SUMMARY
SAGA (Spt-Ada-Gcn5 acetyltransferase) and ATAC (Ada-two-A-containing) are two related coactivator complexes, sharing the same histone acetyltransferase (HAT) subunit. The HAT activities of SAGA and ATAC are required for metazoan development, but the role of these complexes in RNA polymerase II transcription is less understood. To determine whether SAGA and ATAC have redundant or specific functions, we compare the effects of HAT inactivation in each complex with that of inactivation of either SAGA or ATAC core subunits in mouse embryonic stem cells (ESCs). We show that core subunits of SAGA or ATAC are required for complex assembly and mouse ESC growth and self-renewal. Surprisingly, depletion of HAT module subunits causes a global decrease in histone H3K9 acetylation, but does not result in significant phenotypic or transcriptional defects. Thus, our results indicate that SAGA and ATAC are differentially required for self-renewal of mouse ESCs by regulating transcription through different pathways in a HAT-independent manner.
In brief
Fischer et al. analyze the role of SAGA and ATAC coactivator complexes that share a histone acetyltransferase (HAT) subunit in mouse embryonic stem cells (ESCs). The authors separate the core and HAT activities of these complexes to show that they play non-redundant roles in maintaining ESCs, through HAT-independent pathways.
Graphical Abstract
INTRODUCTION
Mouse embryonic stem cells (ESCs), derived from the inner cell mass of blastocysts, can self-renew seemingly endlessly and can differentiate into most cell lineages (Evans and Kaufman, 1981; Martello and Smith, 2014; Martin, 1981). The molecular mechanisms underlying the self-renewal and pluripotency of mouse ESCs are dependent on key pluripotency transcription factors (TFs). These include core pluripotency TFs such as Oct4 (encoded by Pou5f1) and Sox2 and other naive-specific pluripotency TFs, such as Nanog, Tfcp2l1, Klf4, and Esrrb (Martello and Smith, 2014; Young, 2011). Recent findings indicate that the physiology of mouse ESCs is also dependent on several transcriptional coactivator complexes (Acharya et al., 2017; Festuccia et al., 2017; Li et al., 2012; Seruggia et al., 2019; Young, 2011). Coactivators contain enzymatic activities to either deposit or remove post-translational modifications of histone proteins or to mobilize core nucleosomes through ATP-dependent remodeling functions. Within the cell, these complexes facilitate RNA polymerase II (Pol II) transcription by modulating the chromatin environment and thereby enabling the access of the transcription machinery to the template DNA (Kouzarides, 2007; Li et al., 2007; Sainsbury et al., 2015; Young, 2011).
SAGA (Spt-Ada-Gcn5 acetyltransferase) and ATAC (Ada-two-A-containing) are two related coactivators, sharing the same histone acetyltransferase (HAT) enzyme, either Gcn5 (Kat2a) or its paralog Pcaf (Kat2b) (Helmlinger and Tora, 2017). Gcn5 and Pcaf are the vertebrate orthologs of yeast Gcn5, required for histone H3 acetylation, and are incorporated within SAGA or ATAC in a mutually exclusive manner. The association of Gcn5 or Pcaf with three adaptor proteins, Sgf29, Tada3, and either Tada2a or Tada2b, is required for full acetyltransferase activity and substrate specificity (Riss et al., 2015; Balasubramanian et al., 2002; Nagy et al., 2010; Yang et al., 1996). Tada2a specifically incorporates Gcn5/Pcaf within the ATAC HAT module, whereas Tada2b recruits Gcn5/Pcaf to the SAGA HAT module.
Loss of the catalytic subunit Gcn5 causes defects in mesoderm formation and early embryonic lethality, whereas Pcaf null embryos are viable and fertile (Bu et al., 2007; Koutelou et al., 2020; Xu et al., 2000; Yamauchi et al., 2000). Studies of SAGA and ATAC HAT functions in Drosophila revealed that Ada2a or Ada2b mutants have lethal phenotypes, but at different stages of development, suggesting that Ada2a and Ada2b have different functions in development. Such differences may be explained by differential recruitment of the two complexes at distinct sets of genes where their HAT activity would be required for target gene activation (Krebs et al., 2011; Pankotai et al., 2010).
The contribution of other functions of SAGA and ATAC complexes to mammalian development and transcription is less well characterized. Outside of their HAT module, SAGA and ATAC are composed of different and unrelated subunits, suggesting a functional diversity of these two complexes. SAGA is organized in four functional modules and was highly conserved throughout evolution. In addition to its HAT module, SAGA contains a histone deubiquitination (DUB) module, a core structural module, and a TF-binding module (Grant et al., 1997; Helmlinger et al., 2020; Henry et al., 2003; Soffers and Workman, 2020). Recent cryoelectron microscopy studies revealed that the yeast SAGA complex is organized around the core central module of SAGA, proposed to be important for TATA-box binding protein (TBP) delivery at promoters (Papai et al., 2020; Wang et al., 2020). In Saccharomyces cerevisiae, the SAGA complex cannot assemble in the absence of the core subunits Spt20, Spt7, or Ada1, suggesting that most SAGA functions are lost when these critical structural subunits are missing (Han et al., 2014; Lee et al., 2011; Sterner et al., 1999; Wu and Winston, 2002).
In contrast to SAGA, ATAC is exclusively found in metazoans and contains six other subunits (Yeats2, Zzz3, Atac2, Mbip, Wdr5, and Nc2β), in addition to the HAT module (Helmlinger et al., 2020; Spedale et al., 2012). The structural organization of ATAC and the functions of these additional subunits are less well known. In mammalian cells, SAGA or ATAC were shown to regulate different groups of genes, particularly genes involved in stress-induced signaling pathways (Krebs et al., 2011; Nagy et al., 2010; Spedale et al., 2012; Wang and Dent, 2014). However, recent genome-wide studies in human cells imply ATAC in the transcription regulation of genes involved in house-keeping functions, such as ribosome protein genes (RPGs) (Mi et al., 2017, 2018). As these studies used different experimental approaches and different cellular models, it is still unclear whether SAGA and ATAC have overlapping or specific functions in Pol II transcription and to which extent their functions rely on their shared HAT activities.
To better understand the role of SAGA and ATAC in mammalian transcription, we aimed at a comprehensive and comparative analysis of these two complexes in a single cell type. Using CRISPR-Cas9-mediated genome editing in mouse ESCs, we individually targeted different subunits specific to each complex as well as HAT subunits shared between SAGA and ATAC. These cell lines allowed us to show that, while loss of core subunits of SAGA and ATAC severely affected mouse ESC growth and self-renewal capacities, loss of HAT subunits did not cause apparent phenotypes. Genome-wide analysis of newly synthesized RNA revealed that loss of SAGA and ATAC subunits predominantly affected different sets of genes. Thus, our data suggest that SAGA and ATAC play important, but distinct, roles in the maintenance of mouse ESC self-renewal mainly through non-redundant, HAT-independent functions.
RESULTS
Inactivation of the core, but not the HAT, module of SAGA alters mouse ESC proliferation and self-renewal
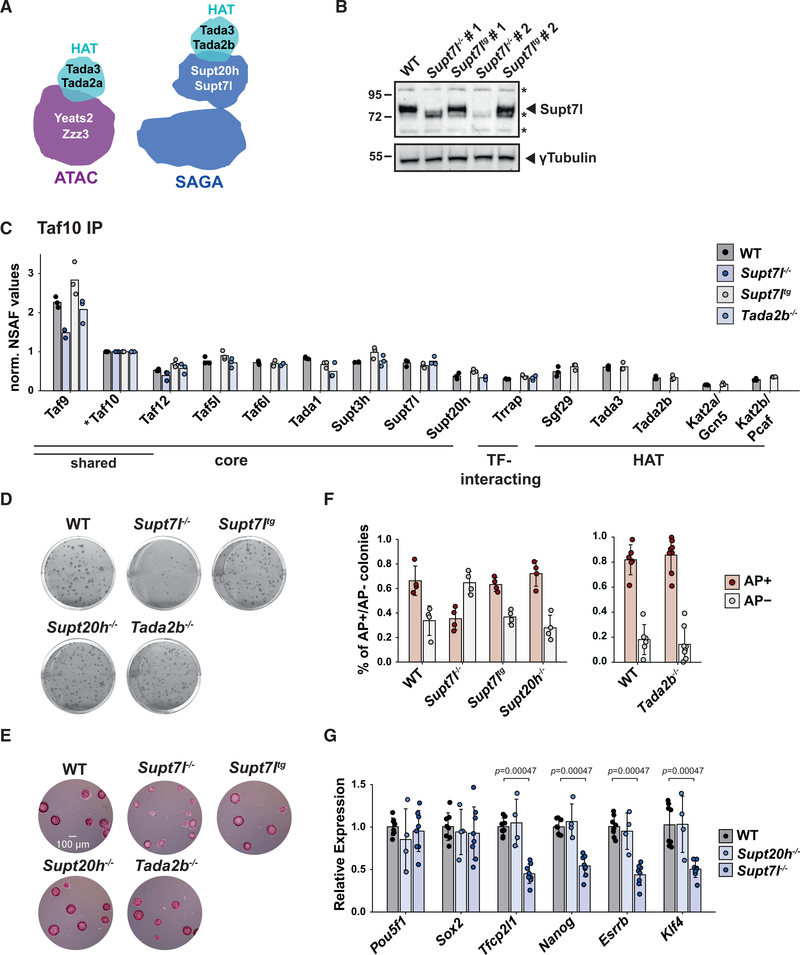
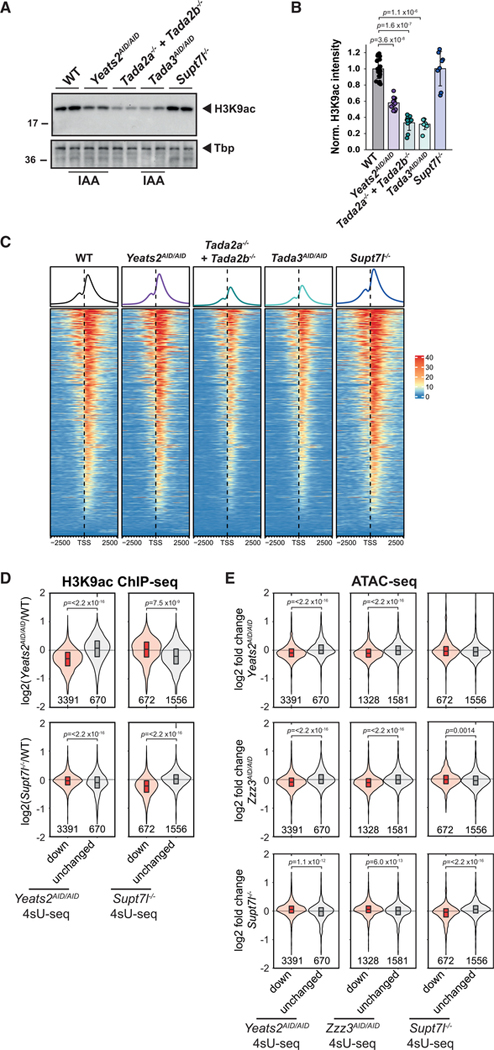
To understand the role of SAGA and ATAC in mouse ESCs, we systematically inactivated genes encoding subunits of different functional modules of these complexes, using CRISPR-Cas9 genome editing. We first targeted genes encoding subunits of the HAT (Tada2b) and core (Supt7l or Supt20h) modules of SAGA (Figures 1A and S1A). In S. cerevisiae, the orthologs of Supt7l and Supt20h are required for SAGA assembly, whereas the loss of Ada2, orthologous to Tada2b, specifically disrupts the HAT module (Han et al., 2014; Lee et al., 2011; Sterner et al., 1999; Wu and Winston, 2002). In the obtained homozygous mutant ESCs, Supt7l, Supt20h, or Tada2b mRNA analyses confirmed the deletion of the targeted out-of-frame exon and the resulting degradation of mRNA containing a premature stop codon (Figures S1B and S1C). Western blot analysis for Supt7l, for which a specific antibody was available, revealed undetectable levels of Supt7l (Figure 1B). These results indicate that these three SAGA subunits are not essential for mouse ESC survival, when cultured in medium containing fetal calf serum (FCS), leukemia inhibitory factor (LIF), and two inhibitors of mitogen-activated protein kinase kinase (MEK)/extracellular signal-regulated kinase (ERK) and glycogen synthase kinase 3 beta (GSK3b) pathways (hereafter referred to as FCS+LIF+2i medium).
Figure 1. Inactivation of the core module of SAGA alters mouse ESC proliferation and self-renewal.
(A) The HAT modules incorporate within SAGA or ATAC through Tada2a or Tada2b, respectively.
(B) Western blot analyses of two independent Supt7l−/− and Supt7ltg cell lines compared with wild-type (WT) cells. gTubulin serves as loading control. *, unspecific bands.
(C) Mass spectrometry analyses of SAGA complexes purified from Supt7l−/−, Supt7ltg, Tada2b−/−, and WT cells. *, bait protein. Shared indicates subunits shared between SAGA and TFIID. norm., normalized; NSAF, normalized spectral abundance factor.
(D) Representative images of clonal assays of SAGA mutant cells cultured in FCS+LIF medium and stained by crystal violet.
(E) Alkaline phosphatase (AP) staining on clonal assays of SAGA mutant lines cultured in FCS+LIF+2i medium. Scale bar represents 100 μm.
(F) Quantification of AP staining as shown in (D). Numbers of AP+ and AP− colonies were normalized to the total number of colonies.
(G) Total mRNA levels of pluripotency factors in WT, Supt20h−/−, and Supt7l−/− cells, normalized to RNA polymerase III genes (Rpph1 and Rn7sk) and to WT cells. For (F) and (G), the statistical test performed is Wilcoxon rank-sum test with Benjamini-Hochberg correction for multiple testing. Error bars show mean ± standard deviation (SD) of at least 4 biological replicates, using at least two independent clones. Only statistically significant (<0.05) results are indicated. See also Figure S1.
To determine how the loss of Supt7l, Supt20h, or Tada2b affects SAGA assembly, we purified SAGA complexes from nuclear extracts that were first depleted for TFIID, which shares subunits with SAGA. Depleted extracts were then subjected to a second immuno-purification using an anti-Taf10 antibody and analyzed by mass spectrometry or western blotting. Complexes purified from Supt7l−/− cells do not contain any SAGA subunits, with the exception of remaining TFIID subunits (Taf9, Taf10, and Taf12), suggesting a disorganization of SAGA in the absence of Supt7l (Figures 1C and S1D). By contrast, the loss of Supt20h in ESCs is less deleterious, as all subunits of the SAGA core module are found in amounts comparable with that of wild-type cells (Figures S1D and S1E). Finally, in Tada2b−/− cells, we observed that the whole HAT module dissociated from an intact core SAGA complex (Figures 1C and S1D).
To assess the role of the SAGA core and HAT modules in ESC proliferation, we performed clonal assays in FCS+LIF and FCS+LIF+2i medium and observed that the loss of Supt7l affects colony formation of mouse ESCs (Figures 1D, 1E, and S1F). Expression of a Supt7l transgene in Supt7l−/− ESCs (Supt7ltg) fully rescues the growth phenotype observed in a Supt7l null background, indicating a direct effect of the loss of Supt7l (Figures 1B, 1D, 1E, S1F, and S1G). By contrast, inactivation of Supt20h does not affect ESC growth, which appears similar to that of wild-type cells. Importantly, inactivation of Tada2b does not cause any defect in ESC proliferation, suggesting that the growth defect observed upon loss of Supt7l results from HAT-independent activities of the SAGA complex (Figures 1D, 1E, and S1F). In agreement, growth curve analyses based on the numbers of viable cells in FCS+LIF+2i medium further suggested a slight growth defect in Supt7l−/− cells, but not in Tada2b−/− cells (Figure S1H).
To further investigate the impact of SAGA on ESC self-renewal, we performed clonal assays in FCS+LIF medium coupled with alkaline phosphatase (AP) staining and quantified the proportion of undifferentiated colonies having high levels of AP (AP positive) and that of differentiated colonies that remain unstained (AP negative). These analyses indicated that the capacity of ESCs to self-renew is reduced upon Supt7l inactivation, but is restored upon re-expression of Supt7l in Supt7ltg cells (Figure 1F). By contrast, we found that inactivation of Supt20h or Tada2b has no effect on the number of AP-positive undifferentiated colonies (Figure 1F). We further quantified the steady-state mRNA levels of several pluripotency factors in Supt7l−/− cells and observed reduced expression of four of the six factors analyzed, namely Tfcp2l1, Nanog, Esrrb, and Klf4. By contrast, Pou5f1 (encoding Oct4) and Sox2 mRNA levels are unaffected (Figure 1G). In agreement with unaffected self-renewal observed in Supt20h−/− cells, mRNA levels for all tested pluripotency factors do not change compared with wild-type cells (Figure 1G).
Overall, our results demonstrate that the SAGA complex disassembles in the absence of Supt7l, ultimately leading to defects in mouse ESC growth and self-renewal. Inactivation of Supt20h had milder effects on SAGA subunit composition with core SAGA complexes still assembling. Importantly, SAGA function in ESC physiology appears mostly independent of its HAT activity.
ATAC core subunits are required for ESC survival
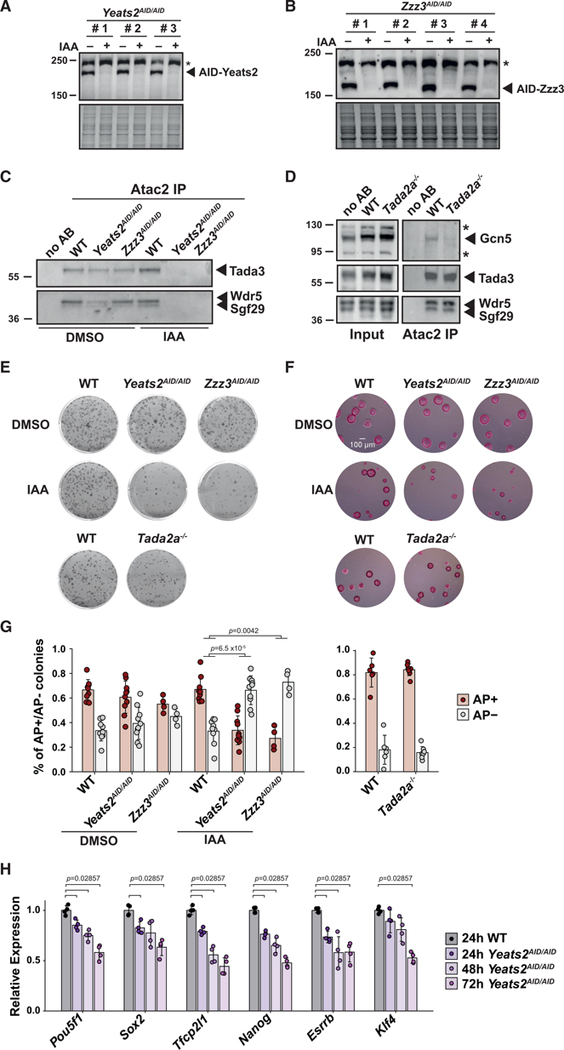
To compare the role of ATAC and SAGA complexes in ESCs, we further inactivated genes encoding subunits of the HAT and central core module of ATAC. We therefore targeted Tada2a, which specifically anchors the HAT module within ATAC, as well as Yeats2 and Zzz3, which were shown to be important for ATAC-mediated histone acetylation and gene expression (Mi et al., 2017, 2018; Wang et al., 2008) (Figures 1A and S2A). Homozygous inactivation of Tada2a was confirmed by mRNA analyses in the obtained Tada2a−/− cell lines (Figures S2B and S2C). However, we could not isolate any homozygous Yeats2 or Zzz3 mutant clones, suggesting that ATAC functions, but not its Tada2a-dependent HAT activity, are essential for mouse ESC survival and cannot be compensated by SAGA (Figure S2B). Therefore, we turned to an inducible degradation system and introduced the auxin-inducible degron (AID) sequence on both alleles of the Yeats2 (Yeats2AID/AID) or Zzz3 (Zzz3AID/AID) genes in an ESC line expressing the Tir1 protein (Natsume et al., 2016). The levels of the AID-Yeats2 and AID-Zzz3 fusion proteins are undetectable after 24 h of auxin (indole-3-acetic acid [IAA]) treatment (Figures 2A and 2B).
Figure 2. The ATAC complex is required for mouse ESC survival.
(A and B) Western blot analyses of protein extracts prepared from ESC lines expressing AID-Yeats2 or AID-Zzz3. Three Yeats2AID/AID clones (A) and four Zzz3AID/AID clones (B) were treated for 24 h with (+) or without (−) auxin (IAA). *, unspecific bands.
(C and D) ATAC was purified from nuclear extracts of WT, Yeats2AID/AID, and Zzz3AID/AID cells treated for 24 h with IAA or not (DMSO) (C) or WT and Tada2a−/− cells (D) by using anti-Atac2 antibodies and peptide elution. Beads incubated without antibody (no AB) are shown as control.
(E) Clonal assays of ATAC mutant cell lines compared with WT cells in FCS+LIF medium and stained with crystal violet. AID cell lines were treated with either DMSO or IAA.
(F) Clonal assays of cell lines in FCS+LIF+2i medium and stained with AP. Scale bar represents 100 μm.
(G) Quantification of AP staining in clonal assays as shown in (E), normalized as in Figure 1F.
(H) mRNA levels of pluripotency factors in Yeats2AID/AID mutant and WT cells upon 24–72 h of IAA treatment were normalized as in Figure 1G. Statistical test performed is Wilcoxon rank-sum test with Benjamini-Hochberg correction for multiple testing for (G) and two-sided Wilcoxon-Mann- Whitney test for (H). Error bars show mean ± SD of at least 4 biological replicates, using at least two independent clones. Only statistically significant (<0.05) results are indicated. See also Figure S2.
To determine ATAC complex integrity upon loss of Tada2a or depletion of Yeats2 or Zzz3, we performed immuno-purification experiments using antibodies targeting ATAC-specific subunits Atac2 or Mbip. Western blot analyses showed that Atac2 copurifies with Tada3, Wdr5, or Sgf29 from wild-type cells and from Yeats2AID/AID or Zzz3AID/AID cell lines in the absence of auxin treatment, indicating that the fusion of the AID sequence to Yeats2 or Zzz3 has no major effect on complex assembly (Figure 2C). By contrast, upon auxin-induced depletion of AID-Yeats2 or AID-Zzz3, the tested subunits are no longer detected in the Atac2-containing complexes, showing that the loss of Yeats2 or Zzz3 affects ATAC organization (Figure 2C). Mass spectrometry analyses of Mbip-associated complexes similarly indicate that ATAC is disassembled upon depletion of Yeats2 or Zzz3 with undetectable levels of all HAT subunits (Figure S2D). By contrast, the characterization of complexes immuno-purified from Tada2a−/− and parental cells revealed that the core ATAC module is not affected and that subunits of the HAT module are still present, but in reduced amount when Tada2a is missing (Figures 2D and S2D). As the lack of Tada2a within ATAC is expected to dramatically affect the catalytic activities of Gcn5/Pcaf (Balasubramanian et al., 2002; Grant et al., 1997; Ringel et al., 2015; Riss et al., 2015), we quantified H3K9 acetylation (H3K9ac) in cells lacking Tada2a or upon depletion of Yeats2 or Zzz3. H3K9ac levels are decreased in auxin-treated Yeats2AID/AID or Zzz3AID/AID cells, in agreement with the observed dissociation of the HAT module from ATAC (Figure S2E). In Tada2a−/− cells, H3K9ac levels are decreased to a similar extent, indicating a dramatic loss of the ATAC HAT activity upon loss of Tada2a (Figure S2E).
To assess the impact of ATAC inactivation on mouse ESC growth, we performed clonal assays with or without auxin in the Yeats2AID/AID, Zzz3AID/AID, and control cell lines cultured either in FCS+LIF or FCS+LIF+2i medium. In the absence of auxin, the size of colonies is comparable between all cell lines. Upon continuous auxin treatment for six days, the size of Yeats2AID/AID or Zzz3AID/AID colonies is reduced compared with that of control cells (Figures 2E, 2F, and S2F). By contrast, we could not detect any growth defect in Tada2a−/− ESCs, indicating that the effects observed upon Yeats2 or Zzz3 depletion are mostly HAT independent (Figures 2E, 2F, and S2F). In addition, growth curve analyses based on the number of viable cells cultured in FCS+LIF+2i further show reduced proliferation rates upon depletion of Yeats2 or Zzz3, but not upon loss of Tada2a, in agreement with the smaller colony area measured in clonal assays (Figures S2F and S2G).
Further AP staining on cells cultured in FCS+LIF in the absence of auxin revealed that the proportion of differentiated and undifferentiated cells is similar in Yeats2AID/AID, Zzz3AID/AID, Tada2a−/−, and wild-type cells (Figure 2G). By contrast, upon depletion of Yeats2 or Zzz3, the fraction of undifferentiated AP-positive colonies is strongly reduced, compared with that of control cells (Figure 2G). In agreement with impaired self-renewal, mRNA levels of all tested pluripotency factors progressively declined upon depletion of AID-Yeats2 (Figure 2H). As earlier studies suggested a role for ATAC subunits in cell-cycle regulation (Fournier et al., 2016; Guelman et al., 2009; Orpinell et al., 2010), we further analyzed cell-cycle distribution in ATAC mutant cells. Depletion of Yeats2 or Zzz3 led to an increase in the proportion of cells in G1 after 48 h of auxin treatment, whereas no differences could be detected upon 24 h of auxin treatment (Figure S2H).
As shown for SAGA, depletion of core ATAC subunits reduces the number of undifferentiated, pluripotent ESCs, indicating that both SAGA and ATAC have important functions to maintain the ESC state. Importantly, these functions appear to be mostly independent of their HAT activities.
SAGA and ATAC have HAT-independent functions, crucial to maintaining the ESC state
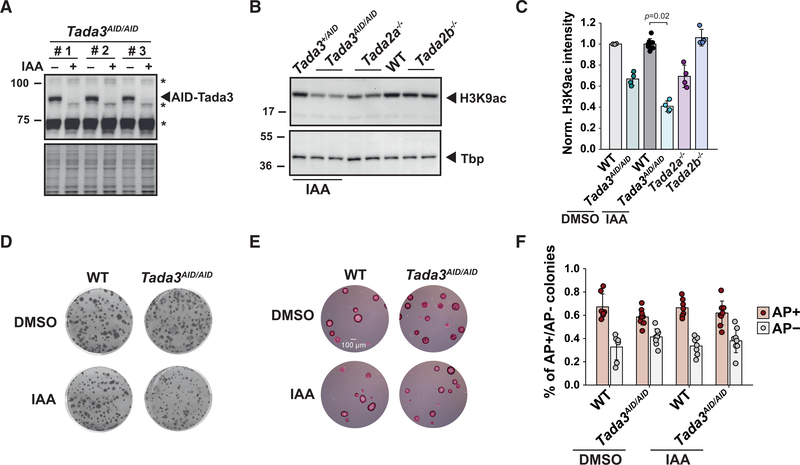
Next, we asked whether the HAT activities of SAGA and ATAC compensate each other and whether this compensation would explain the lack of phenotype observed in either Tada2b−/− or Tada2a−/− deletion mutants. For this, we inactivated Tada3 to supress the HAT activities of both complexes simultaneously (Figure 1A). As Tada3 was reported to be required for inner cell mass formation (Mohibi et al., 2012), we generated mouse ESC lines with homozygous insertion of AID sequence at the Tada3 locus (Tada3AID/AID cell line). AID-Tada3 fusion protein was undetectable after 24 h of auxin treatment (Figure 3A). As a proxy for HAT inactivation, we measured the levels of H3K9ac, an in vivo target of Gcn5 and Pcaf. Whereas we observed a mild reduction of H3K9ac levels upon loss of either SAGA (Tada2b−/−) or ATAC (Tada2a−/−) HAT activity, H3K9ac levels are significantly reduced (by about 60%) in Tada3AID/AID cells treated with auxin for 24 h (Figures 3B and 3C). Surprisingly, the AID fusion to Tada3 interferes with Gcn5 and Pcaf enzymatic activities as H3K9ac levels were reduced by 30% in the absence of auxin treatment in Tada3AID/AID cells (Figure 3C).
Figure 3. Loss of the shared acetyltransferase activity of SAGA and ATAC does not affect proliferation or self-renewal of mouse ESCs.
(A) Western blot analyses of protein extracts prepared from three Tada3AID/AID cell lines treated with (+) or without (−) IAA for 24 h. *, unspecific bands.
(B) Western blot analyses of histone H3 lysine 9 acetylation (H3K9ac) levels in extracts prepared from Tada2a−/−, Tada2b−/−, and Tada3AID/AID cells treated for 24 h with IAA. Control cells are either heterozygous Tada3+/AID or WT cells. Tbp serves as loading control.
(C) Quantification of H3K9ac levels normalized to Ponceau staining. Error bars show mean ± SD of at least 4 biological replicates using at least two independent clones. Only statistically significant (p < 0.05) results are indicated. Statistical test performed is Wilcoxon rank-sum test with Benjamini-Hochberg correction for multiple testing.
(D and E) Clonal assays of Tada3AID/AID and WT cells cultured in FCS+LIF (D) or FCS+LIF+2i (E) medium treated with DMSO or IAA and stained with crystal violet (D) or AP (E). Scale bar represents 100 μm.
(F) Quantification of AP staining of clonal assays as shown in (D), normalized as in Figure 1F. Error bars show mean ± SD of 8 biological replicates. At least two independent clones were analyzed per cell line. Statistical test performed is two-sided Wilcoxon-Mann-Whitney test. No statistically significant (p < 0.05) differences were detected. See also Figure S3.
Immuno-purifications of SAGA and ATAC complexes revealed residual AID-Tada3 within both complexes, together with other subunits of the HAT module (Gcn5 or Sgf29), although the fusion protein was undetectable in whole-cell extracts (Figures S3A and S3B). To further characterize the impact of Tada3 depletion on SAGA and ATAC HAT activities, we measured H3K9ac levels in a double Tada2a−/−+Tada2b−/− ESC line. H3K9ac levels are reduced in these double mutant cells to a similar extent than upon depletion of Tada3 (Figure S3C), suggesting a comparable reduction of SAGA and ATAC HAT activities in both cell lines.
As observed for Tada2a−/− or Tada2b−/− cells, clonal assays performed in FCS+LIF or FCS+LIF+2i medium, as well as growth curve analyses in FCS+LIF+2i medium, did not evidence any growth defect upon depletion of Tada3 (Figures 3D, 3E, and S3E). In addition, the proportion of differentiated and undifferentiated ESC colonies is similar in Tada3AID/AID and control cells treated with or without auxin (Figure 3F). These results indicate that the combined loss of SAGA and ATAC HAT activities has no detectable effects on mouse ESC growth and self-renewal. The role of SAGA and ATAC in these processes is therefore related to HAT-independent activities of these complexes. This further suggests that SAGA and ATAC functions required to maintain the ESC state are different, as SAGA and ATAC do not share any other subunit besides their HAT modules.
Newly synthesized RNA quantification reveals non-overlapping roles for SAGA and ATAC in Pol II transcription
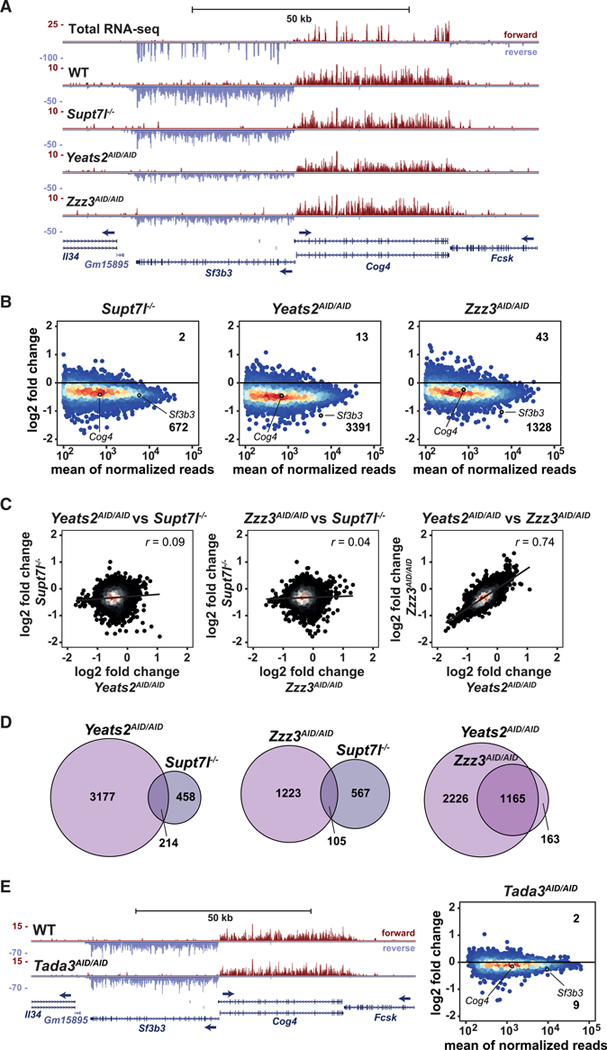
To determine the role of SAGA and ATAC in Pol II transcription in ESCs, we used 4-thiouridine (4sU) labeling of newly synthesized RNA coupled with sequencing of the labeled RNA (4sU-seq) (Rädle et al., 2013; Schwalb et al., 2016). Two independent clones of Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID lines as well as control cells cultured in FCS+LIF+2i were treated for 24 h with auxin prior to 4sU labeling, followed by purification and quantification of the labeled RNA by sequencing. By using 25 min 4sU labeling, we observed throughout all samples a very similar enrichment in in-tronic reads, compared with total RNA sequencing (RNA-seq), indicating an enrichment for unspliced transcripts in a highly reproducible manner (Figure S4A). The efficient purification of newly synthesized RNA was further evidenced when comparing 4sU-seq profiles with that of total RNA-seq at individual representative genes, revealing the presence of mainly unprocessed transcripts as well as unstable transcripts such as upstream antisense RNAs (Figures 4A and S4B).
Figure 4. SAGA and ATAC regulate the expression of different sets of genes.
(A) Genome browser view showing newly synthesized mRNA sequencing coverage of Sf3b3 and Cog4 in Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID cell lines compared with WT cells. The top panel shows total RNA-seq coverage in WT cells. Blue arrows indicate transcription direction.
(B) MA plots showing log2 fold changes (log2 FCs) of newly synthesized mRNA levels in Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID cell lines relative to WT cells against the mean of normalized reads. For each cell line, two independent clones were treated for 24 h with IAA. Numbers of significantly up- and downregulated genes are indicated. An adjusted p value of 0.05 and absolute log2 FC of 0.5 were used as thresholds for significantly affected genes.
(C) Correlation analyses of log2 FCs of newly synthesized mRNA between Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID lines.
(D) Venn diagrams comparing overlaps of significantly downregulated genes between Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID cells.
(E) Left, genome browser view showing newly synthesized mRNA sequencing coverage of Sf3b3 and Cog4 in WT and Tada3AID/AID cells treated with IAA for 24 h. Right, MA plot showing log2 FCs of newly synthesized mRNA levels in Tada3AID/AID cells relative to WT cells treated with IAA. See also Figure S4.
In Supt7l null cells, the newly synthesized mRNA levels of 672 genes (out of 8,201 expressed protein-coding genes) are significantly decreased, when applying a threshold of −0.5 log2 fold change and an adjusted p value below 0.05 (Figure 4B). For many other genes, pre-mRNA levels decrease moderately, without reaching statistical significance. By contrast, only two genes are significantly upregulated in Supt7l−/− cells. Upon depletion of the ATAC subunits, Yeats2 and Zzz3, we observed a significant decrease in newly synthesized mRNA levels for a large number of genes (3,391 and 1,328, respectively), whereas very few genes are upregulated (Figure 4B). Examination of MA plots indicated that the synthesis of a majority of genes is decreased upon Yeats2 or Zzz3 depletion, suggesting that ATAC may have global effects on Pol II transcription (Figure 4B). This mild global effect appears specific to Pol II transcription, as newly synthesized rRNA transcribed by RNA Pol I are unchanged in SAGA or ATAC mutant ESCs (Figure S4C). Importantly, the comparison of gene expression changes induced by Yeats2 or Zzz3 depletion revealed a strong correlation between the two datasets (Pearson correlation coefficient = 0.74), with about 87% of genes significantly downregulated by Zzz3 depletion being also significantly affected upon Yeats2 depletion (Figures 4C and 4D). By contrast, no correlation is observed between SAGA mutant (Supt7l−/−) and both ATAC mutants (Yeats2AID/AID or Zzz3AID/AID) (Figure 4C). When comparing genes significantly downregulated in these different cell lines, we found only little overlap between genes affected upon inactivation of Supt7l and those downregulated by the loss of ATAC core subunits (Figure 4D).
Although these data indicate that SAGA and ATAC predominantly activate the transcription of different sets of genes, it remains unclear to which extent these transcriptional effects are related to their HAT activities. To answer this question, we analyzed RNA synthesis in cells in which Tada3, a subunit shared by the SAGA and ATAC HAT modules, is depleted. Newly synthesized RNA analyses in two independent Tada3AID/AID clones revealed that Tada3 depletion has almost no effect on Pol II transcription, with only 9 genes significantly downregulated upon auxin treatment (Figure 4E). We further analyzed the transcriptional effects of the individual HAT modules of SAGA or ATAC in Tada2a−/− or Tada2b−/− cells, by using qRT-PCR quantification of newly synthesized mRNA levels for selected genes that were found downregulated in the SAGA and ATAC core mutant ESCs. Genes downregulated in Supt7l−/− ESCs are not affected by the loss of Tada2b and genes downregulated upon depletion of ATAC core subunits, Yeats2 or Zzz3, are not affected by the loss of Tada2a (Figures S4D and S4E).
These observations indicate that SAGA and ATAC are important for the expression of different sets of genes and that their effects on mouse ESC self-renewal may be caused by different mechanisms. Importantly, these non-redundant functions for SAGA and ATAC in Pol II transcription in ESCs are mostly independent from their HAT activities. Our observations also suggest that, besides their predominant action on different groups of genes, these two coactivators may have a mild, but broad, effect on Pol II transcription.
SAGA and ATAC preferentially acetylate their target genes
We next asked whether SAGA and ATAC are recruited at genes, which were found downregulated in the corresponding mutant cell lines. As we could not obtain reliable binding profiles of SAGA and ATAC, due to technical limitations, we quantified changes in histone acetylation in mutant ESC lines as a proxy for complex localization. First, we measured H3K9ac levels in extracts from core SAGA (Supt7l−/−) or core ATAC (auxin-treated Yeats2AID/AID) mutant ESCs as well as in cells in which the HAT activities of both complexes are suppressed (auxin-treated Tada3AID/AID and Tada2a−/−+Tada2b−/−). Compared with control cells, H3K9ac levels are reduced by about 40% upon depletion of Yeats2, but are not significantly modified upon loss of Supt7l (Figures 5A and 5B). As expected, the loss of both SAGA and ATAC HAT modules causes a more drastic decrease of H3K9ac, by about 60%–70%, as observed in cells depleted for Tada3 or in cells lacking both Tada2a and Tada2b (Figures 5A and 5B). To determine the genome-wide distribution of SAGA and ATAC HAT activities, we then performed H3K9ac chromatin immunoprecipitation sequencing (ChIP-seq) experiments in these cell lines using spike-in Drosophila chromatin for normalization between the different samples. When looking around the promoters of genes actively transcribed in ESCs (8,201 genes as assessed by our 4sU-seq data), we observed a global decrease of H3K9ac at these promoters upon inactivation of both SAGA and ATAC HAT modules (Figure 5C). Inactivation of either SAGA (Supt7l−/−) or ATAC (auxin-treated Yeats2AID/AID) does not cause a global decrease of H3K9ac signal, detectable by ChIP-seq, in the same genomic regions (Figure 5C). However, in Yeats2-depleted cells, H3K9ac levels are decreased at promoters of genes that were found to be regulated by ATAC, but not at genes regulated by SAGA or at control genes (Figures 5D and S5A). Conversely, in Supt7l−/− cells, reduced H3K9ac is observed at SAGA-regulated promoters, but not at genes regulated by ATAC or at control genes (Figures 5D and S5A). These data together demonstrate that the SAGA HAT activity is reduced at target promoters in Supt7l null cells and that the ATAC HAT activity is decreased at target genes upon depletion of Yeats2, suggesting that each complex is preferentially recruited at these target genes.
Figure 5. H3K9ac and chromatin accessibility are decreased at ATAC- and SAGA-dependent genes.
(A) Representative images of western blot analyses of H3K9ac levels in extracts prepared from WT, Yeats2AID/AID, Tada2a−/−+Tada2b−/−, Tada3AID/AID, and Supt7l−/− cells. AID cells were treated for 24 h with IAA. Tbp serves as loading control.
(B) Quantification of H3K9ac levels normalized to Ponceau staining. Error bars show mean ± SD of at least 6 biological replicates using at least two independent clones.
(C) Heatmap representation of H3K9ac ChIP-seq coverage in WT, Yeats2AID/AID, Tada2a−/−+Tada2b−/−, Tada3AID/AID, and Supt7l−/− cells over promoter regions of 8,201 genes considered as expressed based on 4sU-seq. TSS, transcription start site.
(D) Violin plots showing log2 FC of H3K9ac ChIP-seq coverage at promoters between Yeats2AID/AID (top panels) or Supt7l−/− cells (bottom panels) and WT cells for either genes found significantly downregulated (red) or unchanged (gray, absolute log2 FC < 0.2 and an adjusted p value > 0.2) by 4sU-seq in either Yeats2AID/AID (left panels) or Supt7l−/− cells (right panels). Numbers of genes per category are indicated below each violin graphs.
(E) Violin plots showing log2 FC of ATAC-seq coverage at promoters for genes which were found either downregulated (red) or unchanged (gray) in the respective 4sU-seq experiments. Statistical test performed in (B) is Wilcoxon rank-sum test with Benjamini-Hochberg correction for multiple testing and in (D) and (E) is two-sided Welch t test. Only statistically significant results (p < 0.05) are indicated. See also Figure S5.
We further used ATAC-seq (assay for transposase-accessible chromatin) to compare chromatin accessibility in Supt7l−/− and auxin-treated Yeats2AID/AID or Zzz3AID/AID cell lines with that in control cells. At promoters of genes regulated by ATAC (found downregulated by 4sU-seq upon depletion of Yeats2 or Zzz3), chromatin accessibility is slightly reduced in auxin-treated Yeats2AID/AID or Zzz3AID/AID cells, but not in Supt7l−/− ESCs (Figures 5E and S5B). Conversely, chromatin accessibility is slightly decreased at genes regulated by SAGA (found downregulated by 4sU-seq upon loss of Supt7l) in Supt7l−/− ESCs, but not upon depletion of ATAC core subunits (Figures 5E and S5B).
The mild global decrease of newly synthesized mRNA levels observed upon depletion of Yeats2 or Zzz3 (Figure 4B) suggested a broad recruitment of ATAC to active gene promoters and enhancers. As a further indication of broad ATAC recruitment to active genes, we measured normalized peak intensities in each cell line for twelve different chromatin states previously defined in ESCs (Pintacuda et al., 2017). Upon depletion of Yeats2 or Zzz3, ATAC-seq peaks are decreased exclusively at active promoters and strong enhancers, but not for intergenic regions or insulators (Figures S5C–S5E). Altogether, our 4sU-seq and ATAC-seq data in Yeats2AID/AID and Zzz3AID/AID cells suggest that, in addition to a predominant role on a specific subset of genes, ATAC also has a broad function at the promoter and enhancer regions of most actively transcribed genes.
ATAC regulates the expression of translation-related genes
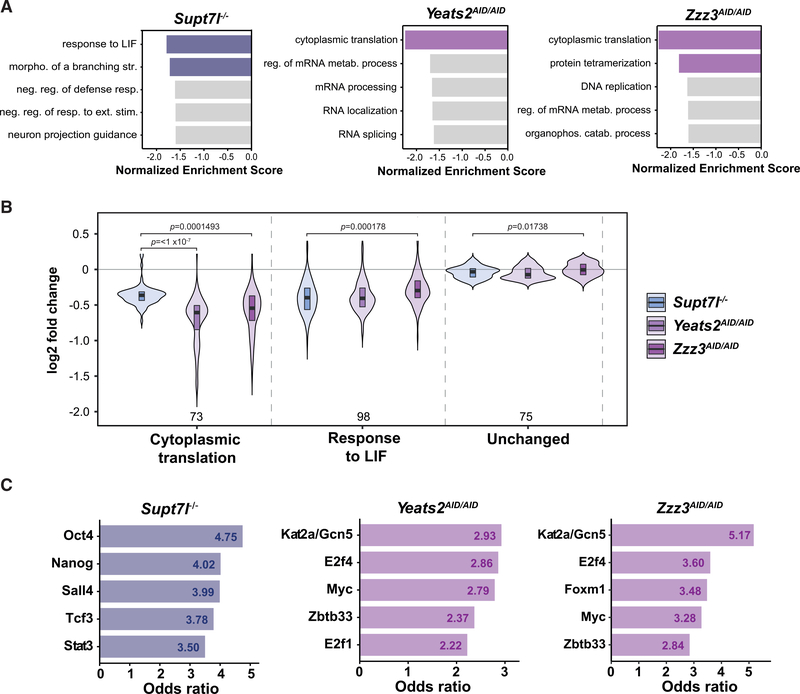
To better understand how SAGA and ATAC differently influence the proliferation and self-renewing capacities of mouse ESCs, we asked which gene categories are affected by the inactivation of each complex. Using gene set enrichment analysis (GSEA), we searched for Gene Ontology (GO) biological processes that are enriched in our newly synthesized 4sU-seq datasets. We identified “cytoplasmic translation” as a GO term that is significantly enriched in the set of genes significantly downregulated upon depletion of either AID-Yeats2 or AID-Zzz3, but not upon loss of Supt7l (Figure 6A). Genes involved in “response to LIF” are enriched in the set of genes downregulated in Supt7l−/− cells, consistent with the phenotype observed in these SAGA mutant cells (Figure 6A). Analyses of these two categories in our different 4sU-seq datasets indicate that even though each complex preferentially regulates different sets of genes, some overlap can be observed between genes regulated by SAGA or ATAC. Indeed, inactivation of ATAC not only impacts translation but also affects the pluripotency network, as genes of the “response to LIF” GO category are downregulated to a similar extent between Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID cells (Figure 6B). Similarly, genes belonging to the “cytoplasmic translation” category are highly regulated by ATAC, but are also modestly affected by SAGA inactivation (Figure 6B). Thus, the two related HAT-containing coactivator complexes SAGA or ATAC appear to be particularly important for the expression of different sets of genes, but also have more moderate effects on many genes belonging to common biological processes.
Figure 6. Gene categories preferentially regulated by SAGA and ATAC.
(A) Gene set enrichment analyses (GSEAs) for Gene Ontology (GO) biological processes based on log2 FCs in newly synthesized RNA levels from Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID cells relative to WT cells. Colored bars represent statistically significant terms (false discovery rate [FDR] < 0.05), while non-significant terms are represented in gray.
(B) Violin plots of log2 FCs comparing the distribution of expression changes of genes belonging to the GO categories “cytoplasmic translation” (73 genes) and “response to LIF” (98 genes) to unchanged genes (75 genes). Statistical test performed is ANOVA test. Only statistically significant results (p < 0.05) are indicated.
(C) Transcription-factor-binding sites from ChEA and Encode ChIP datasets enriched in genes significantly downregulated in Supt7l−/−, Yeats2AID/AID, and Zzz3AID/AID cell lines as identified by Enrichr. Only the first five transcription factors with the highest odds ratio and adjusted p value < 0.05 are shown.
We then asked whether the occurrence of specific TF-binding events could explain the differential recruitment and function of the two complexes on their target genes. For this, we analyzed the significantly downregulated genes identified in the respective mutant cell lines for overlaps with ChIP enrichment analysis (ChEA) and Encode ChIP datasets using Enrichr. TFs enriched in Supt7l-regulated genes include several pluripotency TFs such as Oct4, Nanog, and Sall4, in agreement with our GO analyses (Figures 6A and 6C). Among the Yeats2- and Zzz3-regulated genes, we observed in both cases an enrichment for genes bound by the HAT subunit of SAGA and ATAC, Gcn5 (Kat2a), thereby validating this approach. Myc and the E2f family member E2f4 were also identified as TFs bound at both Yeats2- and Zzz3-regulated genes and are thus potentially recruiting ATAC to regulate the expression of these genes (Figure 6C). Both Myc and E2f4 are involved in cell-cycle regulation, which may partially explain the cell-cycle defects observed upon inactivation of ATAC in mouse ESCs (Fagnocchi and Zippo, 2017; Hsu et al., 2019; Scognamiglio et al., 2016). Thus, the preferential dependence of genes on SAGA, or ATAC inactivation may be dictated by TFs present at each given promoter, eventually favoring the recruitment of either SAGA or ATAC.
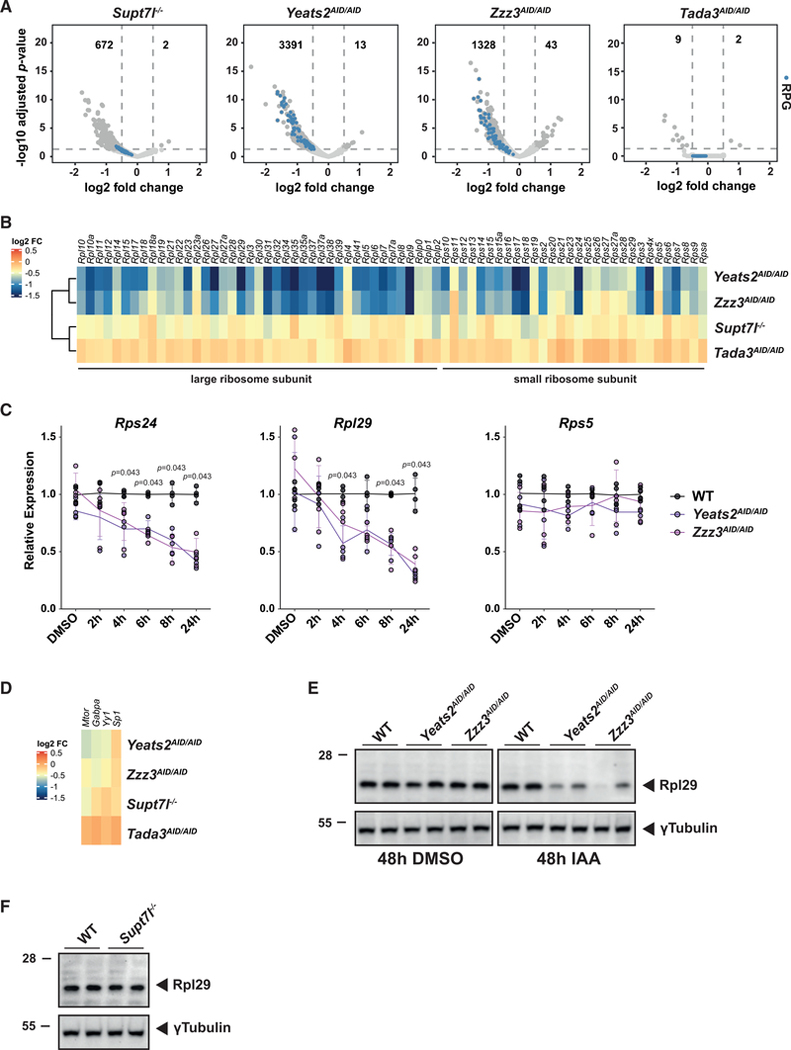
The specific effect of two different ATAC subunits on translation-related genes was further confirmed by the visualization of newly synthesized mRNA levels of RPGs in the different mutant cell lines. Indeed, many, but not all, RPGs are highly and significantly downregulated upon Yeats2 or Zzz3 depletion, but mild changes on few RPGs are seen upon loss of Supt7l (Figures 7A and 7B). In addition, the expression of RPGs is unaltered upon depletion of Tada3, supporting our conclusion that ATAC has HAT-independent functions in Pol II transcription (Figures 7A and 7B). Quantification of nascent mRNA levels for three RPGs (Rps24, Rpl29, and Rps5) in a time-course experiment, revealed a progressive decline of Rps24 and Rpl29 nascent mRNA, detected as early as 4 h of Yeats2 or Zzz3 depletion, whereas Rps5 expression remains largely unchanged after 24 h of auxin treatment, in agreement with our 4sU-seq experiment (Figures 7B and 7C). This observation strongly argues that RPG downregulation is a primary event due to ATAC inactivation. In support of this conclusion, we did not observe any major change in the expression of different TFs, known to control RPG expression (Perry, 2005) (Figure 7D). To further investigate the putative impact of RPG downregulation, we measured Rpl29 protein levels in SAGA and ATAC mutant ESCs. Bulk Rpl29 protein levels decrease upon auxin treatment of two independent Yeats2AID/AID and Zzz3AID/AID cells (Figure 7E), but are unchanged in Supt7l−/− ESCs (Figure 7F). Thus, ATAC, but not SAGA, is particularly important for the expression of RPGs and genes involved in translation, which may explain ATAC essentiality in ESCs.
Figure 7. ATAC is required for the expression of translation-related genes.
(A) Volcano plots representation of differential expression between Supt7l−/−, Yeats2AID/AID, Zzz3AID/AID, Tada3AID/AID, and WT cells. Numbers of significantly misregulated genes are indicated. Ribosome protein genes (RPGs) are highlighted as blue dots.
(B) Heatmap showing log2 FCs observed for all RPGs in Supt7l−/−, Yeats2AID/AID, Zzz3AID/AID, and Tada3AID/AID cell lines.
(C) Intron-containing mRNA levels of three RPGs (Rps24, Rpl29, and Rps5) in Yeats2AID/AID, Zzz3AID/AID, and WT cells upon 2–24 h of IAA treatment were normalized as in Figure 1G. Error bars show SD of 4 biological replicates, using two independent clones. Statistical test performed is Wilcoxon rank-sum test with Benjamini-Hochberg correction for multiple testing.
(D) Heatmap showing log2 FCs observed for four transcription factors involved in the regulation of RPG expression in the indicated cell lines.
(E and F) Western blot analysis of Rpl29 protein levels in two independent Yeats2AID/AID and Zzz3AID/AID cell lines (E) or Supt7l−/− cell lines (F) compared with WT cells. γTubulin serves as loading control.
DISCUSSION
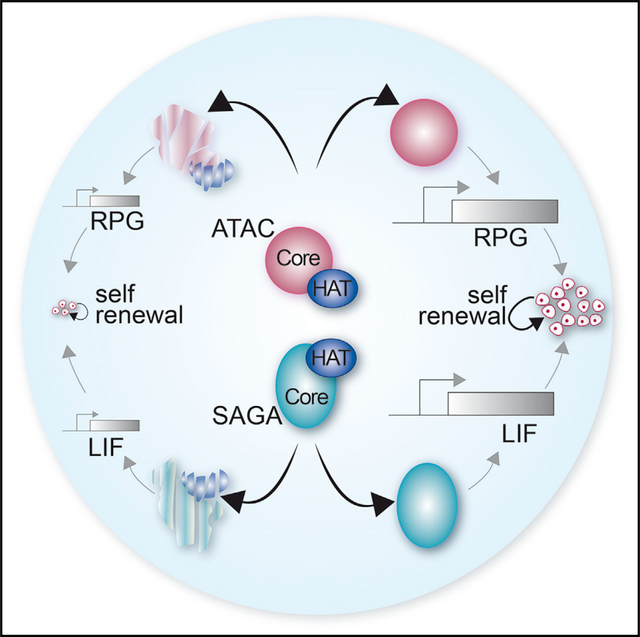
In this study, we demonstrate a key role for the SAGA and ATAC coactivator complexes in the maintenance of ESC self-renewal and growth. The inactivation of the SAGA or ATAC complex significantly affects the expression of distinct gene groups with a varying impact on global Pol II transcription. Importantly, the phenotypes and transcriptional anomalies observed in SAGA and ATAC mutant cells are mostly independent of the activities of their HAT modules. Inactivation of SAGA decreased the expression of genes related to LIF signaling, whereas ATAC depletion affected the expression of genes related to cytoplasmic translation such as RPGs. Thus, each complex makes use of its specific activities to regulate different sets of genes, eventually leading to self-renewal defects of mouse ESCs, in agreement with previous studies in Drosophila or differentiated human cells (Arede et al., 2020; Gamper et al., 2009; Krebs et al., 2011; Nagy et al., 2010; Pankotai et al., 2005, 2010).
SAGA stabilizes the naive pluripotency network, whereas ATAC is required to maintain the whole pluripotency network
A recent study found that the core SAGA subunits Taf5l and Taf6l maintain the self-renewal of mouse ESCs, mainly through acetylation and subsequent expression of SAGA target genes (Seruggia et al., 2019). In our study, the loss of Supt7l or Supt20h, two subunits of the core module of SAGA that were reported to be required for the integrity of SAGA structure in S. cerevisiae (Grant et al., 1997; Sterner et al., 1999; Wu and Winston, 2002), had very different effects in ESCs. Indeed, Supt7l, but not Supt20h, is required for ESC growth and self-renewal as well as for SAGA structural integrity, in agreement with observations made in Schizosaccharomyces pombe, in which spt7Δ cells were severely impaired for growth, whereas deletion of SPT20 showed more modest defects (Helmlinger et al., 2011). These two studies in mouse ESCs concur that several subunits of the core SAGA module, such as Supt7l, Taf5l, or Taf6l are required for SAGA integrity and ESC maintenance.
Using transcriptomic profiling and correlation with binding profiles of pluripotency factors in mouse ESCs, Seruggia et al. (2019) also showed that SAGA activates the expression of Oct4 and c-Myc, and their corresponding regulatory networks. We also observed a link between SAGA and Oct4 as genes downregulated in Supt7l−/− cells are enriched with binding sites for the pluripotency factors Oct4, Sox2, and Nanog. This suggests that SAGA functions as a coactivator for several pluripotency factors and thus regulates the pluripotency network. Reduced expression of naive pluripotency factors (Esrrb, Nanog, Klf4, and Tfcp2l1), but not of core pluripotency regulators (Oct4 and Sox2) in Supt7l−/− cells grown in FCS+LIF medium, indicates that SAGA may directly or indirectly stabilize the naive pluripotency network, in line with increased sporadic differentiation and reduced self-renewal of SAGA mutant ESCs (Martello and Smith, 2014; Navarro, 2018).
By contrast, we found that genes downregulated upon ATAC inactivation were enriched for Myc-bound as well as E2f4-bound genes, two TFs important for cell-cycle progression, in agreement with a previously reported interaction between Yeats2 and E2f4 (Chappell and Dalton, 2013; Chen et al., 2009; Fagnocchi and Zippo, 2017; Hsu et al., 2019; Matsumura et al., 2003). Inactivation of E2f4 or the combined inactivation of c-Myc and n-Myc in mouse ESCs affected cell growth and self-renewal, similarly to our observations in ATAC mutant cell lines (Smith and Dalton, 2010; Varlakhanova et al., 2010). Interestingly, the mRNA levels of all tested pluripotency TFs, including the core pluripotency factors Oct4 and Sox2, were decreased upon depletion of Yeats2 in FCS+LIF medium. This direct or indirect role for ATAC in pluripotency factor expression may explain the self-renewal defects in ATAC mutant cells, whereas the growth defects in ATAC mutant ESCs may be mediated by altered E2f4- and Myc-target gene expression.
SAGA and ATAC have distinct HAT-independent functions needed for Pol II transcription in mouse ESCs
The distinct HAT-independent functions of SAGA and ATAC for Pol II transcription in mouse ESCs were demonstrated through three major observations: (1) whereas inactivation of ATAC HAT module did not affect mouse ESC viability, no Yeats2 or Zzz3 homozygous knockout clones could be obtained, suggesting that the core ATAC is essential for mouse ESC survival; (2) disruption of the HAT module of SAGA or ATAC did not reproduce any of the effects seen upon inactivation or depletion of core subunits of these complexes, although histone acetylation was more dramatically affected in HAT mutants than in individual core mutant cell lines; and (3) newly synthesized RNA analyses in ATAC and SAGA mutant cell lines revealed that SAGA and ATAC affect transcription of distinct sets of genes, although they share the same HAT enzymes.
An earlier study suggested that SAGA regulates expression of its target genes through H3K9ac deposition (Seruggia et al., 2019). Although we could confirm that SAGA core subunits are required to maintain the ESC state as reported by Seruggia et al. (2019), our analyses revealed that inactivation of the SAGA HAT module did not affect ESC self-renewal. Importantly, conclusions by Seruggia et al. (2019) were based on correlation analysis between binding profiles of SAGA subunits and changes in H3K9ac levels at the corresponding promoters revealing that Taf5l- and Taf6l-bound genes show reduced levels of H3K9ac in Taf5l or Taf6l null ESCs. In the same line, in Supt7l−/− ESCs, we observed reduced H3K9ac levels at the promoters of Supt7l-dependent genes. By contrast, inactivation of SAGA and ATAC HAT modules resulted in more dramatic changes in H3K9ac levels at almost all active promoters, without causing comparable phenotypic or transcriptional defects. Thus, the recruitment of SAGA and ATAC at their target genes is revealed by their effects on histone acetylation, but their HAT activity is not crucial for Pol II transcription in mouse ESCs.
The phenotypic and transcriptional differences between mutants of the SAGA core or SAGA HAT modules are in good agreement with data from S. cerevisiae. Indeed, inactivation of yeast SAGA core subunits leads to more severe growth and transcriptional defects than inactivation of its HAT module (Baptista et al., 2017; Helmlinger et al., 2020). Besides their HAT modules, SAGA and ATAC contain 16 and 6 additional subunits, respectively, which could play a role in Pol II transcription. Four SAGA subunits constitute a histone deubiquitination (DUB) module and the yeast ortholog of the SAGA-specific subunit Supt3h may bind and/or deliver TBP at gene promoters. The mammalian SAGA complex additionally possesses a splicing module composed of two subunits (Sf3b3 and Sf3b5), with unknown function within SAGA. The respective role of these different subunits in transcription is still unclear. Similarly, the HAT-independent functions of ATAC remain to be determined and could involve interaction with TBP through its Yeats2/NC2β subunits (Wang et al., 2008) or other factors of the transcription machinery.
Catalytic-independent functions of coactivator complexes were previously demonstrated through the analysis of catalytic mutants of different chromatin-modifying complexes, such as TIP60 and Mll3/4 COMPASS-like complexes (Acharya et al., 2017; Dorighi et al., 2017; Rickels et al., 2017). As such, Mll3/4 COMPASS-like complexes were suggested to be important for the presence of Pol II recruitment at enhancers independently of their histone methylation activities (Dorighi et al., 2017). Earlier studies on Gcn5 null ESCs demonstrated a requirement of the HAT activities of SAGA and ATAC during differentiation of mouse ESCs (Lin et al., 2007; Wang et al., 2018). This suggests that the histone-modifying activities of SAGA and ATAC have a more critical role during differentiation than for ESC self-renewal, consistent with the requirement of Gcn5 catalytic activity during mouse embryonic development (Bu et al., 2007; Xu et al., 2000; Yamauchi et al., 2000). Similarly, catalytic inactivation of the histone-modifying activities of TIP60 did not impair mouse ESC growth or self-renewal, but resulted in defects during mouse embryonic development (Acharya et al., 2017).
Previous findings further highlight the importance of SAGA and ATAC in mouse development. As such, homozygous inactivation of Supt20h in mice was reported to cause severe gastrulation defects with abnormalities in mesoderm migration (Zohn et al., 2006). Inactivation of Atac2, an ATAC-specific subunit, was shown to lead to defects in embryonic development at the post-gastrulation stage (Guelman et al., 2009). Similarly, inactivation of genes encoding catalytic subunits of SAGA, Gcn5, Pcaf, Atxn7l3 or Usp22 resulted in embryonic lethality at stages beyond gastrulation (Bu et al., 2007; Koutelou et al., 2019; Xu et al., 2000; Yamauchi et al., 2000; Wang et al., 2021). These phenotypes do not argue for a crucial role of SAGA or ATAC in the inner cell mass of the blastocyst, from which ESCs are derived. In agreement, no significant growth defects have been observed in mouse ESCs mutant for genes encoding catalytic subunits or the Supt20h subunit of SAGA (Lin et al., 2007; Sussman et al., 2013). However, the role of genes encoding subunits of SAGA or ATAC, which were shown to play a role for ESC growth and self-renewal, i.e., Supt7l, Yeats2 and Zzz3 (this study) and Taf5l and Taf6l (Seruggia et al., 2019), has not yet been investigated in mouse development. Thus, it will be crucial to determine whether the inactivation of these genes affects the peri-implantation development of mouse embryos.
In summary, we generated a large series of mutant ESC lines for SAGA and ATAC subunits, allowing comprehensive and comparative analyses of these two coactivator complexes. Our study allowed the identification of distinct HAT-independent roles for SAGA and ATAC in mouse ESC growth and self-renewal. These findings pave the way to determine the ATAC- and SAGA-specific activities used in mouse ESCs to control their specific gene expression programs.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Didier Devys (devys@igbmc.fr).
Materials availability
Plasmids and cell lines generated in this study are available upon request.
Data and code availability
RNA-seq, ATAC-seq and ChIP-seq data have been deposited at GEO (Edgar et al., 2002) and are publicly available as of the date of publication. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley Data and are publicly available as of the date of publication. The DOIs is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Supt7l | Bethyl Laboratories | Cat#A302–803A; RRID:AB_10630265 |
| Mouse monoclonal anti-γTubulin | Sigma-Aldrich | Cat#T6557; RRID:AB_477584 |
| Rabbit polyclonal anti-Rpl29 | ThermoFisher Scientific | Cat#15799–1-AP; RRID:AB_2878187 |
| Rabbit polyclonal anti-histone H3 lysine 9 acetylation | Abcam | Cat#ab4441; RRID:AB_2118292 |
| Rabbit polyclonal anti-HA tag | Abcam | Cat#ab9110; RRID:AB_307019 |
| Mouse monoclonal anti-Wdr5 | Abcam | Cat#ab56919; RRID:AB_946146 |
| Mouse monoclonal anti-Tbp | In-house | 3TF1–3G3; Brou et al., 1993 |
| Rabbit polyclonal anti-Taf7 | In-house | 3475; Bardot et al., 2017 |
| Mouse monoclonal anti-Taf10 | In-house | 6TA-2B11; Jacq et al., 1994 |
| Mouse monoclonal anti-Taf12 | In-house | 22TA-2A1; Mengus et al., 1995 |
| Rabbit polyclonal anti-Atxn7l3 | In-house | 2325; Zhao et al., 2008 |
| Rabbit polyclonal anti-Supt3h | In-house | 3118; Bardot et al., 2017 |
| Rabbit polyclonal anti-Gcn5 | In-house | 2676; Nagy et al., 2010; Orpinell et al., 2010 |
| Mouse monoclonal anti-Gcn5 | In-house | 2GC-2C11; Brand et al., 2001 |
| Rabbit polyclonal anti-Tada3 | In-house | 2678; Nagy et al., 2010; Orpinell et al., 2010 |
| Rabbit polyclonal anti-Sgf29 | In-house | 2461; Orpinell et al., 2010 |
| Rabbit polyclonal anti-Atac2 | In-house | 2734; Nagy et al., 2010 |
| Rabbit polyclonal anti-Mbip | In-house | 2768; Nagy et al., 2010; Orpinell et al., 2010 |
| Rabbit polyclonal anti-Zzz3 | In-house | 2616; Nagy et al., 2010 |
| Chemicals, peptides, and recombinant proteins | ||
| Indole-3-acetic acid | Sigma-Aldrich | Cat#I3750 |
| 4-thiouridine | Glentham Life Sciences or abcam | Cat#GN6085 or Cat#ab143718 |
| 4-thiouracil | Sigma-Aldrich | Cat#440736 |
| EZ-link HPDP-biotin | ThermoFisher Scientific | Cat#21341 |
| Streptavidin protein coupled to HRP | ThermoFisher Scientific | Cat#21126 |
| Home-made Tn5E54K,L372P transposase | Plasmid provided by Dr. Kim Remans, EMBL Heidelberg, Germany | Hennig et al., 2018 |
| Critical commercial assays | ||
| Alkaline Phosphatase Kit | Vector Laboratories | Cat#SK-5100 |
| TURBO DNA-free™ Kit | ThermoFisher Scientific | Cat#AM1907 |
| μMACS magnetic beads and columns | Miltenyi Biotec | Cat#130–074–101 |
| RNeasy MinElute Cleanup Kit | QIAGEN | Cat#74204 |
| NucleoSpin Gel and PCR Clean-up kit | Macherey-Nagel | Cat#740609.50 |
| RiboPure™ RNA Purification Kit | ThermoFisher Scientific | Cat#AM1926 |
| TruSeq Stranded Total RNA LT Sample Prep Kit with Ribo-Zero Gold | Illumina | Cat#RS-122–2301 or Cat#RS-122–2302 |
| Ribo-Zero Gold rRNA (Yeast) | Illumina | Cat#MRZY1324 |
| MicroPlex Library Preparation kit v2 | Diagenode | Cat#C05010014 |
| SPRIselect beads | Beckman-Coulter | Cat#B23319 |
| AMPure XP beads | Beckman-Coulter | Cat#A63882 |
| Deposited data | ||
| Raw and analyzed data 4sU RNA-seq | This paper | GEO: GSE175905 |
| Raw and analyzed data ATAC-seq | This paper | GEO: GSE175905 |
| Raw and analyzed data H3K9ac ChIP-seq | This paper | GEO: GSE175905 |
| Mass spectrometry proteomics Taf10 IP | This paper | PRIDE: PXD027026 |
| Mass spectrometry proteomics Mbip and Zzz3 IP | This paper | PRIDE: PXD027026 |
| Original western blot images | This paper | Mendeley Data: https://doi.org/10.17632/thjcm8zdpf.1 |
| Experimental models: Cell lines | ||
| Mouse: ES E14tg2a.4 cells (129P2 genetic background) | BayGenomics | N/A |
| Mouse: Supt7l−/− ES E14 cells | This study | N/A |
| Mouse: Supt7ltg ES E14 cells | This study | N/A |
| Mouse: Supt20h−/− ES E14 cells | This study | N/A |
| Mouse: Tada2b−/− ES E14 cells | This study | N/A |
| Mouse: Tada2b−/− ES E14 cells | This study | N/A |
| Mouse: Yeats2AID/AID ES E14 cells N-ter. | This study | N/A |
| Mouse: Tada3AID/AID ES E14 cells N-ter. | This study | N/A |
| Mouse: Zzz3AID/AID ES E14 cells N-ter. | This study | N/A |
| Mouse: Zzz3AID/AID ES E14 cells C-ter. | This study | N/A |
| D. melanogaster: Schneider S2 cells | ATCC | CRL-1963 |
| Experimental models: Organisms/strains | ||
| S. pombe: DHP43 h- | Elías-Villalobos et al., 2019 | N/A |
| S. cerevisiae: Strain background: FY406 | Hirschhorn et al., 1995 | N/A |
| Oligonucleotides | ||
| For sgRNA sequences, see Table S1 | This paper | N/A |
| For primer sequences, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: Supt7l CDS-HygromycinR | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Supt7l for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Supt20h for KO | This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against Tada2b for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Tada2a for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Yeats2 for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Zzz3 for KO | This paper | N/A |
| Plasmid: Tir1-IRES-BirA-NeomycinR | This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against N-ter of Yeats2 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for N-ter AID-tagging of Yeats2: eGFP-P2A-1xFlag-BioTag-AID |
This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against N-ter of Tada3 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for N-ter AID-tagging of Tada3: eGFP-P2A-1xFlag-BioTag-AID |
This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against N-ter of Zzz3 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for N-ter AID-tagging of Zzz3: eGFP-P2A-1xFlag-BioTag-AID | This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against C-ter of Zzz3 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for C-ter AID-tagging of Yeats2: AID-1xFlag-BioTag-P2A-eGFP | This paper | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al., 2012 | https://imagej.net/software/fiji/downloads |
| cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/v1.10/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| htseq-count | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Enrichr | Chen et al., 2013; Kuleshov et al., 2016; Xie et al., 2021 | https://maayanlab.doud/Enrichr/ |
| RTA | Illumina | https://support.illumina.com/content/dam/illumina-support/documents/downloads/software/hiseq/hcs-hd-v3-4-0-install-notes-1000000028070-00.pdf |
| bcl2fastq | Illumina | https://emea.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Encode ATAC-seq pipeline | ENCODE | https://www.encodeproject.org/atac-seq/ |
| MACS2 | Zhang et al., 2008 | https://hbctraining.github.io/Intro-to-ChIPseq/lessons/05_peak_calling_macs.html |
| Homer v4.11.1 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Bedtools | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| Enrichr | Kuleshov et al., 2016 | https://maayanlab.doud/Enrichr/ |
| WebGestalt GEne SeT Analysis Toolkit | Wang et al., 2013 | http://www.webgestalt.org/ |
| chromHMM datasets | Pintacuda et al., 2017 | https://github.com/guifengwei/ChromHMM_mESC_mm10 |
| ATACseqQC | Ou et al., 2018 | https://www.bioconductor.org/packages/release/bioc/html/ATACseqQC.html |
| Proteome Discoverer | ThermoFisher Scientific | https://www.thermofisher.com/fr/fr/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html |
| FlowJo™ | Ashland; Becton, Dickinson and Company | https://www.fiowjo.com/solutions/fiowjo/downloads |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse ESCs
Mouse male ES E14 cells (BayGenomics) were cultured on plates coated with 0.1% gelatine solution in 1x PBS (Dutcher, Cat#P06–20410) using DMEM medium supplemented with 15% fetal calf serum ES-tested (ThermoFisher Scientific, Cat#10270–106), 2 mM ʟ-glutamine (ThermoFisher Scientific, Cat#25030–024), 0.1% β-mercaptoethanol (ThermoFisher Scientific, Cat#31350–010), 100 UI/ml penicillin and 100 μg/ml streptomycin (ThermoFisher Scientific, Cat#15140–122), 0.1 mM non-essential amino acids (ThermoFisher Scientific, Cat#11140–035) and 1,500 U/ml leukemia inhibitory factor (home-made). For medium described as FCS+LIF+2i, 3 μM CHIR99021 (axon medchem, Cat#1386) and 1 μM PD0325901 (axon medchem, Cat#1408) were added freshly to the medium. Cells were grown at 37°C with 5% CO2 levels. Cells were passaged every second day. To induce the auxin-inducible degradation (AID), cells were treated with 500 μM Indole-3-acetic acid (Sigma-Aldrich, Cat#I3750).
Schneider S2 cells
Drosophila melanogaster Schneider S2 cells (CRL-1963, ATCC) were grown in Schneider’s Drosophila medium (ThermoFisher Scientific, Cat#21720–024) containing 10% FCS (heat inactivated) (Sigma Aldrich, Cat#F7524) and 0.5% penicillin and streptomycin at 27°C.
Yeast cells
Schizosaccharomyces pombe cells were grown in autoclaved YES medium (yeast extract, adenine, histidine, uracil, leucine, lysine, 3% glucose) at 32°C. Saccharomyces cerevisiae cells were grown in autoclaved YPD medium (yeast extract, bactopeptone, 2% glucose) at 30°C.
METHOD DETAILS
Plasmid construction
All homologous recombination (HR) templates and plasmids expressing 1 or 2 gRNAs and co-expressing high-fidelity Cas9 (Kleinstiver et al., 2016) fused to eGFP (Cas9-HF-eGFP) were generated by Golden Gate cloning (Engler et al., 2009). For the HR templates, silent mutations were introduced by PCR to prevent Cas9-HF-mediated cleavage of the HR template or the knockin allele. The sequences of the gRNAs for the different constructs are indicated in Table S1. The plasmid containing the mouse Supt7l coding sequence (CDS) for the generation of Supt7ltg cell lines was constructed as follows. The CDS of Supt7l was amplified by PCR from a cDNA bank of mouse embryos (day 9–12) and inserted together with the PGK promoter into a pcDNA3.1 hygro vector (Invitrogen) by replacing the CMV promoter. All plasmids were verified by sequencing.
Generation of stable cell lines
For the generation of Supt7ltg cell lines and the Tir1-BirA stable cell line, the linearized plasmids containing the coding sequences were transfected into either two independent Supt7l−/− cell lines or wild-type ES E14 cells, respectively, using Lipofectamine2000 (ThermoFisher Scientific, Cat#11668019) following manufacturer’s instruction. Antibiotic selection was started 48 hours post-transfection (250 μg/ml hygromycin (Sigma Aldrich, Cat#H0654) or 400 μg/ml geneticin (ThermoFisher Scientific, Cat#11811031)). Selection medium was exchanged every second day for a week. For Supt7ltg cell lines, the polyclonal population was used for subsequent experiments. For Tir1-BirA stable cell lines, monoclonal cell lines were established by colony picking.
Generation of knockout and auxin-inducible degron (AID) cell lines
Mouse ESCs were transfected with the plasmid constructs at a confluency of 70%–80% using Lipofectamine2000 (ThermoFisher Scientific, Cat#11668019) following manufacturer’s instruction. For knock-in (AID and HA-tag) cell lines, donor plasmids were linearized using unique cutter restriction enzymes before transfection and transfected together with the Cas9-containing transient plasmid in a Tir1-BirA expressing cell line. Two to three days after transfection, cells were selected for expression of the fluorescent tags (for knockout cell lines: fusion protein of Cas9 with fluorescent protein, for knock-in cell lines: fusion protein of protein of interest with fluorescent protein) by fluorescence activated cell sorting (FACS). Three to five 96-well plates were seeded with one fluorescent cell per well using the BD FACSAria™ II (BD Biosciences) instrument.
Clonal assays
For clonal assay analyses, 1500 to 3000 cells, which had been adapted to the respective media through at least three passages, were plated in wells of 6-well plates. Medium was changed every other day. On the sixth day, colonies were washed twice with 1x PBS before fixation with 4% Paraformaldehyde (Electron Microscopy Sciences, Cat#15710) for 30 minutes followed by two washes with 1x PBS. To assess the alkaline phosphatase (AP) activity of mouse ESC colonies, Alkaline Phosphatase Kit (Vector Laboratories, Cat#SK-5100) was used following manufacture’s instruction. Colonies were stained with AP for 5–10 minutes. For clonal assay analyses in FCS+LIF medium, an additional staining with crystal violet was performed after AP staining to assess the total number of colonies enabling normalization between replicates. Colonies were stained with 0.1% crystal violet solution for at least 30 minutes.
For clonal assay quantification in FCS+LIF+2i medium, colony areas were measured automatically using ImageJ software. For clonal assay quantification in FCS+LIF medium, crystal violet-stained colonies were counted manually using the ImageJ interface. The number of AP positive colonies was also counted manually using the ImageJ interface, while the number of AP negative colonies were deduced by subtracting the number of AP+ colonies from the total number of colonies. We considered colonies as AP+ colonies if they either stained entirely red or if they possessed a center of red cells surrounded by unstained cells.
Cell cycle analysis
For cell cycle analyses, cells were harvested, washed with 1x PBS and permeabilized with 70% of ice- cold ethanol. Cells were stored at 4°C for up to a week prior to analysis. For propidium iodide staining, permeabilized cells were centrifuged, washed with 1x PBS prior to incubation with 75 μg/ml RNase A and 15 μg/ml propidium iodide (Sigma-Aldrich, Cat#P4170) for at least 30 minutes at room temperature. Samples were filtered and 10,000 to 20,000 cells were analyzed using a BD FACSCelesta™ (BD Biosciences) instrument to determine cell cycle profiles. Data were analyzed using FlowJo™ 10.2. software with manual assignment of the cell cycle phases.
Metabolic labeling
Metabolic labeling of newly synthesized RNA was adapted from previously described protocols (Rabani et al., 2011; Rädle et al., 2013; Schwalb et al., 2016). In brief, the nucleoside analog 4-thiouridine (4sU) (Glentham Life Sciences, Cat#GN6085 or abcam, Cat#ab143718) was added to the cell culture medium at a final concentration of 500 μM for a 25 minutes pulse. After the labeling period, the medium containing 4sU was removed, the cells were washed with ice cold 1x PBS and immediately lysed using TRI® Reagent (Molecular Research Center Inc., Cat#TR 188). Total RNA was extracted following TRI® Reagent manufacturer’s instruction. To remove any potential genomic DNA contamination from the total RNA extracts, the TURBO DNA-free™ Kit (ThermoFisher Scientific, Cat#AM1907) was used following manufacturer’s instructions for rigorous DNase treatment.
To label Drosophila S2 cells, medium containing 4sU at a final concentration of 500 μM was added to the cells during 15 minutes under aluminum foil at room temperature. 4sU-containing medium was removed and 1xPBS was added to collect the cells using a cell scratcher. Cells were centrifuged, flash frozen in aliquots and stored at −80°C. For total RNA extraction, S2 cells were defrozen, lysed using TRI® Reagent (Molecular Research Center Inc., Cat#TR 188) and total RNA was isolated following manufacturer’s instruction.
Yeast cultures were grown to an OD600 of 0.8. 4-thiouracil (Sigma-Aldrich, Cat#440736) was freshly dissolved in DMSO and added to the cultures at a final concentration of 1 mM. Labeling was performed for 6 minutes. After this time period, yeast cells were pelleted, washed with ice-cold 1x PBS and aliquoted before being flash frozen and stored at −80°C. For total RNA extraction, the RiboPure™ RNA Purification Kit (ThermoFisher Scientific, Cat#AM1926) was used following manufacturer’s instruction.
Newly synthesized RNA purification
The purification of newly synthesized RNA was based on previously described protocols (Rabani et al., 2011; Rädle et al., 2013; Schwalb et al., 2016). Labeled, total RNA of spike-in cells (D. melanogaster, S. cerevisiae or S. pombe) was added to labeled, total RNA from mouse ESCs in a ratio 1:5 to 1:10 prior to newly synthesized RNA purification to a final amount of 200–250 μg of total RNA. The RNA was precipitated and resuspended in 130 μL and sonicated using the following program on a Covaris E220 instrument: 1% duty factor, 100 W, 200 cycles per burst, 80 s. Fragment size ranged from 10 kb to 200 bp. For purification, the fragmented total RNA was incubated for 10 minutes at 60°C and immediately chilled on ice for 2 minutes to open secondary RNA structures. The 4sU-labeled RNA was thiol-specific biotinylated by addition of 200 μg EZ-link HPDP-biotin (ThermoFisher Scientific, Cat#21341), biotinylation buffer (10 mM HEPES-KOH pH 7.5 and 1 mM EDTA) and 20% DMSO (Sigma-Aldrich, Cat#D8418). Biotinylation was carried out for 3 hours at 24°C in the dark and with gentle agitations. After incubation, excess of biotin was removed by adding an equal volume of chloroform and centrifugation at 16,000 g for 5 minutes at 4°C. RNA was precipitated from the aqueous phase by adding 0.1 volumes of 5 M NaCl and an equal volume of 100% isopropanol followed by centrifugation at 16,000 g for 30 minutes at 4°C. After washing with 75% ethanol the RNA pellet was resuspended in 100 μL of RNase-free water and denatured for 10 minutes at 65°C followed by immediate chilling on ice for 5 minutes. The samples were incubated with 100 μL of streptavidin-coated μMACS magnetic beads (Miltenyi Biotec, Cat#130–074-101) for 90 minutes at 24°C under gentle agitations. The μMACS columns (Miltenyi Biotec, Cat#130–074-101) were placed on a MACS MultiStand (Miltenyi Biotec) and equilibrated with washing buffer (100 mM Tris-HCl pH 7.5, 10 mM EDTA, 1 M NaCl, 0.1% Tween20) before applying the samples twice to the columns. The columns were then washed one time with 600 μl, 700 μl, 800 μl, 900 μL and 1 mL washing buffer before eluting the newly synthesized RNA with two washes of 100 μL 0.1M DTT. The isolated newly synthesized RNA was recovered using RNeasy MinElute Cleanup Kit (QIAGEN, Cat#74204) following manufacturer’s instruction.
Library preparation and sequencing of total RNA-seq and 4sU-seq
Total RNA-seq libraries were generated from 1 μg of total RNA using TruSeq Stranded Total RNA LT Sample Prep Kit with Ribo-Zero Gold (Illumina, Cat#RS-122–2301 or RS-122–2302) according to the Illumina protocol with the following modifications. Cytoplasmic and mitochondrial ribosomal RNA (rRNA) was removed using Ribo-Zero Gold rRNA (Yeast) (Illumina, Cat#MRZY1324). Following purification, the depleted RNA was fragmented using divalent cations at 94°C for 2 minutes. While, double stranded cDNA synthesis and adaptor ligation were performed according to manufacturer’s instructions, the number of PCR cycles for library amplification was reduced to 10 cycles. After purification using AMPure XP beads (Beckman-Coulter, Cat#A63882), the libraries were sequenced with 1× 50 base pairs on a HiSeq4000 machine (Illumina).
4sU RNA-seq libraries were generated from 15 to 50 ng of purified, newly synthesized RNA using TruSeq Stranded Total RNA LT Sample Prep Kit with Ribo-Zero Gold (Illumina) according to the Illumina protocol with the following modifications. 4sU-labeled RNA was cleaned up using 1.8x RNAClean XP beads and fragmented using divalent cations at 94°C for 1 minutes without depletion of rRNA. While, double stranded cDNA synthesis and adaptor ligation were performed according to manufacturer instructions, the number of PCR cycles for library amplification was reduced to 10 cycles. After purification using SPRIselect beads (Beckman-Coulter, Cat#B23319), the libraries were sequenced with 1× 50 base pairs on a HiSeq4000 machine (Illumina).
Data analysis of total RNA-seq and 4sU-seq
Reads were preprocessed using cutadapt 1.10 (Martin, 2011) in order to remove adaptors and low- quality sequences and reads shorter than 40 bp were removed for further analysis. Remaining reads were mapped to M. musculus, D. melanogaster and S. cerevisiae rRNA sequences for samples VQFR1, VQFR2, VQFR7–12 or M. musculus and S. pombe rRNA sequences for samples VQFR13–18 and VQFR25–28 using bowtie 2.2.8 (Langmead and Salzberg, 2012) and reads mapping to rRNA sequences were removed for further analysis. For samples VQFR1, VQFR2, VQFR7–12, remaining reads were aligned to a hybrid genome composed of mm10, BDGP6 and sacCer3 assemblies of M. musculus, D. melanogaster and S. cerevisiae genome respectively with STAR 2.5.3a (Dobin et al., 2013). For samples VQFR13–18 and VQFR25–28, the hybrid genome was composed of mm10 and ASM294v2 assemblies of M. musculus and S. pombe genome respectively. Gene quantification was performed with htseq-count 0.6.1p1 (Anders et al., 2015), using “union” mode and Ensembl 93 annotations for all organisms except for S. pombe where Ensembl Fungi 41 annotations were used. For 4sU-seq data, ‘type’ option was set to ‘gene’ in order to take also into account reads aligned onto introns. Differential gene expression analysis was performed using DESeq2 1.16.1 (Love et al., 2014) Bioconductor R package on M. musculus counts normalized with size factors computed by the median-of-ratios method proposed by Anders and Huber (Anders and Huber, 2010), on Drosophila melanogaster counts for samples VQFR1, VQFR2, VQFR7–12 or on S. pombe counts for samples VQFR13–18 and VQFR25–28 (using the following options: cooksCutoff = TRUE, independentFiltering = TRUE, alpha = 0.05). P values were adjusted for multiple testing using the Benjamini and Hochberg method (Benjamini et al., 2001). For subsequent data analyses and visualization, genes of the Y chromosomes and mitochondrial chromosome were excluded and only protein-coding genes were considered. Further, a threshold of 100 reads was used to define expressed genes and only genes shared between all 4sU-seq datasets were analyzed. This resulted in the analysis of 8201 protein-coding genes. For the analysis using the Enrichr online interface shown in Figure 6C (https://maayanlab.cloud/Enrichr/) (Kuleshov et al., 2016), a list of significantly downregulated genes was generated (adjusted p-value < 0.05 and log2 fold change < −0.5) and the results for ‘ENCODE and ChEA Consensus TFs from ChIP-X’ were considered following ranking by the odds ratio. For the Gene Set Enrichment Analysis (GSEA) shown in Figure 6A, lists of the 8201 genes which we considered expressed and their log2 fold changes per condition were used with the WebGestalt GEne SeT Analysis Toolkit (http://www.webgestalt.org/) (Wang et al., 2013) and analyzed with Gene Set Enrichment Analysis (GSEA) as method of interest and non-redundant gene ontology biological processes as functional database with the remaining settings set to default with the exception that the minimum number of genes for a category was set to 50. The resulting tables were ranked by the normalized enrichment score.
ATAC-seq
ATAC-seq was preformed using an adaptation of protocols described in Buenrostro et al. (2015) and King and Klose (2017). In short, 100.000 cells were used per sample and centrifuged at 2000 g for 5 minutes at 4°C. Cells were washed with ice-cold 1x PBS and centrifuged at 2000 g for 5 minutes at 4°C. Nuclei were isolated using 50 μL Lysis buffer (10 mM Tris HCl ph 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP-40) and centrifuged immediately at 2000 g for 10 minutes at 4°C. Immediately after removing supernatant pellet was resuspended in 50 μL of TAPS-buffer (10 mM TAPS-NaOH pH 8.5, 5 mM MgCl2, 10% DMF) containing 25 nM adaptor-loaded, home-made Tn5E54K,L372P transposase (plasmids provided by Dr. Kim Remans, EMBL Heidelberg, Germany). The reaction was incubated for 1 hour at 37°C and was immediately purified using NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel, Cat#740609.50). DNA was eluted into a volume of 15 μl.
Library preparation of ATAC-seq
For ATAC-seq libraries, adapters described in Buenrostro et al. (2013, 2015) were used using Phusion High-Fidelity DNA Polymerase (ThermoFisher Scientific, Cat#F530S) following manufacturer’s instruction with 14 cycles. After purification using 1.5x SPRIselect beads (Beckman-Coulter, Cat#B23319), libraries were sequenced with 2× 100 base pairs on a HiSeq4000 machine (Illumina) following Illumina’s instructions.
Data analysis of ATAC-seq
Image analysis and base calling were performed using RTA 2.7.7 and bcl2fastq 2.17.1.14. Data analysis was performed using the Encode ATAC-seq pipeline v1.4.2. Adaptor sequences were removed and low- quality ends were trimmed. Sequence alignment was performed into the mm10 assembly of Mus musculus genome using Bowtie2 (version 2.2.6) (Langmead and Salzberg, 2012) choosing the zero multi-mapping option. Mitochondrial reads were removed. For violin plots shown in Figure 5E, reads were counted over promoter ranges (1500 bp upstream, 500 bp downstream) of genes of the Ensembl 93 database. Peak calling was performed using MACS2 v2.1.1.20160309 (Zhang et al., 2008). The optimal set of IDR peaks were used for the downstream analysis. Peaks from different conditions were merged to form a consensus peak set. The peaks were annotated using annotatePeaks.pl script in Homer software and with Ensembl 94 database. The read coverage for each sample was calculated with multicov function from bedtools program (v2.26.0) (Quinlan and Hall, 2010). Read counts have been normalized across samples with the median-of-ratios method proposed by Love et al. and implemented in the Bioconductor package DESeq2 version 1.16.1 (Love et al., 2014). Differential expression analyses were performed using the DESeq2 package. A multi-factor design was used to control an observed batch effect between day1 (rep1, rep2) and day2 (rep3, rep4). To assign the identified accessibility peaks to chromatin states, the chromHMM 12-states dataset (https://github.com/guifengwei/ChromHMM_mESC_mm10) was used (Pintacuda et al., 2017). Chromatin states analyzed are indicated in the respective graphs. Chromatin states analyzed are indicated in the respective graphs. For the analysis shown in Figure S5D, the ATACseqQC R library was used.
ChIP-seq
Mouse ES E14 and Drosophila S2 cells were washed with 1x PBS, fixed at room temperature with 1% paraformaldehyde in 1x PBS for 10 minutes. Fixation was stopped by adding glycine to a final concentration of 125mM for 10 minutes at room temperature. Cells were washed twice with ice-cold 1x PBS and collected by scrapping. Cells were centrifuged at 2000 g for 5 minutes at 4°C and washed once with ice-cold 1x PBS. Cells were lysed in L1 buffer (50 mM Tris-HCl ph 8.0, 2 mM EDTA pH 8.0, 0.1% NP40, 10% glycerol, 5 mM sodium butyrate and protease inhibitory cocktail) and incubated on ice for 10 minutes before centrifugation at 2000 g for 10 minutes at 4°C. Nuclei were resuspended in L2 buffer (0.5% SDS, 10 mM EDTA ph 8.0, 50 mM Tris-HCl pH 8.0, 5 mM sodium butyrate and protease inhibitory cocktail) and sonicated using a Covaris E210 sonicator to on average 300 bp fragments. Protein-A Sepharose beads were washed twice with TE buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA) and blocked for three hours with 1ug/ul denatured yeast tRNA and 1,5% fish-skin gelatine. Beads were washed twice with TE buffer and stored at 4°C. For ChIP, 49 μg of mouse chromatin was spiked with 1 μg of fly chromatin. The chromatin mix was diluted with chromatin dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl pH 8.0, 5 mM sodium butyrate and protease inhibitory cocktail) to a volume of 800 μl and incubated with 30 μl of blocked bead slurry for 1 hour at 4°C for preclearing. Two μg of anti-H3K9ac antibody (abcam, cat # ab4441) was added to precleared chromatin and incubated overnight at 4°C. 50 μl of bead slurry was added to samples and incubated for two hours at 4°C with overhead shaking. After centrifugation for 2 minutes at 1000 g, beads were washed two times with Low salt washing buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris HCl pH 8.0, 5 mM sodium butyrate and protease inhibitory cocktail) for 10 minutes, two times with High salt washing buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris HCl pH 8.0, 5 mM sodium butyrate and protease inhibitory cocktail) for 10 minutes, two times with LiCl washing buffer (250 mM LiCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0, 5 mM sodium butyrate and protease inhibitory cocktail) for 10 minutes and two times with TE buffer for 10 minutes. Bound fragments were eluted using with freshly prepared elution buffer (1% SDS and 0.1 M sodium bicarbonate) at room temperature. Chromatin was reverse crosslinked by addition of 0.2M NaCl and 50 μg/ml RNase A. Samples were incubated at 37°C for one hour. 20 μg of Proteinase K, 60 mM Tris ph 7.9, 20 mM EDTA was added and samples were incubated at 65°C overnight in a thermomixer. DNA was isolated by Phenol-chloroform purification and resuspended in water.
Library preparation of ChIP-seq
ChIP samples were purified using Agencourt AMPure XP beads (Beckman Coulter, Cat#A63882) and quantified with the Qubit (Invitrogen). ChIP-seq libraries were prepared from 7 to 10 ng of double-stranded purified DNA using the MicroPlex Library Preparation kit v2 (Diagenode, Cat# C05010014), according to manufacturer’s instructions. In the first step, the DNA was repaired and yielded molecules with blunt ends. In the next step, stem-loop adaptors with blocked 5′ ends were ligated to the 5′ end of the genomic DNA, leaving a nick at the 3′ end. The adaptors cannot ligate to each other and do not have single-strand tails, avoiding non-specific background. In the final step, the 3′ ends of the genomic DNA were extended to complete library synthesis and Illumina compatible indexes were added through a PCR amplification (7 cycles). Amplified libraries were purified and size-selected using Agencourt AMPure XP beads to remove unincorporated primers and other reagents. The libraries were sequenced on Illumina Hiseq 4000 sequencer as single-Read 50 base reads following Illumina’s instructions.
Data analysis of ChIP-seq
Image analysis and base calling were performed using RTA 2.7.7 and bcl2fastq 2.17.1.14. Adaptor sequences were removed and low-quality ends were trimmed. Sequence alignment was performed using the mm10 assembly for Mus musculus genome or the BDGP6 assembly for D. melanogaster genome using Bowtie (version 1.0.0) (Langmead and Salzberg, 2012) choosing the zero multi-mapping option. Blacklisted reads for the mouse genome were filtered out using the ENCODE mm10 blacklist v2. For violin plots shown in Figure 5D, reads were counted over promoter ranges (1500 bp upstream, 500 bp downstream) of genes of the Ensembl 93 database. Read counts were normalized across samples by dividing by size factors calculated using the spike-in D. melanogaster reads as follows. Uniquely mapped D. melanogaster reads were divided by black list filtered uniquely mapped mouse reads and then multiplied by 100. Bigwig tracks were generated using Homer software (v4.11.1) and normalized by the D. melanogaster size factors.
RT-qPCR
Reverse Transcription (RT) was performed with 1 – 2 μg total RNA and using 3.2 μg random hexamer primers (ThermoFisher Scientific, Cat#SO142) and Transcriptor Reverse Transcriptase (Roche, Cat#03531287001) following manufacturer’s instruction. For qPCR, the cDNA samples were amplified using LightCycler® 480 SYBR® Green 2x PCR Master Mix I (Roche, Cat#04887352001) and 0.6 μM of forward and reverse primer respectively. The primer pairs used for qPCR are listed in Table S2. The qPCR was conducted using a LightCycler® 480 (Roche). The obtained threshold-values were used to calculate the relative gene expression using the 2−ΔΔCT method and considering the individual primer pair efficiencies (Pfaffl, 2001).
Whole cell protein extraction
For whole cell protein extracts, cells were washed with 1x PBS and harvested using trypsin 0.25% EDTA. After centrifugation at 2,000 g for 3 minutes, the protein pellets were washed twice with ice-cold 1x PBS to remove any remaining FCS. Proteins were then extracted from the collected cells using 1 volume of whole cell extract buffer (50 mM Tris HCl pH 7.9, 25% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 5 mM MgCl2, 600 mM KCl, 0.5% NP40 and 1x protein inhibitor cocktail) and incubated for 10 minutes. The salt concentration was neutralized by addition 2–3 volumes of IP0 buffer (25 mM Tris HCl pH 7.9, 5% glycerol, 5 mM MgCl2, 0.1% NP40, 1 mM DTT and 1x protein inhibitor cocktail) and incubation for 10 minutes. After centrifugation at 12,000 g for 10 minutes at 4°C, the supernatant containing the proteins was collected. The protein concentrations of the extracts were determined using the Coomassie Protein Assay Dye Reagent Concentrate (Bio-Rad, Cat#5000006) and Synergy HTX Multi-Mode Reader (BioTek).
Acidic extraction of histone proteins
Cells were harvested by trypsinization and washed twice with ice-cold 1x PBS. Pellets were resuspended in 5–10 volumes of lysis buffer (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 10 mM N-ethylmaleimide, 5 mM sodium butyrate and protein inhibitor cocktail) and 0.2 M hydrochloric acid was added before incubation on ice for 30 minutes. Extracts were centrifuged for 10 minutes at 11,000 g at 4°C and the supernatant stored at −80°C prior to western blotting.
Western blot analysis
Proteins were separated using 8% to 15% of SDS-PAGE gels prior to blotting onto nitrocellulose membranes. Membranes were blocked in 3% non-fat dry milk for at least 30 minutes at room temperature. The membranes were then incubated overnight with primary antibodies (see Key resources table) in 0.3% non-fat dry milk at 4°C with one exception being Streptavidin-HRP, which was used in 1% BSA. After washing with 1x PBS containing 0.1% Tween20, if required, the membranes were incubated with secondary goat-anti-rabbit or -mouse antibodies conjugated to HRP for 50 minutes at room temperature followed by further three washes. The membranes were developed using the Pierce™ ECL Western Blotting Substrate (ThermoFisher Scientific, Cat#32109) and the ChemiDoc™ Touch Imaging System (Bio-Rad).
Nuclear extraction
To enrich extracts for nuclear proteins, cells were harvested and washed twice with 1x PBS. Cell pellet was resuspended in hypotonic buffer (10 mM Tris-HCl pH 8.0, 1.5 mM MgCl2, 10 mM KCl and 1x protein inhibitor cocktail) and dounced 10 to 20 times using a B dounce homogenizer to isolate the nuclei. After centrifugation at 10,000 g for 10 minutes at 4°C, supernatant was removed and pellet resuspended in high salt buffer (20 mM Tris-HCl pH 8.0, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 450 mM NaCl, 0.1% NP40 and 1x protein inhibitor cocktail). Suspension was homogenized by douncing as described before prior to centrifugation at 10,000 g for 10 minutes at 4°C. The supernatant was kept as nuclear extract and stored at −80°C.
Immunoprecipitation
Protein-A or Protein-G Sepharose beads were washed three times with filtered 1x PBS and two times with IP100 buffer (25 mM Tris-HCl 7.5, 5 mM MgCl2, 10% glycerol, 0,1% NP40, 100 mM KCl, 2 mM DTT and 1x protein inhibitor cocktail). Nuclear extracts were pre-cleared with 1/5 of 50% bead slurry for 2 hours at 4°C with overhead agitation. For antibody binding, the 50% bead slurry was incubated with 1/10 volume of the respective antibody ascites for 2 hours at 4°C with overhead agitation. After incubation, beads were washed three times with IP500 buffer (25 mM Tris-HCl 7.5, 5 mM MgCl2, 10% glycerol, 0,1% NP40, 500 mM KCl, 2 mM DTT and 1x protein inhibitor cocktail) and twice with IP100 buffer before addition of 1/5 volume of the 50% antibody-bead slurry to the pre-cleared nuclear extracts. Nuclear extracts were incubated with beads overnight at 4°C with overhead agitation. After incubation, resins were washed three times with IP500 buffer and twice with IP100 buffer. For anti-Taf10 IPs, anti-Mbip IPs and anti-Zzz3 IPs, complexes were eluted from the beads using two subsequent 0.1 M glycine pH 2.8 elutions at room temperature and with agitation. Importantly, as Taf10 are shared between the SAGA and TFIID complexes and to increase the purification efficiency for SAGA in anti-Taf10 IPs, nuclear extracts were depleted for TFIID prior to anti-Taf10 IP by overnight incubation with beads coated with antibodies targeting the TFIID-specific subunit Taf7. Supernatants depleted for Taf7-containing TFIID were subsequently used for anti-Taf10 IPs. For anti-Atac2 IPs, complexes were eluted from the beads using two subsequent steps of elutions with peptide PI264 at a concentration of 2 mg/ml pH 7.5 for 1 hour each at 4°C with overhead agitation. Eluates were then characterized by western blot or mass spectrometry analyses.
Mass spectrometry
Protein mixtures were precipitated with TCA (Sigma Aldrich, Cat#T0699) overnight at 4°C. Samples were then centrifuged at 14,000 g for 30 minutes at 4°C. Pellet were washed twice with 1 mL cold acetone and centrifuged at 14,000 g for 10 minutes at 4°C. Washed pellet were then urea- denatured with 8 M urea (Sigma Aldrich, Cat#U0631) in Tris-HCl 0.1 mM, reduced with 5 mM TCEP for 30 minutes, and then alkylated with 10 mM iodoacetamide (Sigma Aldrich, Cat#I1149) for 30 minutes in the dark. Both reduction and alkylation were performed at room temperature and under agitation (850 rpm). Double digestion was performed with endoproteinase Lys-C (Wako, Cat#125–05061) at a ratio 1/100 (enzyme/proteins) in 8 M urea for 4 hours, followed by an overnight modified trypsin (Promega, Cat#V5111) digestion at a ratio 1/100 (enzyme/proteins) in 2 M urea. Both Lys-C and Trypsin digestions were performed at 37°C. Peptide mixtures were then desalted on C18 spin-column and dried on Speed-Vacuum before LC-MS/MS analysis. Samples were analyzed using an Ultimate 3000 nano-RSLC (ThermoFisher Scientific) coupled in line with a LTQ-Orbitrap ELITE mass spectrometer via a nano-electrospray ionization source (ThermoFisher Scientific). Peptide mixtures were loaded on a C18 Acclaim PepMap100 trap-column (75 μm ID x 2 cm, 3 μm, 100Å,ThermoFisher Scientific) for 3.5 minutes at 5 μL/min with 2% ACN (Sigma Aldrich, Cat#1207802), 0.1% formic acid (Sigma Aldrich, Cat#94318) in water and then separated on a C18 Accucore nano-column (75 μm ID x 50 cm, 2.6 μm, 150Å, ThermoFisher Scientific) with a 90 minutes linear gradient from 5% to 35% buffer B (A: 0.1% FA in water/ B: 99% ACN, 0.1% FA in water), then a 20 minutes linear gradient from 35% to 80% buffer B, followed with 5 min at 99% B and 5 minutes of regeneration at 5% B. The total duration was set to 120 minutes at a flow rate of 200 nL/min. The oven temperature was kept constant at 38°C. The mass spectrometer was operated in positive ionization mode, in data-dependent mode with survey scans from m/z 350–1500 acquired in the Orbitrap at a resolution of 120,000 at m/z 400. The 20 most intense peaks (TOP20) from survey scans were selected for further fragmentation in the Linear Ion Trap with an isolation window of 2.0 Da and were fragmented by CID with normalized collision energy of 35%. Unassigned and single charged states were rejected. The Ion Target Value for the survey scans (in the Orbitrap) and the MS2 mode (in the Linear Ion Trap) were set to 1E6 and 5E3 respectively and the maximum injection time was set to 100 ms for both scan modes. Dynamic exclusion was used. Exclusion duration was set to 20 s, repeat count was set to 1 and exclusion mass width was ± 10 ppm.
Data analysis of mass spectrometry
Proteins were identified by database searching using SequestHT (ThermoFisher Scientific) with Proteome Discoverer 2.4 software (PD2.4, ThermoFisher Scientific) on Mus musculus database (Swissprot, non-reviewed, release 2019_08_07, 55121 entries). Precursor and fragment mass tolerances were set at 7 ppm and 0.6 Da respectively, and up to 2 missed cleavages were allowed. Oxidation (M) was set as variable modification, and Carbamidomethylation (C) as fixed modification. Peptides were filtered with a false discovery rate (FDR) at 1%, rank 1 and proteins were identified with 1 unique peptide. For the Label-Free Quantification, the protein abundancies were calculated from the average of the peptide abundancies using the TOP N (where N = 3, the 3 most intense peptides for each protein), and only the unique peptide were used for the quantification. Normalized spectral abundance factors (NSAF) were calculated for each protein as follows. To obtain spectral abundance factors (SAF), spectral counts identifying a protein were divided by the protein length represented by the number of amino acids. To calculate NSAF values, the SAF values of each protein were divided by the sum of SAF values of all SAGA subunits. The NSAF values were subsequently normalized to the NSAF values of the bait protein Taf10.
QUANTIFICATION AND STATISTICAL ANALYSIS
Clonal assay quantifications were performed using ImageJ software. Flow cytometry data was analyzed using FlowJo™ 10.2. software. Western blot quantification was performed with ImageLab™ software from Bio-Rad. Statistical analyses were performed using two-sided Wilcoxon-Mann-Whitney test or two-sided Welch t test for two-sample comparisons, or Wilcoxon rank sum test with Benjamini-Hochberg correction for multiple testing or analysis of variance (ANOVA) for multiple-sample comparisons. Details for individual experiments including number of replicates and statistical tests performed can be found in the figure legends. All statistical tests were performed using R (version 4.0.5). Comparisons were considered statistically significant with a p-value below 0.05.
Supplementary Material
Highlights.
SAGA and ATAC are required for mouse ESC growth and self-renewal
SAGA and ATAC predominantly regulate different sets of genes
SAGA and ATAC have a mild global effect on Pol II transcription
SAGA and ATAC effects on Pol II transcription and cell growth are HAT independent
ACKNOWLEDGMENTS
We thank D. Helmlinger and T. Sexton for discussions and critical comments on the manuscript and P. Navarro Gil for advice on mouse ESC analyses. We also thank C. Thibault-Carpentier and B. Jost from the GenomEast platform (France Génomique consortium ANR-10-INBS-0009), S. Bour, N. Jung, C. Birck, C. Ebel, M. Philipps, L. Negroni, B. Morlet, and the IGBMC cell culture facility for their contribution to this work. We thank K. Remans for gift of plasmids encoding the Tn5E54K,L372P transposase. This study was supported by NIH R01 grant (1R01GM131626-01 to L.T.), by the Agence Nationale de la Recherche (AAPG2019 PICen and ANR-PRCI-AAPG2019 EpiCAST to L.T.; ANR-18-CE12-0026 and ANR-20-CE12-0014-02 to D.D.), grant ANR-10-LABX-0030-INRT and a French State fund managed by the ANR under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02 to IGBMC. V.F. was recipient of fellowships by the IdEx-University of Strasbourg international Ph.D. program and by the Fondation pour la Recherche Médicale (FRM) association (FDT201904008368).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPORTING CITATIONS
The following references appear in the supplemental information: Tourigny et al. (2018).
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109598.
REFERENCES
- Acharya D, Hainer SJ, Yoon Y, Wang F, Bach I, Rivera-Pérez JA, and Fazzio TG (2017). KAT-Independent Gene Regulation by Tip60 Promotes ESC Self-Renewal but Not Pluripotency. Cell Rep. 19, 671–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, and Huber W (2010). Differential expression analysis for sequence count data. Genome Biol. 11, R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arede L, Foerner E, Wind S, Kulkarni R, Domingues AF, Kleinwaechter S, Gupta S, Scheer E, Tora L, and Pina C (2020). Unique roles of ATAC and SAGA - KAT2A complexes in normal and malignant hematopoiesis. Bio-Rxiv. 10.1101/2020.05.14.096057. [DOI] [Google Scholar]
- Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, and Tan S (2002). Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277, 7989–7995. [DOI] [PubMed] [Google Scholar]
- Baptista T, Grünberg S, Minoungou N, Koster MJE, Timmers HTM, Hahn S, Devys D, and Tora L (2017). SAGA Is a General Cofactor for RNA Polymerase II Transcription. Mol. Cell 68, 130–143.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardot P, Vincent SD, Fournier M, Hubaud A, Joint M, Tora L, and Pourquié O (2017). The TAF10-containing TFIID and SAGA transcriptional complexes are dispensable for early somitogenesis in the mouse embryo. Development 144, 3808–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, and Golani I (2001). Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284. [DOI] [PubMed] [Google Scholar]
- Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, and Tora L (2001). UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 20, 3187–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly JM, Tora L, and Chambon P (1993). Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 12, 489–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu P, Evrard YA, Lozano G, and Dent SYR (2007). Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol. Cell. Biol. 27, 3405–3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr. Protoc. Mol. Biol. 109, 21.29.1–21.29.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell J, and Dalton S (2013). Roles for MYC in the establishment and maintenance of pluripotency. Cold Spring Harb. Perspect. Med. 3, a014381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H-Z, Tsai S-Y, and Leone G (2009). Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9, 785–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, and Ma’ayan A (2013). Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorighi KM, Swigut T, Henriques T, Bhanu NV, Scruggs BS, Nady N, Still CD 2nd, Garcia BA, Adelman K, and Wysocka J (2017). Mll3 and Mll4 Facilitate Enhancer RNA Synthesis and Transcription from Promoters Independently of H3K4 Monomethylation. Mol. Cell 66, 568–576.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, and Lash AE (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30, 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elías-Villalobos A, Toullec D, Faux C, Séveno M, and Helmlinger D (2019). Chaperone-mediated ordered assembly of the SAGA and NuA4 transcription co-activator complexes in yeast. Nat. Commun. 10, 5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, and Marillonnet S (2009). Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4, e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, and Kaufman MH (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156. [DOI] [PubMed] [Google Scholar]
- Fagnocchi L, and Zippo A (2017). Multiple Roles of MYC in Integrating Regulatory Networks of Pluripotent Stem Cells. Front. Cell Dev. Biol. 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festuccia N, Gonzalez I, and Navarro P (2017). The Epigenetic Paradox of Pluripotent ES Cells. J. Mol. Biol. 429, 1476–1503. [DOI] [PubMed] [Google Scholar]
- Fournier M, Orpinell M, Grauffel C, Scheer E, Garnier J-M, Ye T, Chavant V, Joint M, Esashi F, Dejaegere A, et al. (2016). KAT2A/KAT2B-targeted acetylome reveals a role for PLK4 acetylation in preventing centrosome amplification. Nat. Commun. 7, 13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper AM, Kim J, and Roeder RG (2009). The STAGA subunit ADA2b is an important regulator of human GCN5 catalysis. Mol. Cell. Biol. 29, 266–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. (1997). Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640–1650. [DOI] [PubMed] [Google Scholar]
- Guelman S, Kozuka K, Mao Y, Pham V, Solloway MJ, Wang J, Wu J, Lill JR, and Zha J (2009). The double-histone-acetyltransferase complex ATAC is essential for mammalian development. Mol. Cell. Biol. 29, 1176–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Luo J, Ranish J, and Hahn S (2014). Architecture of the Saccharomyces cerevisiae SAGA transcription coactivator complex. EMBO J. 33, 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, and Tora L (2017). Sharing the SAGA. Trends Biochem. Sci. 42, 850–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Marguerat S, Villén J, Swaney DL, Gygi SP, Bähler J, and Winston F (2011). Tra1 has specific regulatory roles, rather than global functions, within the SAGA co-activator complex. EMBO J. 30, 2843–2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmlinger D, Papai G, Devys D, and Tora L (2020). What do the structures of GCN5-containing complexes teach us about their function? Biochim. Biophys. Acta Gene Regul. Mech. 1864, 194614. [DOI] [PubMed] [Google Scholar]
- Hennig BP, Velten L, Racke I, Tu CS, Thoms M, Rybin V, Besir H, Remans K, and Steinmetz LM (2018). Large-Scale Low-Cost NGS Library Preparation Using a Robust Tn5 Purification and Tagmentation Protocol. G3 (Bethesda) 8, 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry KW, Wyce A, Lo W-S, Duggan LJ, Emre NCT, Kao C-F, Pillus L, Shilatifard A, Osley MA, and Berger SL (2003). Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Bortvin AL, Ricupero-Hovasse SL, and Winston F (1995). A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15, 1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J, Arand J, Chaikovsky A, Mooney NA, Demeter J, Brison CM, Oliverio R, Vogel H, Rubin SM, Jackson PK, and Sage J (2019). E2F4 regulates transcriptional activation in mouse embryonic stem cells independently of the RB family. Nat. Commun. 10, 2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq X, Brou C, Lutz Y, Davidson I, Chambon P, and Tora L (1994). Human TAFII30 is present in a distinct TFIID complex and is required for transcriptional activation by the estrogen receptor. Cell 79, 107–117. [DOI] [PubMed] [Google Scholar]
- King HW, and Klose RJ (2017). The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. eLife 6, e22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, and Joung JK (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Wang L, Schibler AC, Chao H-P, Kuang X, Lin K, Lu Y, Shen J, Jeter CR, Salinger A, et al. (2019). USP22 controls multiple signaling pathways that are essential for vasculature formation in the mouse placenta. Development 146, dev174037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutelou E, Farria AT, and Dent SYR (2020). Complex functions of Gcn5 and Pcaf in development and disease. Biochim. Biophys. Acta Gene Regul. Mech. 1864, 194609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T (2007). Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- Krebs AR, Karmodiya K, Lindahl-Allen M, Struhl K, and Tora L (2011). SAGA and ATAC histone acetyl transferase complexes regulate distinct sets of genes and ATAC defines a class of p300-independent enhancers. Mol. Cell 44, 410–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. (2016). Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44 (W1), W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KK, Sardiu ME, Swanson SK, Gilmore JM, Torok M, Grant PA, Florens L, Workman JL, and Washburn MP (2011). Combinatorial depletion analysis to assemble the network architecture of the SAGA and ADA chromatin remodeling complexes. Mol. Syst. Biol. 7, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, and Workman JL (2007). The role of chromatin during transcription. Cell 128, 707–719. [DOI] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, and Dou Y (2012). The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 11, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Srajer G, Evrard YA, Phan HM, Furuta Y, and Dent SYR (2007). Developmental potential of Gcn5(−/−) embryonic stem cells in vivo and in vitro. Dev. Dyn. 236, 1547–1557. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, and Smith A (2014). The nature of embryonic stem cells. Annu. Rev. Cell Dev. Biol. 30, 647–675. [DOI] [PubMed] [Google Scholar]
- Martin GR (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634–7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. [Google Scholar]
- Matsumura I, Tanaka H, and Kanakura Y (2003). E2F1 and c-Myc in cell growth and death. Cell Cycle 2, 333–338. [PubMed] [Google Scholar]
- Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, and Davidson I (1995). Cloning and characterization of hTAFII18, hTAFII20 and hTAFII28: three subunits of the human transcription factor TFIID. EMBO J. 14, 1520–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang S, Andrews FH, Wang X, Gagea M, Wen H, et al. (2017). YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat. Commun. 8, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Zhang Y, Lyu J, Wang X, Tong Q, Peng D, Xue Y, Tencer AH, Wen H, Li W, et al. (2018). The ZZ-type zinc finger of ZZZ3 modulates the ATAC complex-mediated histone acetylation and gene activation. Nat. Commun. 9, 3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohibi S, Gurumurthy CB, Nag A, Wang J, Mirza S, Mian Y, Quinn M, Katafiasz B, Eudy J, Pandey S, et al. (2012). Mammalian alteration/deficiency in activation 3 (Ada3) is essential for embryonic development and cell cycle progression. J. Biol. Chem. 287, 29442–29456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy Z, Riss A, Fujiyama S, Krebs A, Orpinell M, Jansen P, Cohen A, Stunnenberg HG, Kato S, and Tora L (2010). The metazoan ATAC and SAGA coactivator HAT complexes regulate different sets of inducible target genes. Cell. Mol. Life Sci. 67, 611–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, and Kanemaki MT (2016). Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 15, 210–218. [DOI] [PubMed] [Google Scholar]
- Navarro P (2018). 2i, or Not 2i: The Soliloquy of Nanog-Negative Mouse Embryonic Stem Cells. Stem Cell Rep. 11, 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orpinell M, Fournier M, Riss A, Nagy Z, Krebs AR, Frontini M, and Tora L (2010). The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 29, 2381–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Liu H, Yu J, Kelliher MA, Castilla LH, Lawson ND, and Zhu LJ (2018). ATACseqQC: a Bioconductor package for post-alignment quality assessment of ATAC-seq data. BMC Genomics 19, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Komonyi O, Bodai L, Ujfaludi Z, Muratoglu S, Ciurciu A, Tora L, Szabad J, and Boros I (2005). The homologous Drosophila transcriptional adaptors ADA2a and ADA2b are both required for normal development but have different functions. Mol. Cell. Biol. 25, 8215–8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankotai T, Popescu C, Martín D, Grau B, Zsindely N, Bodai L, Tora L, Ferrús A, and Boros I (2010). Genes of the ecdysone biosynthesis pathway are regulated by the dATAC histone acetyltransferase complex in Drosophila. Mol. Cell. Biol. 30, 4254–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papai G, Frechard A, Kolesnikova O, Crucifix C, Schultz P, and Ben-Shem A (2020). Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 577, 711–716. [DOI] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, et al. (2019). The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP (2005). The architecture of mammalian ribosomal protein promoters. BMC Evol. Biol. 5, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45–e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintacuda G, Wei G, Roustan C, Kirmizitas BA, Solcan N, Cerase A, Castello A, Mohammed S, Moindrot B, Nesterova TB, and Brockdorff N (2017). hnRNPK Recruits PCGF3/5-PRC1 to the Xist RNA B-Repeat to Establish Polycomb-Mediated Chromosomal Silencing. Mol. Cell 68, 955–969.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. (2011). Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat. Biotechnol. 29, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rädle B, Rutkowski AJ, Ruzsics Z, Friedel CC, Koszinowski UH, and Dölken L (2013). Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J. Vis. Exp. 78, 50195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickels R, Herz H-M, Sze CC, Cao K, Morgan MA, Collings CK, Gause M, Takahashi YH, Wang L, Rendleman EJ, et al. (2017). Histone H3K4 monomethylation catalyzed by Trr and mammalian COMPASS-like proteins at enhancers is dispensable for development and viability. Nat. Genet. 49, 1647–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel AE, Cieniewicz AM, Taverna SD, and Wolberger C (2015). Nucleosome competition reveals processive acetylation by the SAGA HAT module. Proc. Natl. Acad. Sci. USA 112, E5461–E5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riss A, Scheer E, Joint M, Trowitzsch S, Berger I, and Tora L (2015). Subunits of ADA-two-A-containing (ATAC) or Spt-Ada-Gcn5-acetyltrasferase (SAGA) Coactivator Complexes Enhance the Acetyltransferase Activity of GCN5. J. Biol. Chem. 290, 28997–29009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury S, Bernecky C, and Cramer P (2015). Structural basis of transcription initiation by RNA polymerase II. Nat. Rev. Mol. Cell Biol. 16, 129–143. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwalb B, Michel M, Zacher B, Frühauf K, Demel C, Tresch A, Gagneur J, and Cramer P (2016). TT-seq maps the human transient transcriptome. Science 352, 1225–1228. [DOI] [PubMed] [Google Scholar]
- Scognamiglio R, Cabezas-Wallscheid N, Thier MC, Altamura S, Reyes A, Prendergast ÁM, Baumgärtner D, Carnevalli LS, Atzberger A, Haas S, et al. (2016). Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell 164, 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seruggia D, Oti M, Tripathi P, Canver MC, LeBlanc L, Di Giammartino DC, Bullen MJ, Nefzger CM, Sun YBY, Farouni R, et al. (2019). TAF5L and TAF6L Maintain Self-Renewal of Embryonic Stem Cells via the MYC Regulatory Network. Mol. Cell 74, 1148–1163.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, and Dalton S (2010). Myc transcription factors: key regulators behind establishment and maintenance of pluripotency. Regen. Med. 5, 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soffers JHM, and Workman JL (2020). The SAGA chromatin-modifying complex: the sum of its parts is greater than the whole. Genes Dev. 34, 1287–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spedale G, Timmers H.Th.M., and Pijnappel WWMP (2012). ATAC-king the complexity of SAGA during evolution. Genes Dev. 26, 527–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Grant PA, Roberts SM, Duggan LJ, Belotserkovskaya R, Pacella LA, Winston F, Workman JL, and Berger SL (1999). Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19, 86–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussman RT, Stanek TJ, Esteso P, Gearhart JD, Knudsen KE, and McMahon SB (2013). The epigenetic modifier ubiquitin-specific protease 22 (USP22) regulates embryonic stem cell differentiation via transcriptional repression of sex-determining region Y-box 2 (SOX2). J. Biol. Chem. 288, 24234–24246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourigny JP, Saleh MM, Schumacher K, Devys D, and Zentner GE (2018). Mediator is essential for small nuclear and nucleolar RNA transcription in yeast. Mol. Cell Biol. 38, 00296–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, and Knoepfler PS (2010). myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation 80, 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, and Dent SY (2014). Functions of SAGA in development and disease. Epigenomics 6, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, El-Saafin F, Ye T, Stierle M, Negroni L, Durik M, Fischer V, Devys D, Vincent SD, and Tora L (2021). Histone H2Bub1 deubiquitylation is essential for mouse development, but does not regulate global RNA polymerase II transcription. Cell Death Differ. 28, 2385–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-L, Faiola F, Xu M, Pan S, and Martinez E (2008). Human ATAC Is a GCN5/PCAF-containing acetylase complex with a novel NC2-like histone fold module that interacts with the TATA-binding protein. J. Biol. Chem. 283, 33808–33815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, and Zhang B (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 41, W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Koutelou E, Hirsch C, McCarthy R, Schibler A, Lin K, Lu Y, Jeter C, Shen J, Barton MC, and Dent SYR (2018). GCN5 Regulates FGF Signaling and Activates Selective MYC Target Genes during Early Embryoid Body Differentiation. Stem Cell Rep. 10, 287–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dienemann C, Stützer A, Urlaub H, Cheung ACM, and Cramer P (2020). Structure of the transcription coactivator SAGA. Nature 577, 717–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P-YJ, and Winston F (2002). Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 22, 5367–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. (2021). Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 1, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Edmondson DG, Evrard YA, Wakamiya M, Behringer RR, and Roth SY (2000). Loss of Gcn5l2 leads to increased apoptosis and mesodermal defects during mouse development. Nat. Genet. 26, ng1000_229. [DOI] [PubMed] [Google Scholar]
- Yamauchi T, Yamauchi J, Kuwata T, Tamura T, Yamashita T, Bae N, Westphal H, Ozato K, and Nakatani Y (2000). Distinct but overlapping roles of histone acetylase PCAF and of the closely related PCAF-B/GCN5 in mouse embryogenesis. Proc. Natl. Acad. Sci. USA 97, 11303–11306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-J, Ogryzko VV, Nishikawa J, Howard BH, and Nakatani Y (1996). A p300/CBP-associated factor that competes with the adenoviral on-coprotein E1A. Nature 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Young RA (2011). Control of the embryonic stem cell state. Cell 144, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. (2008). A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell 29, 92–101. [DOI] [PubMed] [Google Scholar]
- Zohn IE, Li Y, Skolnik EY, Anderson KV, Han J, and Niswander L (2006). p38 and a p38-interacting protein are critical for downregulation of E-cadherin during mouse gastrulation. Cell 125, 957–969. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq, ATAC-seq and ChIP-seq data have been deposited at GEO (Edgar et al., 2002) and are publicly available as of the date of publication. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al., 2019) partner repository and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Original western blot images have been deposited at Mendeley Data and are publicly available as of the date of publication. The DOIs is listed in the key resources table.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this study is available from the lead contact upon request.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Supt7l | Bethyl Laboratories | Cat#A302–803A; RRID:AB_10630265 |
| Mouse monoclonal anti-γTubulin | Sigma-Aldrich | Cat#T6557; RRID:AB_477584 |
| Rabbit polyclonal anti-Rpl29 | ThermoFisher Scientific | Cat#15799–1-AP; RRID:AB_2878187 |
| Rabbit polyclonal anti-histone H3 lysine 9 acetylation | Abcam | Cat#ab4441; RRID:AB_2118292 |
| Rabbit polyclonal anti-HA tag | Abcam | Cat#ab9110; RRID:AB_307019 |
| Mouse monoclonal anti-Wdr5 | Abcam | Cat#ab56919; RRID:AB_946146 |
| Mouse monoclonal anti-Tbp | In-house | 3TF1–3G3; Brou et al., 1993 |
| Rabbit polyclonal anti-Taf7 | In-house | 3475; Bardot et al., 2017 |
| Mouse monoclonal anti-Taf10 | In-house | 6TA-2B11; Jacq et al., 1994 |
| Mouse monoclonal anti-Taf12 | In-house | 22TA-2A1; Mengus et al., 1995 |
| Rabbit polyclonal anti-Atxn7l3 | In-house | 2325; Zhao et al., 2008 |
| Rabbit polyclonal anti-Supt3h | In-house | 3118; Bardot et al., 2017 |
| Rabbit polyclonal anti-Gcn5 | In-house | 2676; Nagy et al., 2010; Orpinell et al., 2010 |
| Mouse monoclonal anti-Gcn5 | In-house | 2GC-2C11; Brand et al., 2001 |
| Rabbit polyclonal anti-Tada3 | In-house | 2678; Nagy et al., 2010; Orpinell et al., 2010 |
| Rabbit polyclonal anti-Sgf29 | In-house | 2461; Orpinell et al., 2010 |
| Rabbit polyclonal anti-Atac2 | In-house | 2734; Nagy et al., 2010 |
| Rabbit polyclonal anti-Mbip | In-house | 2768; Nagy et al., 2010; Orpinell et al., 2010 |
| Rabbit polyclonal anti-Zzz3 | In-house | 2616; Nagy et al., 2010 |
| Chemicals, peptides, and recombinant proteins | ||
| Indole-3-acetic acid | Sigma-Aldrich | Cat#I3750 |
| 4-thiouridine | Glentham Life Sciences or abcam | Cat#GN6085 or Cat#ab143718 |
| 4-thiouracil | Sigma-Aldrich | Cat#440736 |
| EZ-link HPDP-biotin | ThermoFisher Scientific | Cat#21341 |
| Streptavidin protein coupled to HRP | ThermoFisher Scientific | Cat#21126 |
| Home-made Tn5E54K,L372P transposase | Plasmid provided by Dr. Kim Remans, EMBL Heidelberg, Germany | Hennig et al., 2018 |
| Critical commercial assays | ||
| Alkaline Phosphatase Kit | Vector Laboratories | Cat#SK-5100 |
| TURBO DNA-free™ Kit | ThermoFisher Scientific | Cat#AM1907 |
| μMACS magnetic beads and columns | Miltenyi Biotec | Cat#130–074–101 |
| RNeasy MinElute Cleanup Kit | QIAGEN | Cat#74204 |
| NucleoSpin Gel and PCR Clean-up kit | Macherey-Nagel | Cat#740609.50 |
| RiboPure™ RNA Purification Kit | ThermoFisher Scientific | Cat#AM1926 |
| TruSeq Stranded Total RNA LT Sample Prep Kit with Ribo-Zero Gold | Illumina | Cat#RS-122–2301 or Cat#RS-122–2302 |
| Ribo-Zero Gold rRNA (Yeast) | Illumina | Cat#MRZY1324 |
| MicroPlex Library Preparation kit v2 | Diagenode | Cat#C05010014 |
| SPRIselect beads | Beckman-Coulter | Cat#B23319 |
| AMPure XP beads | Beckman-Coulter | Cat#A63882 |
| Deposited data | ||
| Raw and analyzed data 4sU RNA-seq | This paper | GEO: GSE175905 |
| Raw and analyzed data ATAC-seq | This paper | GEO: GSE175905 |
| Raw and analyzed data H3K9ac ChIP-seq | This paper | GEO: GSE175905 |
| Mass spectrometry proteomics Taf10 IP | This paper | PRIDE: PXD027026 |
| Mass spectrometry proteomics Mbip and Zzz3 IP | This paper | PRIDE: PXD027026 |
| Original western blot images | This paper | Mendeley Data: https://doi.org/10.17632/thjcm8zdpf.1 |
| Experimental models: Cell lines | ||
| Mouse: ES E14tg2a.4 cells (129P2 genetic background) | BayGenomics | N/A |
| Mouse: Supt7l−/− ES E14 cells | This study | N/A |
| Mouse: Supt7ltg ES E14 cells | This study | N/A |
| Mouse: Supt20h−/− ES E14 cells | This study | N/A |
| Mouse: Tada2b−/− ES E14 cells | This study | N/A |
| Mouse: Tada2b−/− ES E14 cells | This study | N/A |
| Mouse: Yeats2AID/AID ES E14 cells N-ter. | This study | N/A |
| Mouse: Tada3AID/AID ES E14 cells N-ter. | This study | N/A |
| Mouse: Zzz3AID/AID ES E14 cells N-ter. | This study | N/A |
| Mouse: Zzz3AID/AID ES E14 cells C-ter. | This study | N/A |
| D. melanogaster: Schneider S2 cells | ATCC | CRL-1963 |
| Experimental models: Organisms/strains | ||
| S. pombe: DHP43 h- | Elías-Villalobos et al., 2019 | N/A |
| S. cerevisiae: Strain background: FY406 | Hirschhorn et al., 1995 | N/A |
| Oligonucleotides | ||
| For sgRNA sequences, see Table S1 | This paper | N/A |
| For primer sequences, see Table S2 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: Supt7l CDS-HygromycinR | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Supt7l for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Supt20h for KO | This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against Tada2b for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Tada2a for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Yeats2 for KO | This paper | N/A |
| Plasmid: Cas9-eGFP-sgRNA against Zzz3 for KO | This paper | N/A |
| Plasmid: Tir1-IRES-BirA-NeomycinR | This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against N-ter of Yeats2 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for N-ter AID-tagging of Yeats2: eGFP-P2A-1xFlag-BioTag-AID |
This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against N-ter of Tada3 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for N-ter AID-tagging of Tada3: eGFP-P2A-1xFlag-BioTag-AID |
This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against N-ter of Zzz3 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for N-ter AID-tagging of Zzz3: eGFP-P2A-1xFlag-BioTag-AID | This paper | N/A |
| Plasmid: Cas9-mCherry-sgRNA against C-ter of Zzz3 for AID tagging | This paper | N/A |
| Homologous Recombination Plasmid for C-ter AID-tagging of Yeats2: AID-1xFlag-BioTag-P2A-eGFP | This paper | N/A |
| Software and algorithms | ||
| Fiji | Schindelin et al., 2012 | https://imagej.net/software/fiji/downloads |
| cutadapt | Martin, 2011 | https://cutadapt.readthedocs.io/en/v1.10/ |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| htseq-count | Anders et al., 2015 | https://htseq.readthedocs.io/en/master/ |
| DESeq2 | Love et al., 2014 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Enrichr | Chen et al., 2013; Kuleshov et al., 2016; Xie et al., 2021 | https://maayanlab.doud/Enrichr/ |
| RTA | Illumina | https://support.illumina.com/content/dam/illumina-support/documents/downloads/software/hiseq/hcs-hd-v3-4-0-install-notes-1000000028070-00.pdf |
| bcl2fastq | Illumina | https://emea.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Encode ATAC-seq pipeline | ENCODE | https://www.encodeproject.org/atac-seq/ |
| MACS2 | Zhang et al., 2008 | https://hbctraining.github.io/Intro-to-ChIPseq/lessons/05_peak_calling_macs.html |
| Homer v4.11.1 | Heinz et al., 2010 | http://homer.ucsd.edu/homer/ |
| Bedtools | Quinlan and Hall, 2010 | https://github.com/arq5x/bedtools2 |
| Enrichr | Kuleshov et al., 2016 | https://maayanlab.doud/Enrichr/ |
| WebGestalt GEne SeT Analysis Toolkit | Wang et al., 2013 | http://www.webgestalt.org/ |
| chromHMM datasets | Pintacuda et al., 2017 | https://github.com/guifengwei/ChromHMM_mESC_mm10 |
| ATACseqQC | Ou et al., 2018 | https://www.bioconductor.org/packages/release/bioc/html/ATACseqQC.html |
| Proteome Discoverer | ThermoFisher Scientific | https://www.thermofisher.com/fr/fr/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/multi-omics-data-analysis/proteome-discoverer-software.html |
| FlowJo™ | Ashland; Becton, Dickinson and Company | https://www.fiowjo.com/solutions/fiowjo/downloads |