Abstract
Hepatocellular carcinoma (HCC) is acknowledged to be a fatal malignant cancer around the world. Circular RNAs (circRNAs) function as crucial regulators in the pathological procession of HCC. Here, we elucidated the biological function of a novel circRNA, circNFIX, in HCC tumorigenesis. qRT-PCR was performed to determine the expressions of circNFIX, miR-3064-5p, and HMGA2. circNFIX stability was evaluated after treatment with ribonuclease R. The growth and invasion of HCC cells were assessed by CCK8 and transwell assays. Protein levels were measured by Western blotting. The levels of glutaminolysis metabolites were evaluated by commercial kits. Dual-luciferase report assay, RNA immunoprecipitation (RIP) assay and RNA pull-down assay were performed for validating the interaction between miR-3064-5p and circNFIX/HMGA2. Tumor growth in vivo was detected using xenograft assay. Our results showed that circNFIX was remarkably up-regulated in HCC and was associated with a poor survival. Knockdown of circNFIX repressed proliferation, invasion and glutaminolysis of HCC cells. Moreover, circNFIX directly sponged miR-3064-5p to release HMGA2 expression, and thus conferred the malignant development of HCC. In conclusion, circNFIX serves as a competing endogenous RNA to accelerate HCC progression via regulating miR-3064-5p/HMGA2 axis, suggesting a therapeutic strategy for HCC intervention.
Keywords: Hepatocellular carcinoma, circNFIX, miR-3064-5p, HMGA2, glutaminolysis
Introduction
Hepatocellular carcinoma (HCC) has been regarded as the most common malignancy all over the world [1]. In China, due to the frequent hepatitis B virus infection and significant morbidity of liver cirrhosis, HCC is considered as one of the deadliest cancers [2]. In spite of effective treatments that have been defined, the prognosis is still poor over the last couple of years because of recurrence and metastasis [3]. Therefore, uncovering the pathogenesis is urgently needed for the diagnosis and treatment of HCC.
Circular RNAs (circRNAs), with a closed loop structure, represent a kind of non-coding RNAs. As compared with linear RNAs, circRNAs have greater stability and can resist exonucleases-mediated digestion [4]. Recently, the regulation of circRNAs in tumorigenesis of multiple malignancies has been well documented. For example, circHECTD1 promoted gastric cancer development via sponging miR-1256 to enhance USP5 expression [5]. In HCC, hsa_circ_0004018 restrained malignant growth and metastasis via targeting miR-626/DKK3 axis to inhibit Wnt/β-catenin pathway [6]. CircNFIX (hsa_circ_0049658) has been verified to exert oncogenic roles in glioma [7] and non-small cell lung cancer [8]. However, the expression of circNFIX and its effect on HCC tumorigenesis have not been elucidated. Therefore, circNFIX is the focus in this study.
CircRNAs have been considered to control cancer development by acting as sponges for microRNAs (miRNAs) [9]. MiRNAs are another type of significant non-coding RNAs that repress gene expression via directly binding with 3’ untranslated region (3’-UTR) [10]. The regulation of miRNAs in carcinoma progression has been well accepted. miR-3064-5p expression was reduced in HCC, and an enforced expression of miR-3064-5p presented anti-angiogenic ability [11]. However, the upstream regulatory mechanisms of miR-3064-5p in HCC still need to be explored. Online databases predicted that circNFIX could bind to miR-3064-5p, thus we speculated that circNFIX might regulate HCC progression by targeting miR-3064-5p. Notably, bioinformatic analysis indicated that miR-3064-5p could bind to HMGA2. High mobility group A2 (HMGA2), an important transcription factor of high mobility group A family, regulates gene expression via modification of the chromatin architecture [12]. Mounting evidence has reported that dysregulation of HMGA2 participated in the development of a wide variety of tumors, including osteosarcoma [13], cutaneous squamous cell carcinoma [14], and lung adenocarcinoma [15]. HMGA2 has also been demonstrated to be up-regulated in HCC, which contributed to metastasis of HCC cells [16]. Therefore, the involvement of miR-3064-5p/HMGA2 axis in circNFIX-mediated biological functions in HCC needs to be elucidated.
In this study, the up-regulation of circNFIX was verified in HCC tissues. We further demonstrated that circHECTD1 facilitated HCC cell proliferation, invasion and glutaminolysis by targeting miR-3064-5p/HMGA2 axis, thus driving HCC progression. Our findings uncovered the biological roles of circRNA-miRNA-mRNA network in HCC, which may provide a novel therapeutic target for HCC.
Materials and methods
Clinical specimens
HCC and normal peritumor tissues (n=66) were obtained from He’nan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of He’nan University. All enrolled HCC patients signed written informed consent and did not receive radiotherapy or chemotherapy before surgery. This study was approved by the Ethics Committee of He’nan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of He’nan University (Approval No. Q/ZXYY-ZY-YWB-LL201905201).
Cell lines and culture
Human HCC cell line Hep3B and normal hepatocyte cell line THLE-2 were purchased from American Type Culture Collection (ATCC; Manassas, VA, USA), while HCC cell lines HuH7, MHCC97-H, and LM3 were obtained from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were maintained in high glucose dulbecco’s modified eagle medium (DMEM; BI, Hertzliya Pituach, Israel) with 10% fetal bovine serum (FBS) at 37°C with 5% CO2.
Quantitative reverse transcription PCR (qRT-PCR)
To obtain nuclear and cytoplasmic fractions of HCC cells, a PARIS kit (Thermo Fisher, Franklin Lakes, MA, USA) was selected. Trizol (Thermo Fisher, Franklin Lakes, MA, USA) was used for total RNA purification. cDNA was prepared using PrimeScript RT Master Mix (TaKaRa, Tokyo, Japan). Subsequently, qRT-PCR was performed using the SYBR Premix Ex Taq™ (TaKaRa) with the MyIQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) under the following conditions: an initial denaturation step for 10 min at 95°C and 40 amplification cycles comprising a denaturation step for 15 seconds at 95°C, an annealing step for 30 seconds at 60°C, and finally an elongation step for 30 seconds at 72°C. Relative RNA expression normalized to GAPDH or U6 was calculated by the 2-ΔΔCt. PCR primers are shown in Table 1.
Table 1.
Oligonucleotide primer sets for qPCR
| Name | Sequence (5’¬3’) | Length |
|---|---|---|
| circNFIX F | AGGAGATGCGGACATCAAAC | 20 |
| circNFIX R | GTGAAATACGGGCTCGACTG | 20 |
| NFIX F | TACCAGCAGCGTGTGATGAG | 20 |
| NFIX R | TGATGGTCAGCACGAAGTCC | 20 |
| miR-3064-5p F | CTGGCTGTTGTGGTGTGC | 18 |
| miR-3064-5p R | TGGTGTCGTGGAGTCG | 16 |
| U6 F | CTCGCTTCGGCAGCACA | 17 |
| U6 R | AACGCTTCACGAATTTGCGT | 20 |
| GAPDH F | AATCCCATCACCATCTTCC | 19 |
| GAPDH R | CATCACGCCACAGTTTCC | 18 |
Detection for circNFIX stability
Total RNA was incubated with ribonuclease R (RNase R) at 37°C for 1 h to evaluate circNFIX stability. After treatment, the expression of circNFIX and linear NFIX was assessed by qRT-PCR.
Cell transfection
Small interfering RNA (siRNA) for circNFIX (si-circNFIX), miR-3064-5p mimics and inhibitor were purchased from RIBOBIO (Guangzhou, China). HMGA2 overexpression plasmid was obtained from Sino Biological (Beijing, China). The above fragments were transfected into HCC cells using Lipofectamine 3000 (Thermo Fisher) according to the instructions [17].
Cell counting kit-8 (CCK-8)
The HCC cells inoculated in 96-well plates were added with 10 μL CCK-8 solution (Abcam, Cambridge, UK). After incubation for 2 h, cell viability was detected at 450 nm using a microplate reader (Hidex, Turku, Finland).
Transwell assay
The invasion and migration of HCC cells were determined using Transwell chambers coated with or without Matrigel (BD, Franklin Lakes, NJ, USA). In brief, the suspension of HCC cells (containing 1×105 cells) was added to the upper chamber, while medium containing 10% FBS was added to the lower chamber. The residual cells in the upper chamber were removed after 48 h. The invading or migratory cells were stained with crystal violet and counted. The number of stained cells was counted in five random fields under a light microscope (Olympus, Tokyo, Japan).
Western blotting
Protein samples were acquired using radioimmunoprecipitation buffer (BioTeke Corporation, Beijing, China) and adjusted to the same concentration. Then, separation using sodium dodecyl sulfate-polyacrylamide gel electrophoresis was carried out. Afterwards, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes, followed by probing with primary antibodies against Ki-67 (bs-23102R, 1:500, Bioss, Beijing, China), PCNA (bs-2007R, 1:1000, Bioss), ASCT2 (A6981, 1:1000, ABclonal, Wuhan, China), GLS1 (bs-10341R, 1:500, Bioss), HMGA2 (A2972, 1:500, ABclonal), and β-actin (bs-0061R, 1:5000, Bioss) overnight at 4°C. Subsequently, the secondary antibody (bs-0369M-HRP, 1:5000, Bioss) was applied. An enhanced chemiluminescence reagent (Engreen, Birkenhead, Auckland, New Zealand) was used for visualization of protein bands. The protein bands were quantified by densitometry using ImageJ software (NIH, Bethesda, MD, USA).
Xenograft transplantation in vivo
Female BALB/c nude mice (4 weeks old) were purchased from Slac Jingda Laboratory Animal Co., Ltd., Hunan, China. The HuH7 cells (1×107) stably transfected with sh-circNFIX or sh-NC were subcutaneously injected into the mice (n=6 per group). Tumor volume (mm3) calculated as (width)2 × length/2 was recorded every week. 5 weeks later, the mice were euthanized by pentobarbital overdose (210 mg/kg) and tumor weights were obtained. All animal procedures were approved by the Institutional Animal Care and Use Ethics Committee at the He’nan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, People’s Hospital of He’nan University and carried out according to the National Institutes of Health (NIH) guidelines for the ethical treatment of animals.
Prediction analysis
Online databases circAtlas (http://circatlas.biols.ac.cn/), circBank (http://www.circbank.cn/), and starBase (http://starbase.sysu.edu.cn/index.php) were adopted to predict the potential targets of circNFIX or miR-3064-5p.
Detection of glutamine, glutamate, and α-KG levels
The glutamine assay kit (Abcam), glutamate assay kit (Abcam), and alpha ketoglutarate (α-KG) assay kit (Abcam) were applied to detect glutamine, glutamate, and α-KG levels in HCC cells following the protocol booklets.
Dual-luciferase reporter assay
The wild type (WT) and mutant (MUT) sequences of circNFIX or HMGA2 3’-UTR were subcloned into pGL3 vector (Promega, Madison, WI, USA). HCC cells were co-transfected with the above constructed WT or MUT plasmids together with miR-3064-5p mimics or mimics NC. After transfection for 48 h, luciferase signals were detected using a Dual Luciferase Reporter Gene Assay Kit (Yeasen, Shanghai, China).
RNA pull-down
The lysates of HCC cells were incubated with the biotinylated circNFIX (bio-circNFIX) or bio-NC for 2 h. Then, streptavidin magnetic beads (Thermo Fisher) were used for capturing RNAs complexes, followed by incubation with washing buffer containing proteinase K. miR-3064-5p level from bound RNAs was assessed by qRT-PCR.
RNA immunoprecipitation (RIP)
Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA) was adopted for performing RIP assay. In short, HCC cells were subjected to RIP lysis buffer supplemented with protease and RNase inhibitors to prepare cell lysates. Incubation with magnetic beads with Ago-2 antibody or IgG antibody was carried out at 4°C overnight. Finally, the enrichment of circNFIX and miR-3064-5p in immunoprecipitated RNAs was evaluated by qRT-PCR.
Statistical analysis
The results are presented as mean ± standard deviation (x̅ ± sd). Student’s t test for two group comparison or one-way ANOVA followed by Turdey post hoc test for multiple group comparison was performed using GraphPad prism 8.0, and P<0.05 was set for significant difference.
Results
circNFIX was up-regulated in HCC
First, circNFIX expression in 66 pairs of HCC tissue and normal peritumor tissue was determined by qRT-PCR. The results indicated that circNFIX was highly expressed in HCC samples in comparison to peritumor samples (Figure 1A). Furthermore, HCC patients with high circNFIX level presented lower overall survival rate as compared to those with low circNFIX level (Figure 1B). In line with the increased expression in HCC tissues, circNFIX was up-regulated in HCC cell lines, compared with normal hepatocyte cell line THLE-2 (Figure 1C). Moreover, RNase R degradation assay revealed that RNase R treatment led to reduced expression of linear transcript NFIX, whereas circNFIX level was not changed (Figure 1D). In Hep3B and HuH7 cells, nuclear mass separation assay indicated that circNFIX was mainly located in the cytoplasm (Figure 1E).
Figure 1.
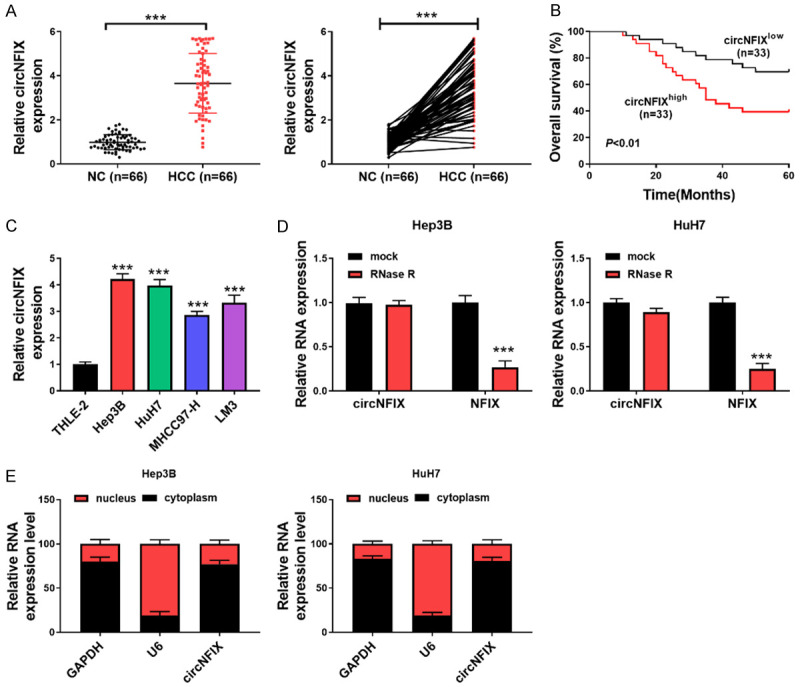
circNFIX was upregulated in HCC and indicates poor survival. A: circNFIX expression in 66 pairs of HCC and normal tissues was assessed by qRT-PCR; B: Overall survival of HCC patients with low or high expression of circNFIX; C: circNFIX expression in multiple cell lines was determined by qRT-PCR; D: qRT-PCR for evaluating expression of circNFIX and NFIX in HCC cells incubated with or without RNase R; E: circNFIX expression in nucleus or cytoplasm of HCC cells was detected by qRT-PCR. ***P<0.001.
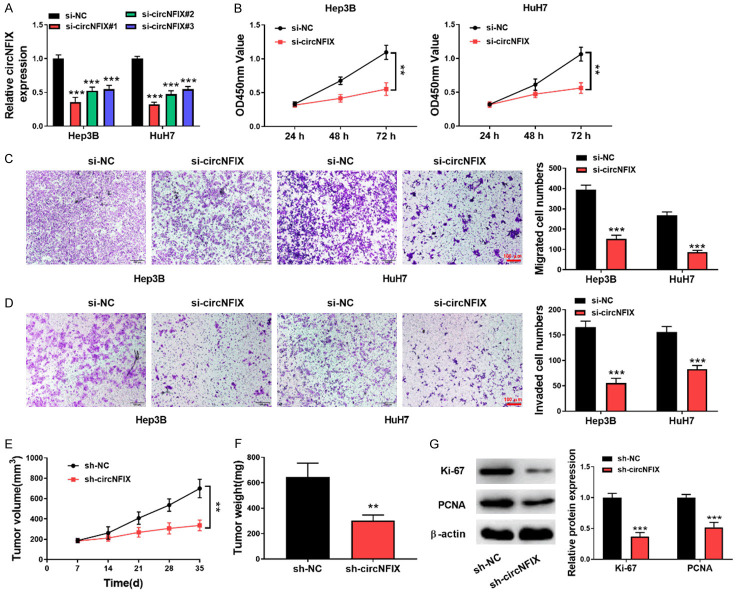
circNFIX depletion repressed HCC cell growth, migration and invasion
To address the function of circNFIX in HCC cell proliferation and metastasis, HCC cells (Hep3B and HuH7) with higher circNFIX expression were transfected with three si-RNAs targeting circNFIX. We found that circNFIX expression was remarkably decreased after transfection with all these three si-RNAs. The highest interference efficiency was found in si-circNFIX #1, which was selected in the subsequent experiments (Figure 2A). CCK8 assay showed that circNFIX silencing slowed down the proliferation of HCC cells (Figure 2B). Additionally, the migrative (Figure 2C) and invasive (Figure 2D) abilities were reduced in circNFIX-silenced HCC cells. Subsequently, the effect of circNFIX on HCC cell growth in vivo was evaluated in nude mice. As shown in Figure 2E, 2F, a smaller volume and weight of xenografts were observed after circNFIX depletion, as compared with sh-NC group. Western blotting analysis indicated that Ki-67 and PCNA protein levels in tumor tissues were reduced after knockdown of circNFIX (Figure 2G). Collectively, circNFIX down-regulation restrained the proliferation and motility of HCC cells.
Figure 2.
Down-regulation of circNFIX repressed HCC cell proliferation and motility. A: qRT-PCR for assessing circNFIX level in HCC cells transfected with si-circNFIX; B: HCC cell proliferation after circNFIX silencing was determined by CCK-8; C, D: The migration and invasion of HCC cells were evaluated by transwell assay (200×); E: The volume of xenograft tumors was measured once a week; F: Tumor weight was detected after the experiment; G: Protein levels of Ki-67 and PCNA in tumor tissues were assessed by Western blotting. **P<0.01; ***P<0.001.
circNFIX knockdown suppressed glutaminolysis of HCC cells
To further examine the regulation of circNFIX in glutaminolysis, a series of glutaminolysis metabolites were detected. As presented in Figure 3A-C, the levels of glutamine, glutamate and α-KG in HCC cells were evidently lowered by silencing circNFIX. In addition, the expression of glutaminolysis-related enzymes ASCT2 and GLS1 was assessed by Western blotting. ASCT2 and GLS1 are pivotal enzymes responsible for glutaminolysis [18,19]. The protein levels of ASCT2 and GLS1 were significantly down-regulated in circNFIX-depleted HCC cells (Figure 3D). Therefore, circNFIX depletion inhibited glutaminolysis of HCC cells.
Figure 3.
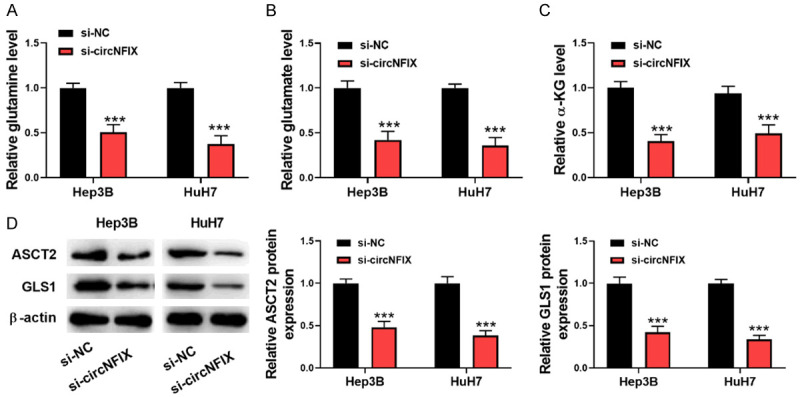
Silencing circNFIX restrains glutaminolysis of HCC cells. A-C: The levels of glutamine, glutamate, and α-KG in HCC cells after transfection with si-circNFIX were detected by commercial kits; D: Western blotting for determining ASCT2 and GLS1 levels in HCC cells. ***P<0.001.
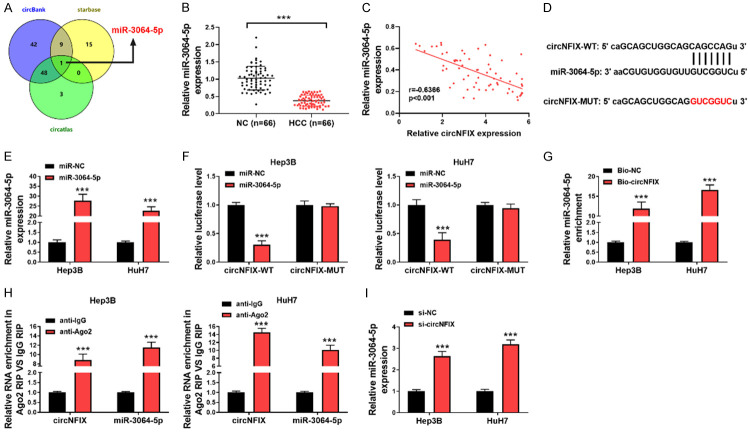
circNFIX binds to miR-3064-5p directly
To further investigate the downstream regulatory mechanism of circNFIX in HCC, three online databases including circBank, starBase and circAtlas were adopted to predict target miRNAs of circNFIX. As presented in Figure 4A, miR-3064-5p was the only candidate according to the intersection results. Moreover, miR-3064-5p was distinctly down-regulated in HCC tissues in comparison to the adjacent tissues (Figure 4B). Accordingly, a negative correlation between circNFIX and miR-3064-5p in HCC tissues was verified (Figure 4C). Figure 4D showed the putative binding sites for miR-3064-5p in circNFIX. Subsequently, miR-3064-5p overexpression strikingly restrained the luciferase activity of circNFIX-WT in HCC cells, but did not change that of circNFIX-MUT (Figure 4E, 4F). Furthermore, RNA pull-down assay revealed that Bio-circNFIX probe resulted in a higher enrichment of miR-3064-5p in HCC cells (Figure 4G). Additionally, RIP assay demonstrated that the enrichment of circNFIX and miR-3064-5p was significantly promoted in Ago2 group, as compared with IgG group (Figure 4H). More importantly, knockdown of circNFIX led to higher miR-3064-5p level in HCC cells (Figure 4I). These data suggested that miR-3064-5p was a direct target of circNFIX.
Figure 4.
circNFIX acted as a sponge for miR-3064-5p. A: Overlap of the predicted target miRNAs of circNFIX by circBank, starBase and circAtlas; B: qRT-PCR for detecting circNFIX level in HCC and normal tissues; C: Correlation analysis between circNFIX and miR-3064-5p expression in HCC tissues; D: Schematic illustration of the predicted complementary sequences for miR-3064-5p in circNFIX; E: Overexpression of miR-3064-5p in HCC cells was validated by qRT-PCR; F: Relative luciferase activity of HCC cells co-transfected with circNFIX-WT or MUT and miR-3064-5p mimics or mimics NC; G: RNA pull-down assay for evaluating the effect of miR-3064-5p enrichment on circNFIX; H: The recruitment of miR-3064-5p by circNFIX was confirmed by anti-Ago2 RIP assay; I: qRT-PCR for detecting miR-3064-5p expression in circNFIX-silence HCC cells. ***P<0.001.
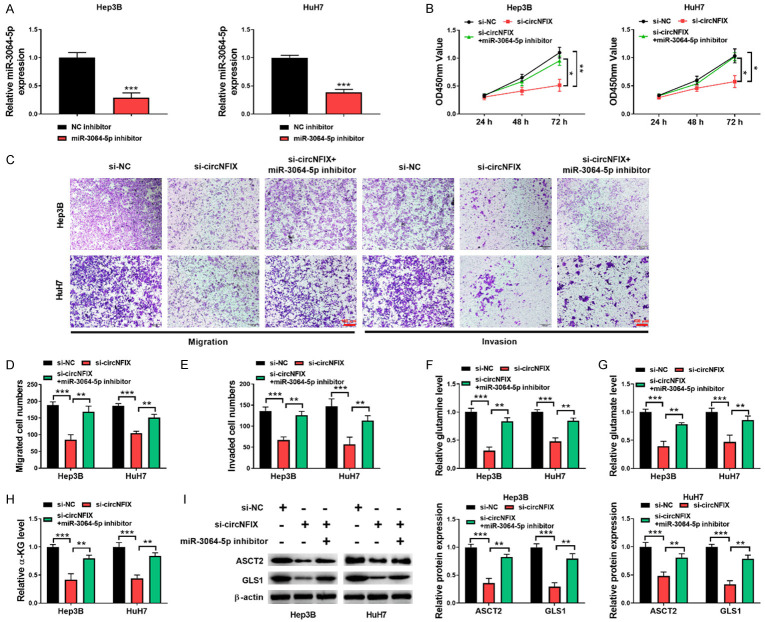
circNFIX knockdown inhibited HCC progression via targeting miR-3064-5p
Next, we determined whether circNFIX regulated HCC development via miR-3064-5p by performing multiple rescue experiments. For this purpose, circNFIX-silenced HCC cells were further transfected with miR-3064-5p inhibitor. The decreased expression of miR-3064-5p in HCC induced by miR-3064-5p inhibitor was confirmed by qRT-PCR (Figure 5A). Functional assays demonstrated that miR-3064-5p inhibitor reversed the inhibitory effect of circNFIX depletion on HCC cell growth (Figure 5B), migration and invasion (Figure 5C-E). Furthermore, inhibition of miR-3064-5p promoted glutaminolysis in circNFIX-silenced HCC cells as evidenced by up-regulating levels of glutaminolysis metabolites glutamine, glutamate and α-KG (Figure 5F-H). Consistently, the decreased ASCT2 and GLS1 levels mediated by si-circNFIX in HCC cells were restored by miR-3064-5p inhibitor (Figure 5I). Taken together, these observations indicated that circNFIX/miR-3064-5p axis participated in the progression of HCC.
Figure 5.
circNFIX depletion played tumor-suppressive roles by sponging miR-3064-5p. A: miR-3064-5p inhibitor-mediated decrease of miR-3064-5p expression was determined by qRT-PCR; B: HCC cell proliferation from different groups was assessed by CCK-8; C-E: The migrative and invasive abilities of HCC cells were detected by transwell assay (200×); F-H: The levels of glutamine, glutamate, and α-KG in HCC cells with various treatments were detected by commercial kits; I: Western blotting for evaluating ASCT2 and GLS1 levels in HCC cells. *P<0.05; **P<0.01; ***P<0.001.
HMGA2 is a downstream target of circNFIX/miR-3064-5p axis
We further explored the downstream effector of circNFIX/miR-3064-5p axis. By bioinformatic analysis, HMGA2 was predicted as the target gene of miR-3064-5p and their complementary sequences are shown (Figure 6A). Further Dual-luciferase assay demonstrated that HMGA2 was the direct target gene of miR-3064-5p in HCC cells (Figure 6B). As assessed by Western blotting, HMGA2 protein level was down-regulated by miR-3064-5p overexpression, while up-regulated by miR-3064-5p suppression (Figure 6C, 6D). Furthermore, silencing of circNFIX-mediated down-regulation of HMGA2 level was effectively counteracted by miR-3064-5p inhibitor (Figure 6E). These data suggested that circNFIX regulated HMGA2 expression by targeting miR-3064-5p in HCC cells.
Figure 6.
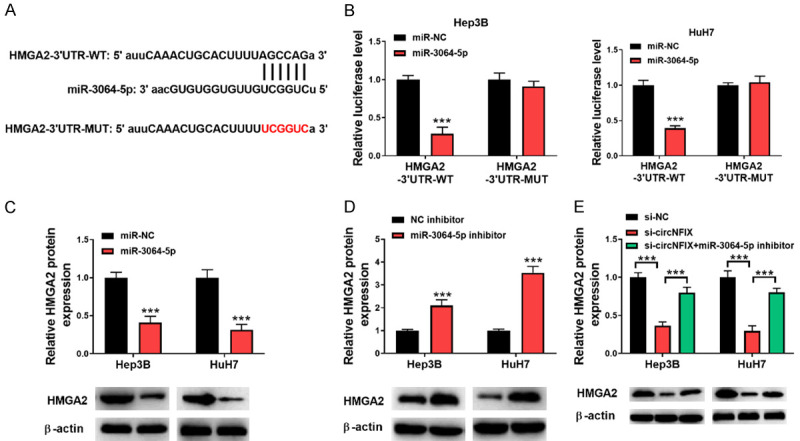
HMGA2 is a target gene of miR-3064-5p. A: HMGA2 3’-UTR containing the miR-3064-5p binding sites were illustrated; B: Relative luciferase activity of HCC cells co-transfected with HMGA2 3’-UTR-WT or MUT and miR-3064-5p mimics or mimics NC; C, D: Protein level of HMGA2 in HCC cells transfected with miR-3064-5p mimics or inhibitor was detected by Western blotting; E: Protein level of HMGA2 in HCC cells transfected with si-circNFIX or combination with miR-3064-5p inhibitor was determined by Western blotting. ***P<0.001.
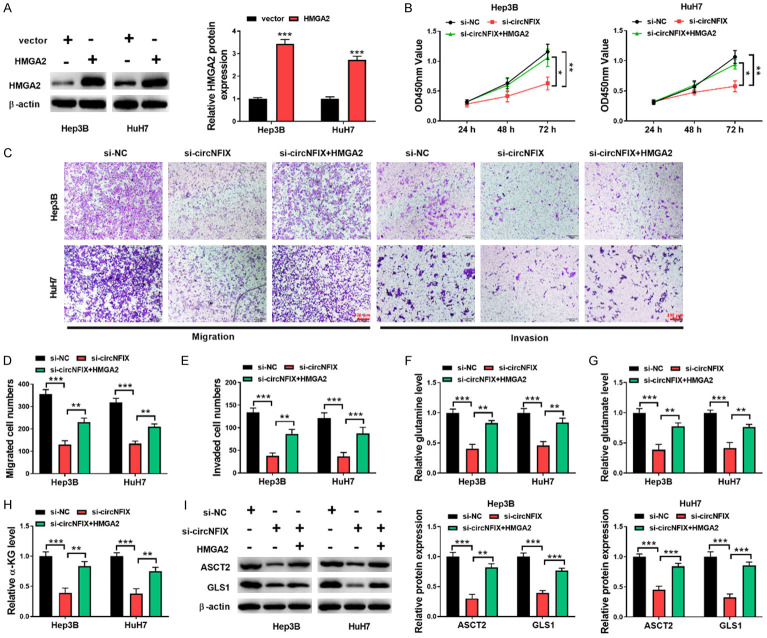
HMGA2 overexpression counteracted the anti-tumor effects of circNFIX silencing on HCC cells
Finally, rescue experiments were carried out to assess the contribution of HMGA2 to circNFIX/miR-3064-5p axis-mediated malignant process of HCC. As shown in Figure 7A, overexpression of HMGA2 in HCC cells was validated by Western blotting assay. In addition, si-circNFIX-induced decreases of proliferative, migratory, and invasive potencies of HCC cells were recovered by HMGA2 overexpression (Figure 7B-E). Moreover, enforced expression of HMGA2 reversed the reduced glutamine, glutamate, and α-KG levels in circNFIX-depleted HCC cells (Figure 7F-H). As expected, the declined expression of ASCT2 and GLS1 owing to circNFIX knockdown was abolished when HMGA2 was overexpressed in HCC cells (Figure 7I). These findings suggested that circNFIX/miR-3064-5p axis affected the growth, motility and glutaminolysis of HCC cells via HMGA2.
Figure 7.
circNFIX knockdown restrained HCC cell progression through the miR-3064-5p/HMGA2 axis. A: Overexpression of HMGA2 in HCC cells was verified by Western blotting assay; B: The proliferation was determined by CCK-8 assay; C-E: Cell migratory and invasive capacities were assessed by transwell assay (200×); F-H: The levels of glutamine, glutamate, and α-KGin in HCC cells were measured by commercial kits; I: Western blotting for assessing ASCT2 and GLS1 levels in HCC cells. *P<0.05; **P<0.01; ***P<0.001.
Discussion
HCC carries a really poor prognosis. A better interpretation of the pathological mechanisms of HCC tumorigenesis may be helpful for seeking effective therapeutic targets. It has been accepted that circRNAs can affect their target gene expression at transcriptional or post-transcriptional level and thus play pivotal biological functions in multiple diseases [20]. Mounting studies have reported that the dysregulation of circRNAs is implicated in the occurrence and development of HCC [21,22]. In this study, a novel circRNA, circNFIX, was identified to be up-regulated in HCC tissues and cells. According to the results, Hep3B and HuH7 cells presented the highest circNFIX expression level among a series of HCC cells. Therefore, Hep3B and HuH7 cells were chosen for transfection with si-circNFIX and adopted in subsequent experiments. Moreover, knockdown of circNFIX repressed glutaminolysis to inhibit the proliferation and metastasis of HCC cells. Mechanistically, circNFIX was demonstrated to interact with miR-3064-5p to enhance HMGA2 expression. These findings implied that circNFIX might function as an oncogene to facilitate malignancy of HCC.
Glutaminolysis has been recognized as an important hallmark in tumor-cell metabolism, which satisfies the bioenergetic needs for the survival and growth of cancer cells [23,24]. The regulation of circRNAs in glutaminolysis during cancer development has been reported. For example, circHMGCS1 has been shown to promote the growth of hepatoblastoma cells via enhancing glutaminolysis [25]. Li et al. reveal that circGSK3B facilitated glutamine metabolism, which accelerated HCC progression [26]. In line with these studies, for the first time, this study also validated the regulation of circNFIX in glutaminolysis during HCC progression. We found that circNFIX reduced the levels of glutaminolysis metabolites and suppressed the expression of ASCT2 and GLS1 enzymes in HCC cells. Thus, glutaminolysis was involved in circNFIX-mediated HCC progression.
It has been widely known that circRNAs, distributed in cytoplasm, can act as sponge of miRNA to affect miRNA level. In human cancers, circRNAs may affect gene expression via sequestering miRNAs, possessing carcinogenic or anti-cancer roles [27]. For instance, Xu et al. suggested that circRNA_0000392 conferred tumorigenesis of colorectal cancer by serving as a sponge of miR-193a-5p [28]. A previous study indicated that circEYA1 drove cervical adenocarcinoma progression by sponging miR-582-3p [29]. In the present study, we demonstrated that circNFIX regulated the progression and glutaminolysis of HCC by targeting miR-3064-5p. The anti-tumor effects of miR-3064-5 have been confirmed in various cancers. As reported, miR-3064-5p inhibition reversed the anti-tumor action of si-circCOL6A3 in gastric cancer cells [30]. In addition, miR-3064-5p inhibition was involved in exosomal circPRRX1-mediated doxorubicin resistance during the advancement of gastric cancer [31]. In HCC, miR-3064-5p was reported to suppress angiogenesis and may be a target for HCC therapy [11]. Our results further verified the function of miR-3064-5p in HCC and showed that inhibition of miR-3064-5p overturned the suppressive effects of si-circNFIX on HCC cell proliferation, migration, invasion and glutaminolysis. Therefore, we concluded that circNFIX depletion slowed down HCC progression by up-regulating miR-3064-5p.
HMGA2 is invariably up-regulated in malignant tumor, which confers the development of many kinds of cancers, such as breast cancer [32], pancreatic cancer [33], colorectal cancer, and so on [34]. Hence, inhibition of HMGA2 may be a crucial therapeutic approach for cancer therapy. Zhao et al. revealed that inhibition of HMGA2 by siRNA-lipoplexes could suppress gastrointestinal cancer cell proliferation and motility [35]. Additionally, enhanced HMGA2 level was reported to correlate with poor prognosis of liver cancer cases [36]. Besides, a previous study showed that panobinostat-mediated HMGA2 suppression could restrain liver cancer cell growth in vitro and in vivo [37]. Our data first demonstrated that HMGA2 was a downstream target gene of miR-3064-5p, and HMGA2 protein level was enhanced by circNFIX-mediated miR-3064-5p inhibition in HCC cells. We also revealed that overexpression of HMGA2 abolished the inhibitory effect of circNFIX depletion on HCC cell malignant progression. Therefore, circNFIX acted as a regulator of HMGA2, and circNFIX/miR-3064-5p/HMGA2 axis played a pivotal role in HCC.
We are aware the fact that there are some limitations in this study. First, the small sample size may result in the statistical limitation of the study. Secondy, circNFIX can exert its function via ceRNA network, and this study only investigated the role of circNFIX/miR-3064-5p/HMGA2 axis. In the future study, more potential target miRNAs of circNFIX and their roles in HCC need to be explored. Also, future research needs to base upon large sample size.
Taken together, our findings suggested that the aberrant up-regulation of circNFIX in HCC tissues and cells could be a biomarker for HCC. In addition, down-regulation of circNFIX restrained HCC malignant progression via regulating miR-3064-5p/HMGA2 axis, uncovering a new mechanism for HCC development (Figure 8). We suggest that circNFIX may be an effective therapeutic target for HCC patients in the future.
Figure 8.
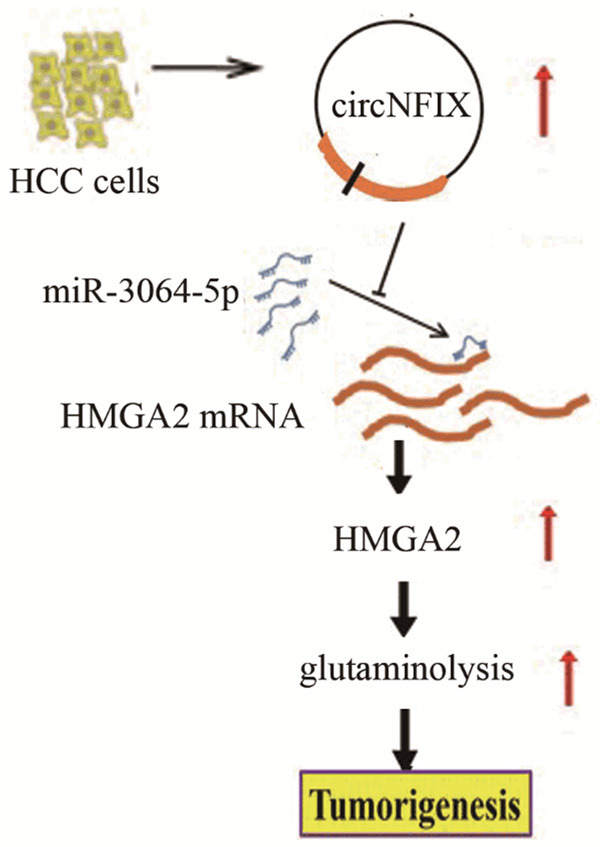
Schematic representation of the proposed mechanism of circNFIX in HCC cells. circNFIX acted as a miR-3064-5p sponge to enhance HMGA2 expression in HCC cells, which facilitated glutaminolysis, thereby promoting HCC tumorigenesis and progression.
Disclosure of conflict of interest
None.
References
- 1.Abouzied MM, Eltahir HM, Abdel Aziz MA, Ahmed NS, Abd El-Ghany AA, Abd El-Aziz EA, Abd El-Aziz HO. Curcumin ameliorate DENA-induced HCC via modulating TGF-beta, AKT, and caspase-3 expression in experimental rat model. Tumour Biol. 2015;36:1763–1771. doi: 10.1007/s13277-014-2778-z. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 5.Cai J, Chen Z, Wang J, Chen X, Liang L, Huang M, Zhang Z, Zuo X. circHECTD1 facilitates glutaminolysis to promote gastric cancer progression by targeting mir-1256 and activating beta-catenin/C-Myc signaling. Cell Death Dis. 2019;10:576. doi: 10.1038/s41419-019-1814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu P, Liang H, Huang X, Zeng Q, Liu Y, Lv J, Ming L. Circular RNA Hsa_circ_0004018 inhibits wnt/beta-catenin signaling pathway by targeting microRNA-626/DKK3 in hepatocellular carcinoma. Onco Targets Ther. 2020;13:9351–9364. doi: 10.2147/OTT.S254997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding C, Wu Z, You H, Ge H, Zheng S, Lin Y, Wu X, Lin Z, Kang D. CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J Exp Clin Cancer Res. 2019;38:506. doi: 10.1186/s13046-019-1483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu J, Zhu Y, Qin Y, Chen Y. CircNFIX acts as a miR-212-3p sponge to enhance the malignant progression of non-small cell lung cancer by up-regulating ADAM10. Cancer Manag Res. 2020;12:9577–9587. doi: 10.2147/CMAR.S272309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, Wei F, Guo C, Wu X, Li X, Li Y, Li G, Zeng Z, Xiong W. Circular RNAs function as cernas to regulate and control human cancer progression. Mol Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, Iliopoulos D. An HNF4alpha-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang P, Ha M, Li L, Huang X, Liu C. MicroRNA-3064-5p sponged by MALAT1 suppresses angiogenesis in human hepatocellular carcinoma by targeting the FOXA1/CD24/Src pathway. FASEB J. 2020;34:66–81. doi: 10.1096/fj.201901834R. [DOI] [PubMed] [Google Scholar]
- 12.Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R, Pierantoni GM, Outwater E, Santoro M, Croce CM, Fusco A. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene. 2002;21:3190–3198. doi: 10.1038/sj.onc.1205428. [DOI] [PubMed] [Google Scholar]
- 13.Zhong J, He C, Xu F, Xu X, Liu L, Xu M, Guo Z, Wang Y, Liao J, Li Y. Lupeol inhibits osteosarcoma progression by up-regulation of HMGA2 via regulating miR-212-3p. J Orthop Surg Res. 2020;15:374. doi: 10.1186/s13018-020-01879-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha W, Hinde A, Xie L, Trager MH, Liu L. Biomarker function of HMGA2 in ultraviolet-induced skin cancer development. Exp Dermatol. 2020;29:1021–1026. doi: 10.1111/exd.14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu L, Ma Y, Zhang H, Lu QJ, Yang L, Jiang GN, Liao WL. HMGA2 regulates circular RNA ASPH to promote tumor growth in lung adenocarcinoma. Cell Death Dis. 2020;11:593. doi: 10.1038/s41419-020-2726-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Y, Li W, Liao H. HMGA2 induces epithelial-to-mesenchymal transition in human hepatocellular carcinoma cells. Oncol Lett. 2013;5:1353–1356. doi: 10.3892/ol.2013.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging Mir-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N, Harvey K, Beith JM, Selinger CI, O’Toole SA, Rasko JE, Holst J. ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple-negative basal-like breast cancer. Oncogene. 2016;35:3201–3208. doi: 10.1038/onc.2015.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katt WP, Cerione RA. Glutaminase regulation in cancer cells: a druggable chain of events. Drug Discov Today. 2014;19:450–457. doi: 10.1016/j.drudis.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 21.Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, Guo J. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8:43878–43888. doi: 10.18632/oncotarget.16709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang G, Liang M, Liu H, Huang J, Li P, Wang C, Zhang Y, Lin Y, Jiang X. CircRNA hsa_circRNA_104348 promotes hepatocellular carcinoma progression through modulating miR-187-3p/RTKN2 axis and activating Wnt/Beta-catenin pathway. Cell Death Dis. 2020;11:1065. doi: 10.1038/s41419-020-03276-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah R, Chen S. Metabolic signaling cascades prompted by glutaminolysis in cancer. Cancers (Basel) 2020;12:2624. doi: 10.3390/cancers12092624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhen N, Gu S, Ma J, Zhu J, Yin M, Xu M, Wang J, Huang N, Cui Z, Bian Z, Sun F, Pan Q. CircHMGCS1 promotes hepatoblastoma cell proliferation by regulating the IGF signaling pathway and glutaminolysis. Theranostics. 2019;9:900–919. doi: 10.7150/thno.29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li K, Cao J, Zhang Z, Chen K, Ma T, Yang W, Yang S, Rao J, Zhang K. Circular RNA circGSK3B promotes cell proliferation, migration, and invasion by sponging mir-1265 and regulating CAB39 expression in hepatocellular carcinoma. Front Oncol. 2020;10:598256. doi: 10.3389/fonc.2020.598256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visci G, Tolomeo D, Agostini A, Traversa D, Macchia G, Storlazzi CT. CircRNAs and fusion-circRNAs in cancer: new players in an old game. Cell Signal. 2020;75:109747. doi: 10.1016/j.cellsig.2020.109747. [DOI] [PubMed] [Google Scholar]
- 28.Xu H, Liu Y, Cheng P, Wang C, Zhou W, Xu Y, Ji G. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39:283. doi: 10.1186/s13046-020-01799-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Zhang Y, Huang Y, Dong X, Xiang Z, Zou J, Wu L, Lu W. circEYA1 functions as a sponge of miR-582-3p to suppress cervical adenocarcinoma tumorigenesis via upregulating CXCL14. Mol Ther Nucleic Acids. 2020;22:1176–1190. doi: 10.1016/j.omtn.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun X, Zhang X, Zhai H, Zhang D, Ma S. A circular RNA derived from COL6A3 functions as a cerna in gastric cancer development. Biochem Biophys Res Commun. 2019;515:16–23. doi: 10.1016/j.bbrc.2019.05.079. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, Ping M, Song B, Guo Y, Li Y, Jia J. Exosomal CircPRRX1 enhances doxorubicin resistance in gastric cancer by regulating MiR-3064-5p/PTPN14 signaling. Yonsei Med J. 2020;61:750–761. doi: 10.3349/ymj.2020.61.9.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu J, Fang X, Long L, Wang S, Qian S, Lyu J. HMGA2 promotes breast cancer metastasis by modulating hippo-YAP signaling pathway. Cancer Biol Ther. 2021;22:5–11. doi: 10.1080/15384047.2020.1832429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo HH, Wang YZ, Zhang ZK, Li MZ, Tian XD, Yang YM. High mobility group AT-Hook 2 promotes tumorigenicity of pancreatic cancer cells via upregulating ANLN. Exp Cell Res. 2020;393:112088. doi: 10.1016/j.yexcr.2020.112088. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Wang J, Wu J. Emerging roles for HMGA2 in colorectal cancer. Transl Oncol. 2020;14:100894. doi: 10.1016/j.tranon.2020.100894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohammadi A, Mansoori B, Savadi P, Khaze V, Minouei M, McMillan NAJ, Hallaj-Nezhadi S, Baradaran B. Targeting of high mobility group A2 by small interfering RNA-loaded nanoliposome-induced apoptosis and migration inhibition in gastrointestinal cancer cells. J Cell Biochem. 2019;120:9203–9212. doi: 10.1002/jcb.28196. [DOI] [PubMed] [Google Scholar]
- 36.Zhao YC, Jiao Y, Li YQ, Fu Z, Yang ZY, He M. Elevated high mobility group A2 expression in liver cancer predicts poor patient survival. Rev Esp Enferm Dig. 2020;112:27–33. doi: 10.17235/reed.2019.6365/2019. [DOI] [PubMed] [Google Scholar]
- 37.Di Fazio P, Montalbano R, Neureiter D, Alinger B, Schmidt A, Merkel AL, Quint K, Ocker M. Downregulation of HMGA2 by the pan-deacetylase inhibitor panobinostat is dependent on Hsa-Let-7b expression in liver cancer cell lines. Exp Cell Res. 2012;318:1832–1843. doi: 10.1016/j.yexcr.2012.04.018. [DOI] [PubMed] [Google Scholar]