Abstract
An in-house reverse transcription (RT)-competitive PCR (RT-cPCR) for the quantitation of human immunodeficiency virus type 1 (HIV-1) RNA in plasma samples was developed and validated. The procedure involves (i) extraction of RNA with spin columns, (ii) ready-to-use bead-mediated RT, (iii) competitive PCR in a microtiter plate, (iv) agarose gel electrophoresis of the reaction products, and (v) densitometric analysis of the digitized image of the gel. Quadruplicate tests and dilution studies showed that the sensitivity and intertest coefficient of variability of the RT-cPCR are comparable to those of the reference AMPLICOR HIV-1 MONITOR test. The results obtained by the two assays with a panel of 45 clinical samples were in good agreement (mean difference, 0.36 ± 0.25 log units). Analysis of 1,982 clinical samples by the in-house RT-cPCR yielded the typical range of plasma HIV-1 RNA levels with the expected inverse correlation between CD4 counts and HIV-1 RNA titers. In addition, testing of plasma from 36 subjects at weeks 0 and 4 with respect to the time of initiation of protease inhibitor therapy detected a significant decrease in HIV-1 viremia. The mean reduction in the HIV-1 RNA level was 0.914 log unit for those receiving saquinavir (P = 0.0210), 1.584 log units for those receiving indinavir (P = 0.0047), and 1.904 log units for those receiving ritonavir (P < 0.0001). The in-house RT-cPCR assay is simple to develop and perform and allows quantitation of HIV-1 RNA in 100 to 200 samples per operator per week. Since the cost is 1/8 to 1/10 of those of reference commercial assays, this procedure could be conveniently used in medium-scale laboratories.
The levels of human immunodeficiency virus type 1 (HIV-1) RNA in plasma are highly predictive of disease progression in individuals with HIV-1 infection (14, 15). Periodic measurement of the plasma HIV-1 RNA load has thus been established as the key laboratory analysis for monitoring the course of HIV-1 infection in patients under treatment with different combinations of antiretroviral compounds (9). Three commercially available methods have been licensed for titration of HIV-1 RNA in clinical samples; these methods are based on target amplification by competitive PCR (AMPLICOR HIV-1 MONITOR test; Roche) or quantitative nucleic acid sequence-based amplification (Q-NASBA; Organon Teknika) and signal amplification by the branched-DNA assay (QUANTIPLEX; Chiron). These systems have been shown to yield equivalent results in comparative studies (5, 20, 23), and all are suitable for clinical use. However, all of these commercial kits are still expensive ($60 to $100 per test), actually limiting full large-scale application in most settings and prompting the development of reliable cost-effective in-house assays (1, 25).
The use of a titrated competitor amplification target for the quantitation of HIV-1 nucleic acid sequences by PCR was first detailed in 1993 (19) and is still considered to be the best approach for quantitative PCR, as shown by its use in the Roche AMPLICOR HIV-1 MONITOR test. We have optimized and clinically evaluated a simplified in-house reverse transcription (RT)-competitive PCR (RT-cPCR) system which combines (i) extraction of RNA from plasma with spin columns, (ii) noncompetitive RT with ready-to-use reaction beads, (iii) four-well competitive PCR with premade reaction mixtures in a microtiter plate, (iv) agarose gel electrophoresis of the reaction products, and (v) densitometric analysis of the digitized image of the gel. Reproducibility studies, parallel analysis of samples titrated by the AMPLICOR HIV-1 MONITOR test, and testing of samples from a large number of patients indicated that the in-house assay is suitable for clinical application and has 1/8 to 1/10 of the cost of commercial systems.
MATERIALS AND METHODS
Construction of competitor HIV-1 DNA and wt HIV-1 RNA standard.
The sequences and relative locations of all primers used in this study are presented in Table 1 and Fig. 1, respectively. A deleted competitor HIV-1 DNA fragment was generated by PCR amplification of a pol sequence of the HIV-1 Z6 genome contained in plasmid pSYC1857 (Perkin-Elmer, Emeryville, Calif.) with the antisense primer LR62 and the sense primer T52. Primer T52 hybridizes with the 3′ terminal 20 bases at positions 3163 to 3182 of the HIV-1 SF2 isolate (GenBank accession no. KO2007) and contains a 27-base 5′ tail that hybridizes to positions 3096 to 3122. PCR amplification with primers LR52 and LR62 generates a 237- or a 197-bp product when wild-type (wt) HIV-1 RNA-derived cDNA or the T52-LR62 competitor DNA, respectively, is used as the template. The concentration of the purified T52-LR62 DNA fragment was estimated by agarose gel electrophoresis and was accurately adjusted by repeated competitive titration of 500 copies of pSYC1857 (as determined by the manufacturer). Competitive amplification and data analysis were essentially the same as described below for clinical samples except that the wt DNA titer was known and the competitor DNA titer was to be determined.
TABLE 1.
Sequences and locations of the primers used for construction of the HIV-1 DNA competitor and in the RT-cPCR assay
| Primera | Sequence (5′-3′) | Positionb |
|---|---|---|
| T52 (+) | CTATCAATACATGGATGATTTGTATGT ATGTGAGGAGCTGAGACAACATCTc | 3163–3182 |
| LR52 (+) | CTATCAATACATGGATGATTTGTATGT | 3096–3122 |
| LR62 (−) | TTCTGTATGTCATTGACAGTCCAGCT | 3307–3332 |
(+) and (−), sense and antisense, respectively.
Coordinates are from HIV-1 isolate SF2 (GenBank accession no. KO2007).
The 27-base 5′ tail (underlined) hybridizes to positions 3096 to 3122.
FIG. 1.
Relative location of HIV-1-specific primers on the HIV-1 pol gene (thick lines). Sense and antisense HIV-1 DNA strands are labeled (+) and (−), respectively. Note that since the sequence of the 5′ tail of primer T52 is identical to that of primer LR52, PCR products obtained with primer pair LR52-LR62 and with primer pair T52-LR62 differ in length but are delimited by the same sequences at the 5′ end (LR52) and the 3′ end (LR62). Primers, target DNA, and PCR products are not drawn to scale.
To construct a wt HIV-1 RNA standard, the wt LR52-LR62 PCR product was inserted into plasmid pCRII by using the TA-Cloning System (Invitrogen, Leek, The Netherlands). The resulting plasmid, pCRII-LR52/LR62, was linearized at a unique SalI site (26 bases from the end of the inserted fragment) and was quantitated by agarose gel electrophoresis. Following in vitro transcription with T7 RNA polymerase, template DNA was digested with DNase I (Promega, Madison, Wis.). The ethanol-precipitated RNA pellet was resuspended in diethylpyrocarbonate (DEPC)-treated water containing 10 mM Tris-HCl (pH 8.5), 10 mM NaCl, and 1 mM EDTA and was quantitated by spectrophotometry. Appropriate single-use aliquots of the wt HIV-1 RNA standard were stored at −70°C and were used in preliminary experiments in order to evaluate the yield and reproducibility of the RNA extraction and RT steps. HIV-1-negative plasma was used to reconstruct clinical samples harboring 103 to 106 copies of wt HIV-1 RNA per ml. These standard samples were subjected in triplicate to RNA extraction and RT followed by competitive PCR in the presence of increasing amounts of the competitor T52-LR62 DNA fragment as described below, except that 2-μl aliquots of cDNA were used as the cPCR template for titration of the samples with 106 copies. The results obtained allowed us to estimate the ratio between the amount of HIV-1 RNA present in the starting material and the amount of HIV-1 cDNA generated. This factor was then used to adjust the actual copy numbers of the competitor DNA to RNA equivalents, i.e., the number of cDNA copies resulting from extraction and RT of equivalent numbers of copies of RNA.
In-house RT-cPCR system.
Plasma RNA was prepared by using the QIAmp Viral RNA kit (Qiagen, Chatsworth, Calif.) according to the manufacturer’s instructions, except that 300 μl of starting material was loaded onto the column and was eluted with 50 μl of preheated DEPC-treated water after incubation at 80°C for 5 min. In addition, 1,000 copies of tobacco mosaic virus (TMV) RNA (Boehringer Mannheim, Mannheim, Germany) per sample were directly added to the virus lysis buffer as an internal control to check that plasma specimens with undetectable HIV-1 RNA levels were suitable for the RT-PCR. The RT step was performed by using Ready-to-Go You-Prime first-strand cDNA synthesis beads (Pharmacia, Uppsala, Sweden), which contain all reaction components except for template RNA and primer. Each bead was reconstituted with 16 μl of DEPC-treated water containing 6 pmol of the HIV-1-specific RT primer LR62 and 3 pmol of the TMV-specific RT primer TMV1 (5′-CATCTTTAGTTGTAGATAAGTTTTTTG-3′). Template RNA was transferred from a heat block (75°C) to a cryobox (0°C), and 20 μl was immediately added to the appropriate tube for RT (30 min at 37°C, followed by 5 min at 94°C to inactivate the enzyme). Each RT run included a blank sample. Following completion of the RT step, four 8-μl aliquots of the RT mixture were used as PCR templates in the presence of increasing amounts (40, 240, 1,440, and 8,640 copies of RNA equivalents) of the competitor T52-LR62 DNA fragment. The amplification reaction mixtures (50 μl) were prepared in a 96-well PCR plate (Corning Costar Corp., Cambridge, Mass.) and contained 50 mM KCl, 10 mM Tris-HCl (pH 8.6), 1.5 mM MgCl2, 0.1% Triton X-100, 100 μM each dATP, dCTP, dGTP, and UTP, 10 pmol of the primer pair LR52-LR62, an additive for direct gel loading (240 μg of tartrazine per ml, 1.5% Ficoll 400) (modified from reference 26), 0.15 U of thermolabile uracyl-N-glycosylase (HK-UNG; Epicentre Technologies, Madison, Wis.), and 1 U of Taq DNA polymerase (Promega). A solid wax layer (DynaWax; Fynnzymes, Espoo, Finland) was used to separate the key reaction components (Taq and primers), providing a synchronous hot start. Each plate allowed competitive quantitation of 23 samples containing cDNA and 1 blank control sample. A Hybaid Touchdown thermal cycler was used with the simulated-tube temperature control algorithm to perform the following program: 30 min at 37°C (UNG-mediated inactivation of possible contaminating U-containing DNA), 15 min at 85°C, and 4 min at 94°C (irreversible inactivation of UNG and first denaturation) and then 44 cycles of annealing at 56°C for 20 s, extension at 72°C for 30 s plus a 3-s increment per cycle, and denaturation at 93°C for 20 s. Ten microliters of the reaction mixtures was then directly loaded onto a 2.4% NuSieve–0.6% Seakem (FMC, Rockland, Maine) agarose gel. Two 12-sample series of products were loaded consecutively with a 10-min delay in order to accommodate the reaction products of a whole plate in two two-comb, 24-well minigels. The gel photograph was digitized with a Hewlett-Packard Scanjet as a 256-level grayscale image in the gel analysis software SigmaGel (Jandel Scientific, Erkrath, Germany). The log of the ratio between the intensity of the competitor band (corrected for the shorter length [in base pairs]) and that of the wt band was plotted against the log of the number of copies of competing DNA added, and linear regression was used to calculate the wt DNA copy number at the equivalence between the corrected competitor and wt band intensities (Fig. 2) (27). Testing of samples containing very low (<1,000 copies/ml) or very high (>750,000 copies/ml) numbers of HIV-1 RNA copies per milliliter could be performed by calculation of the ratio between the amounts of the competitor and the wt PCR products in the first or last lane only, respectively. The results were normalized as the number of HIV-1 RNA copies per milliliter. The efficiency of RNA extraction and RT for samples yielding no wt HIV-1 amplification product was controlled retrospectively by subjecting the residual cDNA (about 2 μl) to a 40-cycle single-round amplification with TMV primers TMV1 and TMV2 (5′-TGGTCTTTCTATGCCCTTGTTTC-3′). These primers amplify a 564-bp region of the TMV genome (GenBank accession no. V01408).
FIG. 2.
Quantitation of a wt HIV-1 RNA standard by RT-cPCR. HIV-1 RNA (5,000 nominal copies) was reverse transcribed with primer LR62. The reaction mixture was split into four aliquots and subjected to amplification with primers LR52 and LR62 in the presence of 40, 240, 1,440, and 8,640 RNA equivalents (eq.) of the competitor (comp) T52-LR62 DNA. The wt target copy number was estimated by plotting the log ratio between wt and competitor intensities (intens) (after correction for the difference in base pairs) versus the log number of competitor target copies. The result for this control experiment is 1,375 × 4 = 5,500 copies.
Specificity, sensitivity, reproducibility, and comparative analysis.
The specificities of the RT and PCR primers were preliminarily checked by testing 50 HIV-1-negative and 50 HIV-1-positive plasma samples in a qualitative RT-PCR (i.e., in a single tube in the absence of competitor DNA). The threshold of sensitivity of the method was determined by testing reconstructed plasma samples containing 1,500 to 100 copies of HIV-1 RNA per ml, with tests conducted eight times to examine the performance at the lower limit of 400, 250, and 100 copies/ml. To determine the intertest coefficient of variation (CV) of the whole assay from RNA extraction to data analysis, 12 plasma samples were divided into four aliquots on receipt and were frozen at −70°C. The different aliquots of these samples were then tested in four totally independent runs. To examine whether repeated freezing-thawing of the RNA obtained by spin column purification affects the quantitative in-house assay, a panel of 50 plasma samples was tested twice with the first and second 20-μl aliquots of the same RNA preparation after one and two freezing-thawing cycles, respectively. To compare the in-house assay with commercial reference tests, another panel of 45 plasma samples containing undetectable to high titers of HIV-1 RNA, as measured by the in-house RT-cPCR procedure, were blindly tested by the Roche AMPLICOR HIV-1 MONITOR test.
AMPLICOR HIV-1 MONITOR test.
The Roche AMPLICOR HIV-1 MONITOR test was performed by following the manufacturer’s instructions (20). This procedure involves (i) treatment of plasma with guanidinium isothiocyanate and precipitation of RNA with isopropanol, (ii) single-step rTth DNA polymerase-mediated RT-PCR with biotinylated primers SK431 and SK462 (gag region) in the presence of one internal competitor RNA (quantification standard [QS]), (iii) separate hybridization of fivefold serial dilutions of the HIV-1 and QS amplification products in specific probe-coated microtiter wells, (iv) enzyme-linked colorimetric detection of amplification products, and (v) calculation of the number of HIV-1 RNA copies on the basis of the optical densities of one QS-containing well and one HIV-1-containing well within a defined range, the dilution factors in the selected wells, and a correction factor given by the manufacturer. The test is carried out in 0.2-ml tubes (RT-PCR) and 12-sample microtiter plates (colorimetric detection) and is expected to be completed in 6 to 7 h.
Clinical samples.
The HIV-1 RNA in a total of 1,982 plasma samples obtained from 720 patients attending different infectious diseases units was quantitated by the in-house RT-cPCR system. Most patients were under treatment with combination therapies that included protease inhibitors. In order to check the capability of the in-house RT-cPCR to detect decreases in HIV-1 RNA load, 72 paired samples were obtained from 36 patients (all of whom had been pretreated with reverse transcriptase inhibitors) at week 0 and 4 with respect to the time of initiation of saquinavir (n = 12), indinavir (n = 12), and ritonavir (n = 12) therapy.
RESULTS
Preliminary testing of 50 HIV-1-negative and 50 HIV-1-positive plasma samples by a qualitative RT-PCR showed that primers LR52 and LR62 specifically amplify the expected 237-bp region of the HIV-1 pol gene. Initial studies were then aimed at defining the yield and the reproducibility of the first noncompetitive steps, i.e., RNA extraction with dedicated spin columns and RT with ready-to-use beads. Triplicate titration of reconstructed plasma samples containing 103 to 106 copies of the wt HIV-1 RNA standard indicated that the ratio between the plasma HIV-1 RNA and the cDNA levels obtained after RNA extraction and RT is fairly constant over the range of numbers of copies considered. Indeed, the mean proportions of HIV-1 RNA copies extracted from plasma and reverse transcribed into cDNA were 35, 43, 37, and 46% for samples containing 103, 104, 105, and 106 HIV-1 RNA copies per ml, respectively. This yielded the conversion of an average of 40% ± 5% of the plasma HIV-1 RNA into cDNA and defined a correction factor of 2.5 for adjustment of the actual number of copies of competitor DNA to RNA equivalents, i.e., the numbers of cDNA copies resulting from extraction and RT of equal numbers of RNA copies in plasma.
Quadruplicate testing of a 12-sample panel in four totally independent assays allowed determination of a mean CV of 41.9%, with a tendency to obtain larger CVs for lower-titer samples (Table 2). The lower limit of sensitivity of the method was then assessed with an accurately titrated plasma sample diluted with HIV-1-negative plasma. When plasma samples with decreasing amounts of RNA equivalent to 1,500, 1,000, 500, and 250 HIV-1 RNA copies per ml were tested, the in-house RT-cPCR assay calculated 1,391, 924, 333, and 202 copies, respectively. However, the wt PCR product could not be detected in none of eight and only three of eight samples with 100 and 250 nominal HIV-1 RNA copies per ml, respectively. Since replicate tests of eight samples nominally containing 400 HIV-1 RNA copies per ml consistently resulted in a measurable wt amplification signal, plasma samples yielding no amplification signal were considered to contain <400 copies/ml. A separate set of experiments demonstrated that inclusion of the TMV control primer in the RT step did not influence either the sensitivity or the performance of the assay (data not shown).
TABLE 2.
Reproducibility of the in-house RT-cPCR assay in quadruplicate independent tests with 12 plasma samples
| Sample | No. of HIV-1 RNA copies/ml
|
CV (%) | ||||
|---|---|---|---|---|---|---|
| Run 1 | Run 2 | Run 3 | Run 4 | Mean ± SD | ||
| A | 557,112 | 230,106 | 569,786 | 388,902 | 436,477 ± 160,392 | 36.7 |
| B | 446,999 | 289,507 | 251,802 | 232,878 | 305,297 ± 97,357 | 31.9 |
| C | 138,086 | 111,417 | 72,225 | 185,099 | 126,707 ± 47,403 | 37.4 |
| D | 100,166 | 63,700 | 102,844 | 148,838 | 103,887 ± 34,883 | 33.6 |
| E | 33,783 | 73,995 | 69,873 | 43,384 | 55,259 ± 19,722 | 35.7 |
| F | 43,528 | 21,814 | 27,068 | 58,193 | 37,651 ± 16,526 | 43.9 |
| G | 30,813 | 45,036 | 43,885 | 20,125 | 34,965 ± 11,810 | 33.8 |
| H | 20,567 | 8,397 | 13,253 | 7,351 | 12,392 ± 6,026 | 48.6 |
| I | 4,300 | 9,473 | 8,499 | 3,210 | 6,371 ± 3,079 | 48.3 |
| L | 3,300 | 5,527 | 4,000 | 1,110 | 3,484 ± 1,836 | 52.7 |
| M | NDa | 640 | 1,530 | ND | 1,085 ± 629 | 58.0 |
| N | ND | ND | ND | ND | NAb | NA |
| Mean ± SD | 41.9 ± 8.9 | |||||
ND, not detectable.
NA, not applicable.
The results obtained in parallel tests of 45 samples by the in-house assay and the AMPLICOR HIV-1 MONITOR test were in good agreement (Table 3). HIV-1 RNA was undetectable by both assays in the same seven samples, and there was a good correlation for the 38 samples in which HIV-1 viremia was detectable (r = 0.86; P < 10−9; Spearman’s correlation). The mean ± standard deviation difference was 0.36 ± 0.25 log unit, and there was no tendency for each test to give values higher than the values given by the other test. The results were different within a maximum of twofold (0.3 log unit) and fourfold (0.6 log unit) for 17 (44.7%) and 28 (73.7%) of these 38 samples, respectively. No result differed by >1 log unit; the largest difference between the AMPLICOR HIV-1 MONITOR test and the in-house assay (+0.96 log unit) was found for sample 7, which was obtained from a subject under triple-combination therapy with zidovudine, lamivudine, and indinavir. However, six of the seven samples in which HIV-1 RNA was undetectable by both tests and another five samples with comparable low levels of HIV-1 RNA (samples 20, 26, 32, 35, and 36) were also obtained from subjects receiving triple-combination therapy. Analysis of the second 20-μl RNA aliquot of the same sample (sample 7) by RT-cPCR confirmed the result obtained previously (32,000 copies/ml), while no residual plasma was available for retesting by the AMPLICOR HIV-1 MONITOR test. A new sample obtained from the same patient 1 month later in the absence of changes in antiretroviral therapy was then tested by RT-cPCR and AMPLICOR HIV-1 MONITOR, and both tests yielded comparable results (43,000 and 39,000 copies/ml, respectively). While this indicated that the different result first obtained by the two assays was not related to a particular virus strain, it was not possible to determine the nature of the occasional factor accounting for the discrepancy.
TABLE 3.
Parallel testing of 45 plasma samples by using the in-house RT-cPCR assay and the AMPLICOR HIV-1 MONITOR test
| Sample | No. of HIV-1 RNA copies/ml
|
CV (%) | Log difference | |
|---|---|---|---|---|
| AMPLICOR | In-house RT-cPCR | |||
| 1 | 750,000 | 830,000 | 7.16 | −0.04 |
| 2 | 500,000 | 650,000 | 18.45 | −0.11 |
| 3 | 380,000 | 660,000 | 38.07 | −0.24 |
| 4 | 320,000 | 140,000 | 55.34 | +0.36 |
| 5 | 310,000 | 360,000 | 10.55 | −0.06 |
| 6 | 250,000 | 81,000 | 72.21 | +0.49 |
| 7 | 200,000 | 22,000 | 113.39 | +0.96 |
| 8 | 120,000 | 120,000 | 0.00 | +0.00 |
| 9 | 100,000 | 87,000 | 9.83 | +0.06 |
| 10 | 100,000 | 77,000 | 18.38 | +0.11 |
| 11 | 90,000 | 240,000 | 64.28 | −0.43 |
| 12 | 85,000 | 37,000 | 55.64 | +0.36 |
| 13 | 60,000 | 12,000 | 94.28 | +0.70 |
| 14 | 42,000 | 130,000 | 72.36 | −0.49 |
| 15 | 40,000 | 84,000 | 50.18 | −0.32 |
| 16 | 30,000 | 6,700 | 89.79 | +0.65 |
| 17 | 29,000 | 6,000 | 92.93 | +0.68 |
| 18 | 20,000 | 9,300 | 51.65 | +0.33 |
| 19 | 20,000 | 5,600 | 79.55 | +0.55 |
| 20 | 19,000 | 7,600 | 60.61 | +0.40 |
| 21 | 18,000 | 4,000 | 90.00 | +0.65 |
| 22 | 15,000 | 26,000 | 37.94 | −0.24 |
| 23 | 10,000 | 47,000 | 91.80 | −0.67 |
| 24 | 10,000 | 44,000 | 89.04 | −0.64 |
| 25 | 9,500 | 46,000 | 93.01 | −0.69 |
| 26 | 8,000 | 6,000 | 20.20 | +0.12 |
| 27 | 6,300 | 6,500 | 2.21 | −0.01 |
| 28 | 4,300 | 21,000 | 93.35 | −0.69 |
| 29 | 4,200 | 17,000 | 85.39 | −0.61 |
| 30 | 3,100 | 5,000 | 33.17 | −0.21 |
| 31 | 3,000 | 2,300 | 18.68 | +0.12 |
| 32 | 2,200 | 800 | 66.00 | +0.44 |
| 33 | 2,100 | 4,200 | 47.14 | −0.30 |
| 34 | 2,100 | 2,400 | 9.43 | −0.06 |
| 35 | 1,600 | 3,100 | 45.13 | −0.29 |
| 36 | 1,300 | 860 | 28.81 | +0.18 |
| 37 | 1,000 | 2,600 | 62.85 | −0.41 |
| 38 | 700 | 1,000 | 24.96 | −0.15 |
| 39 | <400 | <400 | NAa | NA |
| 40 | <400 | <400 | NA | NA |
| 41 | <400 | <400 | NA | NA |
| 42 | <400 | <400 | NA | NA |
| 43 | <400 | <400 | NA | NA |
| 44 | <400 | <400 | NA | NA |
| 45 | <400 | <400 | NA | NA |
| Mean ± SD | 52.47 ± 32.33 | 0.01 ± 0.44 | ||
NA, not applicable.
Since the amount of each RNA preparation obtained from plasma was sufficient for two quantitative RT-cPCR tests, we checked whether comparable results could be obtained by using the amount of RNA remaining after the first assay (i.e., after a second freezing-thawing). When 50 RNA samples first shown to contain 3,300 to 575,310 copies per ml of plasma were retested, the results from the first and second titrations were in excellent agreement, differing by <0.3 and <0.6 log unit for 42 (84%) and 50 (100%) samples, respectively. The mean ± SD difference was 0.25 ± 0.18 log unit, and there was no tendency for the second experiment to yield lower copy numbers than the first experiment (data not shown).
In-house RT-cPCR of 1,982 plasma samples did not produce wt amplification products for 188 (9.5%) samples (number of HIV-1 RNA copies, <400/ml). The standard yield from RNA extraction and RT for these samples was confirmed by successful amplification of the TMV control sequence with residual cDNA. Of the remaining 1,794 samples, the quantities of HIV-1 RNA in 501 (27.9%), 791 (44.1%), and 341 (19.0%) samples were determined by reading the ratio between the amounts of wt and competitor PCR products in four, three, and two lanes, respectively (Fig. 3). A single reading in the first or last lane was available for a total of 161 (9.0%) samples with exceedingly low (<1,000 copies/ml) or high (>750,000 copies/ml) HIV-1 RNA titers, respectively. The mean coefficients of correlation (R2) for the regression lines generated by measurement ratios for four and three lanes were 0.972 and 0.967, respectively. There was a significant inverse correlation between HIV-1 RNA titers and CD4 counts for the whole population analyzed (r = −0.22; P < 10−7; Spearman’s correlation) (Fig. 4). However, when samples were stratified on the basis of CD4 counts, HIV-1 RNA titers spanned at least two orders of magnitude in all groups. To check the capability of the in-house RT-cPCR to monitor decreases in HIV-1 viremia following effective antiretroviral therapy, 72 paired plasma samples obtained from 36 patients at week 0 and 4 with respect to the time of initiation of saquinavir (n = 12), indinavir (n = 12), or ritonavir (n = 12) therapy were analyzed. The baseline HIV-1 RNA titers in the plasma of the three groups examined were not different (mean values, 210,092, 207,970, and 194,555 copies/ml, respectively). This analysis showed a significant decrease in HIV-1 RNA levels in the whole population (P < 0.0001; Wilcoxon’s signed rank test) as well as in each of the separate arms treated with saquinavir (P = 0.0210), indinavir (P = 0.0047), and ritonavir (P < 0.0001) (Fig. 5). However, the mean decrease in the saquinavir group (0.914 log unit) was markedly lower than those in the indinavir (1.584 log units) and ritonavir (1.904 log units) groups.
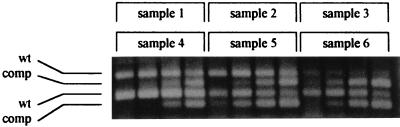
FIG. 3.
Representative quantitative analysis of plasma HIV-1 RNA by the in-house RT-cPCR assay. Competitive amplification products obtained from six clinical samples were loaded as subsequent three-sample series on the same gel with a 10-min delay. Each cDNA was amplified in the presence of 40, 240, 1,440, and 8,640 competitor RNA equivalents. wt and comp, the amplification products generated from wt and competitor templates, respectively. Data analysis for the samples indicated the following numbers of HIV-1 RNA copies per milliliter: 100,166 for sample 1, 138,086 for sample 2, 4,300 for sample 3, 546,999 for sample 4, 30,567 for sample 5, and 33,703 for sample 6.
FIG. 4.
Scatter plot showing the correlation between HIV-1 RNA levels and CD4 counts in the population whose plasma samples were analyzed by the in-house RT-cPCR assay. Samples with undetectable HIV-1 RNA levels (<400 copies/ml) are shown as containing 200 copies/ml.
FIG. 5.
Effect of saquinavir, ritonavir, and indinavir therapy on HIV-1 RNA load in three groups of 12 patients as measured by the in-house RT-cPCR assay. Each line represents a different subject. Weeks refer to the time from the initiation of protease inhibitor therapy. For graphical representation and statistical analysis, samples with <400 HIV-1 RNA copies/ml are considered to contain 200 copies/ml.
DISCUSSION
The in-house HIV-1 RNA quantitation procedure described here was directly adapted from a cPCR system previously used for the titration of DNA targets (28). Although presently replaced by the measurement of HIV-1 RNA levels, robust cPCR procedures were initially developed for the quantitation of HIV-1 DNA by several investigators (7, 8, 13, 15). While the routine clinical use of homemade RNA competitors may pose problems in terms of titration and long-term storage, DNA competitors can be easily and reliably titrated and stored indefinitely (3). Thus, a practical and reliable competitive DNA PCR system can be conveniently used to quantitate HIV-1 RNA, provided that reproducible methods are used for plasma RNA extraction and RT. Effective procedures for both RNA extraction and RT have been assembled in the laboratory and have previously been reported as preliminary steps in successful in-house RT-cPCR assays (16, 17). Commercial RNA spin columns have successfully been evaluated as a means of extraction of RNA from plasma for clinical diagnosis (12, 24). It must be remembered that heparin should not be used as an anticoagulant for blood samples that are to be processed with spin columns, since treatment of the eluate with heparinase may be then required to achieve optimal RT and PCR (12). The use of a commercial ready-to-use system is generally advisable to avoid a number of operator-dependent variables and to increase the uniformity of the reagents, resulting in improved reproducibility. In our hands, commercial RNA spin columns and ready-to-use RT beads proved to fulfill this requirement, resulting in a fairly constant rate of generation of cDNA from the RNA in plasma.
Once the noncompetitive RNA extraction and RT steps have been firmly standardized, there is the need to calculate the rate of cDNA production from plasma RNA. Construction and accurate quantitation of an RNA standard and measurement of the cDNA generated may provide a direct calculation of such a rate. Alternatively, a practical method for estimating the yield from RNA extraction and RT is to test a panel of samples whose HIV-1 RNA titers have been quantitated with a reference commercial kit and adjust empirically the amounts of competitor DNA in order to obtain the same HIV-1 RNA titers. We have been successful with this simple approach when we used a second pair of primers and the appropriate deleted competitor DNA (data not shown).
Comparison with a reference commercial assay is strongly recommended as part of an evaluation of any in-house procedure. The intertest CV calculated for the in-house assays (41.9%) was almost identical to that reported for the Roche AMPLICOR HIV-1 MONITOR test (23). Parallel testing of a panel of plasma samples indicated that the results obtained by the in-house assay and the commercial AMPLICOR HIV-1 MONITOR test are in good agreement. The differences between the two procedures were comparable to those reported in comparative studies of licensed commercial kits (5, 20, 23), and HIV-1 RNA was undetectable in the same set of samples, confirming that the two systems have similar thresholds of sensitivity. Preliminary data suggest that use of the whole RT mixture as the template for a one-tube cPCR in the in-house assay may allow quantitation of samples containing as few as 80 copies/ml, although with a larger intertest variability (70%, as calculated by quintuplicate tests with a reconstructed plasma sample; data not shown). This is similar to what has been recently reported with the new ultrasensitive commercial assays (4, 11, 18, 22) and may be of relevance in light of the need for the monitoring of HIV-1-infected patients whose plasma HIV-1 RNA titer is low. Since the RNA extraction protocol yields material sufficient and suitable for two RTs, a sample with RNA levels below the threshold of 400 copies/ml could be directly retested by using the whole cDNA in a second one-tube cPCR, avoiding the need for a new RNA extraction. A more attractive target is a single system that allows the quantitation of exceedingly low and high HIV-1 RNA levels over a large dynamic range, obviating the need to use an ultrasensitive assay as a second-line test after a negative standard test. Primer labels that increase the sensitivity of the detection phase and procedures for concentration of the RNA in plasma are being investigated in this context.
A clinical field evaluation of the in-house system was carried out by testing a total of 1,982 plasma samples. The HIV-1 RNA levels obtained are in the same range as those commonly reported by reference tests (5, 20, 23). An inverse correlation between CD4 counts and HIV-1 RNA levels with a wide scattering of values was found, in agreement with observations previously obtained by licensed assays (10, 15). Finally, short-term follow-up of the HIV-1 load in patients who were shifted to protease inhibitor therapy demonstrated the capability of the in-house system to monitor antiretroviral treatment in vivo. The extents of the decreases in HIV-1 RNA levels obtained as a result of saquinavir, ritonavir, and indinavir therapy were in agreement with previously reported data (2, 6, 21).
A reliable in-house assay must also be practical in order to be used as a routine means of clinical monitoring. In the in-house procedure described here, RNA extraction and RT are simple to perform and PCR is carried out in a microtiter plate, allowing the convenient use of multichannel pipettes during both preparation and analysis. Hot start of amplification is easily accomplished by multichannel dispensing of wax kept liquid on a heat block inside the laminar flow hood under which the PCR mixture is prepared. An engineered thermolabile UNG is absolutely effective in protecting from carryover without interfering with the generation of PCR products after irreversible inactivation. Notably, multiple ready-to-use plates containing all reagents except cDNA can be stored at 4°C and successfully used up to 2 weeks later, saving most of the time required for each RT-cPCR run. Inclusion of an additive for gel loading (tartrazine-Ficoll) directly in the PCR mixture also significantly speeds the analytic phase. The reaction mixtures are indeed directly loaded with a multichannel pipette and only two two-comb agarose gels are used. Measurement of band intensity requires only a 256-level grayscale optical scanner and minimal densitometric analysis software. Data analysis can be conveniently automated with simple macro commands in popular spreadsheets such as MS Excel, version 5.0 or higher (Microsoft Corp., Redmond, Wash.). The time required to complete a test run is 7 to 8 h, less than 40% of which is hands-on time. In our experience, a single operator may well be capable of measuring the HIV-1 RNA levels in 100 samples per week. This figure can be actually doubled provided that two microcentrifuges and two thermal cyclers are available for RNA extraction and amplification, respectively. An interlaboratory evaluation of the system is in progress.
Assays for the routine medium-scale monitoring of HIV-1 RNA levels in HIV-1-infected subjects should be reliable, practical, and cost-effective. The in-house assay described here is simple to develop and has been shown to be reliable both in reconstruction experiments and for the clinical use in the field. Although not presently available as a complete kit, none of the steps of the procedure requires complex equipment, and all steps can be rapidly learned and routinely used by ordinary laboratory operators. The cost is less than $10 per sample, including the costs for both reagents and disposable supplies. This figure compares with the $60 to $100 per sample required by currently licensed commercial kits. Notably, the cost initially required for additional equipment for data analysis (scanner and densitometric software) translates into only a <10% cost increase for the first 1,000 samples analyzed. This and other in-house systems for the measurement of plasma HIV-1 RNA levels may thus be relevant for increasing both the numbers of subjects analyzed and the frequency of analysis, at least until a significant reduction of the costs of reference methods can be achieved.
ACKNOWLEDGMENTS
This study was supported by Progetto AIDS, Istituto Superiore di Sanità, Ministero della Sanità (9402-15), Rome, Italy. M. Catucci, G. Venturi, and A. De Milito are recipients of AIDS fellowships from the Istituto Superiore di Sanità, Ministero della Sanità.
REFERENCES
- 1.Bagnarelli P, Menzo S, Valenza A, Paolucci S, Petroni S, Scalise G, Sampaolesi R, Manzin A, Varaldo P E, Clementi M. Quantitative molecular monitoring of human immunodeficiency virus type 1 activity during therapy with specific antiretroviral compounds. J Clin Microbiol. 1995;33:16–23. doi: 10.1128/jcm.33.1.16-23.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisset L R, Rothen M, Joller-Jemelka H I, Dubs R W, Grob P J, Opravil M. Change in circulating levels of the chemokines macrophage inflammatory proteins 1 alpha and 11 beta, RANTES, monocyte chemotactic protein-1 and interleukin-16 following treatment of severely immunodeficient HIV-infected individuals with indinavir. AIDS. 1997;11:485–491. doi: 10.1097/00002030-199704000-00012. [DOI] [PubMed] [Google Scholar]
- 3.Clementi M, Menzo S, Bagnarelli P, Manzin A, Valenza A, Varaldo P E. Quantitative PCR and RT-PCR in virology. PCR Methods Appl. 1993;2:191–196. doi: 10.1101/gr.2.3.191. [DOI] [PubMed] [Google Scholar]
- 4.Collins M L, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen L P, Kolberg J, Bushnell S, Urdea M S, Ho D D. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–2984. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coste J, Montes B, Reynes J, Peeters M, Segarra C, Vendrell J P, Delaporte E, Segondy M. Comparative evaluation of three assays for the quantitation of human immunodeficiency virus type 1 RNA in plasma. J Med Virol. 1996;50:293–302. doi: 10.1002/(SICI)1096-9071(199612)50:4<293::AID-JMV3>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 6.Danner S A, Carr A, Leonard J M, Lehman L M, Gudiol F, Gonzales J, Raventos A, Rubio R, Bouza E, Pintado V. A short-term study of the safety, pharmacokinetics, and efficacy of ritonavir, an inhibitor of HIV-1 protease. N Engl J Med. 1995;333:1528–1533. doi: 10.1056/NEJM199512073332303. [DOI] [PubMed] [Google Scholar]
- 7.Ferre F, Marchese A L, Griffin S L, Daigle A E, Richieri S P, Jensen F C, Carlo D J. Development and validation of a polymerase chain reaction method for the precise quantitation of HIV-1 DNA in blood cells from subjects undergoing a 1-year immunotherapeutic treatment. AIDS. 1993;7:s21–s27. doi: 10.1097/00002030-199311002-00006. [DOI] [PubMed] [Google Scholar]
- 8.Gupta P, Ding M, Cottrill M, Rinaldo C, Kingsley L, Wolinsky S, Mellors J. Quantitation of human immunodeficiency virus type 1 DNA and RNA by a novel internally controlled PCR assay. J Clin Microbiol. 1995;33:1670–1673. doi: 10.1128/jcm.33.6.1670-1673.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hammer S M. Advances in antiretroviral therapy and viral load monitoring. AIDS. 1996;10:s1–s11. [PubMed] [Google Scholar]
- 10.Katzenstein D A, Hammer S M, Hughes M D, Gundacker H, Jackson J B, Fiscus S, Rasheed S, Elbeik T, Reichman R, Japour A, Merigan T C, Hirsch M S. The relation of virologic and immunologic markers to clinical outcomes after nucleoside therapy in HIV-infected adults with 200 to 500 CD4 cells per cubic millimeter. N Engl J Med. 1996;335:1091–1098. doi: 10.1056/NEJM199610103351502. [DOI] [PubMed] [Google Scholar]
- 11.Kern D, Collins M, Fultz T, Detmer J, Hamren S, Peterkin J J, Sheridan P, Urdea M, White R, Yeghiazarian T, Todd J. An enhanced-sensitivity branched-DNA assay for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3196–3202. doi: 10.1128/jcm.34.12.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin H J, Tanwandee T, Hollinger F B. Improved methods for quantification of human immunodeficiency virus type 1 RNA and hepatitis C virus RNA in blood using spin column technology and chemiluminescent assays of PCR products. J Med Virol. 1997;51:56–63. [PubMed] [Google Scholar]
- 13.Mallet F, Hebrard C, Livrozet J M, Lees O, Tron F, Touraine J L, Mandrand B. Quantitation of human immunodeficiency virus type 1 DNA by two PCR procedures coupled with enzyme-linked oligosorbent assay. J Clin Microbiol. 1995;33:3201–3208. doi: 10.1128/jcm.33.12.3201-3208.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellors J W, Munoz A, Giorgi J V, Margolick J B, Tassoni C J, Gupta P, Kingsley L A, Todd J A, Saah A J, Detels R, Phair J P, Rinaldo C R., Jr Plasma viral load and CD4+ lymphocytes as prognostic markers of HIV-1 infection. Ann Intern Med. 1997;126:946–954. doi: 10.7326/0003-4819-126-12-199706150-00003. [DOI] [PubMed] [Google Scholar]
- 15.Mellors J W, Rinaldo C R, Jr, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 16.Michael N L, Mo T, Merzouki A, O’Shaughnessy M, Oster C, Burke D S, Redfield R R, Birx D L, Cassol S A. Human immunodeficiency virus type 1 cellular RNA load and splicing patterns predict disease progression in a longitudinally studied cohort. J Virol. 1995;69:1868–1877. doi: 10.1128/jvi.69.3.1868-1877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitra D, Laurence J. Quantitative RT-PCR for human Fas (CD95) expression. BioTechniques. 1997;22:442–446. doi: 10.2144/97223bm14. [DOI] [PubMed] [Google Scholar]
- 18.Mulder J, Resnick R, Saget B, Scheibel S, Herman S, Payne H, Harrigan R, Kwok S A. A rapid and simple method for extracting human immunodeficiency virus type 1 RNA from plasma: enhanced sensitivity. J Clin Microbiol. 1997;35:1278–1280. doi: 10.1128/jcm.35.5.1278-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piatak M, Jr, Luk K C, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–81. [PubMed] [Google Scholar]
- 20.Revets H1, Marissens D, de Wit S, Lacor P, Clumeck N, Lauwers S, Zissis G. Comparative evaluation of NASBA HIV-1 RNA QT, AMPLICOR-HIV monitor, and QUANTIPLEX HIV RNA assay, three methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:1058–1064. doi: 10.1128/jcm.34.5.1058-1064.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schapiro J M, Winters M A, Stewart F, Efron B, Norris J, Kozal M J, Merigan T C. The effect of high-dose saquinavir on viral load and CD4+ T-cell counts in HIV-infected patients. Ann Intern Med. 1996;124:1039–1050. doi: 10.7326/0003-4819-124-12-199606150-00003. [DOI] [PubMed] [Google Scholar]
- 22.Schockmel G A, Yerly S, Perrin L. Detection of low HIV-1 RNA levels in plasma. J Acquired Immune Defic Syndr Hum Retrovirol. 1997;14:179–183. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 23.Schuurman R, Descamps D, Weverling G J, Kaye S, Tijnagel J, Williams L, van Leeuwen R, Tedder R, Boucher C A, Brun-Vezinet F, Loveday C. Multicenter comparison of three commercial methods for quantification of human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 1996;34:3016–3022. doi: 10.1128/jcm.34.12.3016-3022.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shafer R W, Levee D J, Winters M A, Richmond K L, Huang D, Merigan T C. Comparison of QIAamp HCV kit spin columns, silica beads, and phenol-chloroform for recovering human immunodeficiency virus type 1 RNA from plasma. J Clin Microbiol. 1997;35:520–522. doi: 10.1128/jcm.35.2.520-522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trabaud M A, Audoly G, Leriche K, Cotte L, Ritter J, Sepetjan M, Trepo C. Development of a reverse transcriptase PCR-enzyme-linked immunosorbent assay for quantification of human immunodeficiency virus type 1 RNA in plasma: comparison with commercial quantitative assays. J Clin Microbiol. 1997;35:1251–1254. doi: 10.1128/jcm.35.5.1251-1254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittwer C T, Garling D J. Rapid cycle DNA amplification: time and temperature optimization. BioTecheniques. 1991;10:76–83. [PubMed] [Google Scholar]
- 27.Zazzi M, Catucci M, De Milito A, Romano L, Venturi G, Almi P, Gonnelli A, Rubino M, Valensin P E. Zidovudine resistance mutations and human immunodeficiency virus type 1 DNA burden: longitudinal evaluation of six patients under treatment. Infection. 1996;24:419–425. doi: 10.1007/BF01713041. [DOI] [PubMed] [Google Scholar]