Abstract
We have developed a sensitive and quantitative assay using transcription-mediated amplification and hybridization protection assay for the detection of hepatitis B virus (HBV) DNA in serum. The transcription-mediated amplification was carried out in a single tube. The hybridization protection assay was carried out in a microtiter plate with two probes with different specific activities to obtain a broad detection range. As a result, the assay had a detection range of 5 × 103 to 5 × 108 genome equivalents (GE)/ml and good quantitative accuracy on a logarithmic scale. A moderately sized manual assay run can be completed within 5 h. Measurements of the amounts of HBV DNA in clinical samples by the assay showed the amounts under various disease conditions to be widely distributed (more than 5 logs, from approximately 5 × 103 to 5 × 108 GE/ml). It was also shown that the amount of HBV DNA in one chronic hepatitis patient varied widely, with a range of more than 5 logs during long-term monitoring. Our assay has the potential to be used to monitor and determine the prognosis of HBV patients and carriers, especially during interferon treatment.
The detection of the hepatitis B virus (HBV) surface antigen (HBsAg) indicates infection with the hepatitis B virus, while the detection of the HBV core protein, the e antigen (HBeAg), indicates replication activity, and the detection of the e antibody (HBeAb) reveals the nonreplicative phase of infection. However, HBeAg and HBeAb do not serve as rationalized markers in the case of precore mutant HBV infection (3, 5).
The HBV DNA level is a direct measure of the level of viral multiplication, and the detection of HBV DNA has provided important diagnostic and prognostic information (3). Several methods for the detection of HBV DNA have been developed. These methods use advanced amplification and/or detection technologies, some of which have already been applied to products used in clinical laboratories (2, 4, 6–11, 14–21). Hybridization assays without amplification provide quantitative results but lack adequate sensitivity. On the other hand, amplification assays have adequate sensitivity, but the results have been much less quantitative or only qualitative. Furthermore, some of the assays require simplification before they can be introduced into clinical laboratories. HBV DNA amounts can vary widely under different conditions in hepatitis B patients and carriers, and the detection ranges of some assays developed thus far were apparently too narrow to monitor the HBV DNA level (11, 18).
Gen-Probe has developed amplified detection systems for microorganisms using proprietary amplification and detection technologies (1, 12, 13). We describe herein a quantitative amplification system for the detection of HBV DNA based on transcription-mediated amplification (TMA) and hybridization protection assay (HPA). This assay was designed to provide a wide dynamic range in a format that is adaptable to large-volume clinical laboratories.
MATERIALS AND METHODS
HBV quantitative test.
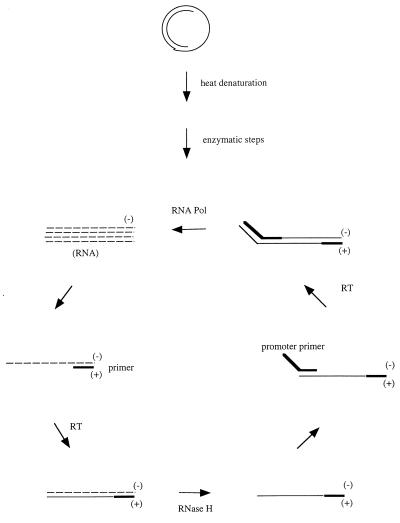
The assay consists of proprietary TMA of target DNA (12, 13) and HPA for detection (1). TMA was originally designed for RNA amplification, but the protocol (heat denaturation of the target in the presence of the primers) is adaptable for DNA amplification as well. The specific mechanisms hypothesized for DNA amplification by TMA can involve different enzymatic pathways, such as generation of the RNA transcript by low-level transcription or by use of the strand displacement activities of enzymes. One of the possible amplification mechanisms is shown in Fig. 1. TMA amplifies RNA or DNA, producing RNA transcripts through double-stranded DNA intermediates by using two primers and two enzymes, i.e., RNA polymerase and reverse transcriptase, resulting in RNA amplicon products. Briefly, the promoter-primer hybridizes to the target DNA after heat denaturation, and the RNA polymerase creates a transcript of the target DNA. A second primer then binds to the transcript, and the reverse transcriptase creates cDNA. The RNA in the resulting RNA-DNA duplex is degraded by the RNase H activity of the reverse transcriptase. The promoter-primer then binds to the cDNA and a new DNA is synthesized by reverse transcriptase, creating a double-stranded DNA molecule. The RNA polymerase recognizes the promoter sequence in the double-stranded DNA and synthesizes a number of RNA transcripts. Each of the newly synthesized RNAs reenters the TMA process and serves as a template for a new round of replication. The RNA amplicons are detected by HPA with amplicon-specific acridinium ester-labeled DNA probes. The acridinium-labeled probe hybridizes to the RNA amplicons. During the selection step, unhybridized probe is hydrolyzed at high pH, while the hybridized probe is protected from hydrolysis and thereby retains the chemiluminescent label. Quantitative detection was carried out by the following procedures. Thirty microliters of sample processing solution prepared with Sample Diluent I, Sample Diluent II, and Primer Reagent and 50 μl of silicone oil were placed in a reaction tube. Ten microliters of the serum sample was added to the reaction tube, below the oil layer. The tube was heated at 95°C for 10 min and was then incubated at 37°C for 10 min. Ten microliters of Neutralization Reagent was added, followed by the addition of 50 μl of reconstituted amplification reagent solution (prepared with the Amplification Reagent, Enzymes, and Reconstitution Buffer), and the reaction mixture was incubated at 37°C for 3 h. Ten microliters of the amplified mixture was transferred to two 96-well microtiter plates for the detection of low and high concentrations, respectively. Thirty microliters of each of the reconstituted probe solutions for the detection of a low or a high concentration was added to the respective microtiter plate, and the plates were incubated at 60°C for 20 min. One hundred microliters of the selection reagent was added to the microtiter plates, and the mixture was incubated at 60°C for 10 min to achieve hydrolysis of the unhybridized probe. The microtiter plates were placed on ice water for 5 min and left at room temperature for 10 min. Chemiluminescence was measured with a plate luminometer. All runs included amplification standards.
FIG. 1.
Proposed mechanism of TMA for HBV DNA. RT, reverse transcriptase; RNA Pol, RNA polymerase.
The amount of HBV DNA present in samples was calculated on the basis of the amounts from a standard curve generated from the amplification standards. Amplification standards contained recombinant double-stranded plasmid DNA containing a partial HBV sequence (nucleotides 248 to 2822). The amplification standards were prepared by serial dilution of the HBV plasmid DNA. The concentrations of HBV DNA in the samples were expressed as the logarithm of the genome equivalent (LGE) per milliliter.
Biochemical and serum markers of HBV.
Serum glutamic oxalacetic transaminase (GOT) and glutamic pyruvic transaminase (GPT) levels were measured by the Scandinavian Society for Clinical Chemistry (SSCC) method (736GOT and 736GPT; Eiken Chemical Co., Ltd., Tokyo, Japan). HBV DNA amounts were also measured by the branched-DNA (bDNA) assay (Quantiplex HBV DNA; Chiron Corp., Emeryville, Calif.). Serum HBeAg and HBeAb levels were measured with an IMx analyzer (IMx HBeAg/HBeAb; Abbott Laboratories, Abbott Park, Ill.). Serum HBsAg levels were measured by reversed passive hemagglutination assay (RPHA; Maiseru HBsAg; Special Immunology Laboratory, Tokyo, Japan) and enzyme immunoassay (EIA; IMx HBsAg; Abbott Laboratories).
Samples.
Serum samples were collected from patients with hepatitis and healthy blood donors. One hundred ninety-two samples were tested: 5 samples from patients with an initial presentation of acute hepatitis B; 7 samples from HBeAg-positive asymptomatic carriers of HBV, 13 samples from HBeAb-positive asymptomatic carriers of HBV, 14 samples from patients with chronic hepatitis B, 3 samples from patients with liver cirrhosis, 9 samples from patients with hepatocellular carcinoma, 14 samples from patients with a low titer of HBsAg (negative by RPHA and positive by EIA), 27 samples from non-B hepatitis patients, and 100 samples from healthy blood donors. Twenty-two samples were also studied to monitor HBV DNA levels in one patient with chronic hepatitis.
RESULTS
Detection range and linearity of quantitation.
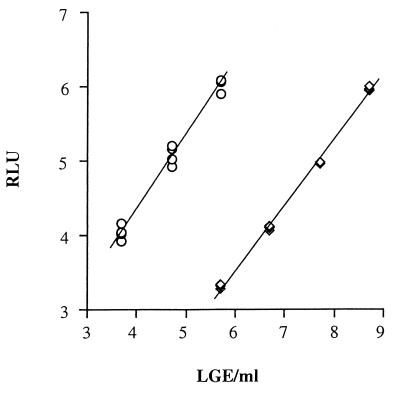
The detection range and linearity of the quantitative HBV DNA test were determined by assaying serial dilutions of the plasmid DNA containing the partial HBV sequence. The plasmid DNA dilutions (3.7 to 8.7 LGE/ml) were amplified by the TMA method, and the amplified products were split into two portions. The amplified products were analyzed by the HPA method with two probe solutions with different specific activities for the detection of high and low concentrations of amplicon. The results showed that the detection limit of the assay was as low as 3.7 LGE/ml and that the detection range of the assay was as broad as 3.7 to 5.7 LGE/ml with the probe solution for the detection of low amplicon concentrations. In addition, the maximal detection limit was as high as 8.7 LGE/ml, and the detection range of the assay was as broad as 5.7 to 8.7 LGE/ml with the probe solution for the detection of high amplicon concentrations (Fig. 2).
FIG. 2.
Standard curve from HBV DNA quantitative kit. ○, low concentration; ◊, high concentration. The horizontal axis indicates the HBV DNA concentration, and the vertical axis indicates chemiluminescence (RLU, relative light units).
Linearity of the assay with sample dilution.
The linearity of the assay was examined by diluting three HBV-positive clinical samples (with Sample Diluent II) followed by TMA amplification and HPA detection. Table 1 indicates the amounts of HBV DNA in the original sample (undiluted) and the diluted samples determined by the assay. The amounts in the diluted samples determined by the assay were essentially equivalent to the expected values, which were calculated from the original amounts (within ±0.1 LGE/ml for each dilution).
TABLE 1.
Linearity of detection of HBV DNA in hepatitis B patients
| Sample no. | Dilution rate (fold) | LGE/ml |
|---|---|---|
| 1 | 1 | 6.2 |
| 10 | 5.3 | |
| 100 | 4.0 | |
| 1,000 | <a | |
| 2 | 1 | 6.7 |
| 10 | 5.8 | |
| 100 | 4.9 | |
| 1,000 | 3.7 | |
| 3 | 1 | 8.0 |
| 10 | 7.0 | |
| 100 | 6.1 | |
| 1,000 | 5.0 |
<, less than 3.7 LGE/ml.
Reproducibilities.
Intra- and interassay reproducibilities were examined with clinical samples. Table 2 shows the overall means of the HBV DNA determinations for the four clinical samples (expressed as LGE per milliliter) and the coefficients of variation for each condition. The intra-assay, interassay, and overall variations ranged from 0.9 to 2.1%, from 0.3 to 1.4%, and from 0.7 to 2.2%, on the logarithmic scale, respectively.
TABLE 2.
Reproducibility of the HBV DNA quantitative kit
| Specimen no. | LGE of HBV DNA/ml (overall mean) | Assay precision (% CVa)
|
||
|---|---|---|---|---|
| Intra-assayb | Interassayc | Overalld | ||
| 1 | 4.1 | 2.1 | 1.4 | 2.2 |
| 2 | 4.8 | 1.1 | 1.3 | 1.6 |
| 3 | 6.4 | 1.1 | 0.7 | 1.1 |
| 4 | 7.6 | 0.9 | 0.3 | 0.7 |
CV, coefficient of variation.
Five determinations, one operator, 1 day.
Five determinations per assay, one operator, 3 days.
Fifteen determinations, one operator, 3 days.
Clinical performance of the assay.
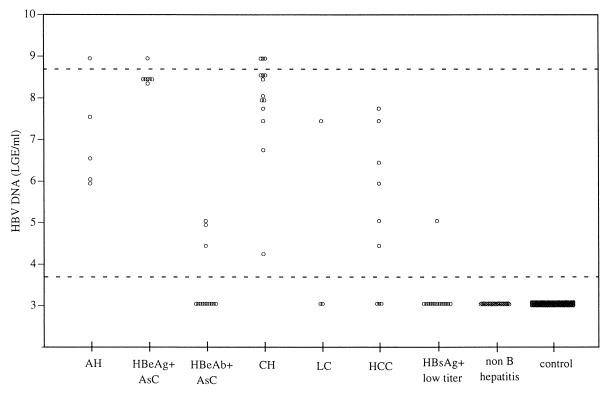
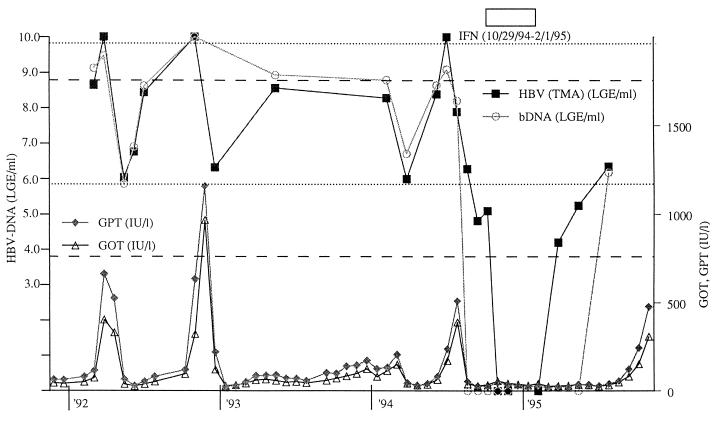
HBV DNA levels in various clinical samples from patients with hepatitis and related diseases were measured by the assay (Fig. 3). HBV DNA amounts in non-B hepatitis patients and healthy controls were below the detection limit (3.7 LGE/ml). Amounts at initial presentation in the acute hepatitis B patients ranged from 6.0 LGE/ml to higher than the upper detection limit (8.7 LGE/ml). In the asymptomatic carriers, the HBV DNA levels approached the upper detection limit (8.7 LGE/ml) in the HBeAg-positive patients, while they ranged from the lower detection limit (3.7 LGE/ml) to 5.0 LGE/ml in the HBeAb-positive patients. Chronic hepatitis patients showed a broad range of HBV DNA levels, from 4.0 LGE/ml to the upper detection limit (8.7 LGE/ml). Liver cirrhosis and hepatocellular carcinoma patients also had broad ranges of HBV DNA levels, from the lower detection limit (3.7 LGE/ml) to nearly 8.0 LGE/ml. Patients with low HBsAg titers (positive by EIA and negative by RPHA) had amounts ranging from the lower detection limit (3.7 LGE/ml) to nearly 5.0 LGE/ml. HBV DNA quantities in serial samples collected from one chronic hepatitis patient over a 4-year period were measured by the assay (Fig. 4). The quantities of HBV DNA varied over a wide range, i.e., more than 5 logs (from the lower to the upper detection limit), during the monitoring period. A transient reduction and a rebound of the HBV DNA levels during and after interferon treatment were clearly demonstrated by the assay. Several sharp increments in the amount of HBV DNA were also detected by the assay 2 to 3 weeks before the appearance of elevated levels of the hepatitis markers (GOT, GPT). The HBV DNA levels determined by TMA-HPA were quite similar to those determined by the bDNA assay over a range of more than 6 LGE/ml, and the former apparently had a higher sensitivity.
FIG. 3.
HBV DNA amounts in patients with various diseases and healthy controls. AH, initial presentation with acute hepatitis; AsC, asymptomatic carrier; CH, chronic hepatitis B; LC, liver cirrhosis; HCC, hepatocellular carcinoma; HBsAg+ low titer, EIA positive and RPHA negative.
FIG. 4.
Monitoring of HBV DNA, GPT, and GOT levels in one chronic hepatitis B patient. The analytical range for the detection of HBV DNA by TMA-HPA is from 3.7 to 8.7 LGE/ml (between the dashed lines). The analytical range for the detection of HBV DNA by the bDNA assay is from 5.8 to 9.8 LGE/ml (between the dotted lines). □, interferon treatment.
DISCUSSION
We have developed an HBV DNA detection system that uses the TMA-HPA technologies (12, 13). The advantages of the TMA-HPA method include isothermal amplification and nonradioactive, single-step differentiation of hybridized and unhybridized probe. The assay procedure involves sample and reagent additions and temperature incubations, but it is not technically demanding. In a moderately sized run (80 specimens and standards), the sample addition and the assay steps can be completed in 1 and 4 h, respectively, when they are done manually. It is possible to incorporate this assay into the normal work routine of clinical laboratories.
Results obtained by serially diluting the HBV DNA standard indicated that our assay has a broad dynamic range of from 3.7 to 8.7 LGE/ml (5 logs) and good quantitative linearity. The detection range of the assay was expanded by using two probe solutions with two different specific activities for the detection of low and high amplicon concentrations. When HBV DNA was extracted from the serum samples by an appropriate method, such as the phenol-chloroform method, guanidine method, or NaI method, and the extracted DNA was used as the sample, it was possible to increase the sensitivity of the assay, and the assay still possessed linearity beyond the current lower limit of 3.7 LGE/ml (data not shown).
Several groups have reported the development of assay systems for the detection of HBV DNA with or without DNA amplification (2, 4, 6–11, 14–21). The assay systems without amplification had lower detection sensitivities (from about 6.0 to 9.0 LGE/ml) and high quantitative accuracies, while the assay systems with amplification had higher sensitivities but lower quantitative accuracies. The detection ranges of both of these assay systems were apparently about 3 logs, regardless of whether amplification was used. As shown from the clinical performance of the assay in Fig. 3, HBV DNA amounts in patients with various disease conditions were widely distributed over more than 5 logs. Furthermore, even HBV DNA levels in a single patient varied markedly (Fig. 4). Two clinical studies have suggested that HBV DNA amounts differ markedly in hepatitis B patients and carriers and that the detection range of some assays was apparently too narrow to monitor the amount of HBV DNA (11, 18). The results reported here suggest that diagnosis of HBV infection and monitoring of patients with HBV infection may require a test not only with adequate sensitivity but also one with a very wide detection range. The quantitative test for the detection of HBV that we have developed has adequate sensitivity and a broad dynamic range for monitoring the condition and prognosis of HBV patients, carriers, and especially, patients undergoing interferon therapy.
REFERENCES
- 1.Arnold L J, Jr, Hammond P W, Wiese W A, Nelson N C. Assay formats involving acridinium-ester-labeled DNA probes. Clin Chem. 1989;35:1588–1594. [PubMed] [Google Scholar]
- 2.Aspinall S, Steele A D, Peenze I, Mphahlele M J. Detection and quantitation of hepatitis B virus DNA: comparison of two commercial hybridization assays with polymerase chain reaction. J Viral Hepatitis. 1995;2:107–111. doi: 10.1111/j.1365-2893.1995.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 3.Baker B L, Bisceglie A M D, Kaneko S, Miller R, Feinstone S M, Waggoner J G, Hoofnagle J H. Determination of hepatitis B virus DNA in serum using the polymerase chain reaction: clinical significance and correlation with serological and biochemical markers. Hepatology. 1991;13:632–636. [PubMed] [Google Scholar]
- 4.Barlet V, Cohard M, Thelu M A, Chaix M J, Baccard C, Zarski J P, Seigneurin J M. Quantitative detection of hepatitis B virus DNA in serum using chemiluminescence: comparison with radioactive solution hybridization assay. J Virol Methods. 1994;49:141–152. doi: 10.1016/0166-0934(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 5.Carman W F, Thomas H C. Genetic variation in hepatitis B virus. Gastroenterology. 1992;102:711–719. doi: 10.1016/0016-5085(92)90125-i. [DOI] [PubMed] [Google Scholar]
- 6.Chen C-H, Wang J-T, Lee C-Z, Sheu J-C, Wang T-H, Chen D-S. Quantitative detection of hepatitis B virus DNA in human sera by branched-DNA signal amplification. J Virol Methods. 1995;53:131–137. doi: 10.1016/0166-0934(95)00007-h. [DOI] [PubMed] [Google Scholar]
- 7.Erhardt A, Schaefer S, Athanassiou N, Kann M, Gerlich W H. Quantitative assay of PCR-amplified hepatitis B virus DNA using a peroxidase-labelled DNA probe and enhanced chemiluminescence. J Clin Microbiol. 1996;34:1885–1891. doi: 10.1128/jcm.34.8.1885-1891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia F, Quiros E, Bernal M C, De Luis B, Leyva A, Piedrola G, Maroto M C. Serum hepatitis B virus DNA detection with S- and C-region-directed probes. J Med Microbiol. 1993;39:473–475. doi: 10.1099/00222615-39-6-473. [DOI] [PubMed] [Google Scholar]
- 9.Garcia F, Jr, Garcia F, Bernal M C, Leyva A, Piedrola G, Maroto M C. Evaluation of enzyme immunoassay for hepatitis B virus DNA based on anti-double-stranded DNA. J Clin Microbiol. 1995;33:413–415. doi: 10.1128/jcm.33.2.413-415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia F G, Quiros E, Bernal M C, Salmeron J, Maroto M C. An enzyme-linked (alkaline phosphatase) oligonucleotide probe for the detection of serum hepatitis B virus DNA. Liver. 1992;12:179–182. doi: 10.1111/j.1600-0676.1992.tb01044.x. [DOI] [PubMed] [Google Scholar]
- 11.Jardi R, Buti M, Rodriguez-Frias F, Cortina M, Esteban R, Guardia J, Pascual C. The value of quantitative detection of HBV-DNA amplified by PCR in the study of hepatitis B infection. J Hepatol. 1996;24:680–685. doi: 10.1016/s0168-8278(96)80263-7. [DOI] [PubMed] [Google Scholar]
- 12.Jonas V, Alden M J, Curry J I, Kamisango K, Knott C A, Lankford R, Wolfe J M, Moore D F. Detection and identification of Mycobacterium tuberculosis directly from sputum sediments by amplification of rRNA. J Clin Microbiol. 1993;31:2410–2416. doi: 10.1128/jcm.31.9.2410-2416.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kacian, D. L., and R. C. Harvey. Application of the transcription-mediated amplification system (TMA) to clinical assays. In Beyond PCR: amplification 90’s, in press. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 14.Kaneko S, Miller R H, Bisceglie A M D, Feinstone S M, Hoofnagle J H, Purcell R H. Detection of hepatitis B virus DNA in serum by polymerase chain reaction. Application for clinical diagnosis. Gastroenterology. 1990;99:799–804. doi: 10.1016/0016-5085(90)90971-3. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko S, Feinstone S M, Miller R H. Rapid and sensitive method for detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J Clin Microbiol. 1989;27:1930–1933. doi: 10.1128/jcm.27.9.1930-1933.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kejian G, Bowden D S. Digoxigenin-labeled probes for the detection of hepatitis B virus DNA in serum. J Clin Microbiol. 1991;29:506–509. doi: 10.1128/jcm.29.3.506-509.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller G H, Huang D P, Shih J W-K, Manak M M. Detection of hepatitis B virus DNA in serum by polymerase chain reaction amplification and microtiter sandwich hybridization. J Clin Microbiol. 1990;28:1411–1416. doi: 10.1128/jcm.28.6.1411-1416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khakoo S I, Soni P N, Brown D, Dusheiko G M. A clinical evaluation of a new method for HBV DNA quantitation in patients with chronic hepatitis B. J Med Virol. 1996;50:112–116. doi: 10.1002/(SICI)1096-9071(199610)50:2<112::AID-JMV2>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 19.Naouv N V, Lau J Y N, Daniels H M, Alexander G J M, Williams R. Detection of HBV-DNA using a digoxigenin-labelled probe. J Hepatol. 1991;12:382–385. doi: 10.1016/0168-8278(91)90844-2. [DOI] [PubMed] [Google Scholar]
- 20.Quint W G V, Heijtink R A, Schirm J, Gerlich W H, Niesters H G M. Reliability of methods for hepatitis B virus DNA detection. J Clin Microbiol. 1995;33:225–228. doi: 10.1128/jcm.33.1.225-228.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaaijer H L, Borg F, Cuypers H T M, Hermus M C A H, Lelie P N. Comparison of methods for detection of hepatitis B virus DNA. J Clin Microbiol. 1994;32:2088–2091. doi: 10.1128/jcm.32.9.2088-2091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]