Abstract
Bcl-w, a member of the Bcl-2 family, is highly expressed in various solid tumor, including lung cancer, suggesting that it is involved in cancer cell survival and carcinogenesis. Solid cancer-induced hypoxia has been reported to increase angiogenesis, growth factor, gene instability, invasion, and metastasis. Despite many studies on the treatment of non-small cell lung cancer (NSCLC) with a high incidence rate, the survival rate of patients has not improved because the cancer cells acquired resistance to treatment. This study investigated the correlation between Bcl-w expression and hypoxia in tumor malignancy of NSCLC. Meanwhile, microRNAs (miRNAs) are involved in a variety of key signaling mechanisms associated with hypoxia. Therefore, we discovered miR-519d-3p, which inhibits the expression of Bcl-w and hypoxia-inducing factor (HIF)-1α, and found that it reduces hypoxia-induced tumorigenesis. Spearman’s correlation analysis showed that the expression levels of miR-519d-3p and Bcl-w/HIF-1α were negatively correlated, respectively. This showed that miR-519d-3p can be used as a diagnostic biomarker and target therapy for NSCLC.
Keywords: Bcl-w, hypoxia, HIF-1α, NSCLC, miR-519d-3p, solid tumor
Graphical abstract
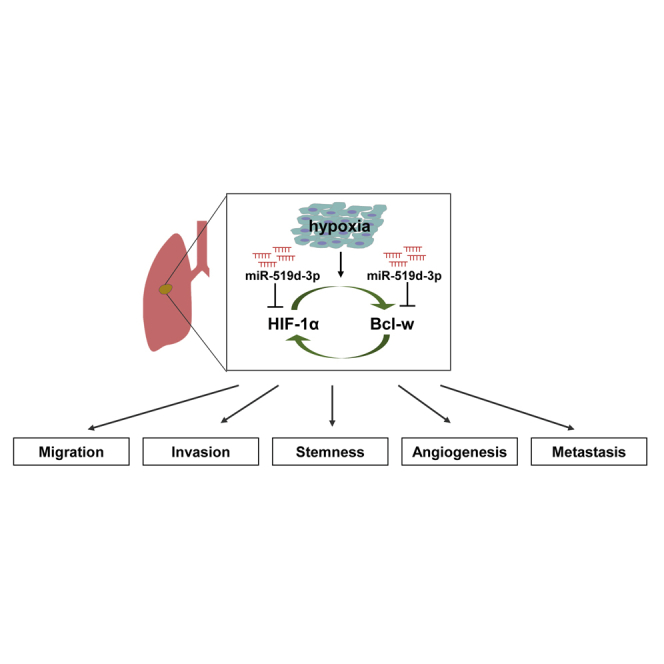
This study investigated the correlation between Bcl-w overexpressed in solid cancer NSCLC and HIF-1α, a hypoxia-related factor, and discovered miR-519d-3p, which inhibited these two factors to suppress hypoxia-induced malignancy. These results showed that miR-519d-3p could be used as a biomarker of diagnosis and targeted therapy for NSCLC.
Introduction
Lung cancer is one of the four most prevalent cancers in the world. About 85% of patients with lung cancer have non-small cell lung cancer (NSCLC), which has a 5-year overall survival rate of <18%.1,2 Despite many studies on the treatment of patients with NSCLC, their overall survival rate has not shown a noticeable increase in the past few years because of metastasis of lung tumor cells.1,3, 4, 5 Therefore, it is imperative to understand the malignant mechanism of NSCLC.
Hypoxia is a common phenomenon in human solid tumors, which are generated by the abnormal proliferation of cancer cells.6, 7, 8 Hypoxia-inducing factor (HIF)-1α is a transcription factor that regulates cellular responses to hypoxia9 and is overexpressed in breast, brain, pharyngeal, cervical, and ovarian cancers.10,11 Increased HIF-1α plays a critical role in cancer malignancy by inducing epithelial-mesenchymal transition (EMT), invasiveness and migration, and angiogenesis as well as metastasis,12,13 eventually leading to a poor prognosis. Therefore, hypoxia in NSCLC is an important factor contributing to patient resistance to treatment and poor survival, suggesting that hypoxia is attractive as a target in the treatment of lung cancer.14, 15, 16
The oncogene Bcl-w, a member of the anti-apoptotic Bcl-2 family,17 is reported to be upregulated in several cancers, including glioblastoma multiforme18, 19, 20 and breast,21 gastric,22 and colorectal23 cancers, leading to tumor progression. It is unclear, however, whether hypoxia and Bcl-w are involved in the mechanism of solid tumor growth and metastasis.
MicroRNAs (miRNAs) are small non-coding RNAs composed of ~22 nucleotides that inhibit target gene expression by binding to the 3′ untranslated region (3′ UTR) of the targeted mRNA.24,25 miRNAs are involved in various cellular processes, including cell proliferation, differentiation, survival, metabolism, inflammation, and angiogenesis.9,26 In particular, it has been reported that dysregulated miRNAs promote tumor malignancy by regulating carcinogenic pathways.26 This study investigated the roles of Bcl-w and HIF-1α in the malignant action of NSCLC. As a result, we describe that the positive feedback loop of Bcl-w and HIF-1α promoted tumorigenicity and metastasis in lung cancer, in vitro and in vivo. It was found that miR-519d-3p, which inhibits Bcl-w and HIF-1α, showed clinical applicability by revealing the mechanism of suppressing cancer malignancy. Recently, in the development of biomarkers for cancer diagnosis and treatment, the focus is on the use of circulating miRNA in the blood, one of the liquid biopsies.27 This study suggested the potential of miR-519d-3p as a target for diagnosis and treatment of lung cancer patients.
Results
High expression of Bcl-w in lung cancer is associated with hypoxia
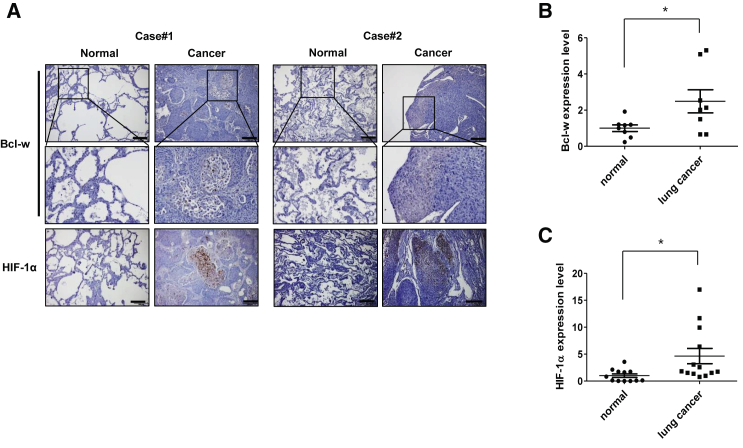
Hypoxia in solid tumor is a very important factor in cancer prognosis and is known to contribute to malignancy by promoting tumor growth, EMT, migration, invasion, and metastasis by increasing HIF-1α expression.12,28,29 We previously reported that Bcl-w, which is highly expressed in solid tumors, including breast cancer21 and glioblastoma multiforme,18, 19, 20 is associated with tumor aggressiveness. To investigate the relationship between Bcl-w and hypoxia in lung cancer, the expression of Bcl-w and HIF-1α was measured in tissue and plasma samples of lung cancer patients. Immunohistochemistry (IHC) staining showed higher expression of Bcl-w and HIF-1α in tumors than in adjacent normal lung tissue (Figure 1A). Bcl-w and HIF-1α mRNA expression was higher in plasma samples from the lung cancer patients than from normal individuals (Figures 1B and 1C). Overall, these results suggested that Bcl-w is associated with hypoxia in lung cancer.
Figure 1.
The level of Bcl-w and HIF-1α expression increases in tissues and plasma of lung cancer patients
(A) Expression of Bcl-w and HIF-1α was confirmed by immunohistochemistry (IHC) staining in tissues of lung cancer patients (scale bars, 200 μm). (B) Bcl-w mRNA levels in the plasma of lung cancer patients were detected by quantitative real-time PCR: normal (n = 8) versus lung cancer (n = 8). (C) Levels of HIF-1α mRNA in the plasma of lung cancer patients were detected by quantitative real-time PCR: normal (n = 12) versus lung cancer (n = 13). The data are presented as mean ± SD. ∗p < 0.05. Student’s t test.
Hypoxic condition in lung cancer induces Bcl-w and HIF-1α expression
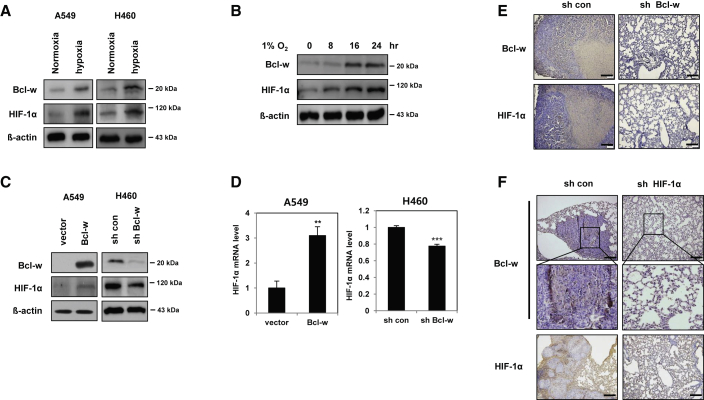
The expression of both of Bcl-w and HIF-1α in A549 and H460 cells was increased in the hypoxic condition induced by 1% O2 (Figure 2A). Bcl-w and HIF-1α expressions were time-dependently increased after treatment with hypoxic condition (1% O2) in H460 cells (Figure 2B). In addition, the expressions of HIF-1α protein and mRNA were upregulated in Bcl-w-overexpressing A549 cells but downregulated in Bcl-w-knockdown H460 cells (Figures 2C and 2D). Moreover, the expression of Bcl-w protein was downregulated in HIF-1α−knockdown H460 cells (Figure S1A). To confirm the above data at the cellular level in experimental animal tissues, H460 cells in which Bcl-w or HIF-1α was knocked down were injected into the tail veins of mice, and the mice were sacrificed 8 weeks later (Figure 2C; Figures S1A and S1B). First, to identify hypoxic regions in the pulmonary tissues of a mouse xenograft model, IHC staining was performed with carbonic anhydrase 9 (CA9) and glucose transporter 1 (GLUT1), known as hypoxia markers.30,31 As a result, both hypoxia markers were highly expressed at the tumor region in pulmonary tissues (Figure S2). In addition, it was confirmed that both Bcl-w and HIF-1α were highly expressed in the region of tumor with high hypoxia marker expression (Figures 2E and 2F). From these two results, it can be suggested that Bcl-w and HIF-1α, which are highly expressed in the hypoxic region of pulmonary tumor, are closely related to each other.
Figure 2.
The expression of Bcl-w and HIF-1α is increased in hypoxic condition
(A) After A549 and H460 cells were incubated for overnight in the hypoxic condition (1% O2), expression of Bcl-w and HIF-1α was determined by western blot analysis. (B) After time-dependent treatment with hypoxic condition (1% O2) in H460 cells, indicated cells were subjected to western blot analysis. β-actin was used for normalization in western blot analysis. (C) A549 and H460 cells were transfected with Bcl-w or shBcl-w, respectively. Bcl-w and HIF-1α expression levels were determined by western blot analysis. (D) HIF-1α mRNA level was quantified in Bcl-w-overexpressing or -knockdown cells by quantitative real-time PCR. (E and F) The pulmonary metastatic tissues were collected 8 weeks after shBcl-w (E) or shHIF-1α (F) H460 cells were injected into the tail vein of mice. The expression of Bcl-w and HIF-1α was detected by immunohistochemistry (IHC) staining (scale bars, 200 μm). The data are presented as mean ± SD. ∗∗p < 0.01; ∗∗∗p < 0.001. Student`s t test.
Bcl-w enhances epithelial-mesenchymal transition, migration, invasion, stemness, and metastasis in lung cancer
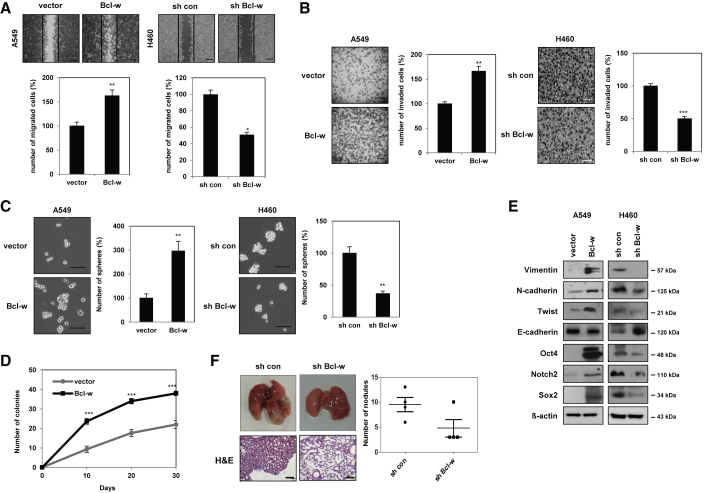
Assessments of the basal levels of expression of Bcl-w in A549 and H460 NSCLC cell lines showed that Bcl-w expression was higher in H460 cells than in A549 cells (Figure S3). Thus, the role of Bcl-w in lung cancer was investigated in Bcl-w-overexpressing A549 cells and Bcl-w-knockdown H460 cells (Figure 2C). The migratory and invasive abilities were increased in Bcl-w-overexpressing A549 cells but inhibited in Bcl-w-knockdown H460 cells (Figures 3A and 3B). Because cancer stem-like cells are one of the major causes of tumor formation,32,33 their presence was assessed by sphere formation and anchorage-independent cell growth assays. In addition, the effect of Bcl-w on the maintenance of stemness was assessed by western blot analysis of the expression of cancer stem-like cell markers. Overexpression of Bcl-w increased the number of spheres, anchorage-independent cell growth, and the level of cancer stem-like cell markers compared with the negative control group. Conversely, knockdown of Bcl-w reduced sphere formation ability, spheroid formation in three-dimensional culture, and expression of stemness-related factors such as Oct4, Notch2, and Sox2 (Figures 3C–3E). Additionally, to investigate the effect of Bcl-w on mesenchymal traits in lung cancer, the expression of EMT-related markers was confirmed by western blot analysis and immunofluorescence (IF) staining. Overexpression of Bcl-w increased mesenchymal makers, including Vimentin, N-cadherin, and Twist, and decreased epithelial markers, such as E-cadherin (Figure 3E; Figures S4A and S4B). Conversely, knockdown of Bcl-w reduced the expression of mesenchymal markers (Figure 3E). Angiogenesis, which creates new blood vessels in cancer tissue, is important for cancer progression.34 To assess the effect of Bcl-w on angiogenesis, tube formation analysis was performed in human umbilical vein endothelial cells (HUVECs). Overexpression of Bcl-w increased the ability to form tubes, whereas knockout of Bcl-w decreased this ability in HUVECs (Figure S5A). The levels of expression of the angiogenesis markers angiopoietin-2 (Ang2) and vascular endothelial growth factor (VEGF)35 were upregulated in Bcl-w-overexpressing A549 cells but decreased in Bcl-w-knockdown H460 cells (Figure S5B). These results showed that Bcl-w enhanced lung cancer cell migration, invasion, stemness, mesenchymal traits, and angiogenesis, consistent with our previous results showing that Bcl-w, which is highly expressed in various cancers, is involved in tumorigenesis.18, 19, 20, 21, 22,36 To verify the tumorigenic actions of Bcl-w, we used BEAS-2B cells, which were overexpressed with Bcl-w. Bcl-w-overexpressed BEAS-2B cells increased the properties of EMT, migration, invasion, angiogenesis, and stemness by western blot analysis, migration, invasion, and sphere formation assays (Figures S6A–S6D). These findings confirmed that Bcl-w promoted the cell migration, invasion, EMT, and sphere formation ability in normal BEAS-2B cells as well as in lung cancer cells.
Figure 3.
Bcl-w promotes EMT, migration, invasion, stemness maintenance, and metastasis in vitro and in vivo
(A and B) Indicated cells were analyzed for wound healing assay (A) (scale bars, 200 μm) and invasion assay with Matrigel-coated Transwells (B) (scale bars, 100 μm). (C) The stemness in indicated cells was shown through sphere forming assay (scale bars, 100 μm). (D) The stemness of Bcl-w was detected by anchorage-independent cell growth assay in A549 cells. (E) The level of indicated proteins was detected by western blot analysis in indicated cells. β-actin was used for normalization in western blot analysis. (F) Bcl-w-knockdown H460 cells were injected into the tail vein in BALB/c nude mice (n = 4, 1 × 106 cells/mouse). After 8 weeks, lung was harvested and subjected to H&E staining and metastatic nodule counting in lung tissue. The data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Student`s t test.
Next, to confirm the effects of Bcl-w on the metastatic mechanism of lung cancer, Bcl-w-knockdown cells were injected into the tail veins of nude mice and the number of nodules was counted. It was confirmed that Bcl-w short hairpin RNA (shRNA) significantly reduced lung metastatic nodule formation compared with the control (Figure 3F), indicating that Bcl-w is a potent factor involved in the mechanism of lung metastasis.
A positive-feedback loop between Bcl-w and HIF-1α induces lung cancer malignancy
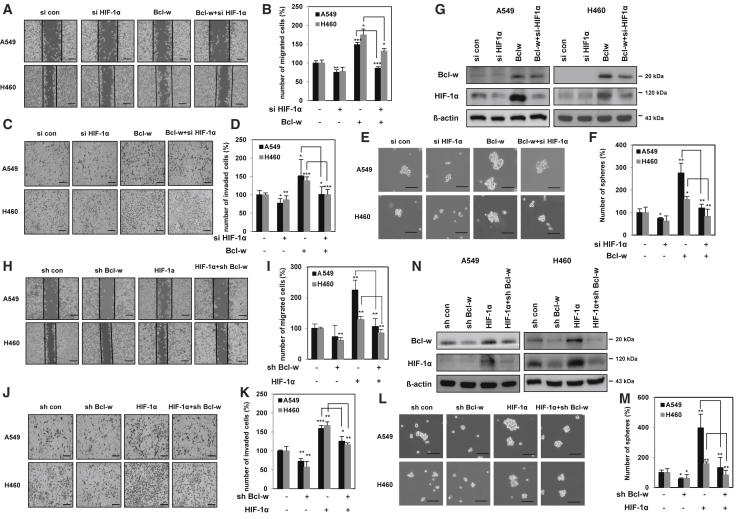
To investigate the relationship between Bcl-w and HIF-1α in NSCLC, Bcl-w-overexpressing A549 and H460 cells were transfected with HIF-1α or negative control small interfering RNA (siRNA). Wound healing, invasion, sphere formation, and western blot analyses were performed to investigate the signaling mechanisms by which both Bcl-w and HIF-1α induced the tumorigenic phenotype. Knockdown of HIF-1α reduced Bcl-w-induced cancer cell motility, invasiveness, stemness, the expression of oncogenic factors such as Vimentin, N-cadherin, Notch2, Sox2, and Oct4 (Figures 4A–4F; Figure S7A), and HIF-1α expression (Figure 4G). On the other hand, knockdown of Bcl-w suppressed HIF-1α-induced migratory and invasive abilities, stemness maintenance, and the expression of these phenotype-related factors and Bcl-w (Figures 4H–4N; Figure S7B) in A549 and H460 cells. These findings suggested that Bcl-w and HIF-1α promoted pulmonary malignancy by positively increasing the expression of each other.
Figure 4.
The positive-feedback loop between Bcl-w and HIF-1α leads to lung cancer malignancy
(A–G) After A549 and H460 cells were transfected with Bcl-w-overexpressing vector or siRNA against HIF-1α (siHIF-1α), indicated cells were analyzed by wound healing assay (A and B) (scale bars, 200 μm), invasion assay using Matrigel-coated Transwells (C and D) (scale bars, 100 μm), sphere forming assay (E and F) (scale bars, 100 μm), and western blot analysis (G). (H–N) After A549 and H460 cells were transfected with HIF-1α-overexpressing vector or shRNA against Bcl-w (shBcl-w), indicated cells were analyzed by wound healing assay (H and I) (scale bars, 200 μm), invasion assay using Matrigel-coated Transwells (J and K) (scale bars, 100 μm), sphere forming assay (L and M) (scale bars, 100 μm), and western blot analysis (N). β-actin was used for normalization in western blot analysis. The data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Student`s t test.
Meanwhile, HIF-1α is known to regulate the expression of several genes as a transcription factor. Therefore, it was investigated whether HIF-1α as a transcription factor affects the transcriptional regulation of Bcl-w. Since CoCl2 induces artificial hypoxia in cells, prevents HIF-1α degradation, and induces its stabilization,37,38 chromatin immunoprecipitation (ChIP) assay was performed after treatment of H460 cells with CoCl2. The chromatin fraction was pulled down with HIF-1α antibody, and a PCR fragment corresponding to the predicted HRE site was detected in the Bcl-w promoter (Figure S8). These results suggested that HIF-1α increased the expression of Bcl-w by acting as a transcription factor of Bcl-w.
A miR-519d-3p that directly inhibits Bcl-w and HIF-1α expression was identified
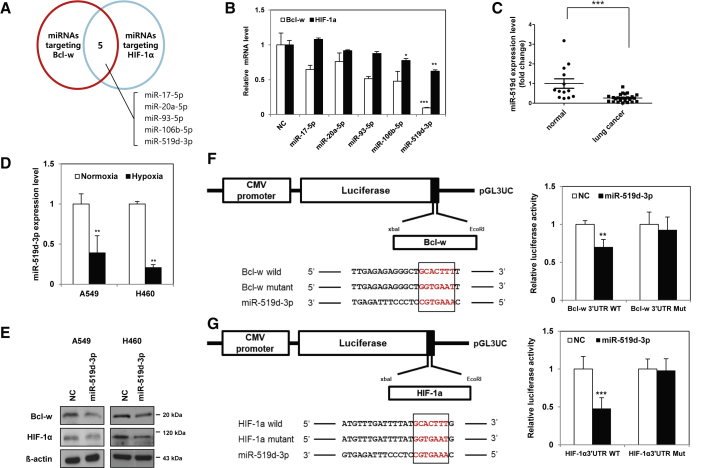
miRNAs inhibit the expression of target proteins by suppressing the translation of mRNAs.39 Therefore, we used miRNAs as a tool to suppress the malignancy of NSCLC, and for more effective control, we tried to discover miRNAs that simultaneously downregulate the expression of both Bcl-w and HIF-1α. Two independent target prediction sites, TargetScan40 and miRDB,41,42 were used to identify miRNAs that inhibit the expression of Bcl-w and HIF-1α (Figure 5A). Five candidate miRNAs were selected with two databases. miR-519d-3p suppressed the expression of the target mRNAs, Bcl-w and HIF-1α, more effectively than other candidate miRNAs (Figure 5B). In addition, as a result of searching The Cancer Genome Atlas (TCGA) dataset and comparing the expression levels of five candidate miRNAs in lung cancer patients, only miR-519d-3p was less expressed in lung cancer patient samples than in the normal group (Figures S9A–S9E). As a result of verifying of public database results using patient samples, the expression of miR-519d-3p was found to be lower in plasma of lung cancer patients than in that of the normal control group (Figure 5C). As a result of testing the correlation with hypoxia induced in NSCLC, the expression of miR-519d-3p decreased in hypoxia condition (Figure 5D).
Figure 5.
MiR-519d-3p directly targets Bcl-w and HIF-1α
(A) Using target prediction sites TargetScan and miRDB, candidate miRNAs that simultaneously regulate the expression of Bcl-w and HIF-1α are shown in a Venn diagram. (B) To select 1 of 5 candidate miRNAs, the mRNA expression levels of Bcl-w and HIF-1α were compared after each miRNA overexpression by quantitative real-time PCR. (C) Clinical association of miR-519d-3p expression in lung cancer patients was analyzed in plasma: normal (n = 13) versus lung cancer (n = 21). (D) After A549 and H460 cells were incubated overnight in hypoxia (1% O2), the expression of miR-519d-3p was measured by quantitative real-time PCR. (E) The protein expression levels of Bcl-w and HIF-1α predicted as targets of miR-519d-3p were confirmed by western blot analysis. β-actin was used for normalization in western blot analysis. (F) After co-transfection with either wild-type (WT) or mutant type (MT) vector of Bcl-w 3′ UTR in the presence or absence of miR-519d-3p mimic, luciferase activities were examined in H460 cells. (G) After co-transfection with either WT or MT vector of HIF-1α 3′ UTR in the presence or absence of miR-519d-3p mimic, luciferase activities were measured. The data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Student`s t test.
Bcl-w and HIF-1α were confirmed to be direct targets of miR-519d-3p by measuring protein expression levels and luciferase activity. Protein levels of Bcl-w and HIF-1α were significantly reduced in A549 or H460 cells transfected with miR-519d-3p compared with the negative control. (Figure 5E). Luciferase activity was decreased in H460 cells transfected with miR-519d-3p and wild-type Bcl-w or HIF-1α, whereas activity in cells transfected with miR-519d-3p and mutant Bcl-w or HIF-1α was unchanged (Figures 5F and 5G). These results confirmed that Bcl-w and HIF-1α are direct targets of miR-519d-3p.
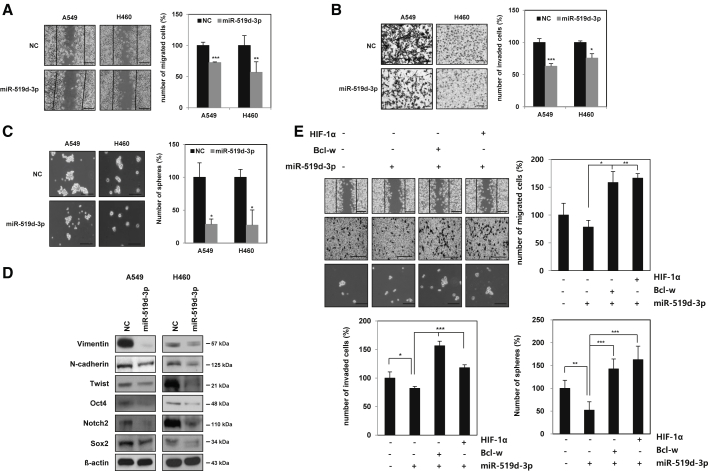
MiR-519d-3p reduces mesenchymal traits, mobility, invasiveness, and maintenance of stemness by decreasing Bcl-w and HIF-1α expression
To investigate the role of miR-519d-3p in the mechanism of malignancy in lung cancer cells, a miR-519d-3p mimic was overexpressed in A549 and H460 cells. Overexpression of miR-519d-3p inhibited migratory, invasive, and sphere formation activities and the expression of mesenchymal and cancer stem-like cell marker proteins (Figures 6A–6D). In particular, the expression of invasion-related matrix metalloproteinases MMP-2 and MMP-9 was analyzed by using western blotting to identify the enzymes involved in miR-519d-3p reducing invasiveness (Figure 6B). Since MMP-2 and MMP-9 are enzymes secreted to the outside of the cell, their expressions were observed in the conditioning medium (CM) of A549 and H460 cells overexpressing miR-519d-3p, respectively. As a result, miR-519d-3p decreased the expression of MMP-2 and MMP-9 (Figure S10). Taken together, it was confirmed that miR-519d-3p was found to reduce invasiveness by reducing the expression of MMP-2 and MMP-9.
Figure 6.
MiR-519d-3p represses EMT, mobility, invasiveness, and stemness maintenance by suppressing Bcl-w or HIF-1α
A549 and H460 cells were transfected with either negative control or miR-519d-3p mimic. (A–C) Transfected cells were analyzed by wound healing assay (A) (scale bars, 100 μm), invasion assay with Matrigel-coated Transwells (B) (scale bars, 100 μm), and sphere forming assay (C) (scale bars, 100 μm). (D) Mesenchymal and cancer stem-like cell marker proteins were identified by western blot analysis. β-actin was used for normalization in western blot analysis. (E) H460 cells were co-transfected with miR-519d-3p or negative control along with Bcl-w or HIF-1α vector. Transfected cells were determined by wound healing assay (scale bars, 100 μm), invasion assay with Matrigel-coated Transwells (scale bars, 100 μm), and sphere forming assay (scale bars, 100 μm). The data are presented as mean ± SD. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. Student`s t test.
The relationship between miR-519d-3p and targeted Bcl-w or HIF-1α on the malignant phenotype of lung cancer cells was investigated using miR-519d-3p mimic and Bcl-w- or HIF-1α-overexpressing vector. Cancer cell motility, invasiveness, and sphere-forming ability reduced by miR-519d-3p were increased by overexpression of Bcl-w or HIF-1α (Figure 6E). In contrast, cell motility, invasiveness, and sphere-forming ability increased by miR-519d-3p inhibitor were suppressed by Bcl-w or HIF-1α siRNA (Figures S11A–S11D). Overall, it was shown that miR-519d-3p is involved in the mechanism of tumor suppression by directly inhibiting the expression of its target proteins, Bcl-w and HIF-1α.
miR-519d-3p inhibited cancer malignancy in vivo and verified clinical applicability
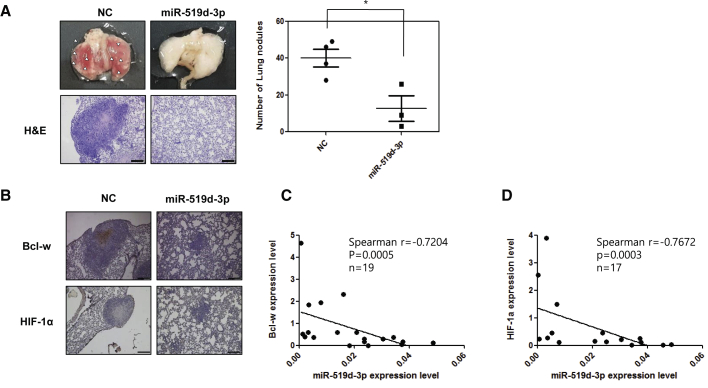
To confirm the efficacy of miR-519d-3p on the metastasis mechanism by the in vivo study, H460 cells overexpressing this miRNA were injected into the tail veins of NOD/SCID mice and the mice were sacrificed 7 weeks later. The number of pulmonary nodules was lower in mice injected with cells overexpressing miR-519d-3p than in mice injected with the control group (Figure 7A, top). Hematoxylin and eosin (H&E) staining of lung tissue confirmed that the sizes of the lung metastatic nodules were reduced in the miR-519d-3p overexpression group compared with the control group (Figure 7A, bottom). IHC staining found that the levels of expression of Bcl-w and HIF-1α were decreased in lung tissue of the miR-519d-3p overexpression group compared with the control group (Figure 7B). These results suggested that metastatic activity was enhanced in tumors with increased Bcl-w expression due to hypoxia. To verify the clinical applicability of these findings, Spearman’s correlation between Bcl-w/HIF-1α and miR-519d-3p in plasma of lung cancer patients was investigated. Negative correlations were observed between Bcl-w and miR-519d-3p and between HIF-1α and miR-519d-3p in plasma of lung cancer patients (Figures 7C and 7D). Because miRNAs circulate in blood vessels,27 miR-519d-3p, whose expression was suppressed in plasma of lung cancer patients, has shown potential as a diagnostic marker for lung cancer.
Figure 7.
miR-519d-3p negatively correlates with its targets Bcl-w and HIF-1α in plasma of lung cancer patients, respectively
Bcl-w and HIF-1α are regulated by miR-519d-3p. (A and B) The miR-519d-3p-overexpressed H460 cells were injected into the tail vein of NOD/SCID mice (n = 4, 1 × 106 cells/mouse), and mice were sacrificed 7 weeks later to obtain lung tissue. (A) The number of metastatic nodules of the obtained lung tissue was counted and graphed. The obtained lung tissue was stained with hematoxylin and eosin (H&E) staining. (B) The expression of Bcl-w and HIF-1α was confirmed by immunohistochemistry (IHC) staining (scale bars, 200 μm). (C and D) The expression patterns of Bcl-w (C) and HIF-1α (D) and miR-519d-3p in plasma of lung cancer patients were analyzed by Spearman’s correlation analysis. The data are presented as mean ± SD. ∗p < 0.05. Student`s t test.
Finally, our study found that a positive feedback loop between Bcl-w and HIF-1α induced by hypoxia promoted EMT, mobility, invasion, stemness, and metastasis. In addition, miR-519d-3p, which suppresses the expression of these two factors, was discovered as a way to overcome this phenomenon, and it was proposed as a factor to suppress the mechanism of cancer malignancy and metastasis induced by hypoxia in lung cancer.
Discussion
Bcl-w is an oncogene highly expressed in various solid tumors, such as glioblastoma multiforme18, 19, 20 and breast,21 gastric,22 and colorectal23 cancers. Solid tumor-derived hypoxia allows cancer cells to adapt to the tumor microenvironment, thereby inducing angiogenesis, growth factor signal transduction, gene instability, tissue invasion, and metastasis, as well as the expression of various genes related to HIF-1α, leading to acquiring tumor malignancy.43, 44, 45
This study investigated the correlation between hypoxia and Bcl-w expression in NSCLC tissues and the effects of these two factors on the mechanism of tumorigenesis. To show that Bcl-w is associated with hypoxia, the expressions of Bcl-w and HIF-1α were measured in tissues and plasma of lung cancer patients, with the results showing that the expression of both factors increased (Figure 1). The association between Bcl-w expression and hypoxia in lung cancer was further verified at the cellular level and in an animal model. First, it was described that the expression of Bcl-w and HIF-1α increased in lung cancer cells, A549 and H460, that were exposed to hypoxia containing 1% oxygen (Figures 2A–2D). The expression of Bcl-w and HIF-1α was suppressed in lung cancer tissues obtained after injection of H460 cells that suppressed Bcl-w or HIF-1α expression into mouse veins (Figures 2E and 2F). These results confirmed the hypothesis of the link between hypoxia and Bcl-w.
HIF-1α has been reported to be involved in the progression of various cancers, including lung cancer,46,47 and increased expression of HIF-1α is associated with poor prognosis.47 Bcl-w is also increased in various cancers, including breast,21 gastric,22 and colorectal23 cancers and glioblastoma multiforme,18, 19, 20 suggesting that Bcl-w contributes to tumorigenicity. Investigation of the relationship between Bcl-w and HIF-1α in malignancy showed that both induced tumorigenic properties and metastasis by increasing the expression of each other via a positive feedback loop (Figures 2, 3, and 4; Figures S2–S8).
The relationship between the expression of the Bcl-2 family and HIF-1α has been revealed in several studies. It has been reported that the expression of HIF-1α is increased and stabilized by the BH4 domain of Bcl-2 to induce the expression of the target gene VEGF,48 thereby increasing angiogenesis in melanoma cells.49 In addition, HIF-1α directly binds to the promoter of Bcl-xL to regulate its expression, and these two factors are positively expressed in various cancers.50,51 Bcl-xL is known to act as a resistance factor in hypoxia-induced apoptosis.50,51 In this study, it was confirmed that HIF-1α functions as a factor that regulates the transcription of Bcl-w (Figure S8). Both HIF-1α and Bcl-w increased each other’s expression by forming a positive feedback loop, contributing to malignancy of NSCLC.
Recent studies have shown that miRNAs are involved in various key signaling mechanisms associated with hypoxia.52 Hypoxia-induced miR-210 has shown involvement in angiogenesis, cell cycle regulation, mitochondrial metabolism, DNA damage repair, and tumor growth, suggesting that this miRNA has excellent therapeutic potential.52, 53, 54 The present study found that miR-519d-3p decreased hypoxia-induced tumorigenesis by inhibiting the expression of Bcl-w and HIF-1α (Figures 5, 6, and 7). miR-519d-3p has been reported to be a tumor suppressor miRNA targeting MMP2, Twist, and X-linked inhibitor of apoptosis protein (XIAP), inhibiting cell proliferation, invasion, and migration of trophoblast cells, ovarian cancer, and gastric cancer,55, 56, 57 supporting our results.
Circulating tumor DNA (ctDNA), non-coding RNA (ncRNAs), miRNAs, proteins, and exosomes are present in body fluids, including blood. Because miRNAs are secreted into the blood and circulate throughout the body, they can be detected in the blood.58 It is known that miRNAs circulate in the blood and act as messengers that communicate between tumor and the surrounding microenvironment.59 Therefore, recently, the potential of miRNA as a biomarker for cancer diagnosis and treatment using liquid biopsy has emerged.27,59
In NSCLC, miRNA is also being developed as a biomarker. As an example, the let-7 family functions as a tumor suppressor that suppresses the expression of RAS and MYC, and low expression in NSCLC patients has a poor prognosis.60 In addition, miR-141 and miR-200c are strongly upregulated in patients with NSCLC and are associated with low overall survival.61 The increased levels of MiR-486 and miR-150 in plasma of NSCLC patients suggest a potential as biomarkers for early diagnosis and prognosis.62
The correlations between plasma expression of Bcl-w/HIF-1α and miR-519d-3p were therefore analyzed in lung cancer patients. Spearman’s correlation analysis disclosed that Bcl-w and HIF-1α each showed a negative correlation with miR-519d-3p (Figures 7C and 7D). Overall, miR-519d-3p in lung cancer has shown potential as a biomarker for cancer diagnosis and targeted treatment.
Materials and methods
Cell culture
H460 and A549 were obtained from the Korea Cell Line Bank (KCLB). HUVECs and BEAS-2B were purchased from CEFO Bio (Seoul, Korea) and American Type Culture Collection (ATCC, Manassas, VA, USA), respectively. H460 and BEAS-2B cells were cultured in RPMI 1640 (Corning, Corning, NY, USA). A549 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning, Corning, NY, USA). Both were supplemented with 10% fetal bovine serum (Genken, Los Alamitos, CA, USA) and penicillin-streptomycin (Corning, Corning, NY, USA). HUVECs were cultured in Endothelial Cell Growth Medium (PromoCell, Heidelberg, Germany). Cells were incubated in 5% CO2 at 37°C. For maintaining the hypoxic condition, cells were incubated in a hypoxia chamber, which was maintained with 1% O2, 5% CO2 and balanced with N2.
Plasmid, miRNA mimic, and transfection
pGL3-UC vector was provided by V. Narry Kim.63 pGL3-UC vector was used for luciferase reporter assay. The wild-type or mutant type fragment of the target gene was introduced into the pGL3-UC vector with xbaI (NEB, Ipswich, MA, USA) and EcoRI (NEB, Ipswich, MA, USA).
Synthetic miRNA mimics or inhibitors were synthesized by Genolution (Seoul, Korea). RNA duplexes were designed from the sequences of miR-519d-3p (5′-CAAAGUGCCUCCCUUUAGAGUG-3′), miR-17-5p (5′-CAAAGUGCUUACAGUGCAGGUAG-3′), miR-20a-5p (5′-UAAAGUGCUUAUAGUGCAGGUAG-3′), miR-93-5p (5′-CAAAGUGCUGUUCGUGCAGGUAG-3′), and miR-106b-5p (5′-UAAAGUGCUGACAGUGCAGAU-3′). All siRNAs and shRNA against Bcl-w were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). shRNA against HIF-1α was obtained from Sigma-Aldrich (St. Louis, MO, USA). pcDNA-3.1 vector containing Bcl-w or HIF-1α was used for production of overexpressing cells. Plasmid, miRNA, siRNA, and shRNA were transfected into cells with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), following the manufacturer’s instructions. After transfection for 24 or 48 h, the expression of Bcl-w, HIF-1α, and miR-519d-3p was confirmed by quantitative real-time PCR and western blot analysis, respectively.
Total RNA isolation and quantitative real-time PCR
Total RNA was isolated in cell lysate with TRIzol reagent (Favorgen Biotech, Taiwan). cDNA synthesis was performed with the SensiFAST cDNA Synthesis Kit (Bioline, London, UK) and the Mir-X miRNA first-strand synthesis kit (Clontech Laboratories, Palo Alto, CA, USA) and amplified with the SimpliAmp thermal cycler (Applied Biosystems, Foster City, CA, USA). Quantitative real-time PCR was performed in a LightCycler 96 (Roche, Basel, Switzerland) with Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The use of the kit in this experiment followed the manufacturer’s instructions.
Primer sequences were as follows: 5′-TGCTCATCAGTTGCCACTTC-3′ (forward) and 5′-TCCTCACACGCAAATAGCTG-3′ (reverse) for HIF-1α; 5′-GGACAAGTGCAGGAGTGGAT-3′ (forward) and 5′-GTCCTCACTGATGCCCAGTT-3′ (reverse) for Bcl-w;64 5′-CATCTCTGCCCCCTCTGCTGA-3′ (forward) and 5′-GGATGACCTTGCCCACAGCCT-3′ (reverse) for GAPDH.
Luciferase reporter assay
H460 cells were seeded in 24-well plates (2.5 × 104 cells/well) and were co-transfected with reporter plasmid (200 ng), pRL-CMV-Renilla plasmid (2 ng), and miR-519d-3p with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). After 48 h, luciferase activity was determined with a dual-luciferase reporter assay system kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Firefly luciferase was normalized with Renilla. All experiments were performed in triplicate.
Western blot analysis
Primary antibodies were Bcl-w (Abcam, 1:1,000 dilution), HIF-1α (Novus, 1:500 dilution), Vimentin (Cell Signaling, 1:1,000 dilution), N-cadherin (Abcam, 1:500 dilution), Twist (Abcam, 1:500 dilution), E-cadherin (BD Biosciences, 1:1,000 dilution), Oct4 (Abcam, 1:1,000 dilution), Notch2 (Cell Signaling, 1:1,000 dilution), Sox2 (Santa Cruz Biotechnology, 1:1,000 dilution), Ang2 (Santa Cruz Biotechnology, 1:500 dilution), VEGF (Abcam, 1:500 dilution), and β-actin (Santa Cruz Biotechnology, 1:2,000 dilution). Protein was extracted with lysis buffer (10 mM Tris-HCl with pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 0.1% SDS) containing protease and phosphatase inhibitors (Roche, Basel, Switzerland) and quantified by Bradford assay (Bio-Rad, Hercules, CA, USA). Quantified protein was separated by SDS-PAGE and transfer to polyvinylidene fluoride (PVDF) membrane (Millipore, Burlington, MA, USA). The membrane was blocked with 5% skim milk for 30 min. Primary antibodies were incubated for overnight at 4°C, and secondary antibodies were incubated for 1 h at room temperature. All procedures were washed with 0.1% TBS-T (Tris Buffered Saline with Tween 20). Samples were normalized with β-actin.
Transwell invasion and wound healing assay
Invasion assays used Transwell chambers with an 8 μm pore (Corning, Corning, NY, USA). The Transwell chamber was coated with Matrigel (BD Biosciences, San Jose, CA, USA). Cells (2.5 × 104 cells/well), which were suspended in a medium containing 0.1% BSA, were placed in the upper Transwell chamber. Complete medium was added to the lower chamber. After incubation for 16 h, the cells that migrated to the lower surface of the filter were fixed with methanol and stained with crystal violet. The stained cells were counted under a microscope (Olympus, Tokyo, Japan). For the wound healing analysis, the cells were seeded in a 6-well plate so as to be 95%–100% confluent. After incubation of the scratched cells for 16–24 h, cells that have moved to the scratched space are counted under a microscope (Olympus, Tokyo, Japan).
Sphere forming assay
The indicated cells were resuspended in DMEM-F12 (Gibco, Waltham, MA, USA) containing B27 (Gibco, Waltham, MA, USA) and grown for 7–10 days. Spheres with a diameter > 20 μm were counted under an inverted microscope (Olympus, Tokyo, Japan).
Immunofluorescence staining
A549 cells, which were seeded on glass coverslips, were fixed with 4% paraformaldehyde solution, permeabilized with 0.1% Triton X-100, and blocked with 1% BSA. All procedures were washed with PBS. The cells were incubated with antibody against E-cadherin (BD Biosciences, 1:500 dilution), Bcl-w (Santa Cruz, 1:250 dilution), and Vimentin (Thermo Scientific, 1:500) at 4°C overnight. The nuclei were stained with DAPI in mounting solution (Vector Laboratories, Burlingame, CA, USA). The images were determined with laser-scanning confocal microscope LSM710 (Zeiss, Oberkochen, Germany).
Chromatin immunoprecipitation assay
CoCl2-treated cells were subjected to a cross-linking reaction by addition of formaldehyde/glycine. After washing with PBS and lysing of the harvested cells, they were subjected to sonication. The supernatant was added with HIF-1α antibody (Bethyl Laboratories, Montgomery, TX, USA) or normal immunoglobulin G (IgG) (Santa Cruz Biotechnology, Dallas, TX, USA) and Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Dallas, TX, USA). The reaction product is washed with low NaCl, high NaCl, and LiCl buffer, and then NaHCO3 and SDS are added to elute the complex. To obtain free DNA, reverse cross-linking of the protein-DNA complex is performed by adding NaCl, RNase, and proteinase K to the complex. DNA fragment is used as a PCR template. Primer for Bcl-w detection: (forward) 5′-GCCCCGAGATGTCCCGAAGTCC-3′, (reverse) 5′-GGCCTGCAAAGGGCCACTACTCAG-3′.
Animal experiments
Six-week-old female BALB/c nude mice were used for the mouse xenograft experiments. Human lung cancer cells, H460 cells, with shBcl-w or shHIF-1α (1 × 106 cells/mouse) were subjected to intravenous injection. After 8 weeks, mice were sacrificed and pulmonary nodules were counted. Lung tissues were formalin fixed and paraffin embedded for H&E and IHC.
Another experiment used 6-week-old female NOD/SCID mice. H460 cells with negative control or miR-519d-3p mimic (1 × 106 cells/mouse) were injected into the tail vein. After 7 weeks, mice were sacrificed and pulmonary nodules were counted. Lung tissues were formalin fixed and paraffin embedded for H&E and IHC. These studies were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the Korea Institute of Radiological and Medical Sciences (KIRAMS).
Hematoxylin and eosin and immunohistochemistry staining
Lung tissues were fixed in 4% formaldehyde and embedded in paraffin. The paraffin section was deparaffinized with xylene and rehydrated with ethanol. For H&E, the slides were stained with H&E (Thermo Scientific, Waltham, MA, USA). In IHC staining, after antigen retrieval, the sections were blocked for endogenous peroxidase activity with 3% H2O2. Bcl-w (Abcam, 1:500 dilution) and HIF-1α (Novus, 1:250 dilution) were incubated overnight at 4°C. After washing with TBS-T (TBS with Tween 20), Vectastain ABC kit and DAB (3,3′-diaminobenzidine; Vector Laboratories, Burlingame, CA, USA) were used according to manufacturer`s protocols. The signal was detected with cellSens (Olympus, Tokyo, Japan).
Clinical specimens
Human specimens were provided from the Radiation Tissue Resources Bank of Korea Cancer Center Hospital and the KIRAMS Radiation Biobank (KRB). Lung cancer tissues and adjacent lung tissues were obtained from 30 patients, respectively. Plasma was provided in 20 samples from normal and lung cancer patients, respectively. All samples used in this experiment have Institutional Review Board (IRB) approval (K-1608-002-048) in KIRAMS.
Statistical analysis
GraphPad software was used for all data analysis. All data are expressed as mean ± SD. Statistical calculations were performed with Student’s t test. The Spearman correlation coefficient was used to verify the relationships between Bcl-w or HIF-1α and miR-519d-3p in plasma of lung cancer patients.
Acknowledgments
This study was supported by grants of the National Research Foundation of Korea and the Korea Institute of Radiological and Medical Sciences (KIRAMS), funded by Ministry of Science and ICT (MSIT) in the Republic of Korea (Nos. NRF-2021R1A2C2005966(50698-2021), NRF-2017M2A2A7A01018542(50035-2019), and 50531-2021).
Author contributions
I.H.B. supervised the work; J.Y.C. performed research and analyzed data; I.H.B. and J.Y.C. designed the experiments and drafted the manuscript; I.H.B., J.Y.C., H.J.S., R.-K.K., M.Y.C., and S.-J.L. designed and performed animal experiments. All authors discussed the results and commented on the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2021.06.015.
Supplemental information
References
- 1.Ceppi P., Mudduluru G., Kumarswamy R., Rapa I., Scagliotti G.V., Papotti M., Allgayer H. Loss of miR-200c expression induces an aggressive, invasive, and chemoresistant phenotype in non-small cell lung cancer. Mol. Cancer Res. 2010;8:1207–1216. doi: 10.1158/1541-7786.MCR-10-0052. [DOI] [PubMed] [Google Scholar]
- 2.Zappa C., Mousa S.A. Non-small cell lung cancer: current treatment and future advances. Transl. Lung Cancer Res. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coello M.C., Luketich J.D., Litle V.R., Godfrey T.E. Prognostic significance of micrometastasis in non-small-cell lung cancer. Clin. Lung Cancer. 2004;5:214–225. doi: 10.3816/CLC.2004.n.002. [DOI] [PubMed] [Google Scholar]
- 4.Herbst R.S., Heymach J.V., Lippman S.M. Lung cancer. N. Engl. J. Med. 2008;359:1367–1380. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei L., Song X.-R., Sun J.-J., Wang X.-W., Xie L., Lv L.-Y. Lysyl oxidase may play a critical role in hypoxia-induced NSCLC cells invasion and migration. Cancer Biother. Radiopharm. 2012;27:672–677. doi: 10.1089/cbr.2012.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaGory E.L., Giaccia A.J. The ever-expanding role of HIF in tumour and stromal biology. Nat. Cell Biol. 2016;18:356–365. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaupel P., Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 8.Schito L., Semenza G.L. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 9.Huang X., Ding L., Bennewith K.L., Tong R.T., Welford S.M., Ang K.K., Story M., Le Q.T., Giaccia A.J. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu X., Kang Y. Hypoxia and hypoxia-inducible factors: master regulators of metastasis. Clin. Cancer Res. 2010;16:5928–5935. doi: 10.1158/1078-0432.CCR-10-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Semenza G.L. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 12.Yang M.-H., Wu M.-Z., Chiou S.-H., Chen P.-M., Chang S.-Y., Liu C.-J., Teng S.C., Wu K.J. Direct regulation of TWIST by HIF-1α promotes metastasis. Nat. Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 13.Hsu Y.L., Hung J.Y., Chang W.A., Lin Y.S., Pan Y.C., Tsai P.H., Wu C.Y., Kuo P.L. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36:4929–4942. doi: 10.1038/onc.2017.105. [DOI] [PubMed] [Google Scholar]
- 14.Ren W., Mi D., Yang K., Cao N., Tian J., Li Z., Ma B. The expression of hypoxia-inducible factor-1α and its clinical significance in lung cancer: a systematic review and meta-analysis. Swiss Med. Wkly. 2013;143:w13855. doi: 10.4414/smw.2013.13855. [DOI] [PubMed] [Google Scholar]
- 15.Salem A., Asselin M.-C., Reymen B., Jackson A., Lambin P., West C.M.L., O’Connor J.P.B., Faivre-Finn C. Targeting hypoxia to improve non–small cell lung cancer outcome. J. Natl. Cancer Inst. 2018;110:14–30. doi: 10.1093/jnci/djx160. [DOI] [PubMed] [Google Scholar]
- 16.Wilson W.R., Hay M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer. 2011;11:393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 17.Lin C.J.-F., Gong H.-Y., Tseng H.-C., Wang W.-L., Wu J.-L. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 18.Kim S., Choi J.Y., Seok H.J., Park M.-J., Chung H.Y., Bae I.H. miR-340-5p suppresses aggressiveness in glioblastoma multiforme by targeting Bcl-w and Sox2. Mol. Ther. Nucleic Acids. 2019;17:245–255. doi: 10.1016/j.omtn.2019.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee W.S., Woo E.Y., Kwon J., Park M.-J., Lee J.-S., Han Y.-H., Bae I.H. Bcl-w enhances mesenchymal changes and invasiveness of glioblastoma cells by inducing nuclear accumulation of β-catenin. PLoS ONE. 2013;8:e68030. doi: 10.1371/journal.pone.0068030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chung H.J., Choi Y.E., Kim E.S., Han Y.-H., Park M.-J., Bae I.H. miR-29b attenuates tumorigenicity and stemness maintenance in human glioblastoma multiforme by directly targeting BCL2L2. Oncotarget. 2015;6:18429–18444. doi: 10.18632/oncotarget.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim E.S., Choi J.Y., Hwang S.J., Bae I.H. Hypermethylation of miR-205-5p by IR governs aggressiveness and metastasis via regulating Bcl-w and Src. Mol. Ther. Nucleic Acids. 2019;14:450–464. doi: 10.1016/j.omtn.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bae I.H., Park M.-J., Yoon S.H., Kang S.W., Lee S.-S., Choi K.-M., Um H.D. Bcl-w promotes gastric cancer cell invasion by inducing matrix metalloproteinase-2 expression via phosphoinositide 3-kinase, Akt, and Sp1. Cancer Res. 2006;66:4991–4995. doi: 10.1158/0008-5472.CAN-05-4254. [DOI] [PubMed] [Google Scholar]
- 23.Wilson J.W., Nostro M.C., Balzi M., Faraoni P., Cianchi F., Becciolini A., Potten C.S. Bcl-w expression in colorectal adenocarcinoma. Br. J. Cancer. 2000;82:178–185. doi: 10.1054/bjoc.1999.0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rupaimoole R., Ivan C., Yang D., Gharpure K.M., Wu S.Y., Pecot C.V., Previs R.A., Nagaraja A.S., Armaiz-Pena G.N., McGuire M. Hypoxia-upregulated microRNA-630 targets Dicer, leading to increased tumor progression. Oncogene. 2016;35:4312–4320. doi: 10.1038/onc.2015.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu J., Sun T., Wang H., Chen Z., Wang S., Yuan L., Liu T., Li H.R., Wang P., Feng Y. MiR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell. 2016;29:49–60. doi: 10.1016/j.ccell.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu M.-Z., Cheng W.-C., Chen S.-F., Nieh S., O’Connor C., Liu C.-L., Tsai W.W., Wu C.J., Martin L., Lin Y.S. miR-25/93 mediates hypoxia-induced immunosuppression by repressing cGAS. Nat. Cell Biol. 2017;19:1286–1296. doi: 10.1038/ncb3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigeyasu K., Toden S., Zumwalt T.J., Okugawa Y., Goel A. Emerging role of microRNAs as liquid biopsy biomarkers in gastrointestinal cancers. Clin. Cancer Res. 2017;23:2391–2399. doi: 10.1158/1078-0432.CCR-16-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maxwell P.H., Dachs G.U., Gleadle J.M., Nicholls L.G., Harris A.L., Stratford I.J., Hankinson O., Pugh C.W., Ratcliffe P.J. Hypoxia-inducible factor-1 modulates gene expression in solid tumors and influences both angiogenesis and tumor growth. Proc. Natl. Acad. Sci. USA. 1997;94:8104–8109. doi: 10.1073/pnas.94.15.8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ryan H.E., Poloni M., McNulty W., Elson D., Gassmann M., Arbeit J.M., Johnson R.S. Hypoxia-inducible factor-1α is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- 30.Tanaka N., Kato H., Inose T., Kimura H., Faried A., Sohda M., Nakajima M., Fukai Y., Miyazaki T., Masuda N. Expression of carbonic anhydrase 9, a potential intrinsic marker of hypoxia, is associated with poor prognosis in oesophageal squamous cell carcinoma. Br. J. Cancer. 2008;99:1468–1475. doi: 10.1038/sj.bjc.6604719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoskin P.J., Sibtain A., Daley F.M., Wilson G.D. GLUT1 and CAIX as intrinsic markers of hypoxia in bladder cancer: relationship with vascularity and proliferation as predictors of outcome of ARCON. Br. J. Cancer. 2003;89:1290–1297. doi: 10.1038/sj.bjc.6601260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aponte P.M., Caicedo A. Stemness in cancer: stem cells, cancer stem cells, and their microenvironment. Stem Cells Int. 2017;2017:5619472. doi: 10.1155/2017/5619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Medema J.P. Cancer stem cells: the challenges ahead. Nat. Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 34.Pezzella F., Harris A.L., Tavassoli M., Gatter K.C. Blood vessels and cancer much more than just angiogenesis. Cell Death Discov. 2015;1:15064. doi: 10.1038/cddiscovery.2015.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mazzieri R., Pucci F., Moi D., Zonari E., Ranghetti A., Berti A., Politi L.S., Gentner B., Brown J.L., Naldini L., De Palma M. Targeting the ANG2/TIE2 axis inhibits tumor growth and metastasis by impairing angiogenesis and disabling rebounds of proangiogenic myeloid cells. Cancer Cell. 2011;19:512–526. doi: 10.1016/j.ccr.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 36.Bae I.H., Yoon S.H., Lee S.B., Park J.K., Ho J.-N., Um H.-D. Signaling components involved in Bcl-w-induced migration of gastric cancer cells. Cancer Lett. 2009;277:22–28. doi: 10.1016/j.canlet.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 37.Bae S., Jeong H.J., Cha H.J., Kim K., Choi Y.M., An I.S., Koh H.J., Lim D.J., Lee S.J., An S. The hypoxia-mimetic agent cobalt chloride induces cell cycle arrest and alters gene expression in U266 multiple myeloma cells. Int. J. Mol. Med. 2012;30:1180–1186. doi: 10.3892/ijmm.2012.1115. [DOI] [PubMed] [Google Scholar]
- 38.Nag S., Resnick A. Stabilization of hypoxia inducible factor by cobalt chloride can alter renal epithelial transport. Physiol. Rep. 2017;5:e13531. doi: 10.14814/phy2.13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W., Fong M.Y., Min Y., Somlo G., Liu L., Palomares M.R., Yu Y., Chow A., O’Connor S.T., Chin A.R. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. 26267216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W., Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019;20:18. doi: 10.1186/s13059-019-1629-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen Y., Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;48(D1):D127–D131. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harris A.L. Hypoxia--a key regulatory factor in tumour growth. Nat. Rev. Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 44.Joseph J.P., Harishankar M.K., Pillai A.A., Devi A. Hypoxia induced EMT: A review on the mechanism of tumor progression and metastasis in OSCC. Oral Oncol. 2018;80:23–32. doi: 10.1016/j.oraloncology.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 45.D’Ignazio L., Rocha S. Hypoxia Induced NF-κB. Cells. 2016;5:10. doi: 10.3390/cells5010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim M.-C., Hwang S.-H., Kim N.-Y., Lee H.-S., Ji S., Yang Y., Kim Y. Hypoxia promotes acquisition of aggressive phenotypes in human malignant mesothelioma. BMC Cancer. 2018;18:819. doi: 10.1186/s12885-018-4720-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keith B., Johnson R.S., Simon M.C. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer. 2011;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trisciuoglio D., Gabellini C., Desideri M., Ragazzoni Y., De Luca T., Ziparo E., Del Bufalo D. Involvement of BH4 domain of bcl-2 in the regulation of HIF-1-mediated VEGF expression in hypoxic tumor cells. Cell Death Differ. 2011;18:1024–1035. doi: 10.1038/cdd.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iervolino A., Trisciuoglio D., Ribatti D., Candiloro A., Biroccio A., Zupi G., Del Bufalo D. Bcl-2 overexpression in human melanoma cells increases angiogenesis through VEGF mRNA stabilization and HIF-1-mediated transcriptional activity. FASEB J. 2002;16:1453–1455. doi: 10.1096/fj.02-0122fje. [DOI] [PubMed] [Google Scholar]
- 50.Park S.Y., Billiar T.R., Seol D.W. Hypoxia inhibition of apoptosis induced by tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) Biochem. Biophys. Res. Commun. 2002;291:150–153. doi: 10.1006/bbrc.2002.6421. [DOI] [PubMed] [Google Scholar]
- 51.Dong Z., Wang J. Hypoxia selection of death-resistant cells. A role for Bcl-X(L) J. Biol. Chem. 2004;279:9215–9221. doi: 10.1074/jbc.M312225200. [DOI] [PubMed] [Google Scholar]
- 52.Shen G., Li X., Jia Y.F., Piazza G.A., Xi Y. Hypoxia-regulated microRNAs in human cancer. Acta Pharmacol. Sin. 2013;34:336–341. doi: 10.1038/aps.2012.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan Y., Song X., Sun W., Wang Y., Liu B. Effect of Hypoxia-Induced MicroRNA-210 Expression on Cardiovascular Disease and the Underlying Mechanism. Oxid. Med. Cell. Longev. 2019;2019:4727283. doi: 10.1155/2019/4727283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X., Le Q.-T., Giaccia A.J. MiR-210--micromanager of the hypoxia pathway. Trends Mol. Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding J., Huang F., Wu G., Han T., Xu F., Weng D., Wu C., Zhang X., Yao Y., Zhu X. MiR-519d-3p suppresses invasion and migration of trophoblast cells via targeting MMP-2. PLoS ONE. 2015;10:e0120321. doi: 10.1371/journal.pone.0120321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pang Y., Mao H., Shen L., Zhao Z., Liu R., Liu P. MiR-519d represses ovarian cancer cell proliferation and enhances cisplatin-mediated cytotoxicity in vitro by targeting XIAP. OncoTargets Ther. 2014;7:587–597. doi: 10.2147/OTT.S60289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yue H., Tang B., Zhao Y., Niu Y., Yin P., Yang W., Zhang Z., Yu P. MIR-519d suppresses the gastric cancer epithelial-mesenchymal transition via Twist1 and inhibits Wnt/β-catenin signaling pathway. Am. J. Transl. Res. 2017;9:3654–3664. [PMC free article] [PubMed] [Google Scholar]
- 58.Kosaka N., Iguchi H., Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101:2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mensah M., Borzi C., Verri C., Suatoni P., Conte D., Pastorino U., Orazio F., Sozzi G., Boeri M. MicroRNA based liquid biopsy: The experience of the plasma miRNA signature classifier (MSC) for lung cancer screening. J. Vis. Exp. 2017;128:e56326. doi: 10.3791/56326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han Y., Li H. miRNAs as biomarkers and for the early detection of non-small cell lung cancer (NSCLC) J. Thorac. Dis. 2018;10:3119–3131. doi: 10.21037/jtd.2018.05.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tejero R., Navarro A., Campayo M., Viñolas N., Marrades R.M., Cordeiro A., Ruíz-Martínez M., Santasusagna S., Molins L., Ramirez J., Monzó M. miR-141 and miR-200c as markers of overall survival in early stage non-small cell lung cancer adenocarcinoma. PLoS ONE. 2014;9:e101899. doi: 10.1371/journal.pone.0101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li W., Wang Y., Zhang Q., Tang L., Liu X., Dai Y., Xiao L., Huang S., Chen L., Guo Z. MicroRNA-486 as a Biomarker for Early Diagnosis and Recurrence of Non-Small Cell Lung Cancer. PLoS ONE. 2015;10:e0134220. doi: 10.1371/journal.pone.0134220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park S.-Y., Lee J.H., Ha M., Nam J.-W., Kim V.N. miR-29 miRNAs activate p53 by targeting p85 α and CDC42. Nat. Struct. Mol. Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 64.Yosef R., Pilpel N., Tokarsky-Amiel R., Biran A., Ovadya Y., Cohen S., Vadai E., Dassa L., Shahar E., Condiotti R. Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 2016;7:11190. doi: 10.1038/ncomms11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.