Abstract
By using recombinase-mediated cassette exchange, a method that allows integration of single copies of different constructs at the same predetermined chromosomal location, several expression cassettes have been integrated at a randomly chosen locus in the genome of mouse erythroleukemia cells. The cassettes studied contain the human β-globin promoter fused to lacZ coding sequences either alone or linked to DNase I-hypersensitive site HS2, HS3, or HS234 (a large locus control region fragment containing HS2, HS3, and HS4) of the human β-globin locus control region. Analysis of expression of these cassettes revealed mosaic expression patterns reminiscent of, but clearly different from, position effect variegation. Further investigations demonstrated that these mosaic expression patterns are caused by dynamic activation and inactivation of the transcription unit, resulting in oscillations of expression. These oscillations occur once in every few cell cycles at a rate specific for the enhancer present at the locus. DNase I sensitivity studies revealed that the chromatin is accessible and that DNase-hypersensitive sites were present whether or not the transcription unit is active, suggesting that the oscillations occur between transcriptionally competent and transcriptionally active chromatin conformations, rather than between open and closed chromatin conformations. Treatment of oscillating cells with trichostatin A eliminates the oscillations only after the cells have passed through late G1 or early S, suggesting that these oscillations might be caused by changes in histone acetylation patterns.
It has long been known that expression of transfected or translocated genes is not always stable and is subject to position effects. In metazoans, the two best-studied such phenomena are heterochromatin-induced position effect variegation (PEV) (24), in which transgenes are clonally inactivated in a fraction of a cell population, and extinction of transgenes in cell culture (50) and transgenic animals (15, 35, 41, 47). We have recently developed a novel method called recombinase-mediated cassette exchange (RMCE) that allows integration of single copies of different expression cassettes at the same predetermined chromosomal locations (1). This method is ideally suited to study position effects and gene regulation because interesting chromosomal locations can be identified and then revisited by insertion of different expression cassettes.
To study position effects and means to prevent them, we have used as a model components of the human β-globin gene cluster. This cluster is located on chromosome 11 and contains five genes that are sequentially expressed during development and are regulated by the locus control region (LCR) (14, 34, 44). The LCR is located 6 to 20 kb upstream of the ɛ-globin gene and consists of five DNase I-hypersensitive sites (HSs) first reported by Tuan et al. (48). The importance of the LCR for regulation of the globin genes was inferred from studies of naturally occurring deletions of the LCR that are associated with transcriptional silencing of the locus and with a switch from early to late replication and a change of chromatin conformation (13). More recent analyses of experimentally induced deletions of HS1 to HS5 on mouse chromosomes (6) and HS2 to HS5 on human chromosomes (40) revealed, however, that the locus was still sensitive to DNase I in the absence of the LCR and that in the case of the murine locus, expression was decreased but not abolished by the deletion. Taken together, these data suggest that the LCR, as currently defined, acts as an enhancer but is not required to render the chromatin sensitive to DNase I at its native location.
Transfection and transgenic experiments show that the LCR confers high-level expression on the globin genes. When multiple copies of LCR-driven cassettes are integrated at ectopic loci, expression is often found to be copy number dependent and site of integration independent (19). On the basis of these experiments, it has been proposed that the LCR differs from other enhancers in having a dominant chromatin-opening activity (19).
No single HS of the LCR is indispensable for transcriptional activation of the globin genes, since deletions of individual HSs introduced at the endogenous mouse locus (8, 26), in yeast artificial chromosomes (3, 38), or in cosmids containing large fragments encompassing the entire human locus (35) are associated with relatively mild phenotypes. These results have led many authors to the conclusion that the LCR acts as a holocomplex composed of all of the HSs. Nevertheless, HS2, HS3, HS4, and, to a lesser extent, HS1 have enhancer activity when tested individually in cell culture and in transgenic mice (reviewed in reference 18).
In a previous study (1), RMCE was used to investigate the function of some of the HSs constituting the LCR by integrating five expression cassettes in the genome of mouse erythroleukemia (MEL) cells at the same randomly chosen chromosomal location. This chromosomal location was termed RL1, for random locus 1. The cassettes studied included the β-globZ gene (the human β-globin promoter fused to lacZ coding sequences) alone or driven by HS2, HS3, or HS234 (a large LCR fragment containing HS2, HS3, and HS4) of the LCR (Fig. 1a). This revealed that expression of these five cassettes occurred in mosaic patterns reminiscent of PEV (1).
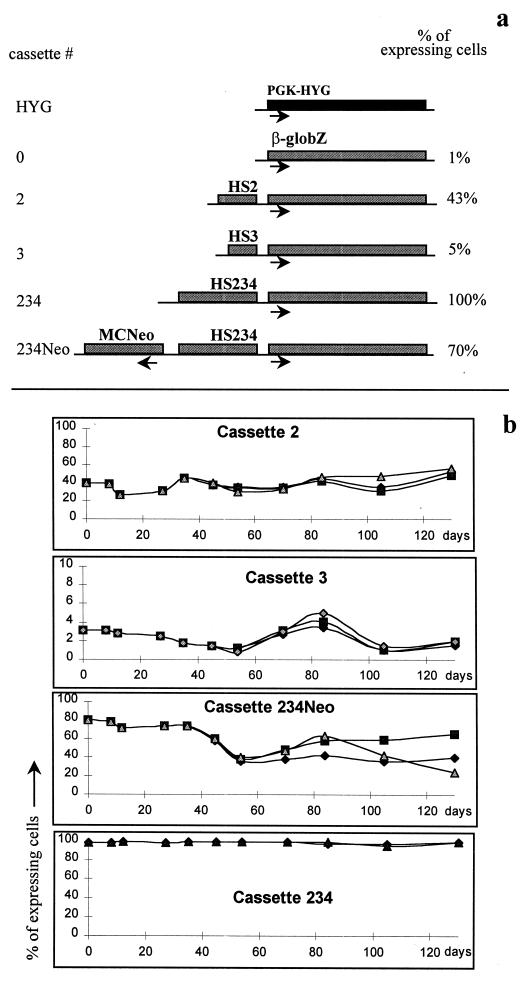
FIG. 1.
Mosaic expression patterns at RL1 in the presence of various cassettes. (a) Cassette integrated at RL1. The HYG cassette contains the PGK-HYG gene and the Lox sites that are required for RMCE. All other cassettes were exchanged with the PGK-HYG gene by RMCE. Cassette 0 contains the β-globZ gene and no enhancer. Cassettes 2, 3, and 234 contain the β-globZ gene linked, respectively, to HS2, HS3, and HS234 (a large LCR fragment containing HS2, HS3, and HS4). In all cassettes, orientation of the HS relative to the β-globin promoter is similar to that of the natural locus. When cassette 234Neo is at RL1, two genes are present: β-globZ, linked to HS234, and MCNeopA, a selectable marker. The direction of transcription is indicated below the genes. Numbers at the right indicate percentages of cells expressing β-globZ 1 month after integration of the cassette (1). The proportion of expressing cells was dependent on the enhancer present at the locus. (b) Mosaic expression patterns are stable over time. Cells with cassettes 2, 3, 234, and 234Neo inserted at RL1 were maintained in culture for 4 months and monitored every 1 to 3 weeks by FACS-Gal. After the first month of incubation, each culture was split into three identical subcultures, and each triplicate subculture was monitored for 3 more months. The percentage of expressing cells over 120 days was fairly constant.
These mosaic expression patterns appeared particularly interesting because the proportion of expressing cells was dependent on the HS and on the number of genes present at the locus. In the present study this phenomenon has been further investigated and found to be clearly different from PEV and to be caused by dynamic activation and inactivation of β-globZ resulting in oscillations of expression. The oscillations occur once every few cell cycles at a rate specific to the enhancer present at the locus. We have also found that treatment of the cells with trichostatin A (TSA) (51) eliminates the oscillations in a cell cycle-dependent manner. Oscillations at a second locus were also observed.
MATERIALS AND METHODS
Plasmids.
Construction of the expression cassettes used in this study was previously described (1). Fragments containing HS2, HS3, and HS4 are fragments 7764-9172, 4273-5122, and 951-2199, respectively, of GenBank file HUMHBB. The β-globin promoter extends from −344 to +44 relative to the cap site. The MCNeopA gene was obtained from Stratagene (La Jolla, Calif.).
Cell line construction.
Generation of the cell lines used in this study is described in detail elsewhere (1). Briefly, RL1 was constructed by electroporation of a DNA fragment containing the phosphoglycerate kinase (PGK)-hygromycin (HYG) reporter flanked by two Lox sites differing by a point mutation in their spacer regions. The transfected fragment also contained the coding sequence of the Neo gene (but no promoter) located just 3′ from the cassette defined by the Lox sites. One clone containing a single copy of this fragment (as determined by Southern blotting) was then used as the target cell line in which we integrated the four cassettes depicted in Fig. 1. These integrations were performed via RMCE using the preintegrated mutated Lox sites and a gene trap system based on the activation of the Neo gene by integration of the MC promoter next to the Neo coding sequence. In a secondary step, this reconstituted MCNeo selectable marker was excised via FLP-mediated recombination. The end result of these manipulations was therefore the integration of four different cassettes at the same chromosomal locus (RL1) in the absence of any selectable marker. Integration of cassette 3 at RL3 was performed similarly, except that the locus was initially generated with a plasmid containing the cytomegalovirus-HYTK selectable marker and that the second step (excision of the selectable marker) was precluded by the use of cell sorting to select for cells that had undergone RMCE. Details of this improved RMCE procedure will be published elsewhere (7).
Cell culture.
MEL cells (clone 745 [DS19]) were grown at 37°C and 7% CO2 in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Benzidine staining and induction of differentiation with dimethyl sulfoxide (DMSO) or hemin were performed as previously described (1).
Long-term culture.
To standardize the culture conditions as much as possible, the same batch of serum was used throughout the experiments and cells were fed at fixed times, three times a week, by the same person. Absence of cross-contamination was routinely verified by taking advantage of specific properties of each cell line used in this study: controls had a HYG gene at RL1 and were the only cells that were hygromycin resistant; cells with cassette 234Neo at RL1 were the only G418-resistant cells; cells with cassette 3 are not inducible by hemin (1). At the end of each long-term experiment, all cultures were induced to differentiate with DMSO, with over 95% of the cells turning β-galactoridase (β-Gal) positive, demonstrating that loss of the chromosome carrying RL1 was not occurring at an appreciable frequency.
Subclones were obtained by depositing cells at a density of one cell per well in 96-well plates, using a Becton Dickinson (San Jose, Calif.) FACSStar flow cytometer. Under these conditions, MEL cells have a plating efficiency of 50 to 60% and a single colony could be visually detected after 7 days of incubation in about half of the wells. The precision of the flow cytometer was evaluated by sorting cells at a density of two or four cells per well. As expected, one or two visible colonies grew in most wells. This demonstrates the accuracy of the sorting procedure and confirms that the subclones obtained were truly the progeny of single cells.
FACS-Gal (fluorescence-activated cell sorting–β-Gal) analyses were performed as described elsewhere (10). The experiments represented in Fig. 2b were performed three times on three different days.
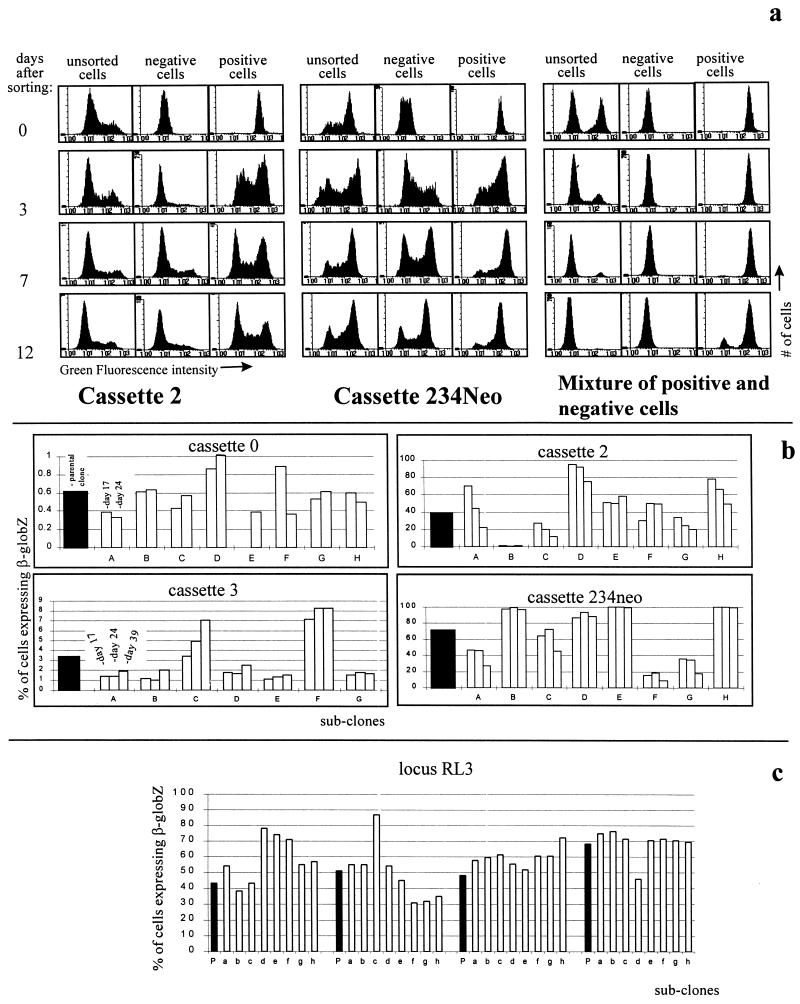
FIG. 2.
Mosaic expression patterns are caused by oscillations between activation and inactivation of the β-globZ gene. (a) Demonstration of oscillation by flow cytometry. Cells with cassettes 2 and 234Neo at RL1, as well as an artificial mixture of 100% negative cells and 100% positive cells, were sorted into populations of cells >99% negative and >99% positive for β-globZ expression and were monitored by FACS-Gal. In the case of cassettes 2 and 234Neo, the unsorted populations remained unchanged, while both the negative and positive populations returned to mosaic expression patterns similar to those of their parental populations within a few days, suggesting that expression of the β-globZ gene oscillates between activation and inactivation. (b) Demonstration of oscillations by subcloning. Subclones of cells with cassettes 0, 2, 3, and 234Neo at RL1 were monitored by FACS-Gal for up to 40 days. Black bars represent percentages of cells expressing β-globZ before subcloning; white bars represent percentages of cells expressing β-globZ 17, 24, or 39 days after subcloning for each of eight subclones (A to H). Almost all of the subclones displayed mosaic expression patterns similar to those of the parental populations, demonstrating the existence of oscillations. (c) Oscillations at RL3. Cassette 3 was inserted by RMCE at RL3. Subclones were then generated from four independent clones, and expression was analyzed by FACS-Gal. As at RL1, the progeny of individual cells display mosaic expression patterns demonstrating that the cells oscillate between activation and inactivation at this locus too. P, parental clones; a to h, eight independent subclones.
TSA incubations.
Cells (5 × 104/ml) were incubated and fed every 3 days for 16 days with medium containing 5 or 10 nM TSA. After day 16 of incubation, all cultures were less than 0.5% benzidine positive, confirming that these concentrations of TSA did not induce differentiation. All experiments were performed in duplicate.
HS mapping studies.
HS mapping studies were performed as described by Dhar et al. (5). Briefly, 2 × 106 to 10 × 106 nuclei were incubated with 0.5, 1, or 2 U of RQ1 DNase I (Promega, Madison, Wis.) for 10 min at 37°C, and the DNA was purified by extraction with organic solvents and analyzed by Southern blotting after digestion with EcoRI, using the lacZ coding sequence as a probe.
General DNase I sensitivity studies.
Partially DNase I-digested genomic DNA was obtained exactly as described above. DNase I sensitivity of the lacZ coding sequence was then determined relative to the mouse β-globin major (β-major) gene (DNase I-sensitive control) and to the Cγ2b gene from the immunoglobulin heavy-chain locus (DNase I-resistant control). The LacZ probe was a 2-kb Not I fragment from plasmid pCMVbeta (Clontech, Palo Alto, Calif.) that contains the lacZ coding sequence. The β-major probe was a 0.45-kb Sau3A/XbaI fragment located just 3′ of the β-major gene coding sequence (fragment E in reference 25). The immunoglobulin heavy-chain region probe was a 1.4-kb fragment located 5′ of the Cy2b gene (pBR 1.4 probe in reference 33). The probes were used either sequentially or simultaneously. Similar results were obtained in both cases. The intensity of each band was quantified on a phosphorimager. At least two independent series of DNase I digests were performed for each cell line tested.
Cell cycle analysis.
Cells were separated according to size by centrifugal elutriations as described elsewhere (2) except that the cells were not labeled with bromodeoxyuridine. Briefly, 108 exponentially growing MEL cells were resuspended in 20 ml of Hanks’ medium containing 5% fetal bovine serum and loaded into an elutriation rotor spinning at 2,000 rpm, and cells of increasing sizes were incrementally recovered by collecting 50-ml fractions after the flow rate of the peristaltic pump was increased by a fixed amount. The quality of the separation was monitored by FACS by measuring the DNA content of citrate nuclei with the help of propidium iodide by the procedure of Krishan (32).
β-Gal half-life.
MEL cells containing cassette 234 at RL1 (1) were incubated with either cycloheximide (20 or 60 μg/ml) or actinomycin D (5 or 30 μg/ml) (11, 12, 27, 30, 42), aliquots were taken every 2 h, and LacZ expression was evaluated by chemiluminescence on protein extract as described elsewhere (1). For both drugs, similar kinetics of β-Gal decay were obtained with both concentrations tested. All experiments were performed in duplicate. Both drugs totally blocked cell division at both concentrations tested, since no increase in cell number was detectable at any point after addition of the drug.
RESULTS
We previously reported (1) that when two genes are cointegrated at RL1 (cassette 234Neo [Fig. 1a and reference 1]), one of the genes can be expressed in all cells whereas the other displays mosaic expression patterns. This observation contrasted with clonally inherited PEVs observed in Drosophila and with silencing of globin regulatory elements in transgenic mice, since these phenomena often involve silencing of a large chromatin region (28, 50a) and a closed chromatin conformation (16). To investigate the differences between PEV and the mosaic expression patterns at RL1, we have assessed the mode of epigenetic inheritance and the long-term stability of expression at RL1.
Mosaic expression patterns are stable.
The stability of expression of cassettes 2, 3, 234, and 234Neo (Fig. 1a) inserted at RL1 was assessed by monitoring the proportion of cells expressing β-globZ via FACS-Gal, a highly sensitive method to detect β-Gal activity in single cells (10). For each of the four cassettes, three independently derived clones were studied. A total of 12 clones were therefore studied. As shown in Fig. 1b, the percentages of expressing cells characteristic of each cassette did not vary significantly over a period of 4 months. To confirm these results a different set of 12 subclones was studied. Similar results were obtained, except that one of the three clones containing cassette 2 was subject to gradual extinction and one of the three clones containing cassette 234Neo was subject to gradual activation (data not shown). In summary, 22 of the 24 clones studied exhibited mosaic expression patterns that were stable for at least 200 cell cycles and that were specific for the enhancer present at the locus.
Mosaic expression patterns result from oscillation of expression.
The mode of epigenetic inheritance of β-globZ expression at RL1 was then assessed by sorting expressing and nonexpressing cells and by monitoring the two sorted populations by FACS-Gal (Fig. 2a). We tested cells with cassettes 2, 3, and 234Neo at RL1 and a 1:1 mixture of control 100% negative cells (PGK-HYG at RL1) and control 100% positive cells (cassette 234 at RL1 [1]). For all cassettes tested, populations of cells that were over 99% either expressing or nonexpressing returned to the mosaic expression pattern of the unsorted populations within a few days. This finding demonstrates that these mosaic expression patterns are not clonally inherited and that expression at RL1 is dynamically regulated: active RL1 loci are constantly being inactivated, and inactive RL1 loci are constantly being activated.
In the case of the mixture of controls, the proportion of positive cells in the unsorted mixture declines gradually to almost zero because this particular negative control grows faster than the positive control. Because of the imperfections of the sorting procedure, this growth advantage led to a gradual increase in the proportion of β-globZ-negative cells in the positive population. In contrast to the situation with cassette 2 or 234Neo at RL1, however, the negative population remained 100% negative. This graphically illustrates that the results observed with cassettes 2, 3, and 234 cannot be caused by differential growth rates of the β-Gal-positive and -negative cells.
To confirm the existence of the oscillations, subclones were generated, expanded, and monitored by FACS-Gal for 2 months (Fig. 2b). Almost all of the subclones with cassette 0, 2, 3, or 234Neo integrated at RL1 exhibited mosaic expression patterns similar to those of their parental cell populations. This finding definitively establishes that the mosaic expression patterns at RL1 are caused by oscillations since the progenies of individual cells are mixed populations of expressing and nonexpressing cells.
To assess the generality of transcriptional oscillations, cassette 3 was introduced by RMCE at a second random locus in MEL cells (RL3), and expression and epigenetic inheritance were analyzed. FACS-Gal analysis on four clones with a single copy of cassette 3 at RL3 revealed that 40 to 60% of the cells were expressing LacZ, suggesting that RL3 was more permissive for expression than RL1 since when the same cassette is integrated at the latter locus, only 2 to 4% of the cells express LacZ. To determine if the mosaic expression patterns at RL3 were caused by transcriptional oscillations, we derived eight subclones from each of the four clones containing cassette 3 integrated at RL3 and evaluated expression by FACS. All 32 subclones thus analyzed exhibited mosaic expression patterns similar to those of the parental clones (Fig. 2c). Since each of the subclones originated from a single cell, we concluded that the mosaic expression at RL3 was caused by oscillations similar to the ones at RL1.
To better interpret these results, we measured the half-life of β-Gal as described in Materials and Methods. After inhibition of protein synthesis, the calculated half-life of β-Gal was about 9 + 2 h. After inhibition of transcription, the calculated half-life was 11 + 2 h. Since this half-life is about as long as a MEL cell cycle, it follows that many cell divisions must be required for a β-Gal-positive cell to turn FACS-Gal negative (after the gene has been switched off). Oscillations between activation and inactivation are therefore less frequent than once per cell cycle, since otherwise all of the cells would appear to be expressing at all times.
HS2 and HS3 are formed whether the RL1 locus is transcriptionally active or inactive.
Active globin genes reside in open chromatin domains characterized by early replication, DNase I sensitivity, hypomethylation, and histone hyperacetylation. In contrast, inactive globin genes in nonerythroid cells, or in erythroid cells with an LCR deletion, reside in closed chromatin domains characterized by late replication, DNase I resistance, hypermethylation, and hypoacetylation (13, 21, 23). The oscillations described here could occur either between the open and closed chromatin structures defined above or between two different states of active chromatin. To address this question, cells with cassette 2 at RL1 were sorted into expressing and nonexpressing populations, nuclei were extracted, and the presence of HS2 was assessed by limited DNase I digestion of the nuclei (Fig. 3A). We found that HS2 was present in both the expressing and nonexpressing cells. Interestingly, an additional HS mapping at the promoter was detected in the sorted expressing cells but not in the sorted nonexpressing cells. HS mapping was also performed with unsorted cells containing HS3 at RL1. This revealed that HS3 formed at this locus in a significant proportion of the cells (Fig. 3B). Since less than 3% of the cells containing this enhancer express the lacZ gene, we conclude that HS3 also forms in nonexpressing cells.
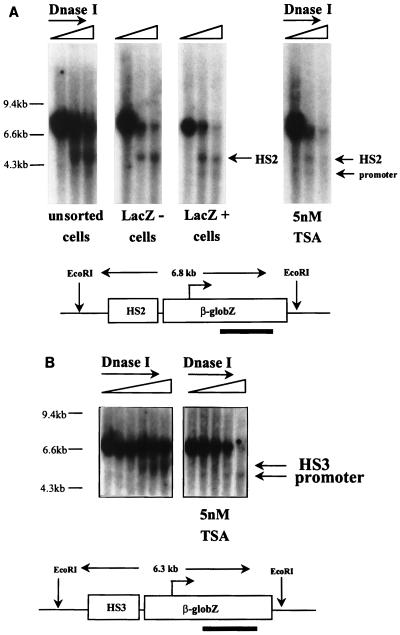
FIG. 3.
HS mapping. (A) HS2. Cells with HS2 integrated at RL1 were sorted into expressing and nonexpressing populations, and formation of HS2 was assessed by limited DNase I digestion and hybridization of EcoRI-digested genomic DNA with a lacZ coding sequence probe. HS2 was found in both expressing and nonexpressing cells. A second HS that maps at the promoter was present in the expressing cells. For the assay on the right, cells with HS2 integrated at RL1 were cultured in the presence of 5 nM TSA for 14 days, and DNase I analysis was performed as described above. HS2 was still present after TSA treatment. The promoter HS could also be detected. (B) HS3. Unsorted cells with HS3 integrated at RL1 were tested as described above before or after treatment with TSA for 2 weeks. This assay revealed that HS3 forms in these cells and that an additional HS mapping at the promoter is induced by TSA.
General DNase I sensitivity.
To determine if the chromatin at the RL1 locus is more sensitive to DNase I when LacZ is expressed than when it is silent, we partially digested nuclei from expressing and nonexpressing sorted populations of cells with HS2 at RL1 with increasing concentrations of DNase I and extracted genomic DNA. After digestion with BamHI, the genomic DNA was hybridized with three probes that we had previously shown to be devoid of any detectable HS (data not shown): (i) a fragment of the lacZ coding sequence, (ii) a fragment of the β-major gene (which is known to be accessible to DNase I in MEL cells), and (iii) a fragment of the Cγ2b gene of the immunoglobulin heavy-chain locus (which is transcriptionally silent in MEL cells and therefore presumably in a chromatin region that is less accessible to DNase I). As expected, the β-major fragment faded out with lower DNase I concentrations than the immunoglobulin fragment, demonstrating that in these cells, the chromatin at the β-globin locus is more accessible to DNase I than the chromatin at the immunoglobulin locus. For all three cell populations tested (sorted expressing, sorted nonexpressing, and unsorted), the fragment containing the lacZ coding sequences was at least as accessible to DNase I as the β-major fragment (Fig. 4A). Similar experiments with unsorted cells containing HS3 integrated at RL1 revealed that in this case also, the lacZ sequences were as accessible to DNase I as the β-major region (Fig. 4B). Since when cassette 3 is at RL1 only a few percent of the cells are expressing, this result suggests that in the presence of HS3, the locus is accessible to DNase I, whether or not it is transcribed.
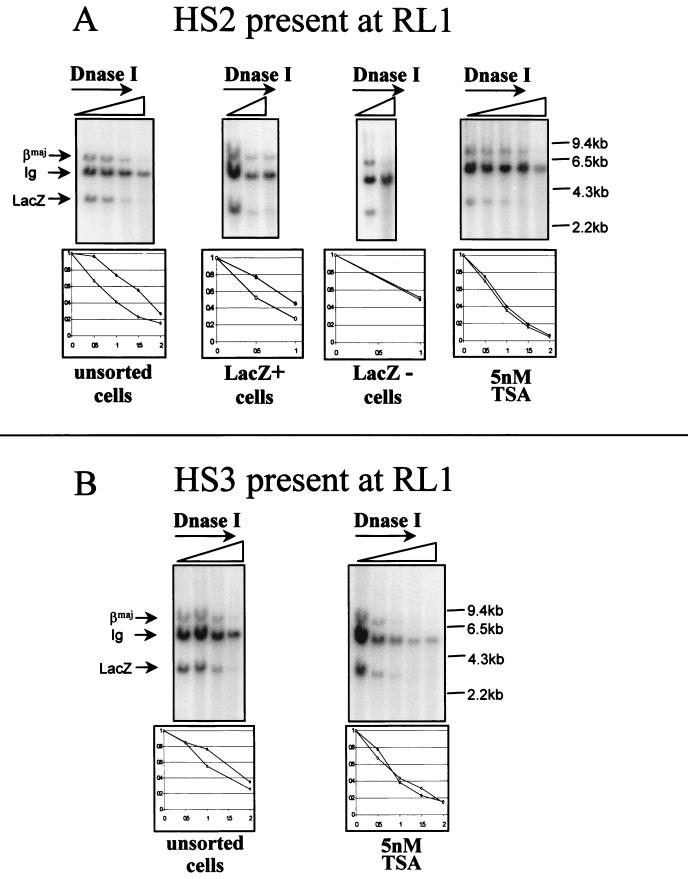
FIG. 4.
General DNase I sensitivity. (A) HS2. Nuclei were incubated with increasing concentrations of DNase I, and genomic DNA was extracted, digested with BamHI, and hybridized with three probes specific for the β-major gene (βmaj; DNase I-sensitive control), the region 5′ of the Cγ2b gene (DNase I-resistant control), and the lacZ coding sequences. Representative autoradiograms obtained in assays using unsorted, sorted expressing, and sorted nonexpressing cells and cells treated with 5 nM TSA for 2 weeks are shown. Intensities of each band were quantified by phosphorimaging. The plot below each autoradiogram represents the ratio of the intensity of the LacZ band to the intensity of the Cγ2b band (●) and the ratio of the intensity of the β-major band to the intensity of the Cγ2b band (○) for each concentration of DNase I used. All ratios were normalized to the value obtained when no DNase I was added. This study clearly demonstrated that in the presence of cassette 2, the RL1 locus is as sensitive to DNase I as the β-globin gene regardless of whether the gene is expressed and whether TSA was used. Ig, immunoglobulin. (B) HS3. Unsorted cells (untreated or treated with 5 nM TSA for 2 weeks) containing HS3 at locus RL1 were examined for general DNase I sensitivity as described above. This assay revealed that in the presence of this HS too, the chromatin was as accessible to DNase I as the β-major gene.
TSA progressively eliminates the oscillations.
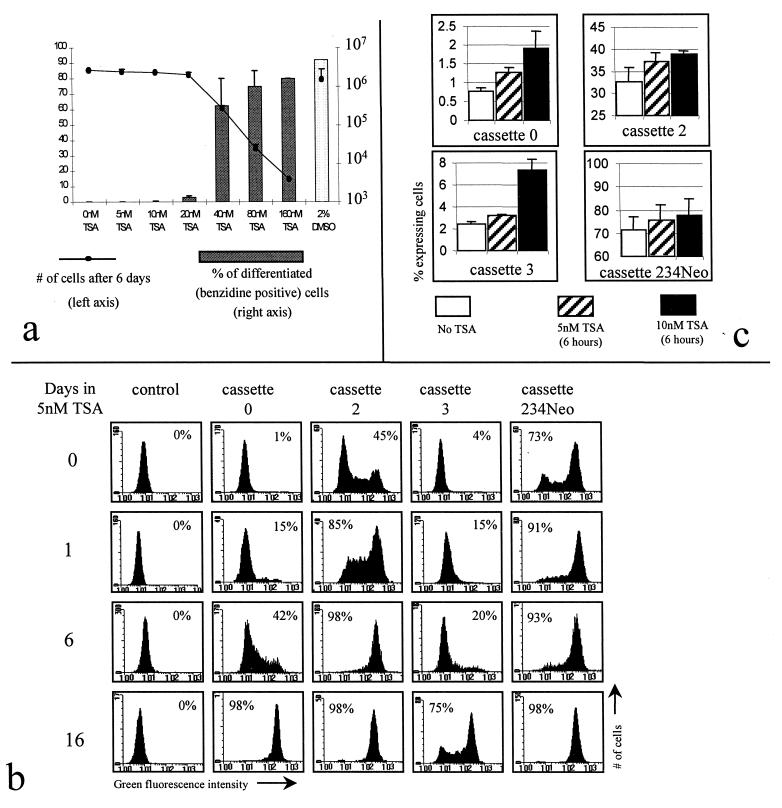
A possible role of histone acetylation in the generation of oscillations was investigated by using TSA, a potent and specific inhibitor of histone deacetylase (52). Since TSA is a known inducer of MEL cell differentiation (53) and since we had previously shown that oscillations at RL1 are eliminated when MEL cells differentiate (1), an evaluation of a potential effect of TSA directly on the RL1 locus, rather than through differentiation, required finding concentrations of TSA that do not induce differentiation. To find such concentrations, dose-response experiments were performed by incubating MEL cells with increasing amounts of TSA, or with 2% DMSO, for 6 days (Fig. 5a). This revealed that concentrations below 20 nM do not induce differentiation of our MEL cell strain. Further experiments revealed that even after a 2-month-long incubation of MEL cells in 5 or 10 nM TSA, the percentage of benzidine-positive cells was always less than 2%. Concentrations of 5 and 10 nM were therefore chosen to test the effect of TSA on the oscillations. At these concentrations, no cytotoxicity was observed.
FIG. 5.
Enhancer-dependent oscillations are eliminated by a histone deacetylase inhibitor. (a) TSA dose response. Cells (5 × 104/ml) were incubated with increasing amounts of TSA, or with 2% DMSO, for 6 days. The cells were then counted (line graph), and the percentage of cells induced to differentiate was evaluated by benzidine staining (bar graph). Concentrations below 20 nM did not induce differentiation and were not cytotoxic. (b) TSA eliminates oscillations of β-globZ. MEL cells with cassettes 0, 2, 3, and 234 at RL1 were incubated with 5 nM TSA, and β-globZ expression was monitored by FACS-Gal. Regardless of the cassettes at RL1, the proportion of expressing cells gradually increased. (c) Minimum response times to TSA. Cells with cassette 0, 2, 3, or 234Neo were incubated with 5 nM TSA for 6 h and tested by FACS-Gal. An increase in the percentage of positive cells could be clearly detected after 6 h.
Cells with cassette 0, 2, 3, or 234Neo at RL1 were grown in the presence of 5 and 10 nM TSA for 2 weeks, and the proportion of cells expressing LacZ was periodically monitored by FACS-Gal. At the end of each experiment, all cells were stained with benzidine to verify that differentiation had not occurred. As shown in Fig. 5b and c, the mosaic expression patterns were eliminated or greatly attenuated in the presence of TSA, regardless of the cassette integrated at RL1. Slightly faster kinetics of induction were observed when the cells were incubated with 10 nM TSA (data not shown). Less than 2% benzidine-positive cells were observed after the 2 weeks of culture in 5 or 10 nM TSA.
To determine the effect of TSA at the chromatin level, DNase I HS mapping and general DNase I sensitivity studies were performed with cells grown in the presence of 5 nM TSA for 2 to 4 weeks (Fig. 3 and 4). As expected, after TSA treatment, HS2 and HS3 were still present. In both cases, an additional site mapping at the β-globin promoter could also be detected. Presence of an HS at the promoter demonstrates that TSA treatment affects expression at the transcriptional level. Regardless of the enhancer present at RL1, the chromatin was about as accessible to DNase I before and after treatment with TSA.
To determine whether TSA activation of gene expression at locus RL1 is direct or indirect, the minimum time required for detection of an increase in the proportion of expressing cells was determined. The response time was found to be 6 h when 5 or 10 nM TSA was used (Fig. 4B) and 1 to 3 h when 75 nM TSA (data not shown) was used. While quite short, these response times do not completely rule out that TSA activates expression indirectly.
Activation by TSA requires passage through late G1 or early S.
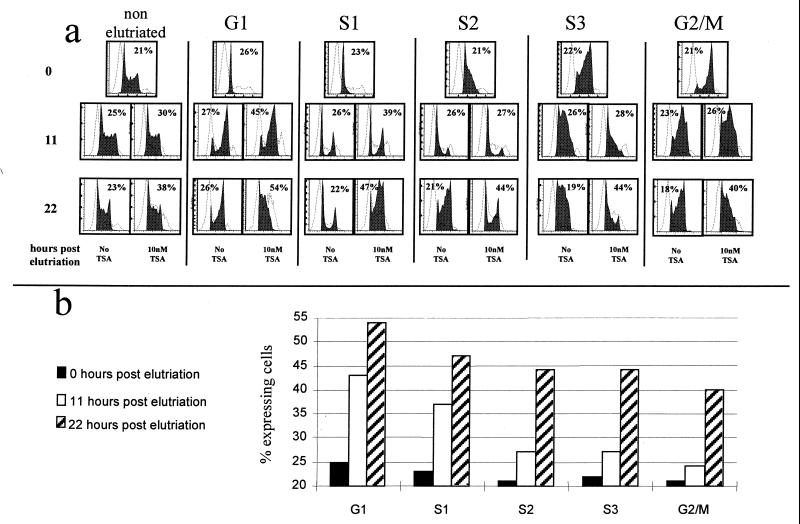
To determine if activation by TSA occurs throughout the cell cycle or only at a particular stage, cells with cassette 2 at RL1 were fractionated into five cell cycle fractions (G1, S1, S2, S3, and G2/M) by centrifugal elutriation. Each fraction was then cultured in the presence of 10 nM TSA, and the kinetics of activation of β-globZ was monitored for about two cell cycles (Fig. 6). When TSA was added to the G1 fraction, the percentage of cells expressing β-globZ increased by 80% in 11 h and doubled in 24 h. When TSA was added in G2/M, the response was delayed: the percentage of expressing cells was almost unchanged after 11 h and increased by 80% in 22 h. Results with the S1 to S3 fractions were intermediate. Yoshida et al. have reported that inhibition of histone deacetylation by TSA can be detected after only 30 min of incubation (52). The delay response to TSA that we observed is therefore best explained by the hypothesis that histone hyperacetylation affects transcription only after passage through late G1 or more likely after replication of the locus. The finding of an 80% increase in the proportion of expressing cells after 11 h in the G1 fraction confirms that the response to TSA starts occurring after a relatively short time.
FIG. 6.
TSA acts only in G1 or early S. (a) Cells with HS2 at RL1 were elutriated into five cell cycle fractions (first row), and these five synchronized fractions were incubated for 11 (second row) and 22 (third row) h with or without TSA. Each fraction was monitored by FACS for its position in the cell cycle (black histogram; x axis = DNA content and y axis = cell number) and by FACS-Gal to determine the proportion of expressing cells (dotted histograms; x axis = level of β-globZ expression and y axis = cell number). The percentage of expressing cells determined by FACS-Gal is shown in the upper corner of each diagram. (b) The percentage of expressing cells is plotted for each cell fraction at each time point. The cells in G1 and S1 respond earlier than the other cells to TSA.
These experiments also show that TSA significantly slows the cell cycle: after 22 h, cells grown with TSA were several hours behind cells grown without TSA (Fig. 6a).
DISCUSSION
Summary of the results.
Expression at locus RL1 is dynamically regulated and oscillates at a rate set by the LCR fragments present at the locus. In the presence of cassette 2 or 3 at RL1, HS2 and HS3 can be detected whether the locus is actively transcribed or not. Similarly, in the presence of cassettes 2 and 3, the chromatin is accessible to DNase I whether the locus is transcribed or not. Treatment with TSA progressively eliminates the oscillations, but passage through late G1 or early S phase is required before TSA affects expression.
DNase I sensitivity studies.
The chromatin of the human β-globin gene locus is preferentially DNase I sensitive in erythroid cells (20, 22). Since this general DNase I sensitivity is established prior to the onset of gene transcription and affects active and inactive globin genes as well as nontranscribed flanking sequences, the chromatin of the β-globin locus can adopt at least three different conformations: a closed conformation present in nonerythroid cells, a transcriptionally competent conformation in immature erythroid cells that do not transcribe the globin genes, and a transcribed conformation in mature erythroid cells.
By analogy to the endogenous locus, transcriptional oscillations at RL1 could be caused by oscillations between the transcribed chromatin conformation and either the closed or transcriptionally competent conformation. The findings that (i) HS2 and HS3 are present in nontranscribing cells and (ii) the chromatin is in a conformation accessible to DNase I regardless of transcription clearly support the second alternative. These cells should therefore prove useful as a model system to study the structure of these different types of open chromatin.
The results also imply that at this locus, enhancers affect the frequency of the transition from transcriptionally competent to transcriptionally active chromatin rather than the transition from closed chromatin to transcriptionally competent chromatin since the proportion of expressing cells at RL1 is dependent on the HS. Recently, Reik et al. (40) deleted HS2, HS3, and HS4 from the human β-globin endogenous locus by homologous recombination in DT40 cells and transferred the mutated chromosome into MEL cells. This deletion was associated with a complete inactivation of transcription of the locus. However, the remaining sites, HS1 and HS5, were still formed, and the chromatin of the locus remained preferentially DNase I sensitive. The dissociation between transcription and chromatin structure defined by sensitivity to DNase I is analogous to the findings reported here. One explanation for this dissociation would be that in MEL cells, the β-globin promoter itself has the capacity to open up the chromatin.
The facts that the chromatin is sensitive to DNase I and that the HSs are present regardless of transcription also confirm that the oscillations are distinct from PEV, since the latter type of position effects is associated with a closed chromatin conformation (16).
Presence of an HS is believed to be due to a disrupted nucleosome or to the presence of a nucleoprotein complex that displaced the nucleosome (17). Our results clearly show that the presence of such a complex on the enhancer is not sufficient to activate transcription. The finding that when the cells are actively transcribing an HS can also be detected at the promoter suggests that during active transcription, nucleoprotein complexes are present at the enhancer and at the promoter. Whether these two complexes interact via looping of the intervening sequences or via some other mechanism is not clear at this point. We cannot exclude that the nucleoprotein complex present at HS2 when the cells are expressing is different from that present when they are silent.
The fact that when cassette 0 is at RL1, a small but clearly detectable fraction of the cells are expressing suggests that formation of the nucleoprotein complex at the promoter does not require the presence of an LCR fragment. However, the observation that the proportion of expressing cells depends on the type of enhancer present at RL1 suggests that enhancers can affect the frequency at which the nucleoprotein complex forms at the promoter.
Role of enhancers in the generation of oscillations.
Previous reports had suggested that in K562 cells, HS2 affects transcription by decreasing the probability of permanent gene silencing, a process that might be caused by progressive heterochromatinization (49, 50). The oscillations in MEL cells are clearly different from permanent gene silencing since they result from dynamic activation and inactivation of the transcription unit. Nevertheless, as discussed above, the data reported here also suggest that enhancers affect the probability of expression. However, in contrast with K562 cells, when expression ceases to occur, the silencing is not permanent. Differences between these two sets of results could be due to the use of different cell lines, integration sites, regulatory elements, or experimental strategies in the two studies.
Role of histone acetylation.
Results of the TSA experiments suggest that histone acetylation plays a role in the generation of these oscillations. However, one cannot definitively conclude that the effect of TSA is direct because this compound probably has global effects on chromatin structure.
A striking feature of activation by TSA is its progressivity even when synchronized populations are tested. TSA does not activate transcription at RL1 in all cells at the same time but rather increases the probability that a gene will be active after each replication. Histone hyperacetylation after inhibition of histone deacetylase with TSA is believed to occur rapidly (52). Therefore, the progressive activation of β-globZ in the presence of TSA cannot be explained by a slow increase in the amount of hyperacetylated histones in the cells. This suggests that TSA affects the transmission of the epigenetic modifications that are required for active transcription after each cell cycle.
The finding that activation by TSA requires passage through the late G1 or early S phase of the cell cycle is also compatible with a role of histone acetylation in the transmission of the epigenetic modifications that are required for transcription.
Mechanism of generation of oscillations.
Whether or not the cells express the β-globZ gene, they have the same genomic structure at locus RL1. By definition the oscillations are therefore caused by an epigenetic phenomenon. Two factors could influence these oscillations: either the concentration of critical transcription factors varies more or less randomly in the cells or the fine chromatin structure of the locus differs in a semiinheritable manner in expressing versus nonexpressing cells. As discussed by Struhl (45), several histone deacetylases and histone acetyltransferases either are intrinsically part of the polymerase II transcription machinery or are able to interact with it. This complicates this classical distinction between trans- and cis-acting factors because these recent observations demonstrate that the chromatin structure is not a passive matrix on which transcription is initiated but is modified by the transcription machinery itself. Consequently, active transcription and chromatin structure are inextricably linked.
In this context, the results obtained with TSA support models in which the establishment and/or the maintenance of an active transcription unit depends on histone acetylation and its inheritance after each replication. Since the rate of oscillation is set by enhancers, these results also indirectly suggest that one role of enhancers may be to act as nucleation sites for histone acetyltransferases. Further studies are required to decisively demonstrate these points.
Transcriptional oscillations might be quite frequent.
The observation of oscillations at both of the two random loci that we tested (RL1 and RL3) and the fact that oscillations might account for the mosaic expression patterns observed in several lymphocytic and fibroblastic cell lines (9, 29, 31, 39) suggest that oscillations are not limited to one locus or to MEL cells.
Milot et al. (35) have observed “transcription timing” position effects in transgenic mice containing cassettes driven by mutated LCRs. These position effects were caused by the fact that at any moment in time only a fraction of the erythroid cells were transcribing the transgene (35). Although in these mice all cells contained globin mRNA, this transcription pattern might be related to the oscillations in MEL cells since a more pronounced reduction in the frequency of transgene transcription in these mice might result in mosaic expression patterns. When HS234 is at RL1, all cells are expressing (Fig. 1a). This could be due either to the lack of oscillation in the presence of this larger enhancer or to the fact that in the presence of HS234 the oscillations are so rapid that detectable amounts of β-Gal remain in the cells between oscillations. If the latter alternative is correct, the pattern of expression in these cells would be similar to the one found for the transgenic lines with transcription timing position effects described by Milot et al.
Possible biological consequences of the oscillations.
Although our results involve small regulatory elements artificially introduced at random chromosomal sites, it is tempting to speculate that expression of natural genes can also oscillate. A recent report that transcription in primary muscle fibers is pulsed demonstrates that this is indeed the case (36). Oscillations of expression of receptors, transcription factors, or other regulatory proteins would have profound consequences for many biological phenomena. For instance, oscillations of genes involved in the response to differentiation and proliferation signals in stem cells could explain the stochastic behaviors observed during hematopoiesis (37, 46): complex cycles of inactivation and activation of cytokine receptors could spontaneously and reversibly produce daughter cells with different levels of sensitivity to cytokines and therefore produce apparently stochastic differentiation patterns. More generally, oscillations of expression could be the molecular mechanism of the heterogeneity and apparent randomness of the response of numerous biological systems to endogenous or exogenous signaling molecules.
Finally, these findings have important implications for gene therapy, since extinction of provirus (43) and integrated adeno-associated virus sequences (4) is a common problem. Extinctions of adeno-associated virus sequences are reversible by treatment with TSA (4) and therefore might be caused by unregulated oscillations. Overcoming these extinctions will require a better understanding of the cause and role of oscillations.
ACKNOWLEDGMENTS
We are very grateful to Ronald Nagel for ongoing support of this project; to Ilia Rochlin, Antonietta Marmorato, and Maria Martinovski for excellent technical help; and to E. Klein-Bouhassira, A. Eisen, S. Fiering, A. Skoultchi, C. Schildkraut, and M. Rapport for critical review of the manuscript.
This work was supported by grants NIH HL38655 and HL 55435 from the NIH and by grant-in-aid 95015110 from the American Heart Association.
REFERENCES
- 1.Bouhassira E E, Westerman K, Leboulch P. Transcriptional behavior of LCR enhancer elements integrated at the same chromosomal locus by recombinase-mediated cassette exchange. Blood. 1997;90:3332–3344. [PubMed] [Google Scholar]
- 2.Braunstein J D, Schildkraut C L. The β-major and β-minor globin genes in murine erythroleukemia cells replicate during the same early interval of the S phase. Biochem Biophys Res Commun. 1984;123:108–113. doi: 10.1016/0006-291x(84)90386-3. [DOI] [PubMed] [Google Scholar]
- 3.Bungert J, Dave U, Lim K C, Lieuw K H, Shavit J A, Liu Q H, Engel J D. Synergistic regulation of human β-globin gene switching by locus control region elements HS3 and HS4. Genes Dev. 1995;9:3083–3096. doi: 10.1101/gad.9.24.3083. [DOI] [PubMed] [Google Scholar]
- 4.Chen W Y, Bailey E C, McCune S L, Dong J Y, Townes T M. Reactivation of silenced, virally transduced genes by inhibitors of histone deacetylase. Proc Natl Acad Sci USA. 1997;94:5798–5803. doi: 10.1073/pnas.94.11.5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhar V, Nandi A, Schildkraut C L, Skoultchi A I. Erythroid-specific nuclease-hypersensitive sites flanking the human β-globin domain. Mol Cell Biol. 1990;10:4324–4333. doi: 10.1128/mcb.10.8.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epner E, Reik A, Cimbora D, Telling A, Bender M A, Fiering S, Enver T, Martin D I, Kennedy M, Keller G, Groudine M. The β-globin LCR is not necessary for an open chromatin structure or developmentally regulated transcription of the native mouse β-globin locus. Mol Cell. 1998;2:447–455. doi: 10.1016/s1097-2765(00)80144-6. [DOI] [PubMed] [Google Scholar]
- 7.Feng, Y. Q., J. Seibler, R. Alami, A. Eisen, S. N. Fiering, and E. E. Bouhassira. Site-specific chromosomal integration in mammalian cells: highly efficient CRE recombinase-mediated cassette exchange. Submitted for publication. [DOI] [PubMed]
- 8.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin DIK, Enver T, Ley T T, Groudine M. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 9.Fiering S, Northrop J P, Nolan G P, Mattila P S, Crabtree G R, Herzenberg L A. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 10.Fiering S N, Roederer M, Nolan G P, Micklem D R, Parks D R, Herzenberg L A. Improved FACS-Gal: flow cytometric analysis and sorting of viable eukaryotic cells expressing reporter gene constructs. Cytometry. 1991;12:291–301. doi: 10.1002/cyto.990120402. [DOI] [PubMed] [Google Scholar]
- 11.Flamigni F, Marmiroli S, Caldarera C M, Guarnieri C. Effect of sodium arsenite on the induction and turnover of ornithine decarboxylase activity in erythroleukemia cells. Cell Biochem Funct. 1989;7:213–217. doi: 10.1002/cbf.290070310. [DOI] [PubMed] [Google Scholar]
- 12.Flamigni F, Marmiroli S, Guarnieri C, Caldarera C M. Stabilization of ornithine decarboxylase in erythroleukemia cells depleted of ATP. Biochem Biophys Res Commun. 1989;163:1217–1222. doi: 10.1016/0006-291x(89)91107-8. [DOI] [PubMed] [Google Scholar]
- 13.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 14.Fraser P, Grosveld F. Locus control regions, chromatin activation and transcription. Curr Opin Cell Biol. 1998;10:361–365. doi: 10.1016/s0955-0674(98)80012-4. [DOI] [PubMed] [Google Scholar]
- 15.Garrick D, Fiering S, Martin DIK, Whitelaw E. Repeat-induced gene silencing in mammals. Nat Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- 16.Garrick D, Sutherland H, Robertson G, Whitelaw E. Variegated expression of a globin transgene correlates with chromatin accessibility but not methylation status. Nucleic Acids Res. 1996;24:4902–4909. doi: 10.1093/nar/24.24.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gross D S, Garrard W T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1995;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- 18.Grosveld F, Antoniou M, Berry M, De Boer E, Dillon N, Ellis J, Hanscombe O, Hurst J, Imam A, et al. The regulation of human globin gene switching. Philos Trans R Soc Lond Ser B. 1993;339:183–291. doi: 10.1098/rstb.1993.0015. . [Review.] [DOI] [PubMed] [Google Scholar]
- 19.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 20.Groudine M, Eisenman R, Gelinas R, Weintraub H. Developmental aspects of chromatin structure and gene expression. Prog Clin Biol Res. 1983;134:159–182. [PubMed] [Google Scholar]
- 21.Groudine M, Forrester W C, Novak U, Epner E. Replication and activation of the human β-globin gene domain. Prog Clin Biol Res. 1989;316A:15–35. [PubMed] [Google Scholar]
- 22.Groudine M, Kohwi-Shigematsu T, Gelinas R, Stamatoyannopoulos G, Papayannopoulou T. Human fetal to adult hemoglobin switching: changes in chromatin structure of the β-globin gene locus. Proc Natl Acad Sci USA. 1983;80:7551–7555. doi: 10.1073/pnas.80.24.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebbes T R, Clayton A L, Thorne A W, Crane-Robinson C. Core histone hyperacetylation co-maps with generalized DNase I sensitivity in the chicken β-globin chromosomal domain. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Henikoff S. Position effect and related phenomena. Curr Opin Genet Dev. 1992;2:907–912. doi: 10.1016/s0959-437x(05)80114-5. . (Review.) [DOI] [PubMed] [Google Scholar]
- 25.Hofer E, Darnell J E., Jr The primary transcription unit of the mouse β-major globin gene. Cell. 1981;23:585–593. doi: 10.1016/0092-8674(81)90154-9. [DOI] [PubMed] [Google Scholar]
- 26.Hug B A, Wesselschmidt R L, Fiering S, Bender M A, Epner E, Groudine M, Ley T J. Analysis of mice containing a targeted deletion of β-globin locus control region 5′ hypersensitive site 3. Mol Cell Biol. 1996;16:2906–2912. doi: 10.1128/mcb.16.6.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanova E, Pironcheva G, Djondjurov L. Turnover of the major polypeptides of 40-S monomer particles. Eur J Biochem. 1981;113:569–573. doi: 10.1111/j.1432-1033.1981.tb05100.x. [DOI] [PubMed] [Google Scholar]
- 28.Karpen G H. Position-effect variegation and the new biology of heterochromatin. Curr Opin Genet Dev. 1994;4:281–291. doi: 10.1016/s0959-437x(05)80055-3. . (Review.) [DOI] [PubMed] [Google Scholar]
- 29.Kerr W G, Nolan G P, Herzenberg L A. In situ detection of transcriptionally active chromatin and genetic regulatory elements in individual viable mammalian cells. Immunology. 1989;2(Suppl.):74–78. [PubMed] [Google Scholar]
- 30.Khochbin S, Principaud E, Chabanas A, Lawrence J J. Early events in murine erythroleukemia cells induced to differentiate. Accumulation and gene expression of the transformation-associated cellular protein p53. J Mol Biol. 1988;200:55–64. doi: 10.1016/0022-2836(88)90333-6. [DOI] [PubMed] [Google Scholar]
- 31.Ko M S, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishan A. Rapid flow cytofluorometric analysis of mammalian cell cycle by propidium iodide staining. J Cell Biol. 1975;66:188–193. doi: 10.1083/jcb.66.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang R B, Stanton L W, Marcu K B. On immunoglobulin heavy chain gene switching: two gamma 2b genes are rearranged via switch sequences in MPC-11 cells but only one is expressed. Nucleic Acids Res. 1982;10:611–630. doi: 10.1093/nar/10.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin DIK, Fiering S, Groudine M. Regulation of β-globin gene expression: straightening out the locus. Curr Opin Genet Dev. 1996;6:488–495. doi: 10.1016/s0959-437x(96)80072-4. [DOI] [PubMed] [Google Scholar]
- 35.Milot E, Strouboulis J, Trimborn T, Wijgerde M, Deboer E, Langeveld A, TanUn K, Vergeer W, Yannoutsos N, Grosveld F, Fraser P. Heterochromatin effects on the frequency and duration of LCR-mediated gene transcription. Cell. 1996;87:105–114. doi: 10.1016/s0092-8674(00)81327-6. [DOI] [PubMed] [Google Scholar]
- 36.Newlands S, Levitt L K, Robinson C S, Karpf A B, Hodgson V R, Wade R P, Hardeman E C. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogawa M. Differentiation and proliferation of hematopoietic stem cells. Blood. 1993;81:2844–2853. [PubMed] [Google Scholar]
- 38.Peterson K R, Clegg C H, Navas P A, Norton E J, Kimbrough T G, Stamatoyannopoulos G. Effect of deletion of 5′HS3 or 5′HS2 of the human β-globin locus control region on the developmental regulation of globin gene expression in β-globin locus yeast artificial chromosome transgenic mice. Proc Natl Acad Sci USA. 1996;93:6605–6609. doi: 10.1073/pnas.93.13.6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reddy S, DeGregori J V, von Melchner H, Ruley H E. Retrovirus promoter-trap vector to induce lacZ gene fusions in mammalian cells. J Virol. 1991;65:1507–1515. doi: 10.1128/jvi.65.3.1507-1515.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reik A, Telling A, Zitnik G, Cimbora D, Epner E, Groudine M. The locus control region is necessary for gene expression in the human β-globin locus but not the maintenance of an open chromatin structure in erythroid cells. Mol Cell Biol. 1998;18:5992–6000. doi: 10.1128/mcb.18.10.5992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson G, Garrick D, Wilson M, Martin DIK, Whitelaw E. Age-dependent silencing of globin transgenes in the mouse. Nucleic Acids Res. 1996;24:1465–1471. doi: 10.1093/nar/24.8.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rousseau D, Khochbin S, Gorka C, Lawrence J J. Regulation of histone H1(0) accumulation during induced differentiation of murine erythroleukemia cells. J Mol Biol. 1991;217:85–92. doi: 10.1016/0022-2836(91)90613-b. [DOI] [PubMed] [Google Scholar]
- 43.Sadelain M. Genetic treatment of the haemoglinopathies: recombinations and new combinations. Br J Haemat. 1997;98:247–253. doi: 10.1046/j.1365-2141.1997.2313048.x. [DOI] [PubMed] [Google Scholar]
- 44.Stamatoyannopoulos G, Nienhius A W. Hemoglobin switching. In: Stamatoyanoupoulos G, editor. The molecular basis of blood diseases. W. B. Philadelphia, Pa: Saunders; 1994. pp. 107–156. [Google Scholar]
- 45.Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 46.Suda T, Suda J, Ogawa M. Single-cell origin of mouse hematopoietic colonies expressing multiple lineages in variable combinations. Proc Natl Acad Sci USA. 1983;80:6689–6693. doi: 10.1073/pnas.80.21.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutherland HGE, Martin DIK, Whitelaw E. A globin enhancer acts by increasing the proportion of erythrocytes expressing a linked transgene. Mol Cell Biol. 1997;17:1607–1614. doi: 10.1128/mcb.17.3.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tuan D, Solomon W, Li Q, London I M. The “β-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walters M C, Fiering S, Eidemiller J, Magis W, Groudine M, Martin DIK. Enhancers increase the probability but not the level of gene expression. Proc Natl Acad Sci USA. 1995;92:7125–7129. doi: 10.1073/pnas.92.15.7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walters M C, Magis W, Fiering S, Eidemiller J, Scalzo D, Groudine M, Martin DIK. Transcriptional enhancers act in cis to suppress position-effect variegation. Genes Dev. 1996;10:185–195. doi: 10.1101/gad.10.2.185. [DOI] [PubMed] [Google Scholar]
- 50a.Weiler K S, Wakimoto B T. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida M, Horinouchi S, Beppu T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays. 1995;17:423–430. doi: 10.1002/bies.950170510. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Antibiot. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 53.Yoshida M, Nomura S, Beppu T. Effects of trichostatins on differentiation of murine erythroleukemia cells. Cancer Res. 1987;47:3688–3691. [PubMed] [Google Scholar]