintain silencing at the Xi (Brown and Willard 1994; Rack et al. 1994), the impact of XIST loss on Xi chromatin is less well explored. A recent report (Lee et al. 2019) used genome engineering to systematically delete portions of the endogenous human XIST locus in a female cell line derived from a patient with chronic myelogenous leukemia (Lozzio and Lozzio 1975). This study assigned an important role for exon-5 in maintaining proper silencing and showed that exon-6 was important for the maintenance of the XIST cloud. However, chromatin features at the Xi were not possible to examine as H3K27me3 and H2AK119Ub1 were not robustly detected at the Xi (Lee et al. 2019). A likely explanation for this is because Xi chromatin and silencing is commonly disrupted in female cancer (Chaligne and Heard 2014), including changes to H3K27me3 distribution (Chaligne et al. 2015). In non-cancerous human cells, distinct multi-megabase non-overlapping bands of H3K9me3 and spatially separate bands of H3K27me3, H2AK119Ub1 and macroH2A1 can be observed at the Xi at metaphase (Chadwick 2007; Chadwick 2020; Chadwick and Willard 2002; Chadwick and Willard 2004; Darrow et al. 2016; Nozawa et al. 2013; Smith et al. 2004) that remain spatially separate at interphase. Here we use the CRISPR/Cas9 system (Ran et al. 2013) to delete the XIST promoter from the Xi in non-cancerous human cells in order to assess the impact that loss of XIST expression has on the organization and maintenance of chromatin and to evaluate changes to Xi gene expression.
Materials and Methods
Cells and Culture
hTERT-RPE1 (RPE1) are 46, X,t(X:10) female retinal pigment epithelial cells derived from telomerase immortalization of primary RPE-340 cells (Bodnar et al. 1998). These cells were originally obtained from Clontech Laboratories Inc. (Cat. No. C4000-1) but are now available through the American Type Tissue Culture Collection (CRL-4000). The X:10 translocation is the Xa in all cells (Darrow et al. 2016). RPE1 cells were cultured in DMEM-F12 (Genesee Scientific Cat. No. 25-503) supplemented with 1x Penicillin-Streptomycin-Glutamine (Thermo Fisher Scientific Cat. No. 10378-016), 1x Non-Essential Amino Acids (Thermo Fisher Scientific Cat. No. 11140-050), 0.23% volume per volume (v/v) sodium bicarbonate and 10% (v/v) fetal bovine serum (Thermo Fisher Scientific Cat. No. 16000-069).
Antibodies
Antibodies raised against human macroH2A1 were described previously (Chadwick and Willard 2001). Mouse monoclonal to HP1-gamma (HP1γ) (Cat. No. sc-398562), ATRX (Cat. No. sc-55584) and CTCF (Cat. No. sc-271474) were obtained from Santa Cruz Biotechnology. Rabbit polyclonal antibodies to EZH2 (Cat. No. 07-400), H3K4me2 (Cat. No. 07-030) and H3K9me3 (Cat. No. 07-523) were obtained from Millipore Sigma. Mouse monoclonal antibodies to H3K27me2/3 (Cat. No. 39535) were obtained from Active Motif. Rabbit monoclonal antibodies to H2AK119Ub1 (Cat. No. 8240S) was obtained from Cell Signaling Technology. Rabbit polyclonal antibodies to SMCHD1 (Cat. No. A302-872A) were obtained from Bethyl Laboratories Inc. Alexa Fluor 555 direct-labeled goat anti-rabbit (A21429) and goat anti-mouse (A21424), and Alexa Fluor 488 direct-labeled goat anti-rabbit (A11034) and goat anti-mouse (A11029) were obtained from Thermo Fisher Scientific.
XIST Targeting
Sequences up and downstream of the human XIST minimal promoter and A-repeats in exon-1 Hendrich et al. 1997) were used to identify suitable guide-RNA (gRNA) target sites using the Optimized CRISPR Design (http://crispr.mit.edu. The chosen gRNA sequences (Fig S1a and Table S1) were designed to be compatible with cloning into pSpCas9(BB)-2A-GFP (pX458) (Ran et al. 2013). pSpCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang (Addgene plasmid # 48138; http://n2t.net/addgene:48138; RRID:Addgene_48138). Clones were sequence verified before combinations of upstream (US) and downstream (DS) gRNAs were tested for deletion capability by transient transfection into 293 cells using Lipofectamine 3000 (Thermo Fisher Scientific, Cat. No. L3000008). Forty-eight hours post-transfection, genomic DNA was isolated from gRNA and mock transfected cells using the Nucleospin Tissue Kit from Machery-Nagel (Cat. No. 740952). Genomic DNA was used for Polymerase Chain Reaction (PCR) with primer combinations flanking the deletion site. The most efficient pair of gRNAs were chosen (Fig S1b) and transfection grade plasmid DNA was isolated using the Qiagen midi-prep DNA isolation kit (Cat. No. 12945). Introduction of the gRNA pair into RPE1 cells was achieved by Nucleofection (Lonza). Seventy- two hours later, single GFP positive cells were directly sorted into single wells of four 96-well plates using a BD FACSAria SORP flow cytometer. Colonies were expanded 3–4 weeks later and assessed for the presence or absence of the desired deletion by PCR (Fig S1c). All standard PCR was performed with HotStarTaq Plus (Qiagen, Cat. No. 203605).
Metaphase Chromosomes
Metaphase chromosomes were prepared essentially as described previously (Chadwick and Willard 2002). Briefly, a 1:400 dilution of KaryoMAX Colcemid (Thermo Fisher Scientific, Cat. No. 15212-012) was added to media of actively dividing cells and incubated for one hour at 37°C 5% CO2. The media was collected and transferred to a 50ml tube. Cells were washed with phosphate buffered saline (PBS) which was added to the media in the 50ml tube. Cells were detached with TrypLE (Thermo Fisher Scientific, Cat. No. 12605-010) and collected using the media/PBS mixture, before counting followed by briefly pelleting the cells. Chromosomes were attached to slides by centrifugation using a Shandon Cytospin 4 (Thermo Fisher Scientific), spinning at 1,900 × rpm for 10 minutes. Post-spin, samples were fixed for 10 minutes in 1x PBS supplemented with 0.1% (v/v) Triton X-100 and 3.7% formaldehyde, before washing twice in 1x PBS. Samples were then subjected to immunofluorescence as described below.
Immunofluorescence
The preparation of microscope slides with immobilized metaphase chromosomes for immunofluorescence is described above. For interphase analysis, cells were grown on standard microscope slides overnight in 100mm dishes. Slides were removed from the dishes before rinsing briefly with 1x PBS. Cells were fixed for 10 minutes in 1x PBS supplemented with 0.1% (v/v) Triton X-100 and 3.7% formaldehyde, before washing twice in 1x PBS. Samples were blocked for 15–30 minutes in a humidified chamber with 3% bovine serum albumin (BSA) in 1x PBS containing 0.1% (v/v) Tween-20 (PBS-T) overlaid with cover glass. Cover glass were tipped off before rinsing for 2 minutes in 1x PBS. An appropriate dilution of primary antibody/antibodies in PBS-T containing 1% (v/v) BSA were applied to samples followed by cover glass and incubation in a humidified chamber for at least 1 hour. Samples were washed three times for 2 minutes each in 1x PBS before applying a 1:200 dilution of direct-labeled secondary antibodies in PBS-T supplemented with 1% (v/v) BSA. Samples were washed three times for 2 minutes each in 1x PBS before fixing once more as described above. Vectashield with 4,6-diaminidino-2-phenylindole (DAPI) (Vector Laboratories, Cat. No. H-1200) was applied along with cover glass.
RNA Fluorescence In Situ Hybridization
For RNA fluorescence in situ hybridization (FISH), cells grown overnight on slides were washed and fixed as described above for immunofluorescence before proceeding to dehydrate through 70% and 100% ethanol (v/v) for 2 minutes each before air drying. A direct-labeled fluorescent red probe for the X-linked microsatellite DXZ4 and a direct-labeled green XIST probe were prepared as described previously (Tremblay et al. 2011). One microgram of labeled DNA was ethanol precipitated with 25 micrograms of human Cot-1 DNA (Sigma-Aldrich, Cat. No. 11581074001) before resuspending in 0.1ml of Hybrisol VII (MP Biomedicals, Cat. No. 11RIST13901). A 1:1 mixture of the two probes were denatured at 72°C for 5 minutes before incubation at 37°C for 60 minutes. The repeat-blocked probe was then applied to the slides and sealed with rubber cement under cover glass before incubating at 37°C overnight. After removal of the cover glass, slides were washed twice at room temperature in 50% formamide, 2x sodium citrate sodium chloride (SSC), followed by 3 minutes at 37°C in 50% formamide 2x SSC and one wash of 3 minutes at 37°C in 2x SSC before addition of Vectashield with DAPI (Vector Laboratories, Cat. No. H-1200) and cover glass. A negative control in which slides were pre-treated with RNaseA in PBS for 30 minutes prior to hybridization was included, to confirm that signals were only obtained when RNA was present.
Fluorescence Microscopy
Images were collected using a Delta Vision pDV deconvolution system controlled by softWoRx 7 (GE Healthcare). Images were exported as 24-bit RGB TIFF files and assembled into figures using Adobe Photoshop CC 2018 (Adobe Systems). Line profiles were generated from deconvolved images using the Arbitrary Profile tool in softWoRx with Constant Min/Max settings. Data was extracted as symbolic link (SYLK) files and line profiles generated in Microsoft Excel. Fluorescent intensity measurements were achieved using the Edit Polygon tool in softWoRx using Enhanced Ratio deconvolved images. A total of 20 images were collected for each sample using a grid pattern, capturing images of the first 20 nuclei encountered. Data for each channel was extracted as SYLK files. Total fluorescence and relative to DAPI was calculated in Microsoft Excel. Violin plots were generated in R using package ggplot2 (Wickham 2016).
Standard and Quantitative Reverse Transcription PCR
Total RNA was isolated from cells using the Machery Nagel Nucleospin RNA kit (Cat. No 740955.250). DNaseI (NEB, Cat. No. M030L) treated RNA was used to make complementary DNA (cDNA) using random primers (NEB, Cat. No. S1330S) and M-MuLV reverse transcriptase (NEB, Cat. No. M0253L). The cDNA was used for standard or quantitative RT-PCR (qRT-PCR). All qRT-PCR was performed on a CFX96 (Bio-Rad) using SYBR Green PCR master mix (Thermo Fisher Scientific, Cat. No. 4334973). All oligonucleotides for PCR and qRT-PCR were obtained from Eurofins Genomics. Oligonucleotides used for RT-PCR and qRT-PCR can be found in Table S2.
Single Nucleotide Polymorphism Analysis
X-linked putative expressed single nucleotide polymorphisms (SNPs) in RPE1 cells were obtained from phased SNP and RNA-Seq data (Darrow et al. 2016). Primer pairs were designed flanking the SNP sites (Table S2) and used for PCR with RPE1 genomic DNA. Primers were removed from PCR products using the Monarch PCR and DNA Cleanup Kit (NEB, Cat. No. T1030L) and submitted to Eurofins Genomics for Sanger sequencing with either the forward or reverse primer. Sequence chromatograms were examined using Sequencher 5.4.6 (Gene Codes) to validate the presence of the SNP. PCR was then repeated using DNA isolated from a somatic cell hybrid in which the RPE1 Xa is the only human chromosome, in order to confirm which SNP allele corresponded to the Xa and Xi respectively, as done previously (Sun and Chadwick 2018). Where PCR primers were colinear between the genomic and cDNA sequence, these primers were used to amplify the SNP interval from cDNA. If one or both primers resided in intronic sequences, new cDNA-specific primers were designed in order to genotype the cDNA (Table S2). Sequence analysis of cDNA was used to determine mono versus bi-allelic expression.
Statistical Analysis
Expression of XIST relative to GAPDH was determined using the delta-delta-Ct method. Relative expression was measured on three separate occasions using three independent RNA extractions and cDNA preparations, each assessed by qRT-PCR performed in duplicate. Significant differences were calculated using unpaired t tests. Fluorescence intensity data was assessed using One-Way ANOVA and Tukey honestly significant difference procedure.
Results
Deletion of the human XIST minimal promoter and A-repeats
Little is known about the impact that XIST loss has on the organization and maintenance of chromatin in non-cancerous human cells. Here we sought to begin to address this by using genome engineering to delete the XIST promoter from the Xi. We followed a similar strategy to what we have used previously to delete the macrosatellite DXZ4, an enhancer within the IL1RAPL1 gene and the promoter of the IL1RAPL1 gene (Darrow et al. 2016; Sun and Chadwick 2018). The minimal promoter of both the human and mouse XIST/Xist genes have been described previously (Hendrich et al. 1997; Martindale et al. 2000) and encompass less than 1kb upstream of the transcription start site of a large first exon (Fig1a). In addition to searching for a suitable guide RNA (gRNA) target site in this upstream region, we decided to select a second gRNA downstream of the transcription start site in order to encompass the X-conserved repeats, that are one of several highly conserved repeat elements in XIST (Nesterova et al. 2001) and this particular repeat is now more commonly referred to as the A-repeats. The rationale for this is that these were shown to boost the activity of the minimal promoter 3-fold (Hendrich et al. 1997), and others have reported a role for these repeats in increasing Xist expression and proper splicing of Xist (Colognori et al. 2019; Royce-Tolland et al. 2010; Wang et al. 2019b; Zhao et al. 2008). Three gRNAs located upstream (US1, 2 and 3) and three gRNAs located downstream (DS1, 2 and 3) of the minimal promoter and A-repeats were selected (FigS1a). Combinations of up and downstream gRNAs were introduced into 293 cells before harvesting DNA and assessing for the presence of the desired deletions by PCR across the interval. The desired deletion could be detected for most gRNA combinations. The gRNA pair US2 and DS3 along with screening primers F2 and R1 were selected (FigS1b). The chosen gRNAs were then introduced into the hTERT immortalized diploid female cell line RPE1 (Bodnar et al. 1998). These cells carry an Xq28:10 translocation on the Xa and the karyotypically normal X is the Xi in 100% of cells (Chadwick and Willard 2003), allowing cytological distinction between the two X chromosomes at metaphase and by allele-specific assignment of X-linked SNPs. RPE1 cells Nucleofected with the gRNA pair were first sorted as single cells by FACS based on the presence of GFP that is co-expressed from the CRISPR/Cas9-gRNA construct (Ran et al. 2013). Clones were expanded before the presence of a deletion was assessed by PCR (FigS1c). A single clone (XIST-76) was identified in the first targeting attempt, a second clone was identified from a second independent targeting attempt (XIST-75) and two other clones were identified from a third targeting attempt (XIST-51 and XIST-92). Notably, XIST-75 grew considerably slower than the other clones, which impacted the ability to perform some of the desired downstream experiments. To confirm the deletion, the breakpoint was amplified by PCR and the product sequenced. These data confirmed that the screening PCR product was indeed the desired deletion of XIST and defined the extent of the deletion as 2,240 or 2,241bp for each clone (FigS2).
Fig.1.

XIST targeting. (a) Schematic map showing the structure of the XIST gene locus corresponding to bases 73,040,491 – 73,072,558 of the human X-chromosome (GRCh37/hg19). The open boxes represent the location and size of exons 1–8, joined by introns. The shaded interval overlapping the 5’ end of exon-1 is expanded in part-b. (b) Schematic map corresponding to the promoter region and 5’ end of the first exon of human XIST, corresponding to 73,071,053 – 73,074,052 of the human X-chromosome (GRCh37/hg19). The location of the gRNA target sites is indicated, as is the location of the oligonucleotides used for PCR across the deleted interval. The thick black horizontal line corresponds to the location of the minimal promoter (Hendrich et al. 1997). The open rectangle represents the 5’ end of XIST exon-1, and the location of the A-repeats is indicated by the shaded area.
XIST expression analysis in deletion mutants
In order to further characterize the clones and determine whether the deletion had occurred on the Xa or Xi, clones were assessed by RT-PCR with primers encompassing the A-repeats and therefore within the deleted region (Fig2a and Table S2). Products of the expected size were detected for genomic DNA which is colinear with the mRNA for this region as well as for parental RPE1 and XIST-92 cDNA, but not for the other three clones or the negative controls (Fig2b). These data suggest that the deletion is on the Xa for XIST-92 but Xi for XIST-51, XIST-75 and XIST-76. To confirm the assignment of the deletion to the Xa or Xi, qRT-PCR was performed between exon-1 and exon-2 of XIST (Fig2a). The level of XIST RNA in XIST-92 was comparable to parental RPE1, whereas no significant product could be detected for XIST-51, XIST-75 and XIST-76. To further validate these observations, RNA FISH was performed using a probe encompassing the complete XIST locus and a second probe to the microsatellite DXZ4 that is expressed from the Xa in >90% of RPE1 nuclei (Tremblay et al. 2011). Consistent with the RT-PCR and qRT-PCR data, an XIST RNA cloud was detected in RPE1 and XIST-92 (Fig2d), but not in XIST-51 and XIST-76. The DXZ4 RNA FISH positive control was detected in all clones. Based on these observations, we conclude that the XIST promoter and A-repeats are deleted from the Xa in clone XIST-92, whereas it is deleted from the Xi in XIST clones 51, 75 and 76.
Fig.2.

Expression of XIST RNA. (a) Schematic map showing the structure of the XIST gene locus corresponding to bases 73,040,491 – 73,072,558 of the human X-chromosome (GRCh37/hg19). The open boxes represent the location and size of exons 1–8, joined by introns. The region targeted for deletion covering the minimal promoter and 5’ end of XIST exon-1 is indicated. The location of primer pairs used for RT-PCR and qRT-PCR are indicated beneath the gene structure. (b) Inverted image of an ethidium bromide stained agarose gel showing the results of RT-PCR analysis of XIST exon-1 (top gel) and the GAPDH control (bottom gel). The molecular weight marker is loaded in the first lane and the size of various bands in base pairs is indicated. The water lane corresponds to a no-template control and DNA corresponds to genomic DNA from RPE1 cells that is co-linear with cDNA. Samples of cDNA prepared with (+RT) or without (−RT) reverse transcriptase are indicted above the gel for parental RPE1 and deletion positive clones. (c) Expression of XIST RNA in cDNA from the samples indicated (x-axis) relative to GAPDH (y-axis) as determined by qRT-PCR with primers spanning exons 1 and 2 of XIST. Columns correspond to the mean from three biological replicates, each performed as two technical replicates. The error bars correspond to the standard deviation of the mean. The indicated p-values were derived from performing unpaired t tests. (d) Images showing the results of RNA FISH with direct-labeled XIST (green) and DXZ4 (red) probes. The white-arrows indicate the location of the XIST cloud, whereas the white-arrowheads indicate the location of the DXZ4 signal. Nuclei are counter stained with DAPI (blue). The white horizontal scale bars in the bottom left corners correspond to 5 microns.
Changes in chromatin features at the Xi in XIST deletion mutants
Next, we sought to characterize the chromatin in these clones by indirect immunofluorescence. Given that most genes on the Xi are transcriptionally silent, the euchromatin associated covalent histone modification H3K4me2 is largely absent from the Xi and provides a robust signal to delineate the chromosome territory at interphase, typically as a void in the staining pattern within which is an intensely staining focus (Boggs et al. 2002) that corresponds to the euchromatic Xi-copy of DXZ4 (Chadwick 2008; Chadwick and Willard 2002).
Immunofluorescence to the multi-functional zinc-finger protein CCCTC-binding factor (CTCF) (Arzate-Mejia et al. 2018) results in an Xi-pattern that is very similar to H3K4me2 (Chadwick and Willard 2003) (Fig3a). Therefore, our mouse anti-CTCF antibody could be used to define the interphase Xi in combination with other rabbit primary antibodies, while the rabbit anti-H3K4me2 could be used with mouse primary antibodies, avoiding the need for direct-labeling.
Fig.3.
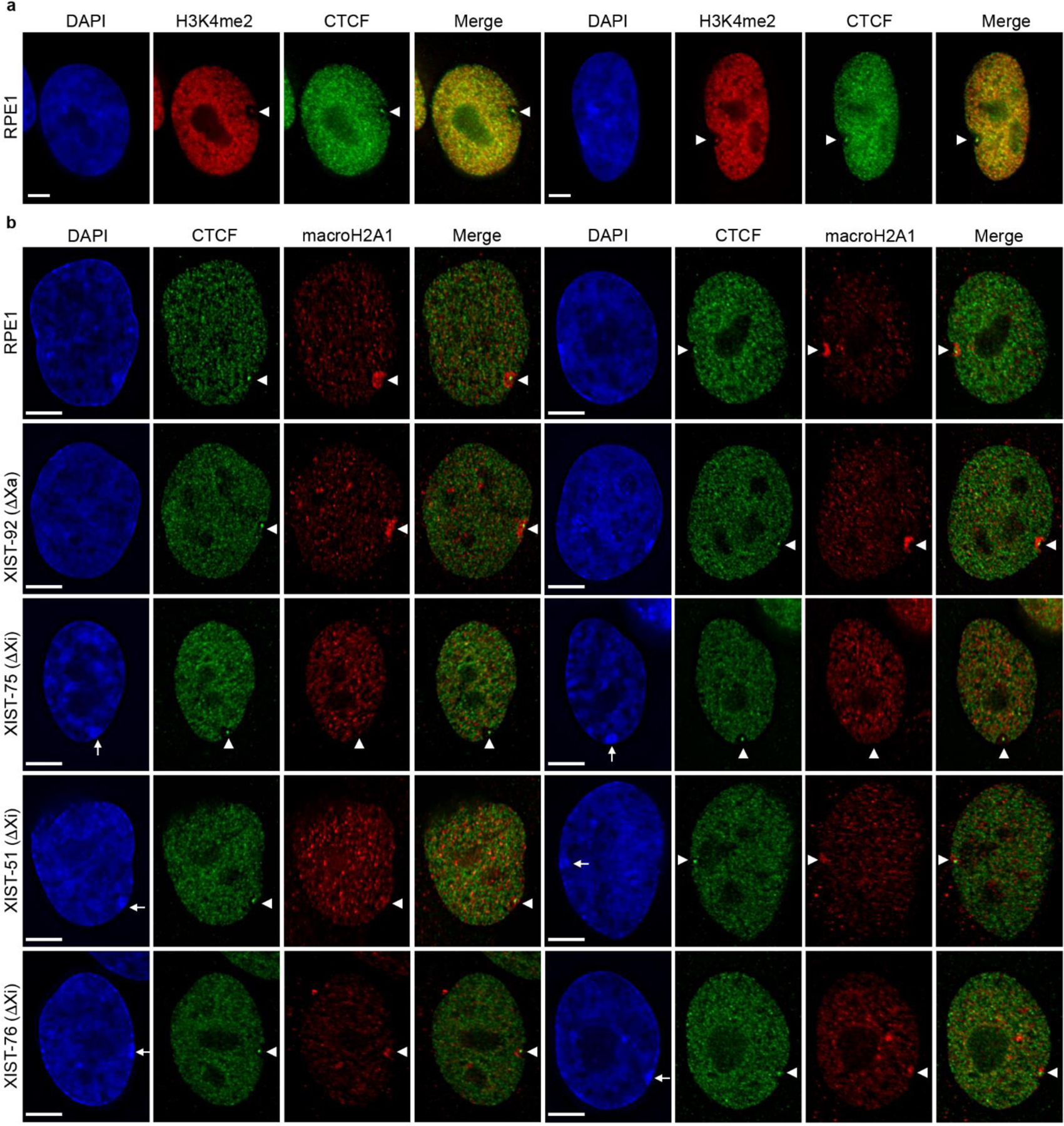
Chromatin characterization in XIST deletion mutants. (a) Distribution of H3K4me2 (red) and CTCF (green) in RPE1 cells by indirect immunofluorescence. Overlap of signals in the merged images appear yellow/orange. White arrowheads indicate the location of the intense H3K4me2 and CTCF signals at DXZ4 on the Xi. (b) Distribution of macroH2A1 (red) and CTCF (green) in RPE1 cells and XIST deletion mutants as indicated to the left of each row of images. White arrowheads indicate the location of the Xi territory and white arrows point to the DAPI dense Barr body in XIST Xi mutants. Nuclei are counter stained with DAPI (blue). The white horizontal scale bars in the bottom left corners of the DAPI images correspond to 5 microns.
Initially we examined the distribution of the histone variant macroH2A1 (Chadwick and Willard 2003), which is enriched within the territory of the Xi (Costanzi and Pehrson 1998) in what is referred to as a macrochromatin body (MCB). As expected, (Costanzi and Pehrson 1998), an MCB was readily detected in RPE1 and XIST-92 cells (Fig3b). In contrast, the MCB was substantially smaller and reduced in intensity in XIST-51 and XIST-76 and was not observed in XIST-75 (Fig3b). In all cases the DXZ4-associated CTCF focus and void was readily observed, coinciding with the DAPI-dense Barr body (Barr and Bertram 1949). The absence of an MCB combined with general nuclear distribution of macroH2A1 in XIST-75 is consistent with observations made in mouse somatic cells in which Xist was deleted post-XCI using the Cre-LoxP system (Csankovszki et al. 1999). However, the detection of an MCB, albeit reduced in size, in XIST-51 and XIST-76 indicate that in human cells, macroH2A1 can associate with the Xi territory in the absence of XIST.
Next, we investigated the robust Xi marker Structural maintenance of chromosomes flexible hinge domain containing 1 (SMCHD1) (Blewitt et al. 2008). In XIST-92, SMCHD1 signal at the Xi looked indistinguishable from RPE1 (Fig4a). However, similar to XIST knockdown experiments in RPE1 (Nozawa et al. 2013), SMCHD1 signal was negatively impacted in XIST-51 and XIST-76. The overall intensity of SMCHD1 at the Xi was reduced, but not absent, while intense SMCHD1 signals at other non-Xi nuclear foci remained unchanged (Fig4a). In contrast, SMCHD1 at the Xi in XIST-75 was very obvious and appeared unchanged. These data suggest that unlike in mouse where deletion of Xist post XCI or dissociation of Xist from the Xi using locked nucleic acids (Jansz et al. 2018; Wang et al. 2019a; Wang et al. 2018) results in loss of Smchd1 at the Xi, SMCHD1 can remain associated with the Xi territory to a varying extent in humans in the absence of XIST (Fig4a) (Nozawa et al. 2013).
Fig.4.
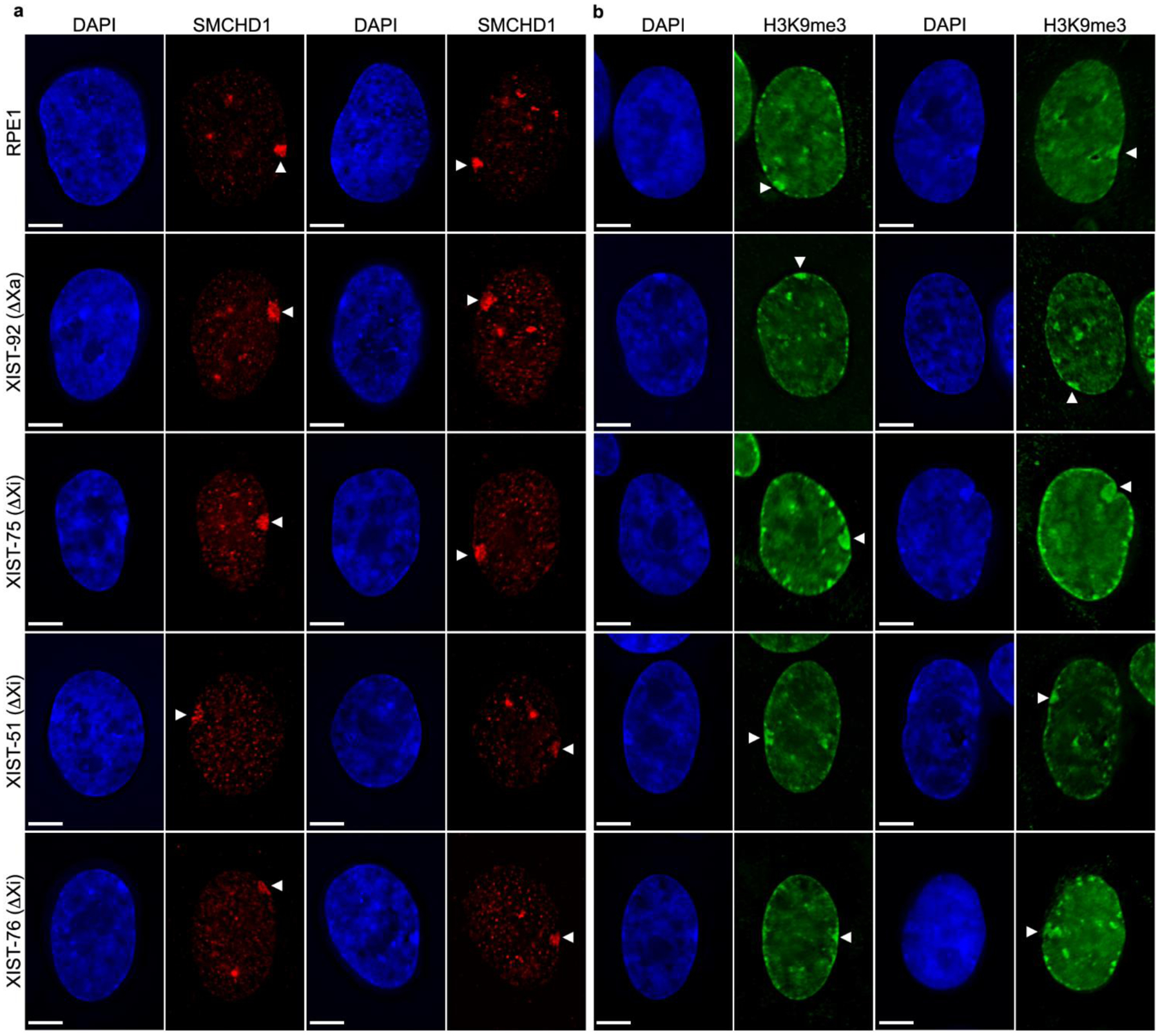
Heterochromatin characterization in XIST mutants. (a) Distribution of SMCHD1 (red), and (b) H3K9me3 (green) by indirect immunofluorescence in RPE1 and XIST mutants as indicated to the left of each row. White arrowheads point to the territory of the Xi. Nuclei are counter stained with DAPI (blue). The white horizontal scale bars in the bottom left corners correspond to 5 microns.
H3K9me3 is a prominent marker of the Xi in RPE1 (Chadwick and Willard 2004; Nozawa et al. 2013). This modification was obvious at the Xi regardless of whether the XIST promoter was deleted from the Xa or Xi (Fig4b). However, in the XIST-75 mutants, the H3K9me3 signal was substantially larger than that observed in parental cells and other XIST mutants. Close examination of the parental cells and all mutants revealed that the DAPI dense Barr body signal appeared to mimic the H3K9me3 signal. This can be more readily observed through line profiles (Fig5a). In the representative examples shown in Fig5a, the fluorescence profile for DAPI and H3K9me3 are very similar, and clearly show the more extensive and intense Barr body and H3K9me3 territory in XIST-75.
Fig.5.
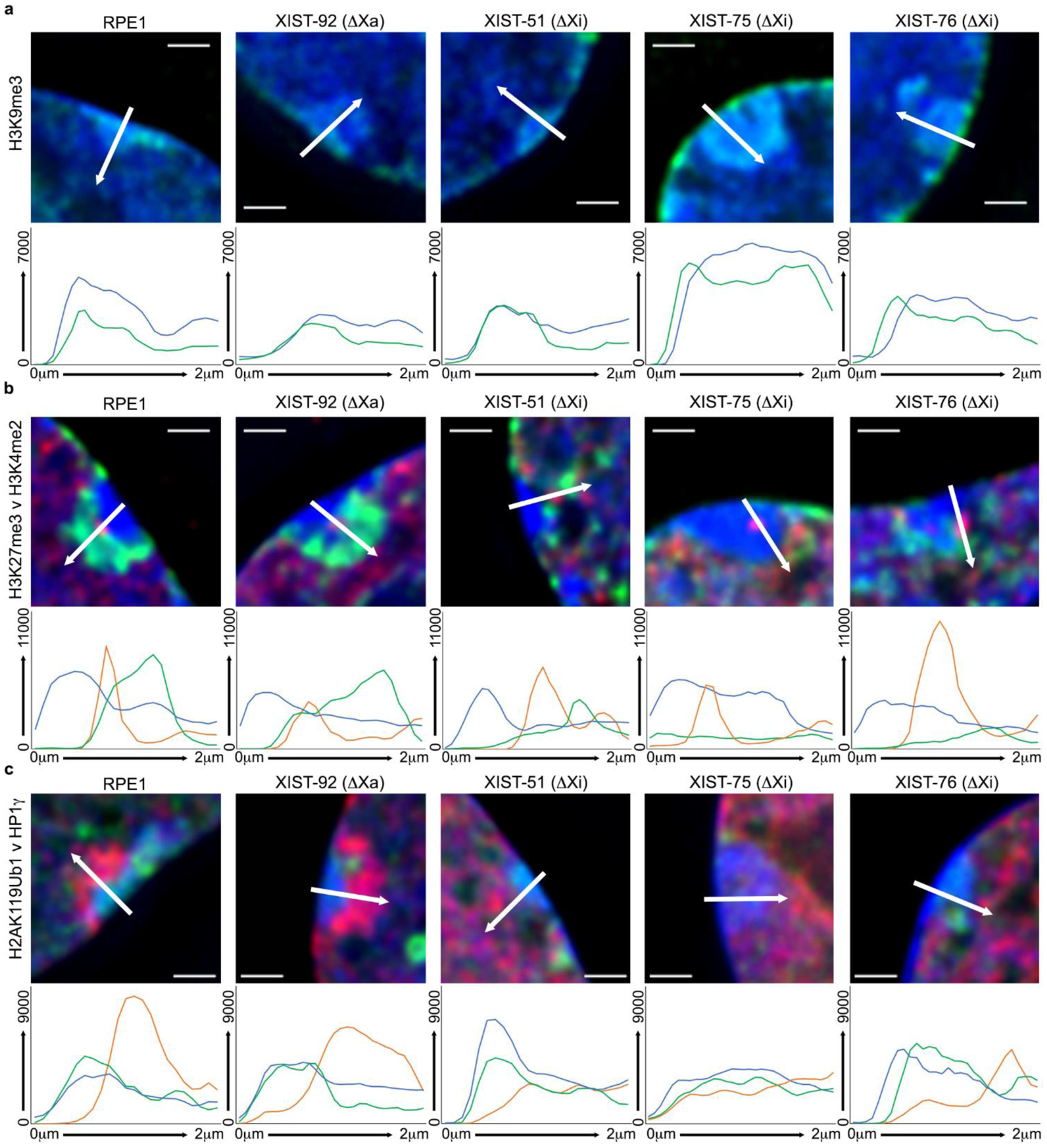
Arbitrary line profiles across the Xi territory for (a) H3K9me3 (green), (b) H3K27me3 (green) and H3K4me2 (red), and (c) H2AK119Ub1 (red) and HP1γ (green). Examples shown are the cells indicated above each image. Beneath each image is the arbitrary line profile based on the white-arrow shown in the image that spans 2 microns. The line colors correspond to those in the associated image. The y-axis is camera fluorescence intensity value and the x-axis is distance along the arrow from 0–2 microns. Nuclei are counter stained with DAPI (blue) and merged with the immunofluorescence signals. The white horizontal line in each image corresponds to 1 micron.
The H3K27me3 histone methyltransferase Enhancer of Zeste (EZH2) (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002) is responsible for this modification at the Xi (Plath et al. 2003; Silva et al. 2003), and is initially assisted by the activity of Alpha Thalassemia/Mental Retardation Syndrome X-Linked (ATRX) (Sarma et al. 2014), and ATRX can be detected at the Xi in somatic cells (Baumann and De La Fuente 2009). Consistent with these data, we observed both EZH2 and ATRX at the Xi in RPE1 cells (Fig6a). Notably, EZH2 association with the Xi was not common, which is likely consistent with its reported transient association with the Xi territory (Plath et al. 2003; Silva et al. 2003). Interestingly, the ATRX signal was spatially distinct from the EZH2 signal with no obvious overlap (Fig6a, right two panels), suggesting that while ATRX may assist the initial loading of EZH2 onto the Xi during the establishment of XCI, it may not be performing a similar role during maintenance. Like RPE1 cells, both EZH2 (Fig6b) and ATRX (Fig6c) were observed at the Xi in XIST-92 cells. Of the Xi XIST promoter mutants, only XIST-51 and XIST-76 showed occasional EZH2 signals at the Xi comparable to RPE1 (Fig6b), but no EZH2 signals were observed in the vicinity of the Xi in XIST-75. In contrast, ATRX was observed at the Xi in both Xa and Xi XIST promoter mutants (Fig6c).
Fig.6.
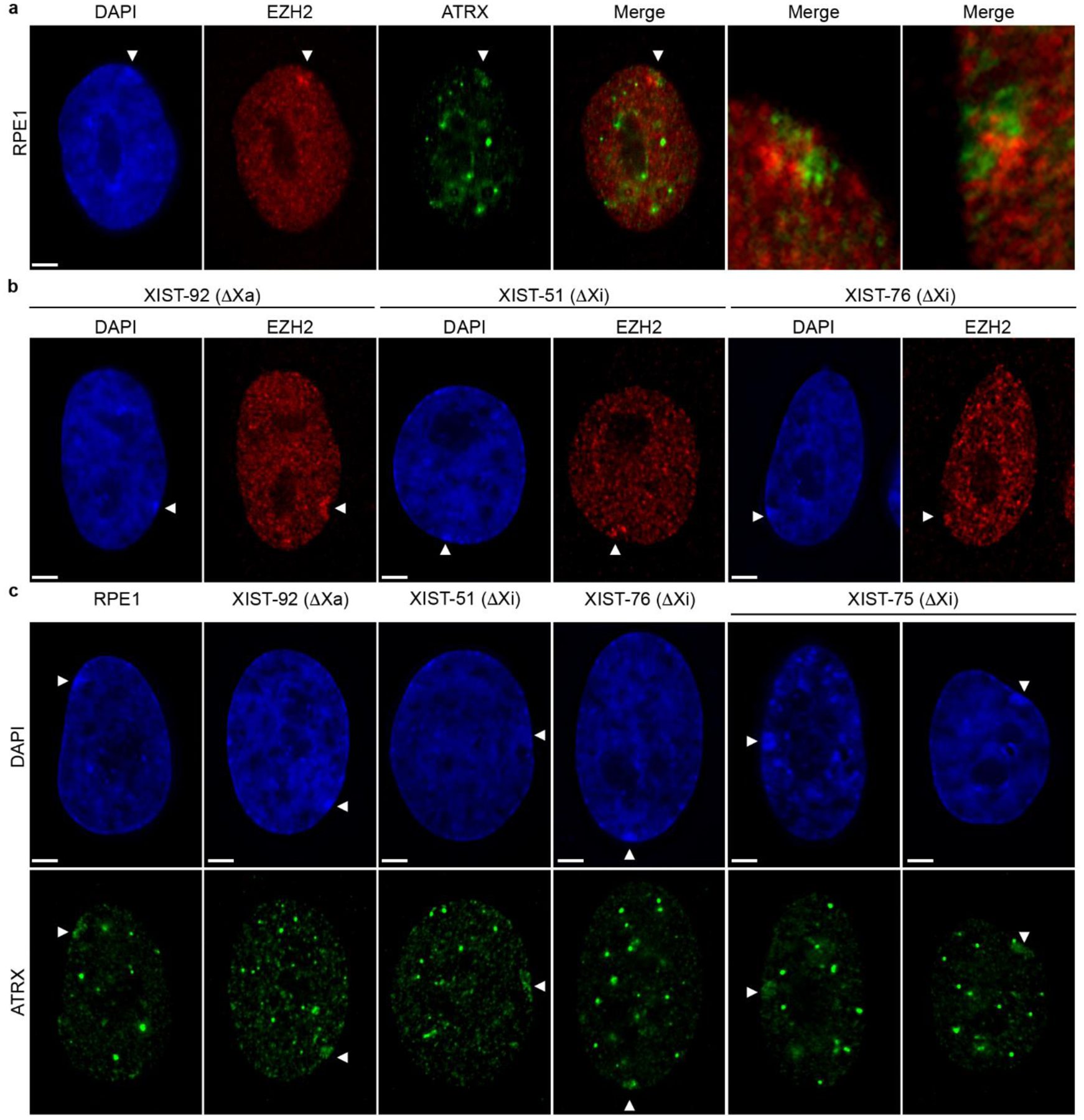
Distribution of ATRX and EZH2 at the Xi. (a) Distribution of EZH2 (red) and ATRX (green) at the Xi in RPE1 cells by indirect immunofluorescence. The two images on the far right are zoomed in 4x on the distribution of the two proteins at the Xi in two independent samples. (b) Indirect immunofluorescence showing the association of EZH2 (red), and (c) ATRX (green) with the Xi in the XIST deletion mutants indicated. White arrowheads indicate the location of the Xi. Nuclei are counter stained with DAPI (blue). The white horizontal scale bars in the bottom left corners of the DAPI images correspond to 5 microns.
We next examined H3K27me3, in combination with H3K4me2 to highlight the void and DXZ4 enriched focus to demarcate the territory of the Xi. As expected, RPE1 and the Xa XIST promoter mutant XIST-92 showed a clear enrichment of H3K27me3 nestled within the H3K4me2 void of the Xi (Fig7a). However, the Xi XIST promoter mutants XIST-51 an XIST-76, like macroH2A1 (Fig3b), showed a considerably smaller H3K27me3 signal compared to parental cells, whereas XIST-75 showed no H3K27me3 signal (Fig7a), which is consistent with the inability to detect EZH2 at the Xi in this clone. Additionally, close examination of the H3K4me2 void in XIST-75 revealed the occasional appearance of a second, albeit weaker H3K4me2 focus. Given that the CTCF void and focus in this clone was comparable to all other clones (Fig3b), these cells were re-stained with CTCF and H3K4me2. While the DXZ4 associated CTCF and H3K4me2 signals clearly overlapped, the new H3K4me2 signal was weaker and did not overlap CTCF (Fig7b). The DNA sequence that underlies this acquired H3K4me2 signal is currently unknown. While the H3K27me3 signal was generally smaller in the Xi XIST promoter mutants XIST-51 and XIST-76, we noticed that the H3K4me2 void was frequently not as clearly defined and that a single intense H3K4me2 focus that typically coincides with DXZ4 was often accompanied by other nearby intense foci not seen in RPE1 or XIST-92 (Fig7c). Consistent with the bi-partite organization of the Xi chromatin (Chadwick and Willard 2004), the H3K27me3 territory in RPE1 and XIST-92 is oriented toward the nuclear interior and is not overlapping the most DAPI-dense portion of the Barr body based upon line profiles (Fig5b). Line profiles across the Xi more readily reveal the reduced intensity (XIST-51 and XIST-76) or absence (XIST-75) of H3K27me3 at the Xi in the mutants lacking XIST (Fig5b).
Fig.7.

Chromatin changes at the Xi. (a) Indirect immunofluorescence showing the distribution of H3K4me2 (red) and H3K27me3 (green) in RPE1 or XIST mutants (indicated to the left of each row). White arrowheads indicate the location of the Xi. Nuclei are counter stained with DAPI (blue). The white horizontal scale bars in the bottom left corner of the DAPI images correspond to 5 microns. (b) Close up images of the Xi territory in XIST Xi deletion mutant XIST-75 showing the distribution of H3K4me2 (red) and CTCF (green). The white arrowheads point to the prominent H3K4me2/CTCF signal associated with DXZ4 and the additional weaker H3K4me2 signal. (c) Distribution of H3K27me3 (green) and H3K4me2 (red) at the Xi in RPE1 and XIST deletion mutants 92, 51 and 76. The white arrowheads point to the territory of the Xi.
Finally, we examined the distribution of Heterochromatin Protein 1 gamma (HP1γ) and H2AK119Ub1 at the Xi. Previously we have reported the presence of all three HP1 paralogs at the Xi in several different human cell types, including TERT immortalized RPE1 and a variety of primary cells (Chadwick 2020; Chadwick and Willard 2003; Chadwick and Willard 2004). HP1 has also been observed at the Xi in vole (Shevchenko et al. 2018; Shevchenko et al. 2009). Given that H3K9me3 serves as a binding site for the chromodomain of HP1 (Bannister et al. 2001; Jacobs et al. 2001; Lachner et al. 2001), and that H3K9me3 is not lost from the Xi in our Xi XIST promoter mutants (Fig4b), we anticipated that HP1γ would also be retained at the Xi, which was indeed the case for parental cells and all mutants (Fig8). As expected, the HP1γ signal in XIST-75 was considerably larger, consistent with the more extensive H3K9me3 signal and Barr body (Fig5c). H2AK119Ub1 enrichment at the Xi is established by the PRC1 complex (de Napoles et al. 2004; Fang et al. 2004; Smith et al. 2004). The combination of HP1γ with H2AK119Ub1 reliably reveal the bi-partite structure of the Xi (Chadwick and Willard 2004), with HP1γ oriented more toward the nuclear periphery and H2AK119Ub1 oriented more toward the interior, as was observed for RPE1 and XIST-92 (Fig8, top two rows). The separation of the two territories can be seen more clearly through line profiles (Fig5c, first two panels). Given the lack of EZH2 association and H3K27me3 at the Xi in the XIST-75 clone (Fig7a), the absence of H2AK119Ub1 signal at the Xi in this clone was anticipated (Fig8, middle row and Fig5c). However, it was somewhat surprising that we did not detect H2AK119Ub1 at the Xi in either of the Xi XIST promoter mutants (Fig8, bottom two rows and Fig5c). Among the chromatin proteins and covalent histone modifications examined in this study, this was the only Xi associated feature that was consistently absent in all three of the Xi promoter mutants.
Fig.8.

Chromatin changes at the Xi. Indirect immunofluorescence showing the distribution of HP1g (green) and H2AK119Ub1 (red) in RPE1 and XIST deletion mutants (indicated to the left of each row). Nuclei are counter stained with DAPI (blue). White arrowheads point to the location of the Xi. The white horizontal scale bar in the bottom left corner of the DAPI images correspond to 5 microns.
In order to strengthen the observations described above, we sought to quantitate the changes in H2AK119Ub1, H3K27me3 and H3K9me3 in the various mutant clones. Consistent with our interpretation of the images, H2AK119Ub1 signal at the Xi was significantly reduced to a comparable extent in all three Xi XIST promoter mutants, but not the Xa promoter mutant (Fig9a). Like H2AK119Ub1, H3K27me3 was also significantly reduced at the Xi in all three Xi XIST promoter deletion mutants (Fig9b). Above, we reported that H3K27me3 was reduced, but not completely absent from the XIST Xi promoter mutants XIST-51 and XIST-76, whereas H3K27me3 was not obvious in XIST-75. In agreement with this interpretation, the H3K27me3 signal at the Xi was significantly higher in XIST-51 (p=0.002) and XIST-76 (p=0.012) relative to XIST-75. Finally, although H3K9me3 was readily detected at the Xi in all mutants, the signal appeared more intense in Xi promoter mutant XIST-75. Correspondingly, only this mutant showed a significant increase in H3K9me3 at the Xi relative to parental cells and all other mutants (p=<0.001) (Fig9c).
Fig.9.

Violin plots showing data for fluorescence relative to DAPI at the Xi for (a) H2AK119ub1, (b) H3K27me3 and (c) H3K9me3. Samples are listed along the x-axis and the ratio of fluorescence for the indicated marker relative to DAPI is on the y-axis. The median is the black-horizontal line within the white box, which itself indicates the interquartile range. Each plot represents data from 20 Xi for each sample. The p-values relative to RPE1, calculated using One-Way ANOVA and Tukey honestly significant difference, are indicated above corresponding samples. Plots were created using R software and ggplot2 package.
Metaphase analysis of Xi chromatin in XIST deletion mutants
To more readily assess the changes observed at the interphase Xi for H3K4me2, H3K27me3 and H2AK119Ub1, we prepared cytospun metaphase chromosomes, which allow for the assessment of changes to chromatin banding at the Xi (Chadwick and Willard 2004; Darrow et al. 2016; Nozawa et al. 2013; Smith et al. 2004). Metaphase RPE1 Xi show consistent banding for all of the above modifications and H3K9me3 (Chadwick 2007). Due to the slow growth of XIST-75, it was not possible to isolate metaphase chromosomes in sufficient quantity for analysis, therefore we focused on Xi XIST promoter deletion mutants XIST-51 and XIST-76. Initially we examined H3K27me3 and H3K4me2 given that the interphase H3K27me3 signal for these mutants was reduced and H3K4me2 signals were elevated (Fig7). As expected for parental RPE1, H3K27me3 occupied 4–5 large reproducible bands (Chadwick and Willard 2002) and H3K4me2 was predominantly at Xq23-24 where it is enriched at DXZ4 with weak signals at the tip of Xp (Chadwick and Willard 2002) (Fig10a, top row). In contrast, both Xi XIST promoter mutants (XIST-51 and 76) showed a reduction in H3K27me3 bands, accompanied with the gain of several intense H3K4me2 bands with most change occurring on Xp (Fig10a, second and third row). Collectively these gains and losses account for the changes in signals observed at interphase. Also consistent with interphase staining, we observed prominent non-overlapping alternating bands of H3K9me3 and H2AK119Ub1 at the Xi in parental RPE1 cells (Fig10b, top row), whereas H2AK119Ub1 signals were severely depleted at the Xi in XIST-51 while H3K9me3 bands appeared unaffected.
Fig.10.
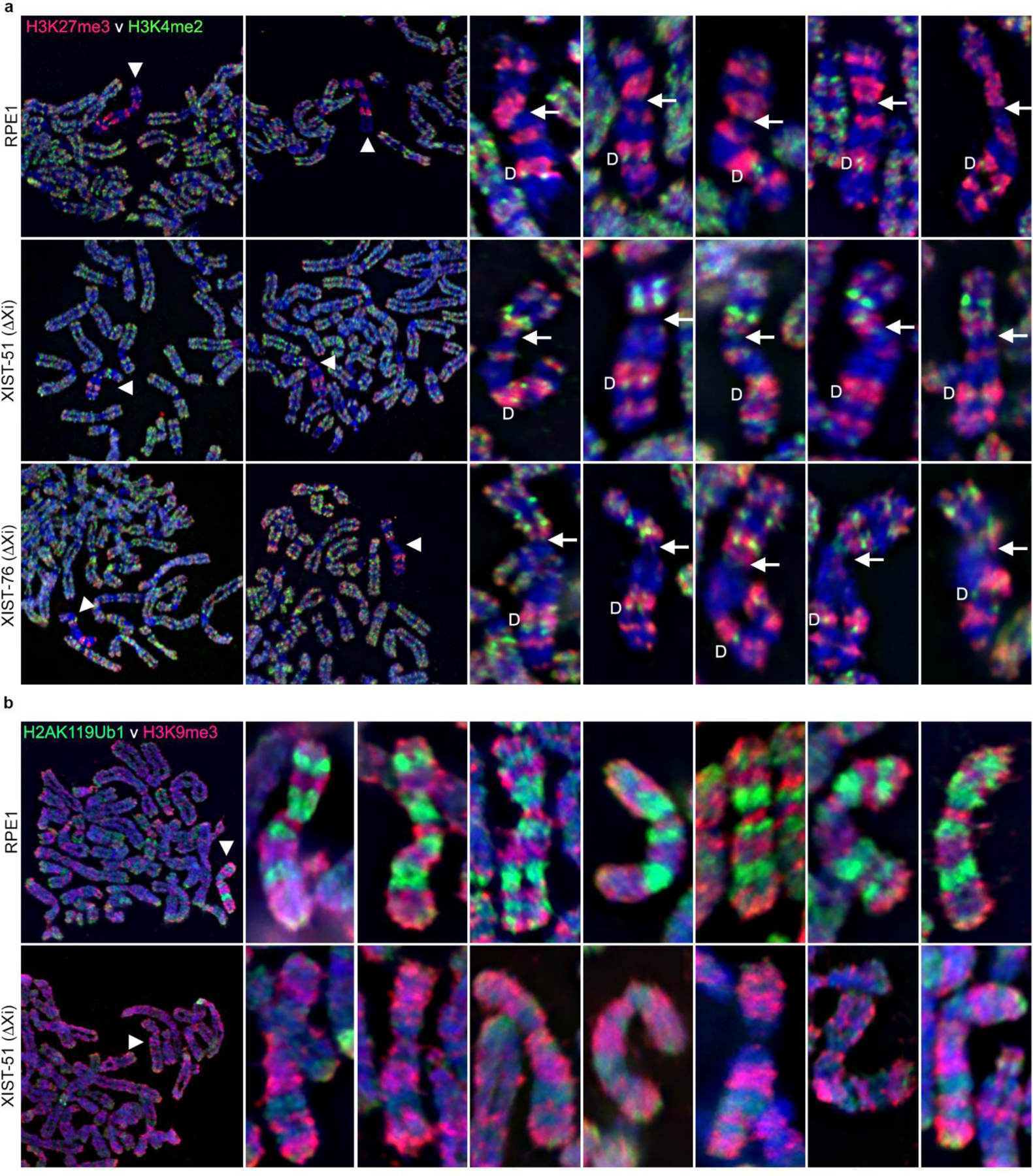
Covalent histone modifications at the metaphase Xi. (a) Indirect immunofluorescence showing the metaphase distribution of H3K27me3 (red) and H3K4me2 (green) merged with DAPI (blue) in RPE1 cells and Xi XIST deletion mutants XIST-51 and XIST-76. The two panels on the left show partial metaphase spreads with the location of the Xi indicated by the white arrowhead. To the right are 5 additional zoomed in images of the Xi for RPE1 or the XIST mutant clones. The location of the DXZ4-associated H3K4me2 signal is indicated by the white “D” to the left of the metaphase Xi. The approximate location of the centromere in indicated by the white-arrow located to the right of the metaphase Xi. For ease of comparison, the metaphase Xi has been oriented vertically, with Xp immediately above the arrow, while Xq is below. (b) Metaphase chromosomes, showing the distribution of H3K9me3 (red) and H2AK119Ub1 (green) by indirect immunofluorescence, merged with DAPI (blue). the Top row shows a partial metaphase spread on the far left and seven zoomed in Xi immediately to the right for RPE1, whereas the bottom row shows the same but for the XIST mutant XIST-51.
Stable silencing at the Xi in XIST deletion mutants
Our analysis of chromatin maintenance at the Xi in the absence of XIST reveals that the most prominent impact occurs to Xp (Fig10). The more pronounced instability of chromatin in this interval probably reflects that the ancestral portion of the X chromosome upon which XCI evolved is Xq (Graves 1995; Lahn and Page 1999), whereas Xp was acquired more recently. Consistent with the evolution of the X whereby proximal Xp was likely acquired first followed by distal portions of Xp as the most recent addition (Lahn and Page 1999), outside the pseudoautosomal regions (Helena Mangs and Morris 2007), more genes escape XCI and are thus biallelically expressed from Xp (Carrel et al. 2003; Cotton et al. 2013; Tukiainen et al. 2017). Given the reduced heterochromatin and acquired euchromatin signals on Xp in our mutants, we looked for changes in expression from the Xi. RPE1 cells are clonal, and as such, the same X chromosome is the Xi in all cells. Using data for known SNPs residing within expressed portions of X-linked genes (Helena Mangs and Morris 2007), we selected 24 X-linked genes for analysis, with the majority located on Xp (Fig11). To confirm the presence of the SNP, we first amplified genomic DNA from RPE1 cells (Table S2) and sequenced the PCR product, confirming that all did indeed contain informative SNPs (Table S3). Next, we amplified cDNA from RPE1 and Xi XIST promoter mutants XIST-51 and XIST-76. Of all the genes analyzed, only TCEANC showed expression from the Xa and Xi in all three samples, whereas FRMPD4 showed expression from the Xi in the deletion mutants only (Fig11). These data are consistent with maintenance of the human Xi in the absence of XIST post XCI (Brown and Willard 1994; Rack et al. 1994) and reports in mouse where loss of Xist alone in somatic cells results in heterochromatin changes and gains in euchromatin in the absence of appreciable gain of expression from the Xi (Adrianse et al. 2018; Diaz-Perez et al. 2006), but is more prone to reactivation (Adrianse et al. 2018; Diaz-Perez et al. 2006). Nevertheless, while loss of XIST is tolerated for maintenance of XCI, recent reports highlight the important role that retention of Xist plays in suppressing tumor susceptibility (Yang et al. 2020; Yildirim et al. 2013) and sex-linked autoimmune disease (Syrett et al. 2019).
Fig.11.

X-linked allele-specific gene expression analysis. At the top is a horizontal ideogram of the human X chromosome with Xp on the left and Xq on the right. The centromere is represented by the grey oval at the intersection of the two arms. Immediately beneath this are indicated the approximate location of all genes assessed that were verified to contain an expressed SNP. The bottom image shows partial chromatograms of Sanger dideoxy sequence traces for the gene examples indicated. The colored bases at top correspond to the trace signals and the white-S on a black background indicates the location of the SNP, which is further highlighted by the downward facing black arrowhead. The top row shows sequence data obtained from sequencing PCR products derived from RPE1 genomic DNA (gDNA). The following three rows show sequence data obtained from PCR products derived from cDNA samples for the templates indicated to the left. The asterisks indicate samples in which bi-allelic expression can be detected.
Discussion
Here we report our observations regarding the organization of chromatin at the human Xi and associated impact on expression from the chromosome upon knock-out of XIST through CRISPR/Cas9 genome engineering. Most functional studies in mice involve knocking out part or all of Xist. With the exception of site-specific recombination-based approaches (such as Cre-Lox) whereby a desired sequence can be excised, and the impact monitored shortly thereafter, the main limitation of a knock-out approach is that engineered cells have undergone numerous cell divisions before clones emerge, that can then be verified as knockouts, and the overall effect investigated. Approaches to assess the immediate impact of Xist/XIST loss could utilize short-interfering RNA (siRNA) to knock-down Xist/XIST as has been done previously in hTERT-RPE1 cells (See below) (Nozawa et al. 2013). Although siRNA-based approaches are impeded by the inability to completely eliminate a target RNA, they have been reported to be effective at improving somatic cell nuclear transfer in mice (Matoba et al. 2011) and pigs (Zeng et al. 2016), with further improvement when short-hairpin based approaches are used (Yang et al. 2019). Alternatively, approaches that strip Xist from the Xi without reducing Xist RNA levels have proved effective, such as short protein-nucleic-acid (PNA) molecules that showed loss of macroH2A1 at the Xi after a 72-hour incubation (Beletskii et al. 2001), or short locked-nucleic-acid (LNA) molecules that stripped cis-association of Xist from the chromosome within 2 hours of Nucleofection accompanied by loss of EZH2 at the Xi (Sarma et al. 2010). Nevertheless, a knock-out approach has the advantage that upon cloning, all cells are impacted equally with permanent loss of the target gene. Although not assessed in the current study, knock-out clones provide the added advantage that the long-term impact XIST loss has on the Xi could be assessed by culturing cells for extended periods and monitoring change to chromatin features over time.
In this study we describe the impact XIST loss has on Xi chromatin and expression in three independent knock-out clones, which is summarized in Table 1. Something that became apparent early in this study is the importance of isolating and assessing biological replicates, because one of the three mutant clones (XIST-75) possessed some chromatin changes that were unique to this clone, and overall conclusions could have been quite different had only one clone been assessed. Furthermore, XIST-75 grew considerably slower than the other two clones. Why this clone differs slightly could reflect off-target impacts of the CRISPR/Cas9 system. Nevertheless, several chromatin changes were consistent between this clone and the other two, which highlight the shared impact XIST loss has.
Table 1.
Summary of observations made from assessing the Xi XIST deletion mutants.
| Feature Assessed | Observations in Xi Deletion Mutants |
|---|---|
| Barr Body |
|
| CTCF |
|
| macroH2A1 |
|
| SMCHD1 |
|
| H3K9me3 |
|
| HP1g |
|
| EZH2 |
|
| ATRX |
|
| H3K4me2 |
|
| H3K27me3 |
|
| H2AK119Ub1 |
|
| Gene expression |
|
Previously, we reported that CTCF is largely absent from the Xi with the exception of association with the macrosatellite DXZ4 (Horakova et al. 2012), resulting in an intense focus within a void in staining pattern corresponding the Xi territory (Chadwick 2008; Chadwick and Willard 2003). Consistent with a report in mouse (Minajigi et al. 2015) where loss of Xist did not significantly alter CTCF association with the Xi, we did not detect obvious change in the distribution of this protein. One possible explanation could reflect retention of SMCHD1 at the Xi in all three of our mutants, because in mouse, SMCHD1 has been shown to bind to many sites that correspond to CTCF sites, with the two proteins acting antagonistically (Chen et al. 2015). As such, retention of SMCHD1 could prevent CTCF binding. While loss of Xist in mouse through deletion (Wang et al. 2019a; Wang et al. 2018) or displacement from the Xi via LNA (Jansz et al. 2018) results in loss of SMCHD1 enrichment at the Xi, at least in human cells loss of XIST is not sufficient to dissociate SMCHD1. One possible explanation for this is that SMCHD1 at the Xi in hTERT-RPE1 cells associates with both the H3K27me3 and H3K9me3 Xi territories (Nozawa et al. 2013). Given that in XIST-51 and XIST-76 we see no significant change in H3K9me3 coupled with a significant reduction in H3K27me3 at the Xi, this may account for the slight reduction in Xi SMCHD1 signal in these clones. Likewise, in XIST-75 we see even lower H3K27me3 levels combined with a significant increase in H3K9me3 levels, which may account for why Xi SMCHD1 signal appears unchanged in this mutant. The two groups of mutants also differ to some extent with regards to the association of the histone variant macroH2A1 that is normally enriched at the Xi (Costanzi and Pehrson 1998). In XIST-51 and XIST-76, the macroH2A1 MCB is diminished, but not absent from the Xi, whereas a MCB could not be detected in XIST-75. In this situation, XIST-75 is comparable to data in mouse where knocking XIST from the Xi with PNAs (Beletskii et al. 2001) or cre-mediated excision of Xist from the Xi in somatic cells (Csankovszki et al. 1999) results in loss of macroH2A1.
Interestingly, transient knockdown of XIST in hTERT-RPE1 cells through a 72-hour exposure to XIST siRNAs resulted in reduced detection of the Barr body and a substantial reduction of cells showing H3K9me3 enrichment at the Xi (from >90% of cells treated with a control siRNA to <28% of cells treated with an XIST siRNA) (Nozawa et al. 2013). The knockouts reported here do not show a reduction in H3K9me3, and in fact levels increased significantly in XIST-75. Consistent with the retention of H3K9me3, we did not see a reduction in HP1γ at the Xi, or loss of ATRX, which associates with H3K9me3 via it’s ADD domain, an interaction which is enhanced by HP1 (Dhayalan et al. 2011; Eustermann et al. 2011; Iwase et al. 2011). Perhaps this reflects differences in short-term and long-term impacts of XIST loss on the Xi, whereby H3K9me3 levels recover at the Xi over time?
Deletion of XIST had a significant effect on levels of H3K27me3 at the Xi, which is comparable to the effect seen when Xist is deleted from mouse somatic cells (Colognori et al. 2019). The detection of significantly lower levels of H3K27me3 at the Xi in XIST-51 and XIST-76, is consistent with the occasional detection of EZH2 which is known to transiently associate with the Xi (Plath et al. 2003; Silva et al. 2003). Furthermore, the reduction, but not loss of H3K27me3 at the Xi in these clones, is similar to the reduced detection of H3K27me3 at the Xi from >85% of cells to <22% in hTERT-RPE1 cells treated with XIST siRNA (Nozawa et al. 2013). It is possible that the reduced macroH2A1 signal at the Xi in these clones corresponds to the same genomic interval where H3K27me3 is retained. The exception once again is XIST-75 where H3K27me3 was not apparent and EZH2 was not observed at the Xi.
The Xi chromatin change that was consistent for all three XIST deletion mutants was the significant reduction in H2AK119Ub1. The deletion of Xist in mouse embryonic fibroblasts results in reduced H2AK119Ub1 (Colognori et al. 2019), and targeting of PRC1 to the Xi has been shown to be dependent upon Xist (Plath et al. 2004; Schoeftner et al. 2006), but independent of H3K27me3 (Schoeftner et al. 2006; Tavares et al. 2012), which may explain why H2AK119Ub1 is absent in our mutants, but H3K27me3 can be detected. Furthermore, the reduced SMCHD1 signal we observe in XIST-51 and XIST-76 may be in part due to the loss of H2AK119Ub1, as SMCHD1 association with the Xi has been reported to be dependent on H2AK119Ub1 (Jansz et al. 2018). The fact that H2AK119Ub1 appears to be reduced to general nuclear levels here, yet SMCHD1 is retained at the Xi in XIST-51 and XIST-76 (at reduced levels), but unaffected in XIST-75, may reflect the retention of high levels of H3K9me3, where SMCHD1 is also associated (Nozawa et al. 2013).
Examination of euchromatin signatures in the mutants showed no appreciable change in XIST-75 with the exception of the occasional gain of an additional H3K4me2 focus within the hypo-H3K4me2 void defining the Xi territory. Interestingly, we have observed a similar phenomenon at the Xi in hTERT-RPE1 cells when the H3K9me3 histone methyltransferase SETDB1 is deleted (Sun and Chadwick 2018). In these mutants, the Xi territory had increased in size which was linked to the acquired H3K4me2 focus that corresponded to activation of an enhancer within the IL1RAPL1 gene. What the DNA sequence is that is associated with the acquired H3K4me2 Xi signal in XIST-75 is presently unknown, but it has not escaped our notice that the Xi territory also appears larger in this mutant, and therefore perhaps this gain of signal may also correspond to activation of the same enhancer cluster. Both XIST-51 and XIST-76 occasionally showed more extensive H3K4me2 signal within the territory of the Xi, which is consistent with observations made when Xist is deleted in mouse (Diaz-Perez et al. 2006; Zhang et al. 2007). While euchromatin signals were acquired (predominantly on Xp), expression analysis from the Xi revealed that only one gene of twenty-four showed any appreciable reactivation. These data are consistent with the Xi possessing multiple redundant layers of heterochromatin that lock-in gene silencing, and are in agreement with the report that the XIC is not required to maintain silencing at the Xi (Brown and Willard 1994; Rack et al. 1994), but clearly XIST has an important role in ensuring that some of those silencing layers are properly organized and preserved.
Supplementary Material
Acknowledgements
We thank R. Didier (Florida State University, College of Medicine) for assistance with flow cytometry, F. Zhang (Broad Institute of MIT) for the gift of pSpCas9(BB)-2A-GFP (Addgene plasmid # 48138) and H. F. Willard for the use of the RPE1 Xa somatic cell hybrid. This work was supported by the National Institute of General Medical Sciences at the National Institutes of Health [grant number GM117003 to B.P.C.]
Abbreviations
- ATCC
American Type Culture Collection
- ATRX
Alpha Thalassemia/Mental Retardation Syndrome X-Linked
- BSA
Bovine serum albumin
- cDNA
Complementary DNA
- CTCF
CCCTC-Binding Factor
- DAPI
4,6-diaminidino-2-phenylindole
- EZH2
Enhancer of Zeste 2
- FBS
Fetal bovine serum
- FISH
Fluorescence in situ hybridization
- gRNA
Guide RNA
- H2AK119Ub1
Histone H2A mono ubiquitylated at lysine 119
- H3K4me2
Histone H3 di-methylated at lysine 4
- H3K9me3
Histone H3 tri-methylated at lysine 9
- H3K27me3
Histone H3 tri-methylated at lysine 27
- HP1γ
Heterochromatin protein 1 gamma
- LNA
Locked nucleic acid
- MCB
Macrochromatin body
- PBS
Phosphate buffered saline
- PBS-T
Phosphate buffered saline supplemented with Tween-20
- PCR
Polymerase chain reaction
- PNA
Protein nucleic acid
- PX458
pSpCas9(BB)-2A-GFP
- qRT-PCR
Quantitative reverse transcription PCR
- RT-PCR
Reverse transcription PCR
- siRNA
Short interfering RNA
- SMCHD1
Structural maintenance of chromosomes flexible hinge domain containing 1
- SSC
Sodium citrate sodium chloride
- v/v
Volume per volume
- Xa
Active X chromosome
- Xi
Inactive X chromosome
- XIC
X inactivation center
- XIST
X Inactive specific transcript
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Conflicts of interest/Competing interests
The authors declare that they do not have any conflicts of interest or competing interests.
Ethics approval
N/A
Availability of data and material
Constructs will be submitted to Addgene for distribution.
Code availability
N/A
References
- Adrianse RL et al. (2018) Perturbed maintenance of transcriptional repression on the inactive X-chromosome in the mouse brain after Xist deletion Epigenetics Chromatin 11:50 doi: 10.1186/s13072-018-0219-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzate-Mejia RG, Recillas-Targa F, Corces VG (2018) Developing in 3D: the role of CTCF in cell differentiation Development 145 doi: 10.1242/dev.137729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain Nature 410:120–124. [DOI] [PubMed] [Google Scholar]
- Barr ML, Bertram EG (1949) A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis Nature 163:676–677 [DOI] [PubMed] [Google Scholar]
- Baumann C, De La Fuente R (2009) ATRX marks the inactive X chromosome (Xi) in somatic cells and during imprinted X chromosome inactivation in trophoblast stem cells Chromosoma 118:209–222 doi: 10.1007/s00412-008-0189-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beletskii A, Hong YK, Pehrson J, Egholm M, Strauss WM (2001) PNA interference mapping demonstrates functional domains in the noncoding RNA Xist Proc Natl Acad Sci U S A 98:9215–9220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blewitt ME et al. (2008) SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation Nat Genet 40:663–669 doi: 10.1038/ng.142 [DOI] [PubMed] [Google Scholar]
- Bodnar AG et al. (1998) Extension of life-span by introduction of telomerase into normal human cells Science 279:349–352 [DOI] [PubMed] [Google Scholar]
- Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD (2002) Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes Nat Genet 30:73–76. [DOI] [PubMed] [Google Scholar]
- Borsani G et al. (1991) Characterization of a murine gene expressed from the inactive X chromosome Nature 351:325–329 doi: 10.1038/351325a0 [DOI] [PubMed] [Google Scholar]
- Brockdorff N et al. (1992) The product of the mouse Xist gene is a 15kb inactive X-specific transcript containing no conserved ORF and located in the nucleus Cell 71:515–526. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Kay G, Smith S, Keer JT, Hamvas RM, Brown SD, Rastan S (1991) High-density molecular map of the central span of the mouse X chromosome Genomics 10:17–22 doi: 10.1016/0888-7543(91)90478-w [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF (1991a) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome Nature 349:38–44. [DOI] [PubMed] [Google Scholar]
- Brown CJ et al. (1991b) Localization of the X inactivation centre on the human X chromosome in Xq13 Nature 349:82–84 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Willard HF (1994) The human X-inactivation centre is not required for maintenance of X-chromosome inactivation Nature 368:154–156. [DOI] [PubMed] [Google Scholar]
- Cao R et al. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing Science 298:1039–1043 [DOI] [PubMed] [Google Scholar]
- Carrel L, Nickel G, Trevarthen K, Dunn J, Willard HF (2003) A comprehensive X inactivation profile of the human X chromosome. Am J Hum Genet 73:S94 [Google Scholar]
- Carrel L, Willard HF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females Nature 434:400–404 [DOI] [PubMed] [Google Scholar]
- Chadwick BP (2007) Variation in Xi chromatin organization and correlation of the H3K27me3 chromatin territories to transcribed sequences by microarray analysis Chromosoma 116:147–157 [DOI] [PubMed] [Google Scholar]
- Chadwick BP (2008) DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts Genome Res 18:1259–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP (2020) Characterization of chromatin at structurally abnormal inactive X chromosomes reveals potential evidence of a rare hybrid active and inactive isodicentric X chromosome Chromosome research : an international journal on the molecular, supramolecular and evolutionary aspects of chromosome biology 28:155–169 doi: 10.1007/s10577-019-09621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF (2001) Histone H2A variants and the inactive X chromosome: identification of a second macroH2A variant Hum Mol Genet 10:1101–1013. [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF (2002) Cell cycle-dependent localization of macroH2A in chromatin of the inactive X chromosome J Cell Biol 157:1113–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF (2003) Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome Hum Mol Genet 12:2167–2178 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF (2004) Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome Proc Natl Acad Sci U S A 101:17450–17455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaligne R, Heard E (2014) X-chromosome inactivation in development and cancer FEBS Lett 588:2514–2522 doi: 10.1016/j.febslet.2014.06.023 [DOI] [PubMed] [Google Scholar]
- Chaligne R et al. (2015) The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer Genome Res 25:488–503 doi: 10.1101/gr.185926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K et al. (2015) Genome-wide binding and mechanistic analyses of Smchd1-mediated epigenetic regulation Proc Natl Acad Sci U S A 112:E3535–3544 doi: 10.1073/pnas.1504232112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Baldry SE, Thorogood NP, Lawrence JB, Brown CJ (2007) Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible Proc Natl Acad Sci U S A 104:10104–10109 doi: 10.1073/pnas.0610946104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB (1996) XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure J Cell Biol 132:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colognori D, Sunwoo H, Kriz AJ, Wang CY, Lee JT (2019) Xist Deletional Analysis Reveals an Interdependency between Xist RNA and Polycomb Complexes for Spreading along the Inactive X Mol Cell 74:101–117 e110 doi: 10.1016/j.molcel.2019.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR (1998) Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals Nature 393:599–601. [DOI] [PubMed] [Google Scholar]
- Cotton AM, Ge B, Light N, Adoue V, Pastinen T, Brown CJ (2013) Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome Genome Biol 14:R122 doi: 10.1186/gb-2013-14-11-r122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R (1999) Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation Nat Genet 22:323–324. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites Cell 111:185–196. [DOI] [PubMed] [Google Scholar]
- Darrow EM et al. (2016) Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture Proc Natl Acad Sci U S A 113:E4504–4512 doi: 10.1073/pnas.1609643113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M et al. (2004) Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation Dev Cell 7:663–676 [DOI] [PubMed] [Google Scholar]
- Dhayalan A et al. (2011) The ATRX-ADD domain binds to H3 tail peptides and reads the combined methylation state of K4 and K9 Hum Mol Genet 20:2195–2203 doi: 10.1093/hmg/ddr107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Perez SV et al. (2006) A deletion at the mouse Xist gene exposes trans-effects that alter the heterochromatin of the inactive X chromosome and the replication time and DNA stability of both X chromosomes Genetics 174:1115–1133 doi:genetics.105.051375 [pii] 10.1534/genetics.105.051375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustermann S et al. (2011) Combinatorial readout of histone H3 modifications specifies localization of ATRX to heterochromatin Nat Struct Mol Biol 18:777–782 doi: 10.1038/nsmb.2070 [DOI] [PubMed] [Google Scholar]
- Fang J, Chen T, Chadwick B, Li E, Zhang Y (2004) Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X inactivation J Biol Chem 279:52812–52815 [DOI] [PubMed] [Google Scholar]
- Graves JA (1995) The evolution of mammalian sex chromosomes and the origin of sex determining genes Philos Trans R Soc Lond B Biol Sci 350:305–311; discussion 311–302 doi: 10.1098/rstb.1995.0166 [DOI] [PubMed] [Google Scholar]
- Helena Mangs A, Morris BJ (2007) The Human Pseudoautosomal Region (PAR): Origin, Function and Future Curr Genomics 8:129–136 doi: 10.2174/138920207780368141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich BD, Plenge RM, Willard HF (1997) Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation Nucleic Acids Res 25:2661–2671 doi:gka431 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horakova AH, Moseley SC, McLaughlin CR, Tremblay DC, Chadwick BP (2012) The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome Hum Mol Genet doi:dds270 [pii] 10.1093/hmg/dds270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S et al. (2011) ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome Nat Struct Mol Biol 18:769–776 doi: 10.1038/nsmb.2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs SA et al. (2001) Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3 EMBO J 20:5232–5241 doi: 10.1093/emboj/20.18.5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansz N et al. (2018) Smchd1 Targeting to the Inactive X Is Dependent on the Xist-HnrnpK-PRC1 Pathway Cell Rep 25:1912–1923 e1919 doi: 10.1016/j.celrep.2018.10.044 [DOI] [PubMed] [Google Scholar]
- Kelsey AD et al. (2015) Impact of flanking chromosomal sequences on localization and silencing by the human non-coding RNA XIST Genome Biol 16:208 doi: 10.1186/s13059-015-0774-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein Genes Dev 16:2893–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins Nature 410:116–120. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC (1999) Four evolutionary strata on the human X chromosome Science 286:964–967 [DOI] [PubMed] [Google Scholar]
- Lee HJ et al. (2019) En bloc and segmental deletions of human XIST reveal X chromosome inactivation-involving RNA elements Nucleic Acids Res 47:3875–3887 doi: 10.1093/nar/gkz109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppig KA, Brown CJ, Bressler SL, Gustashaw K, Pagon RA, Willard HF, Disteche CM (1993) Mapping of the distal boundary of the X-inactivation center in a rearranged X chromosome from a female expressing XIST Hum Mol Genet 2:883–887 doi: 10.1093/hmg/2.7.883 [DOI] [PubMed] [Google Scholar]
- Loda A, Heard E (2019) Xist RNA in action: Past, present, and future PLoS Genet 15:e1008333 doi: 10.1371/journal.pgen.1008333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozzio CB, Lozzio BB (1975) Human chronic myelogenous leukemia cell-line with positive Philadelphia chromosome Blood 45:321–334 [PubMed] [Google Scholar]
- Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature 190:372–373. [DOI] [PubMed] [Google Scholar]
- Martindale DW, Wilson MD, Wang D, Burke RD, Chen X, Duronio V, Koop BF (2000) Comparative genomic sequence analysis of the Williams syndrome region (LIMK1-RFC2) of human chromosome 7q11.23 Mamm Genome 11:890–898 [DOI] [PubMed] [Google Scholar]
- Matoba S et al. (2011) RNAi-mediated knockdown of Xist can rescue the impaired postimplantation development of cloned mouse embryos Proc Natl Acad Sci U S A 108:20621–20626 doi: 10.1073/pnas.1112664108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minajigi A et al. (2015) A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation Science doi: 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex Cell 111:197–208. [DOI] [PubMed] [Google Scholar]
- Nesterova TB et al. (2001) Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence Genome Res 11:833–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS et al. (2013) Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway Nat Struct Mol Biol 20:566–573 doi: 10.1038/nsmb.2532 [DOI] [PubMed] [Google Scholar]
- Patrat C, Ouimette JF, Rougeulle C (2020) X chromosome inactivation in human development Development 147 doi: 10.1242/dev.183095 [DOI] [PubMed] [Google Scholar]
- Peters AH, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N, Jenuwein T (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin Nat Genet 30:77–80. [DOI] [PubMed] [Google Scholar]
- Plath K et al. (2003) Role of histone H3 lysine 27 methylation in X inactivation Science 300:131–135 [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B (2004) Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome J Cell Biol 167:1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack KA et al. (1994) Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia Hum Mol Genet 3:1053–1059 doi: 10.1093/hmg/3.7.1053 [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system Nat Protoc 8:2281–2308 doi: 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royce-Tolland ME et al. (2010) The A-repeat links ASF/SF2-dependent Xist RNA processing with random choice during X inactivation Nat Struct Mol Biol 17:948–954 doi: 10.1038/nsmb.1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K et al. (2014) ATRX directs binding of PRC2 to Xist RNA and Polycomb targets Cell 159:869–883 doi: 10.1016/j.cell.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Levasseur P, Aristarkhov A, Lee JT (2010) Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome Proc Natl Acad Sci U S A 107:22196–22201 doi: 10.1073/pnas.1009785107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S et al. (2006) Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing EMBO J 25:3110–3122 doi: 10.1038/sj.emboj.7601187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko AI et al. (2018) Impact of Xist RNA on chromatin modifications and transcriptional silencing maintenance at different stages of imprinted X chromosome inactivation in vole Microtus levis Chromosoma 127:129–139 doi: 10.1007/s00412-017-0650-9 [DOI] [PubMed] [Google Scholar]
- Shevchenko AI, Pavlova SV, Dementyeva EV, Zakian SM (2009) Mosaic heterochromatin of the inactive X chromosome in vole Microtus rossiaemeridionalis Mamm Genome 20:644–653 doi: 10.1007/s00335-009-9201-x [DOI] [PubMed] [Google Scholar]
- Silva J et al. (2003) Establishment of histone h3 methylation on the inactive x chromosome requires transient recruitment of eed-enx1 polycomb group complexes Dev Cell 4:481–495 [DOI] [PubMed] [Google Scholar]
- Smith KP, Byron M, Clemson CM, Lawrence JB (2004) Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: enrichment in gene rich bands Chromosoma 113:324–335 [DOI] [PubMed] [Google Scholar]
- Sun Z, Chadwick BP (2018) Loss of SETDB1 decompacts the inactive X chromosome in part through reactivation of an enhancer in the IL1RAPL1 gene Epigenetics Chromatin 11:45 doi: 10.1186/s13072-018-0218-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrett CM et al. (2019) Altered X-chromosome inactivation in T cells may promote sex-biased autoimmune diseases JCI Insight 4 doi: 10.1172/jci.insight.126751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares L et al. (2012) RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3 Cell 148:664–678 doi: 10.1016/j.cell.2011.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay DC, Moseley S, Chadwick BP (2011) Variation in Array Size, Monomer Composition and Expression of the Macrosatellite DXZ4 PLoS ONE 6:e18969 doi: 10.1371/journal.pone.0018969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukiainen T et al. (2017) Landscape of X chromosome inactivation across human tissues Nature 550:244–248 doi: 10.1038/nature24265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Colognori D, Sunwoo H, Wang D, Lee JT (2019a) PRC1 collaborates with SMCHD1 to fold the X-chromosome and spread Xist RNA between chromosome compartments Nature communications 10:2950 doi: 10.1038/s41467-019-10755-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Jegu T, Chu HP, Oh HJ, Lee JT (2018) SMCHD1 Merges Chromosome Compartments and Assists Formation of Super-Structures on the Inactive X Cell 174:406–421 e425 doi: 10.1016/j.cell.2018.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhong Y, Zhou Y, Tanaseichuk O, Li Z, Zhao JC (2019b) Identification of a Xist silencing domain by Tiling CRISPR Sci Rep 9:2408 doi: 10.1038/s41598-018-36750-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H (2016) ggplot2 : Elegant Graphics for Data Analysis Use R!,. doi: 10.1007/978-3-319-24277-4 [DOI]
- Yang L, Yildirim E, Kirby JE, Press W, Lee JT (2020) Widespread organ tolerance to Xist loss and X reactivation except under chronic stress in the gut Proc Natl Acad Sci U S A 117:4262–4272 doi: 10.1073/pnas.1917203117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X et al. (2019) Improvement of developmental competence of cloned male pig embryos by short hairpin ribonucleic acid (shRNA) vector-based but not small interfering RNA (siRNA)-mediated RNA interference (RNAi) of Xist expression J Reprod Dev 65:533–539 doi: 10.1262/jrd.2019-070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT (2013) Xist RNA is a potent suppressor of hematologic cancer in mice Cell 152:727–742 doi: 10.1016/j.cell.2013.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F et al. (2016) Effects of RNAi-mediated knockdown of Xist on the developmental efficiency of cloned male porcine embryos J Reprod Dev 62:591–597 doi: 10.1262/jrd.2016-095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT (2007) Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing Cell 129:693–706 doi:S0092–8674(07)00452–7 [pii] 10.1016/j.cell.2007.03.036 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome Science 322:750–756 doi:322/5902/750 [pii] 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


