Abstract
RNA polymerase (Pol) III transcription is abnormally active in fibroblasts that have been transformed by simian virus 40 (SV40). This report presents evidence that two separate components of the general Pol III transcription apparatus, TFIIIB and TFIIIC2, are deregulated following SV40 transformation. TFIIIC2 subunits are expressed at abnormally high levels in SV40-transformed cells, an effect which is observed at both protein and mRNA levels. In untransformed fibroblasts, TFIIIB is subject to repression through association with the retinoblastoma protein RB. The interaction between RB and TFIIIB is compromised following SV40 transformation. Furthermore, the large T antigen of SV40 is shown to relieve repression by RB. The E7 oncoprotein of human papillomavirus can also activate Pol III transcription, an effect that is dependent on its ability to bind to RB. The data provide evidence that both TFIIIB and TFIIIC2 are targets for activation by DNA tumor viruses.
A broad range of transformed cell types display abnormally elevated levels of RNA polymerase (Pol) III transcripts (reviewed in references 53 and 54). This was first discovered with murine fibroblast lines that have been transformed by simian virus 40 (SV40) (5, 36, 42). When different SV40-transformed clones are compared, those which most efficiently induce tumors in nude mice also display the highest abundance of Pol III transcripts, whereas lower levels are detected in the less tumorigenic lines (36, 60). A tight link between transformation and Pol III induction is suggested by analyses of two cell lines that were transformed with temperature-sensitive mutants of the SV40 oncoprotein large T antigen; these cells down-regulate Pol III products at the nonpermissive temperature while reverting to normal morphology and phenotype (36). Large T antigen has also been shown to activate Pol III transcription in transient transfection assays (9, 28).
The activity of a general Pol III factor called TFIIIC2 has been shown to be greater in the SV40-transformed cell lines SV3T3 Cl38 and SV3T3 Cl49 than in the untransformed parental 3T3 line A31 (60). TFIIIC2 is a large DNA-binding factor that is required for the expression of most class III genes (reviewed in reference 53). It can relieve chromatin-mediated repression of Pol III transcription and has been shown to display histone acetyltransferase activity (18). TFIIIC2 can be detected in at least two forms, distinguishable by their differential migrations in electrophoretic mobility shift assays (EMSAs) (8, 15, 17, 43, 60). Whereas the high-mobility form predominates in untransformed A31 cell extracts, the SV3T3 derivatives contain a greater proportion of TFIIIC2 in the slowly migrating form (60). Chromatographic fractionation of HeLa cells has revealed that the low-mobility form is transcriptionally active, whereas the higher-mobility species can bind DNA but is unable to support transcription (15, 17). Following purification, the inactive form was found to be missing a 110-kDa subunit, which was named TFIIICβ (17, 43). The abundance of TFIIICβ in HeLa cells has been shown to increase substantially following serum stimulation or adenovirus infection, two conditions which activate TFIIIC2 (43). In contrast, the level of TFIIICα, which is the DNA-binding subunit of TFIIIC2, remains constant under these conditions (43).
The large T antigen of SV40 can bind and neutralize RB, the protein product of the retinoblastoma susceptibility gene (11, 13, 31). Mutations in T antigen that interfere with RB binding also abrogate its transforming activity (11, 13, 31). Gene disruption experiments have shown that endogenous RB represses Pol III transcription approximately fivefold in murine fibroblasts (61). The increased synthesis of tRNA and 5S rRNA that is observed in RB-knockout cells correlates with the deregulation of the general Pol III factor TFIIIB (24). TFIIIB is required for the expression of all class III genes; it is recruited via TFIIIC2 and then serves to position the polymerase over the initiation site (reviewed in reference 53). TFIIIB activity has been shown to be inhibited specifically by recombinant RB in vitro (7, 24). Furthermore, immunoprecipitation, cofractionation, and pull-down experiments have demonstrated that TFIIIB interacts with both endogenous and recombinant RB (7, 24). The ability of large T antigen to neutralize RB therefore raises the possibility that TFIIIB is released from repression following SV40 transformation. We present evidence that this is the case. We also demonstrate that SV40-transformed Cl38 and Cl49 cells express much higher levels of TFIIICα and TFIIICβ than the untransformed parental A31 line. The data suggest that at least two components of the general Pol III transcription apparatus become activated following transformation by SV40.
MATERIALS AND METHODS
Tissue culture.
SV3T3 Cl38 and Cl49 cell lines were generated by infection of BALB/c 3T3 A31 cells with SV40 (wt830 strain) and selected by focus formation in low serum (35). All cell lines were grown in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml and were harvested when subconfluent.
Antibodies and Western blotting.
Antibodies used were C-15 (Santa Cruz Biotechnology) and G99-549 (PharMingen) against RB, M-19 (Santa Cruz) against TAFI48, 330 and 128 against BRF (1, 4), SL30 against TATA-binding protein (TBP) (27), clone 46 (Transduction Laboratories) against TFIIICβ, and Ab4 against TFIIICα (38). Western immunoblot analysis was performed as previously described (56).
Reverse transcription-PCR (RT-PCR) analysis.
RNA was extracted from subconfluent A31, Cl38, and Cl49 cells by using TRI reagent (Sigma) according to the manufacturer’s specifications. Reverse transcription reactions were performed for 1 h at 42°C, using 3 μg of RNA, 200 ng of random hexamers (Promega), and 400 U of Superscript II reverse transcriptase (Life Technologies) in a total volume of 40 μl of 1× First Strand Buffer (Life Technologies) containing 10 mM dithiothreitol (DTT) and 0.5 mM each deoxynucleoside triphosphate.
PCRs were carried out in a PTC-100 programmable thermal controller (MJ Research Inc.); 2 μl of cDNA was amplified with 20 pmol of either glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers (5′-TCCACCACCCTGTTGCTGTA-3′ and 5′-ACCACAGTCCATGCCATCAC-3′) to give a 452-bp product, TFIIICβ primers (5′-CCAGAAGGGGTCTCAAAAGTCC-3′ and 5′-CTTTCTTCAGAGATGTCAAAGG-3′) to give a 303-bp product, TFIIICα primers (5′-TCCAGCGAGACCTTCACACC-3′ and 5′-GGATTGAGTGTTGCTGGGCT-3′) to give a 144-bp product, or BRF primers (5′-AAATTCTGTGAGCCTCTTCCGTAGTG-3′ and 5′-AGACCCATGCTTGTACATTCCACG-3′) to give a 260-bp product. Amplification reaction mixtures contained 0.5 U of Taq DNA polymerase (Promega) in a total volume of 20 μl of 1× Taq DNA polymerase buffer (Promega) containing 1.5 mM MgCl2 and 0.2 mM each deoxynucleoside triphosphate. PCR was performed under the following cycling parameters: for GAPDH, (i) 94°C for 3 min, (ii) 18 cycles of 95°C for 30 s, 66°C for 40 s, and 72°C for 1 min, (iii) 72°C for 5 min; for TFIIICβ, (i) 94°C for 3 min, (ii) 6 cycles of 95°C for 1 min, 66°C for 40 s, and 72°C for 40 s, (iii) 22 cycles of 95°C for 1 min, 62°C for 40 s, and 72°C for 40 s, (iv) 72°C 5 min; for TFIIICα, (i) 95°C for 3 min, (ii) 30 cycles of 94°C for 20 s, 62°C for 30 s, and 72°C for 30 s, (iii) 72°C 10 min; for BRF, 95°C for 2 min, 26 cycles of 95°C for 1 min, 60°C for 30 s, and 72°C for 1 min, (iv) 72°C 5 min. Reaction products were resolved on a 2% agarose gel and visualized by ethidium bromide staining. Gels were scanned, and the intensity of each band was quantitated by using a UVP image analysis system and Adobe Photoshop 4.0 software.
Plasmids.
pVAI contains the adenovirus VAI gene (10). pCAT (Promega) contains the chloramphenicol acetyltransferase (CAT) gene driven by the SV40 promoter and enhancer. Expression constructs encoding the E7 oncoprotein of human papillomavirus type 16 (HPV-16) and its mutant derivatives Δ21-35, PRO2, and GLY24 have been described elsewhere (21).
Transfection assays.
Cell lines were transiently transfected by the calcium phosphate precipitation method. DNA precipitates were left on the plates overnight, and then the cells were washed with phosphate-buffered saline and cultured for 24 h before harvesting. Total RNA was extracted by using TRI reagent (Sigma) according to the manufacturer’s instructions and then analyzed by primer extension using both VAI-specific (5′-CACGCGGGCGGTAACCGCATG-3′) and CAT-specific (5′-CGATGCCATTGGGATATATCA-3′) labeled primers. Primer extension reactions were conducted as previously described (61).
Transcription.
Pol III transcription was carried out as described previously (59) except that pBR322 was not included and the incubations were for 60 min at 30°C.
Preparation of extracts and protein fractions.
Whole-cell extracts were prepared from exponentially growing cells by the method of Manley et al. (29). Nuclear extracts were purchased from the Computer Cell Culture Center (Mons, Belgium). PC-B is the 0.1 to 0.35 M KCl step fraction from a phosphocellulose column and contains both TFIIIB and Pol III (37). PC-C is the 0.35 to 0.6 M KCl step fraction from a phosphocellulose column and contains both TFIIIC and Pol III (37). PC-D is the 0.6 to 1.0 M KCl step fraction from a phosphocellulose column and contains TFIID (37). The CHep-1.0 fraction was generated as described previously (56) by loading PC-C onto heparin-Sepharose CL-6B in BC buffer (25 mM Tris-HCl [pH 7.9], 10% glycerol, 10 mM β-mercaptoethanol) plus 100 mM KCl (BC-100). The column was washed with BC-280 and eluted with BC-1000 to produce the CHep-1.0 fraction containing TFIIIC. The A25(0.15) fraction containing TFIIIB was generated as described previously (56) by applying PC-B to an A25 DEAE-Sephadex column in buffer A (20 mM HEPES-KOH [pH 7.9], 20% glycerol, 5 mM MgCl2, 3 mM DTT, 0.2 mM phenylmethylsulfonyl fluoride) plus 50 mM (NH4)2SO4. After extensive washing with this buffer, the A25(0.15) fraction containing TFIIIB was eluted in buffer A plus 150 mM (NH4)2SO4. Mono Q gradient-purified TFIIIB was prepared as described previously (27).
Recombinant glutathione S-transferase (GST) and GST-RB fusion proteins were expressed in bacteria and purified on glutathione-agarose as previously described (24). GST-RB contains residues 379 to 928 of RB. Immunoaffinity-purified recombinant SV40 large T antigen was a generous gift from David Lane.
Immunoprecipitation.
Whole-cell extract (150 μg) was incubated at 4°C on an orbital shaker with 20 μl of protein A-Sepharose beads carrying equivalent amounts of prebound immunoglobulin G. Samples were then pelleted, supernatants were removed, and the beads were washed five times with 150 μl of LDB buffer (20 mM HEPES-KOH [pH 7.9], 17% glycerol, 100 mM KCl, 12 mM MgCl2, 0.1 mM EDTA, 2 mM DTT). The bound material was analyzed by Western blotting. In the experiments shown in Fig. 5, reticulocyte lysate (15 μl) containing BRF translated in the presence of [35S]Met and [35S]Cys was mixed with nuclear extract (150 μg) during immunoprecipitation. In this case, the precipitated material was analyzed by autoradiography rather than Western blotting.
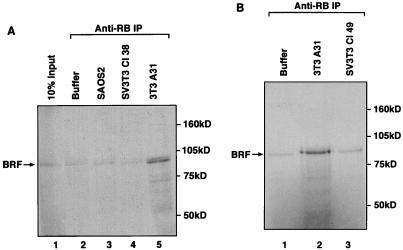
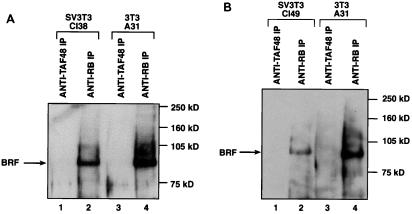
FIG. 5.
The BRF subunit of TFIIIB coimmunoprecipitates with endogenous RB present in A31 extracts, but this interaction is compromised in extracts of Cl38 and Cl49 cells. (A) Reticulocyte lysate (15 μl) containing in vitro-translated BRF was immunoprecipitated (IP) with anti-RB antibody G3-245 in the presence of buffer alone (lane 2) or 150 μg of whole-cell extract prepared from SAOS2 (lane 3), Cl38 (lane 4), or A31 (lane 5) cells. Proteins retained after extensive washing were resolved on an SDS–7.8% polyacrylamide gel and then visualized by autoradiography. Lane 1 shows 10% of the input reticulocyte lysate containing in vitro-translated BRF. (B) Reticulocyte lysate (15 μl) containing in vitro-translated BRF was immunoprecipitated with anti-RB antibody C-15 in the presence of buffer alone (lane 1) or 150 μg of whole-cell extract prepared from A31 (lane 2) or Cl49 (lane 3) cells. Proteins retained after extensive washing were resolved on an SDS–7.8% polyacrylamide gel and then visualized by autoradiography.
RESULTS
TFIIIC2 subunits are more abundant in SV3T3 Cl38 and Cl49 cell extracts than in parental 3T3 A31 cell extracts.
Previous studies have shown that the SV40-transformed cell lines Cl38 and Cl49 express high levels of Pol III transcripts relative to the untransformed parental line A31 (36, 60). Nuclear run-on assays demonstrated that this up-regulation occurs at the transcriptional level (60). EMSA analysis revealed that the DNA-binding activity of TFIIIC2 is significantly higher in Cl38 and Cl49 cell extracts than in extracts of the A31 cells (60). This is a specific effect, since Sp1 activity is not elevated in the transformed cell lines (60). In addition, the Cl38 and Cl49 extracts were found to contain a greater proportion of TFIIIC2 in the slowly migrating transcriptionally active form (60). It was proposed that these changes might account for the abnormally high rates of Pol III transcription that are observed in these SV3T3 cell lines (60).
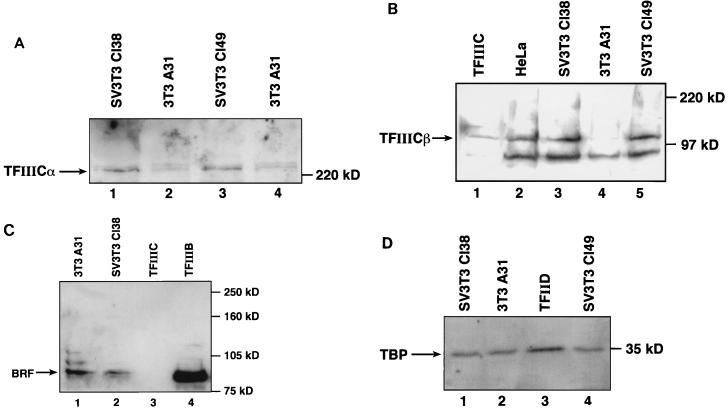
Since the time of this previous analysis, the two forms of TFIIIC2 have been purified to homogeneity and shown to differ in the presence of a 110-kDa subunit that is found only in the active form and is referred to as TFIIICβ (17, 43). The availability of antibodies that recognize TFIIICβ (43) and the ∼240-kDa DNA-binding subunit TFIIICα (20, 26) has provided the opportunity to investigate further the changes in TFIIIC2 that accompany SV40 transformation. We therefore carried out Western blot analyses to compare the levels of these subunits in A31, Cl38, and Cl49 cell extracts. The level of TFIIICα was found to be higher in Cl38 and Cl49 cell extracts than in A31 cell extracts (Fig. 1A), consistent with the increased DNA-binding activity detected by EMSA following SV40 transformation (60). A striking change was also detected with the TFIIICβ subunit, which is substantially more abundant in the Cl38 and Cl49 cell extracts than in A31 cell extracts (Fig. 1B). This increase in TFIIICβ may account for the higher proportion of TFIIIC2 that is found in the transcriptionally active form following SV40 transformation (60).
FIG. 1.
The SV40-transformed cell lines Cl38 and Cl49 express elevated levels of TFIIIC2 subunits. (A) Whole-cell extracts (57 μg of protein) prepared from Cl38 (lane 1), A31 (lanes 2 and 4), and Cl49 (lane 3) cells were resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western immunoblotting with anti-TFIIICα antibody Ab4. Lanes 2 and 4 show extracts prepared from two separate batches of A31 cells. (B) CHep-1.0 fraction (15 μg of protein) containing TFIIIC2 (lane 1) and whole-cell extracts (38 μg of protein) prepared from HeLa (lane 2), Cl38 (lane 3), A31 (lane 4), and Cl49 (lane 5) cells were resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western immunoblotting with anti-TFIIICβ monoclonal antibody clone 46. (C) Whole-cell extracts (50 μg of protein) prepared from A31 (lane 1) and Cl38 (lane 2) cells, CHep-1.0 (12 μg of protein) TFIIIC2 fraction (lane 3), and A25(0.15) (1.6 μg of protein) TFIIIB fraction (lane 4) were resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western immunoblotting with anti-BRF antibody 330. (D) Whole-cell extracts (50 μg of protein) prepared from Cl38 (lane 1), A31 (lane 2), and Cl49 (lane 4) cells and PC-D (5.6 μg of protein) TFIID fraction (lane 3) were resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western immunoblotting with anti-TBP antibody SL30.
To investigate the specificity of these effects, we also examined the abundance of two subunits of TFIIIB. Human TFIIIB has been shown to contain TBP and an associated polypeptide of ∼90 kDa called TFIIIB90 or human BRF (27, 30, 40, 45, 51, 57). Neither of these subunits was found to be significantly more abundant in Cl38 cell extracts than in A31 cell extracts (Fig. 1C and D). We conclude that the up-regulation of TFIIICα and TFIIICβ in the SV40-transformed cells is a specific phenomenon.
SV40-transformed Cl38 and Cl49 cells express elevated levels of mRNA encoding TFIIICα and TFIIICβ.
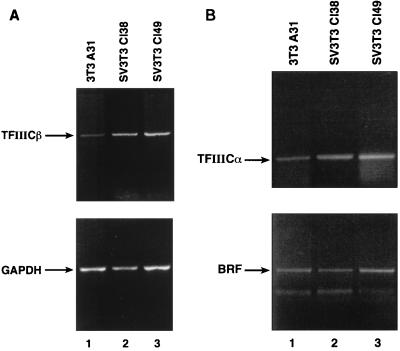
To determine the level at which TFIIICα and TFIIICβ become up-regulated, we carried out semiquantitative RT-PCR analyses of their mRNAs. The transcript encoding TFIIICβ was found to be significantly more abundant in Cl38 and Cl49 cells than in A31 cells (Fig. 2A, top). Again, this is a specific phenomenon, since the mRNA encoding GAPDH is expressed at similar levels in these three cell lines (Fig. 2A, bottom). The TFIIICα transcript is also more abundant in Cl38 and Cl49 cells than in A31 cells (Fig. 2B, top). However, the mRNA for BRF shows little or no increase following SV40 transformation (Fig. 2B, bottom), consistent with the Western blotting data. After normalization to the level of GAPDH mRNA, the levels of the BRF transcript in Cl38 and Cl49 cells were 90 and 114%, respectively, of that seen in A31 cells. In contrast, expression of TFIIICα transcript was elevated four- to fivefold, whereas that of TFIIICβ was elevated seven- to eightfold in Cl38 and Cl49 cells, relative to the parental A31 cells. The observed specific increases in the mRNAs encoding TFIIICα and TFIIICβ may therefore account for the elevated abundance of these subunits in the SV40-transformed cell lines.
FIG. 2.
The SV40-transformed cell lines Cl38 and Cl49 express elevated levels of transcripts encoding TFIIICα and TFIIICβ. (A) cDNAs generated by reverse transcription of 3 μg of RNA from A31 (lane 1), Cl38 (lane 2), and Cl49 (lane 3) cells were PCR amplified by using TFIIICβ (top) and GAPDH (bottom) primers. Amplification products were resolved on a 2% agarose gel. (B) cDNAs generated by reverse transcription of 3 μg of RNA from A31 (lane 1), Cl38 (lane 2), and Cl49 (lane 3) cells were PCR amplified by using TFIIICα (top) and BRF (bottom) primers. Amplification products were resolved on a 2% agarose gel.
SV40 large T antigen can activate Pol III transcription by overcoming the repressive effects of RB.
The transforming protein of SV40 is its large T antigen, and this has been shown to be sufficient to stimulate Pol III transcription in transient transfection assays (9, 28). The ability of T antigen to transform cells is dependent on its capacity to bind and inactivate RB (11, 13, 31). Since RB is a potent repressor of Pol III transcription in murine fibroblasts (61), we investigated whether T antigen can release Pol III from repression by RB (Fig. 3). When recombinant T antigen was added to reactions that had been reconstituted with partially purified factors, it caused no increase in the level of VAI transcription by Pol III (lanes 1 to 3). However, a different response was obtained after a recombinant GST-RB fusion protein had been added to depress transcription. In the presence of this added RB, titration of T antigen resulted in a significant stimulation of VAI expression (lanes 4 to 6). The fact that T antigen increases Pol III transcription in the presence of excess RB but not in its absence suggests that its stimulatory effect in this system results from its ability to neutralize RB.
FIG. 3.
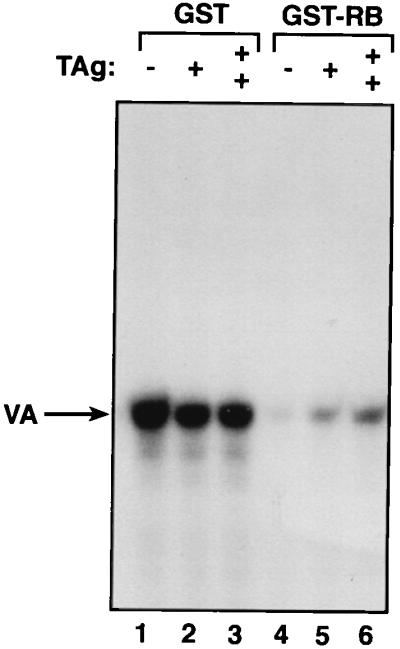
SV40 large T antigen can counteract the repressive effect of RB on Pol III transcription. Template pVAI (250 ng of DNA) was transcribed by using 2 μl of Mono Q-purified TFIIIB and 2 μl of CHep-1.0 fraction in the presence of 100 ng of either GST (lanes 1 to 3) or GST-RB (lanes 4 to 6) and 20 ng (lanes 2 and 5) or 40 ng (lanes 3 and 6) of immunoaffinity-purified recombinant large T antigen (TAg).
TFIIIB activity is abnormally elevated in the SV40-transformed cell lines Cl38 and Cl49.
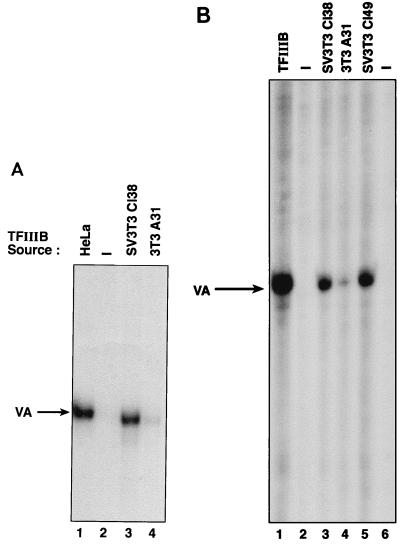
Previous studies have shown that the general Pol III factor TFIIIB is a target for repression by RB (7, 24). Since the T antigen present in SV40-transformed cells might be expected to counteract the effects of RB, we investigated whether this results in an increase in TFIIIB activity. Extracts of 3T3 A31 cells and SV3T3 Cl38 cells were subjected to chromatography on phosphocellulose. Equivalent TFIIIB fractions were then compared for their abilities to reconstitute transcription when mixed with a fraction containing TFIIIC and Pol III from HeLa cells (Fig. 4A). No transcription was observed with this system in the absence of added TFIIIB (lane 2). However, a robust transcription signal was obtained following the addition of TFIIIB fractions derived from HeLa or Cl38 cells (lanes 1 and 3, respectively). In contrast, an equal amount of the equivalent TFIIIB fraction derived from 3T3 A31 cells gave much lower levels of Pol III transcription (lane 4).
FIG. 4.
Cl38 and Cl49 cell extracts contain higher TFIIIB activity than A31 cell extracts. (A) Template pVAI (250 ng of DNA) was transcribed by using 2 μl of HeLa-derived PC-C fraction and buffer alone (lane 2) or PC-B fraction (6.4 μg of protein) derived from HeLa (lane 1), Cl38 (lane 3), or A31 (lane 4) cell extracts. (B) Template pVA1 (500 ng DNA) was transcribed by using 4 μl of HeLa-derived PC-C fraction supplemented with 10 ng of recombinant TBP and 6 μl of heat-treated Mono Q-purified TFIIIB (lane 1), buffer alone (lanes 2 and 6), or 7.5 μg of heat-treated extract from Cl38 (lane 3), A31 (lane 4), or Cl49 (lane 5) cells.
The reduced activity of A31 cell-derived TFIIIB fractions could conceivably result from differential recovery or inactivation during chromatography. We therefore also conducted complementation assays to measure TFIIIB activity in unfractionated extracts. These assays make use of the differential sensitivity of Pol III transcription factors to inactivation by mild heat treatment. When an extract is heated at 47°C for 15 min, TFIIIC and TBP become inactivated whereas the other components of the Pol III machinery are not compromised (17, 41, 57, 58). The activity of TFIIIB in an unfractionated extract can therefore be assayed by heat treating the extract and then measuring its ability to reconstitute transcription when added to a complementing fraction containing TFIIIC, TBP, and Pol III (1, 24, 55, 56). Using such assays, we found that A31 cell extracts contain significantly less TFIIIB activity than extracts of Cl38 or Cl49 cells (Fig. 4B). We conclude that SV40 transformation of A31 cells is accompanied by an increase in TFIIIB activity, even though the levels of TBP and BRF do not increase.
The interaction between RB and TFIIIB is compromised following SV40 transformation.
Previous genetic analyses have shown that TFIIIB is subject to repression by RB in murine fibroblasts (24). Coimmunoprecipitation experiments were therefore carried out to test whether the physical interaction between RB and TFIIIB is diminished following SV40 transformation. The BRF subunit of the TFIIIB complex was synthesized by using a reticulocyte lysate in the presence of [35S]cysteine and [35S]methionine. The radiolabeled protein was then incubated with whole-cell extracts under conditions which allow complex assembly (4). The mixtures were subjected to immunoprecipitation using a monoclonal antibody that is specific for RB, and the precipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography (Fig. 5A). Significant amounts of radiolabeled BRF were coprecipitated with RB from an A31 cell extract (lane 5). This was not due to cross-reaction with the antibody, since only background levels of BRF were detected when buffer was used in place of the whole-cell extract (lane 2). Similar background levels were also obtained using extracts of SAOS2 osteosarcoma cells (lane 3), which express an inactive truncated mutant form of RB (39). Furthermore, very little BRF was coimmunoprecipitated with RB when it was mixed with an extract of SV3T3 Cl38 cells (lane 4), despite the fact that Cl38 cells contain full-length RB that is no less abundant than the RB present in A31 cells (25). A similar analysis using a different RB antibody revealed that the interaction between BRF and RB is also compromised in SV3T3 Cl49 cell extracts (Fig. 5B). Identical results were obtained in assays using a third antibody against a different region of RB (44). In contrast, irrelevant control antibodies failed to coprecipitate BRF (44).
The previous experiments suggest that the BRF subunit of TFIIIB is much less able to associate with cellular RB following SV40 transformation. As these results were obtained in studies using exogenous radiolabeled BRF, we also carried out coimmunoprecipitation analyses to compare the extent to which endogenous TFIIIB is bound by RB in 3T3 and SV3T3 cells. Extracts of A31 and Cl38 cells were immunoprecipitated with either an anti-RB antibody or an irrelevant control antibody against the TAFI48 subunit of the Pol I factor SL1. Immunoprecipitated material was then resolved by SDS-PAGE and probed for the presence of BRF by Western blotting (Fig. 6A). Substantial amounts of endogenous BRF were found to coprecipitate with RB from A31 cell extracts, an effect that was specific since none was detected in the control immunoprecipitates. In contrast, significantly less BRF was coprecipitated with RB from the Cl38 cell extracts. Similarly, anti-RB immunoprecipitates from Cl49 cell extracts contained much less BRF than those obtained in parallel from A31 cell extracts (Fig. 6B). These results reinforce the data obtained in assays using radiolabeled BRF and indicate that the interaction between endogenous RB and TFIIIB is diminished following SV40 transformation.
FIG. 6.
The association between endogenous RB and endogenous TFIIIB is greater in A31 cells than in Cl38 and Cl49 cells. (A) Cl38 (lanes 1 and 2) and A31 (lanes 3 and 4) cell extracts (150 μg of protein) were immunoprecipitated (IP) with anti-TAFI48 antibody M-19 (lanes 1 and 3) or anti-RB antibody C-15 (lanes 2 and 4). Precipitated material was resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western blotting with anti-BRF antiserum 128. (B) Cl49 (lanes 1 and 2) and A31 (lanes 3 and 4) cell extracts (150 μg of protein) were immunoprecipitated with anti-TAFI48 antibody M-19 (lanes 1 and 3) or anti-RB antibody C-15 (lanes 2 and 4). Precipitated material was resolved on an SDS–7.8% polyacrylamide gel and then analyzed by Western blotting with anti-BRF antiserum 128.
The E7 oncoprotein of HPV-16 can also activate Pol III transcription.
The data described above suggest that T antigen can activate TFIIIB by releasing it from the repressive influence of RB. The RB-binding site within T antigen has been mapped to an LXCXE motif, where X can be any amino acid (11, 64, 65). HPV-16 encodes an oncoprotein called E7 that has also been shown to bind and neutralize RB by means of an LXCXE motif (12, 32, 48). We therefore tested whether E7 resembles T antigen in being able to activate Pol III transcription.
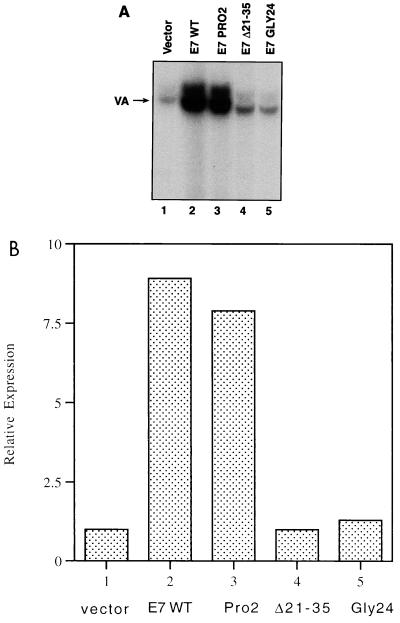
The untransformed murine fibroblast cell line NIH 3T3 was transfected with expression vectors encoding either wild-type E7 or several mutant forms of E7 (Fig. 7). Primer extension analysis was used to determine the levels of Pol III transcription of a cotransfected VAI gene; a cotransfected control plasmid, in which the CAT gene is driven by the SV40 early promoter, was used to normalize for transfection efficiency. Specific VAI expression was stimulated substantially by the wild-type E7 protein (lane 2) relative to empty vector (lane 1). The ability of E7 to activate VAI transcription was severely compromised by deletion of residues 21 to 35, which removes the LXCXE motif that is located between amino acids 22 and 26 (lane 4). Indeed, substitution of the C24 residue to glycine is sufficient to abrogate the ability of E7 to stimulate VAI expression (lane 5). In contrast, mutant PRO2, which carries a proline substitution in its second residue and has an intact LXCXE motif, remains fully capable of activating Pol III transcription (lane 3). The PRO2 mutant is significant, because it has lost the ability to transform although it can still bind and neutralize RB (21). The fact that the PRO2 mutant can activate VAI expression indicates that the effect of E7 on Pol III transcription is not an indirect consequence of cell transformation. These E7 proteins are all expressed at comparable levels in NIH 3T3 cells (21). We conclude that E7 can activate Pol III transcription in vivo in a manner that is dependent on its RB-binding LXCXE motif.
FIG. 7.
Transfected HPV E7 activates Pol III transcription in vivo in a manner that is dependent on its RB-binding domain. NIH 3T3 cells were transfected with pVAI (2 μg), pCAT (2 μg), and 6 μg of empty vector (lane 1) or vector encoding wild-type E7 (E7 WT; lane 2), PRO2 mutant E7 (lane 3), Δ21-35 mutant E7 (lane 4), or GLY24 mutant E7 (lane 5). (A) VAI primer extension products. (B) VAI transcription levels that have been normalized to the levels of CAT expression to correct for transfection efficiency and are expressed relative to the value obtained for the empty vector control (designated 1.0).
DISCUSSION
Abnormally high levels of Pol III transcripts are observed in SV40-transformed cells (5, 36, 42, 60). The present study has provided evidence that more than one mechanism contributes to this effect and that two distinct Pol III factors, TFIIIB and TFIIIC2, are both targeted for activation by SV40.
The deregulation of TFIIIC2 in SV3T3 cells was first detected by EMSA, which revealed an overall increase in DNA-binding activity and a greater proportion of the transcriptionally active form (60). These observations can be explained by our new data which show that SV40 transformation is accompanied by an increase in the abundance of both TFIIICα and TFIIICβ, two of the principal components of the TFIIIC2 complex. Cross-linking has demonstrated that the TFIIICα subunit is responsible for contacting promoter DNA (3, 17, 63). In contrast, the presence of TFIIICβ is not required for DNA binding but is necessary for transcriptional activity (17, 43). Both the α and β subunits have been reported to display histone acetyltransferase activity (18). TFIIIC2 also contains three other subunits (17, 63), but the cloning of cDNAs encoding these subunits has not yet been reported and their roles have yet to be established. In the absence of molecular reagents that recognize the uncharacterized components of TFIIIC2, we cannot determine whether these are also up-regulated in SV3T3 cells. However, it seems probable that they would be, in order to allow an overall increase in the level of functional TFIIIC2. Our RT-PCR analyses have demonstrated that the transcripts encoding TFIIICα and TFIIICβ are more abundant in Cl38 and Cl49 cells than they are in the parental A31 cells. This is likely to account, at least in part, for the elevated levels of the TFIIICα and TFIIICβ polypeptides.
SV40 transformation of A31 cells also results in the activation of TFIIIB. In contrast to TFIIIC2, this does not appear to result from an increase in the abundance of TFIIIB subunits. Thus, Western blotting reveals little or no change in the levels of TBP and BRF when A31 extracts are compared with Cl38 and Cl49 extracts. Human TFIIIB is incompletely characterized at present but is believed to contain at least one additional subunit besides TBP and BRF (27, 30, 46, 47). We cannot exclude the possibility that the abundance of one or more uncharacterized component(s) of the complex becomes elevated in response to SV40 transformation. However, it is not necessary to invoke such a contingency in order to explain the observed activation of TFIIIB. Both biochemical and genetic data have shown that TFIIIB is subject to negative regulation in untransformed murine fibroblasts through a physical interaction with RB (24, 61). Coimmunoprecipitation analyses reveal that much less of the TFIIIB in SV3T3 cells is associated with RB (Fig. 5 and 6). The higher specific activity of TFIIIB that is observed in Cl38 and Cl49 cells (Fig. 4) is therefore likely due, at least in part, to a partial release from repression by RB. This interpretation is supported strongly by the fact that recombinant SV40 T antigen can stimulate Pol III transcription in a partially purified system in an RB-dependent fashion (Fig. 3). These data are consistent with a model in which T antigen counteracts the inhibitory influence of RB on TFIIIB.
This study has described two distinct mechanisms which allow SV40-transformed cells to maintain abnormally high levels of Pol III transcription, namely, the increased abundance of TFIIIC2 and the release of TFIIIB from repression by RB. Given the complexity of gene regulation by SV40, it is entirely possible that additional levels of control are also involved. For example, T antigen has been shown to bind and neutralize the tumor suppressor p53, and this capability is important for the transformation of some cell types (22, 48, 65). Among its many functions, p53 has been found to interact with TFIIIB and repress Pol III transcription (4, 6). Inactivation of p53 might therefore provide an additional mechanism whereby T antigen is able to deregulate TFIIIB. However, this may not have a major effect in the 3T3 system investigated here, since p53 levels are extremely low in unstressed A31 cells (25).
Damania et al. have reported recently that T antigen can be found in a complex with TFIIIB, a conclusion based on both cofractionation and coimmunoprecipitation experiments with extracts prepared from HeLa cells that had been infected with a recombinant adenovirus which overexpresses SV40 large T antigen (9). The authors suggested that binding to TFIIIB may allow T antigen to activate Pol III transcription directly, although no evidence was presented to establish this model (9). Our experiments with partially purified factors are not consistent with a direct effect on TFIIIB, since T antigen was found to activate Pol III transcription only in the presence of added RB (Fig. 3). It is possible that T antigen acts directly on TFIIIB under different experimental conditions. However, we have been unable to detect T antigen associated with TFIIIB in extracts of SV3T3 cells. Thus, T antigen is not detected in anti-BRF immunoprecipitates, nor is BRF detected in anti-T immunoprecipitates from Cl38 or Cl49 cells, even though both cell lines express high levels of T antigen (44). We can therefore find no evidence that T antigen acts directly on TFIIIB in the SV3T3 cell system.
The up-regulation of both TFIIIB and TFIIIC2 is likely to ensure that SV40-transformed cells maintain high rates of Pol III transcription. At present it is not clear which target is of greater importance in determining the overall level of class III gene expression. Add-back experiments carried out previously with crude fractions have suggested that TFIIIC activity is limiting in extracts prepared from A31, Cl38, and Cl49 cells (60). If TFIIIC is also limiting for Pol III transcription in vivo, then the up-regulation of this factor may make the principal contribution to the activation observed in SV3T3 cells. However, much of the complexity of the situation is likely to be concealed when asynchronous cell populations are analyzed. Thus, experiments with synchronized HeLa populations have revealed that alternative Pol III factors can be limiting during different phases of the cell cycle (55). Whereas TFIIIC activity limits the rate of VAI expression in extracts of S- or G2-phase HeLa cells, TFIIIB is the limiting factor in extracts of cells that were harvested during early G1 phase (55). Under these circumstances, stimulation of TFIIIB or TFIIIC alone might influence the transcriptional output only during a restricted interval of the cell cycle. If a similar situation pertains in 3T3 cells, then the activation of both TFIIIB and TFIIIC might allow SV40 to sustain elevated rates of Pol III transcription throughout interphase.
Like SV40, adenovirus appears able to use more than one mechanism to stimulate the production of Pol III products. Adenovirus infection results in a marked increase in TFIIIC2 activity (15, 16, 43, 62). Western blotting revealed that the infected cells express elevated levels of TFIIICβ, although TFIIICα was not induced under these circumstances (43). Induction of TFIIICβ requires the adenoviral transforming protein E1A (43). In addition to its effect on TFIIICβ, E1A has also been shown to relieve Pol III transcription from repression by RB, both in vitro and in vivo (61). Adenovirus therefore resembles SV40 in employing at least two distinct mechanisms to activate the Pol III machinery. The present study has provided evidence to suggest that a third DNA tumor virus, HPV, can also stimulate Pol III transcription by overcoming the repressive influence of RB. We have shown that the E7 oncoprotein of HPV activates VAI expression in transfected fibroblasts (Fig. 7). This capacity is abolished by mutations that specifically prevent E7 from binding and neutralizing RB. Previous work has shown that hepatitis B virus and human T-cell leukemia virus type 1 can also stimulate Pol III transcription by activating TFIIIB (2, 14, 19, 34, 49, 50). It therefore seems that TFIIIB is a frequent target for transforming viruses (reviewed in reference 54). High rates of Pol III transcription are a prerequisite of rapid growth, in order to sustain the production of tRNA and 5S rRNA. It has therefore been suggested that repression of the Pol III system may provide a mechanism for restraining cell growth (23, 33, 52). The fact that this system is targeted by two unrelated tumor suppressors, RB and p53, provides strong support for this contention. Additional support is provided by the range of oncogenic viruses that have evolved mechanisms to stimulate Pol III transcription.
ACKNOWLEDGMENTS
We are extremely grateful to K. Vousden and R. Watson for the E7 expression vectors, D. Lane for recombinant T antigen, N. Hernandez for antibody SL30, and Y. Shen and A. Berk for antibodies Ab2 and Ab4.
This work was funded by grant SP2314/0101 to R.J.W. from the Cancer Research Campaign and grant 98-46 to R.J.W. from the Association for International Cancer Research. J.E.S. and Z.A.F-E. are supported by postgraduate studentships from the Biotechnology and Biological Sciences Research Council and the Medical Research Council, respectively. R.J.W. is a Jenner Research Fellow of the Lister Institute of Preventive Medicine.
REFERENCES
- 1.Alzuherri H M, White R J. Regulation of a TATA-binding protein-associated factor during cellular differentiation. J Biol Chem. 1998;273:17166–17171. doi: 10.1074/jbc.273.27.17166. [DOI] [PubMed] [Google Scholar]
- 2.Aufiero B, Schneider R J. The hepatitis B virus X-gene product trans-activates both RNA polymerase II and III promoters. EMBO J. 1990;9:497–504. doi: 10.1002/j.1460-2075.1990.tb08136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boulanger P A, Yoshinaga S K, Berk A J. DNA-binding properties and characterization of human transcription factor TFIIIC2. J Biol Chem. 1987;262:15098–15105. [PubMed] [Google Scholar]
- 4.Cairns C A, White R J. p53 is a general repressor of RNA polymerase III transcription. EMBO J. 1998;17:3112–3123. doi: 10.1093/emboj/17.11.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey M F, Singh K, Botchan M, Cozzarelli N R. Induction of specific transcription by RNA polymerase III in transformed cells. Mol Cell Biol. 1986;6:3068–3076. doi: 10.1128/mcb.6.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesnokov I, Chu W-M, Botchan M R, Schmid C W. p53 inhibits RNA polymerase III-directed transcription in a promoter-dependent manner. Mol Cell Biol. 1996;16:7084–7088. doi: 10.1128/mcb.16.12.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu W-M, Wang Z, Roeder R G, Schmid C W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 8.Clark M E, Dasgupta A. A transcriptionally active form of TFIIIC is modified in poliovirus-infected HeLa cells. Mol Cell Biol. 1990;10:5106–5113. doi: 10.1128/mcb.10.10.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damania B, Mital R, Alwine J C. Simian virus 40 large T antigen interacts with human TFIIB-related factor and small nuclear RNA-activating protein complex for transcriptional activation of TATA-containing polymerase III promoters. Mol Cell Biol. 1998;18:1331–1338. doi: 10.1128/mcb.18.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean N, Berk A J. Ordering promoter binding of class III transcription factors TFIIIC1 and TFIIIC2. Mol Cell Biol. 1988;8:3017–3025. doi: 10.1128/mcb.8.8.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeCaprio J A, Ludlow J W, Figge J, Shew J-Y, Huang C-M, Lee W-H, Marsilio E, Paucha E, Livingston D M. SV40 large tumour antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54:275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 12.Dyson N, Howley P M, Munger K, Harlow E. The human papillomavirus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 13.Ewen M E, Ludlow J W, Marsilio E, DeCaprio J A, Millikan R C, Cheng S H, Paucha E, Livingston D M. An N-terminal transformation-governing sequence of SV40 large T antigen contributes to the binding of both p110Rb and a second cellular protein, p120. Cell. 1989;58:257–267. doi: 10.1016/0092-8674(89)90840-4. [DOI] [PubMed] [Google Scholar]
- 14.Gottesfeld J M, Johnson D L, Nyborg J K. Transcriptional activation of RNA polymerase III-dependent genes by the human T-cell leukemia virus type 1 Tax protein. Mol Cell Biol. 1996;16:1777–1785. doi: 10.1128/mcb.16.4.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoeffler W K, Kovelman R, Roeder R G. Activation of transcription factor IIIC by the adenovirus E1A protein. Cell. 1988;53:907–920. doi: 10.1016/s0092-8674(88)90409-6. [DOI] [PubMed] [Google Scholar]
- 16.Hoeffler W K, Roeder R G. Enhancement of RNA polymerase III transcription by the E1A gene product of adenovirus. Cell. 1985;41:955–963. doi: 10.1016/s0092-8674(85)80076-3. [DOI] [PubMed] [Google Scholar]
- 17.Kovelman R, Roeder R G. Purification and characterization of two forms of human transcription factor IIIC. J Biol Chem. 1992;267:24446–24456. [PubMed] [Google Scholar]
- 18.Kundu T K, Wang Z, Roeder R G. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwee L, Lucito R, Aufiero B, Schneider R J. Alternate translation initiation on hepatitis B virus X mRNA produces multiple polypeptides that differentially transactivate class II and III promoters. J Virol. 1992;66:4382–4389. doi: 10.1128/jvi.66.7.4382-4389.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagna G, Kovelman R, Sukegawa J, Roeder R G. Cloning and characterization of an evolutionarily divergent DNA-binding subunit of mammalian TFIIIC. Mol Cell Biol. 1994;14:3053–3064. doi: 10.1128/mcb.14.5.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam E W-F, Morris J D H, Davies R, Crook T, Watson R J, Vousden K H. HPV16 E7 oncoprotein deregulates B-myb expression: correlation with targeting of p107/E2F complexes. EMBO J. 1994;13:871–878. doi: 10.1002/j.1460-2075.1994.tb06330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane D P, Crawford L V. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278:261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 23.Larminie C G C, Alzuherri H M, Cairns C A, McLees A, White R J. Transcription by RNA polymerases I and III: a potential link between cell growth, protein synthesis and the retinoblastoma protein. J Mol Med. 1998;76:94–103. doi: 10.1007/s001090050196. [DOI] [PubMed] [Google Scholar]
- 24.Larminie C G C, Cairns C A, Mital R, Martin K, Kouzarides T, Jackson S P, White R J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997;16:2061–2071. doi: 10.1093/emboj/16.8.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larminie, C. G. C., and R. J. White. Unpublished observations.
- 26.L’Etoile N D, Fahnestock M L, Shen Y, Aebersold R, Bertc A J. Human transcription factor IIIC box B binding subunit. Proc Natl Acad Sci USA. 1994;91:1652–1656. doi: 10.1073/pnas.91.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobo S M, Tanaka M, Sullivan M L, Hernandez N. A TBP complex essential for transcription from TATA-less but not TATA-containing RNA polymerase III promoters is part of the TFIIIB fraction. Cell. 1992;71:1029–1040. doi: 10.1016/0092-8674(92)90397-u. [DOI] [PubMed] [Google Scholar]
- 28.Loeken M, Bikel I, Livingston D M, Brady J. Transactivation of RNA polymerase II and III promoters by SV40 small t antigen. Cell. 1988;55:1171–1177. doi: 10.1016/0092-8674(88)90261-9. [DOI] [PubMed] [Google Scholar]
- 29.Manley J L, Fire A, Cano A, Sharp P A, Gefter M L. DNA-dependent transcription of adenovirus genes in a soluble whole-cell extract. Proc Natl Acad Sci USA. 1980;77:3855–3859. doi: 10.1073/pnas.77.7.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran E. A region of SV40 large T antigen can substitute for a transforming domain of the adenovirus E1A products. Nature. 1988;334:168–170. doi: 10.1038/334168a0. [DOI] [PubMed] [Google Scholar]
- 32.Munger K, Werness B A, Dyson N, Phelps W C, Harlow E, Howley P M. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumour suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nasmyth K. Another role rolls in. Nature. 1996;382:28–29. doi: 10.1038/382028a0. [DOI] [PubMed] [Google Scholar]
- 34.Piras G, Dittmer J, Radonovich M F, Brady J N. Human T-cell leukaemia virus type I Tax protein transactivates RNA polymerase III promoter in vitro and in vivo. J Biol Chem. 1996;271:20501–20506. doi: 10.1074/jbc.271.34.20501. [DOI] [PubMed] [Google Scholar]
- 35.Rigby P W J, Chia W, Clayton C E, Lovett M. The structure and expression of the integrated viral DNA in mouse cells transformed by Simian virus 40. Proc R Soc Lond Ser B. 1980;210:437–450. doi: 10.1098/rspb.1980.0145. [DOI] [PubMed] [Google Scholar]
- 36.Scott M R D, Westphal K-H, Rigby P W J. Activation of mouse genes in transformed cells. Cell. 1983;34:557–567. doi: 10.1016/0092-8674(83)90388-4. [DOI] [PubMed] [Google Scholar]
- 37.Segall J, Matsui T, Roeder R G. Multiple factors are required for the accurate transcription of purified genes by RNA polymerase III. J Biol Chem. 1980;255:11986–11991. [PubMed] [Google Scholar]
- 38.Shen Y, Igo M, Yalamanchili P, Berk A J, Dasgupta A. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol Cell Biol. 1996;16:4163–4171. doi: 10.1128/mcb.16.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shew J-Y, Lin B T-Y, Chen P-L, Tseng B Y, Yang-Feng T L, Lee W-H. C-terminal truncation of the retinoblastoma gene product leads to functional inactivation. Proc Natl Acad Sci USA. 1990;87:6–10. doi: 10.1073/pnas.87.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simmen K A, Bernues J, Lewis J D, Mattaj I W. Cofractionation of the TATA-binding protein with the RNA polymerase III transcription factor TFIIIB. Nucleic Acids Res. 1992;20:5889–5898. doi: 10.1093/nar/20.22.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmen K A, Bernues J, Parry H D, Stunnenberg H G, Berkenstam A, Cavallini B, Egly J M, Mattaj I W. TFIID is required for in vitro transcription of the human U6 gene by RNA polymerase III. EMBO J. 1991;10:1853–1862. doi: 10.1002/j.1460-2075.1991.tb07711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh K, Carey M, Saragosti S, Botchan M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985;314:553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- 43.Sinn E, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of a TFIIIC2 subunit (TFIIICβ) whose presence correlates with activation of RNA polymerase III-mediated transcription by adenovirus E1A expression and serum factors. Genes Dev. 1995;9:675–685. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- 44.Sutcliffe, J. E., and R. J. White. Unpublished observations.
- 45.Taggart A K P, Fisher T S, Pugh B F. The TATA-binding protein and associated factors are components of pol III transcription factor TFIIIB. Cell. 1992;71:1015–1028. doi: 10.1016/0092-8674(92)90396-t. [DOI] [PubMed] [Google Scholar]
- 46.Teichmann M, Dieci G, Huet J, Ruth J, Sentenac A, Seifart K H. Functional interchangeability of TFIIIB components from yeast and human cells in vitro. EMBO J. 1997;16:4708–4716. doi: 10.1093/emboj/16.15.4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teichmann M, Seifart K H. Physical separation of two different forms of human TFIIIB active in the transcription of the U6 or the VAI gene in vitro. EMBO J. 1995;14:5974–5983. doi: 10.1002/j.1460-2075.1995.tb00286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vousden K H. Regulation of the cell cycle by viral oncoproteins. Semin Cancer Biol. 1995;6:109–116. doi: 10.1006/scbi.1995.0014. [DOI] [PubMed] [Google Scholar]
- 49.Wang H-D, Trivedi A, Johnson D L. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signalling pathway. Mol Cell Biol. 1997;17:6838–6846. doi: 10.1128/mcb.17.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang H-D, Yuh C-H, Dang C V, Johnson D L. The hepatitis B virus X protein increases the cellular level of TATA-binding protein, which mediates transactivation of RNA polymerase III genes. Mol Cell Biol. 1995;15:6720–6728. doi: 10.1128/mcb.15.12.6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White R J. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]
- 53.White R J. RNA polymerase III transcription. New York, N.Y: Springer-Verlag; 1998. [Google Scholar]
- 54.White R J. Transcription factor IIIB: an important determinant of biosynthetic capacity that is targeted by tumour suppressors and transforming proteins. Int J Oncol. 1998;12:741–748. [PubMed] [Google Scholar]
- 55.White R J, Gottlieb T M, Downes C S, Jackson S P. Cell cycle regulation of RNA polymerase III transcription. Mol Cell Biol. 1995;15:6653–6662. doi: 10.1128/mcb.15.12.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.White R J, Gottlieb T M, Downes C S, Jackson S P. Mitotic regulation of a TATA-binding-protein-containing complex. Mol Cell Biol. 1995;15:1983–1992. doi: 10.1128/mcb.15.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White R J, Jackson S P. Mechanism of TATA-binding protein recruitment to a TATA-less class III promoter. Cell. 1992;71:1041–1053. doi: 10.1016/0092-8674(92)90398-v. [DOI] [PubMed] [Google Scholar]
- 58.White R J, Jackson S P, Rigby P W J. A role for the TATA-box-binding protein component of the transcription factor IID complex as a general RNA polymerase III transcription factor. Proc Natl Acad Sci USA. 1992;89:1949–1953. doi: 10.1073/pnas.89.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.White R J, Stott D, Rigby P W J. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- 60.White R J, Stott D, Rigby P W J. Regulation of RNA polymerase III transcription in response to Simian virus 40 transformation. EMBO J. 1990;9:3713–3721. doi: 10.1002/j.1460-2075.1990.tb07584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 62.Yoshinaga S, Dean N, Han M, Berk A J. Adenovirus stimulation of transcription by RNA polymerase III: evidence for an E1A-dependent increase in transcription factor IIIC concentration. EMBO J. 1986;5:343–354. doi: 10.1002/j.1460-2075.1986.tb04218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoshinaga S K, L’Etoile N D, Berk A J. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–10731. [PubMed] [Google Scholar]
- 64.Zalvide J, DeCaprio J A. Role of pRb-related proteins in simian virus 40 large-T-antigen-mediated transformation. Mol Cell Biol. 1995;15:5800–5810. doi: 10.1128/mcb.15.10.5800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu J, Rice P W, Gorsch L, Abate M, Cole C N. Transformation of a continuous rat embryo fibroblast cell line requires three separate domains of simian virus 40 large T antigen. J Virol. 1992;66:2780–2791. doi: 10.1128/jvi.66.5.2780-2791.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]