Abstract
A stable, human sporadic vestibular schwannoma cell line is not currently available. By using a lentivirus-mediated transfection from a 41-year-old sporadic vestibular schwannoma patient, primary schwannoma cells were obtained, cultured and immortalized using the hHERT gene. The NF2 gene of the resulting JEI-001 cell line contains a specific Exon 5 mutation. The schwannoma cell origin of this cell line was confirmed using STR techniques and immunocytochemistry. A comparison between the primary tumor tissue and JEI-001 revealed a common mutation of the NF2 gene, which indicated that the JEI-001 cell line had retained most of its original tumor characteristics. The JEI-001 cell line was found to be non-tumorigenic in nude mice, but certain growth features had been altered, resulting in changes such as independence from the Schwann cell growth factors and a higher proliferation rate. This was the first known study to establish cell lines immortalized from human sporadic vestibular schwannoma that had different characteristics from that of HEI-193. This is a novel model system that can be used for the study of NF2 gene functions, in order to elaborate on the biological features of sporadic vestibular schwannoma, even including familial NF2 tumors, and to further explore the molecular pathogenesis and develop new adjuvant therapies.
Keywords: Sporadic vestibular schwannoma, characterization, establishment, immortalization
Introduction
Vestibular schwannoma, a benign tumor, arises from the nerve sheath of the VIII cranial nerve and accounts for approximately 6% of all intracranial tumors, including 90% lesions of the cerebellopontine angle (CPA) [1,2]. Vestibular schwannomas consist of 96% sporadic vestibular schwannomas and 4% neurofibromatosis type 2 (NF2)-related vestibular schwannomas [3,4]. Neurofibromatosis type 2 is a multiple neoplasia syndrome that results from a mutation in the NF2 tumor suppressor gene. Bilateral vestibular schwannomas are the distinctive feature of NF2, otherwise affected people can develop schwannomas in other cranial, spinal, and peripheral nerves [5]. NF2 mutations/merlin loss-driven tumorigenesis are widely believed to cause either sporadic or NF2-related vestibular schwannomas. Even though animal-derived or NF2-related schwannomas have been described previously, no stable cell line originating from human sporadic vestibular schwannoma has been available thus far. The main obstacle to generating immortalized benign tumor cell lines is replicative senescence. This results from low telomerase activity and to a certain extent, the progressive shortening of telomeres at each cell cycle, in comparison with malignant tumors that show high telomerase activity, which leads to limitless growth [6,7]. Benign tumor cells and non-neoplastic cells have been used to construct immortalized cell lines of several types of cells through transfection of the gene for human telomerase reverse transcriptase (hTERT) [8].
In this study, we established an immortalized cell line derived from sporadic vestibular schwannoma created using lentivirus-mediated hHERT gene transduction.
Materials and methods
Ethics statement
All experimental protocols were approved by the Research Ethics Review Committee of Shanghai Jiaotong University (Document serial number: SH9H-2019-T179-2). The methods and experimental protocols in the human samples used in the present study were carried out in accordance with the approved guidelines and regulations, and the study participant signed the informed consent form. It conformed to the provisions of the Declaration of Helsinki.
Material from the patient
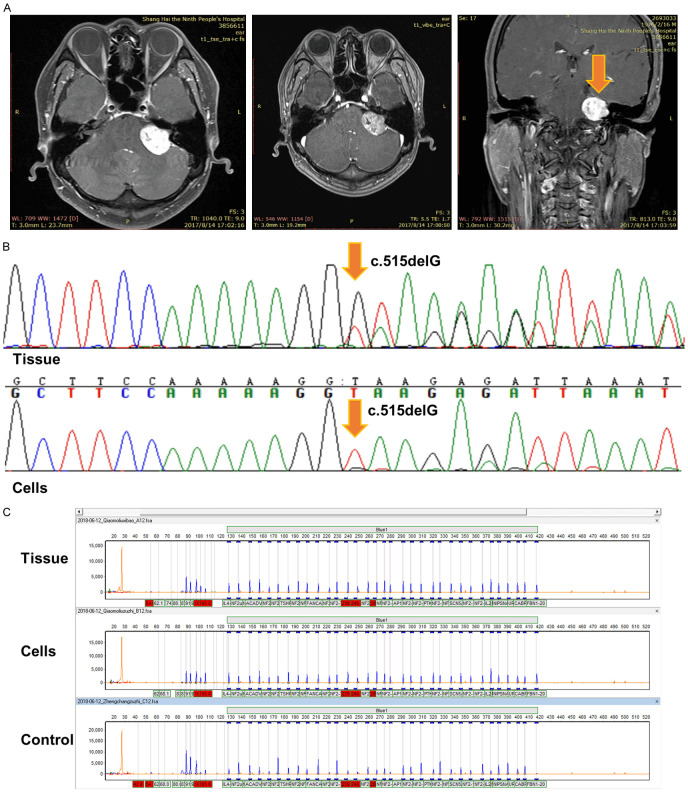
Vestibular schwannoma tissue was obtained from a 41-year-old male sporadic vestibular schwannoma patient who underwent the tumor excision after providing a written informed consent, at Shanghai Ninth People’s Hospital, Shanghai Jiaotong University School of Medicine. The patient had only one unilateral sporadic vestibular schwannoma on the left side. He had persistent tinnitus for 3 years and hearing loss during recent months and was diagnosed using MR (Figure 1A).
Figure 1.
The clinical information and the genetic characteristics of this sporadic vestibular schwannoma patient. A. MRI of a forty-one-year-old sporadic vestibular schwannoma patient. B. The tumor tissue and cells were both confirmed to contain a specific NF2 heterozygous mutation (c.515delG) at Exon 5 using Sanger sequencing. C. No exonic deletions were found in the tissue or the cells using MLPA.
Primary sporadic vestibular schwannoma cell culture
The tissue was taken to the laboratory at 4°C after being placed in DMEM media. The tumor tissue was minced to around 1 mm3 and digested at 37°C for 2 hours with 0.125% enzymes-EDTA (Gibco) + 0.1% collagenase type II (Gibco). Then it was washed with fetal bovine serum (FBS) (Gibco) and phosphate-buffered saline (PBS, PH = 7.4 7.4; Gibco), and plated into 10 cm plates in a modified DMEM/high glucose medium (1 × N-2 supplement (Gibco) + insulin (Sigma) 5 µg/ml + 10% FBS + 1 × P/S (Proteintech)) at 37°C with 5% CO2. The tumor cells that had not attached were transferred onto new plates after each block of 12 hours during the first 3 days, to remove the fibroblasts and the other non-targeted cells. S-100 was also used to ensure that more than 90% of the target cells remained.
Immortalization of the primary culture
On a 60 mm laminin-coated plate with DMEM media (90% DMEM/high glucose medium + 10% FBS) 5 × 105 cells were plated. A lentivirus-mediated human telomerase reverse transcriptase gene (hHERT) was added after the cells had attached. Two ug/mL of puromycin was used to select the transfected cells that contained puromycin resistance. Passage 0 was regarded as the resulting drug resistant cells, which were harvested after the culture medium was changed 5 times with puromycin. The name JEI-001 (for Ear Institute Shanghai Jiaotong University School of Medicine) was given to the cell line.
Immunofluorescence analysis
The cells were cultured on slides. Twenty-four hours later, paraformaldehyde was applied for 15 min to fix the plated cells. The slides were then permeabilized in Triton X-100 and blocked with 10% normal goat serum. An immunocytochemical analysis was performed using antibodies specific for S100 (#ab868, Abcam) and NGFR (#A11050, Abclonal). The nuclear counterstaining was conducted using 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) (#C1002, Beyotime). The slides were observed using a Zeiss fluorescence microscope (Zeiss, Germany) after three rounds of washing using PBS and covered with regular Permount.
Immunocytochemical analysis
The cells were cultured on slides. Twenty-four hours later, paraformaldehyde was applied for 15 min to fix the plated cells. 3% H2O2 was used for 15 min in RT. The slides were then blocked using 10% normal goat serum for 30 min. The immunocytochemical analysis was performed using antibodies specific for CNPase (#ab6319, Abcam), MBP (#ab7349, Abcam), and collagen type IV (#GB13362, Servicebio). The cells were stained using a DAB-system kit (#DAB4033, Maxim) and hematoxylin (#BA4041, Baso). The slides were observed under a microscope (Olympus, Japan).
Immunoblotting analysis
The immunoblotting analysis was performed using antibodies specific for Merlin (#HPA003097, Sigma-Aldrich), and the β-actin antibody (#AA128, Beyotime) was used to ensure equal loading of the total protein.
RNA sequencing
The total RNAs from each cell line were quantified using a NanoDrop ND-1000 instrument. The total RNAs were extracted using oligo (dT) magnetic beads. A KAPA-Stranded RNA-Seq Library Prep Kit (Illumina) was used to construct the sequencing libraries after a quantitative analysis and a quality inspection. The sequencing was carried out using an Illumina HiSeq 4000 Sequencing System for 150 cycles. After the data preprocessing, significant changes in the gene and transcript expressions were then calculated using Ballgown software.
DNA sequencing
For the NF2 gene message measurement and the comparison between the JEI-001 cells and the patient tumor tissue, DNA samples were prepared using a TIANamp Genomic DNA Kit (Tiangen), following the protocols provided by the manufacturer. In order to sequence the entire coding region, 17 primers were synthesized for PCR product direct DNA sequencing. The manufacturer’s instructions (Takara Biotechnology) were followed for the sequencing reaction procedure, and after the reactions were run, 3730 XL (Applied Biosystems) and DNA sequencing analysis 5.2.0 software were used for the analysis. In order to determine the exonic deletions, a Multiplex Ligation-Dependent Probe Amplification Analysis (MLPA) kit (MRC-Holland) was used.
Exosome Isolation
In order to isolate the exosomes, the cells were ultracentrifuged. First, the liquid samples were centrifuged for 5 min at 500 × g, then the cells were eliminated from the samples. Next, the supernatants were transferred into new polycarbonate tubes, which were centrifuged for 10 min at 2000 × g. Then, the supernatants were collected and transferred into new polycarbonate tubes to eliminate debris from the samples. In order to eliminate the shed microvesicles, the centrifugation was conducted for 30 min at 10,000 × g (sMV, 200-1000 nm). Then, the supernatants were collected and filtered using 0.22 μm membrane filters (Merck Millipore). The centrifugation was conducted for 2 h at 100,000 × g. For the RNA isolation, exosome pellets were washed once with 1 × PBS and centrifuged for 2 h at 100,000 × g for a second time. The exosomes were resuspended in 1 × PBS and stored at -80°C for further use, and then they were viewed using transmission electron microscopy (Tecnai G2 Spirit 120 KV).
Karyotype analysis
The chromosomes of the JEI-001 cells were identified using a karyotype analysis [9]. First, the cell cultures of the 30th passage of JEI-001 in the exponential phase of growth were treated with colchicine (0.2 μg/ml; Sigma) at 37°C for 1-2 hours. Then, the cells were Trypsinized and collected. After centrifugation (10 min at 1,000 × g), the precipitates of the cells were incubated in a salt solution (0.075 M potassium chloride) at 37°C for 40 min and then fixed in methanol/ice-cold acetic acid (v/v = 3:1) for 10 min in RT. Subsequently, the fixed cells were harvested using centrifugation (10 min at 1,000 × g) and resuspended in PBS three times. The cell suspension was added on a slide, dried for 6 hours, and stained with Giemsa (sigma). The metaphase chromosomes were observed under a microscope (Leica, H1210).
Label-free quantitative proteomics using LC-MS/MS [10]
A Q-Exactive mass spectrometer (Thermo Finnigan) and a nano-UPLC (EASY-nLC1200) were used to analyze approximately 2 μg of peptides from each sample, after separation. A reversed-phase column (100 μm, ID × 15 cm, Reprosil-Pur 120 C18-AQ, 1.9 μm, Dr. Math) was used for the separation. H2O with 80% ACN, 0.1% FA and 0.1% FA, 2% ACN were used as the mobile phases for phase B and phase A, respectively. A sample separation was conducted using a 120 min gradient at a 300 nL/min flow rate. Gradient B: 8 to 30%, for 92 min, 30 to 40% for 20 min, followed by 40 to 100%, 100%, 100 to 2% and 2% for 2 min each. The data dependent acquisition was performed using the positive mode, and an Orbitrap analyzer was used in the positive mode at a m/z range of 350-1600 and a resolution of 70,000 (@200 m/z) for MS1, while the resolution was set to 17,500 for MS2 using the dynamic first mass. The automatic gain control (AGC) target for MS1 was set at 3.0 E + 6 with max IT 50 ms, and 5.0 E + 4 for MS2 with a max IT of 100 ms. HCD with a normalized collision energy (NCE) of 27% and an isolation window of 2 m/z were used to fragment the top 20 most intense ions. The dynamic exclusion time window used was 30 s.
Results
Establishment of a cell line
Vestibular schwannoma tissue was used to culture schwannoma cells from a 41 year old sporadic vestibular schwannoma patient (Figure 1A), which had been confirmed to contain a specific NF2 heterozygous mutation (c.515delG) at Exon5 through Sanger sequencing (Figure 1B). The primary cells were used for immortalization through lentivirus-mediated hHERT gene transduction. The target cell line was harvested using a puromycin selection. The cell line was designed as JEI-001 for the Ear Institute Shanghai Jiaotong University School of Medicine and was propagated for over 30 passages. A further analysis detected exonic deletions through a Multiplex Ligation-Dependent Probe Amplification Analysis (MLPA) and found no exonic deletions in either the tumor tissue or the JEI-001 (Figure 1C).
Cell morphology
Several days after the lentiviral transduction of the primary cells, they were found to be atypical and with a differentiated morphology. The cells appeared to be of an irregular polygon shape and were growing in a disorderly manner. These cells were observed to be overlapped and piled up at a high density on the plates where they grew. These results indicated that the cells were gradually immortalized (primary cells, P0, P10, P30) (Figure 2A).
Figure 2.
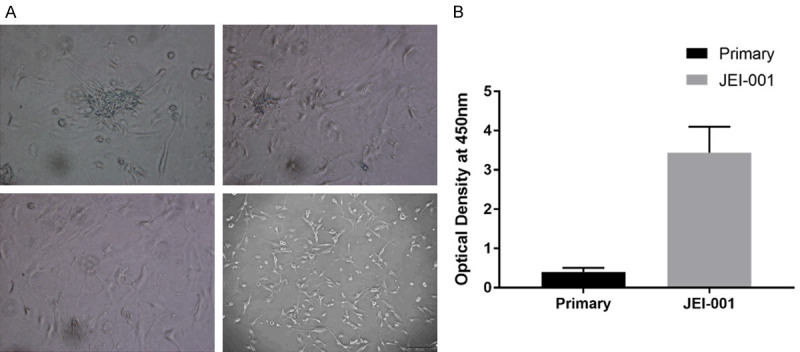
The proliferation ability of JEI-001. A. The cells had gradually become immortalized: primary cells, P0, P10, P30. B. The CCK8 results showed the significantly different growth speeds of the primary cells and JEI-001 (P < 0.05).
Growth rate
A large increase in the growth rate of the transduced cells was observed. The rate of the JEI-001 cell proliferation was determined by measuring the optical density value at 450 nm. The CCK8 results show the growth rates of the primary cells and of JEI-001 (Figure 2B).
Short tandem repeat (STR) analysis
A search for multiple alleles in the cell lines of the ATCC, DSMZ, JCRB and RIKEN cell banks did not find any similar cell lines. The harvested cells and tissue were found to be of the same origin. The test results are shown in Table 1.
Table 1.
STR results for the tumor tissue and JEI-001
| Marker | Tissue | Cell line | ||
|---|---|---|---|---|
|
|
|
|||
| Allele 1 | Allele 2 | Allele 1 | Allele 2 | |
| 01-D5S818 | 10 | 12 | 10 | 12 |
| 02-D13S317 | 8 | 11 | 8 | 11 |
| 03-D7S820 | 10 | 12 | 10 | 12 |
| 04-D16S539 | 9 | 12 | 9 | 12 |
| 05-VWA | 15 | 19 | 15 | 19 |
| 06-TH01 | 9 | 9 | ||
| 07-AMEL | X | Y | X | Y |
| 08-TPOX | 11 | 12 | 11 | 12 |
| 09-CSF1PO | 10 | 14 | 10 | 14 |
| 10-D12S391 | 17 | 18 | 17 | 18 |
| 11-FGA | 22 | 23 | 22 | 23 |
| 12-D2S1338 | 19 | 20 | 19 | 20 |
| 13-D21S11 | 29 | 29 | ||
| 14-D18S51 | 14 | 14 | ||
| 15-D8S1179 | 15 | 16 | 15 | 16 |
| 16-D3S1358 | 16 | 17 | 16 | 17 |
| 17-D6S1043 | 11 | 20 | 11 | 20 |
| 18-PENTAE | 16 | 23 | 16 | 23 |
| 19-D19S433 | 15 | 15.2 | 15 | 15.2 |
| 20-PENTAD | 9.1 | 11 | 9.1 | 11 |
Karyotype analysis
A karyotype analysis was carried out on the 30th passage JEI-001. The 30th passage of JEI-001 was found to have a diploid chromosome complement of 2n = 46, consisting of a pair of sex chromosomes (X, Y) and 22 autosomal pairs (Figure 3A). No chromosome abnormalities were found in the 30th passage of JEI-001.
Figure 3.
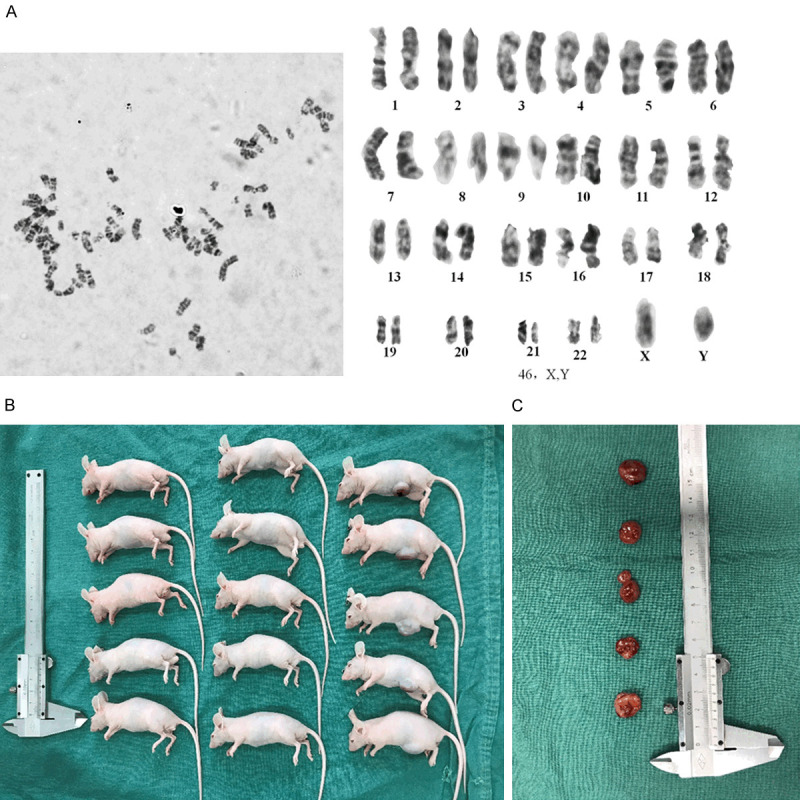
A karyotype analysis and the tumorigenicity assays of JEI-001. A. No chromosome abnormalities were found in the 30th passage JEI-001 cells. B and C. JEI-001 showed no tumorigenic transformation.
Tumorigenicity assays
No tumorigenic transformations were detected in the JEI-001 cells. The tumorigenicity assays were conducted by injecting the JEI-001 cells, as well as PBS as a negative control, and HEI-193 as the positive control. After 3 weeks, tumors were observed at the injected sites of all 6 BALB/c nude mice subcutaneously injected with 1*106 HEI-193 cells, but no tumors were found in the mice injected with JEI-001 cells or PBS, indicating that the JEI-001 cells do not produce tumorigenic transformations (Figure 3B and 3C).
Detection of the tumor biomarkers
There are no specific markers for schwannoma since it is a benign tumor. The schwannoma cell origin was confirmed by identifying its Schwann cell characteristics, as previously described [11].
The results of the immunocytochemical staining for the certain biomarkers of JEI-001 cells are shown below. 2’3’-cyclic nucleotide 3’-phosphohydrolase (CNPase), S100, NGFR and Type IV collagen were found to be positive (Figure 4A-D), and the myelin basic protein (MBP) was negative (Figure 4E), clearly indicating that over 99% of the JEI-001 cells were derived from the vestibular schwannoma tissue. In addition, we detected the expression of the key protein merlin, and significantly lower expressions of merlin were noted in JEI-001 and HEI-193 than the expression in HSC (Figures 4F, S1), which verified that JEI-001 shares the same trend of merlin expression with HEI-193 and a deficiency of merlin leading to tumorigenesis in schwannomas.
Figure 4.
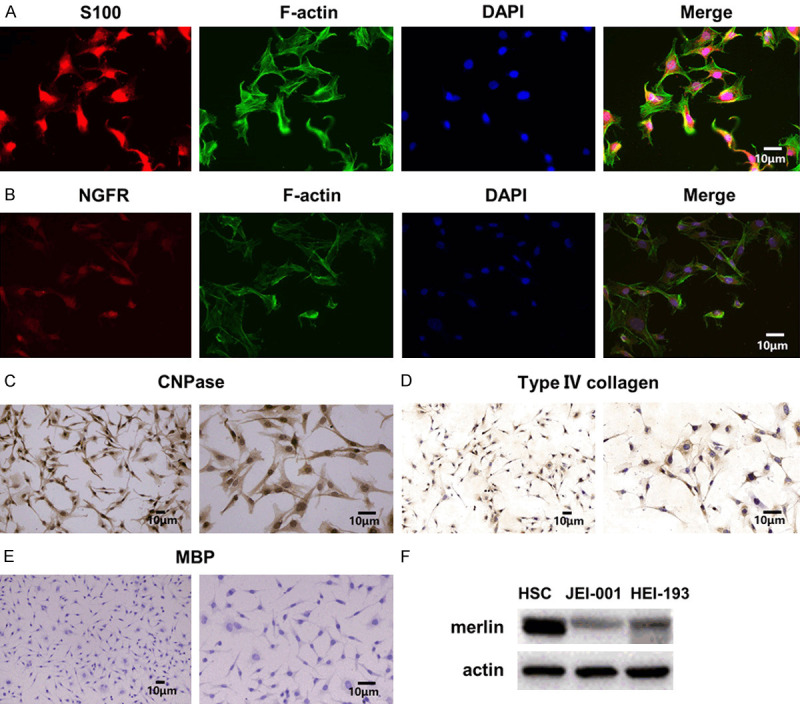
Characteristics of the JEI-001 cells. A-D. Immunocytochemical staining of the JEI-001 cells for S100 (20 ×), NGFR (10 ×, 20 ×), CNPase (10 ×, 20 ×), Type VI collagen (10 ×, 20 ×) produced positive results. E. Immunocytochemical staining of the JEI-001 cells for MBP produced a negative result (10 ×, 20 ×). F. A lower expression of merlin was noted in JEI-001 than in the normal Schwann cells.
RNA-seq analysis between JEI-001 and HEI-193
We examined the different gene expression levels between the JEI-001 and HEI-193 cell lines using RNA sequencing analysis. We sequenced the transcripts, and 5795 differential expressed genes (DEGs) were detected in a test of the two cell lines (Table S1).
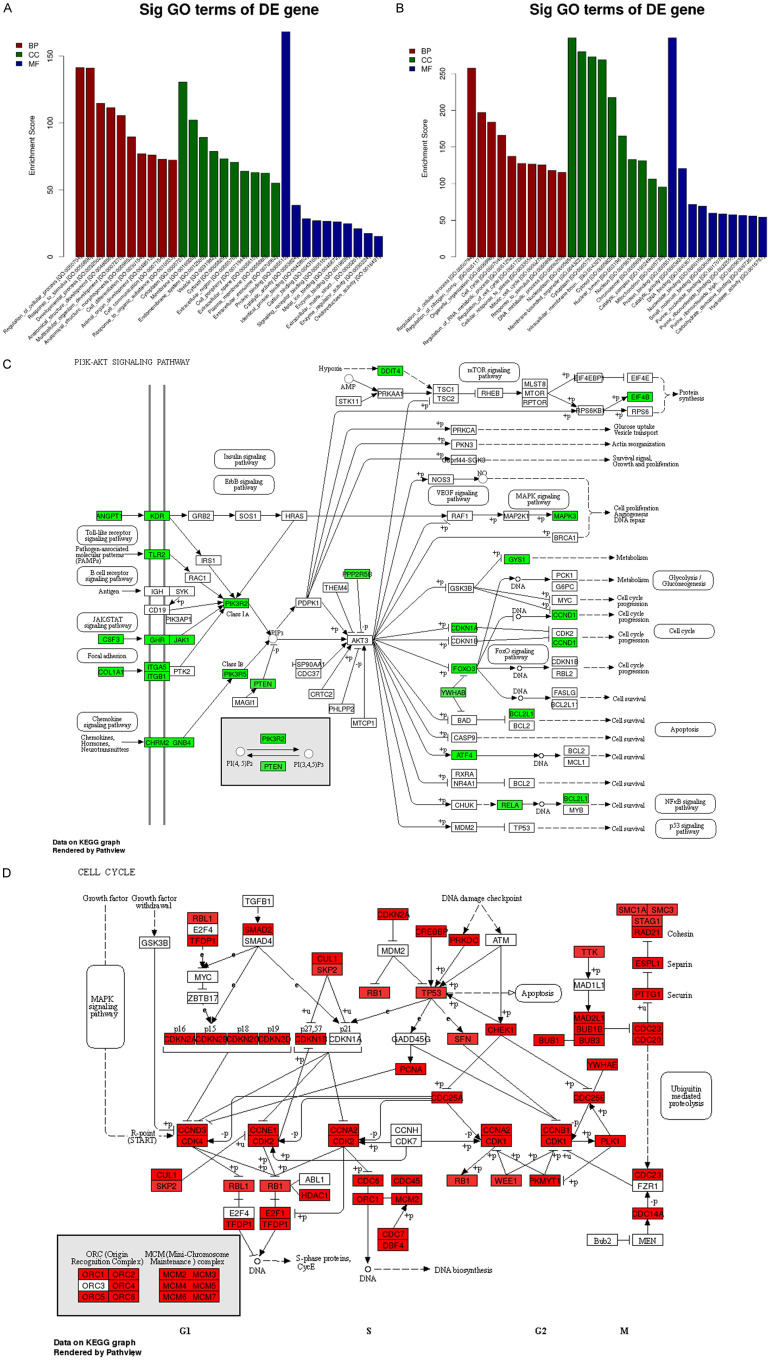
A GO analysis (http://www.geneontology.org/) was applied to search for significantly enriched GO terms which were made on the biological process (BP), the cellular component (CC), and the molecular function (MF) for the DEGs. Prediction terms with a P value < 0.05 were selected and ranked by enrichment score (-log 10 (P-value)). According to the results, 3528 BP terms, 357 CC terms, and 600 MF terms were found to be down-regulated. In contrast, 3969 BP terms, 640 CC terms, and 786 MF terms were found to be up-regulated in JEI-001 compared with HEI-193. The top 10 generally changed GO terms in the JEI-001 group classified by BP, CC, MF, and ranked by enrichment score are listed in Figure 5A, 5B.
Figure 5.
The RNA-seq analysis between JEI-001 and HEI-193. A. A GO analysis of the differentially down-regulated genes. B. A GO analysis of the differentially up-regulated genes. C. The PI3K-Akt signaling pathway, green color = down-regulated genes at ≥1.5 folds. D. Cell cycle, red color = up-regulated genes at ≥1.5 folds.
Our GO analysis showed (Figure 5A) that in the down-regulated genes the most enriched BP terms were primarily about cells, such as “Regulation of the cellular process (GO: 0050794)”, “Multicellular organism development (GO: 0007275)”, “Cell differentiation (GO: 0030154)”, “Cell communication (GO: 0007154)”, and “Animal organ development (GO: 0048513)”. While the GO CC terms were ranked by their enrichment scores, the mainly enriched terms may be closely related to the membrane. The represented terms were “Membrane (GO: 0016020)”, “Endomembrane system (GO: 0012505)”, and “plasma membrane (GO: 0005886)”. The most enriched MF terms were primarily about binding function such as “Protein binding (GO: 0005515)”, “Identical protein binding (GO: 0042802)”, “Cation binding (GO: 0043169)”, “Signaling receptor binding (GO: 0005102)”, “Metal ion binding (GO: 0046872)”, and “Enzyme binding (GO: 0019899)”.
Furthermore, a KEGG pathway analysis was made, and the pathways (P < 0.05) were selected and ranked by their enrichment scores (Figure 5C). Our KEGG pathway analysis showed that the down-regulated genes were mainly enriched in the pathways like the ECM receptor interaction, the P53 signaling pathway, the PI3K-Akt signaling pathway, and the HIF-1 signaling pathway. In the PI3k-Akt signaling pathway, the gene expressions of the JEI-001 genes (JAK1, PTEN, CCND1) were down-regulated. This indicates that the different expression levels were between JEI-001 and HEI-193 in PI3K-Akt, previously known as one of the most remarkable pathways in the merlin-deficiency schwannomas.
Figure 5B shows that most of the enriched and remarkable up-regulated BP terms were also related to the cellular process, for instance, “Regulation of cellular process (GO: 0050794)”, “Cell cycle (GO: 0007049)”, “Regulation of cell cycle (GO: 0051726)”, and “Mitotic cell cycle (GO: 0000278)”. As for the GO CC terms ranked by enrichment score, the mainly enriched terms were correlated with the intracellular matrix, such as “Nucleoplasm (GO: 0005654)”, “Cytoplasm (GO: 0005737)”, and “Cytosol (GO: 0005829)”. The most enriched MF terms were mainly about binding functions, such as “Protein binding (GO: 0005515)”, “DNA binding (GO: 0003677)”, “Nucleotide binding (GO: 0000166)”, “Small molecule binding (GO: 0036094)”, and “Purine nucleotide binding (GO: 0017076)”.
In the same way, our KEGG pathway analysis showed that the up-regulated genes were primarily enriched in the pathways containing spliceosome, the cell cycle, and proteasome. In the cell cycle, the gene expressions of the JEI-001 genes (CDK1, CDK2, RB1, and CHEK1) were up-regulated (Figure 5D). In the mini-chromosome maintenance complex, MCM2/MCM3/MCM4/MCM5/MCM6/MCM7 were also significantly up-regulated.
Identification of the exosomal proteins
In this study, the HEI-193 and JEI-001 exosomes were isolated using the gold-standard ultracentrifugation method (Figure 6A). Table S2 provides the protein information in detail. The volcano plots (Figure 6B) show the statistically significant changes in the differences between the HEI-193 and JEI-001 proteins and their magnitudes. The obtained integrated DEGs were analyzed using the DAVID database for the gene ontology (GO) functional annotation and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.
Figure 6.
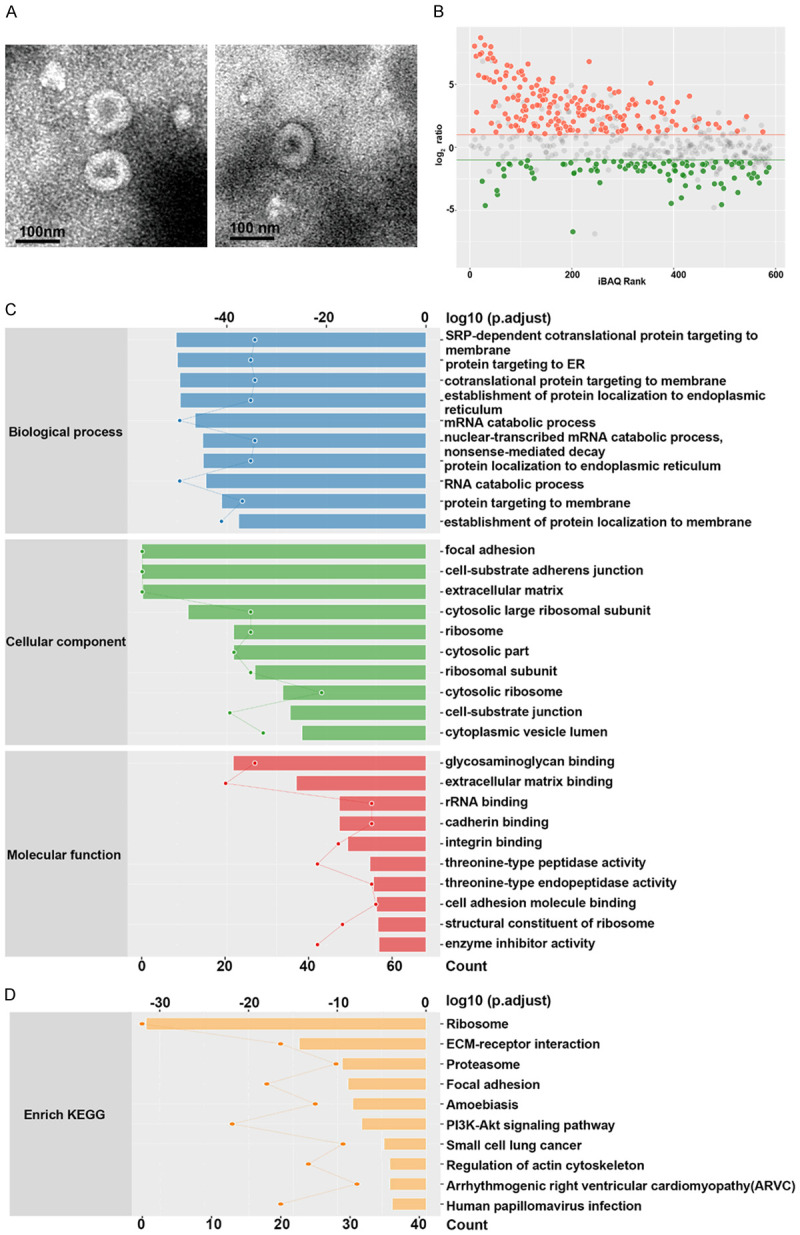
Identification of the exosomal proteins between JEI-001 and HEI-193. A. The exosomes of HEI-193 and the JEI-001 cells were isolated. B. Statistically significant and magnitude of change differences between the exosomal proteins of HEI-193 and JEI-001 cells are presented as volcano plots. C. The enrichment analysis of the gene ontology (GO) was performed to compare the differentially expressed proteins between these two lines of schwannoma cells. D. An enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) was performed to compare the differentially expressed proteins between these two lines of schwannoma cells.
The key biological processes and the potential pathways that may be used to discriminate JEI-001 from the HEI-193 cells were identified by conducting an enrichment analysis of the gene ontology (GO) biological processes on the differentially expressed proteins between the two schwannoma cell lines. The biological processes that were significantly represented included an SRP-dependent co-translational protein targeting the membrane, a protein targeting ER, a co-translational protein targeting the membrane, the establishment of protein localization to the endoplasmic reticulum, protein localization to the endoplasmic reticulum, and protein targeting to the membrane, which are mainly related with protein targeting. The cellular components of the GO enrichment analysis showed that a majority of the differentially expressed proteins were enhanced in the cell-substrate junctions, the cell-substrate adherens junctions, and the focal adhesion, which involve cell cycle regulation and cell junctions (Figure 6C). The differences between the proteins that participate in molecular function were relatively mild. We also demonstrated the most significant enriched pathways the DEGs analyzed by the KEGG analysis (Figure 6D). The DEGs were enriched in the ribosomes, the ECM receptor interactions, proteasome, and the focal adhesion PI3K-akt signaling pathways, etc.
Discussion
Using the lentivirus-mediated hHERT gene transduction technique, we established an immortalized cell line from a patient with sporadic vestibular schwannoma and a specific NF2 gene mutation. For vestibular schwannoma, only two kinds of related cell lines, such as RT4-D6P2T and HEI-193, which can be used for mechanism exploration, have been reported. However, the former involves schwannoma cells of rat origin and the latter is from a neurofibromatosis type 2 (NF2) patient. It is undeniable that in most cases NF2 patients present a much more severe phenotype than sporadic vestibular schwannomas, which may result in different mechanisms that are still obscure.
It is necessary to establish stable immortalized schwannoma cell lines [12], which may carry different NF2 alterations. The ideal method of studying schwannomas in vitro in molecular biology is to use a primary cell culture that best takes on its characteristics. However, the limited lifespan of benign tumors leads to slow growth of the primary cells, growth which may completely stop after several passages. For this reason, schwannoma cells from sporadic patients are even harder to enrich than the cells from NF2 patients. The latter usually carries a biallelic inactivation of NF2 based on the “two-hit” theory, which may result in stronger proliferation, but the former one usually contains only a single inactivation of NF2. Moreover, most studies on this topic have related the genotype of NF2 to different clinical phenotypes [13], such as onset latency, growth speed, and even tumor burden, with no conclusion yet. Indeed, JEI-001 is the first cell line derived from a sporadic vestibular schwannoma patient. The NF2 gene encodes a tumor suppressor protein called merlin, which regulates a series of complex signal transduction pathways. Merlin suppresses tumorigenesis by inhibiting E3 ubiquitin ligase CRL4 (DCAF1) in the nucleus [14]. The deficiency of merlin activates the multiple signal transduction pathways such as RAS-GTPase [15] and mTOR [16], and it also severely affects the synthesis and release of cytokines such as vascular endothelial growth factor (VEGF), platelet growth factor (PGF) and insulin growth factor (IGF), leading to tumorigenesis [17,18]. Here we further determined that the expression levels of the cell lines from sporadic vestibular schwannoma and from NF2 patients presented in different ways (Figures 5, 6), which were considered as the main points of sporadic vestibular schwannoma formation. In addition, the source of the primary cells makes it difficult to obtain monoclonal cells free from contamination by other stoma cells. Additionally, the dangerous location in CPA of the tumors leads to the availability of only finite volumes of tumor specimens. The quality and reproducibility of any related research studies may be influenced by the above mentioned elements. Therefore, this newly established cell line can be used for the in vitro study of human vestibular schwannoma cells.
In this study, a primary Schwann cell culture was used to explore the immortalization of vestibular schwannoma cells. For the delivery of the immortalization agent, a lentiviral mediated gene transfer technique was used. The lentivirus was found to have relatively stable transduction efficiency after being integrated into the target cell genome. The hTERT gene has been found to extend normal human cell life spans and stabilize telomere length, which remains repressed for the adult somatic cells directly associated with tumorigenesis. Ectopic hTERT expression maintained the telomere length and restored the telomerase activity of the JEI-001 cells. A two-stage model was used to clarify the mortal cell cycle. When mortality stage 1 (M1) proceeded to mortality stage 2 (M2), the cells underwent senescence and death, or overcame M1 to achieve their immortal proliferation capacity [19]. Previously, methods that introduced the human papillomavirus type 16 E6 or E7 genes alone or in combination (E6/E7) were used to change the telomerase activity and obtain cell lines, such as HEI-193 [11].
During the culturing process, the primary cells and the JEI-001 cell line were both proved to contain a specific mutation (c.515delG), which is the same as the tumor tissue (but not the same as the blood sample, data not shown). The results indicate that the characteristics of the original tumor are conserved.
The presence of the protein markers, S100, NGFR, CNPase, and Type IV collagen confirmed the Schwann cell origin of JEI-001. S100 has been used as the classic marker for schwannomas, and its B-type has been found to be prevalently expressed in human Schwann cells [20]. NGFR (p75) is another marker used to discriminate Schwann cells from fibroblasts [20,21], which were also found to be positive in this study. Type IV collagen and 2’3’-cyclic nucleotide 3’-phosphohydrolase (CNPase) were used to identify its origin. Type IV collagen is secreted by Schwann cells as a major extracellular matrix component [22], and CNPase is related to biogenesis, early growth, and the differentiation of myelin, and its stains are positive in all schwannoma cultures [11].
As for JEI-001 tumorigenicity in animals, the same genetic situation has been confirmed for benign tumor formation based on past research. In order to identify the biological differences between the JEI-001 and HEI-193 cells, we examined the different gene expression levels and identified their exosomal proteins, and significant differences were found in the processes such as protein targeting, cell cycle regulation, and cell junction, which are closely correlated with the function of merlin as a membrane protein. This indicated that it may be questionable to use HEI-193 for the study of sporadic vestibular schwannoma. In previous studies, the p53-MDM2 pathway has been reported to regulate the growth of vestibular schwannoma, but the usage of inhibitors presented a few differences, which made us reconsider whether it is the use of HEI-193 that made the sporadic vestibular schwannomas obscure, while the primary cells still presented with poor repeatability [23,24].
In conclusion, this is the first study to prove that immortalization using a lentivirus-mediated hHERT transduction can be used for the establishment of a human sporadic schwannoma cell line. The JEI-001 cell line was found to be a stable and benign immortalized schwannoma cell line that originated from Schwann cells and contained the NF2 gene mutation. Studies on the mutagenesis and tumorigenicity of the schwannoma cells and the genotype phenotype correlation of NF2 and NF2 gene function may benefit from the establishment of this cell line, by providing a novel model system for elaborating the biological features of sporadic vestibular schwannoma, even for familial NF2 tumors, and for further exploring its molecular pathogenesis and new adjuvant therapies.
Acknowledgements
The authors would like to thank KangChen Bio-tech (Shanghai, China) for the Proteomics experiments. This work was supported by the National Natural Science Foundation of China (Grant No. 81870713 to Zhaoyan Wang, Grant No. 81970872 to Hao Wu, Grant No. 81600815 to Yongchuan Chai, and Grant No. 81700900 to Weidong Zhu), and the Shanghai Municipal Science and Technology Commission (Grant No. 14DZ2260300 to Yongchuan Chai). The funding sources were not involved in the design of the study, the collection, analysis, and interpretation of the data, the writing the manuscript, or the decision to submit the manuscript for publication.
Disclosure of conflict of interest
None.
Figure S1
Table S1
Table S2
References
- 1.Pathmanaban ON, Sadler KV, Kamaly-Asl ID, King AT, Rutherford SA, Hammerbeck-Ward C, McCabe MG, Kilday JP, Beetz C, Poplawski NK, Evans DG, Smith MJ. Association of genetic predisposition with solitary schwannoma or meningioma in children and young adults. JAMA Neurol. 2017;74:1123–1129. doi: 10.1001/jamaneurol.2017.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrilli AM, Fuse MA, Donnan MS, Bott M, Sparrow NA, Tondera D, Huffziger J, Frenzel C, Malany CS, Echeverri CJ, Smith L, Fernandez-Valle C. A chemical biology approach identified PI3K as a potential therapeutic target for neurofibromatosis type 2. Am J Transl Res. 2014;6:471–493. [PMC free article] [PubMed] [Google Scholar]
- 3.Cutfield SW, Wickremesekera AC, Mantamadiotis T, Kaye AH, Tan ST, Stylli SS, Itineang T. Tumour stem cells in schwannoma: a review. J Clin Neurosci. 2019;62:21–26. doi: 10.1016/j.jocn.2018.12.022. [DOI] [PubMed] [Google Scholar]
- 4.Evans DG. Neurofibromatosis 2. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, Amemiya A, editors. GeneReviews((R)) Seattle (WA): 1993. [Google Scholar]
- 5.Asthagiri AR, Parry DM, Butman JA, Kim HJ, Tsilou ET, Zhuang Z, Lonser RR. Neurofibromatosis type 2. Lancet. 2009;373:1974–1986. doi: 10.1016/S0140-6736(09)60259-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puttmann S, Senner V, Braune S, Hillmann B, Exeler R, Rickert CH, Paulus W. Establishment of a benign meningioma cell line by hTERT-mediated immortalization. Lab Invest. 2005;85:1163–1171. doi: 10.1038/labinvest.3700307. [DOI] [PubMed] [Google Scholar]
- 7.Hiyama E, Hiyama K. Telomerase as tumor marker. Cancer Lett. 2003;194:221–233. doi: 10.1016/s0304-3835(02)00709-7. [DOI] [PubMed] [Google Scholar]
- 8.Hao J, Li Z, Zhang C, Yu W, Tang Z, Li Y, Feng X, Gao Y, Liu Q, Huang W, Guo W, Deng W. Targeting NF-kappaB/AP-2beta signaling to enhance antitumor activity of cisplatin by melatonin in hepatocellular carcinoma cells. Am J Cancer Res. 2017;7:13–27. [PMC free article] [PubMed] [Google Scholar]
- 9.Hong HX, Zhang YM, Xu H, Su ZY, Sun P. Immortalization of swine umbilical vein endothelial cells with human telomerase reverse transcriptase. Mol Cells. 2007;24:358–363. [PubMed] [Google Scholar]
- 10.Wang C, Tang Y, Wang Y, Li G, Wang L, Li Y. Label-free quantitative proteomics identifies Smarca4 is involved in vascular calcification. Ren Fail. 2019;41:220–228. doi: 10.1080/0886022X.2019.1591997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung G, Li X, Faudoa R, Xeu Z, Kluwe L, Rhim JS, Slattery W, Lim D. Establishment and characterization of a schwannoma cell line from a patient with neurofibromatosis 2. Int J Oncol. 2002;20:475–482. [PubMed] [Google Scholar]
- 12.Guo J, Grovola MR, Xie H, Coggins GE, Duggan P, Hasan R, Huang J, Lin DW, Song C, Witek GM, Berritt S, Schultz DC, Field J. Comprehensive pharmacological profiling of neurofibromatosis cell lines. Am J Cancer Res. 2017;7:923–934. [PMC free article] [PubMed] [Google Scholar]
- 13.Evans DG, Hartley CL, Smith PT, King AT, Bowers NL, Tobi S, Wallace AJ, Perry M, Anup R, Lloyd SKW, Rutherford SA, Hammerbeck-Ward C, Pathmanaban ON, Stapleton E, Freeman SR, Kellett M, Halliday D, Parry A, Gair JJ, Axon P, Laitt R, Thomas O, Afridi SK, Obholzer R, Duff C, Stivaros SM, Vassallo G, Harkness EF, Smith MJ. Incidence of mosaicism in 1055 de novo NF2 cases: much higher than previous estimates with high utility of next-generation sequencing. Genet Med. 2020;22:53–59. doi: 10.1038/s41436-019-0598-7. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Cooper J, Zhou L, Yang C, Erdjument-Bromage H, Zagzag D, Snuderl M, Ladanyi M, Hanemann CO, Zhou P, Karajannis MA, Giancotti FG. Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases lats1 and 2 in the nucleus. Cancer Cell. 2014;26:48–60. doi: 10.1016/j.ccr.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison H, Sperka T, Manent J, Giovannini M, Ponta H, Herrlich P. Merlin/neurofibromatosis type 2 suppresses growth by inhibiting the activation of Ras and Rac. Cancer Res. 2007;67:520–527. doi: 10.1158/0008-5472.CAN-06-1608. [DOI] [PubMed] [Google Scholar]
- 16.James MF, Han S, Polizzano C, Plotkin SR, Manning BD, Stemmer-Rachamimov AO, Gusella JF, Ramesh V. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Mol Cell Biol. 2009;29:4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plotkin SR, Stemmer-Rachamimov AO, Barker FG 2nd, Halpin C, Padera TP, Tyrrell A, Sorensen AG, Jain RK, di Tomaso E. Hearing improvement after bevacizumab in patients with neurofibromatosis type 2. N Engl J Med. 2009;361:358–367. doi: 10.1056/NEJMoa0902579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamura R, Fujioka M, Morimoto Y, Ohara K, Kosugi K, Oishi Y, Sato M, Ueda R, Fujiwara H, Hikichi T, Noji S, Oishi N, Ogawa K, Kawakami Y, Ohira T, Yoshida K, Toda M. A VEGF receptor vaccine demonstrates preliminary efficacy in neurofibromatosis type 2. Nat Commun. 2019;10:5758. doi: 10.1038/s41467-019-13640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Tang WJ, Shi JB, Liu MM, Liu XH. Therapeutic strategies for targeting telomerase in cancer. Med Res Rev. 2019;40:532–585. doi: 10.1002/med.21626. [DOI] [PubMed] [Google Scholar]
- 20.Monje PV, Sant D, Wang G. Phenotypic and functional characteristics of human schwann cells as revealed by cell-based assays and RNA-SEQ. Mol Neurobiol. 2018;55:6637–6660. doi: 10.1007/s12035-017-0837-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshi N, Hiraki H, Yamaki T, Natsume T, Watanabe K, Suzuki T. Frequent expression of 75 kDa nerve growth factor receptor and phosphotyrosine in human peripheral nerve tumours: an immunohistochemical study on paraffin-embedded tissues. Virchows Arch. 1994;424:563–568. doi: 10.1007/BF00191444. [DOI] [PubMed] [Google Scholar]
- 22.Eldridge CF, Bunge MB, Bunge RP. Differentiation of axon-related Schwann cells in vitro: II. Control of myelin formation by basal lamina. J Neurosci. 1989;9:625–638. doi: 10.1523/JNEUROSCI.09-02-00625.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voltan R. p53 and merlin tumor suppressors: two of a kind. EBioMedicine. 2018;37:23–24. doi: 10.1016/j.ebiom.2018.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Xue L, Huang H, Wang H, Zhang X, Zhu W, Wang Z, Wang Z, Wu H. Synergistic effect of nutlin-3 combined with MG-132 on schwannoma cells through restoration of merlin and p53 tumour suppressors. EBioMedicine. 2018;36:252–265. doi: 10.1016/j.ebiom.2018.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.