Abstract
Malaria is a highly prevalent parasitic disease in regions with tropical and subtropical climates worldwide. Among the species of Plasmodium causing human malaria, P. vivax is the second most prevalent and the most geographically widespread species. A major target of a pre-erythrocytic vaccine is the P. vivax circumsporozoite protein (PvCSP). In previous studies, we fused two recombinant proteins representing three allelic variants of PvCSP (VK210, VK247 and P. vivax-like) to the mumps virus nucleocapsid protein to enhance immune responses against PvCSP. The objective of the present study was to evaluate the protective efficacy of these recombinants in mice challenged with transgenic P. berghei parasites expressing PvCSP allelic variants. Formulations containing Poly (I:C) or Montanide ISA720 as adjuvants elicited high and long-lasting IgG antibody titers specific to each PvCSP allelic variant. Immunized mice were challenged with two existing chimeric P. berghei parasite lines expressing PvCSP-VK210 and PvCSP-VK247. We also developed a novel chimeric line expressing the third allelic variant, PvCSP-P. vivax-like, as a new murine immunization-challenge model. Our formulations conferred partial protection (significant delay in the time to reach 1% parasitemia) against challenge with the three chimeric parasites. Our results provide insights into the development of a vaccine targeting multiple strains of P. vivax.
Subject terms: Protein vaccines, Parasitology
Introduction
Human malaria is caused by five different etiological agents, all belonging to the phylum Apicomplexa and the genus Plasmodium: P. falciparum, P. vivax, P. ovale, P. malariae and P. knowlesi. The latter is a non-human primate (NHP) parasite that causes infections in humans, including severe malaria1. The most prevalent species in the world are P. falciparum and P. vivax, both accounting for 95% of human malaria infections. Despite efforts to eliminate malaria from the world, the resistance of the main etiological agents to antimalarial drugs has increased considerably in recent years2, which has allowed malaria to spread in new areas and re-emerge in places where the disease has previously been considered eradicated3,4.
P. falciparum malaria is often considered the main target, as it causes the highest number of deaths among cases of infection. However, P. vivax malaria also causes severe symptoms and occasionally death5, and it is endemic in different regions of South and Central America, some parts of Africa and much of Asia. In 2019, 75% of malaria cases in the Americas were attributed to P. vivax, which has the highest geographical distribution and highest prevalence in the Americas among the etiological agents6. Research on P. vivax has long been neglected, resulting in limited knowledge of its biology, pathogenesis, and epidemiology compared to P. falciparum. Therefore, P. vivax is a relevant challenge to overcome for the success of malaria eradication programs7.
The main target of pre-erythrocytic malaria vaccines is the circumsporozoite protein (CSP), which covers the sporozoite surface. P. vivax CSP (PvCSP) has two widely recognized variants, VK210 and VK247, which differ mainly in the sequence of repetitive amino acids in their central region. Each variant displays repeating nonapeptides, of which the more prevalent are GDRA(D/A)GQPA and ANGA(G/A) (C/D)QPG, respectively8,9. Nonetheless, several different peptide repeat motifs were described in the central region of both variants, and this genetic polymorphism could have impact on the efficacy of CSP-based vaccines10. A third variant from a parasite that causes P. vivax malaria in humans, called Plasmodium vivax-like, expresses CSP with APGANQ(E/G)GAA repeats (hereafter named PvCSP-P. vivax-like) and was described in endemic regions of Papua New Guinea, Brazil, Indonesia and Madagascar11. Since the P. vivax-like parasite is among the Plasmodium species that infects NHP12,13 and the PvCSP-P. vivax-like sequence is identical to P. simiovale CSP14, human infections with P. vivax-like parasites are commonly reported as cases of zoonoses15,16. However, when analyzing the genotype of parasites causing infections characterized microscopically as P. vivax in humans, a significant proportion of the parasites were the P. vivax-like variant, both in single infections and mixed with other PvCSP allelic variants17–19. Moreover, the prevalence of this P. vivax-like variant is greater in regions of low incidence18, suggesting that if it is neglected, this variant might become an important reservoir of the disease. Other Plasmodium specie that commonly infects NHP and causes zoonotic P. vivax malaria in humans is P. simium, which shares high genetic identity with P. vivax20 and their two main CSP variants (VK210 and VK247) are identical21. Thus, a universal vaccine against all types of P. vivax malaria should include the VK210 and VK247 P. vivax variants and also the P. vivax-like variant.
Previously, we reported the generation of a recombinant protein fusing the repeat domains of the three PvCSP variants (VK210, VK247 and P. vivax-like) in tandem, which contain immunodominant epitopes for B cells, and the conserved C-terminal region of P. vivax CSP (PvCSP-AllCT)22. Additionally, we generated two chimeric recombinant proteins containing the sequence of PvCSP-AllCT fused to the mumps virus nucleocapsid protein to form nucleocapsid-like particles (NLPs) as a strategy to elicit strong and protective immune responses23. These recombinant proteins, NLP-CSPCT and NLP-CSPR (with and without the PvCSP C-terminal region, respectively), were successfully produced in yeast Pichia pastoris and were highly immunogenic in mice when administered with Poly (I:C) adjuvant. Moreover, the immunization of mice with NLP-CSPCT/Poly (I:C) conferred partial protection against intradermal challenge23 with chimeric P. berghei parasites expressing the repetitive region of PvCSP (VK210 allelic variant)24. Although these results were encouraging, the protective efficacy that these recombinant proteins potentially confer against the two other PvCSP allelic variants, VK247 and P. vivax-like, remains to be elucidated.
To investigate whether the two recombinant PvCSP proteins, NLP-CSPR and NLP-CSPCT, can induce protective immune responses when combined with suitable adjuvants, we analyzed protective efficacy by immunizing mice followed by challenge with different chimeric P. berghei parasites expressing the three different PvCSP variants. We have used two of these chimeric parasites in our established immunization-challenge model because they express full-length P. vivax CSP of VK210 and VK247 variants (Pb-PvCSP210, Pb-PvCSP247) on the sporozoite surface25. In this study, we generated a novel chimeric P. berghei parasite line that expresses the PvCSP-P. vivax-like protein in sporozoites (Pb-PvCSP-like G10). In addition, the protective efficacy of these recombinant proteins was analyzed in the presence of Poly (I:C) or Montanide ISA720 as adjuvants.
Materials and methods
Animals and ethics statements
Female inbred C57BL/6 (H-2b) mice were used to assess immunogenicity and protection after challenge. Tuck-ordinary (TO) outbred mice were used for parasite production and transmission. Mice were purchased from Harlan (UK). Female OF1 mice (6–7 weeks; Charles River, NL) were used to generate chimeric P. berghei lines. Immunogenicity and protection studies were performed in accordance with the recommendations of the UK Home Office Animals Act Project License. Procedures were approved by the University of Oxford Animal Care and Ethical Review Committee (PPL P9804B4F1).
Longevity assays were performed in accordance with the terms of the Guide for the Care and Use of Laboratory Animals of the Brazilian National Council of Animal Experimentation (http://www.cobea.org.br/). The protocol (CEUA/FCF no. 74.2016-P53) was approved by the Research Committee on Animal Experimentation of the School of Pharmaceutical Sciences of the University of São Paulo, Brazil.
Experiments for the generation of the chimeric P. berghei lines were granted with a license by the Competent Authority after advice on the ethical evaluation by the Animal Experiments Committee Leiden (AVD1160020171625). All experiments were performed in accordance with the Experiments on Animals Act, the applicable legislation in the Netherlands in accordance with the European guidelines (EU directive no. 2010/63/EU) regarding the protection of animals used for scientific purposes. The experiments were executed in a licensed establishment for the use of experimental animals (LUMC). Mice were housed in individually ventilated cages furnished with autoclaved aspen woodchip, fun tunnel, wood chew block and Nestlets at 21 ± 2 °C under a 12:12 h light–dark cycle with a relative humidity of 55 ± 10%.
This study was carried out in compliance with the ARRIVE (Animal Research: Reporting of In Vivo Experiments) guidelines for animals.
Parasites
The following P. berghei ANKA reference parasite lines were used: (1) 1596cl1 (230p-GIMOPbANKA; RMgm-687, www.pberghei.eu), which contains a positive–negative selectable marker (SM) (human dihydrofolate reductase:: yeast cytosine deaminase and uridyl phosphoribosyl transferase (hdhfr::yfcu)) cassette integrated into the silent 230p gene locus (PBANKA_030600)26; (2) the wild-type (WT) reference line cl15cy1 of P. berghei ANKA27 and the reporter PbANKA parasite line PbGFP-Luccon (676m1cl1). The PbGFP-Luccon parasite expresses a GFP (mutant3) and firefly luciferase (LUC-IAV) fusion protein from the constitutive eef1a promoter and is selectable marker (SM)-free27. The reporter cassette is integrated into the neutral 230p locus (PBANKA_030600). For details of PbGFP-Luccon, see RMgmDB entry #29 (http://www.pberghei.eu/index.php?rmgm=29).
In addition, we used two existing chimeric P. berghei lines in which the P. berghei csp gene was replaced with either the P. vivax csp VK210 allele or the P. vivax VK247 allele (Pb-PvCSP210, https://www.pberghei.eu/index.php?rmgm=4136; Pb-PvCSP247, https://www.pberghei.eu/index.php?rmgm=4137)25.
Generation and genotyping of chimeric P. berghei lines expressing the PvCSP-P. vivax-like protein
To generate the chimeric P. berghei replacement line, we replaced the P. berghei csp coding sequence (CDS; Pbcsp; PBANKA_0403200) with the PvCSP-P. vivax-like CDS (Locus PVU09738, Accession U09738) using a 2-step GIMO transfection protocol25,26,28. In the first step, we deleted the P. berghei csp CDS and replaced it with a positive–negative selectable marker to create a P. berghei csp deletion GIMO line (PbANKA-CSP GIMO). The construct (pL1929) used and the generation of the PbANKA-CSP GIMO line (line 2251cl1) have been described previously29. This construct contains the positive–negative (hdhfr::yfcu) SM cassette and was used to insert both the Pbcsp 5′ and 3′ gene targeting regions (TRs), encompassing the full-length promoter and transcription terminator sequences, respectively, and was transfected into PbGFP-Luccon parasites (676m1cl1) using standard transfection methods30. Transfected parasites were selected in mice by applying positive selection by providing pyrimethamine in the drinking water30. Transfected parasites were cloned using the limiting dilution method31, resulting in the PbANKA-CSP GIMO line (line 2251 cl1). In the second step, we replaced the positive–negative SM in the PbANKA-CSP GIMO genome with the PvCSP-P. vivax-like CDS by GIMO transfection to create the P. berghei chimeric Pb-PvG10 replacement line Pb-PvG10(r). This line was obtained by modifying the construct used in the first step (pL1929); specifically, the hdfhr::yfcu SM cassette was removed and replaced with the PvCSP-P. vivax-like CDS, generating plasmid pL2161. The PvCSP-P. vivax-like CDS was ordered from GeneArt Gene Synthesis—Thermo Fisher Scientific. The pL2161 construct was sequenced to ensure that no mutations were present in the PvCSP-P. vivax-like CDS during the cloning process. The construct was linearized using AflII and SacI restriction enzymes outside of the 5’ and 3’ TRs before transfection. The construct was used to transfect parasites of the PbANKA-CSP GIMO line (line 2251 cl1)26 using standard methods of GIMO transfection to generate a single replacement gene chimeric parasite25,32–34. Transfected parasites were selected in mice by applying negative selection by providing 5-fluorocytosine (5-FC) in the drinking water35. Negative selection results in the selection of chimeric parasites where the hdhfr::yfcu SM in the csp locus on chromosome 4 of the PbANKA-CSP GIMO line is replaced with the PvCSP-P. vivax-like CDS. Selected chimeric parasites were cloned using the limiting dilution method31. Correct integration of the constructs into the genome of chimeric parasites was analyzed by performing a gDNA Southern analysis of pulsed field gel (PFG)-separated chromosomes, as previously described36. This method creates chimeric ‘gene replacement’ P. berghei parasites that lack the Pbcsp CDS but express the PvCSP-P. vivax-like protein (Pb-PvG10(r); line 2710 cl1) under the control of the P. berghei csp regulatory sequences.
The PvCSP-P. vivax-like Gabon Clone G10 CDS (Locus PVU09738, Accession U09738) gene was introduced into the genome as an additional copy of the gene in the neutral 230p locus using the previously described ‘gene insertion/marker out’ (GIMO) technology26,28,37 and the standard GIMO DNA construct pL0043 to generate the chimeric P. berghei additional copy line. This construct contains 5’ and 3’ targeting sequences for the 230p locus, as well as a multiple-cloning site for the integration of transgene expression cassettes. This construct integrates transgenes by double crossover homologous recombination and replaces the positive–negative SM (human dihydrofolate reductase:: yeast cytosine deaminase and uridyl phosphoribosyl transferase (hdhfr::yfcu)) cassette with the transgene expression cassette. The expression cassette contained the PvCSP-P. vivax-like CDS flanked by the 5’ and 3’ promoter and transcription terminator sequences of the P. berghei uis4 gene (PBANKA_0501200), which were amplified from P. berghei ANKA WT genomic DNA36. The coding sequence of the P. vivax CSP-like G10 gene (Locus PVU09738, Accession U09738) was ordered from GeneArt Gene Synthesis—Thermo Fisher Scientific. In addition, a reporter cassette containing GFP::luciferase30 driven by the constitutive P. berghei elongation factor 1 alpha (ef1α) promoter was also cloned into the transgene construct to generate the gene insertion construct pL2163 (PvCSP-Like G10@Pbuis4 + GFP::Luc@Pbeef1a_230p) targeting the neutral 230p locus on chromosome 3. The coding sequence and promoter region of the construct were confirmed by sequencing.
The pL2163 construct was linearized by SacII restriction digestion and introduced into parasites of the GIMO motherline 1596cl1 using standard methods of GIMO transfection26. Transfected parasites were selected in mice through the addition of 5-fluorocytosine (5-FC) to the drinking water35, resulting in negative selection of parasites in which the SM in the 230p locus was replaced by the PvCSP-P. vivax-like expression/reporter cassette. The selected chimeric parasites were cloned using the limiting dilution method31. Primer sequences are listed in Table S1. Correct integration of the PvCSP-P. vivax-like coding sequence (under control of the Pbuis4 promoter) into the genome of clones of the chimeric line (Pb-PvCSP-like G10, 2700 cl1) was analyzed by performing a diagnostic PCR analysis of gDNA and Southern analysis of pulsed field gel (PFG)-separated chromosomes36.
Phenotype and fitness assessment of the chimeric P. berghei lines expressing PvCSP-P. vivax-like protein
Multiplication of blood stages in mice was determined during the cloning period as previously described,38. Feeding of Anopheles stephensi mosquitoes, determination of oocyst production, and sporozoite (spz) collection were performed as described elsewhere36,38. The infectivity of chimeric spz was assessed by determining the T1% period (i.e., the time to reach 1% parasitemia) after an intravenous injection of 1000 spz in the tail vein of inbred BALB/c mice (Harlan, UK).
The expression of the PvCSP-P. vivax-like protein in spz was analyzed by performing an immunofluorescence assay (IFA) using sera from mice immunized with the recombinant proteins (diluted 1:100). As a control, the 3D11 antibody39 recognizing P. berghei CSP was used (diluted 1:1000). Purified spz were fixed with 4% paraformaldehyde in PBS for 20 min on ice, washed three times with PBS and blocked with 20 µl of 10% FCS + 1% BSA in PBS for 30 min at room temperature. Excess blocking medium was removed, followed by the addition of 20–25 µL of primary monoclonal antibody in PBS containing 10% FCS + 1% BSA (blocking medium) for 1–2 h at room temperature or overnight at 4 °C. After the incubation, the primary antibody was removed, and the slides were washed three times with PBS, followed by staining with the secondary antibody (Alexa Fluor-488 goat anti-mouse IgG from Life Technologies, Cat# A-11001) diluted 1:800 in PBS containing 10% FCS + 1% BSA (blocking medium) for 1 h at room temperature. After three washes with PBS, nuclei were stained with 2% Hoechst-33342 (Cell Signaling Technology #4082S) in PBS for 10 min at room temperature, washed twice with PBS and air-dried, followed by the addition of fluorescence mounting medium (Dako, code S3023). Cover slips were mounted onto the slides, and the slides were sealed with nail polish and allowed to dry overnight in the dark as described in a previous study37. The spz were analyzed using a DMI-300B Leica fluorescence microscope in both blue and green channels, and images were processed using ImageJ software.
Purified recombinant proteins obtained after expression in P. pastoris
Expression and purification of recombinant CSP proteins from P. pastoris was carried out as described previously23. Briefly, yeast clones containing the previously selected plasmids of interest were cultured for 24 h at 30 °C under constant stirring (230 rpm) in 40–200 mL of 3% glycerol-containing medium (BMGY). Cells were then harvested by centrifugation, solubilized in 40–200 mL of medium containing 1.0% methanol (BMMY) and cultured at 28 °C with constant stirring (230 rpm). Induction was maintained by the daily addition of 1.0% methanol. After 72–96 h of incubation, the cells were removed by centrifugation, and the supernatants were filtered through 0.45 µm membranes (Millipore). Recombinant proteins were then purified by affinity and ion exchange chromatography using a HisTrap FF column and Q-Sepharose resin, respectively, both coupled to the ÄKTAprime system (GE Healthcare). Fractions containing the highly pure recombinant proteins were collected and dialyzed against PBS.
Mouse immunization protocol
Groups of six female C57BL/6 mice aged 6–8 weeks were subcutaneously (s.c.) immunized thrice with the corresponding formulation of recombinant protein/adjuvant. For each dose, a final volume of 100 µl (10 µg of protein/sterile PBS/adjuvant) was injected at the base of the tail of each mouse. The adjuvants used were Poly (I:C) HMW (InvivoGen, 50 μg per dose per mouse in ratio 50/50 protein/adjuvant) and Montanide ISA720 (Seppic, emulsion in ratio 70/30 protein/adjuvant). The formulations were prepared just before administration.
Antibody measurements
Twelve days after each immunization, blood was collected from the tail vein, and sera were analyzed for the presence of antibodies recognizing each recombinant protein. Antibodies were detected by enzyme-linked immunosorbent assay (ELISA), essentially as described in a previous study22. The recombinant proteins NLP-CSPCT, NLP-CSPR23, yPvCSP-VK210CT, yPvCSP-VK247CT, and yPvCSP-P. vivax-like CT22 were employed as solid phase-bound antigens (200 ng/well). After an overnight incubation at RT, plates were washed with a solution of PBS containing 0.05% Tween-20 (PBS-T) and blocked with a blocking solution (PBS, 5% (w/v) skimmed milk) for 2 h at 37 °C. Serial dilutions of murine polyclonal sera were added to the wells and incubated for 1 h at RT; after washes with PBS-T, peroxidase-labeled goat anti-mouse IgG (Sigma, St. Louis, USA), diluted 1:3000, was added to each well. Reactions were developed with the OPD/acid stop system. Anti-IgG titers were determined based on the highest dilution of sera yielding an A492 greater than 0.1.
Challenge of mice with chimeric sporozoites
Spz of the chimeric parasite lines Pb-PvCSP-210 (2196cl1), Pb-PvCSP247 (2199cl1)25 and Pb-PvCSP-like G10 (2700cl1) were used to challenge immunized mice. Female A. stephensi mosquitoes were used to produce chimeric spz. After 21 days of incubation in a humidified incubator at 19–21 °C on a 12-h day-night cycle and feeding on a fructose-p-aminobenzoic acid (PABA) solution, the mosquitoes were dissected, salivary glands were isolated, and spz were extracted. The total number of spz was determined using a hemocytometer, and 2000 spz were intravenously (i.v.) injected in 100 µL 14 days after the second booster immunization.
Parasitemia analyses
Thin blood smears were prepared daily from day 4 to day 12 after challenge or until the day after mice reached 1% parasitemia. The smears were prepared on glass slides with a drop of blood obtained from mouse tail veins, fixed with methanol and stained for 15 min using 10% Giemsa. The glass slides were observed under a light microscope, and the percentages of parasitized red blood cells were determined. The time required to reach 1% parasitemia (T1%) is a variable calculated by a linear regression equation using the percentage of parasites detected in blood on the first three consecutive days with positive parasitemia. Protection analysis using T1% period as index is a useful tool to assess vaccine efficacy, as the comparative time to reach a determined level of parasitemia reflects the prepatent period and the number of parasites erupting from the liver40.
Statistical analyses
All analyses and graphics were performed/generated using GraphPad Prism version 8.0 (GraphPad Software Inc., La Jolla, CA, USA). IgG Ab titers were compared using one-way analysis of variance (ANOVA). One-way ANOVA was also used to compare normally distributed log-transformed means for the different animal groups. Multiple comparisons were assessed using Tukey’s posttest with a significance level of p < 0.05. Survival curves were compared using a log-rank Mantel-Cox test with a significance level of p < 0.05.
Results
PvCSP-specific antibody responses in mice immunized with the recombinant proteins NLP-CSPCT and NLP-CSPR
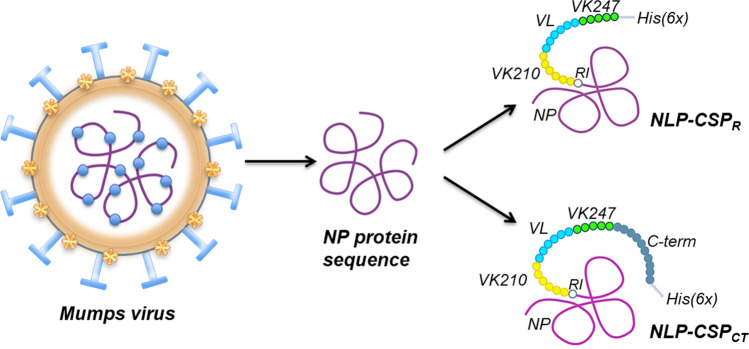
In previous studies, we generated two chimeric recombinant proteins, NLP-CSPCT and NLP-CSPR, fusing domains of the three PvCSP variants (VK210, VK247, and P. vivax-like) to the mumps virus nucleocapsid protein (NP)23. A schematic representation of the mumps virus, the NP protein and the new recombinants is depicted in Fig. 1. Briefly, the strategy used was fusing the malaria antigens to a core-viral protein rather than the surface proteins, thus avoiding the possible interference of immunological memory against mumps virus in the general population23.
Figure 1.
Schematic representation of mumps virus and NLP-CSP proteins. Mumps virus proteins are represented in the left panel. From outside to inside: Hemagglutinin-Neuraminidase protein (light blue) and Fusion protein (gold) in the membrane surface; Matrix protein (orange) and Nucleocapsid protein (purple) with associated RNAs (blue) in the inside. NLP-CSPs proteins are represented in the right panels. NP sequence is represented in purple, PvCSP-RI sequence in withe circle, PvCSP-VK210 repeats in yellow circles, PvCSP-P. vivax-like (VL) in cyan circles, PvCSP-VK247 in green circles, PvCSP-C-terminal sequence in dark grey and the His-tag sequence in light grey.
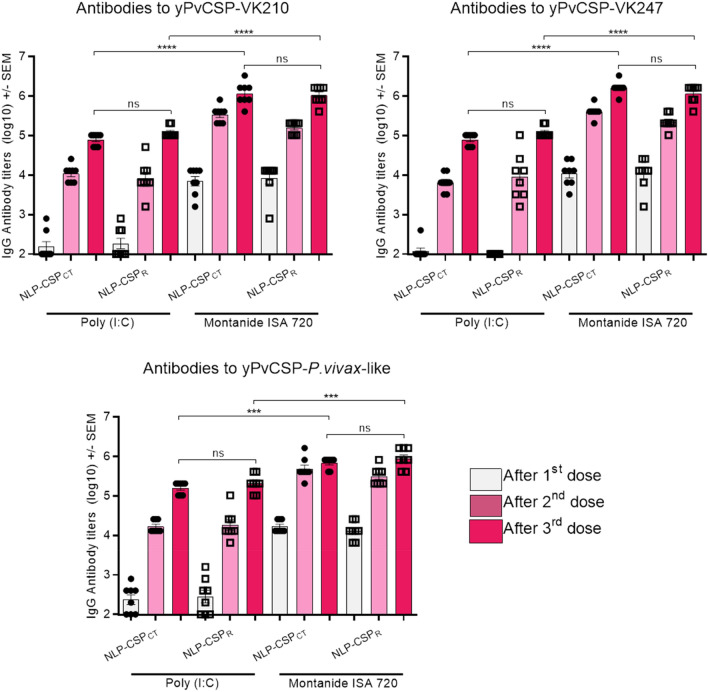
First, we compared the humoral immune response induced by immunizing mice with different vaccine formulations. Groups of six C57BL/6 female mice were immunized with 10 μg of the two recombinant proteins, NLP-CSPCT or NLP-CSPR, mixed with either Poly (I:C) (50 μg/dose) adjuvant or emulsified in Montanide ISA720 (7:3), an oil–water emulsion. Each animal received three immunizations 14 days apart in a homologous prime-boost vaccination regimen (Table 1). The antibody titers against each recombinant PvCSP variant (yPvCSP-VK210, yPvCSP-VK247, yPvCSP-P. vivax-like) were measured by ELISA twelve days after the administration of each dose. As shown in Fig. 2, no statistically significant difference was observed in the antibody titers elicited by the two recombinant proteins (NLP-CSPCT or NLP-CSPR) when combined with the same adjuvant (NLP-CSPCT or NLP-CSPR in the presence of Poly (I:C) for yPvCSP-VK210 p = 0.9795, yPvCSP-VK247 p = 0.9608 and yPvCSP-P. vivax-like p = 0.9994, and NLP-CSPCT or NLP-CSPR in the presence of Montanide ISA 720 for yPvCSP-VK210 p > 0.9999, yPvCSP-VK247 p = 0.9925 and yPvCSP-P. vivax-like p = 0.9994).
Table 1.
Groups of immunized C57BL/6 female mice.
| Group (n = 6 mice/group) | Prime (day 0) | Boost 2x (days 14 and 28) | Adjuvant |
|---|---|---|---|
| G1 | – | – | Poly (I:C) |
| G2 | NLP-CSPCT | NLP-CSPCT | Poly (I:C) |
| G3 | NLP-CSPR | NLP-CSPR | Poly (I:C) |
| G4 | – | – | Montanide ISA720 |
| G5 | NLP-CSPCT | NLP-CSPCT | Montanide ISA720 |
| G6 | NLP-CSPR | NLP-CSPR | Montanide ISA720 |
Figure 2.
Humoral immune response in mice immunized with recombinant proteins. C57BL/6 mice were s.c. immunized with the recombinant NLP-CSPCT and NLP-CSPR proteins in the presence of Poly (I:C) or Montanide ISA 720 adjuvants using the scheme shown in Table 1. IgG antibody titers were determined using ELISA assay at 12 days after the administration of each dose using the individual PvCS proteins (yPvCSP-VK210, yPvCSP-VK247 and yPvCSP-P. vivax-like) as solid-phase bound antigens.
In contrast, significant differences in IgG titers were observed when we compared the effects of the different adjuvants. With the Poly (I:C) adjuvant, the titers of IgG specific for all three variants were detectable (~ 104) in both immunized groups only after the administration of two doses. With the administration of the third dose, IgG titers were greater than 105 in all immunized mice. When the same analysis was performed for groups immunized with Montanide ISA 720, antibody titers reaching 104 levels were detected after the administration of a single dose. Moreover, after the administration of the second and third doses, even higher IgG titers (106) against all three PvCSP variants were detected (Fig. 2).
When antibody titers were compared after the administration of three doses using the same recombinant protein but with a different adjuvant, all groups of mice immunized with Montanide ISA 720 had significantly higher specific IgG titers than mice immunized with Poly (I:C) (NLP-CSPCT to yPvCSP-VK210 p > 0.0001, yPvCSP-VK247 p > 0.0001 and yPvCSP-P. vivax-like p = 0.0004, and NLP-CSPR to yPvCSP-VK210 p > 0.0001, yPvCSP-VK247 p > 0.0001 and yPvCSP-P. vivax-like p = 0.0004).
We also analyzed the longevity of the IgG antibodies. As shown in Suppl. Figure 1, PvCSP-specific IgG titers remained higher than 104 in the Poly (I:C) adjuvant-treated groups and on the order of 105 in the Montanide adjuvant-treated groups for at least 102 days after priming. These responses were antigen-specific, since mice immunized only with either adjuvant did not elicit detectable PvCSP-specific IgG antibodies at any time point analyzed (Suppl. Figure 1).
It is worth mentioning that any potential interference of yeast components in the specificity of the produced antibodies was discarded in previous studies, by using recombinant proteins produced in bacteria as solid phase-bound antigens22,23.
Preclinical immunization-challenge model: generation of a chimeric P. berghei line expressing P. vivax-like CSP in sporozoites (Pb-PvCSP-like G10)
In previous studies, we generated two chimeric P. berghei parasites (Pb-PvCSP210 and Pb-PvCSP247) that express the full-length VK210 and VK247 variants of P. vivax CSP on the spz surface25. These parasites have been used to analyze protective efficacy in mice immunized with different vaccine candidates targeting PvCSP with an immunization-challenge mouse model41–43. To analyze protective immune responses induced by the two recombinant proteins NLP-CSPCT or NLP-CSPR against not only the VK210 and VK247 repeats but also against the P. vivax-like variant, we generated a third chimeric P. berghei parasite that expressed the PvCSP-P. vivax-like protein in spz.
We first generated a chimeric P. berghei ANKA line in which the endogenous P. berghei csp gene was replaced with the PvCSP-P. vivax-like gene using the same GIMO transfection approach that was used to generate the chimeric Pb-PvCSP210 and Pb-PvCSP247 lines (Suppl. Figure 2 and Fig. 4a)25. Two independent clones of this Pb-PvG10(r) line (2710cl1, 2710cl2) produced normal numbers of oocysts. However, in contrast to the other two chimeric lines that express PvCSP-VK210 and PvCSP-VK247, the parasites expressing the PvCSP-P. vivax-like protein did not form visible spz inside oocysts, and only very few salivary gland spz were observed (Table S2). The cause of the lack of sporozoite formation is unclear, but the CSP of different Plasmodium species do not always fully complement the function of the CSP of other species. For example, chimeric P. falciparum replacement lines expressing P. vivax CSP (VK210 and VK247) also have a defect in the formation of spz44. An alternative strategy is the generation of rodent-infectious P. berghei and P. falciparum spz which can be engineered to express CSP proteins on the spz surface from two different Plasmodium species45–47. Based on this observation, we decided to generate chimeric P. berghei spz expressing both the PbCSP and the PvCSP-P. vivax-like protein. In this chimeric line (Pb-PvCSP-like G10, 2700 cl1), the PvCSP-P. vivax-like gene is introduced into the genome as an additional copy of the csp gene in the neutral 230p locus using GIMO transfection (Figs. 3, 4a,b). The PvCSP-P. vivax-like gene is flanked by the 5′ and 3′ promoter and transcription terminator sequences of the P. berghei uis4 gene, which is specifically expressed in spz and liver stages48. Pb-PvCSP-like G10 parasites showed normal asexual blood stage multiplication in mice (data not shown), and both oocyst and sporozoite production in A. stephensi mosquitoes was comparable to wild-type P. berghei parasites (Table S2). The infectivity of chimeric spz, as determined by the length of the T1% period after an intravenous injection of 1000 spz in BALB/c mice, was similar to the T1% period in mice infected with wild-type P. berghei spz (Table S2). These results demonstrate that the chimeric Pb-PvCSP-like G10 parasites produce fully infectious spz that are able to complete liver stage development in mice. The expression of the PvCSP-P. vivax-like protein in Pb-PvCSP-like G10 spz was determined by immunofluorescence analysis using sera from mice immunized with the recombinant proteins. The 3D11 antibody recognizing P. berghei CSP was used as a control. Pb-PvCSP-like G10 spz stained both with the antiserum and the 3D11 antibody (Suppl. Figure 3), demonstrating the expression of both P. berghei CSP and PvCSP-P. vivax-like protein in spz of the Pb-PvCSP-like G10 line.
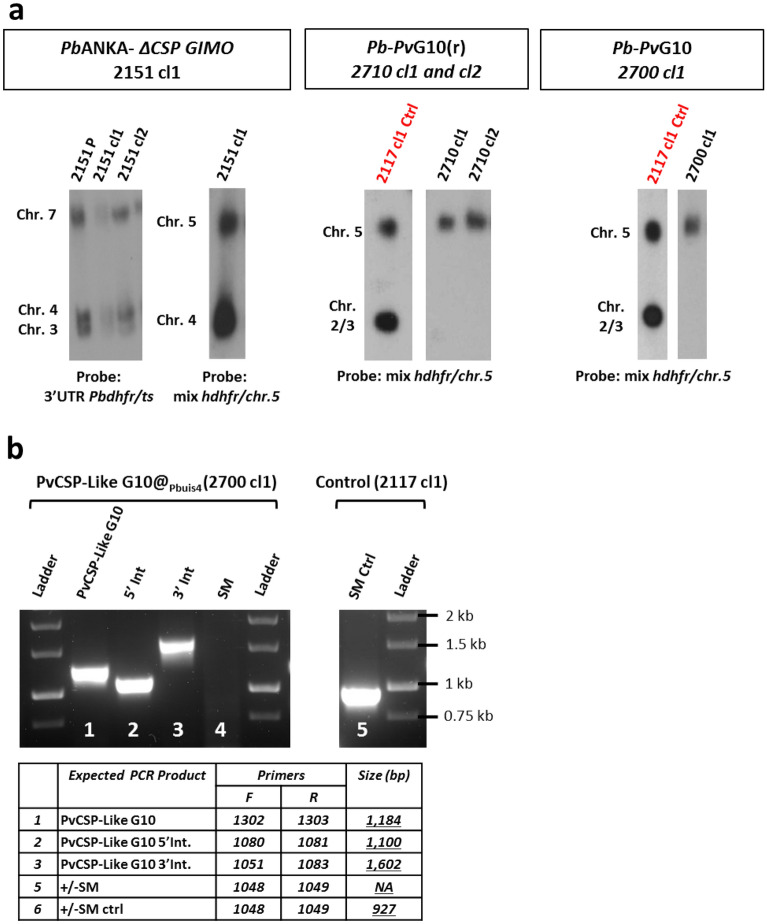
Figure 4.
Genotyping analyses of chimeric P. berghei parasite lines expressing the PvCSP-P. vivax-like protein. Cropped gels and blots are displayed in this figure. Full-length blots/gels are presented in Supplementary Fig. 4. (a) Genotyping analysis of the replacement line Pb-PvG10(r) (2710cl1 and cl2) and its intermediate GIMO mother-line (2151cl1) using a Southern analysis of chromosomes (chr.) separated by pulsed-field gel electrophoresis (PFGE) and diagnostic PCR analysis. Left panel: Hybridization of chr. from line 2151cl1 with the 3′UTR Pbdhfr/ts confirms integration of construct pL1929 into the Pbcsp gene on chr. 4. In addition, this probe hybridizes to the GFP-Luc reporter cassette on chr. 3 and to the endogenous Pbdhfr/ts on chr. 7. The correct integration of the SM is also confirmed by using a mixture of two probes: one recognizing hdhfr and a control probe recognizing chr. 5. Middle panel: The correct integration of the PvCSP-P. vivax-like gene expression construct (pL2161) into the GIMO locus was confirmed by showing the removal of the hdhfr::yfcu selectable marker (SM) cassette in clones of the chimeric parasite line Pb-PvG10(r) (2710cl1 and 2710cl2). The Southern blot was hybridized with a mixture of two probes: one recognizing hdhfr and a control probe recognizing chr. 5. As an additional control (ctrl), parasite line 2117cl1 was used with the hdhfr::yfcu SM integrated into chr. 3. Right panel: Southern analysis of chr. of the ‘additional copy’ chimeric line Pb-PvG10(r) (2700cl1) confirms the correct integration of the expression PvCSP-Like G10@Pbuis4 construct (pL2163) into the GIMO locus (230p on chr. 3), shown as the removal of the hdhfr::yfcu SM cassette in cloned chimeric parasites compared to a control probe recognizing chr. 5. As an additional control (Ctrl), parasite line 2117cl1 is also shown, as it retains hdhfr::yfcu SM in the 230p locus on chr. 3. (b) Genotyping using a diagnostic PCR analysis of the chimeric Pb-PvCSP-like G10 line (2700cl1; left panel) confirms correct integration of the PvCSP-Like@Pbuis4 expression cassette. Correct integration is shown by the absence of the hdhfr::yfcu SM and the presence of the PvCSP-P. vivax-like CDS and the correct integration of the construct into the genome at both the 5′ and 3′ regions (5′int and 3′int; see Fig. 3 for primer locations). The primer sequences used in this study are shown in Table S1, while the expected PCR product sizes and the primer numbers are listed in the table below the PCR analysis. As an additional control (ctrl), parasite line 2117cl1 (right panel) was used to validate the primers used to amplify the hdhfr::yfcu SM that was integrated into chr. 3 but has been removed from 2700cl1.
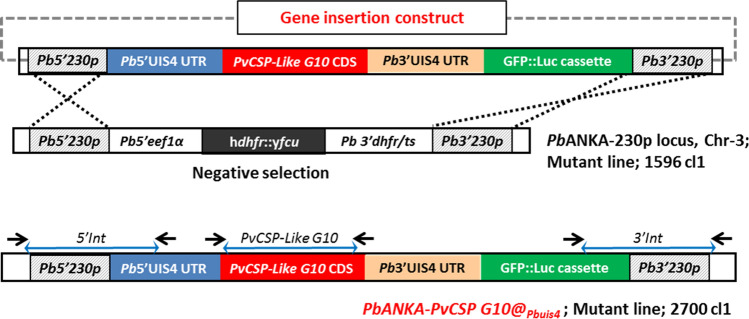
Figure 3.
Strategy to generate a chimeric P. berghei parasite line expressing a PvCSP-P. vivax-like protein as additional CSP. An additional copy line in which the PvCSP-P. vivax-like CDS (PVU09738) gene was introduced into the genome as an additional copy of the gene in the neutral 230p locus. The construct that contains the ‘PvCSP-P. vivax-like gene expression cassette’ was integrated into the 230p locus on chromosome 3 of the P. berghei ANKA GIMO mother line by GIMO transfection using negative selection (5-FC), resulting in the expression of the PvCSP-P. vivax-like gene under the control of the Pbuis4 gene promoter and transcriptional terminator sequences. This construct also expresses a GFP and firefly luciferase (LUC-IAV) fusion protein under control of the constitutive Pbeef1a promoter and is selectable marker (SM)-free. The construct is integrated into the neutral p230p locus by double crossover integration. Black arrows: location of PCR primers used for the diagnostic PCR analysis.
Protective efficacy in mice immunized with different vaccine candidates targeting PvCSP variants using the immunization-challenge mouse model
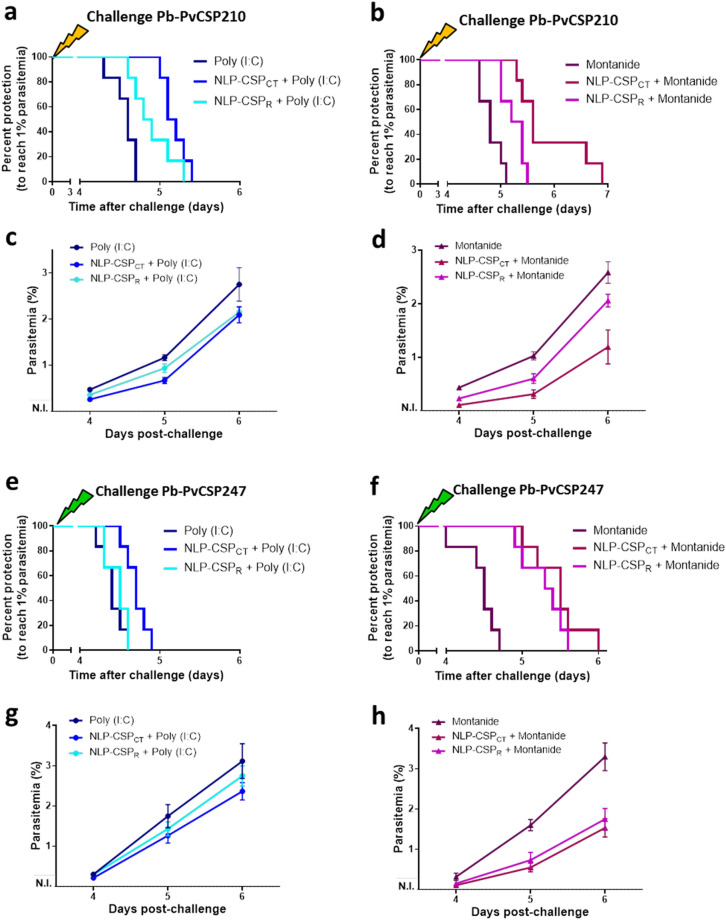
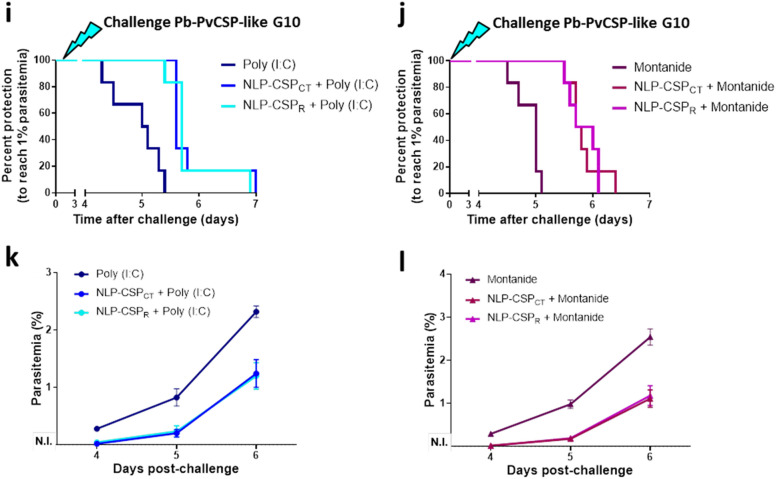
To analyze protective immune responses induced in the mice by immunization with the recombinant proteins NLP-CSPR and NLP-CSPCT, we challenged mice with spz of the three chimeric P. berghei lines, Pb-PvCSP210, Pb-PvCSP247 and Pb-PvCSP-like G10. The results are shown in Fig. 5; Pb-PvCSP210 challenge in the upper panel (Fig. 5a–d), Pb-PvCSP247 challenge in the middle panel (Fig. 5e–h) and Pb-PvCSP-like G10 challenge in the lower panel (Fig. 5i–l).
Figure 5.
T1% period in immunized mice after challenge with Pb-PvCSP transgenic sporozoites. Groups of six C57BL/6 mice were s.c. immunized with the vaccine formulations in the presence of Poly (I:C) or Montanide adjuvants, following the scheme shown in Table 1. Upper panel: Immunized mice were challenged 14 days after the third dose with 2,000 Pb-PvCSPVK210 sporozoites. Percent protection to reach 1% parasitemia (a, b) and follow-up analysis of parasitemia at days 4, 5 and 6 after challenge (c, d) is shown. Middle panel: Immunized mice were challenged 14 days after the third dose with 2,000 Pb-PvCSPVK247 sporozoites. Percent protection to reach 1% parasitemia (e, f) and follow-up analysis of parasitemia at days 4, 5 and 6 after challenge (g, h) is shown. Lower panel: Immunized mice were challenged 14 days after the third dose with 2000 spz of the new chimeric Pb-PvCSP-like G10 parasite. Percent protection to reach 1% parasitemia (i, j) and follow-up analysis of parasitemia at days 4, 5 and 6 after challenge (k, l) is shown. Significant differences in T1% periods (see Table 2) were analyzed by applying the log-rank (Mantel-Cox) test.
Groups of six C57BL/6 mice were immunized as described in Table 1 and challenged 2 weeks after the last immunization by administering an intravenous (i.v.) injection of 2000 spz Pb-PvCSP210. The protective efficacy was assessed by determining the T1% period after challenge (Fig. 5a, b) by following parasitemia (Fig. 5c, d); p values denoting significant differences in the T1% period between control (adjuvant) and immunized mice are depicted in Table 2. With either adjuvant, a statistically significant delay in the T1% was observed in mice immunized with NLP-CSPCT and NLP-CSPR compared to the adjuvant-only groups. However, a significant difference between the protective efficacy of mice immunized with NLP-CSPCT or NLP-CSPR was only observed in the Montanide adjuvant-treated groups (p = 0.0192, Table 2), showing higher protective efficacy of the NLP-CSPCT protein.
Table 2.
Prepatent (T1%) periods and statistical significance of protective efficacy.
| Challenge | Comparison | Poly (I:C) | Montanide ISA720 | ||||
|---|---|---|---|---|---|---|---|
| NLP-CSPCT vs. Adj | NLP-CSPR vs. Adj | NLP-CSPCT vs. NLP-CSPR | NLP-CSPCT vs. Adj | NLP-CSPR vs. Adj | NLP-CSPCT vs. NLP-CSPR | ||
| Pb-PvCSP210 | Median T1% | 5.15 vs. 4.6 | 4.85 vs. 4.6 | 5.15 vs. 4.85 | 5.6 vs. 4.8 | 5.3 vs. 4.8 | 5.6 vs. 5.3 |
| p values | p = 0.0008 | p = 0.0136 | n.s. p = 0.1026 | p = 0.0005 | p = 0.0068 | p = 0.0192 | |
| Pb-PvCSP247 | Median T1% | 4.7 vs. 4.4 | 4.5 vs. 4.4 | 4.7 vs. 4.5 | 5.5 vs. 4.5 | 5.35 vs. 4.5 | 5.5 vs. 5.35 |
| p values | p = 0.0049 | n.s. p = 3797 | p = 0.0136 | p = 0.0005 | p = 0.0005 | n.s. p = 0.2798 | |
| Pb-PvCSP-like G10 | Median T1% | 5.6 vs. 5.05 | 5.7 vs. 5.05 | 5.6 vs. 5.7 | 5.75 vs. 5 | 5.85 vs. 5 | 5.75 vs. 5.85 |
| p values | p = 0.0005 | p = 0.0012 | n.s. p = 0.9949 | p = 0.0007 | p = 0.0007 | n.s. p = 0.9910 | |
Significant differences (p < 0.05) were analyzed by applying the log-rank (Mantel-Cox) test. n = 6 mice/group. n.s.: not significant differences.
Other groups of C57BL/6 mice immunized as described in Table 1 were challenged 2 weeks after the last immunization by administering an i.v. injection of 2000 spz of Pb-PvCSP247. Percent protection to reach T1% and parasitemia are shown in Fig. 5 (e,f and g,h, respectively). Mice immunized with formulations containing the Montanide adjuvant presented a significant delay in the T1% period compared to the adjuvant-only groups (Fig. 5f and Table 2, p = 0.0005). Additionally, a significant delay in the T1% period (p = 0.0049) was observed in mice immunized with NLP-CSPCT protein formulated with the Poly (I:C) adjuvant (Fig. 5e and Table 2).
Finally, a third set of C57BL/6 mice immunized as described in Table 1 were challenged 2 weeks after the last immunization by administering an i.v. injection of 2,000 spz Pb-PvCSP-like G10. Percent protection to reach T1% and parasitemia are shown in Fig. 5 (i, j and k, l, respectively). With either adjuvant, a statistically significant delay in the T1% period in mice immunized with either protein compared to the adjuvant-only groups was observed (p = 0.0007 for Montanide adjuvant-treated groups and p < 0.0015 for Poly (I:C) adjuvant-treated groups, Table 2). As in previous experiments using Pb-PvCSP210 and Pb-PvCSP247 parasites, no significant differences were observed between the naïve (not immunized, infected mice) and adjuvant-only groups (data not shown).
Discussion
The development of an effective vaccine would be an important tool against malaria, as it would provide a cost-effective form of prevention and would help circumvent adaptive strategies both from the vector and parasite.
In addition to the general obstacles to overcome when developing vaccines against parasitic diseases, research groups developing vaccines against P. vivax face other issues. One of them is the formation of hypnozoites, which can cause relapses within months and even years after the primary infection49. Our vaccine formulations were able to elicit high and long-lasting titers of PvCSP-specific antibodies in mice, providing partial protection against challenge with chimeric P. berghei spz expressing the P. vivax CSP variants. Since a significant proportion of sporozoites released in the challenge were prevented from causing an infection, our formulation could hypothetically contribute to the reduction in cases of relapse, as it would prevent the formation of new hypnozoites in the liver49.
Another important obstacle to overcome is that P. vivax does not infect rodents. For this reason, the preclinical evaluation of the protective efficacy of vaccine formulations is mainly restricted to the use of monkeys. In addition to the ethical conflict associated with the use of NHP in such early stage of the vaccine development, these animals must undergo a splenectomy to facilitate the development of parasitemia. This procedure may not provide robust data since organ removal causes immunological changes50. Thus, a strategy that enables the preclinical determination of protective immune responses induced by vaccine formulations against P. vivax malaria is based on the use of chimeric parasites expressing P. vivax proteins that are the targets of the vaccine formulations51. In particular, the use of transgenic parasites in the study of CSP-based vaccine formulations for the pre-erythrocytic phase of infection has allowed the analysis of functional inhibition of the exogenous CSP, expressed in replacement of the endogenous protein. Following this strategy, chimeric P. berghei parasites expressing the P. vivax CSP variants VK210 and VK247 were used to determine the protective efficacy of vaccine formulations consisting of viral vectors carrying P. vivax CSP alleles25,41–43. In this study, we generated for the first time a chimeric P. berghei parasite line expressing the third PvCSP variant, PvCSP-P. vivax-like.
We first attempted to develop a chimeric parasite in which the endogenous P. berghei csp gene was replaced with the P. vivax-like csp gene. Unfortunately, this transgenic parasite failed to produce visible spz inside oocysts, and only very few in the salivary glands of An. stephensi mosquitoes. This failure to complement the function of the endogenous CSP was also observed in transgenic P. falciparum expressing P. vivax CSP as replacement lines44. In addition, the chimeric P. berghei Pb-PvCSP247 line produces significantly less salivary gland spz than the Pb-PvCSP210 line25. To overcome this concern, we generated an “additional copy” chimeric parasite in which the endogenous P. berghei csp gene is maintained and the P. vivax-like csp gene is expressed under the control of the promoter region of the sporozoite- and liver-specific P. berghei gene uis4. This strategy was successfully applied in previous studies, in which chimeric P. berghei spz have been generated by introducing a P. falciparum csp gene as an additional copy into the P. berghei genome. These chimeric P. berghei spz expressed both PbCSP and PfCSP at their surface45–47. Additionally, chimeric P. falciparum spz have been generated by introducing a P. vivax csp gene as an additional copy into the P. falciparum genome. These chimeric P. falciparum spz also expressed both PvCSP and PfCSP at their surface52, similarly to our results.
The development of the first chimeric P. berghei parasite expressing the PvCSP-P. vivax-like protein allows the preclinical determination of protective immunity of vaccines targeting this mostly neglected CSP variant. The genome of P. vivax-like as a malaria-transmitting parasite in apes has been recently published53. Similar to P. knowlesi and P. simium infections in humans, P. vivax-like malaria could currently be considered a zoonotic disease, probably with continuing cross-species exchange of P. vivax between humans and apes in tropical Africa16. This hypothesis is based on not only the shared vector species (An. vinckei, An. moucheti, and An. marshallii) but also their low host specificity and high longevity54. Nevertheless, a significant proportion of this variant was found in P. vivax infections of patients from endemic areas of the Brazilian Amazon, as determined by molecular methods17–19. Consistent with these findings, Soares et al. (2020) recently reported the high (~ 40–60%) prevalence of antibodies against P. vivax-like in patients from three communities in this region55. These results prompted us to propose that human-to-human transmission is very likely. The aim of this work is not to elucidate whether human-infective P. vivax-like is a P. vivax allelic variant or a different species causing zoonosis; however, regardless of the origin and classification, we cannot continue to neglect actions to combat it. Therefore, in this work, we developed the first chimeric P. berghei parasite expressing the PvCSP-P. vivax-like protein, and used these parasites to analyze protective immunity in mice immunized with recombinant proteins representing all three P. vivax CSP variants.
Our recombinant proteins include the repeats of the PvCSP VK210 and VK247 variants and a sequence representing P. vivax-like CSP. Thus, our formulations would be predicted to be effective against a broad spectrum of cases of vivax malaria, caused not only by P. vivax, but P. simium and P. vivax-like as well. This could represent an improvement when compared to formulations such as VMP00156 and Rv2125, which contain only sequences of PvCSP VK210 and VK247, as would confer higher protection against P. vivax-like infections, than that expected to be achieved through cross-reactivity from other CSP allelic variants. Moreover, our immunization data show that our vaccine formulations stimulated the production of high titers of specific antibodies against each of the variants. In previous studies, it was demonstrated the absence of significative cross-reaction or antigenic interference among the PvCSP-repeat sequences in animals immunized with individual (VK210, VK247 and P. vivax-like) recombinant proteins, produced in bacteria57 or yeast22. Besides it, formulations containing the chimeric fusion protein, comprising epitopes of all three different allelic forms, were as immunogenic as the mixture of three individual PvCSP proteins. Thus, the specific response to PvCSP RI, repeats and C-terminal regions combined in our formulations might contribute substantially to enhancing protective efficacy, since specific antibodies against these regions are highly neutralizing41, thus indicating the importance of a universal formulation.
Although our vaccine formulations did not confer sterile protection after challenge with chimeric spz, the significant delay in T1% periods was noteworthy for several reasons, as described below.
i. Each day of delay in blood-stage parasitemia is representative of ~ tenfold fewer sporozoites reaching the liver58, and as previously discussed, this decrease would impact hypnozoite formation, thus preventing relapses49. ii. Due to technical reasons, we used our previously established i.v. challenge system, which does not allow us to consider the effect of specific CSP antibodies that potentially act in the skin. In the case of Plasmodium infections, these types of antibodies were recently shown to contribute significantly to protective effects59,60. iii. In natural infections, most mosquitoes inoculate only ~ 1% of the sporozoites in their salivary glands, with median inocula ranging between ~ 40 and 100 sporozoites61. Supporting this, it was demonstrated that a natural infection with 8 P. berghei-infected mosquitoes is equivalent to i.v. inoculation of 250–500 sporozoites60. Therefore, our challenge system (i.v. inoculation of 2000 spz) is stronger than other strategies using intradermal or s.c. challenge. The variation in the challenge system also explains the apparent discrepancies in protective efficacy comparing our results with previous studies23 (30% mice protected in the s.c. challenge system vs. significant delay in T1% in the i.v. system). In agreement, a similar situation was observed comparing the protective performance of previous PvCSP-based formulations using s.c.22 and i.v.43 challenges, respectively. Taking into account all these facts, the effectiveness of the formulations developed in this study might be even greater when considered in the case of natural infection.
Consistent with a previous study43, a clear positive relation was observed between high titers of CSP-specific antibodies and protection. In a comparison of both recombinant proteins, the delay in the T1% period obtained with NLP-CSPCT was longer than with NLP-CSPR when administered with the same adjuvant. Most likely, antibodies against the C-terminal domain of PvCSP, which is absent in NLP-CSPR, are responsible for the differences, as this region might be important for protection41. The lack of anti-C-terminal antibodies and the lower titers against VK247 repeats in vaccines using Poly (I:C) as an adjuvant reported in previous work23 would explain the lack of protection provided by NLP-CSPR/Poly (I:C) against challenge with Pb-PvCSP247 parasites. By the other hand, the C-terminal region of PvCSP contains predicted T cell epitopes, which could also contribute to protective effect of pre-erythrocytic vaccines40. However, in previous studies, very low levels of PvCSP-specific CD4+ and CD8+ T cell responses were elicited by our PvCSP recombinant proteins22,57. Results from the VMP001 clinical trial also indicate the low contribution of repeats and C-terminal region to induce T cell responses, as only 17% of vaccinated subjects responded to these antigens whereas 90% showed strong cellular responses to the N-terminal region56 (absent in NLP-CSP proteins). Moreover, it was shown that PvCSP short repeat-region peptides, when presented on a VLP, can induce antibodies mediated protection41. Nonetheless, we do not exclude the participation of T cell-mediated immune responses in the protection observed in this study.
Finally, Montanide ISA720 was overall a better adjuvant in terms of IgG titers and protective efficacy. However, phase I clinical trials showed some concerns regarding the safety of Montanide ISA720-adjuvanted vaccines against malaria, particularly high reactogenicity62–64.
For all these reasons, we aim to analyze the mechanisms underlying the observed protection before moving into clinical testing of safety and toxicology. Ongoing research analyzing the specificity of humoral and cellular responses and performing transcriptomic analysis of lymphocytes from immunized mice will provide insights into the pathways that are selectively activated by these formulations and will provide valuable information about the type of immune response that a protective vaccine against vivax malaria should elicit.
Supplementary information
Acknowledgements
This work was supported by funds from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, 2016/26123-0, 2014/18102-7, and 2012/13032-5). AMG received a fellowship from FAPESP, RFM received a fellowship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) and ISS received a fellowship from CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). We would like to thank Professor Shahid M. Khan for supportive advices and knowledge, Erwan Atcheson for helping with experimental challenges and Katia S. Françoso for providing technical support.
Author contributions
A.R.S., I.S.S. and C.J.J. conceived and designed the study. A.M.G., A.M.S., R.F.M. and C.L.C. delineated, performed and analyzed the experiments. K.H. and Y.C.K. performed experiments. A.M.G., A.M.S., R.F.M. and C.J.J. analyzed the data and prepared the figures. C.J.J. contributed reagents and materials. A.M.G., A.M.S., R.F.M., C.J.J., I.S.S. and A.R.S. wrote and edited the manuscript. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Alba Marina Gimenez and Ahmed M. Salman.
Contributor Information
Irene S. Soares, Email: isoares@usp.br
Arturo Reyes-Sandoval, Email: arturo.reyes@ndm.ox.ac.uk.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-96986-1.
References
- 1.Kotepui M, Kotepui KU, Milanez GD, Masangkay FR. Prevalence of severe Plasmodium knowlesi infection and risk factors related to severe complications compared with non-severe P. knowlesi and severe P. falciparum malaria: a systematic review and meta-analysis. Infect. Dis. Poverty. 2020;9:106. doi: 10.1186/s40249-020-00727-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capela R, Moreira R, Lopes F. An overview of drug resistance in protozoal diseases. Int. J. Mol. Sci. 2019 doi: 10.3390/ijms20225748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park JW, Jun G, Yeom JS. Plasmodium vivax malaria: Status in the Republic of Korea following reemergence. Korean J Parasitol. 2009;47(Suppl):S39–50. doi: 10.3347/kjp.2009.47.S.S39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antonio-Nkondjio C, et al. Review of malaria situation in Cameroon: technical viewpoint on challenges and prospects for disease elimination. Parasit. Vectors. 2019;12:501. doi: 10.1186/s13071-019-3753-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahimi BA, et al. Severe vivax malaria: A systematic review and meta-analysis of clinical studies since 1900. Malar. J. 2014;13:481. doi: 10.1186/1475-2875-13-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization. World Malaria Report 2020: 20 Years of Global Progress and Challenges. Geneva, Switzerland (2020).
- 7.Armistead JS, Adams JH. Advancing research models and technologies to overcome biological barriers to Plasmodium vivax control. Trends Parasitol. 2018;34:114–126. doi: 10.1016/j.pt.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnot DE, et al. Circumsporozoite protein of Plasmodium vivax: Gene cloning and characterization of the immunodominant epitope. Science. 1985;230:815–818. doi: 10.1126/science.2414847. [DOI] [PubMed] [Google Scholar]
- 9.Rosenberg R, et al. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–976. doi: 10.1126/science.2672336. [DOI] [PubMed] [Google Scholar]
- 10.Vo TC, et al. Genetic polymorphism and natural selection of circumsporozoite protein in Myanmar Plasmodium vivax. Malar. J. 2020;19:303. doi: 10.1186/s12936-020-03366-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qari SH, et al. Identification of Plasmodium vivax-like human malaria parasite. Lancet. 1993;341:780–783. doi: 10.1016/0140-6736(93)90559-y. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser M, et al. Wild chimpanzees infected with 5 Plasmodium species. Emerg. Infect. Dis. 2010;16:1956–1959. doi: 10.3201/eid1612.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu W, et al. African origin of the malaria parasite Plasmodium vivax. Nat. Commun. 2014;5:3346. doi: 10.1038/ncomms4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Udhayakumar V, Qari SH, Patterson P, Collins WE, Lal AA. Monoclonal antibodies to the circumsporozoite protein repeats of a Plasmodium vivax-like human malaria parasite and Plasmodium simiovale. Infect. Immun. 1994;62:2098–2100. doi: 10.1128/iai.62.5.2098-2100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prugnolle F, et al. Diversity, host switching and evolution of Plasmodium vivax infecting African great apes. Proc. Natl. Acad. Sci. U S A. 2013;110:8123–8128. doi: 10.1073/pnas.1306004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramasamy R. Zoonotic malaria—Global overview and research and policy needs. Front. Public Health. 2014;2:123. doi: 10.3389/fpubh.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machado RL, Povoa MM. Distribution of Plasmodium vivax variants (VK210, VK247 and P. vivax-like) in three endemic areas of the Amazon region of Brazil and their correlation with chloroquine treatment. Trans. R. Soc. Trop. Med. Hyg. 2000;94:377–381. doi: 10.1016/s0035-9203(00)90110-x. [DOI] [PubMed] [Google Scholar]
- 18.Cerutti C, Jr, et al. Epidemiologic aspects of the malaria transmission cycle in an area of very low incidence in Brazil. Malar. J. 2007;6:33. doi: 10.1186/1475-2875-6-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gomes MD, et al. Evaluation of circumsporozoite protein of Plasmodium vivax to estimate its prevalence in Oiapoque, Amapa State, Brazil, bordering French Guiana. Rev. Inst. Med. Trop. Sao Paulo. 2016;58:72. doi: 10.1590/S1678-9946201658072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buery JC, et al. Mitochondrial genome of Plasmodium vivax/simium detected in an endemic region for malaria in the Atlantic Forest of Espirito Santo state, Brazil: Do mosquitoes, simians and humans harbour the same parasite? Malar. J. 2017;16:437. doi: 10.1186/s12936-017-2080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lim CS, Tazi L, Ayala FJ. Plasmodium vivax: Recent world expansion and genetic identity to Plasmodium simium. Proc Natl Acad Sci U S A. 2005;102:15523–15528. doi: 10.1073/pnas.0507413102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gimenez AM, et al. Vaccine containing the three allelic variants of the Plasmodium vivax circumsporozoite antigen induces protection in mice after challenge with a transgenic rodent malaria parasite. Front. Immunol. 2017;8:1275. doi: 10.3389/fimmu.2017.01275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marques RF, et al. Protective malaria vaccine in mice based on the Plasmodium vivax circumsporozoite protein fused with the mumps nucleocapsid protein. Vaccines (Basel) 2020 doi: 10.3390/vaccines8020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espinosa DA, et al. Development of a chimeric Plasmodium berghei strain expressing the repeat region of the P. vivax circumsporozoite protein for in vivo evaluation of vaccine efficacy. Infect. Immun. 2013;81:2882–2887. doi: 10.1128/IAI.00461-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salman AM, et al. Rational development of a protective P. vivax vaccine evaluated with transgenic rodent parasite challenge models. Sci. Rep. 2017;7:46482. doi: 10.1038/srep46482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JW, et al. A novel 'gene insertion/marker out' (GIMO) method for transgene expression and gene complementation in rodent malaria parasites. PLoS ONE. 2011;6:e29289. doi: 10.1371/journal.pone.0029289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janse CJ, Franke-Fayard B, Waters AP. Selection by flow-sorting of genetically transformed, GFP-expressing blood stages of the rodent malaria parasite, Plasmodium berghei. Nat. Protoc. 2006;1:614–623. doi: 10.1038/nprot.2006.88. [DOI] [PubMed] [Google Scholar]
- 28.Salman AM, et al. Generation of transgenic rodent malaria parasites expressing human malaria parasite proteins. Methods Mol Biol. 2015;1325:257–286. doi: 10.1007/978-1-4939-2815-6_21. [DOI] [PubMed] [Google Scholar]
- 29.Triller G, et al. Natural parasite exposure induces protective human anti-malarial antibodies. Immunity. 2017;47:1197–1209. doi: 10.1016/j.immuni.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janse CJ, et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol. Biochem. Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Menard R, Janse C. Gene targeting in malaria parasites. Methods. 1997;13:148–157. doi: 10.1006/meth.1997.0507. [DOI] [PubMed] [Google Scholar]
- 32.Cabral-Miranda G, et al. DOPS adjuvant confers enhanced protection against malaria for VLP-TRAP based vaccines. Diseases. 2018 doi: 10.3390/diseases6040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espinosa DA, et al. The Plasmodium falciparum cell-traversal protein for ookinetes and sporozoites as a candidate for preerythrocytic and transmission-blocking vaccines. Infect. Immun. 2017 doi: 10.1128/IAI.00498-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alves E, et al. Evaluation of Plasmodium vivax cell-traversal protein for ookinetes and sporozoites as a preerythrocytic P. vivax Vaccine. Clin. Vaccine Immunol. CVI. 2017 doi: 10.1128/CVI.00501-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr RY, Philip N, Waters AP. Improved negative selection protocol for Plasmodium berghei in the rodent malarial model. Malar. J. 2012;11:103. doi: 10.1186/1475-2875-11-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Janse CJ, Ramesar J, Waters AP. High-efficiency transfection and drug selection of genetically transformed blood stages of the rodent malaria parasite Plasmodium berghei. Nat. Protoc. 2006;1:346–356. doi: 10.1038/nprot.2006.53. [DOI] [PubMed] [Google Scholar]
- 37.Longley RJ, et al. Comparative assessment of vaccine vectors encoding ten malaria antigens identifies two protective liver-stage candidates. Sci. Rep. 2015;5:11820. doi: 10.1038/srep11820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Annoura T, et al. Assessing the adequacy of attenuation of genetically modified malaria parasite vaccine candidates. Vaccine. 2012;30:2662–2670. doi: 10.1016/j.vaccine.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Yoshida N, Nussenzweig RS, Potocnjak P, Nussenzweig V, Aikawa M. Hybridoma produces protective antibodies directed against the sporozoite stage of malaria parasite. Science. 1980;207:71–73. doi: 10.1126/science.6985745. [DOI] [PubMed] [Google Scholar]
- 40.Reyes-Sandoval A, et al. CD8+ T effector memory cells protect against liver-stage malaria. J. Immunol. 2011;187:1347–1357. doi: 10.4049/jimmunol.1100302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atcheson E, Reyes-Sandoval A. Protective efficacy of peptides from Plasmodium vivax circumsporozoite protein. Vaccine. 2020;38:4346–4354. doi: 10.1016/j.vaccine.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atcheson E, et al. Tailoring a Plasmodium vivax vaccine to enhance efficacy through a combination of a CSP virus-like particle and TRAP viral vectors. Infect. Immun. 2018;86:1. doi: 10.1128/IAI.00114-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.de Camargo TM, et al. Prime-boost vaccination with recombinant protein and adenovirus-vector expressing Plasmodium vivax circumsporozoite protein (CSP) partially protects mice against Pb/Pv sporozoite challenge. Sci. Rep. 2018;8:1118. doi: 10.1038/s41598-017-19063-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marin-Mogollon C, et al. Chimeric Plasmodium falciparum parasites expressing Plasmodium vivax circumsporozoite protein fail to produce salivary gland sporozoites. Malar. J. 2018;17:288. doi: 10.1186/s12936-018-2431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reuling IJ, et al. An open-label phase 1/2a trial of a genetically modified rodent malaria parasite for immunization against Plasmodium falciparum malaria. Sci. Transl. Med. 2020 doi: 10.1126/scitranslmed.aay2578. [DOI] [PubMed] [Google Scholar]
- 46.Mendes AM, et al. Pre-clinical evaluation of a P. berghei-based whole-sporozoite malaria vaccine candidate. NPJ Vaccines. 2018;3:54. doi: 10.1038/s41541-018-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mendes AM, et al. A Plasmodium berghei sporozoite-based vaccination platform against human malaria. NPJ Vaccines. 2018;3:33. doi: 10.1038/s41541-018-0068-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mueller AK, et al. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc. Natl. Acad. Sci. U S A. 2005;102:3022–3027. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White M, Amino R, Mueller I. Theoretical implications of a pre-erythrocytic Plasmodium vivax vaccine for preventing relapses. Trends Parasitol. 2017;33:260–263. doi: 10.1016/j.pt.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Butcher GA. The role of the spleen and immunization against malaria. Trends Parasitol. 2005;21:356–357. doi: 10.1016/j.pt.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 51.Othman AS, et al. The use of transgenic parasites in malaria vaccine research. Expert Rev. Vaccines. 2017;16:1–13. doi: 10.1080/14760584.2017.1333426. [DOI] [PubMed] [Google Scholar]
- 52.Miyazaki Y, et al. Generation of a genetically modified chimeric Plasmodium falciparum parasite expressing Plasmodium vivax circumsporozoite protein for malaria vaccine development. Front. Cell. Infect. Microbiol. 2020;10:591046. doi: 10.3389/fcimb.2020.591046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilabert A, et al. Plasmodium vivax-like genome sequences shed new insights into Plasmodium vivax biology and evolution. PLoS Biol. 2018;16:e2006035. doi: 10.1371/journal.pbio.2006035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Makanga B, et al. Ape malaria transmission and potential for ape-to-human transfers in Africa. Proc. Natl. Acad. Sci. U S A. 2016;113:5329–5334. doi: 10.1073/pnas.1603008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soares IF, et al. Recombinant Plasmodium vivax circumsporozoite surface protein allelic variants: Antibody recognition by individuals from three communities in the Brazilian Amazon. Sci. Rep. 2020;10:14020. doi: 10.1038/s41598-020-70893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lumsden JM, et al. Evaluation of immune responses to a Plasmodium vivax CSP-based recombinant protein vaccine candidate in combination with second-generation adjuvants in mice. Vaccine. 2012;30:3311–3319. doi: 10.1016/j.vaccine.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Teixeira LH, et al. Immunogenicity of a prime-boost vaccine containing the circumsporozoite proteins of Plasmodium vivax in rodents. Infect Immun. 2014;82:793–807. doi: 10.1128/IAI.01410-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamauchi LM, Coppi A, Snounou G, Sinnis P. Plasmodium sporozoites trickle out of the injection site. Cell. Microbiol. 2007;9:1215–1222. doi: 10.1111/j.1462-5822.2006.00861.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aliprandini E, et al. Cytotoxic anti-circumsporozoite antibodies target malaria sporozoites in the host skin. Nat. Microbiol. 2018;3:1224–1233. doi: 10.1038/s41564-018-0254-z. [DOI] [PubMed] [Google Scholar]
- 60.Flores-Garcia Y, et al. Antibody-mediated protection against plasmodium sporozoites begins at the dermal inoculation site. MBio. 2018 doi: 10.1128/mBio.02194-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Graumans W, Jacobs E, Bousema T, Sinnis P. When is a plasmodium-infected mosquito an infectious mosquito? Trends Parasitol. 2020;36:705–716. doi: 10.1016/j.pt.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hu J, et al. Safety and immunogenicity of a malaria vaccine, Plasmodium falciparum AMA-1/MSP-1 chimeric protein formulated in montanide ISA 720 in healthy adults. PLoS ONE. 2008;3:e1952. doi: 10.1371/journal.pone.0001952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Roestenberg M, et al. Safety and immunogenicity of a recombinant Plasmodium falciparum AMA1 malaria vaccine adjuvanted with Alhydrogel, Montanide ISA 720 or AS02. PLoS ONE. 2008;3:e3960. doi: 10.1371/journal.pone.0003960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCarthy JS, et al. A phase 1 trial of MSP2-C1, a blood-stage malaria vaccine containing 2 isoforms of MSP2 formulated with Montanide(R) ISA 720. PLoS ONE. 2011;6:e24413. doi: 10.1371/journal.pone.0024413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.