Abstract
Stromal-epithelial interactions are critical to the morphogenesis, differentiation, and homeostasis of the prostate, but the molecular identity and anatomy of discrete stromal cell types is poorly understood. Using single cell RNA-sequencing, we identified and validated the in situ localization of three smooth muscle subtypes (prostate smooth muscle, pericytes, and vascular smooth muscle) and two novel fibroblast subtypes in human prostate. Peri-epithelial fibroblasts (APOD+) wrap around epithelial structures while interstitial fibroblasts (C7+) are interspersed in extracellular matrix. In contrast, the mouse displayed three fibroblast subtypes with distinct proximal-distal and lobe specific distribution patterns. Statistical analysis of mouse and human fibroblasts showed transcriptional correlation between mouse prostate (C3+) and urethral (Lgr5+) fibroblasts and the human interstitial fibroblast subtype. Both urethral fibroblasts (Lgr5+) and ductal fibroblasts (Wnt2+) in the mouse contribute to a proximal Wnt/Tgfb signaling niche that is absent in human prostate. Instead, human peri-epithelial fibroblasts express secreted WNT inhibitors SFRPs and DKK1, which could serve as a buffer against stromal WNT ligands by creating a localized signaling niche around individual prostate glands. We also identified proximal-distal fibroblast density differences in human prostate that could amplify stromal signaling around proximal prostate ducts. In human Benign Prostatic Hyperplasia, fibroblast subtypes upregulate critical immunoregulatory pathways and show distinct distributions in stromal and glandular phenotypes. A detailed taxonomy of leukocytes in BPH reveals an influx of myeloid dendritic cells, T cells and B cells, resembling a mucosal inflammatory disorder. A receptor-ligand interaction analysis of all cell types revealed a central role for fibroblasts in growth factor, morphogen and chemokine signaling to endothelia, epithelia, and leukocytes. These data are foundational to the development of new therapeutic targets in benign prostatic hyperplasia.
Keywords: Prostate, prostatic urethra, stroma, fibroblast, smooth muscle, single cell RNA sequencing, benign prostatic hyperplasia
Introduction
Stromal signaling is essential at all stages of prostate development and homeostasis. The highly inductive urogenital sinus mesenchyme (UGM), composed primarily of fibroblastic cells, instructs prostate budding and differentiation during fetal development [1]. Stromal signaling niches high in Tgfβ [2] and Wnt [3] signaling have been described as maintaining quiescence of epithelium in the urethra-proximal region of adult mouse prostate. We previously characterized heterogeneity in epithelia of proximal prostate [4], but it is unclear whether discrete fibroblast subtypes are responsible for localized signaling niches in proximal and distal regions.
The mouse prostate has multiple lobes while the human prostate is comprised of zones enclosed by a capsule [5]. The restriction of Benign Prostatic Hyperplasia (BPH) to the peri-urethral transition zone led McNeal to propose that the unique stroma in this region undergoes an ‘embryonic reawakening’ to drive epithelial hyperplasia [6]. Although studies in mouse prostate suggest a unique proximal niche, a similar niche in human prostate has not been described.
BPH is characterized by discrete histologic phenotypes comprised of mixed epithelia and stroma (glandular nodules) or predominantly stromal cells (stromal nodules) [6]. Early studies speculated that stromal nodules form first and then recruit epithelial structures [7], but the cellular composition and molecular alterations underlying these unique phenotypes are unknown.
To generate an atlas of stromal cell types in normal human prostate, single cell RNA-sequencing (scRNA-seq) with subsequent in situ validation of cellular subtype location was performed on specimens from healthy young organ donors. Cells from mouse prostate and urethra were also profiled by scRNA-seq to determine orthologous fibroblasts that traverse the putative stem cell niche in the urethra and proximal prostate.
Chronic inflammation in BPH is associated with higher prostate volume, higher risk of urinary retention and higher risk of symptomatic progression [8–11]. Here, we provide an objective taxonomy of immune/inflammatory cell types in BPH. Receptor-ligand interaction analyses of all cell types reveal actionable interactions between fibroblasts, epithelia, endothelia, and leukocytes. The identification of orthologous stromal cell types in human and mouse prostate and new targetable cellular interactions in BPH are critical to modelling prostate homeostasis and disease.
Materials and methods
Human tissue processing
Prostate specimens were obtained from organ donors whose families were consented at the Southwest Transplant Alliance under IRB STU 112014–033. Organ donors with BPH were excluded from the control cohort. BPH specimens were obtained from patients undergoing simple prostatectomy at UT Southwestern Medical Center (Table S1). Fresh human tissue samples were dissected into portions for digestion of fresh tissue or fixation in 10% formalin (Fisher Scientific, Waltham, MA, USA). To obtain single cells, a 4 h enzymatic digestion at 37 °C was performed as described previously [12].
Mouse tissue handling and processing
All animal work described in this manuscript was approved by the UT Southwestern Institutional Animal Care and Use Committee. Male C57BL/6J mice (8–12 weeks of age) were obtained from the UT Southwestern Mouse Breeding core or Jackson laboratories (Bar Harbor, ME, USA, #000664). Dissected mouse tissues were fixed overnight in 10% formalin (Fisher Scientific) and paraffin embedded or digested into single cells as described previously [4].
Single cell RNA-Sequencing
Single cells were loaded into the 10x Genomics (Pleasanton, CA, USA) chromium controller. Single cell data were analyzed as previously described [4,12]. Sequencing metrics for all human and mouse samples are provided in Table S1. The bulk and scRNA-seq data from human and mouse prostate tissues were deposited into the GEO SuperSeries GSE172357 and GSE173096. Analyzed data from scRNA-seq experiments can be found at http://strandlab.net/sc.data. Code used for the single cell analysis is publicly accessible [13].
Immunohistochemistry and immunofluorescence
Immunohistochemistry and immunofluorescence were performed as described previously [4]. For primary and secondary antibody information see supplementary material, Table S2.
RNA in situ hybridization
RNA in situ hybridization was performed on paraffin sections of human and mouse tissue using the RNAscope 2.5 HD Assay kit from Advanced Cell Diagnostics (Newark, CA, USA) following the manufacturer’s instructions. For RNAscope probe information see supplementary material, Table S2.
Fibroblast to smooth muscle ratio calculation
Fibroblasts and smooth muscle cells were fluorescently labeled with antibodies to decorin and desmin, respectively. Three magnified images each from the proximal and distal regions were obtained from transverse sections of individual prostates (n=4). Fibroblast and smooth muscle cell densities were calculated as percentages of the image area covered by labeled pixels. Fibroblast to smooth muscle ratio for each inset was obtained by dividing fibroblast density by smooth muscle density. Average of three insets were obtained for proximal and distal regions of each prostate.
Data and software availability:
To increase rigor, reproducibility, and transparency, raw RNA-seq data generated as part of this study were deposited into the GUDMAP consortium database and are fully accessible at https://doi.org/10.25548/17-DRBC. The bulk and scRNA-seq data from human and mouse prostate tissues were deposited into the GEO SuperSeries GSE172357 and GSE173096. Analyzed data from human and mouse scRNA-seq experiments can be explored at http://strandlab.net/sc.data, where gene expression can be investigated in the cell type clusters identified in this study.
For an extended description of methods see Supplementary materials and methods.
Results
Molecular identification and localization of stromal cell subtypes in normal human prostate
We set out to build a hierarchical taxonomy of prostate stroma and map the spatial distribution of individual cell types in situ. First, major epithelial, immune, and stromal lineages were identified from 3 normal prostate scRNA-seq datasets (Figure 1A, supplementary material, Table S1). To reveal heterogeneity in fibromuscular stroma, fibroblast and smooth muscle cells were subsetted for re-clustering (Figure 1B). Three muscle and two novel fibroblast subtypes were identified and found to localize to distinct anatomical regions.
Figure 1. Identification and validation of fibromuscular stroma subtypes in the human prostate.
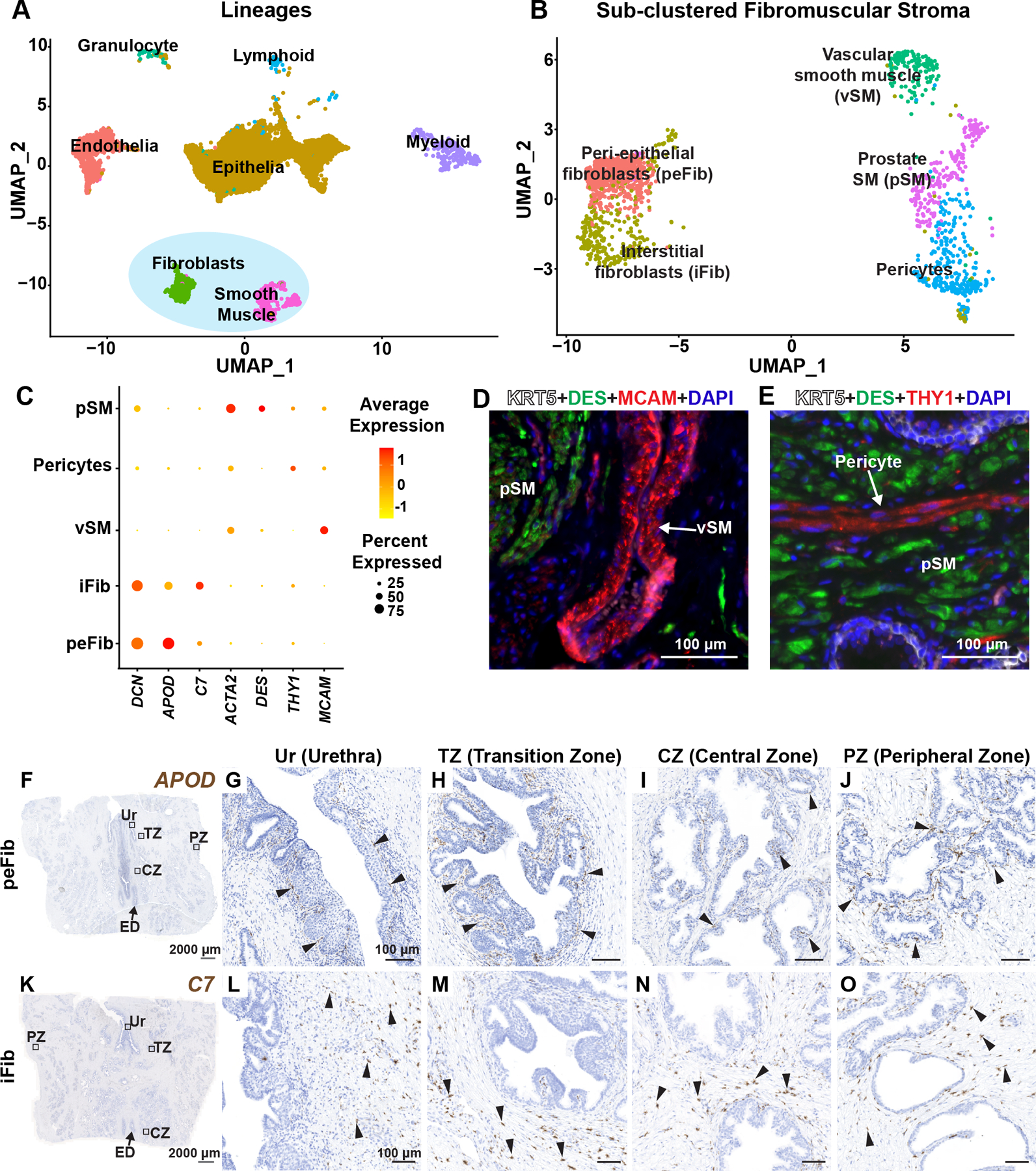
(A) Clustering of major cell lineages in normal human prostate samples (n=3 prostate donors). (B) Subsetted fibromuscular stroma from normal human prostate samples (n=3 prostate donors). (C) Dot plot of selected differentially expressed marker genes across each fibromuscular stromal subtype. (D) Section through prostate tissue labeled with fluorescent antibodies to the prostate smooth muscle marker DES (in green), vascular smooth muscle marker MCAM (in red) and basal epithelial marker KRT5 (in white). (E) Section through prostate tissue labeled with fluorescent antibodies to the prostate smooth muscle marker DES (in green), Pericyte marker THY1 (in red) and basal epithelial marker KRT5 (in white). DAPI (in blue) labeled nuclei. (F) Section through prostate tissue from organ donor labeled with probes to the peri-epithelial fibroblast marker APOD (in brown). Magnified images of distinct anatomical locations from (F) corresponding to (G) Urethra, (H) Transition zone, (I) Central zone and (J) Peripheral zone. (K) Section through prostate tissue from organ donor labeled with probes to the interstitial fibroblast marker C7 (in brown). Magnified images of distinct anatomical locations from (K) corresponding to (L) Urethra, (M) Transition zone, (N) Central zone and (O) Peripheral zone. Black arrowheads indicate individual fibroblasts. Grey scale bar represents 2000 microns. Black scale bar represents 100 microns. See also supplementary material, Figure S1.
We looked at differentially expressed genes for each fibromuscular stromal cluster to identify marker genes for in situ validation (supplementary material, Table S3). In the muscle lineage, we identified a vascular smooth muscle cluster that highly expresses MCAM and BCAM (Figure 1C, supplementary material, Table S3). We validated the vascular smooth muscle cluster in situ by immunostaining for MCAM to show vascular smooth muscle localization around large blood vessels in prostate (Figure 1D). Pericytes are characterized by high expression of THY1 and RGS5 (Figure 1C, supplementary material, Table S3) and can be visualized around small diameter blood vessels by staining with a THY1 antibody (Figure 1E). Prostate smooth muscle cells highly express DES and ACTG2 (Figure 1C, supplementary material, Table S3) and are distributed in muscle bundles around prostate glands (Figure 1D,E).
We discovered two novel fibroblast clusters in prostate that could be validated in situ. We identified a ‘peri-epithelial’ fibroblast cluster, marked by expression of APOD, PTGDS, PTGS2 and MMP2 (Figure 1C, supplementary material, Table S3). APOD+ fibroblasts (termed ‘peri-epithelial’ fibroblasts, or peFibs) lie in a narrow band adjacent to the epithelium of urethra, ejaculatory ducts, and prostate glands (Figure 1F–J, supplementary material, Figure S1B). We identified another fibroblast marked by expression of C7, CCK, PCOLCE2 and GSN (Figure 1C, supplementary material, Table S3). C7+ fibroblasts (termed ‘interstitial fibroblasts, or iFib) localized to interstitial spaces between prostate glands. Interstitial fibroblasts are widely distributed throughout all prostate anatomical zones but not around ejaculatory ducts (Figure 1K–O, supplementary material, Figure S1C).
Gene sets related to the cyclooxygenase pathway, regulation of lipoprotein metabolic process and positive regulation of fibroblast growth factor production were enriched in peFibs. In iFibs, gene sets related to mast cell cytokine production, transforming growth factor beta 3 production and regulation of antigen processing and presentation were among the top 20 significantly enriched gene sets (supplementary material, Figure S1D,E and Table S3).
Distinct spatial localization of fibroblast subtypes in mouse prostate and urethra
The molecular identity of stromal fibroblasts that maintain the relatively quiescent and castration-resistant proximal epithelial compartment is of particular clinical relevance in benign and malignant disease [4]. We analyzed 5,617 fibromuscular stromal cells from prostate and urethra (from n=4 mice each) (supplementary material, Table S1). We observed a smooth muscle population and three distinct fibroblast populations (Figure 2A). We observed a novel ‘urethral’ fibroblast cluster with cells largely coming from urethra samples. A ‘prostate’ fibroblast cluster was largely comprised of cells from prostate samples. ‘Ductal’ fibroblasts were also largely from prostate samples although a small contribution came from urethra samples (Figure 2B). All three fibroblast clusters expressed varying degrees of Decorin (Dcn) mRNA with low expression of Acta2 (Figure 2C). Mouse prostate and urethral fibroblast clusters show the highest correlation to human interstitial fibroblasts while none of the three mouse fibroblasts correlate strongly with human peri-epithelial fibroblasts (Figure 2D).
Figure 2. Identification and validation of fibromuscular stroma subtypes in the mouse prostate and prostatic urethra.
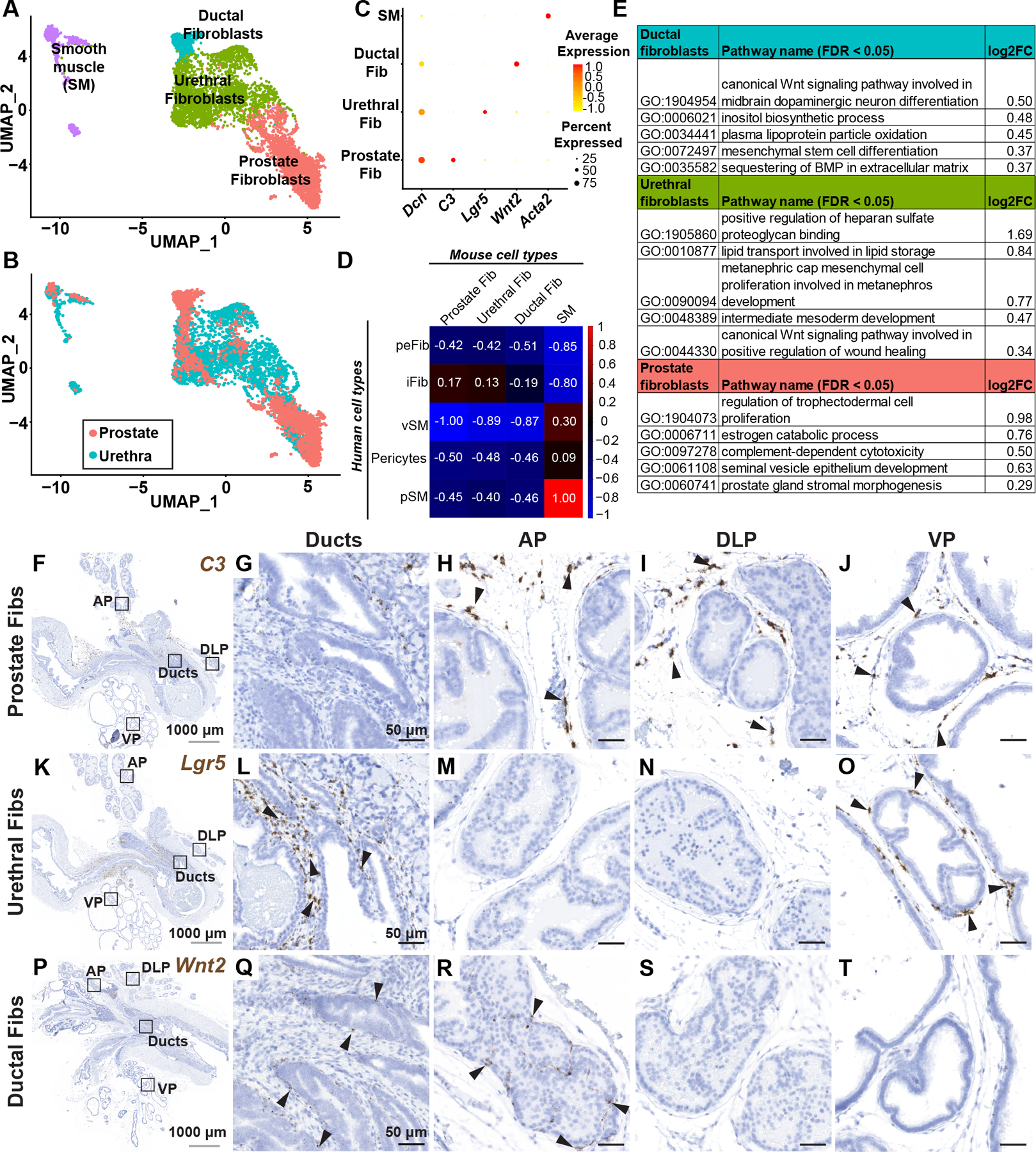
(A) Clustering of subsetted fibromuscular stroma from adult mouse prostates (n=4 mice) and prostatic urethras (n=4 mice). (B) Subsetted fibromuscular stroma from adult mice displaying contribution from prostate and prostatic urethra samples. (C) Dot plot of selected differentially expressed marker genes across each fibromuscular stromal subtype. (D) Pearson correlation of human (genes converted to mouse orthologs) and mouse fibromuscular stromal subtypes. Correlation coefficients are globally scaled between 1 and −1. (E) Quantitative Set Analysis for Gene Expression (QuSAGE) performed on mouse fibroblast subtypes. Selected significantly upregulated gene sets (false discovery rate < 0.05) displayed with log2 fold-change values. (F) Sagittal section through adult mouse lower urinary tract labeled with probes to the prostate fibroblast marker C3. (G–J) Magnified regions corresponding to distinct anatomical locations in (F). (K) Sagittal section through adult mouse lower urinary tract labeled with probes to the urethral fibroblast marker Lgr5. (L–O) Magnified regions corresponding to distinct anatomical locations in (K). (P) Sagittal section through adult mouse lower urinary tract labeled with probes to the ductal fibroblast marker Wnt2. (Q–T) Magnified regions corresponding to distinct anatomical locations in (P). Black arrowheads indicate individual fibroblasts. Grey scale bar represents 1000 microns. Black scale bar represents 50 microns. See also supplementary material, Figure S2.
‘Prostate’ fibroblasts are marked by expression of C3, Ebf1, Gpx3, Sult1e1 and Igf1 (Figure 2C, supplementary material, Table S4) and were enriched for gene sets involved in estrogen catabolic process, complement-dependent cytotoxicity and prostate gland stromal morphogenesis (Figure 2E, supplementary material, Table S4). C3+ prostate fibroblasts localize to loose matrix between glands in distal prostate lobes. C3+ prostate fibroblasts were largely absent from the proximal ducts and peri-urethral region (Figure 2F–J, supplementary material, Figure S2H–J).
‘Urethral’ fibroblasts highly express Lgr5, Apoe, Osr1, Sfrp2 and Mfap4 (Figure 2C, supplementary material, Table S4) and gene sets involved in lipid transport, canonical Wnt signaling in wound healing and regulation of heparan sulfate proteoglycan binding (Figure 2E, supplementary material, Table S4). In situ hybridization with an Lgr5 probe showed urethral fibroblasts enriched around proximal ducts (Figure 2K–O, supplementary material, Figure S2K–M). The distribution of Lgr5+ urethral fibroblasts in the ventral lobe is interesting in light of the unique secretory profile and gene expression pattern in ventral prostate [14].
‘Ductal’ fibroblasts were marked by expression of Wnt2, Rorb, Wif1, Ifitm1 and Srd5a2 (Figure 2C, supplementary material, Table S4). Ductal fibroblasts were enriched for gene sets involved in canonical Wnt signaling, inositol biosynthetic process, mesenchymal stem cell differentiation and sequestering of BMP in extracellular matrix (Figure 2E, supplementary material, Table S4). Wnt2+ ductal fibroblasts were preferentially localized around proximal prostate ducts and ejaculatory ducts and also present in proximal and intermediate segments of the anterior prostate lobe (Figure 2P–T, supplementary material, Figure S2N–P).
Fibroblast distribution and signaling niches in mouse and human prostate
Studies in mouse have shown that proximal prostate ducts are relatively quiescent and resistant to castration due to paracrine Tgfβ and Wnt signaling from local stroma [2,3]. In human prostate, we did not observe a unique proximal-enriched fibroblast in the transition zone. Instead, the two fibroblasts distributed in distinct layers relative to epithelial structures and this was observed throughout the prostate and prostatic urethra (Figure 3A). In contrast, mouse fibroblast subtypes had distinct proximal-distal and lobe-specific distribution patterns (Figure 3B). Ductal and urethral fibroblasts were discretely localized in the proximal region and show differential expression of several Wnt pathway genes. Sfrp2, Wnt10a, Wnt2, Wnt6, Wif1 and Fzd1 were upregulated in ductal fibroblasts while urethral fibroblasts showed high expression of Wnt5a, Wnt4, Lgr5 and Sfrp2 (supplementary material, Figure S3A). Ductal fibroblasts had the highest expression of Wnt ligands followed by urethral fibroblasts (supplementary material, Figure S3B). Previous studies have shown that Wnt signaling upregulates Tgfβ ligands in mouse prostate stromal cells, which act to maintain dormancy of proximal epithelia [2,3]. Tgfb2 is upregulated in urethral fibroblasts while ductal fibroblasts have higher expression of Bmp2, Bmp4 and Bmp7 (supplementary material, Figure S3C).
Figure 3. Fibroblast density and distribution in the prostate.
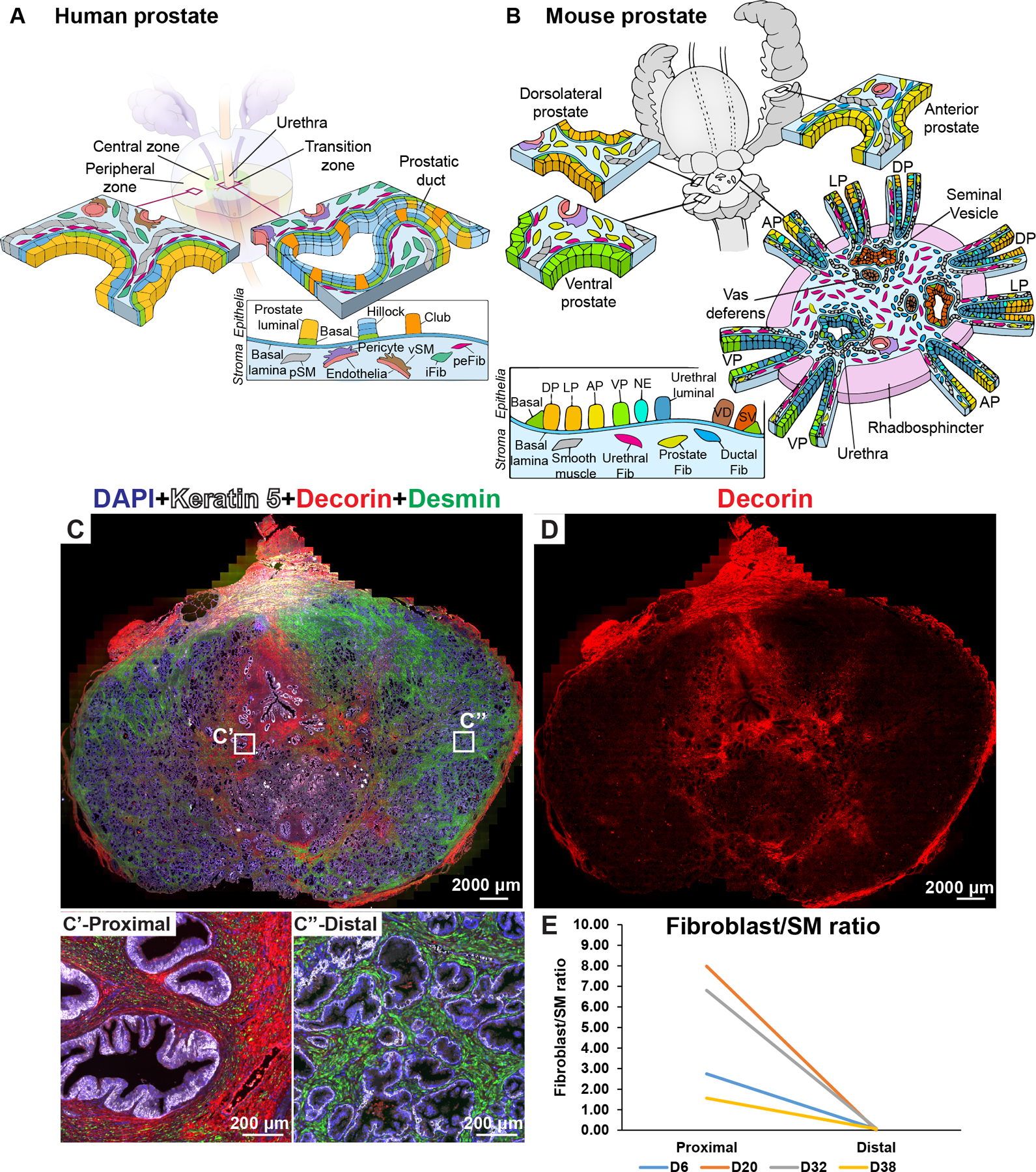
(A) Illustration depicting stromal subtypes in the human prostate. (B) Illustration depicting distribution of mouse stromal subtypes across the prostate lobes and prostatic urethra. (C) Whole mount section through a young normal prostate labeled with antibodies to Desmin (in green, labels prostate smooth muscle), Decorin (in red, labels fibroblasts) and Keratin 5 (in white, labels basal epithelial cells). DAPI (in blue) labeled nuclei. Magnified insets from (C) correspond to the proximal region (C’) and the distal region (C”) of the prostate. (D) Red channel corresponding to Decorin immunostaining isolated from image (C) to visualize fibroblast density differences. (E) Fibroblast to smooth muscle ratio in the proximal and distal region from four donor prostates. See also supplementary material, Figure S3, S4.
In the human, secreted WNT inhibitors SFRP1, SFRP2, SFRP4 and DKK1 were found to be upregulated in peri-epithelial fibroblasts (supplementary material, Figure S4A and Table S3). TGFB1 expression was also upregulated in peri-epithelial fibroblasts (supplementary material, Figure S4B and Table S3). To test for the existence of a proximal WNT and TGFβ signaling niche in human prostate, we performed bulk RNA sequencing on flow sorted fibroblasts from proximal transition zone (TZ) and distal peripheral zone (PZ) of normal human prostate (supplementary material, Figure S4C and Table S5).
The modest differences in WNT and TGFβ pathway genes expression between fibroblasts from proximal and distal regions (supplementary material, Figure S4) led us to speculate that fibroblast density differences could be maintaining a proximal-distal signaling gradient (rather than discrete urethral and ductal fibroblasts like in the mouse). The ratio of fibroblasts (DCN+) to smooth muscle (DES+) was consistently higher in proximal compared to distal regions of normal prostate due to the increased density of interstitial fibroblasts (Figure 3C–E, Figure 1K,L). In hyperplastic prostates, fibroblasts predominate in stromal nodules, while the stromal composition of glandular nodules had a greater proportion of smooth muscle cells and resembled distal prostate (supplementary material, Figure S5A,B).
Changes in fibroblast distribution and gene expression in BPH
We calculated the composition of fibromuscular stromal sub-types across discrete stromal and glandular phenotypes. The proportion of interstitial fibroblasts was consistently higher than peri-epithelial fibroblasts in BPH (Figure 4A,B).
Figure 4. Fibroblasts in human benign prostatic hyperplasia (BPH).
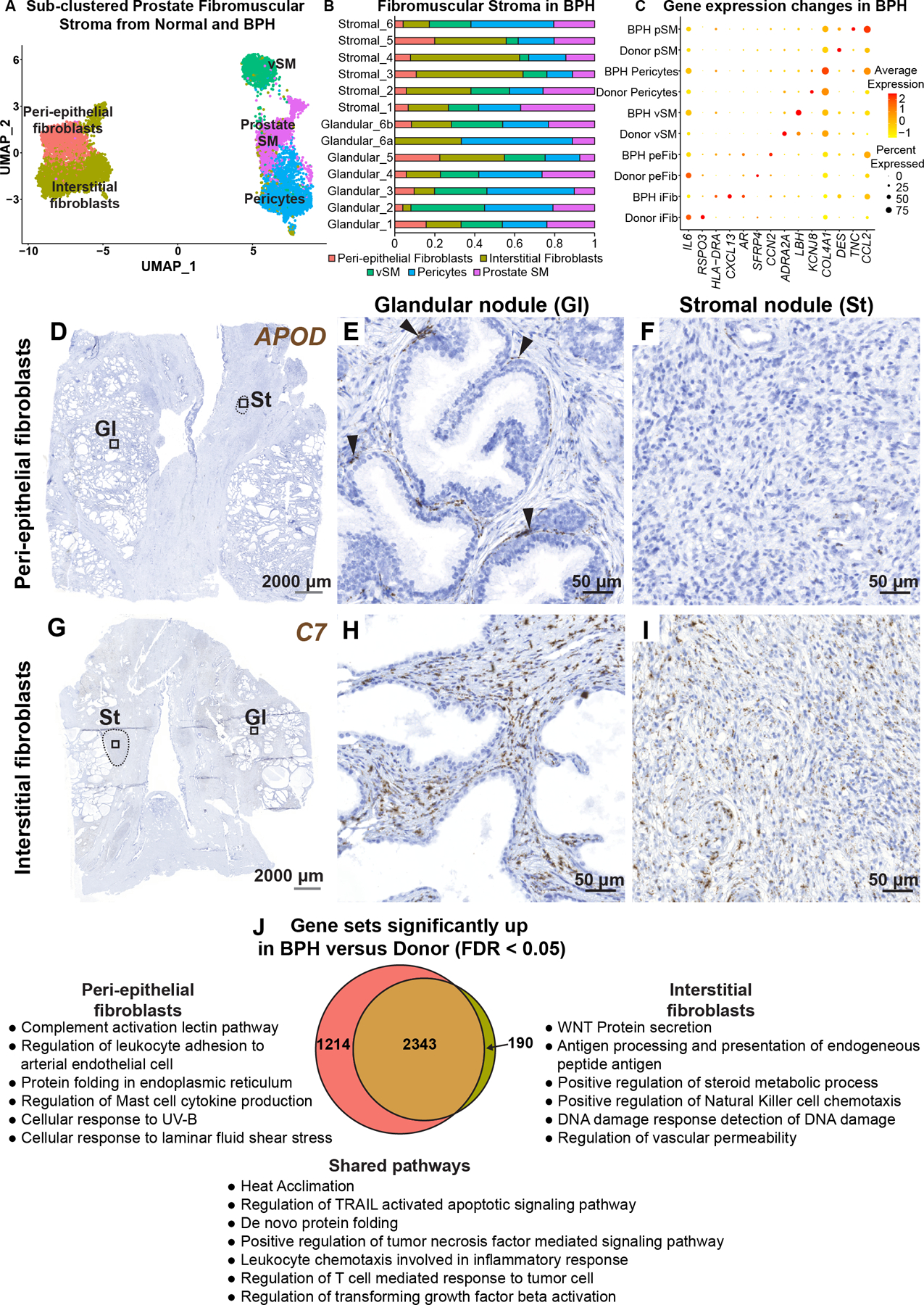
(A) Subsetted fibromuscular stroma from normal human prostate samples (n=3 prostate donors) and BPH samples (n=6 BPH patients). (B) Fibromuscular stromal composition of glandular (n=6) and stromal samples (n=6) from six BPH patients. (C) Dot plot of selected differentially regulated genes (padj < 0.05) in BPH versus normal. (D) Section through BPH tissue labeled with probes to the peri-epithelial fibroblast marker APOD (in brown). Magnified images of distinct histologic phenotypes from (D) corresponding to (E) glandular nodule and (F) stromal nodule. Black arrowheads indicate APOD+ fibroblasts in the peri-epithelial compartment. (G) Section through BPH tissue labeled with probes to the interstitial fibroblast marker C7 (in brown). Magnified images of distinct histologic phenotypes from (G) corresponding to (H) glandular nodule and (I) stromal nodule. Grey scale bar represents 2000 microns. Black scale bar represents 50 microns. Black dotted lines outline stromal nodules. (J) Quantitative Set Analysis for Gene Expression (QuSAGE) performed for each fibroblast subtype comparing donor (normal) and BPH (diseased). Significant gene sets (false discovery rate < 0.05) were subsetted to obtain unique and shared gene sets upregulated by the fibroblast subtypes in BPH. See also supplementary material, Figure S5.
In BPH interstitial fibroblasts, we observed upregulation of AR, CXCL13 and the MHC Class II molecules HLA-DRA and HLA-DPA1 while IL6, CXCL8, RSPO3, and FGF2 were downregulated. Peri-epithelial fibroblasts in BPH showed upregulation of VEGFA and LIF as well as the extracellular matrix related genes COL8A1, CCN2 and ELN while SFRP4, FZD10 and HGF were downregulated. Prostate smooth muscle cells in BPH displayed reduced expression of normal smooth muscle genes such as DES and SMTN and upregulated myofibroblast marker TNC [15]. Pericytes in BPH displayed reduced expression of normal marker genes such as KCNJ8 and increased expression of collagen genes including COL4A2, COL4A1, COL6A3 and COL5A2. Vascular smooth muscle cells displayed reduced expression of the normal marker ADRA2A and increased expression of LBH and ACTG2 (Figure 4C, supplementary material, Table S6).
APOD+ peri-epithelial and C7+ interstitial fibroblasts were present in glandular nodules with similar localization as observed in normal prostate. C7+ interstitial fibroblasts were highly enriched in stromal nodules while APOD+ peri-epithelial fibroblasts were largely absent from these regions (Figure 4D–I).
PAGE4, a tissue-specific regulator of WNT/beta catenin signaling [16], was significantly enriched in fibroblasts in the BPH scRNA-seq dataset (supplementary material, Table S6). To validate the single cell data, we performed RT-qPCR on sorted fibroblasts from normal and BPH specimens, which showed increased PAGE4 mRNA expression in BPH samples. Immunohistochemistry with an antibody to PAGE4 also demonstrated high expression in both glandular and stromal nodules (supplementary material, Figure S5C–I).
A total of 2,343 gene sets were found upregulated in common from analysis of interstitial and peri-epithelial fibroblasts in BPH (supplementary material, Table S6). Upregulated gene sets include those involved in Leukocyte chemotaxis, Regulation of T cell response to tumor cell, and Regulation of TGFβ activation. Interstitial fibroblasts and peri-epithelial fibroblasts upregulated 190 and 1,214 unique gene sets, respectively. Examples of pathways enriched in peri-epithelial fibroblasts included Complement activation and Regulation of leukocyte adhesion to arteriolar endothelial cells. Examples of pathways enriched in interstitial fibroblasts included WNT protein secretion and Antigen processing and presentation (Figure 4J).
Fibroblast interactions with endothelia and epithelia
We queried the CellPhoneDB database of receptor ligand interactions to obtain a map of mechanistic interactions between cell types in prostate, which could serve as a map for finding new druggable targets in BPH (Figure 5A, supplementary material, Table S7). Interstitial and peri-epithelial fibroblasts had the largest number of interactions with endothelial cells. Interstitial fibroblasts produced COL4A4, which is predicted to bind integrin complexes α1β1 and α2β1 on endothelial cells. Different matrix-integrin interactions, including COL27A1–α1β1 and LAMC1–α2β1, are predicted between peri-epithelial fibroblasts and endothelial cells. In addition, both fibroblast types secrete the angiogenic factors VEGFA and FGF2, which are predicted to bind to FLT1 (VEGFR1) and FGFR1, respectively (Figure 5B, supplementary material, Table S7). Moreover, VEGFA was significantly enriched in peri-epithelial fibroblasts in BPH, which could explain the increased angiogenesis observed in BPH [17] (supplementary material, Table S6).
Figure 5. Human fibroblast interactions with endothelial and epithelial cells.
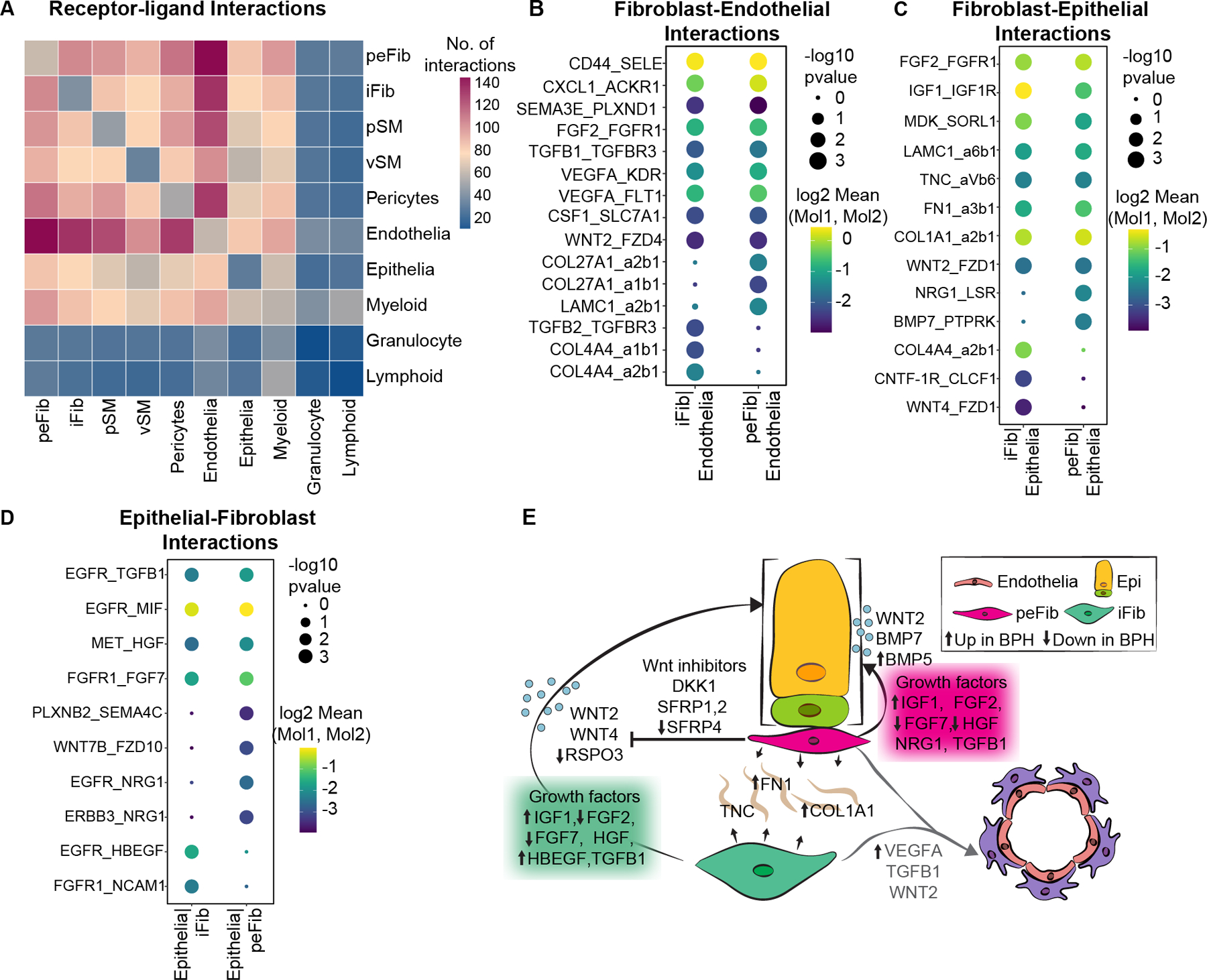
(A) Heatmap of predicted interactions from CellPhoneDB. (B) Dot plot of predicted interactions between fibroblast subtypes and endothelial cells. (C) Dot plot of predicted interactions from fibroblasts to epithelial cells. (D) Dot plot of predicted interactions from epithelial cells to fibroblasts. (E) Illustration depicting signals from fibroblasts to epithelial and endothelial cells. Up and down arrows indicate genes upregulated or downregulated respectively in the fibroblast subtype in BPH.
Epithelial-fibroblast interactions such as Wnt/beta catenin and Bmp signaling are critical for prostate development and homeostasis [18,19]. Interstitial fibroblasts produce WNT4, which is predicted to bind to FZD1 receptors on epithelial cells. Peri-epithelial fibroblasts express a different morphogen, BMP7, predicted to interact with the Protein Tyrosine Phosphatase Receptor PTPRK. In addition, peri-epithelial fibroblasts express NRG1, which interacts with EGFR and ERBB3 receptors on epithelial cells. Another morphogen secreted by both fibroblasts is WNT2, which is predicted to interact with FZD1 receptors on epithelial cells. Both fibroblasts secrete several well-known growth factors including FGF2, IGF1, FGF7 and HGF, which signal to cognate receptor tyrosine kinases on epithelial cells. In contrast to previous reports, many of these growth factors are decreased in BPH, which suggests they play a role in normal homeostasis. Fibroblasts also secrete several matrix proteins including FN1, LAMC1, TNC and COL1A1, which are predicted ligands for integrin complexes on epithelial cells (Figure 5C,D, supplementary material, Table S7). FN1 is significantly increased in BPH peri-epithelial fibroblasts whereas COL1A1 is increased in both fibroblast subtypes in BPH (supplementary material, Table S6, Figure 5E).
Immune cell subtypes and interactions with prostate fibroblasts
Chronic inflammation in BPH is associated with higher prostate volume, higher risk of urinary retention and higher risk of symptomatic progression [8–11], but a detailed taxonomy of infiltrating leukocytes is lacking. Using scRNA-seq data from BPH samples, we hierarchically classified leukocytes under lymphoid, myeloid, and granulocytic lineages (Figure 6A). BPH samples were predominantly comprised of lymphoid and myeloid cells with a smaller proportion of granulocytic cells (Figure 6B). Using CellPhoneDB, we found that interstitial and peri-epithelial fibroblasts had more significant interactions with myeloid cells compared to granulocytes and lymphoid cells (Figure 5A, supplementary material, Table S7). The T cell chemoattractant CXCL12, secreted by both fibroblasts, could be a key player in BPH inflammation as it is predicted to interact with CXCR3 and CXCR4 on lymphoid cells [20]. In addition, TGFB1, TGFB3, MDK, CSF1, CSF3 and MIF are secreted by fibroblasts and predicted to interact with TGFβ receptors on myeloid cells (Figure 6C, supplementary material, Table S7).
Figure 6. Immune subtypes and interactions with fibroblasts in BPH.
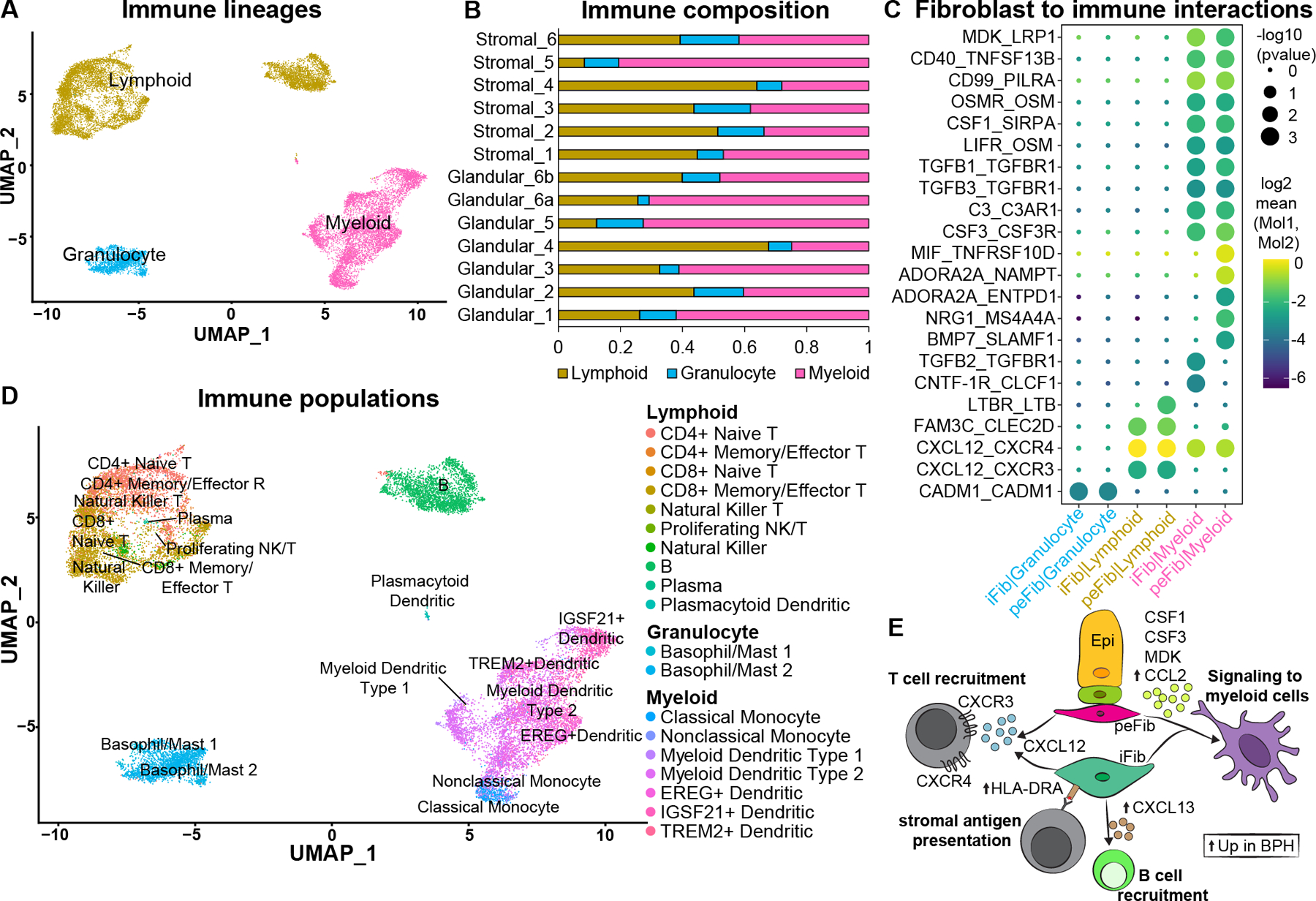
(A) Subsetted leukocytes from normal human prostate samples (n=3 prostate donors) and BPH samples (n=6 BPH patients) with major immune lineages identified. (B) Immune composition of glandular (n=6) and stromal samples (n=6) from six BPH patients. (C) Dot plot of predicted interactions from fibroblasts to immune cells. (D) Immune subtypes in the prostate identified using Travaglini et al [21]. (E) Illustration depicting signals from fibroblasts to immune cells. Up arrows indicate genes upregulated in the fibroblast subtype in BPH. See also supplementary material, Figures S6, S7.
In order to create a more detailed taxonomy of the leukocytic infiltrate based on transcriptional identities rather than individual markers, we used a well annotated scRNA-seq study from the lung as a reference for leukocyte identification [21] (Figure 6D and supplementary material, Figure S6A, Table S8). Within the lymphoid lineage, we identified CD4 and CD8 naïve and memory/effector subtypes, B cells, Natural Killer T, Natural Killer, Proliferating NK/T, plasma and plasmacytoid dendritic cells. Detailed receptor-ligand interactions for immune subtypes showed that CXCL13, which is significantly enriched in interstitial fibroblasts in BPH (supplementary material, Table S6, Figure 4C), can recruit B cells through the CXCR5 receptor [22] (supplementary material, Figure S7A, Table S7). Within the lymphoid lineage, CD4+ and CD8+ T cells were skewed towards high IFNγ expression (Figure S7B,C), a type 1 polarization profile of chronic inflammation that is implicated in mucosal immune disorders such as Crohn’s disease [23]. Mast cells are the predominant granulocytes identified in the prostate and were consistently present across all BPH tissue regions. In the myeloid lineage, we identified several dendritic cell subtypes as well as classical and nonclassical monocytes. Myeloid dendritic cells are the predominant myeloid cell type in BPH samples (Figure S6). Our data suggest dendritic cells may be a key component of the immune response in BPH and could represent an underappreciated therapeutic opportunity.
Discussion
Our study provides the first comparative atlas of human and mouse prostate fibroblasts at the single cell level. Using a single cell transcriptomic approach combined with spatial validation and functional prediction, we identified two novel fibroblast subtypes in human prostate. We extended stromal cell characterization in mouse prostate [24] by including the urethra, which enabled the identification of two spatially and molecularly distinct proximal fibroblast populations (Wnt2+ ductal and Lgr5+ urethral) in addition to distal prostate fibroblasts marked by C3 expression. We provide an overview of alterations in fibroblast organization and gene expression in BPH and a detailed taxonomy of infiltrating immune cells. Predicted receptor-ligand interactions establish a central role for fibroblasts in coordinating signals to endothelia, epithelia, and immune cells.
We found two distinct layers of fibroblasts in human prostate. APOD+ peri-epithelial fibroblasts are in close apposition to prostate, urethral and ejaculatory duct epithelium. Peri-epithelial fibroblasts express a distinct signaling program from interstitial fibroblasts, which could be driven by their proximity to epithelial cells. Several genes expressed in peri-glandular fibroblasts, including APOD [25–27], IL6 [28], CXCL8 [29,30], SOD3 [31] and PTGS2 [32,33] (supplementary material, Table S3) are associated with inflammation, cellular stress, aging and senescence. These data suggest a potential protective role for peri-epithelial fibroblasts in managing cellular stress in epithelia of the prostate, urethra, and ejaculatory ducts. C7+ interstitial fibroblasts appear to carry out conventional fibroblast roles through expression of the WNT signaling regulator RSPO3, matrix proteins such as FBLN1, GSN, MFAP4 and growth factors such as IGF1 and FGF2. These data provide cell type-specific context for previous observations of growth factor expression in prostate fibroblasts [34,35]. Although growth factor expression in general was decreased per cell (supplementary material, Table S6), the proportional increase in fibroblasts in BPH versus normal could explain the overall gain of growth factor expression seen in previous bulk tissue experiments.
We identified two proximal fibroblast populations in the mouse after profiling both the prostate and urethra. Lgr5+ urethral fibroblasts are abundant around the urethra and proximal ducts and extend into the ventral prostate (Figure 3B). Wnt2+ ductal fibroblasts are much fewer in number and predominantly localize around the ejaculatory ducts, proximal prostate ducts, and the anterior prostate. Wnt2 expression in the adult mirrors developmental Wnt2, which is highly expressed around anterior prostate buds and the Wolffian ducts (ejaculatory duct precursors). Wnt2+ ductal fibroblasts are the major source of Wnt and R-spondins in the prostate and urethra and appear to represent a specialized stromal compartment encasing the anterior prostate and ejaculatory ducts that are situated in close proximity. Lgr5+ urethral fibroblasts selectively express Tgfb2, Wnt4 and Wnt5a, which are expressed at low levels in ductal and prostate fibroblasts. Since the proximal Wnt niche was described based on Wnt5a and Tgfb2 [3] expression, the Lgr5+ urethral fibroblast cluster appears to be the predominant cell type in this niche.
C3+ prostate fibroblasts, present in the sparse matrix around mouse prostate glands, represent a more conventional fibroblast, with expression of extracellular matrix proteins like Col3a1, Gsn, Mfap5 and Lum (supplementary material, Table S4). Similar to human C7+ interstitial fibroblasts, C3+ prostate fibroblasts are interspersed in the spaces between prostate glands. Indeed, the C3+ prostate and Lgr5+ urethral fibroblasts correlated most strongly with human interstitial fibroblasts. Peri-epithelial fibroblasts appear to have no anatomical or molecular equivalent in mouse prostate. For mouse studies, our data suggest that C3+ prostate fibroblasts are likely the best candidate for modeling the role of human interstitial fibroblasts in prostate homeostasis and disease.
A proximal-specific fibroblast in human prostate was not identified. Bulk RNA-sequencing of all fibroblasts in the proximal versus distal region revealed a handful of WNT ligand genes that are differentially expressed between the proximal and distal region. We speculate that the increased density of interstitial fibroblasts observed in the proximal region could create a proximal-distal signaling gradient important for maintaining quiescence of proximal ducts. We did observe a significant enrichment of secreted Wnt pathway inhibitors, SFRP1, SFRP2, SFRP4 and DKK1, in APOD+ peri-epithelial fibroblasts compared to C7+ interstitial fibroblasts (supplementary material, Table S3). Higher expression of WNT pathway inhibitors in peri-epithelial fibroblasts, which lie in close proximity to epithelial cells, could result in a localized WNT signaling gradient by blocking activity of WNT ligands emanating from interstitial fibroblasts (Figure 5E). The higher density of interstitial fibroblasts in the transition zone compared to the peripheral zone could create an environment conducive to the emergence of BPH and prostate cancer in discrete anatomical locations.
BPH fibroblasts showed increased expression of immunomodulatory gene programs. Interstitial fibroblasts displayed increased expression of the chemokine CXCL13 (supplementary material, Table S6), which is a key regulator of B cell recruitment [22]. Previous studies have suggested that stromal cells in BPH can act as antigen presenting cells (APCs) [36], and also actively recruit immune cells through expression of chemoattractants such as CCL2, a chemokine that was significantly enriched in BPH fibroblasts (supplementary material, Table S6). The expression of MHC Class II molecule, HLA-DRA, is increased in BPH interstitial fibroblasts and could participate in antigen presentation or cytokine production [37] (Figure 6E). These data suggest an immunomodulatory role for stromal fibroblasts in BPH. Of note, we did not observe evidence of a ‘myofibroblast’ (as defined by co-expression of decorin in fibroblasts and alpha actin in smooth muscle) in either homeostasis or BPH and suggest this phenotype could be in large part an artifact of cell culture conditions.
Chronic inflammation has long been implicated in BPH progression [8–11], but the lack of an objective classification of infiltrating leukocytes has hindered a comprehensive categorization of the type of inflammatory/immune disorder that BPH represents. While our data confirm that stromal cells can act as antigen presenting cells through increased expression of MHC Class II molecules, the most striking feature of our leukocyte taxonomy was the abundance of myeloid-derived dendritic cells, which are professional antigen presenting cells that control the polarization of T cells [23]. The common expression of markers such as CD68 and CD163 in both macrophages and myeloid derived dendritic cells likely led to the mischaracterization of dendritic cells as macrophages in immunohistochemical studies [11,38]. It has been suggested previously that stromal antigen presentation and IL6/IL8 expression regulate TH1-polarization of CD4+ T helper cells, making BPH an immune inflammatory disease [39]. Our data actually demonstrate reduced stromal IL6 and CXCL8/IL8 expression in BPH (supplementary material, Table S6), but high expression of IFNγ in both CD4+ and CD8+ T cells (Figure S7B,C). The Type I polarization of T cells and the abundance of dendritic and B cells are reminiscent of chronic mucosal immune disorders such as Crohn’s disease and could represent a unique therapeutic opportunity. Weaknesses of these data include the fact that the BPH samples used are from late-stage disease and may not represent the initial inflammatory response to prostate growth. This could explain the relatively high abundance of myeloid versus lymphoid cell types compared to previous studies. In addition, while we validated some of the scRNA-seq data by immunohistochemistry, the differences between RNA and protein expression can be pronounced for certain genes and further validation of these data are needed.
Together, our cell type-specific data provide new insights into the etiology of human BPH by identifying key interactions of specialized fibroblasts with the surrounding microenvironment. By creating a cellular atlas of orthologous fibroblasts in the mouse prostate, we enable the means to test the role of cell type specific genes in prostate disease.
Supplementary Material
Figure S1. Human prostate fibromuscular stroma
Figure S2. Distribution of mouse prostate fibromuscular subtypes
Figure S3. Signaling in mouse fibroblasts
Figure S4. Signaling in human prostate fibroblasts
Figure S5. Fibroblasts in BPH
Figure S6. Immune subtype composition in BPH
Figure S7. Cellular interactions and cytokine expression in immune cells
Figure S7. Cellular interactions and cytokine expression in immune cells
Supplementary materials and methods
Table S1. Sample metrics and usage by figure
Table S2. Details of key reagents
Table S3. Human fibromuscular stroma differentially expressed genes and Qusage results
Table S4. Mouse fibromuscular stroma differentially expressed genes and Qusage results
Table S5. TZ versus PZ Bulk RNA-sequencing DESeq2 analysis
Table S6. Donor versus BPH differentially expressed genes and Qusage results
Table S7. CellPhoneDB results showing significant means of receptor-ligand interactions
Table S8. Leukocyte DEGs
Acknowledgments
We thank the families of organ donors at the Southwest Transplant Alliance for their commitment to basic science research. We acknowledge the assistance of the University of Texas Southwestern Tissue Resource (5P30CA142543), UT Southwestern Children’s Research Institute Flow Cytometry Core and the UT Southwestern Medical Center Whole Brain Microscopy Core Facility, RRID:SCR_017949. Financial support came from NIH R01 DK115477 (DWS), U54DK104310 (DWS and CMV), U01DK110807, R01 ES001332 and R01DK099328 (CMV), AUA research scholar award 659333 (DBJ); and the generous donations of the Smith, Penson and Harris families to the UTSW Department of Urology (CGR).
Footnotes
No conflicts of interest were declared
Data availability statement
Bulk and scRNA-seq data from human and mouse prostate tissues were deposited into the GEO SuperSeries GSE172357 and GSE173096 and into the GUDMAP database at https://doi.org/10.25548/17-DRBC.
References
* References 40–58 are cited only in supplementary materials
- 1.Boufaied N, Nash C, Rochette A, et al. Identification of genes expressed in a mesenchymal subset regulating prostate organogenesis using tissue and single cell transcriptomics. Sci Rep 2017; 7: 16385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salm SN, Burger PE, Coetzee S, et al. TGF-{beta} maintains dormancy of prostatic stem cells in the proximal region of ducts. The Journal of cell biology 2005; 170: 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wei X, Zhang L, Zhou Z, et al. Spatially Restricted Stromal Wnt Signaling Restrains Prostate Epithelial Progenitor Growth through Direct and Indirect Mechanisms. Cell Stem Cell 2019; 24: 753–768 e756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Joseph DB, Henry GH, Malewska A, et al. Urethral luminal epithelia are castration-insensitive cells of the proximal prostate. The Prostate 2020; 80: 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ittmann M. Anatomy and Histology of the Human and Murine Prostate. Cold Spring Harb Perspect Med 2018; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McNeal JE. Origin and evolution of benign prostatic enlargement. Investigative urology 1978; 15: 340–345. [PubMed] [Google Scholar]
- 7.Franks LM. Benign nodular hyperplasia of the prostate; a review. Ann R Coll Surg Engl 1953; 14: 92–106. [PMC free article] [PubMed] [Google Scholar]
- 8.Nickel JC, Roehrborn CG, O’Leary MP, et al. The relationship between prostate inflammation and lower urinary tract symptoms: examination of baseline data from the REDUCE trial. European urology 2008; 54: 1379–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Silverio F, Gentile V, De Matteis A, et al. Distribution of inflammation, pre-malignant lesions, incidental carcinoma in histologically confirmed benign prostatic hyperplasia: a retrospective analysis. European urology 2003; 43: 164–175. [DOI] [PubMed] [Google Scholar]
- 10.Roehrborn CG KS, Noble WD, Slawin KM, McVary KT, Kusek JW. The impact of acute or chronic inflammation in baseline biopsy on the risk of progression in the MTOPS study. Eur Urol Suppl 2005; 4: Abstract 10. [Google Scholar]
- 11.Torkko KC, Wilson RS, Smith EE, et al. Prostate Biopsy Markers of Inflammation are Associated with Risk of Clinical Progression of Benign Prostatic Hyperplasia: Findings from the MTOPS Study. The Journal of urology 2015; 194: 454–461. [DOI] [PubMed] [Google Scholar]
- 12.Henry GH, Malewska A, Joseph DB, et al. A Cellular Anatomy of the Normal Adult Human Prostate and Prostatic Urethra. Cell reports 2018; 25: 3530–3542 e3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henry G, Strand D. Strand Lab analysis of single-cell RNA sequencing v3.0.02020; DOI: 10.5281/zenodo.4684652 [last accessed 13 May 2021] [DOI]
- 14.Economides KD, Capecchi MR. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development 2003; 130: 2061–2069. [DOI] [PubMed] [Google Scholar]
- 15.Katoh D, Kozuka Y, Noro A, et al. Tenascin-C Induces Phenotypic Changes in Fibroblasts to Myofibroblasts with High Contractility through the Integrin alphavbeta1/Transforming Growth Factor beta/SMAD Signaling Axis in Human Breast Cancer. The American journal of pathology 2020; 190: 2123–2135. [DOI] [PubMed] [Google Scholar]
- 16.Koirala S, Klein J, Zheng Y, et al. Tissue-Specific Regulation of the Wnt/beta-Catenin Pathway by PAGE4 Inhibition of Tankyrase. Cell reports 2020; 32: 107922. [DOI] [PubMed] [Google Scholar]
- 17.Deering RE, Bigler SA, Brown M, et al. Microvascularity in benign prostatic hyperplasia. The Prostate 1995; 26: 111–115. [DOI] [PubMed] [Google Scholar]
- 18.Mehta V, Abler LL, Keil KP, et al. Atlas of Wnt and R-spondin gene expression in the developing male mouse lower urogenital tract. Dev Dyn 2011; 240: 2548–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta V, Schmitz CT, Keil KP, et al. Beta-catenin (CTNNB1) induces Bmp expression in urogenital sinus epithelium and participates in prostatic bud initiation and patterning. Developmental biology 2013; 376: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith EC, Teixeira AM, Chen RC, et al. Induction of megakaryocyte differentiation drives nuclear accumulation and transcriptional function of MKL1 via actin polymerization and RhoA activation. Blood 2013; 121: 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Travaglini KJ, Nabhan AN, Penland L, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020; 587: 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rupprecht TA, Plate A, Adam M, et al. The chemokine CXCL13 is a key regulator of B cell recruitment to the cerebrospinal fluid in acute Lyme neuroborreliosis. J Neuroinflammation 2009; 6: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neurath MF, Finotto S, Glimcher LH. The role of Th1/Th2 polarization in mucosal immunity. Nature medicine 2002; 8: 567–573. [DOI] [PubMed] [Google Scholar]
- 24.Kwon OJ, Zhang Y, Li Y, et al. Functional Heterogeneity of Mouse Prostate Stromal Cells Revealed by Single-Cell RNA-Seq. iScience 2019; 13: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dassati S, Waldner A, Schweigreiter R. Apolipoprotein D takes center stage in the stress response of the aging and degenerative brain. Neurobiol Aging 2014; 35: 1632–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganfornina MD, Do Carmo S, Lora JM, et al. Apolipoprotein D is involved in the mechanisms regulating protection from oxidative stress. Aging Cell 2008; 7: 506–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muffat J, Walker DW, Benzer S. Human ApoD, an apolipoprotein up-regulated in neurodegenerative diseases, extends lifespan and increases stress resistance in Drosophila. Proceedings of the National Academy of Sciences 2008; 105: 7088–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Montero P, Londoño-Vallejo A, Vernot JP. Senescence-associated IL-6 and IL-8 cytokines induce a self- and cross-reinforced senescence/inflammatory milieu strengthening tumorigenic capabilities in the MCF-7 breast cancer cell line. Cell Commun Signal 2017; 15: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Shalmanova L, Hammad A, et al. Induction of IL-8(CXCL8) and MCP-1(CCL2) with oxidative stress and its inhibition with N-acetyl cysteine (NAC) in cell culture model using HK-2 cell. Transpl Immunol 2016; 35: 40–46. [DOI] [PubMed] [Google Scholar]
- 30.Campbell LM, Maxwell PJ, Waugh DJ. Rationale and Means to Target Pro-Inflammatory Interleukin-8 (CXCL8) Signaling in Cancer. Pharmaceuticals (Basel) 2013; 6: 929–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wert KJ, Velez G, Cross MR, et al. Extracellular superoxide dismutase (SOD3) regulates oxidative stress at the vitreoretinal interface. Free Radic Biol Med 2018; 124: 408–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SR, Park JH, Lee ME, et al. Selective COX-2 inhibitors modulate cellular senescence in human dermal fibroblasts in a catalytic activity-independent manner. Mech Ageing Dev 2008; 129: 706–713. [DOI] [PubMed] [Google Scholar]
- 33.Onodera Y, Teramura T, Takehara T, et al. Reactive oxygen species induce Cox-2 expression via TAK1 activation in synovial fibroblast cells. FEBS Open Bio 2015; 5: 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giri D, Ittmann M. Interleukin-8 is a paracrine inducer of fibroblast growth factor 2, a stromal and epithelial growth factor in benign prostatic hyperplasia. The American journal of pathology 2001; 159: 139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giri D, Ittmann M. Interleukin-1alpha is a paracrine inducer of FGF7, a key epithelial growth factor in benign prostatic hyperplasia. The American journal of pathology 2000; 157: 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penna G, Fibbi B, Amuchastegui S, et al. Human benign prostatic hyperplasia stromal cells as inducers and targets of chronic immuno-mediated inflammation. J Immunol 2009; 182: 4056–4064. [DOI] [PubMed] [Google Scholar]
- 37.Ohyama H, Nishimura F, Meguro M, et al. Counter-antigen presentation: fibroblasts produce cytokines by signalling through HLA class II molecules without inducing T-cell proliferation. Cytokine 2002; 17: 175–181. [DOI] [PubMed] [Google Scholar]
- 38.Robert G, Descazeaud A, Nicolaiew N, et al. Inflammation in benign prostatic hyperplasia: a 282 patients’ immunohistochemical analysis. Prostate 2009; 69: 1774–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kramer G, Mitteregger D, Marberger M. Is benign prostatic hyperplasia (BPH) an immune inflammatory disease? Eur Urol 2007; 51: 1202–1216. [DOI] [PubMed] [Google Scholar]
- *40.Henry G, Mathews J, Gesell J, et al. BICF Cellranger mkfastq Analysis Workflow2019; DOI: 10.5281/zenodo.2652611 [last accessed 13 May 2021] [DOI]
- *41.Henry G, Mathews J, Malladi V. BICF Cellranger count Analysis Workflow2019; DOI: 10.5281/zenodo.2652622 [last accessed 13 May 2021]. [DOI]
- *42.Landini G, Randell DA, Fouad S, et al. Automatic thresholding from the gradients of region boundaries. J Microsc 2017; 265: 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *43.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019; 20: 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *44.Stuart T, Butler A, Hoffman P, et al. Comprehensive Integration of Single-Cell Data. Cell 2019; 177: 1888–1902 e1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *45.van den Brink SC, Sage F, Vertesy A, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods 2017; 14: 935–936. [DOI] [PubMed] [Google Scholar]
- *46.Aran D, Looney AP, Liu L, et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019; 20: 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *47.Cunningham F, Achuthan P, Akanni W, et al. Ensembl 2019. Nucleic Acids Res 2019; 47: D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *48.Finak G, McDavid A, Yajima M, et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 2015; 16: 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *49.Yaari G, Bolen CR, Thakar J, et al. Quantitative set analysis for gene expression: a method to quantify gene set differential expression including gene-gene correlations. Nucleic Acids Res 2013; 41: e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *50.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics 2000; 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *51.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005; 102: 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *52.Law M, Shaw DR. Mouse Genome Informatics (MGI) Is the International Resource for Information on the Laboratory Mouse. Methods Mol Biol 2018; 1757: 141–161. [DOI] [PubMed] [Google Scholar]
- *53.Efremova M, Vento-Tormo M, Teichmann SA, et al. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc 2020; 15: 1484–1506. [DOI] [PubMed] [Google Scholar]
- *54.Kim D, Paggi JM, Park C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019; 37: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *55.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *56.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- *57.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *58.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Human prostate fibromuscular stroma
Figure S2. Distribution of mouse prostate fibromuscular subtypes
Figure S3. Signaling in mouse fibroblasts
Figure S4. Signaling in human prostate fibroblasts
Figure S5. Fibroblasts in BPH
Figure S6. Immune subtype composition in BPH
Figure S7. Cellular interactions and cytokine expression in immune cells
Figure S7. Cellular interactions and cytokine expression in immune cells
Supplementary materials and methods
Table S1. Sample metrics and usage by figure
Table S2. Details of key reagents
Table S3. Human fibromuscular stroma differentially expressed genes and Qusage results
Table S4. Mouse fibromuscular stroma differentially expressed genes and Qusage results
Table S5. TZ versus PZ Bulk RNA-sequencing DESeq2 analysis
Table S6. Donor versus BPH differentially expressed genes and Qusage results
Table S7. CellPhoneDB results showing significant means of receptor-ligand interactions
Table S8. Leukocyte DEGs


