Abstract
Complex multicellular organisms require quantitative and qualitative assessments on each of their constitutive cell types to ensure coordinated and cooperative behavior towards overall functional proficiency. Cell competition represents one of the operating arms of such quality control mechanisms and relies on fitness comparison among individual cells. However, what is exactly included in the fitness equation for each cell type is still uncertain. Evidence will be discussed to suggest that the ability of the cell to integrate and collaborate within the organismal community represents an integral part of the best fitness phenotype. Thus, under normal conditions, cell competition will select against the emergence of altered cells with disruptive behavior towards tissue integrity and/or tissue pattern formation. On the other hand, the winner phenotype prevailing as a result of cell competition does not entail, by itself, any degree of growth autonomy. While cell competition per se should not be considered as a biological driving force towards the emergence of the neoplastic phenotype, it is possible that the molecular machinery involved in the winner/loser interaction could be hijacked by evolving cancer cell populations.
Keywords: Cell competition, Cell cooperation, Cell fitness, Cancer, Cell clones, Aging
Transition from unicellularity to multicellularity
The appearance of multicellular organisms represents a major evolutionary transition of life on earth. Available evidence indicates that, in spite of its complex nature, such process has occurred independently several times in diverse microbial lineages and contexts [1]. In fact, while the transient, reversible assembly of unicellular organisms in cooperating groups is of relatively common occurrence, the emergence of a stably interacting and interdependent cell communities relies on the appearance of new genotypic/phenotypic traits acting as “ratchets,” i.e., limiting the possibilities for reversal towards the unicellular status [2]. This process is typically associated with features reinforcing cohesion and including cell specialization with division of labor and the ability to communicate at the group level in order to coordinate and integrate cell behavior at the new level of individuality [3]. Mechanisms limiting conflict and favoring mutual dependence and cooperative interaction must be incorporated in the birth of the new organismal community [4].
The need to count: quantity control mechanisms
Division of labor entails an additional fundamental requirement in order for a cell community to be successful: the need to count. If indeed different cell types perform different essential and complementary tasks towards maintaining organismal fitness, it follows that there must be a more or less fixed balance in the quantitative output of different tasks and hence among different cell types performing those tasks [5]. The ability to control tissue mass is in fact an essential and defining facet of complexity in multicellular organisms, although the underlying biological and molecular mechanisms are still largely elusive [5,6]. Regeneration of mammalian liver constitutes one of the best examples to illustrate this concept [7]. Within minutes after partial surgical hepatectomy, a finely orchestrated series of events takes place in the host (including the residual liver), leading, under normal conditions, to complete recovery of the original organ mass within days or weeks, depending on the species; at which point the regenerative process subsides [7,8]. Furthermore, such organ mass control also operates in the reverse direction. Following hypertrophy and/or hyperplasia, the liver returns to its reference size through enforcement of regression mechanisms, including cell deletion by apoptosis [9,10]. These findings point to the existence of fine rheostats, each specific for a given tissue, that are continuously monitoring parameters likely related to functional proficiency, be it cell number, size or a combination thereof [11]. Very little is known about sensors and their location(s); what appears to be essential for the implementation of these mechanisms is an extensive connectedness both within and among tissue types, in order to support survival of the organism as a whole [6]. Conditions such as the age-associated benign prostatic hyperplasia (BPH) would appear to challenge the paradigm of tissue mass control. However, for a tissue that is exquisitely hormone responsive it is not surprising that its steady state mass may vary according to age-related fluctuations in estrogen and androgen signalling [12].
The need to assess fitness: quality-control mechanisms
Albeit still poorly understood, the ability to control tissue size in multicellular organisms has long been recognized and intensely investigated over the past several decades. By contrast, the idea that such multicellular communities might also be endowed with the capacity to assess qualitative parameters and, under given conditions, select for the fittest, is relatively novel. This idea is encompassed in the ever-expanding concept of cell competition [13]. Almost 50 years ago, Morata and Ripoll reported a phenomenon that is considered as one of the first documented accounts of cell competition [14]. While Drosophila homozygous mutants for ribosomal proteins (Minute) are lethal, heterozygous Minute flies develop normally into adults, albeit at slower rate compared to WT counterparts. However, if Minute heterozygous cells are induced in the context of a WT tissue background, the former are selectively deleted to the advantage of the latter phenotype [14]. It is important to emphasize that the paradigm indwelled in cell competition is not simply one of a growth advantages, such as proposed, for example, during tissue repopulation by transplanted cells 15, 16, 17. The salient feature is that 1 cell type (the winner) actually outstrips the other (the loser), leading to its clearance, and this occurs by means of a direct comparison of their relative fitness. As such, cell competition is now recognized as a basic biological process of quality control at cellular level that plays a fundamental role during development and in maintaining tissue integrity throughout adult life [13,18,19]. A notable example is the involvement of the Hippo pathway in the selection of high-quality pluripotent stem cells in mouse epiblast. Accumulation of YAP protein and activation of TEAD are in fact required for a strong expression of factors associated with pluripotency. Cells displaying low TEAD activity are eliminated through cell competition, thereby introducing a stringent quality control mechanism over survival of pluripotent stem cells [20]. Besides TEAD, additional transcription factors that have been repeatedly implicated in cell competition include MYC, p53, NF-κB, and STAT; however, their mechanistic role in the process is yet to be elucidated, if any [21]. On the other hand, the cell metabolic efficiency is likely to represent a basic parameter for the regulation of cell competition, as suggested by the Minute mutant phenotype referred to above. In fact, defects in protein synthesis have been associated to a loser phenotype also in mammalian systems [22]. Similarly, the winner phenotype displayed by Myc-overexpressing over wild type cells is dependent on the increased glucose uptake and increased rate of glycolysis in the former compared to the latter [23]. Furthermore, decreased mitochondrial function, including the inability to preserve membrane potential, defective ATP production and reduced mitophagy, lead to reduced fitness in a cell competition scenario [24].
The biological bases of cell competition
While apparently simple and straightforward, the concept of cell competition does indeed add a considerable layer of complexity to the organization and functioning of multicellular organisms [25]. A series of basic questions immediately arise and need to be addressed. First and paramount: what is cell fitness? Secondly, how do cells compare their fitness level? Furthermore, which cell types are involved in the comparison? Does cell competition in vivo operate within specific tissue boundaries (i.e. only among homotypic cells)? In an attempt to define the biological perimeter of this phenomenon, a series of “hallmarks” have been proposed to characterize cell competition [26]. They include: (1) a context-dependency effect, i.e. a fitness gradient must exist for the winner vs. loser cell phenotype to emerge; (2) a mechanistic coupling between proliferation of winner and clearance of loser cells; (3) maintenance of overall tissue size, i.e. cell competition occurs under homeostatic control mechanisms; (4) importantly, the process is enforced within defined developmental compartments [26,27]. A relevant aspect that emerges from the above is that cell competition is enacted at a high hierarchical level, beyond that of single cells, allowing tissues and/or organisms to implement quality control strategies in order to maintain functional proficiency of their constituent cells. Furthermore, if cell competition is based on assessment of relative fitness, then it can possibly operate only among homotypic cells, i.e., within the same tissue type, as it would make little biological sense to compare “fitness” and instigate competition between a glomerular and a tubular epithelial cell in the kidney, as an example [25,28]. The latter consideration highlights the central theme alluded to earlier and facing any research efforts aimed at unravelling the intimate nature of cell competition: what is meant by cell fitness? In an elegant review, Di Gregorio et al defined fitness as “the ability of a cell to thrive in a given environment, an ability determined by a number of parameters, including cell-cycle length, transcriptional output, signaling activity, and metabolic rate” [29]. This definition undoubtedly addresses main aspects of competition from the perspective of single cell confrontation. On the other hand, little or no reference is made (1) to any tissue-specific function that might enter the competition battleground or, most critically, (2) to a fundamental attribute that competing cells must express in a multicellular community, i.e. the ability to cooperate towards the common good [3,4,30]. As discussed in the preceding paragraphs, multicellular organisms, by definition, require a high degree of intercellular cooperation to maintain homeostasis. However, cellular traits selected during evolution to multicellularity may represent a fitness cost to the individual cell [31], implying that any cell that loses such traits would gain a selective advantage over more cooperative counterparts [31]. Thus, cooperative capacity must be in position to override, in a cell competition scenario, the emergence of phenotypic traits that are advantageous to the single cell but would be detrimental to organismal fitness [30] (Fig. 1). As an example, non-cooperative, polarity-deficient and oncogenic cells are eliminated from eye‐antennal imaginal epithelium of Drosophila by normal surrounding cells, thereby exerting a protective anti‐tumor effect via cell competition that favors cooperation 32, 33, 34.
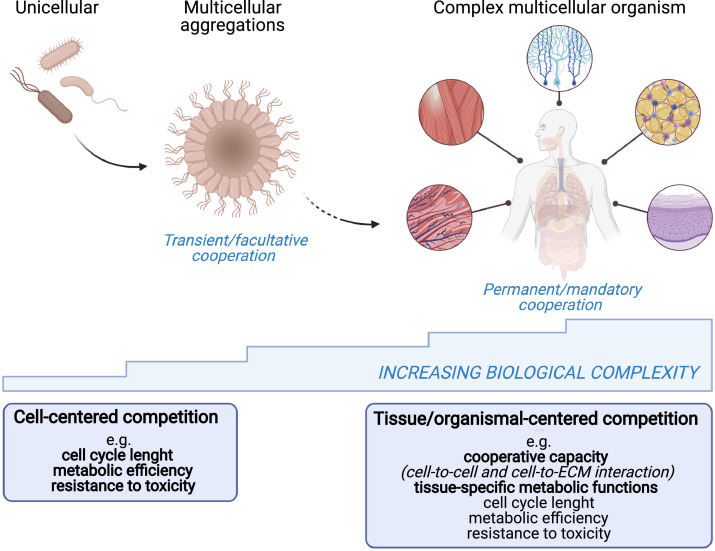
Fig. 1.
While the unit of selection in unicellular organisms is the single cell [97], multicellular organisms function as an interdependent community and cell competition must therefore be included in a higher-order scenario of cell cooperation. Cooperative capacity and tissue-specific metabolic functions become essential components of the fitness equation, although this occur at the expense of basic parameters such as cell cycle length or resistance to toxicity.
The emergence of cell clones in normal tissues
The advent of deep targeted and whole-genome sequencing has revealed the widespread, pervasive presence of clonal growths in normal adult human tissues, with increasing frequency as we age 35, 36, 37, 38. They were first reported in bone marrow-derived cells and referred to as clonal hematopoiesis of indeterminate potential (CHIP), with single clones contributing to a sizeable proportion of cells in peripheral blood [35,39, 40, 41]. Up to 20% of healthy individuals aged 60 to 69 years were found to harbor such clones, and this is probably an underestimate [42]; in 1 study, as many as 18% of blood cells were found to originate from a single clone [35]. It soon became clear that this finding is not unique to bone marrow-derived cells. Phenotypically normal skin also contains a patchwork of genetically altered clones [36], whose size and genotype relates to location [37]; similarly, human esophageal epithelium is progressively colonized by mutant clonal populations compatible with a normal tissue architecture [43,44] and clonal expansions of genetically altered cells were reported to occur in normal human endometrial epithelium [45] human urinary bladder [46,47], and colorectal epithelium [48,49]. Furthermore, mutations conferring increased clonal fitness upon phenotypically normal hepatocytes have been described during the evolution of human liver cirrhosis [50]. The last, most extreme and intriguing arrival to the club of clonally populated tissues is the placenta, which was found to be a mosaic of extensively mutated clones, yet performing its normal function [51].
There is now a general consensus that positive selection, as opposed to random drift, is the main biological force driving this phenomenon [52,53]. It is also clear that mutant clones expand within the normal tissue boundaries, implying that their growth is balanced by a corresponding loss of surrounding counterparts and is therefore well within homeostatic control [53] (Fig. 2). Accordingly, a mechanism of cell competition is likely to form the bases for the selective growth of such clones, although a formal demonstration of its occurrence has been difficult to obtain. Experimental studies in mice have shed light on the dynamics of clonal selection in a mutagenized tissue landscape [53,54]. Skin progenitor cells carrying a heterozygous p53 gain-of-function mutation display a competitive advantage over wild type counterparts, with a reduced tendency to differentiate during migration to the upper layers of the epidermis; however, the mutant phenotype is progressively attenuated and the skin remains phenotypically normal [54]. Similarly, the esophageal mucosa mice treated with a mutagen 1 year earlier was extensively populated by clones carrying alterations in Notch1, Notch2, Trp53, Cul3 and Arid1a genes, despite being >98% histologically normal [53]. In agreement with previous reports [55], it was found that cell division of Notch1-mutant progenitors is strongly biased towards persistence of the proliferative phenotype and against differentiation. In addition, expanding mutant cells induce differentiation of surrounding wild-type counterparts, leading to their progressive clearance from the tissue. Ultimately, notch1-defective clones reinstate their attitude to differentiate and a seemingly new homeostasis is achieved in a phenotypically normal esophageal mucosa [53]. Thus, selection of winner cell phenotypes emerges as a fundamental mechanism shaping the clonal landscape of tissues exposed to mutagenic events, either at genetic and/or epigenetic level.
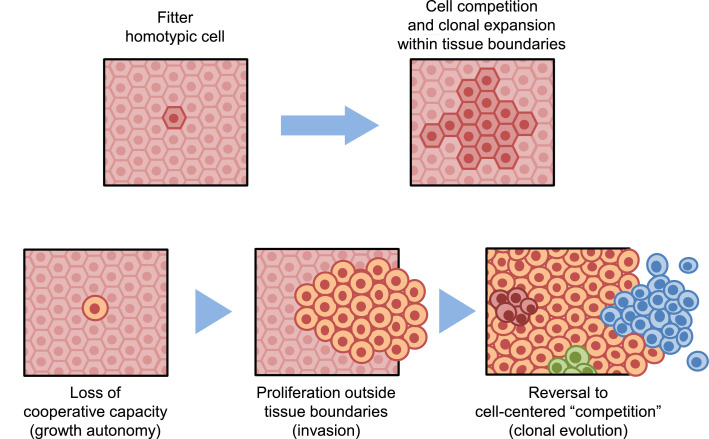
Fig. 2.
Upper panels: a single cell with a relatively higher fitness expands at the expense of neighboring homotypic cells (i.e., cells of the same tissue type). The higher fitness of the rare cell could result from a relative loss in surrounding counterparts, as it occurs in aging [70,71]. Clones maintain cooperative capacity and expand within tissue boundaries.
Lower panels: a single cell with a relatively higher fitness and loss of cooperative capacity expands forming a focal proliferative lesion with histological alterations. These cells display growth autonomy (uncontrolled growth) and revert back to cell-centered competition, fueling clonal evolution.
On the other hand, a similar biological paradigm appears to be at play during normal tissue development. When cells expressing high levels of Myc are induced in mouse epiblast, they expand selectively and cause apoptotic clearance of surrounding cells with lower Myc expression, in the absence of developmental defects [56,57]. Most critically, endogenous Myc levels were reported to be heterogeneous in normal mouse epiblast cells during development, leading to continuous competition towards selection of cells with highest Myc expression [56].
Are we at the peak of fitness? Why do “mutant” clones win over surrounding cells?
The latter considerations point to a core issue in the field of cell competition and, more specifically, to the central question regarding the biological significance of clonal proliferations in aged tissues. If indeed such clones are positively selected because of a higher fitness compared to surrounding tissue counterparts, 2 possibilities can be envisioned: (1) winner cells have improved their fitness above normal level, or (2) surrounding (loser) cells have incurred a decrease in their level of fitness. Although the prevalent view is that clonal expansions result from a gain-of-function alteration in the genotype of constituent cells, recent evidence highlights the complex dynamics of competition-based cell turnover in normal tissues and the essential role played by local environmental constraints [58].
Both evolutionary and developmental processes converge towards achieving near-maximum fitness in tissue composition in multicellular organisms, as exemplified by the above-mentioned selection of highest Myc expression levels in mouse epiblast [56,59]. This implies that any deviation from a normal phenotype is likely to result in a relative loss of fitness in the affected cell, followed by its removal by the surrounding neighbors [60,61]. However, such interpretation is apparently at odds with the common finding of mutant cell clones in normal aged tissues referred to above. If indeed Notch1-defective progenitors are able to outcompete their normal counterparts in aged esophageal mucosa [43,44], it is legitimate to ask why this does not occur earlier during development and, most strikingly, why it did not occur during evolution. The fact that Notch1 mutants are of so common occurrence during a single lifetime of any individual [43,44] rules out de facto the possibility that such genotype has not been available for (positive) evolutionary selection. Thus, it would appear that genotypes (such as Notch1 mutants) that are positively selected in phenotypically normal tissues of aged individuals or mutagenized animals [53] are not inherently fitter than their wild type counterparts; rather, their competitive advantage emerges under specific tissue environments bearing permissive alterations. Which highlights a seemingly trivial and therefore often overlooked additional attribute of cell competition: the winner and loser phenotypes are not related to absolute features of the confronting cells, but are heavily dependent on external environmental cues. As a notable example, p53 mutant cells outcompete normal epithelial cells in the esophagus of mice exposed to oxidative stress induced by low dose ionizing radiation (LDIR); however, such competitive advantage is countered when LDIR is coupled with antioxidant treatment [62]. Similarly, high fat diet-induced inflammation provides a competitive advantage to intestinal RasV12-mutant cells, which is attenuated following treatment with aspirin [63]. Moreover, caerulein-induced chronic inflammation was able to suppress competition-dependent apical elimination of RasV12 cells from mouse pancreatic epithelium [64]. These results indicate that a pro-inflammatory microenvironment can affect the outcome of cell competition to favor accumulation of altered cells over those with a normal phenotype. Along the same lines, intriguing findings were reported in a recent study analyzing the clonal landscape of normal human bronchial mucosa of tobacco smokers. While smoking imposes a high mutational burden on the majority of bronchial epithelial cells, a subpopulation of mitotically quiescent cells remains relatively free of mutagenic events and is able to replenish large segments of bronchial mucosa upon smoking cessation [65]. It is suggested that the expansion of such normal cell population, with a near-normal genotype, which remained quiescent during smoking, relates to its better fitness under conditions of smoking cessation. Conversely, mutant cells such as those with aberrant NOTCH or TP53 signaling, which are positively selected by smoking, were no longer favored after smoke exposure had stopped [65]. The fundamental message emerging from this type of studies is that selection of specific genotypes, including those with pre-neoplastic potential, is context-dependent; moreover, re-normalizing an altered tissue landscape might represent a viable strategy to counter the expansion of mutant clones, including those on the path to neoplastic disease 66, 67, 68. It is noteworthy that caloric restriction, which is known to delay aging and age-associated chronic diseases, including cancer, was reported to increase competitiveness of healthy stem cells and decrease retention of mutant cells in mouse intestine, thereby improving overall tissue fitness 66, 67, 68, 69.
Several years ago, we described the increased clonogenic potential of the liver microenvironment in aged rats compared to that of the young animal [70] and more recently we reported a cell-autonomous decrease of in vivo proliferative potential in hepatocytes isolated from old vs. young donor rats [71]. Since proliferative fitness is an important functional attribute of the normal hepatocyte (albeit, by no means, the only one!), its age-related decline can contribute to the emergence of clones that have gained a relative competitive advantage. Interestingly, clonal expansions were reported during the evolution of human chronic liver disease, and clone size was directly related to the extent of tissue damage [50]. Furthermore, cell clones harbored recurrent mutations in critical genes (including PKD1, KMT2D, and ARID1A) that were related to an increase of hepatocyte proliferative fitness in vivo [50]. In another study, loss of arid1a function was shown to accelerate liver regeneration after injury, while its overexpression impaired the healing process [72]. Once again, it remains an open question why arid1a loss of function was not selected for during development or evolution, given its apparent positive effect on hepatocyte proliferative fitness. A similar argument applies to super-competitor phenotypes that have been described in previous classical reports, including cells over-expressing dMyc, STAT or Yorki in the Drosophila wing disc 73, 74, 75, 76: these mutants are all able to proliferate faster and display a winner phenotype vis a vis their wild type counterparts. Therefore, as with previous examples, it is intriguing to consider why such super-competitors were not successfully selected during evolutionary phases and/or during normal developmental processes.
Cooperation and the fitness equation
Functional analysis of clonal dynamics during cell turnover in health and disease would require a comprehensive understanding of biological properties included and evaluated in the “fitness phenotype” in any specific tissue. While the full details of the fitness equation have yet to be resolved, important hints begin to emerge from the studies referred to above. Along this line, it is noteworthy that both esophageal progenitor Notch1 mutants [53,55] and hepatocyte Arid1a mutants [72] express a phenotype that is relatively refractory towards differentiation and/or functional maturation. As noted above, Notch1-defective progenitors are more likely to remain within the proliferative compartment rather than differentiate and migrate to the upper layers of the esophageal epithelium [55]; on the other hand, Arid1a-mutant hepatocytes display globally attenuated, lineage-specific transcriptional activities, as exemplified by a generalized decrease in the expression of cytochrome p450 enzymes [72]. Thus, although these mutants display a winner phenotype when confronted with neighboring wild type cells within aged, mutagenized or diseased tissue environments, their overall fitness does not stand out as highest in a normal developmental context, possibly as a consequence of defective differentiation/maturation potential referred to above. The latter processes are indeed central to the assembling and maintenance of tissues in multicellular organisms, as part of the mutual dependence and cooperative interactions characterizing such complex communities [4]. Therefore, it would be expected that such properties, ultimately related to the ability to contribute to normal tissue pattern formation, are meticulously screened and positively selected by mechanisms overlooking the dynamics of cell competition [77]. As already mentioned, in Drosophila epithelium, cells with mutations in cell polarity genes, such as scribble (scrib) or discs large (dlg), which are disruptive of tissue architecture and potentially tumorigenic, are eliminated from the tissue when surrounded by wild-type cells [78], through a mechanism that involves cell competition via secretion of fibroblast growth factor 21 by loser cells [34]. As another example, anterior-posterior tissue patterning in zebrafish is tightly regulated via cell competition-mediated elimination of unfit cells upsetting the correct establishment of morphogen gradients [79].
This type of evidence indicates that the ability to communicate, integrate and cooperate in a complex system represents an integral part of the equation that defines the operational boundaries of cell competition during normal tissue development and homeostasis in multicellular organisms [30]. Such consideration bears obvious relevance to the role of this process in the context of neoplastic disease. If indeed cell competition is a facet of quality control mechanisms aimed at achieving and preserving the fittest cell phenotypes in any tissue, it comes as a corollary that it can act as a powerful barrier against the emergence and overgrowth of altered/dysfunctional cells, including (pre)-neoplastic ones [77,80]. A paradigmatic example of such mechanism is possibly represented by what has been referred to as EDAC (epithelial defense against cancer): in the words of the proponents, it is a cell competition-based “intrinsic anti-tumor activity within the epithelial cell society to reduce the risk of oncogenesis” [81]. Hyperinsulinemia was reported to enhance epithelial carcinogenesis via inhibition of cell competition [82]. Furthermore, given the constitutive role of cell cooperation in the birth of multicellular organisms, such process will tend to select against any cell autonomous phenotype, including growth autonomy of neoplastic cells. Although the presence of super-competitor phenotypes, such as conferred by myc overexpression, may suggest otherwise [83], it is important to point out that myc-driven growth of winner cells is still under the constraints of tissue homeostatic mechanisms [56,73,76]. Similarly, expansion of Notch1- or Arid1a-mutant clones, which are able to out-compete normal surrounding counterparts, occurs at the expense of neighboring cells and does not disrupt overall tissue architecture [43,44,72]. This strongly speaks against the existence of a direct link between the winner phenotype associated with cell competition per se and uncontrolled neoplastic proliferation [54,84]. On the other hand, recent studies describing the behavior of initiated cells in genetic mouse models of intestinal carcinogenesis are in apparent contrast with the above conclusion 85, 86, 87. It was reported that crypt stem cells expressing mutant Kras, Pik3ca or Apc are able to release mediators that increase the rate of differentiation of wild type stem cells, leading to their gradual and complete deletion. Parallel expansion of mutant clones results in adenoma formation. Importantly, inhibition of such a paracrine effect prevents both the loss of wild type stem cells and the competitive outgrowth of mutant cells 85, 86, 87. The latter finding is particularly significant: it indicates that, at least at the initial stages, the selective expansion of Kras, Pik3ca or Apc mutants is not cell- autonomous, but it depends on the clearance of neighboring wild type stem cells, i.e., it still occurs within the boundaries of tissue homeostatic control mechanisms. In this respect, it is similar to the behavior of Notch1-mutant progenitors in the esophageal mucosa referred to above: they expand selectively by inducing differentiation of surrounding wild-type counterparts [53]. However, while Notch1-defective clones eventually differentiate forming a seemingly normal esophageal mucosa [53], Kras, Pik3ca and Apc mutants form discrete focal lesions (adenomas), with altered tissue architecture, that can further progress to overt neoplasia. Thus, growth pattern, rather than growth per se, emerges as a critical differential attribute of early lesions with pre-neoplastic potential, thereby dissociating the super-competitive phenotype per se from the risk of neoplastic disease [88].
On the other hand, the molecular machinery mediating cell competition might be hijacked during invasive and metastatic cancer growth, as suggested by some studies [83,89, 90, 91, 92]. For example, cell competition in the context of cancer cell heterogeneity shapes the dynamics of clonal evolution during tumor progression [92,93] (Fig. 2), a principle that informs the newly proposed paradigm of adaptive therapy for the treatment of cancer [94]. Heterotypic cell competition between neoplastic cells and surrounding stromal cells has also been described as a mechanism supporting invasion: elevated levels of c-Myc and YAP in cancer cells was positively correlated with caspase-3 positive apoptotic cells in the adjacent stroma [95].
Concluding remarks
Humans have roughly 300 different cell types by histological criteria [96]. Both quantitative and qualitative assessments are required on each cell type to ensure their coordinated and cooperative behavior towards functional proficiency of the organism as a whole. Cell competition represents one of the operating arms of such quality control mechanisms overseeing fitness level in individual cells. Central to any analysis on its possible role in any process, including neoplastic development, is therefore a comprehensive and workable definition of cell fitness, the cumulative phenotype encompassing salient functional properties specific for each cell type. We have argued that such fitness equation, i.e., the basis for cell competition, must include the ability of the cell to communicate and integrate in a large community, the society of cells constituting multicellular organisms. Furthermore, cell competition operates within defined tissue boundaries dictated by precise developmental programs. Thus, under normal conditions, it represents an effective barrier against the emergence of altered cell phenotypes disruptive of tissue integrity and/or tissue pattern formation. Moreover, cell competition does it entail per se any degree of growth autonomy, in that the growth of winner cells is always counterbalanced by clearance of loser neighbors. Based on these considerations, cell competition does not stand as a biological driving force fueling the emergence of the neoplastic phenotype, although it is possible that the molecular machinery involved in the winner/loser interaction could be hijacked by expanding cancer cell populations.
Author contributions
FM and SC contributed to the writing of the manuscript; EL conceived and finalized the content of the manuscript
Footnotes
Funding: This work was funded in part by Fondazione di Sardegna (FM and EL).
Conflicts of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Grosberg R.K., Strathmann R.R. The evolution of multicellularity: a minor major transition? Ann Rev Ecol Evol Syst. 2007;38:621–654. doi: 10.1146/annurev.ecolsys.36.102403.114735. [DOI] [Google Scholar]
- 2.Libby E., Conlin P.L., Kerr B., Ratcliff W.C. Stabilizing multicellularity through ratcheting. Philos Trans R Soc Lond B Biol Sci. 2016;371 doi: 10.1098/rstb.2015.0444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.West S.A., Fisher R.M., Gardner A., Kiers E.T. Major evolutionary transitions in individuality. PNAS. 2015;112:10112–10119. doi: 10.1073/pnas.1421402112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Queller D.C., Strassmann J.E. Beyond society: the evolution of organismality. Philos Trans R Soc Lond B Biol Sci. 2009;364:3143–3155. doi: 10.1098/rstb.2009.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu F.-X., Zhao B., Guan K.-L. hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hariharan I.K. Organ size control: lessons from drosophila. Dev Cell. 2015;34:255–265. doi: 10.1016/j.devcel.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michalopoulos G.K., Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol. 2021;18:40–55. doi: 10.1038/s41575-020-0342-4. [DOI] [PubMed] [Google Scholar]
- 8.Michalopoulos G.K. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384–1392. doi: 10.1002/hep.28988. [DOI] [PubMed] [Google Scholar]
- 9.Columbano A., Ledda-Columbano G.M., Coni P.P., Faa G., Liguori C., Santa Cruz G., Pani P. Occurrence of cell death (apoptosis) during the involution of liver hyperplasia. Lab Invest. 1985;52:670–675. [PubMed] [Google Scholar]
- 10.Grasl-Kraupp B., Rossmanith W., Ruttkay-Nedecky B., Müllauer L., Kammerer B., Bursch W., Schulte-Hermann R. Levels of transforming growth factor beta and transforming growth factor beta receptors in rat liver during growth, regression by apoptosis and neoplasia. Hepatology. 1998;28:717–726. doi: 10.1002/hep.510280318. [DOI] [PubMed] [Google Scholar]
- 11.Romaker D., Kumar V., Cerqueira D.M., Cox R.M., Wessely O. MicroRNAs are critical regulators of tuberous sclerosis complex and mTORC1 activity in the size control of the Xenopus kidney. PNAS. 2014;111:6335–6340. doi: 10.1073/pnas.1320577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho C.K.M., Habib F.K. Estrogen and androgen signaling in the pathogenesis of BPH. Nat Rev Urol. 2011;8:29–41. doi: 10.1038/nrurol.2010.207. [DOI] [PubMed] [Google Scholar]
- 13.Kim W., Jain R. Picking winners and losers: cell competition in tissue development and homeostasis. Trends Genet. 2020;36:490–498. doi: 10.1016/j.tig.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morata G., Ripoll P. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol. 1975;42:211–221. doi: 10.1016/0012-1606(75)90330-9. [DOI] [PubMed] [Google Scholar]
- 15.Genovese P., Schiroli G., Escobar G., Tomaso T.D., Firrito C., Calabria A., Moi D., Mazzieri R., Bonini C., Holmes M.C. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra M.P., Marongiu F., Sini M., Laconi E. Hepatocyte senescence in vivo following preconditioning for liver repopulation. Hepatology. 2012;56:760–768. doi: 10.1002/hep.25698. [DOI] [PubMed] [Google Scholar]
- 17.Chandler R.J., Venturoni L.E., Liao J., Hubbard B.T., Schneller J.L., Hoffmann V., Gordo S., Zang S., Ko C.-W., Chau N. Promoterless, nuclease-free genome editing confers a growth advantage for corrected hepatocytes in mice with methylmalonic acidemia. Hepatology. 2020 doi: 10.1002/hep.31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker N.E. Emerging mechanisms of cell competition. Nat Rev Gene. 2020;21:683–697. doi: 10.1038/s41576-020-0262-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto M., Sasaki H. Cell competition controls differentiation in mouse embryos and stem cells. Curr Opin Cell Biol. 2020;67:1–8. doi: 10.1016/j.ceb.2020.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Hashimoto M., Sasaki H. Epiblast formation by TEAD-YAP-dependent expression of pluripotency factors and competitive elimination of unspecified cells. Developmental Cell. 2019;50:139–154. doi: 10.1016/j.devcel.2019.05.024. e5. [DOI] [PubMed] [Google Scholar]
- 21.Lawlor K., Pérez-Montero S., Lima A., Rodríguez T.A. Transcriptional versus metabolic control of cell fitness during cell competition. Semin Cancer Biol. 2020;63:36–43. doi: 10.1016/j.semcancer.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver E.R., Saunders T.L., Tarlé S.A., Glaser T. Ribosomal protein L24 defect in Belly spot and tail (Bst), a mouse Minute. Development. 2004;131:3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de la Cova C., Senoo-Matsuda N., Ziosi M., Wu D.C., Bellosta P., Quinzii C.M., Johnston L.A. Supercompetitor status of drosophila myc cells requires p53 as a fitness sensor to reprogram metabolism and promote viability. Cell Metab. 2014;19:470–483. doi: 10.1016/j.cmet.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yadav A.K., Srikrishna S. scribble (scrib) knockdown induces tumorigenesis by modulating Drp1-Parkin mediated mitochondrial dynamics in the wing imaginal tissues of Drosophila. Mitochondrion. 2019;44:103–110. doi: 10.1016/j.mito.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Marongiu F., Laconi E. Cell competition in liver carcinogenesis. World J Hepatol. 2020;12:475–484. doi: 10.4254/wjh.v12.i8.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston L.A. Socializing with MYC: cell competition in development and as a model for premalignant cancer. Cold Spring Harb Perspect Med. 2014;4 doi: 10.1101/cshperspect.a014274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leevers S.J., McNeill H. Controlling the size of organs and organisms. Curr Opin Cell Biol. 2005;17:604–609. doi: 10.1016/j.ceb.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Walderich B., Singh A.P., Mahalwar P., Nüsslein-Volhard C. Homotypic cell competition regulates proliferation and tiling of zebrafish pigment cells during colour pattern formation. Nat Commun. 2016;7:11462. doi: 10.1038/ncomms11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Gregorio A., Bowling S., Rodriguez T.A. Cell Competition and its role in the regulation of cell fitness from development to cancer. Dev Cell. 2016;38:621–634. doi: 10.1016/j.devcel.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Nelson P., Masel J. Intercellular competition and the inevitability of multicellular aging. Proc Natl Acad Sci U S A. 2017;114:12982–12987. doi: 10.1073/pnas.1618854114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baillon L., Basler K. Reflections on cell competition. Semin Cell Dev Biol. 2014;32:137–144. doi: 10.1016/j.semcdb.2014.04.034. [DOI] [PubMed] [Google Scholar]
- 32.Igaki T., Pastor-Pareja J.C., Aonuma H., Miura M., Xu T. Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF signaling in drosophila. Dev Cell. 2009;16:458–465. doi: 10.1016/j.devcel.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohsawa S. Elimination of oncogenic cells that regulate epithelial homeostasis in drosophila. Dev Growth Differ. 2019;61:337–342. doi: 10.1111/dgd.12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa M., Kawarazaki Y., Fujita Y., Naguro I., Ichijo H. FGF21 induced by the ASK1-p38 pathway promotes mechanical cell competition by attracting cells. Curr Biol. 2021;31:1048–1057. doi: 10.1016/j.cub.2020.11.052. e5. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martincorena I., Roshan A., Gerstung M., Ellis P., Van Loo P., McLaren S., Wedge D.C., Fullam A., Alexandrov L.B., Tubio J.M. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fowler J.C., King C., Bryant C., Hall M.W.J., Sood R., Ong S.H., Earp E., Fernandez-Antoran D., Koeppel J., Dentro S.C. Selection of oncogenic mutant clones in normal human skin varies with body site. Cancer Discov. 2021;11:340–361. doi: 10.1158/2159-8290.CD-20-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kakiuchi N., Ogawa S. Clonal expansion in non-cancer tissues. Nat Rev Cancer. 2021;21:239–256. doi: 10.1038/s41568-021-00335-3. [DOI] [PubMed] [Google Scholar]
- 39.Steensma D.P., Bejar R., Jaiswal S., Lindsley R.C., Sekeres M.A., Hasserjian R.P., Ebert B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fey M.F., Liechti-Gallati S., von Rohr A., Borisch B., Theilkäs L., Schneider V., Oestreicher M., Nagel S., Ziemiecki A., Tobler A. Clonality and X-inactivation patterns in hematopoietic cell populations detected by the highly informative M27 beta DNA probe. Blood. 1994;83:931–938. [PubMed] [Google Scholar]
- 41.Busque L., Patel J.P., Figueroa M., Vasanthakumar A., Provost S., Hamilou Z., Mollica L., Li J., Viale A., Heguy A. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Acuna-Hidalgo R., Sengul H., Steehouwer M., van de Vorst M., Vermeulen S.H., Kiemeney L.A.L.M., Veltman J.A., Gilissen C., Hoischen A. Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am J Hum Genet. 2017;101:50–64. doi: 10.1016/j.ajhg.2017.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martincorena I., Fowler J.C., Wabik A., Lawson A.R.J., Abascal F., Hall M.W.J., Cagan A., Murai K., Mahbubani K., Stratton M.R. Somatic mutant clones colonize the human esophagus with age. Science. 2018;362:911–917. doi: 10.1126/science.aau3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yokoyama A., Kakiuchi N., Yoshizato T., Nannya Y., Suzuki H., Takeuchi Y., Shiozawa Y., Sato Y., Aoki K., Kim S.K. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565:312–317. doi: 10.1038/s41586-018-0811-x. [DOI] [PubMed] [Google Scholar]
- 45.Suda K., Nakaoka H., Yoshihara K., Ishiguro T., Tamura R., Mori Y., Yamawaki K., Adachi S., Takahashi T., Kase H. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 2018;24:1777–1789. doi: 10.1016/j.celrep.2018.07.037. [DOI] [PubMed] [Google Scholar]
- 46.Li R., Du Y., Chen Z., Xu D., Lin T., Jin S., Wang G., Liu Z., Lu M., Chen X. Macroscopic somatic clonal expansion in morphologically normal human urothelium. Science. 2020;370:82. doi: 10.1126/science.aba7300. [DOI] [PubMed] [Google Scholar]
- 47.Lawson A.R.J., Abascal F., Coorens T.H.H., Hooks Y., O'Neill L., Latimer C., Raine K., Sanders M.A., Warren A.Y., Mahbubani K.T.A. Extensive heterogeneity in somatic mutation and selection in the human bladder. Science. 2020;370:75. doi: 10.1126/science.aba8347. [DOI] [PubMed] [Google Scholar]
- 48.Nicholson A.M., Olpe C., Hoyle A., Thorsen A.-S., Rus T., Colombé M., Brunton-Sim R., Kemp R., Marks K., Quirke P. Fixation and spread of somatic mutations in adult human colonic epithelium. Cell Stem Cell. 2018;22:909–918. doi: 10.1016/j.stem.2018.04.020. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee-Six H., Olafsson S., Ellis P., Osborne R.J., Sanders M.A., Moore L., Georgakopoulos N., Torrente F., Noorani A., Goddard M. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574:532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 50.Zhu M., Lu T., Jia Y., Luo X., Gopal P., Li L., Odewole M., Renteria V., Singal A.G., Jang Y. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell. 2019;177:608–621. doi: 10.1016/j.cell.2019.03.026. e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coorens T.H.H., Oliver T.R.W., Sanghvi R., Sovio U., Cook E., Vento-Tormo R., Haniffa M., Young M.D., Rahbari R., Sebire N. Inherent mosaicism and extensive mutation of human placentas. Nature. 2021;592:80–85. doi: 10.1038/s41586-021-03345-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall M.W.J., Jones P.H., Hall B.A. Relating evolutionary selection and mutant clonal dynamics in normal epithelia. J R Soc Interface. 2019;16 doi: 10.1098/rsif.2019.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colom B., Alcolea M.P., Piedrafita G., Hall M.W.J., Wabik A., Dentro S.C., Fowler J.C., Herms A., King C., Ong S.H. Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat Gene. 2020;52:604–614. doi: 10.1038/s41588-020-0624-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murai K., Skrupskelyte G., Piedrafita G., Hall M., Kostiou V., Ong S.H., Nagy T., Cagan A., Goulding D., Klein A.M. Epidermal tissue adapts to restrain progenitors carrying clonal p53 mutations. Cell Stem Cell. 2018;23:687–699. doi: 10.1016/j.stem.2018.08.017. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alcolea M.P., Greulich P., Wabik A., Frede J., Simons B.D., Jones P.H. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat Cell Biol. 2014;16:615–622. doi: 10.1038/ncb2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Clavería C., Giovinazzo G., Sierra R., Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500:39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- 57.Sancho M., Di-Gregorio A., George N., Pozzi S., Sánchez J.M., Pernaute B., Rodríguez T.A. Competitive interactions eliminate unfit embryonic stem cells at the onset of differentiation. Dev Cell. 2013;26:19–30. doi: 10.1016/j.devcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Higa K.C., DeGregori J. Decoy fitness peaks, tumor suppression, and aging. Aging Cell. 2019;18:e12938. doi: 10.1111/acel.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreno E., Rhiner C. Darwin's multicellularity: from neurotrophic theories and cell competition to fitness fingerprints. Curr Opin Cell Biol. 2014;31:16–22. doi: 10.1016/j.ceb.2014.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanimura N., Fujita Y. Epithelial defense against cancer (EDAC) Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 61.Kon S., Ishibashi K., Katoh H., Kitamoto S., Shirai T., Tanaka S., Kajita M., Ishikawa S., Yamauchi H., Yako Y. Cell competition with normal epithelial cells promotes apical extrusion of transformed cells through metabolic changes. Nat Cell Biol. 2017;19:530–541. doi: 10.1038/ncb3509. [DOI] [PubMed] [Google Scholar]
- 62.Fernandez-Antoran D., Piedrafita G., Murai K., Ong S.H., Herms A., Frezza C., Jones P.H. Outcompeting p53-mutant cells in the normal esophagus by redox manipulation. Cell Stem Cell. 2019;25:329–341. doi: 10.1016/j.stem.2019.06.011. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sasaki A., Nagatake T., Egami R., Gu G., Takigawa I., Ikeda W., Nakatani T., Kunisawa J., Fujita Y. Obesity suppresses cell-competition-mediated apical elimination of RasV12-transformed cells from epithelial tissues. Cell Rep. 2018;23:974–982. doi: 10.1016/j.celrep.2018.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sato N., Yako Y., Maruyama T., Ishikawa S., Kuromiya K., Tokuoka S.M., Kita Y., Fujita Y. The COX-2/PGE2 pathway suppresses apical elimination of RasV12-transformed cells from epithelia. Commun Biol. 2020;3:132. doi: 10.1038/s42003-020-0847-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yoshida K., Gowers K.H.C., Lee-Six H., Chandrasekharan D.P., Coorens T., Maughan E.F., Beal K., Menzies A., Millar F.R., Anderson E. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature. 2020;578:266–272. doi: 10.1038/s41586-020-1961-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cadoni E., Marongiu F., Fanti M., Serra M., Laconi E. Caloric restriction delays early phases of carcinogenesis via effects on the tissue microenvironment. Oncotarget. 2017;8:36020–36032. doi: 10.18632/oncotarget.16421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marongiu F., Serra M.P., Sini M., Angius F., Laconi E. Clearance of senescent hepatocytes in a neoplastic-prone microenvironment delays the emergence of hepatocellular carcinoma. Aging (Albany NY) 2014;6:26–34. doi: 10.18632/aging.100631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Serra M., Marongiu F., Pisu M.G., Serra M., Laconi E. Time-restricted feeding delays the emergence of the age-associated, neoplastic-prone tissue landscape. Aging (Albany NY) 2019;11:3851–3863. doi: 10.18632/aging.102021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bruens L., Ellenbroek S.I.J., Suijkerbuijk S.J.E., Azkanaz M., Hale A.J., Toonen P., Flanagan D.J., Sansom O.J., Snippert H.J., van Rheenen J. Calorie restriction increases the number of competing stem cells and decreases mutation retention in the intestine. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pasciu D., Montisci S., Greco M., Doratiotto S., Pitzalis S., Pani P., Laconi S., Laconi E. Aging is associated with increased clonogenic potential in rat liver in vivo. Aging Cell. 2006;5:373–377. doi: 10.1111/j.1474-9726.2006.00230.x. [DOI] [PubMed] [Google Scholar]
- 71.Serra M.P., Marongiu F., Marongiu M., Contini A., Laconi E. Cell-autonomous decrease in proliferative competitiveness of the aged hepatocyte. J Hepatol. 2015;62:1341–1348. doi: 10.1016/j.jhep.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 72.Sun X., Chuang J.-C., Kanchwala M., Wu L., Celen C., Li L., Liang H., Zhang S., Maples T., Nguyen L.H. Suppression of the SWI/SNF component Arid1a promotes mammalian regeneration. Cell Stem Cell. 2016;18:456–466. doi: 10.1016/j.stem.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moreno E., Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 74.de la Cova C., Abril M., Bellosta P., Gallant P., Johnston L.A. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 75.Rodrigues A.B., Zoranovic T., Ayala-Camargo A., Grewal S., Reyes-Robles T., Krasny M., Wu D.C., Johnston L.A., Bach E.A. Activated STAT regulates growth and induces competitive interactions independently of Myc, Yorkie, Wingless and ribosome biogenesis. Development. 2012;139:4051–4061. doi: 10.1242/dev.076760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ziosi M., Baena-López L.A., Grifoni D., Froldi F., Pession A., Garoia F., Trotta V., Bellosta P., Cavicchi S., Pession A. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Morata G. Cell competition: a historical perspective. Dev Biol. 2021;476:33–40. doi: 10.1016/j.ydbio.2021.02.012. [DOI] [PubMed] [Google Scholar]
- 78.Kanda H., Igaki T. Mechanism of tumor-suppressive cell competition in flies. Cancer Sci. 2020;111:3409–3415. doi: 10.1111/cas.14575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akieda Y., Ogamino S., Furuie H., Ishitani S., Akiyoshi R., Nogami J., Masuda T., Shimizu N., Ohkawa Y., Ishitani T. Cell competition corrects noisy Wnt morphogen gradients to achieve robust patterning in the zebrafish embryo. Nat Commun. 2019;10:4710. doi: 10.1038/s41467-019-12609-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji Z., Chuen J., Kiparaki M., Baker N. Cell competition removes segmental aneuploid cells from Drosophila imaginal disc-derived tissues based on ribosomal protein gene dose. Elife. 2021;10 doi: 10.7554/eLife.61172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kon S., Fujita Y. Cell competition-induced apical elimination of transformed cells, EDAC, orchestrates the cellular homeostasis. Dev Biol. 2021;476:112–116. doi: 10.1016/j.ydbio.2021.03.015. [DOI] [PubMed] [Google Scholar]
- 82.Sanaki Y., Nagata R., Kizawa D., Léopold P., Igaki T. Hyperinsulinemia drives epithelial tumorigenesis by abrogating cell competition. Dev Cell. 2020;53:379–389. doi: 10.1016/j.devcel.2020.04.008. e5. [DOI] [PubMed] [Google Scholar]
- 83.Rhiner C., Moreno E. Super competition as a possible mechanism to pioneer precancerous fields. Carcinogenesis. 2009;30:723–728. doi: 10.1093/carcin/bgp003. [DOI] [PubMed] [Google Scholar]
- 84.Aktipis C.A., Boddy A.M., Gatenby R.A., Brown J.S., Maley C.C. Life history trade-offs in cancer evolution. Nat Rev Cancer. 2013;13:883–892. doi: 10.1038/nrc3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yum M.K., Han S., Fink J., Wu S.-H.S., Dabrowska C., Trendafilova T., Mustata R., Chatzeli L., Azzarelli R., Pshenichnaya I. Tracing oncogene-driven remodelling of the intestinal stem cell niche. Nature. 2021;594:442–447. doi: 10.1038/s41586-021-03605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Neerven S.M., de Groot N.E., Nijman L.E., Scicluna B.P., van Driel M.S., Lecca M.C., Warmerdam D.O., Kakkar V., Moreno L.F., Vieira Braga F.A. Apc-mutant cells act as supercompetitors in intestinal tumour initiation. Nature. 2021;594:436–441. doi: 10.1038/s41586-021-03558-4. [DOI] [PubMed] [Google Scholar]
- 87.Flanagan D.J., Pentinmikko N., Luopajärvi K., Willis N.J., Gilroy K., Raven A.P., Mcgarry L., Englund J.I., Webb A.T., Scharaw S. NOTUM from Apc-mutant cells biases clonal competition to initiate cancer. Nature. 2021;594:430–435. doi: 10.1038/s41586-021-03525-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Altered growth pattern, not altered growth per se, is the hallmark of early lesions preceding cancer development. Histol Histopathol. 2008:101–106. doi: 10.14670/HH-24.101. [DOI] [PubMed] [Google Scholar]
- 89.Fujita Y. Flower power as human cancer cells compete with normal cells. Nature. 2019;572:181–182. doi: 10.1038/d41586-019-02161-y. [DOI] [PubMed] [Google Scholar]
- 90.Madan E., Pelham C.J., Nagane M., Parker T.M., Canas-Marques R., Fazio K., Shaik K., Yuan Y., Henriques V., Galzerano A. Flower isoforms promote competitive growth in cancer. Nature. 2019;572:260–264. doi: 10.1038/s41586-019-1429-3. [DOI] [PubMed] [Google Scholar]
- 91.Madan E., Peixoto M.L., Dimitrion P., Eubank T.D., Yekelchyk M., Talukdar S., Fisher P.B., Mi Q.-S., Moreno E., Gogna R. Cell competition boosts clonal evolution and hypoxic selection in cancer. Trends Cell Biol. 2020;30:967–978. doi: 10.1016/j.tcb.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 92.Parker T., Madan E., Gupta K., Moreno E., Gogna R. Cell competition spurs selection of aggressive cancer cells. Trends Cancer. 2020;6:732–736. doi: 10.1016/j.trecan.2020.03.008. [DOI] [PubMed] [Google Scholar]
- 93.Parker T.M., Henriques V., Beltran A., Nakshatri H., Gogna R. Cell competition and tumor heterogeneity. Semin Cancer Biol. 2020;63:1–10. doi: 10.1016/j.semcancer.2019.09.003. [DOI] [PubMed] [Google Scholar]
- 94.Strobl M.A.R., West J., Viossat Y., Damaghi M., Robertson-Tessi M., Brown J.S., Gatenby R.A., Maini P.K., Anderson A.R.A. Turnover modulates the need for a cost of resistance in adaptive therapy. Cancer Res. 2021;81:1135–1147. doi: 10.1158/0008-5472.CAN-20-0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Di Giacomo S., Sollazzo M., de Biase D., Ragazzi M., Bellosta P., Pession A., Grifoni D. Human cancer cells signal their competitive fitness through MYC activity. Sci Rep. 2017;7:12568. doi: 10.1038/s41598-017-13002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bornholdt S., Kauffman S. Ensembles, dynamics, and cell types: Revisiting the statistical mechanics perspective on cellular regulation. J Theor Biol. 2019;467:15–22. doi: 10.1016/j.jtbi.2019.01.036. [DOI] [PubMed] [Google Scholar]
- 97.Ra G., S A., Ky T., Js B. Integrating genetic and nongenetic drivers of somatic evolution during carcinogenesis: the biplane model. Evol Appl. 2020;13 doi: 10.1111/eva.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]