Abstract
Gouty arthritis is a very familiar inflammatory arthritis. Controlling inflammation is the key to preventing gouty arthritis. However, colchicine, the most highly represented drug used in clinical practice, has strict contraindications owing to some severe side effects. Curcumin (Cur), a natural anti-inflammatory drug, has demonstrated good safety and efficacy. However, the rapid degradation, poor aqueous solubility, and low bioavailability of Cur limit its therapeutic effect. To strengthen the effectiveness and bioavailability of Cur. Cur loaded tetrahedral framework nucleic acids (Cur-TFNAs) were synthesized to deliver Cur. Compared with free Cur, Cur-TFNAs exhibit a preferable drug stability, good biocompatibility (CCK-8 assay), ease of uptake (immunofluorescence), and higher tissue utilization (in vivo biodistribution). Most importantly, Cur-TFNAs present better anti-inflammatory effect than free Cur both in vivo and in vitro experiments through the determination of inflammation-related cytokines expression. Therefore, we believe that Cur-TFNAs have great prospects for the prevention of gout and similar inflammatory diseases.
Keywords: Tetrahedral framework DNA nanoparticle, Oxidative stress, NF-κB signaling pathway, Macrophage, Gouty arthritis
Graphical abstract
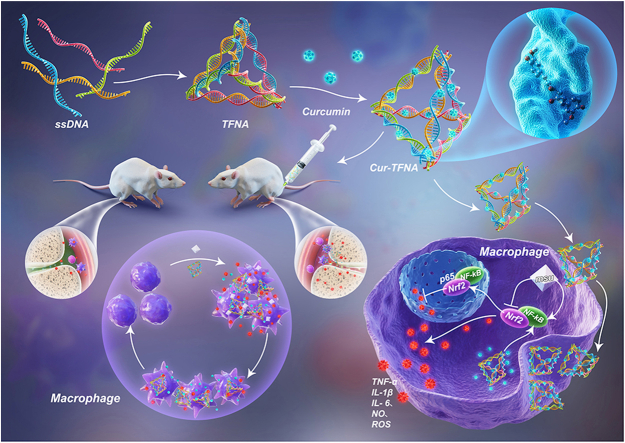
Schematic illustration of treating acute gouty arthritis with Cur loaded TFNAs. Cur-TFNAs were absorbed by inflammatory RAW264.7 cells and inhibited the activation of NF-κB, which decreased the levels of ROS, NO and inflammatory factors (IL-6, IL-1β and TNF-α), thus affecting the progression of acute gouty arthritis.
Highlights
-
•
The drug curcumin system based on DNA nanostructures (Cur-TFNAs) were developed to obtain a novel nanomaterial with high water solubility, large encapsulation efficiency, sustained drug release and excellent drug stability.
-
•
Based on cellular uptake and in vivo drug imaging, Cur-TFNAs improve the retention of Cur in cells and tissues.
-
•
Cur-TFNAs prevent acute gouty arthritis through mediating anti-inflammatory and antioxidant responses of macrophage.
1. Introduction
Gout, a complex and common inflammatory arthritis in adults, is caused by the accumulation of monosodium urate (MSU) crystals and affects approximately 3–6% of the adult population in some western countries [1,2]. When MSU crystals are deposited in the joint, an acute gout attack may occur, which manifests as extreme pain, swelling, fever, and difficulty in moving the affected joints. Macrophages are considered to be important cells that initiate and drive inflammation caused by MSU crystals [3,4]. Namely, macrophages phagocytize MSU crystals and release considerable inflammatory-related factors such as interleukin (IL)-6, IL-1β and tumor necrosis factor (TNF)-α, which recruit neutrophils to initiate an inflammatory cascade [5]. Many mechanisms have been proposed to solve acute gout. And, the very prominent one is the anti-inflammatory response of macrophages to MSU crystals [6]. Given these mechanisms, anti-inflammatory drugs are the top option to control acute inflammation quickly and effectively. At present, nonsteroidal anti-inflammatory drugs (NSAIDs), corticosteroids, and colchicine are usually used to block the acute gouty attacks and can effectively relieve inflammatory symptoms well in general. While their systemic use has severe side effects [7]. Colchicine easily causes a wide range of gastrointestinal toxicities, damages liver metabolism and liver intestinal circulation, and slows drug metabolism, simultaneously,it has dose-dependent toxicity. In addition, NSAIDs have similar side effects [8]. Hence, other effective and safe anti-inflammatory drugs are being explored to prevent acute gout.
Cur, a natural polyphenol compound, is derived from the Curcuma longa and has numerous physiological and pharmacological characteristics, such as anticancer, antibacterial, antioxidant, anti-rheumatism, and anti-inflammatory activities [9,10]. Most importantly, it has minimal acute or chronic toxicity and can be used for a long time [9]. The use of Cur has been documented for treating various common diseases, including inflammatory diseases, tumor, wound infection, etc. [11] Cur has been used as a conventional drug for treating arthritis. Past studies have reported that Cur can upregulate heme oxygenase-1 (HO-1) and nuclear factor E2-related factor 2 (Nrf2) to scavenge reactive oxygen species (ROS), showing excellent antioxidant properties to protect tissues [12,13]. Furthermore, Cur can inhibit the activation of the NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome, which caused by nuclear factor-κ-gene binding (NF-κB) activation. By inhibiting the activation of NF-κB, Cur play an antitumor and anti-inflammatory role [14,15]. Cur has been verified to play a non-negligible role in various chronic inflammatory diseases and possesses many advantages, such as easy achievement, low cost, and pharmacological safety. Thus, it is easily accepted by most people.
Unfortunately, the clinical application of Cur is not ideal. Cur has limited bioavailability owing to rapid degradation and elimination, extremely poor aqueous solubility, inadequate tissue absorption, which severely restricts its therapeutic efficacy [[16], [17], [18]]. Therefore, the difficulty and hot spot of research is to improve stability, cellular uptake, and bioavailability of Cur through delivery systems. To date, metal complexes, polypeptides, solid lipids, and biodegradable polylactic glycolic acid (PLGA) nanoparticles have been used for Cur delivery [[18], [19], [20], [21]]. However, they all have some disadvantages, such as the slow efficiency of the drug release and the toxicity [22,23]. Therefore, it is urgent to develop a delivery system with high biocompatibility. As a drug carrier, DNA nanostructures have become a hot research object, ascribed to their incomparable advantages in biocompatibility, structural stability, and programmability [[24], [25], [26]]. DNA nanomaterials can carry oligonucleotides and small molecule drugs through complementary hybridization, direct covalent extension, and insertion into DNA duplex, which are not only simple in synthesis but also high in yield [27]. Compared with other complex DNA structures, TFNAs are considered to be one of the easiest to synthesize with higher stability [25]. TFNAs are not only easier to be absorbed by cells through simple endocytosis due to the unique spatial structure, but also stable for 48 h in living cells [28]. Moreover, TFNAs have successfully carried a variety of small molecular materials, peptides, DNA enzymes, and other molecules through electrostatic adsorption [[29], [30], [31], [32]]. Hence, it is very desirable to select TFNAs to deliver Cur to obtain better stability, and higher bioavailability of Cur-TFNAs. Therefore, in view of the excellent anti-inflammatory effect and increased biological safety, Cur-TFNAs were firstly explored the anti-inflammatory ability in vitro and then applied to gout in vivo.
2. Material and methods
2.1. Synthesis of Cur-TFNAs
Four prepared ssDNAs were dissolved at the same concentration in enzyme-free water. Then, ssDNAs and TM buffer (50 mM MgCl2 and 10 mM Tris-HCl, pH 8.0) were mixed. TFNAs were successfully synthesized by heating (95 °C, 10 min) and then cooling (4 °C, 20 min) [33]. Cur was dissolved in DMSO in advance. Cur (40 μM–160 μM) was then added to the TFNA solution. According to previous experiments, the final concentration of TFNAs is 200 nM [34]. Based on previous research, Cur-loaded TFNAs was synthesized by shaking the mixture for 6 h at ambient temperature [35]. Then, the residual Cur was removed by centrifugation with a 30 kDa ultrafiltration tube. The absorbance of Cur was detected at a wavelength of 425 nm. The encapsulation and binding of Cur in Cur-TFNAs were further studied by spectral analysis. The encapsulation efficiency of Cur was calculated according to the following formula:
2.2. Characterization of Cur-TFNAs
For verification of the successful synthesis of Cur-TFNAs as described previously, transmission electron microscopy (TEM) and polyacrylamide gel electrophoresis (PAGE) were selected to measure the shape and size of the nanoparticles. The zeta potential and average size were determined to analyze the size and stability of Cur-TFNAs by dynamic light scattering (DLS).
2.3. In vitro release kinetics
The synthesized Cur-TFNAs (40 μM, 50 mL) was added into a dialysis bag (30 kDa; Solarbio, Beijing, China). The dialysis bag was put into a 50 mL release medium, namely, PBS (pH 7.4, 1% (v/v) Tween-80; 37 °C, 150 rpm). Next, 2 mL of solution was separately taken out at the pre-designed time points to obtain the OD at a wavelength of 425 nm.
2.4. Study on solubility and stability of Cur
The equivalent Cur-TFNAs and Cur were dissolved in PBS (0.01 M, pH 7.4) to observe the water solubility of the preparation [36]. Natural Cur and Cur-TFNAs with a final fixed concentration of about 40 μM were incubated in a 150 rpm rotating oscillator at 37 °C for 6 h. Besides, 200 μL of the solution was taken from the total solution at the pre-designed time points of 1 h–6 h. The Cur content was determined according to the above method.
2.5. Uptake of Cy5-loaded Cur-TFNAs by RAW264.7 cells
After adherent growth, the RAW264.7 cells were incubated with 20 μM Cur, Cy5-loaded TFNAs (Cy5-TFNAs, 100 nM) and Cy5-loaded Cur-TFNAs (Cy5-Cur-TFNAs, 20 μM) in 12-well plates with cell slides for 4 h. After rinsing cells thrice with PBS, the cells were fixed in cold polyoxymethylene for 15 min. After washing the cells again, the nuclei were stained with DAPI for 10 min. Finally, the images were obtained by confocal microscopy (N-SIM, Nikon, Tokyo, Japan). In addition, the results of cell entry by a flow cytometer (FC500 Beckman, IL, USA).
2.6. ELISA
To measure the contents of TNF-α and IL-6 in the cell supernatant, we used ELISA kits (MULTI SCIENCES, Hangzhou, China). The supernatants were collected. After washing the plate 6 times, the samples and the diluted standard product were added to the plates and incubated at 37 °C for 1 h. Subsequently, after washing the plate, 50 μL enzyme labeling reagent was added, and the samples were incubated again for 30 min. Finally, after washing once again, the developer and TMB terminator were added at an interval of 10–30 min. The results were obtained in 15 min.
2.7. Immunofluorescence
After being treated as described before, the cells were soaked in cold 4% paraformaldehyde solution for 15 min and 0.5% Triton X-100 for 10 min. After rinsing them again, the samples were blocked in goat serum for 1 h. Then, after washing them again, the samples were incubated in a diluted antibody against CD68 (1:100), Nrf2 (1:250), IL-1β (1:50), iNOS (1:500), TNF-α (1:500), IL-6 (1:500), and HO-1 (1:250) overnight at 4 °C. On the second day, after washing them again, the samples were incubated with the relevant secondary antibody (1:500) for 1 h. Next, the samples were washed again, which was followed by immersion in FITC phalloidin for 20 min and DAPI for 10 min to stain the cytoskeleton and nucleus. Finally, immunofluorescence images were obtained by confocal laser microscopy.
2.8. Western blotting
Western blotting was performed to quantify the protein levels of TNF-α, IL-6, IL-1β, iNOS, Nrf2, HO-1, and NF-κB p65. The total proteins of RAW264.7 cells were extracted and prepared to obtain protein samples as described before [37]. Briefly, the concentrations of proteins were detected, the loading buffer was added, and then the protein samples were mixed and boiled. Next, after isolating of proteins and transferring and blocking of membrane, the protein strips were immersed in the primary antibody at 4 °C overnight. On the second day, after rewarming them for 1 h, the strips were incubated with the relevant secondary antibody (1:2000; Beyotime, Shanghai, China). TBST was used to rinse the strips thrice (15 min/times) throughout the whole process. On the last, the strips were exposed.
2.9. Establishment of the Gout model in mice
Adult CD-1 mice (male, 18–22 g, 6–8weeks old) were provided by Dossy (Chengdu, China). Under chloral hydrate anesthesia, the right ankle joints of mice were injected with MSU crystal (0.5 mg, 20 μL PBS), and the left ankle joints of mice were only injected with 20 μL PBS. To determine the preventive effect, the ankle joints were pretreated with normal saline (NS), Cur, TFNAs, and Cur-TFNAs 1 h before the injection of MSU crystals. The ankle joint diameter was measured with a vernier caliper at the beginning and 24 h later. This study meets the requirements of the ethics committee of the Department of Experimental Animal Science, Sichuan University.
2.10. In vivo biodistribution of Cur and Cur-TFNAs
To determine the distribution of Cur and Cur-TFNAs in vivo, adult CD-1 mice (male, 18–22 g, 6–8weeks old) were selected. According to previous studies, the mice were anesthetized with isoflurane and given Cur and Cur-TFNAs, respectively, at the same level as that injected into the ankle joint of the mice [19]. Images were obtained using an IVIS (Bio Real Quick View 3000, Austria).
2.11. Statistical analysis
The data contains mean ± standard deviation (SD). The data of two groups were statistically significant (P < 0.05) through a one-tailed Student's t-test (SPSS 19.0; IBM, Armonk, NY).
3. Results and discussion
3.1. Synthesis and characterization of Cur-TFNAs
Cur was widely used since it was discovered. So far, the transition of Cur from basic experiments to clinical application has not achieved great success [36]. On the one hand, Cur shows low structural stability and is easy to be hydrolyzed or biotransformation without the protection of the drug delivery system; on the other hand, because of its hydrophobic property, Cur has very low water solubility and cannot be easily absorbed by cells and tissues [18,25]. Cur requires the help of drug carriers. However, owing to cationic surface charges, these organic carrier materials are inherently cytotoxic [38]. Although inorganic materials have been greatly improved in drug delivery, they also have inherent cytotoxicity [39,40]. In addition of some inherent toxicity and low biocompatibility, these organic or inorganic carrier materials also have some nonnegligible disadvantages. As a natural material in all organisms, DNA nanostructures have fully proved excellent biocompatibility [41]. Due to the clear structure and high addressing attribute, TFNAs have high cargo capacity. Compared with organic or inorganic nanomaterials, TFNAs can precisely control the valence and position of molecules [42]. Besides, TFNAs are the simplest and relatively stable DNA nanostructures among most of DNA origami materials [43]. Therefore, TFNAs were chosen to deliver Cur.
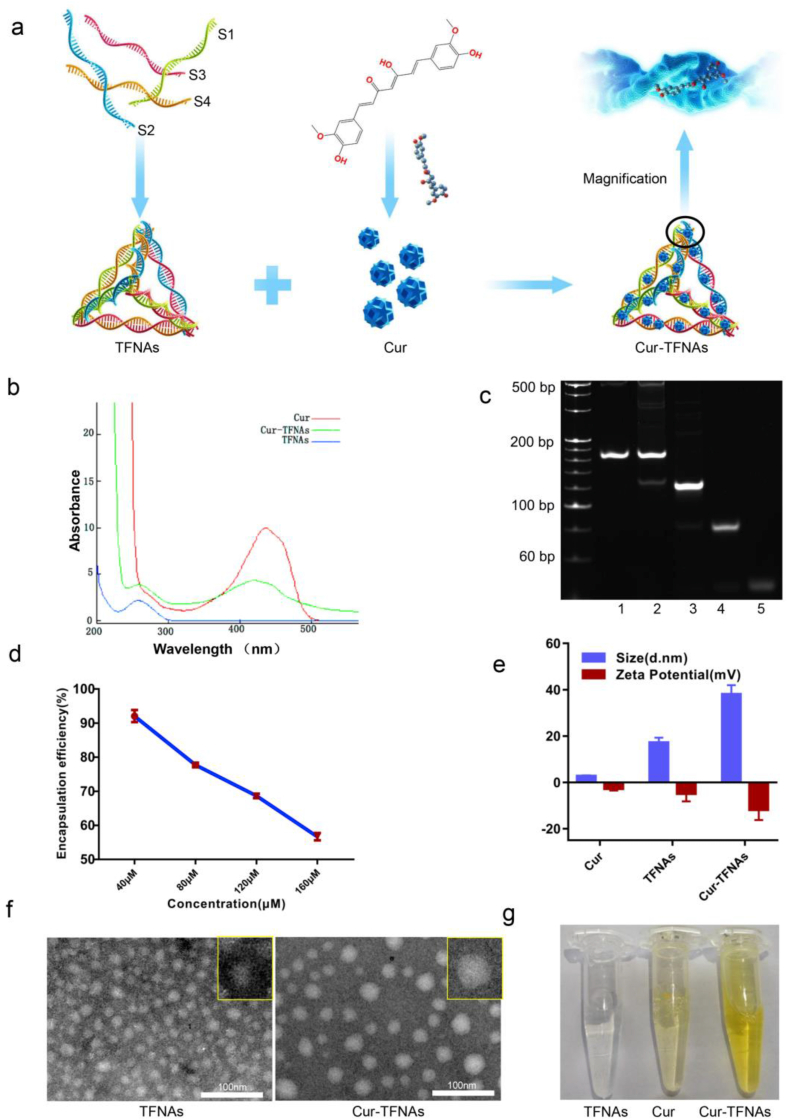
First of all, TFNAs were composed of four paired ssDNAs according to the principle of base pairing, as shown in Table 1. By incubation with TFNAs, Cur was loaded onto TFNAs to form Cur-TFNAs, as shown in Fig. 1a. Cur and its analogs have been confirmed to bind to DNA through thymine O2 and guanine N7 [44]. Then, the photophysical properties were determined to confirm the successful encapsulation and binding of Cur in TFNAs. As illustrated in Fig. 1b, by scanning the Ultraviolet–Visible absorbance spectra of Cur, it revealed that the natural Cur in DMSO solution exhibits an obvious high absorption peak near 425 nm. The absorbance peak value of Cur-TFNAs was close to that of natural Cur, which confirmed the successful encapsulation of Cur in TFNAs [36]. Next, the PAGE result (Fig. 1c) was obtained to verify the synthesis of the material. It indicated that TFNAs were not only successfully fabricated, but not affected by Cur. To determine the proportion of Cur-carrying TFNAs, the encapsulation efficiency of different proportions of components was measured, as shown in Fig. 1d. Based on the previous experiments, the concentration of TFNAs was fixed at 200 nM. It can be seen that the encapsulation efficiency of the loading of Cur onto TFNAs shows a declining curve. And the relatively best encapsulation efficiency (92.08 ± 0.89%) achieved for the loading of 40 μM Cur onto 200 nM TFNAs. Considering that the encapsulation efficiency of 40 μM Cur to 200 nM TFNAs is significantly better than that of other components (77.74%, 68.65%, and 56.67%). We chose 40 μM Cur and 200 nM TFNAs to synthesize Cur-TFNAs for the subsequent experiments [45].
Table 1.
Base sequence of ssDNAs.
| DNA | Base sequence |
|---|---|
| S1 | 5′-ATTTATCACCCGCCATAGTAGACGTATCACC-3′ |
| AGGCAGTTGAGACGAACATTCCTAAGTCTGAA-3′ | |
| S2 | 5′-ACATGCGAGGGTCCAATACCGACGATTACA-3′ |
| GCTTGCTACACGATTCAGACTTAGGAATGTTCG-3′ | |
| S3 | 5′-ACTACTATGGCGGGTGATAAAACGTGTAGCA-3′ |
| AGCTGTAATCGACGGGAAGAGCATGCCCATCC-3′ | |
| S4 | 5′-ACGGTATTGGACCCTCGCATGACTCAACTGC-3′ |
| CTGGTGATACGAGGATGGGCATGCTCTTCCCG-3′ | |
| Cy5-S1 | 5′-Cy5-ATTTATCACCCGCCATAGTAGACGTATCAC-3′ |
| CAGGCAGTTGAGACGAACATTCCTAAGTCTGAA-3′ |
Fig. 1.
Synthesis and characteristics of TFNAs and Cur-TFNAs. (a) A structure diagram showing the composition of Cur-TFNAs. (b) Ultraviolet–Visible absorbance spectra of native Cur and Cur-TFNAs at a fixed concentration of 40 μM. (c) PAGE graph showing the successful synthesis of TFNAs (1: Cur-TFNAs, 2: TFNAs, 3: S1+S2+S3, 4: S1+S2, 5: S1). (d) The curve of encapsulation efficiency of Cur loaded on TFNAs (n = 4). (e) Molecular size and zeta potential of Cur, TFNAs, and Cur-TFNAs measured by DLS (n = 4). (f) TEM image showing successfully synthesized TFNAs, and Cur-TFNAs. (g) Images of TFNAs, Cur, and Cur-TFNAs.
Then, we detected the size and zeta potential of Cur, TFNAs, and Cur-TFNAs by DLS. As shown in Fig. 1e, the approximate size of TFNAs and Cur-TFNAs was respectively 17.17 ± 2.17 nm and 38.04 ± 3.95 nm; especially, it indicated a single peak size distribution with narrow monodisperse. Besides, the zeta potentials show the stability of nanoparticles [46]. After integration, Cur-TFNAs manifested a larger negative zeta potential, which not only contributes to the permeability and retention of Cur in tissues but also affects the stability of Cur [47]. Then, the morphology of TFNAs and Cur-TFNAs were characterized by TEM. According to the TEM images (Fig. 1f), both TFNA (~10 nm) and Cur-TFNA (~20 nm) present the discrete spherical profile with a single size distribution. Not only that, the difference visible to the naked eye between synthetic Cur-TFNAs and free Cur is shown in Fig. 1g. Cur-TFNAs appeared as a clear yellow liquid, while free Cur appeared as a lightly yellow liquid, and insoluble Cur could also be observed. The color of the Cur-TFNAs was significantly different from that of Cur. In particular, compared with the precipitation of Cur alone, due to its low water solubility, there is no solid matter precipitated in the Cur-TFNA solution, and the Cur-TFNAs are well dispersed in the aqueous solution [36].
3.2. The stability and cellular uptake of Cur-TFNAs
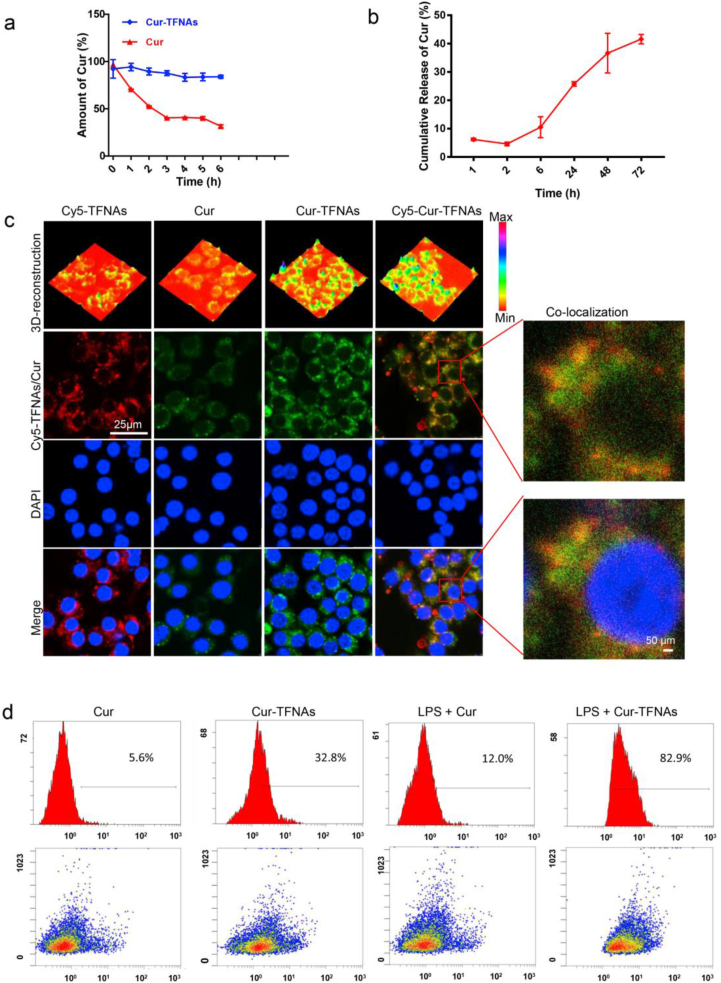
Previous results have confirmed that TFNAs increase the stability and bioavailability of Cur, leading to an increase in the uptake of Cur by macrophages. In addition, because of their special and highly ordered structure, TFNAs can protect molecular from degradation of the enzymes and increase the stability of small molecular [31]. Therefore, the drug stability experiment was further detected. As illustrated in Fig. 2a, the Cur-TFNAs and Cur were placed in PBS (pH:7.4, 37 °C, and 150 rpm) [36]; less than 20% of Cur in Cur-TFNAs was degraded in the first 6 h; approximately 60% Cur alone has been rapidly degraded in the first 3 h. Next, we measured the release of Cur from Cur-TFNAs. It can be observed from Fig. 2b that Cur slowly releases from Cur-TFNAs, and the release rate is basically maintained at about 40%. At present, although the mechanisms of DNA protecting Cur are not clear, it is undeniable that slow-release can further avoid the rapid degradation of Cur. We speculate that the combination of Cur and TFNAs avoids the structure of Cur completely exposed, thus preventing Cur from decomposition or biotransformation.
Fig. 2.
The stability and cellular uptake of Cur-TFNAs. (a) Stability of Cur (native and Cur-TFNAs in PBS (37 °C, pH 7.4; n = 4). (b) In vitro release kinetics of Cur from Cur-TFNAs formulation in PBS (37 °C, pH 7.4; n = 4). (c) Uptake of Cy5-TFNAs, Cur and cy5-Cur-TFNAs in RAW264.7 cells (Cur: green; Cy5-TFNAs: red; nuclear: blue). Scale bars are 25 μm and 50 μm. (d) Cellular uptakes of Cur and Cur-TFNAs in RAW264.7 cells detected by flow cytometry. Statistical analysis: *p < 0.05, **p < 0.01, and ***p < 0.001.
Then, to further determine whether TFNAs help more Cur enter RAW264.7 cells, immunofluorescence and flow cytometry were used to monitor cellular uptake. The autofluorescence of Cur and fluorescence of Cy5 labeled TFNAs were used to visually observe the entry of materials. As shown in Fig. 2c, the fluorescence of Cur-TFNAs was significantly stronger than the fluorescence of Cur. Nevertheless, there was almost no difference in Cy5 fluorescence between the Cy5-loaded TFNAs (Cy5-TFNAs) and Cy5-loaded Cur-TFNAs (Cy5-Cur-TFNAs). Besides, most of Cur and Cy5-TFNAs exhibited co-localization. Combined with the case that the Cy5 fluorescence was not changed, the results suggested that TFNAs successfully carried Cur and increased the uptake of Cur to RAW264.7 cells. Then, the flow cytometry was used to detect the uptake of Cur to further verify this conclusion. As illustrated in Fig. 2d, the result was consistent with the expected result, suggesting that the uptake level of Cur-TFNAs (~32.8% or ~82.9%) was higher than that of Cur alone (~5.6% or ~12.0%) no matter in normal RAW264.7 cells or LPS induced inflammatory RAW264.7 cells.
3.3. Anti-inflammatory effects of Cur-TFNAs via regulating NF-κB signaling pathway
MSU crystals can induce the development of gout by stimulating the activation of the NLRP3 inflammasome and the secretion of pro-inflammatory factors [4]. Therefore, according to previous experiments, we can establish an in vitro model of the LPS-induced activation of the NLRP3 inflammasome in macrophages [48]. First of all, to ensure that these drugs used in the experiment are nontoxic toward cells, we used the CCK-8 assay kit to measure the cell activity. Fig. S1a and S1b, Supporting information show that Cur-TFNAs within the concentration range (10 μM, 15 μM, and 20 μM) were nontoxic. Moreover, 20 μM Cur, 100 nM TFNAs, and 20 μM Cur-TFNAs did not decrease cell viability after LPS treatment. In addition, TFNAs can be degraded by lysosomes, showing good biological safety [49].
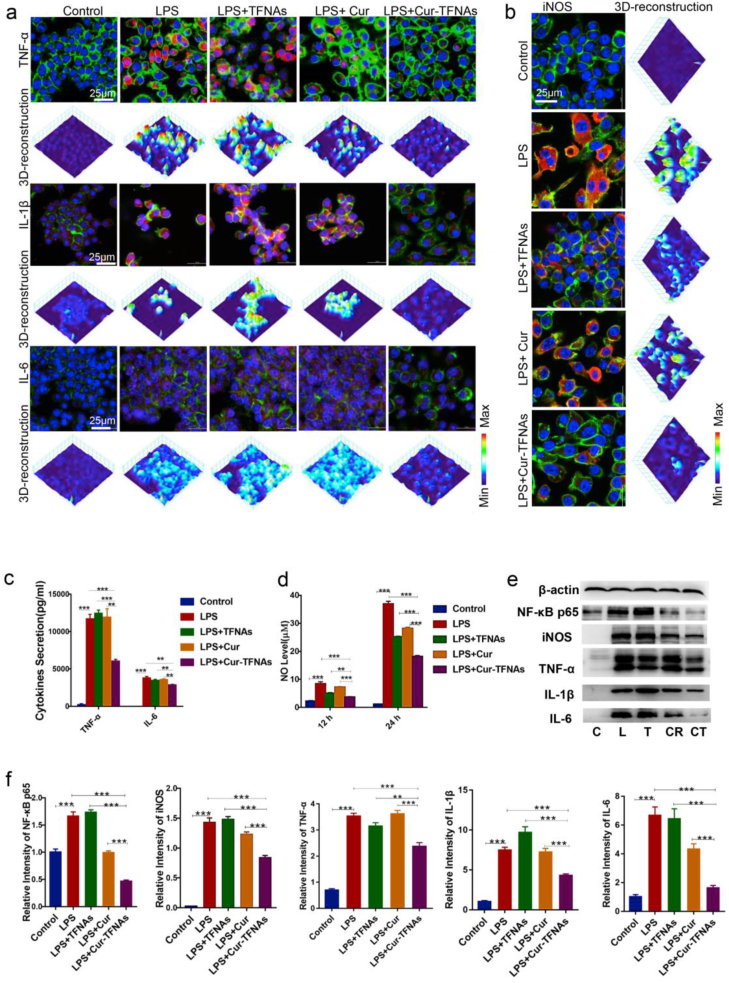
Then, the expression of inflammatory cytokines was measured. After preincubated with drugs (20 μM Cur, 100 nM TFNAs, and 20 μM Cur-TFNAs) for 1 h and treated by LPS for 24 h, the RAW264.7 cells produced a lot of IL-1β, IL-6, and TNF-α. As shown in Fig. 3a, the expressions of inflammatory cytokines were also detected by immunofluorescence. The protein expressions all showed the same trend; namely, under inflammation, the expression of inflammatory cytokines extremely decreased in Cur-TFNA group. Besides, In the TFNA group and Cur group, there was almost no effect. Then, the secretion of IL-6 and TNF-α was further analyzed to verify the anti-inflammatory effect of Cur-TFNAs. According to the determination of ELISA (Fig. 3c), there were slight effects between the TFNA group and Cur group, which were considerably lower than that in the Cur-TFNA group.
Fig. 3.
Anti-inflammatory effects of Cur-TFNAs via regulating the NF-κB signaling pathway. (a) and (b) Immunofluorescence micrographs of LPS-treated RAW264.7 cells (cytoskeleton: green; nucleus: blue; iNOS, IL-1β, IL-6, and TNF-α: red; 3D-reconstruction: 3D reconstruction of fluorescence microscopic images based on fluorescence intensity of iNOS, IL-1β, IL-6, and TNF-α). Scale bars are 25 μm. (c) ELISA detection of the inflammatory cytokines (TNF-α, IL-6; n = 4). (d) The NO level of RAW264.7 cells after treatment of Cur, TFNAs, Cur-TFNAs, and LPS for 12 h and 24 h (n = 4). (e) Western blotting analysis of the NF-κB p65, iNOS, IL-1β, IL-6, and TNF-α expression level (C: Control, L: LPS, T: LPS + TFNAs, CR: LPS + Cur, CT: LPS + Cur-TFNAs; β-actin was used as an internal control). (f) The relative protein expression intensity of NF-κB p65, iNOS, IL-1β, IL-6, and TNF-α (n = 4). Statistical analysis: *p < 0.05, **p < 0.01, and ***p < 0.001.
In addition to the secretion of considerable inflammatory factors, much NO is also produced to affect the development of inflammation when macrophages are activated by inflammation. Hence, the NO level was detected. First, to confirm the optimal concentration of Cur-TFNAs, 10 μM, 20 μM, and 30 μM Cur-TFNAs were used in the experiment. The results (Fig. S1c, Supporting information) showed that the Cur-TFNAs at these concentrations all had the effect of reducing NO levels, but 20 μM Cur-TFNAs had the most obvious effect. Then, to compare the effect of Cur and Cur-TFNAs, NO levels were measured at 12 h and 24 h. As shown in Fig. 3d, the effect of the Cur-TFNAs was much greater than that of the TFNAs and Cur, both of which showed a statistically significant difference.
To further confirm whether Cur-TFNAs play an anti-inflammatory role by blocking the NF-κB, we measured the protein expressions of related signaling pathways. NF-κB is composed of heterodimers of p65 (REL), p52 and p50 proteins. In the light of the prevalent NF-κB activated model, activation of NF-κB needs the degradation of IκBs, which allows the heterodimers (p65 and p50) of NF-κB transfer from cytoplasm to the nucleus [50,51]. Compared with p50, p65 mutation is more lethal [52]. Therefore, by measuring the increased nucleoprotein expression of NF-κB p65, we can prove that LPS-induced inflammation successfully activated the NF-κB [20,53]. Additionally, the inhibition of NF-κB is a major regulatory step of inhibiting of iNOS, thus restraining excessive production of NO [54,55]. iNOS, which is also known as NOS2, is a dimeric enzyme mainly located in macrophages and neutrophils. It is of the same subtype as other NO synthases (NOS), which convert l-arginine and oxygen into NO [56,57]. As shown in Fig. 3b, after LPS treatment for 24 h, the fluorescence intensity of iNOS in macrophages was evidently higher than that of the control group. In the TFNA group and the Cur group, protein expression was decreased slightly, but in the Cur-TFNA group, the fluorescence intensity of iNOS decreased significantly. Consequently, a further determination of western blotting was used to confirm our conjecture. The expression of TNF-α, NF-κB p65, IL-6, iNOS, and IL-1β was further determined. As shown in Fig. 3e and f, the protein expressions were all increased after LPS treatment and then decreased with Cur-TFNA treatment. The results in the Cur group were not very stable, and the protein expression was decreased to some extent, but the gene expression did not change (Fig. S3, Supporting information). In contrast, Cur-TFNAs had a very clear inhibitory effect on both the gene and protein expressions of NF-κB p65, and iNOS. Therefore, Cur-TFNAs showed not only a clear anti-inflammatory effect but also enhancement of overall stability, which was consistent with the results of the characterization measurement of Cur-TFNAs.
3.4. Antioxidant effects of Cur-TFNAs via regulating Nrf2 signaling pathway
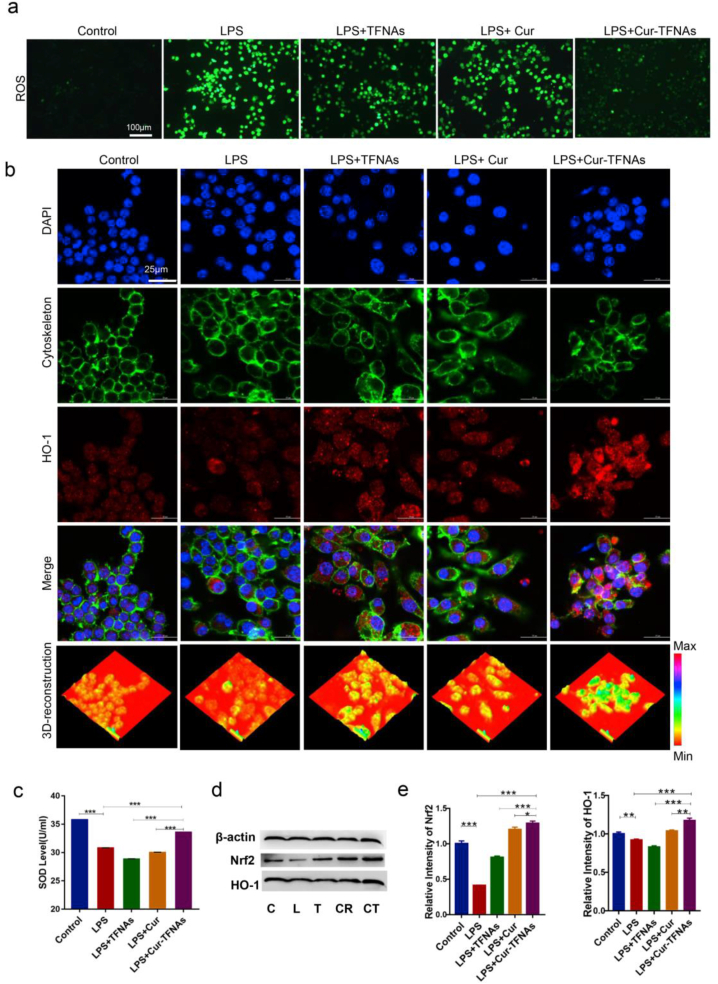
Studies show that ROS are also involved in the regulation of inflammatory signals, which generally occurs through the activation of NF-κB by inflammatory agonists [58]. To confirm the oxidative damage in RAW264.7 cells, the immunofluorescence of ROS was determined. As shown in Fig. 4a, macrophages produced a lot of ROS with LPS treatment for 24 h, but after TFNAs, Cur and Cur-TFNAs pretreatment for 1 h, only the Cur-TFNA group showed significantly decreased fluorescence. In addition, the superoxide dismutase (SOD) content reflected the reduced expression of ROS, indicating the significantly increased antioxidative effects of the Cur-TFNAs (Fig. 4c).
Fig. 4.
Antioxidant effects of Cur-TFNAs via regulating the Nrf2 signaling pathway. (a) Immunofluorescence images of ROS in RAW264.7 cells (ROS: green). Scale bars are 100 μm. (b) Immunofluorescence micrographs of LPS-treated RAW264.7 cells (cytoskeleton: green; nucleus: blue; HO-1: red; 3D-reconstruction: 3D reconstruction of fluorescence microscopic images based on fluorescence intensity of HO-1). Scale bars are 25 μm. (c) The SOD level of RAW264.7 cells were treated with TFNAs, Cur, Cur-TFNAs, and LPS (n = 3). (d) Western blotting analysis of the Nrf2 and HO-1 expression level (C: Control, L: LPS, T: LPS + TFNAs, CR: LPS + Cur, CT: LPS + Cur-TFNAs; β-actin was used as an internal control). (e) The relative protein expression intensity of Nrf2 and HO-1 (n = 4). Statistical analysis: *p < 0.05, **p < 0.01, and ***p < 0.001.
In addition, activation of the Nrf2 can contribute to relative antioxidant activity [57]. Hence, the protein expressions of related signaling pathways were measured to confirm that Cur-TFNAs play an antioxidation role by activating the Nrf2 signaling pathway. When Nrf2 is activated, antioxidant genes then were upregulated to play a further role. Among them, HO-1 is upregulated to participate in antioxidation and anti-inflammatory responses [37]. As shown in Fig. 4b, d, 4e, and Fig S2, Supporting information, the expression of the HO-1 and Nrf2 proteins were respectively detected by western blotting and immunofluorescence technologies to confirm this view. The expression of Nrf2 was suppressed after LPS treatment, while the expression of Nrf2 was improved when the cells were preincubated with drugs, especially Cur-TFNAs. It was demonstrated that the expression of HO-1 was as described above, namely, the drug prevented HO-1 from being inhibited. This effect was most obvious in the Cur-TFNA group.
3.5. Prevention by Cur-TFNAs of acute gouty arthritis
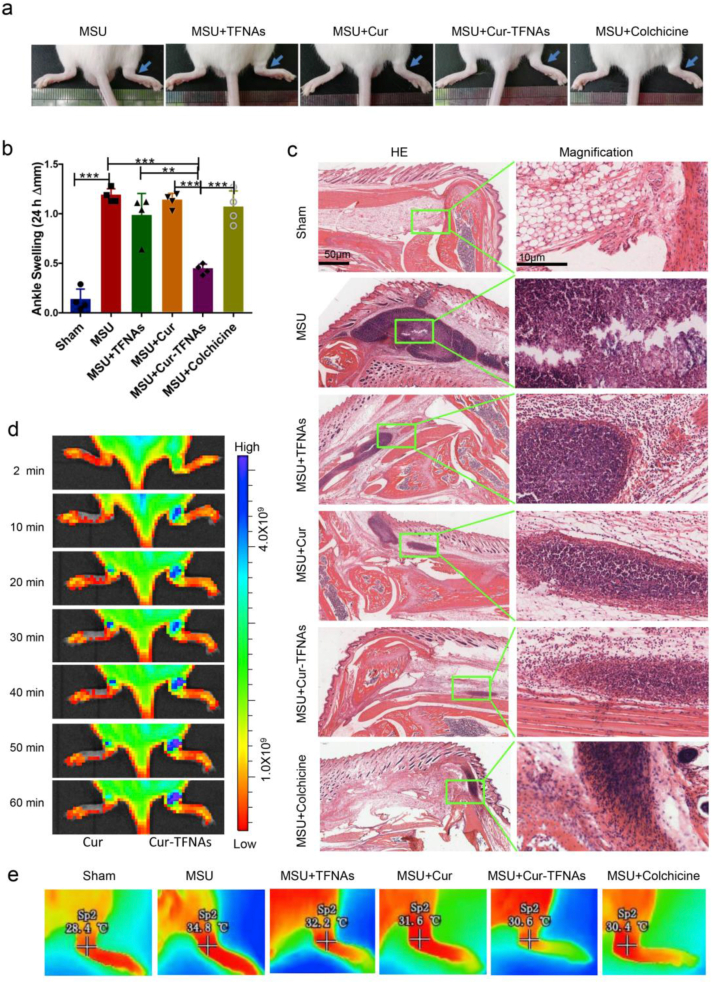
Gouty arthritis, an acute inflammatory reaction, is especially marked by difficulty in moving the joints and extreme pain. Blocking the acute attack is the most urgent to reduce the pain of patients. Usually, for these diseases with the repeated onset of acute inflammation, the curative effect of drugs that can be effectively released by the drug delivery system is better than that of drugs alone. And this kind of treatment can reduce the drug dose, thereby reducing unnecessary side effects [59]. At present, NSAIDs, corticosteroids, and colchicine can effectively relieve inflammatory symptoms well in general [7]. However, NSAIDs are inexpensive but not applicable to many gouty patients due to comorbidity, age, renal impairment or concomitant drug therapy [60]. The colchicine might be more available to most patients, but can lead to the substantial gastrointestinal disturbance [8]. While the Cur-TFNAs have high biological safety and the effect of Cur-TFNAs is equal to colchicine. Although some biologic agents also used to acute gouty arthritis, these drugs are extremely expensive [61]. Compared with these drugs, Cur-TFNAs have the advantages of simple synthesis, suitable price, high biological safety and no less anti-inflammatory effect than colchicine. In consideration of these studies that have demonstrated the anti-inflammatory ability of Cur-TFNAs in vitro, acute gout was induced in mice to explore whether Cur-TFNAs can block the onset of acute gouty arthritis. Then, 20 μL MSU suspension was injected into the tissues near the right ankle joint to simulate acute gouty arthritis, while 20 μL PBS was injected into the left side as the control. Next, we investigated the role of Cur-TFNAs in the model of gouty arthritis induced by MSU. Above all, the degree of ankle joint swelling can be seen in Fig. 5a. After successful modeling, compared with the left joint, obvious swelling of the right joint appeared. Besides, the 50 μL PBS and 50 μL drugs (40 μM Cur, 200 nM TFNAs, 40 μM Cur-TFNAs, and 40 μM colchicine) were respectively given near the joint 1 h in advance to prevent acute gouty arthritis. Then, further quantitative analysis of the degree of joint swelling to suggest the effect of these drugs more intuitively. It can be seen from Fig. 5b that the joint swelling after the modeling decreased gradually in the Cur-TFNA group, and the least degree of joint swelling occurred 24 h later. There was almost no joint swelling in the control group within 24 h. Meanwhile, other groups exhibited significant joint swelling, which did not decrease evidently at 24 h. In addition, temperature performance of the ankles 24 h later is shown in Fig. 5e, and the result is consistent with that of the joint swelling.
Fig. 5.
In vivo biodistribution and anti-inflammatory effect of Cur-TFNAs on acute gouty arthritis. (a) and (b) Joint swelling in mice (left ankle: PBS, right ankle: MSU + TFNAs/Cur/Cur-TFNAs/Colchicine; n = 4). Statistical analysis: *p < 0.05, **p < 0.01, and ***p < 0.001. (c) Photomicrographs stained by HE. Scale bars are 10 μm and 50 μm. (d) IVIS image of ankles treated with Cur and the Cur-TFNAs (left ankle: Cur, right ankle: Cur-TFNAs). (e) Thermal image of ankles.
Then, because the recruitment of leukocytes to the joint is a pathological marker of gouty arthritis, HE staining was detected to determine leukocyte infiltration [7]. As shown in Fig. 5c, there was obvious inflammatory infiltration and focal necrosis in the gout model group, while in the other groups, inflammation was reduced with different levels after the corresponding drug treatment. Moreover, there was no apparent difference between Cur and TFNAs, and their effects were inferior in that of both the Cur-TFNA group and the colchicine group. As a clinical marker drug, there was no significant difference in the effect of colchicine and the Cur-TFNA group on inflammation, which further confirms the good anti-inflammatory ability of Cur-TFNAs. According to the measurement of in vitro material-related properties, this result may be because TFNAs increase the stability of Cur, contributing to boosting its utilization in tissues.
The pure Cur and Cur-TFNAs containing Cur were injected into the ankle joints to detect the drug distribution in vivo. Fluorescence imaging of Cur in the ankle joints was performed by in vivo fluorescence imaging system (IVIS). As shown in Fig. 5d, the fluorescence intensity of Cur was weaker than the fluorescence intensity of Cur-TFNAs. Moreover, the fluorescence of Cur in Cur-TFNAs always increased gradually in the first 1 h while Cur alone increased in the first 40 min and then decreased gradually. Cur alone disappeared almost completely at 60 min while Cur in Cur-TFNAs did not decrease at 60 min. This result may be due to the higher solubility of Cur-TFNA solution. It also indicates that the Cur-TFNAs not only have better stability but also are easier to retain in the injection site than free Cur [19].
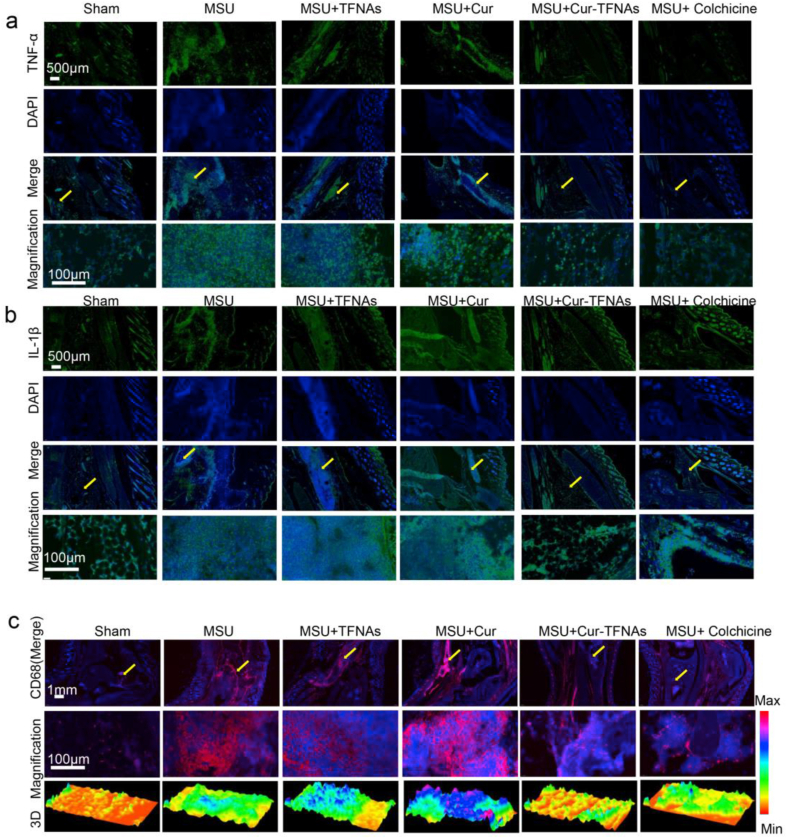
Gout is characterized by the accumulation of MSU crystals in the joint, which induces both acute and chronic inflammation and eventually leads to unbearable pain and inflammatory events [1]. In the process of inflammation, macrophages play a central role in the production of pain by the excretion of TNF-α and IL-1β and some simultaneous activation of nociceptors [62]. Hence, the expressions of TNF-α and IL-1β near the ankle joint were further measured. According to Figs. 6a and 7b, more cells expressed more TNF-α and IL-1β in the gout group. The expressions of inflammatory factors in the Cur-TFNA group and colchicine group are lower than those in the Cur group and TFNA group. More importantly, it shows that the infiltration of CD68-labeled macrophages induced by inflammation around the joint in the Cur-TFNA group was remarkably reduced, compared with the Cur group and the TFNA group (Fig. 6c) [63]. Hence, combined with the results of in vivo and in vitro studies, we confirmed that TFNAs can increase the stability, and bioavailability of Cur, contributing to an increase in the anti-inflammatory ability of macrophages in vitro and an improvement of the ability to resist acute gouty arthritis.
Fig. 6.
Influence on inflammatory cytokines and infiltration of activated macrophages of Cur-TFNAs. (a), (b) and (c) Immunofluorescence micrographs of tissue around the ankle joint (nuclear: blue; IL-1β and IL-6: green; CD68: red; magnification: yellow arrow; 3D: 3D reconstruction of fluorescence microscopic images based on fluorescence intensity of CD68). Scale bars are100 μm, 500 μm, and 1 mm.
4. Conclusion
In summary, it is the first study to comprehensively and systematically analyze the advantages of TFNAs delivering Cur, that is, Cur-TFNAs show better drug stability, and bioavailability. According to the drug distribution in vivo and in vitro, it elucidated that TFNAs not only increased the uptake and utilization of Cur in cells and tissues but also kept Cur in tissues for a long time due to its sustained and stable slow release. Simultaneously, this study confirmed that Cur-TFNA as a composite material has a superior anti-inflammatory effect on acute gouty arthritis. In addition, considering the excellent biosafety and low toxicity of TFNA, we believe that TFNA is a very promising drug delivery system and Cur-TFNA is an encouragingly anti-inflammatory drug that can be used to prevent and treat inflammatory diseases, not just acute gouty arthritis. Finally, because the pathogenesis of gout attacks is complex actually and sometimes related to genetic and environmental factors. Therefore, abundant clinical studies may be needed to further determine the clinical efficacy of Cur-TFNA.
CRediT authorship contribution statement
Mei Zhang: Conceptualization, Investigation, Methodology, Project administration, Writing – original draft. Xiaolin Zhang: Data curation, Formal analysis, Investigation, Writing – review & editing. Taoran Tian: Methodology, Investigation, Software. Qi Zhang: Data curation, Methodology, Resources. Yuting Wen: Data curation, Investigation, Writing – review & editing. Junyao Zhu: Data curation, Methodology, Software. Dexuan Xiao: Resources, Methodology, Writing – review & editing. Weitong Cui: Writing – review & editing. Yunfeng Lin: Project administration, Funding acquisition, Supervision, Writing – review & editing.
Declaration of competing interest
There is no conflict to declare.
Acknowledgement
This study was funded by the National Key R&D Program of China [2019YFA0110600] and National Natural Science Foundation of China [81970916, 81671031].
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioactmat.2021.06.003.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Dalbeth N., Merriman T.R., Stamp L.K., Gout Lancet. 2016;388:2039–2052. doi: 10.1016/S0140-6736(16)00346-9. [DOI] [PubMed] [Google Scholar]
- 2.Chen-Xu M., Yokose C., Rai S.K., Pillinger M.H., Choi H.K. Contemporary prevalence of gout and hyperuricemia in the United States and decadal trends: the national health and nutrition examination survey, 2007-2016. Arthritis Rheum. 2019;71:991–999. doi: 10.1002/art.40807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin W.J., Walton M., Harper J. Resident macrophages initiating and driving inflammation in a monosodium urate monohydrate crystal–induced murine peritoneal model of acute gout. Arthritis Rheum. 2009;60:281–289. doi: 10.1002/art.24185. [DOI] [PubMed] [Google Scholar]
- 4.Martinon F., Pétrilli V., Mayor A., Tardivel A., Tschopp J. Gout-associated uric acid crystals activate the nalp3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- 5.Cronstein B.N., Sunkureddi P. Mechanistic aspects of inflammation and clinical management of inflammation in acute gouty arthritis. J. Clin. Rheumatol. 2013;19:19–29. doi: 10.1097/RHU.0b013e31827d8790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terkeltaub R., Zachariae C., Santoro D., Martin J., Peveri P., Matsushima K. Monocyte-derived neutrophil chemotactic factor/interleukin-8 is a potential mediator of crystal-induced inflammation. Arthritis Rheum. 1991;34:894–903. doi: 10.1002/art.1780340716. [DOI] [PubMed] [Google Scholar]
- 7.Dalbeth N., Choi H.K., Joosten L.A.B., Khanna P.P., Matsuo H., Perez-Ruiz F., Stamp L.K., Gout Nat Rev Dis Primers. 2019;5:69. doi: 10.1038/s41572-019-0115-y. [DOI] [PubMed] [Google Scholar]
- 8.Conaghan P.G., Day R.O. Risks and benefits of drugs used in the management and prevention of gout. Drug Saf. 1994;11:252–258. doi: 10.2165/00002018-199411040-00004. [DOI] [PubMed] [Google Scholar]
- 9.Maheshwari R.K., Singh A.K., Gaddipati J., Srimal R.C. Multiple biological activities of curcumin: a short review. Life Sci. 2006;78:2081–2087. doi: 10.1016/j.lfs.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Basnet P., Skalko-Basnet N. Curcumin: an anti-inflammatory molecule from a curry spice on the path to cancer treatment. Molecules. 2011;16:4567–4598. doi: 10.3390/molecules16064567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh S. From exotic spice to modern drug? Cell. 2007;130:765–768. doi: 10.1016/j.cell.2007.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Yang C., Zhang X., Fan H., Liu Y. Curcumin upregulates transcription factor nrf2, ho-1 expression and protects rat brains against focal ischemia. Brain Res. 2009;1282:133–141. doi: 10.1016/j.brainres.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Krausz A.E., Adler B.L., Cabral V., Navati M., Doerner J., Charafeddine R.A., Chandra D., Liang H., Gunther L., Clendaniel A. Curcumin-encapsulated nanoparticles as innovative antimicrobial and wound healing agent. Nanomedicine. 2015;11:195–206. doi: 10.1016/j.nano.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun H., Ren X., Xiong X., Chen Y., Zhao M., Wang J., Zhou Y., Han Y., Chen Q., Li Y., Kang Y., Zhu G. Nlrp3 inflammasome activation contributes to vsmc phenotypic transformation and proliferation in hypertension. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karlstetter M., Lippe E., Walczak Y., Moehle C., Aslanidis A., Mirza M., Langmann T. Curcumin is a potent modulator of microglial gene expression and migration. J. Neuroinflammation. 2011;8:125. doi: 10.1186/1742-2094-8-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anand P., Kunnumakkara A.B., Newman R.A., Aggarwal B.B. Bioavailability of curcumin: problems and promises. Mol. Pharm. 2007;4:807–818. doi: 10.1021/mp700113r. [DOI] [PubMed] [Google Scholar]
- 17.Szwed A., Miłowska K. The role of proteins in neurodegenerative disease. Postepy Hig. Med. Dosw. 2012;66:187–195. doi: 10.5604/17322693.991446. [DOI] [PubMed] [Google Scholar]
- 18.Tiwari S.K., Agarwal S., Seth B., Yadav A., Nair S., Bhatnagar P., Karmakar M., Kumari M., Chauhan L.K.S., Patel D.K. Curcumin-loaded nanoparticles potently induce adult neurogenesis and reverse cognitive deficits in alzheimer's disease model via canonical wnt/β-catenin pathway. ACS Nano. 2014;8:76–103. doi: 10.1021/nn405077y. [DOI] [PubMed] [Google Scholar]
- 19.Pu H.-L., Chiang W.-L., Maiti B., Liao Z.-X., Ho Y.-C., Shim M.S., Chuang E.-Y., Xia Y., Sung H.-W. Nanoparticles with dual responses to oxidative stress and reduced ph for drug release and anti-inflammatory applications. ACS Nano. 2014;8:1213–1221. doi: 10.1021/nn4058787. [DOI] [PubMed] [Google Scholar]
- 20.Wang J., Wang H., Zhu R., Liu Q., Fei J., Wang S. Anti-inflammatory activity of curcumin-loaded solid lipid nanoparticles in il-1β transgenic mice subjected to the lipopolysaccharide-induced sepsis. Biomaterials. 2015;53:475–483. doi: 10.1016/j.biomaterials.2015.02.116. [DOI] [PubMed] [Google Scholar]
- 21.Banerjee S., Chakravarty A.R. Metal complexes of curcumin for cellular imaging, targeting, and photoinduced anticancer activity. Acc. Chem. Res. 2015;48:2075–2083. doi: 10.1021/acs.accounts.5b00127. [DOI] [PubMed] [Google Scholar]
- 22.Allen T.M., Mumbengegwi D.R., Charrois G.J. Anti-cd19-targeted liposomal doxorubicin improves the therapeutic efficacy in murine b-cell lymphoma and ameliorates the toxicity of liposomes with varying drug release rates. Clin. Canc. Res. 2005;11:3567–3573. doi: 10.1158/1078-0432.CCR-04-2517. [DOI] [PubMed] [Google Scholar]
- 23.Wang A.Z., Langer R., Farokhzad O.C. Nanoparticle delivery of cancer drugs. Annu. Rev. Med. 2012;63:185–198. doi: 10.1146/annurev-med-040210-162544. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y., Sun Y., Li S., Liu M., Qin X., Chen X., Lin Y. Tetrahedral framework nucleic acids deliver antimicrobial peptides with improved effects and less susceptibility to bacterial degradation. Nano Lett. 2020;20:3602–3610. doi: 10.1021/acs.nanolett.0c00529. [DOI] [PubMed] [Google Scholar]
- 25.Zhang T., Tian T., Zhou R., Li S., Ma W., Zhang Y., Liu N., Shi S., Li Q., Xie X., Ge Y., Liu M., Zhang Q., Lin S., Cai X., Lin Y. Design, fabrication and applications of tetrahedral DNA nanostructure-based multifunctional complexes in drug delivery and biomedical treatment. Nat. Protoc. 2020;15:2728–2757. doi: 10.1038/s41596-020-0355-z. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y., Ma W., Zhu Y., Shi S., Li Q., Mao C., Zhao D., Zhan Y., Shi J., Li W., Wang L., Fan C., Lin Y. Inhibiting methicillin-resistant staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 2018;18:5652–5659. doi: 10.1021/acs.nanolett.8b02166. [DOI] [PubMed] [Google Scholar]
- 27.Tian T., Xiao D., Zhang T., Li Y., Shi S., Zhong W., Gong P., Liu Z., Li Q., Lin Y. A framework nucleic acid based robotic nanobee for active targeting therapy. Adv. Funct. Mater. n/a. 2020:2007342. doi: 10.1002/adfm.202007342. [DOI] [Google Scholar]
- 28.Walsh A.S., Yin H., Erben C.M., Wood M.J., Turberfield A.J. DNA cage delivery to mammalian cells. ACS Nano. 2011;5:5427–5432. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 29.Fu W., You C., Ma L., Li H., Ju Y., Guo X., Shi S., Zhang T., Zhou R., Lin Y. Enhanced efficacy of temozolomide loaded by a tetrahedral framework DNA nanoparticle in the therapy for glioblastoma. ACS Appl. Mater. Interfaces. 2019;11:39525–39533. doi: 10.1021/acsami.9b13829. [DOI] [PubMed] [Google Scholar]
- 30.Hu Q., Li H., Wang L., Gu H., Fan C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2018;119:6459–6506. doi: 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- 31.Meng L., Ma W., Lin S., Shi S., Li Y., Lin Y. Tetrahedral DNA nanostructure-delivered dnazyme for gene silencing to suppress cell growth. ACS Appl. Mater. Interfaces. 2019;11:6850–6857. doi: 10.1021/acsami.8b22444. [DOI] [PubMed] [Google Scholar]
- 32.Shi S., Fu W., Lin S., Tian T., Li S., Shao X., Zhang Y., Zhang T., Tang Z., Zhou Y. Targeted and effective glioblastoma therapy via aptamer-modified tetrahedral framework nucleic acid-paclitaxel nanoconjugates that can pass the blood brain barrier. Nanomedicine. 2019;21:102061. doi: 10.1016/j.nano.2019.102061. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y., Liu Y., Zhang B., Shi S., Zhang T., Zhao D., Tian T., Li Q., Lin Y. Erythromycin loaded by tetrahedral framework nucleic acids are more antimicrobial sensitive against escherichia coli (e. Coli) Bioactive Materials. 2021;6:2281–2290. doi: 10.1016/j.bioactmat.2020.12.027. https://www.sciencedirect.com/science/article/pii/S2452199X20303595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie X., Shao X., Ma W., Zhao D., Shi S., Li Q., Lin Y. Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures. Nanoscale. 2018;10:5457–5465. doi: 10.1039/c7nr09692e. [DOI] [PubMed] [Google Scholar]
- 35.Sirong S., Yang C., Taoran T., Songhang L., Shiyu L., Yuxin Z., Xiaoru S., Tao Z., Yunfeng L., Xiaoxiao C. Effects of tetrahedral framework nucleic acid/wogonin complexes on osteoarthritis. Bone Res. 2020;8:6. doi: 10.1038/s41413-019-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohanty C., Sahoo S.K. The in vitro stability and in vivo pharmacokinetics of curcumin prepared as an aqueous nanoparticulate formulation. Biomaterials. 2010;31:6597–6611. doi: 10.1016/j.biomaterials.2010.04.062. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M., Zhu J., Qin X., Zhou M., Zhang X., Gao Y., Zhang T., Xiao D., Cui W., Cai X. Cardioprotection of tetrahedral DNA nanostructures in myocardial ischemia-reperfusion injury. ACS Appl. Mater. Interfaces. 2019;11:30631–30639. doi: 10.1021/acsami.9b10645. [DOI] [PubMed] [Google Scholar]
- 38.Lv H., Zhang S., Wang B., Cui S., Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Contr. Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Chen N., He Y., Su Y., Li X., Huang Q., Wang H., Zhang X., Tai R., Fan C. The cytotoxicity of cadmium-based quantum dots. Biomaterials. 2012;33:1238–1244. doi: 10.1016/j.biomaterials.2011.10.070. [DOI] [PubMed] [Google Scholar]
- 40.De Jong W.H., Hagens W.I., Krystek P., Burger M.C., Sips A.J., Geertsma R.E. Particle size-dependent organ distribution of gold nanoparticles after intravenous administration. Biomaterials. 2008;29:1912–1919. doi: 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- 41.Ko S., Liu H., Chen Y., Mao C. DNA nanotubes as combinatorial vehicles for cellular delivery. Biomacromolecules. 2008;9:3039–3043. doi: 10.1021/bm800479e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J., Fan C., Pei H., Shi J., Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater. 2013;25:4386–4396. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 43.Lv Y., Hu R., Zhu G., Zhang X., Mei L., Liu Q., Qiu L., Wu C., Tan W. Preparation and biomedical applications of programmable and multifunctional DNA nanoflowers. Nat. Protoc. 2015;10:1508–1524. doi: 10.1038/nprot.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gupta S.C., Prasad S., Kim J.H., Patchva S., Webb L.J., Priyadarsini I.K., Aggarwal B.B. Multitargeting by curcumin as revealed by molecular interaction studies. Nat. Prod. Rep. 2011;28:1937–1955. doi: 10.1039/c1np00051a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mohanty C., Acharya S., Mohanty A.K., Dilnawaz F., Sahoo S.K. Curcumin-encapsulated mepeg/pcl diblock copolymeric micelles: a novel controlled delivery vehicle for cancer therapy. Nanomed. 2010;5:433–449. doi: 10.2217/nnm.10.9. [DOI] [PubMed] [Google Scholar]
- 46.Misra R., Acharya S., Dilnawaz F., Sahoo S.K. Sustained antibacterial activity of doxycycline-loaded poly(d,l-lactide-co-glycolide) and poly(epsilon-caprolactone) nanoparticles. Nanomedicine. 2009;4:519–530. doi: 10.2217/nnm.09.28. [DOI] [PubMed] [Google Scholar]
- 47.Sahoo S.K., Panyam J., Prabha S., Labhasetwar V. Residual polyvinyl alcohol associated with poly (d,l-lactide-co-glycolide) nanoparticles affects their physical properties and cellular uptake. J. Contr. Release. 2002;82:105–114. doi: 10.1016/s0168-3659(02)00127-x. [DOI] [PubMed] [Google Scholar]
- 48.He H., Jiang H., Chen Y., Ye J., Wang A., Wang C., Liu Q., Liang G., Deng X., Jiang W., Zhou R. Oridonin is a covalent nlrp3 inhibitor with strong anti-inflammasome activity. Nat. Commun. 2018;9:2550. doi: 10.1038/s41467-018-04947-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S., Tian T., Zhang T., Cai X., Lin Y. Advances in biological applications of self-assembled DNA tetrahedral nanostructures. Mater. Today. 2019;24:57–68. doi: 10.1002/smll.201101576. [DOI] [Google Scholar]
- 50.Natoli G., Chiocca S. Nuclear ubiquitin ligases, nf-κb degradation, and the control of inflammation. Sci. Signal. 2008;1:pe1. doi: 10.1126/stke.11pe1. http://stke.sciencemag.org/content/1/1/pe1.abstract [DOI] [PubMed] [Google Scholar]
- 51.Creus K.K., De Paepe B., De Bleecker J.L. Idiopathic inflammatory myopathies and the classical nf-κb complex: current insights and implications for therapy. Autoimmun. Rev. 2009;8:627–631. doi: 10.1016/j.autrev.2009.02.026. https://www.sciencedirect.com/science/article/pii/S1568997209000433 [DOI] [PubMed] [Google Scholar]
- 52.Beg A.A., Sha W.C., Bronson R.T., Baltimore D. Constitutive nf-kappa b activation, enhanced granulopoiesis, and neonatal lethality in i kappa b alpha-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 53.Wang D., Veena M.S., Stevenson K., Tang C., Ho B., Suh J.D., Duarte V.M., Faull K.F., Mehta K., Srivatsan E.S. Liposome-encapsulated curcumin suppresses growth of head and neck squamous cell carcinoma in vitro and in xenografts through the inhibition of nuclear factor κb by an akt-independent pathway. Clin. Canc. Res. 2008;14:6228–6236. doi: 10.1158/1078-0432.CCR-07-5177. [DOI] [PubMed] [Google Scholar]
- 54.Aktan F. Inos-mediated nitric oxide production and its regulation. Life Sci. 2004;75:639–653. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 55.Bogdan C. Nitric oxide and the immune response. Nat. Immunol. 2001;2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 56.Nathan C. Nitric oxide as a secretory product of mammalian cells. Faseb. J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 57.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 2015;36:161–178. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 58.Canty T.G., Jr., Boyle E.M., Jr., Farr A., Morgan E.N., Verrier E.D., Pohlman T.H. Oxidative stress induces nf-kappab nuclear translocation without degradation of ikappabalpha. Circulation. 1999;100 doi: 10.1161/01.cir.100.suppl_2.ii-361. II-361-II-364. [DOI] [PubMed] [Google Scholar]
- 59.Stubelius A., Sheng W., Lee S., Olejniczak J., Guma M., Almutairi A. Disease-triggered drug release effectively prevents acute inflammatory flare-ups, achieving reduced dosing. Small. 2018;14 doi: 10.1002/smll.201800703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gambaro G., Perazella M.A. Adverse renal effects of anti-inflammatory agents: evaluation of selective and nonselective cyclooxygenase inhibitors. J. Intern. Med. 2003;253:643–652. doi: 10.1046/j.1365-2796.2003.01146.x. [DOI] [PubMed] [Google Scholar]
- 61.Rees F., Hui M., Doherty M. Optimizing current treatment of gout. Nat. Rev. Rheumatol. 2014;10:271–283. doi: 10.1038/nrrheum.2014.32. [DOI] [PubMed] [Google Scholar]
- 62.Hainer B.L., Matheson E., Wilkes R.T. Diagnosis, treatment, and prevention of gout. Am. Fam. Physician. 2014;90:831–836. [PubMed] [Google Scholar]
- 63.Boumans M., Houbiers J., Verschueren P., Ishikura H., Westhovens R., Brouwer E., Rojkovich B., Kelly S., den Adel M., Isaacs J., Jacobs H., Gomez-Reino J., Holtkamp G., Hastings A., Gerlag D., Tak P. Safety, tolerability, pharmacokinetics, pharmacodynamics and efficacy of the monoclonal antibody ask8007 blocking osteopontin in patients with rheumatoid arthritis: a randomised, placebo controlled, proof-of-concept study. Ann. Rheum. Dis. 2012;71:180–185. doi: 10.1136/annrheumdis-2011-200298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.