Abstract
Background: Hypoxia/reoxygenation (H/R)-mediated apoptosis and inflammation are major causes of tissue injury in acute myocardial infarction (AMI). Exploring the underlying mechanisms of cardiomyocyte injury induced by H/R is important for AMI treatment. Circular RNAs have been demonstrated to paly vital roles in the pathogenesis of AMI. Our study aimed to explore the function of circular RNA UBXN7 (circUBXN7) in regulating H/R-induced cardiomyocyte injury. Methods: H/R-treated H9c2 cells and a mouse model of AMI were used to investigate the function of circUBXN7 in H/R damage and AMI. The expressions of circUNXN7, miR-622 and MCL1 were analyzed by RT-qPCR. CCK-8 was used for examining cell viability. Cell apoptosis was evaluated with caspase 3 activity and Annexin V/PI staining. MCL1, Bax, Bcl-2 and cleaved-caspase 3 were examined with western blot. ELISA was used to examine the secretion of IL-6, TNF-α and IL-1β. Results: CircUBXN7 was downregulated in patients and mice with AMI, as well as in H/R-treated cells. Overexpression of circUBXN7 mitigated H/R-mediated apoptosis and secretion of inflammatory factors including IL-6, TNF-α and IL-1β. CircUBXN7 suppressed cell apoptosis and inflammatory reaction induced by H/R via targeting miR-622. MiR-622 targeted MCL1 to restrain its expression in H9c2 cells. Knockdown of MCL1 abrogated circUBXN7-mediated alleviation of apoptosis and inflammation after H/R treatment. Conclusion: CircUBXN7 mitigates cardiomyocyte apoptosis and inflammatory reaction in H/R injury by targeting miR-622 and maintaining MCL1 expression. Our study provides novel potential therapeutic targets for AMI treatment.
Keywords: circular RNA UBXN7, hypoxia/reoxygenation, apoptosis, inflammatory response, miR-622-MCL1 axis
Introduction
Acute myocardial infarction (AMI) is a common and fatal cardiovascular disease worldwide [1,2]. AMI is caused by the necrosis of myocardial cells in response to prolonged ischemia which commonly occurs when the coronary arteries are blocked by thrombus formation, leading to tissue injury [3,4]. Reperfusion of infarcted coronary arteries restores blood flow and oxygen supply and is the most common and effective treatment for AMI, dramatically reducing the mortality rate in recent years [5]. However, reperfusion therapy also causes further cellular and tissue injury, a condition called ischemia/reperfusion (I/R) or hypoxia/reoxygenation (H/R) injury [6,7]. H/R-induced myocardial cell damage mainly triggers apoptosis and inflammatory response [8]. Therefore, exploring the regulatory mechanisms underlying myocardial cell apoptosis and inflammatory reaction in H/R damage is important for understanding the pathogenesis of AMI and developing novel therapeutic targets for the management of H/R injury and AMI.
Non-coding RNAs, such as microRNAs (miRNAs) and circular RNAs (circRNAs), have been demonstrated to exert important functions in regulating cardiac cell apoptosis and inflammatory reaction in H/R injury [9,10]. Zhang and colleagues reported that suppression of miR-92a attenuated H/R-induced myocardiocyte apoptosis through increasing Smad7 expression [11]. CircRNAs are a class of covalently closed non-coding RNAs, which play important roles in physiological and pathological conditions including apoptosis, proliferation, cancers and cardiovascular diseases. Mechanistically, circRNAs act as miRNA sponges to reduce miRNA abundance and thus rescue miRNA-mediated suppression of downstream targets [12-14]. In recent years, the roles of circRNAs in the regulation of H/R damage and myocardial infarction have been increasingly elucidated. Geng et al. demonstrated that circRNA Cdr1as enhanced myocardial cell apoptosis and myocardial infarction through sponging miR-7a and subsequently suppressing the expression of PARP and SP1 [15]. CircMACF1 was reported to alleviate myocardial apoptosis and AMI by functioning as a miR-500b-5p sponge and increasing EMP1 expression [16]. Liu and colleagues found that circUBXN7 worked as a miR-1247-3p sponge to upregulate B4GALT3, thereby suppressing cell proliferation, invasion and growth in bladder cancer [17]. The roles and underlying regulatory mechanisms of circUBXN7 in H/R injury and AMI are still unknown.
Myeloid cell leukemia 1 (MCL1) belongs to the anti-apoptotic Bcl-2 family and plays central roles in regulating cell apoptosis. It has been demonstrated that MCL1 suppressed cell apoptosis by sequestering pro-apoptotic Bak or interfering with mitochondrial activation [18,19]. Importantly, MCL1 is highly expressed in myocardium, and loss of MCL1 caused mitochondrial dysfunction in myocytes and accelerated severe heart failure [20]. Although the expression of MCL1 is high in the heart, its roles in H/R injury and myocardial infarction have not been reported yet. In this study, we investigated the function of circUBXN7 in H/R-induced myocardiocyte apoptosis and inflammatory response, as well as the underlying regulatory mechanisms, in order to provide potential therapeutic targets for the treatment of H/R injury and AMI.
Materials and methods
Cell culture and treatment
Rat H9c2 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in Dulbecco’s Modified Eagle’s medium (ThermoFisher, Waltham, MA, USA) containing 10% fetal bovine serum (Sigma, St. Louis, MO, USA). Actinomycin D, a widely used inhibitor of transcription, was used to treat cells for inhibiting transcription, and the stability and half-life of CircUBXN7 and linear UBXN7 mRNA were examined. For actinomycin D treatment, H9c2 cells were plated and treated with actinomycin D (Sigma) at 2 µg/mL for 0, 4, 8 and 12 hours. Cells were harvested and RNA was extracted for assessing the stability of circUBXN7 and linear UBXN7 mRNA. For H/R treatment, cells were kept under a hypoxic condition (95% N2 and 5% CO2) for 2 hours and then reoxygenated for 12 hours.
Cell transfection
CircUBXN7 was cloned into pcDNA 3.1 vectors for its overexpression. MiR-622 mimics and siRNA against MCL1 (si-MCL1) were obtained from RiboBio (Guangzhou, China). Empty pcDNA 3.1 vectors, si-NC and miR-NC were used as controls. H9c2 cells were transfected with circUBXN7, pcDNA3.1 mock vector, pcDNA3.1 mock vector+miR-NC, circUBXN7+miR-NC, circUBXN7+miR-622 mimics, si-NC, si-MCL1, pcDNA3.1 vector+si-NC, circUBXN7+si-NC or circUBXN7+si-MCL1 using Lipo 2000 reagents (ThermoFisher) following the instructions. After 48 hours, cells were harvested for subsequent experiments.
CircRNA expression data from GSE160717
The circRNA expression data from three AMI patients and three normal controls were downloaded from Gene Expression Omnibus (GEO) Database with accession number GSE160717 [21]. Differentially expressed circRNAs between AMI patients and controls were analyzed with a volcano plot (Padj < 0.05). The clinical information of patients and normal controls was summarized in the previous publication [21].
Real-time quantitative PCR (RT-qPCR)
Total RNA was extracted from mouse myocardial tissues and H9c2 cells using TRI Reagent (Sigma). RNA was quantified and reversely transcribed into cDNA. For miRNAs, miRNAs were extracted using the miRcute miRNA kit (TIANGEN, Beijing, China), and the miRcute miRNA First-strand cDNA synthesis kit (TIANGEN) was used to synthesize the first-strand cDNA. Subsequently, the relative expressions of circUBXN7, UBXN7, miR-622 and MCL1 were examined using SYBR green dye (Toyobo, Osaka, Japan) by real-time quantitative PCR. The quantitative PCR reaction system (20 µL) included SYBR green dye (10 µL), ddH2O (6.5 µL), cDNA template (3 µL) and paired primers (0.5 µL, final concentration: 0.3 µM). The quantitative PCR conditions were as follows: initial denaturation at 95°C for 3 min; 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and elongation at 72°C for 15 s. The final cycle was followed by extension at 72°C for 3 min. 2-ΔΔCt method was used for calculation. RT-qPCR primers used in our study are shown in Table 1.
Table 1.
RT-qPCR primers in this study
| Genes | Sequence |
|---|---|
| circUBXN7 | 5’-CCACCCATTGATTTGATGC-3’ |
| 5’-CCGTCGTCTTTTAGGAGCAC-3’ | |
| Linear UBXN7 | 5’-GGATAGCCGCTCAGATGAAG-3’ |
| 5’-TTTGTGGGGAGACTTTCTGG-3’ | |
| MCL1 | 5’-TGCTTCGGAAACTGGACATTAAA-3’ |
| 5’-ATGGGTCATCACTCGAGAAAAAG-3’ | |
| U6 | 5’-AGAGAAGATTAGCATGGCCCCTG-3’ |
| 5’-AGTGCAGGGTCCGAGGTATT-3’ | |
| GAPDH | 5’-CAGGAGGCATTGCTGATGAT-3’ |
| 5’-GAAGGCTGGGGCTCATTT-3’ |
RNase R digestion
5 µg of total RNA was digested with RNase R (Sigma) at 4 U/µg for 20 minutes at 37°C. Subsequently, RNA was purified, and the abundance of circUBXN7 and linear UBXN7 mRNA were examined with RT-qPCR.
A mouse model of AMI
The mouse model of AMI was established as previously described [22]. Briefly, 16 male C57BJ/6L mice (8 to 10-week old) were obtained from animal center of Fuwai Central China Cardiovascular Hospital, Heart Center of He’nan Provincial People’s Hospital and divided into two groups: sham and AMI. After being anesthetized by inhaling 2% isoflurane, the hearts of mice were exposed, and the left anterior descending coronary artery (LAD) was located and ligated using a suture. The AMI was determined by the ST elevation with electrocardiography. Sham mice received the same procedure except ligation. Mice were kept in original cages and euthanatized by adjusting CO2 flow into cages at 3 L/min until death. Subsequently, the hearts were quickly excised and stored in liquid nitrogen for further RT-qPCR analysis. Animal procedures in this study were conducted following the National Institutes of Health guidelines, which were approved by the Institutional Animal Care and Use Committee of Fuwai Central China Cardiovascular Hospital, Heart Center of He’nan Provincial People’s Hospital.
Dual-luciferase reporter assay
Luciferase reporters were constructed via cloning wildtype (WT) or mutant (MUT) potential binding sites for miR-622 in circUBXN7 or the 3’UTR of MCL1 into the pmirGLO vectors (Promega, Madison, WI, USA). H9c2 cells were co-transfected with miR-622 mimics and WT or MUT luciferase reporters (circUBXN7-WT, circUBXN7-MUT, MCL1-3’UTR-WT and MCL1-3’UTR-MUT). NC mimic (miR-NC) was used as the control. After 48 hours, cells were harvested, and the activity of luciferase was assessed using the Dual-Glo system (Promega).
Cell counting kit-8 (CCK-8) assay
CCK-8 was ordered from Dojindo (Rockville, MD, USA) and used for analyzing cell viability following the manual. In brief, H9c2 cells were transfected as indicated and treated with H/R. Untransfected and untreated cells were used as controls. After H/R treatment, culture media were replaced. CCK-8 regent was added, and cells were subsequently incubated for 4 hours. The absorbance at 450 nm was recorded.
Cell apoptosis analysis
Annexin V-FITC/PI kit was obtained from Solarbio (Beijing, China). H9c2 cells were transfected as indicated and treated with H/R. Untransfected and untreated cells were used as controls. After H/R treatment, H9c2 cells were suspended and stained with FITC-Annexin V and PI for 20 minutes. 400 µL of phosphate buffered saline was added, and cells were analyzed using a flow cytometer in an hour.
Enzyme-linked immunosorbent assay (ELISA)
H9c2 cells were transfected as indicated and treated with H/R. Untransfected and untreated cells were used as controls. After H/R treatment, culture supernatants were harvested, and the concentrations of IL-6, TNF-α and IL-1β were examined using commercial kits following the manufacturer’s recommendation. Rat IL-1 beta and IL-6 ELISA kits were obtained from Abcam (Cambridge, UK). Rat TNF-alpha ELISA was purchased from RayBiotech Life (Peachtree Corners, GA, USA).
Caspase 3 activity
H9c2 cells were transfected as indicated and treated with H/R. Untransfected and untreated cells were used as controls. After H/R treatment, cells were pelleted and lysed on ice for 15 minutes, and the supernatants were collected after centrifugation. Reaction buffer and DEVD-pNA substrate were added directly into each sample and incubated for 2 hours. The absorbance at 405 nm was recorded. Caspase-3 Activity Assay Kit (Colorimetric) was bought from Novus Biologicals (Centennial, CO, USA).
RNA pull-down assay
The interaction between circUBXN7 and miR-622 or miR-3140-3p in H9c2 cells was determined by RNA pull-down using a magnetic RNA-Protein pull-down kit (ThermoFisher). Cells were lysed, and the supernatants were collected. Then, biotin-conjugated circUBXN7 probes were mixed with the supernatants and incubated for 4 hours at 4°C with rotation. Samples were subsequently mixed with streptavidin-conjugated magnetic beads and incubated at 4°C for 2 hours with rotation. RNA was recovered from the complex pulled down by beads, and the level of miR-622 and miR-3140-3p was examined by RT-qPCR.
Western blot
H9c2 cells were transfected as indicated and treated with H/R. Untransfected and untreated cells were used as controls. H9c2 cells were lysed, and the supernatants were harvested prior to protein quantification with the BCA kit (ThermoFisher). 40 μg of protein was loaded, electrophoresed and transferred to PVDF membranes (GE, Waukesha, WI, USA). After block, membranes were incubated with primary antibodies against cleaved-caspase 3 (1:500, Abcam), Bcl-2 (1:800, Abcam), Bax (1:2000, Abcam), MCL1 (1:800, ThermoFisher) and GAPDH (1:5000, SantaCruz, Dallas, TX, USA) for 12 hours. Next day, membranes were rinsed and incubated with HRP-conjugated secondary antibodies (Abcam, 1:5000) for 1 hour. Subsequently, signals were visualized with ECL substrates (Bio-Rad, Hercules, CA, USA). The ImageJ software was used to analyze the intensity of bands.
Statistical analysis
Results were from three independent assays and shown as mean ± standard deviation. The variance of multiple groups was analyzed with one-way analysis of variance (ANOVA). Comparisons of means between two groups were performed by Tukey’s post hoc test. ’A Venn diagram was applied to predict potential targets of circUBXN7. P < 0.05 was statistically significant.
Results
CircUBXN7 was downregulated in AMI and H/R-treated H9c2 cells
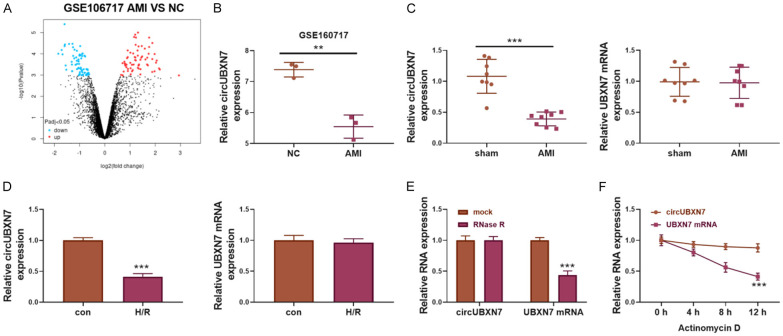
To investigate the roles of circRNAs in AMI, differentially expressed circRNAs between AMI patients and controls were analyzed with a circRNA array (GEO accession: GSE160717, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160717, Figure 1A). AMI patients showed reduced expression of circUBXN7 compared to normal controls (Figure 1B). We then established a mouse model of AMI and found that circUBXN7 expression was decreased in mice with AMI (Figure 1C). The reduced expression of circUBXN7 was also observed in H/R-treated H9c2 cells, a cell model of myocardial H/R injury (Figure 1D). However, the expression of linear UBXN7 was not affected in AMI mice and H/R-treated cells (Figure 1C, 1D). As RNase R can digest linear RNAs but not circular RNAs due to its exoribonuclease activity [23], we treated total RNA with RNase R or mock. The RT-qPCR assay showed that circUBXN7, but not linear UBXN7, was resistant to RNase R treatment (Figure 1E). Furthermore, circUBXN7 was stable in response to actinomycin D treatment with a half-life over 12 hours compared to the shorter half-life of linear UBXN7 (Figure 1F). These results demonstrated that circUBXN7 was a circular RNA, and it was downregulated in mice with AMI and H/R-treated cardiomyocytes, suggesting that circUBXN7 might be implicated in H/R damage and AMI.
Figure 1.
CircUBXN7 was downregulated in patients and mice with AMI and H/R-treated cells. A. The volcano plot was applied for analyzing differentially expressed circRNAs between AMI patients and controls (GSE160717, AMI patients = 3, normal controls = 3). B. RT-qPCR analysis of circUBXN7 expression in AMI patients and normal controls (GSE160717, AMI patients = 3, normal controls = 3). C. RT-qPCR analysis of the expression of circUBXN7 and UBXN7 in AMI and sham mice (AMI mice = 8, sham mice = 8). D. RT-qPCR analysis of the expression of circUBXN7 and UBXN7 in H/R-treated or control H9c2 cells (n = 3). E. CircUBXN7 and linear UBXN7 mRNA were detected after RNase R digestion (n = 3). Mock was used as a control. F. CircUBXN7 and linear UBXN7 mRNA were detected after actinomycin D treatment for 0, 4, 8 and 12 hours (n = 3). **P < 0.01 and ***P < 0.001.
Overexpression of circUBXN7 suppressed H9c2 cell apoptosis and inflammatory reaction induced by H/R
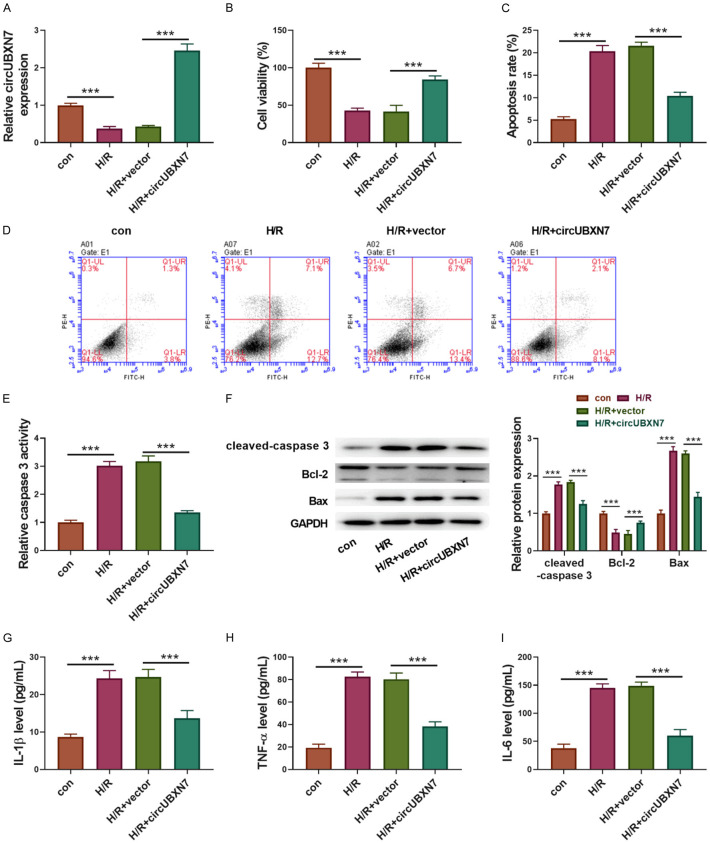
CircUBXN7 was overexpressed in H2c9 cells to examine its function in H/R. CircUBXN7 expression was reduced upon H/R treatment, but it was dramatically enhanced through overexpression of circUBXN7 in H/R-treated H9c2 cells (Figure 2A). H/R obviously impaired H9c2 cell viability, which was largely improved by overexpression of circUBXN7 (Figure 2B). Compared to control cells, H/R-treated H9c2 cells exhibited increased apoptotic cell ratio and caspase 3 activity, which were rescued by overexpression of circUBXN7 (Figure 2C-E). In addition, H/R treatment promoted the cleavage of caspase 3 and pro-apoptotic Bax expression, but suppressed anti-apoptotic Bcl-2 expression in H9c2 cells, all of which were reversed by overexpression of circUBXN7 (Figure 2F). Moreover, H/R-treated H9c2 cells showed much higher secretion of IL-1β, TNF-α and IL-6 than controls. However, overexpression of circUBXN7 significantly inhibited the production of these inflammatory factors in H9c2 cells (Figure 2G-I). Collectively, these results implied that overexpression of circUBXN7 alleviated H/R-induced myocardiocyte apoptosis and inflammatory reaction.
Figure 2.
Overexpression of circUBXN7 inhibited H9c2 cell apoptosis and inflammatory reaction induced by H/R. (A) RT-qPCR analysis of circUBXN7 expression (n = 3). (B) Cell viability analysis with CCK-8 (n = 3). (C, D) Cell apoptosis analysis (n = 3). (E) The relative caspase 3 activity (n = 3). (F) Cleaved-caspase 3, Bcl-2 and Bax were examined by western blotting. IL-1β (G), TNF-α (H) and IL-6 (I) were detected with ELISA (n = 3). H9c2 cells were transfected with circUBXN7 or vector and treated with H/R, which were divided into four groups: con, H/R, H/R+vector and H/R+circUBXN7. ***P < 0.001.
CircUBXN7 targeted miR-622 in cardiomyocytes
To elucidate the mechanism by which circUBXN7 regulates cardiomyocyte apoptosis and inflammation, cytoplasmic and nuclear fractions of H9c2 cells were separated, and then the levels of 18s rRNA (cytoplasmic internal reference), U6 snRNA (nuclear internal reference) and circUBXN7 were examined with RT-qPCR. Similar to 18s rRNA, the level of circUBXN7 in the cytoplasmic fraction was higher than that in the nuclear fraction, indicating that circUBXN7 was mainly localized in the cytoplasm (Figure 3A). Arising evidences have demonstrated that circRNAs act as miRNA sponges [12,24]. By bioinformatics using circBank (http://www.circbank.cn/), starBase (http://starbase.sysu.edu.cn/) and circinteractome (https://circinteractome.nia.nih.gov/), miR-622 or miR-3140-3p was predicted to interact with circUBXN7 (Figure 3B). The RNA pull-down assay confirmed that miR-622, not miR-3140-3p, could be enriched by the circUBXN7 probe (Figure 3C), suggesting that circUBXN7 targeted miR-622 in H9c2 cells. Next, we overexpressed miR-622 in H9c2 cells (Figure 3D). The luciferase assay showed that overexpression of miR-622 suppressed the luciferase activity in cells transfected with wildtype circUBXN7 reporters, but not in cells transfected with mutated circUBXN7 reporters (Figure 3E, 3F). Furthermore, we analyzed the expression of miR-622 in AMI and sham mice and observed an increased miR-622 expression in AMI mice (Figure 3G). The expression of miR-622 was negatively correlated with circUBXN7 expression in AMI (Figure 3H). In addition, miR-622 was dramatically upregulated in H/R-treated H9c2 cells, which was reversed by overexpression of circUBXN7 (Figure 3I). Therefore, miR-622 was identified as a novel target of circUBXN7 in H9c2 cells.
Figure 3.
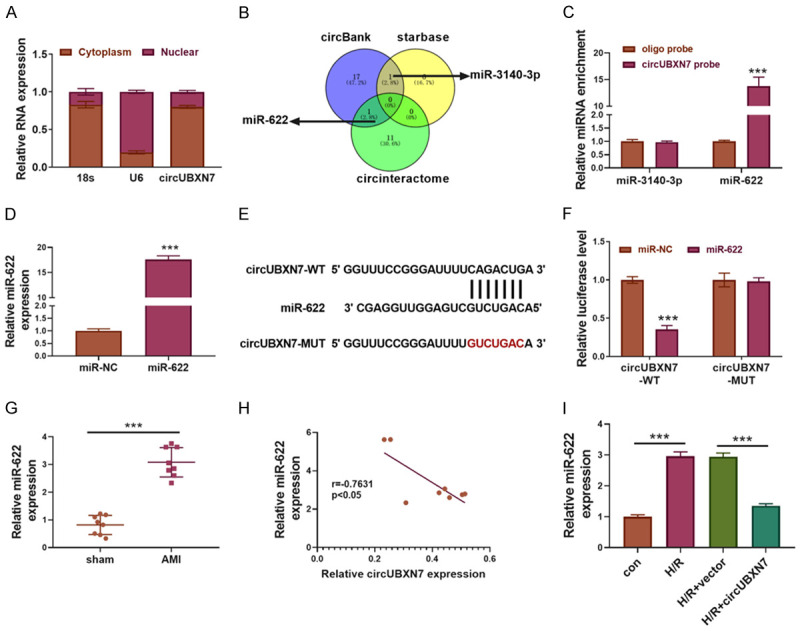
CircUBXN7 targeted miR-622 in H9c2 cardiomyocytes. A. The expression of 18s rRNA, U6 snRNA and circUBXN7 in cytoplasm and nucleus of H9c2 cells (n = 3). B. A Venn diagram was applied to predict potential targets of circUBXN7: miR-622 and miR-3140-3p. C. Enrichment of miR-3140-3p and miR-622 by circUBXN7 probe (n = 3). An oligo probe was used as a control. D. RT-qPCR analysis of miR-622 expression (n = 3). E. Predicted and mutated binding sites for miR-622 in circUBXN7. F. The interaction between circUBXN7 and miR-622 was analyzed by luciferase activity (n = 3). G. RT-qPCR analysis of miR-622 expression in sham and AMI mice (AMI mice = 8, sham mice = 8). H. Correlation analysis of the expression of circUBXN7 and miR-622 in AMI mice (n = 8). I. RT-qPCR analysis of the relative expression of miR-622 in con, H/R, H/R+vector and H/R+circUBXN7 groups (n = 3). ***P < 0.001.
CircUBXN7 inhibited apoptosis and inflammatory reaction through suppressing miR-622 expression in H/R-treated H9c2 cells
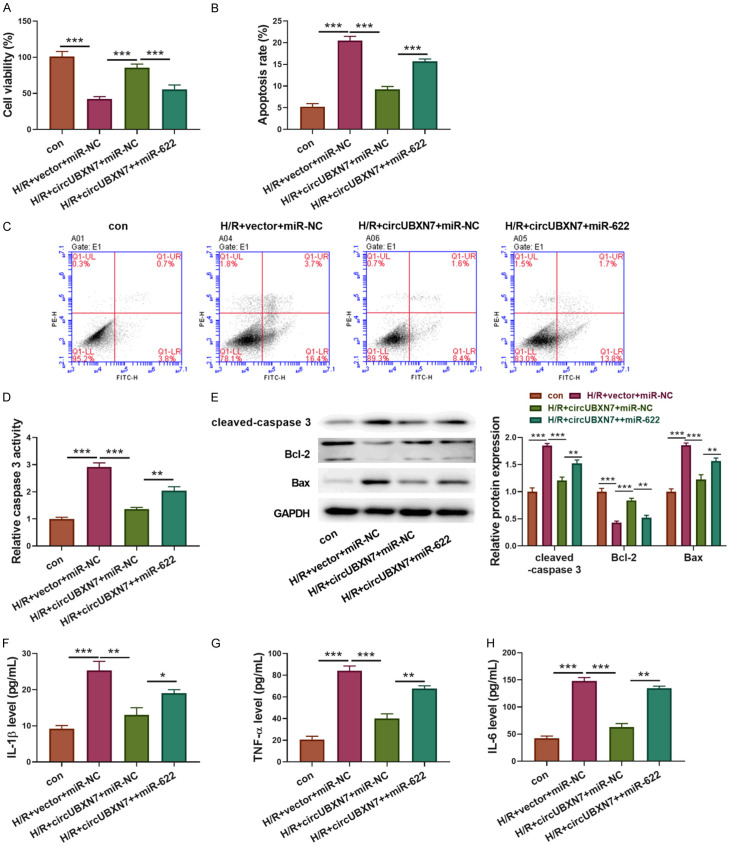
To investigate whether circUBXN7-mediated suppression of H9c2 cell apoptosis and inflammatory response was dependent on miR-622 inhibition, H9c2 cells were transfected with circUBXN7 alone or in combination with miR-622 mimics. The empty vector and NC mimics were used as controls. Reduced H9c2 cell viability caused by H/R treatment was improved by overexpression of circUBXN7, but it was abrogated by concomitant overexpression of miR-622 (Figure 4A). In consistent with cell viability, the reduced apoptotic cell rate and caspase 3 activity in circUBXN7 overexpressing cells were reversed by overexpression of miR-622 (Figure 4B-D). Moreover, overexpression of circUBXN7 inhibited the expression of cleaved-caspase 3 and Bax and promoted Bcl-2 expression in H/R-treated cells, but concomitant overexpression of miR-622 reversed these effects (Figure 4E). Besides, overexpression of circUBXN7 prohibited H/R-induced secretion of IL-6, TNF-α and IL-1β, which was abolished by concomitant overexpression of miR-622 (Figure 4F-H). Taken together, these data indicated that circUBXN7 inhibited apoptosis and inflammation induced by H/R via suppressing miR-622 expression.
Figure 4.
CircUBXN7 inhibited apoptosis and inflammation through suppressing miR-622 expression in H/R-treated cells. (A) Cell viability analysis with CCK-8 (n = 3). (B, C) Cell apoptosis analysis (n = 3). (D) The caspase 3 activity was examined by recording the absorbance at 405 nm (n = 3). (E) Cleaved-caspase 3, Bcl-2 and Bax were examined by western blotting. IL-1β (F), TNF-α (G) and IL-6 (H) were detected with ELISA (n = 3). H9c2 cells were transfected with circUBXN7 alone or in combination with miR-622 mimics and treated with H/R, which were divided into four groups: con, H/R+vector+miR-NC, H/R+circUBXN7+miR-NC and H/R+circUBXN7+miR-622. *P < 0.05, **P < 0.01 and ***P < 0.001.
MiR-622 targeted MCL1 to inhibit its expression in H9c2 cells
To further explore potential downstream targets of miR-622, a potential binding site for miR-622 in the 3’UTR of MCL1 was predicted through bioinformatics (Figure 5A). Wildtype and mutated MCL1 reporters for luciferase assays were constructed (Figure 5A). Overexpression of miR-622 reduced the luciferase activity in cells transfected with wildtype MCL1 reporter, but not in those transfected with mutated MCL1 reporter (Figure 5B). Moreover, MCL1 expression was suppressed by miR-622 mimics in H9c2 cells (Figure 5C). Compared to control cells, MCL1 expression was significantly reduced after H/R treatment in H9c2 cells, but overexpression of circUBXN7 partially increased MCL1 expression (Figure 5D). Concomitant overexpression of miR-622 reversed this effect (Figure 5D). Additionally, MCL1 expression in AMI mice was lower than that in sham mice (Figure 5E). These results suggested that miR-622 targeted MCL1 to suppress its expression in H9c2 cells.
Figure 5.
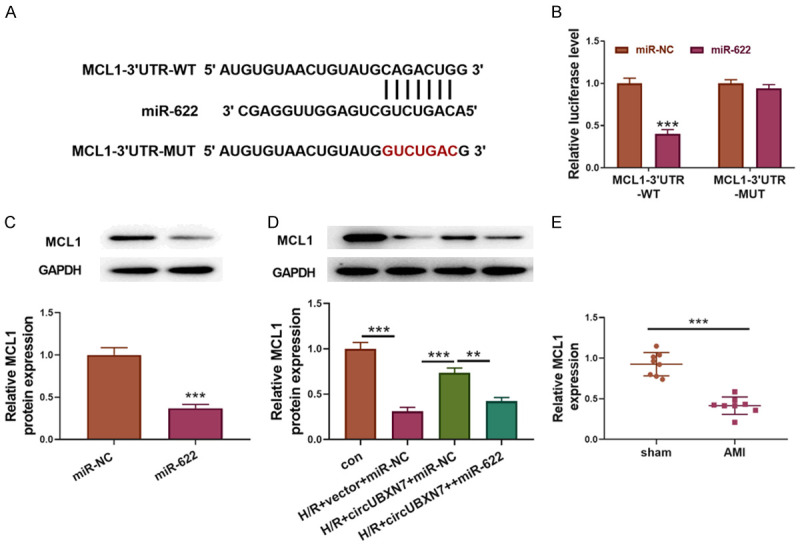
MiR-622 targeted MCL1 to inhibit its expression in H9c2 cells. A. Wildtype and mutated binding sites for miR-622 in the 3’UTR of MCL1. B. The interaction between miR-622 and MCL1 was analyzed by luciferase activity (n = 3). C. MCL1 expression was examined through western blotting (n = 3). D. Protein levels of MCL1 in con, H/R+vector+miR-NC, H/R+circUBXN7+miR-NC and H/R+circUBXN7+miR-622 groups were examined through western blotting (n = 3). E. RT-qPCR analysis of MCL1 expression in sham and AMI mice (AMI mice = 3, sham mice = 3). **P < 0.01 and ***P < 0.001.
Knockdown of MCL1 abrogated circUBXN7-mediated alleviation of apoptosis and inflammatory reaction induced by H/R
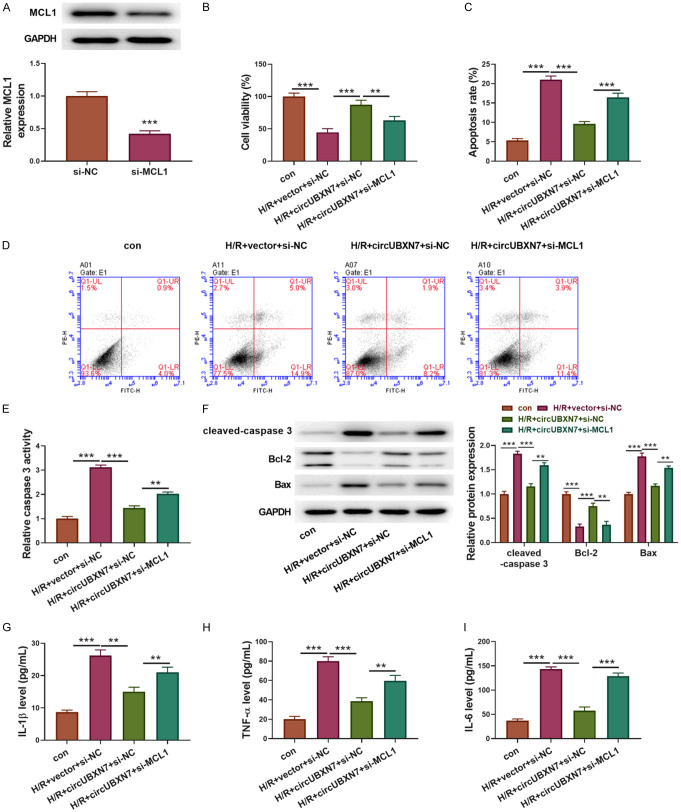
Finally, we examined whether knockdown of MCL1 can reverse circUBXN7 overexpression-mediated effects on cell apoptosis and inflammatory reaction after H/R treatment. To this end, we silenced MCL1 in circUBXN7-overexpressing H9c2 cells by si-MCL1 transfection (Figure 6A). In consistence with miR-622 overexpression, knockdown of MCL1 also reversed the anti-apoptotic effect of circUBXN7 in H/R-induced H9c2 cells (Figure 6B). In addition, circUBXN7 overexpression-mediated suppression of H/R-induced cell apoptosis and caspase 3 activity were also reversed by knockdown of MCL1 (Figure 6C-E). Cleaved-caspase 3 and Bax were downregulated, and Bcl-2 was upregulated in H/R-treated H9c2 cells with circUBXN7 overexpression. Concomitant knockdown of MCL1 enhanced the expression of cleaved-caspase 3 and Bax but inhibited Bcl-2 expression (Figure 6F). On the other hand, H/R-induced secretion of IL-1β, TNF-α and IL-6 was suppressed by overexpression of circUBXN7, and concomitant knockdown of MCL1 abrogated these effects (Figure 6G-I). Collectively, these results indicated that circUBXN7 targeted miR-622 and promoted MCL1 expression to mitigate H/R-mediated cardiomyocyte apoptosis and inflammatory reaction.
Figure 6.
Knockdown of MCL1 reversed circUBXN7-mediated alleviation of apoptosis and inflammation in H/R-treated cells. (A) The protein level of MCL1 in H9c2 cells transfected with si-NC or si-MCL1 was detected by western blotting (n = 3). (B) Cell viability was examined with CCK-8 (n = 3). (C, D) Cell apoptosis analysis (n = 3). (E) The caspase 3 activity was examined by recording the absorbance at 405 nm (n = 3). (F) Cleaved-caspase 3, Bcl-2 and Bax were examined through western blotting. IL-1β (G), TNF-α (H) and IL-6 (I) were determined with ELISA (n = 3). H9c2 cells were transfected with circUBXN7 alone or in combination with si-MCL1 and treated with H/R, which were divided into four groups: con, H/R+vector+si-NC, H/R+circUBXN7+si-NC and H/R+circUBXN7+si-MCL1. **P < 0.01 and ***P < 0.001.
Discussion
AMI is one of the leading causes of death globally and affects millions of patients each year, leading to irreversible injury to the heart muscle because of restrained oxygen supply [25,26]. To save lives of AMI patients, reperfusion of infarcted arteries needs to be done timely [5,27]. However, reperfusion paradoxically results in H/R injury and further heart damage and even death of AMI patients [28]. Understanding the regulatory mechanism of H/R injury is crucial for improving the prognosis and treatment of AMI. In this study, we first reported that circUBXN7 was downregulated in AMI patients and mice and H/R-treated H9c2 cells, and its overexpression attenuated cell apoptosis and inflammation induced by H/R. Moreover, miR-622 was identified as a novel target of circUBXN7 in cardiomyocytes, and miR-622 targeted MCL1 to suppress its expression. Overexpression of miR-622 or knockdown of MCL1 inhibited circUBXN7-mediated alleviation of apoptosis and secretion of inflammatory factors.
In recent years, apoptosis and inflammatory reaction induced by H/R have been believed to be two of vital elements in the progression of cardiac damage induced by H/R [29,30]. The dysregulated apoptosis and inflammation in H/R damage are closely associated with abnormal gene expression and function [31-33]. CircRNAs are circular single-stranded RNAs and ubiquitously expressed in eukaryotic cells, serving central roles in regulating gene expression and protein translation by acting as miRNA or RNA-binding protein decoys [34]. Intriguingly, circRNAs have been reported to be implicated in regulating various pathological processes including H/R injury. Sun et al. identified several abnormally expressed circRNAs in myocardial H/R injury and indicated potential roles of circRNAs in the process of H/R injury [35]. The expression of circFoxo3 was upregulated in H/R-treated cardiomyocytes and the hearts with I/R damage, and knockdown of circFoxo3 suppressed H/R injury by interacting with Foxo3 [36]. CircUBXN7 was reported to suppress bladder cancer cell growth and invasion, but its role in cardiovascular system and diseases is unclear [17]. Here, we reanalyzed a circRNA array (GSE160717) and found that circUBXN7 was downregulated in AMI patients. Furthermore, we first reported the low expression of circUBXN7 in mice with AMI and H/R-treated cardiomyocytes, and overexpression of circUBXN7 reduced cell apoptosis and inflammation in H/R injury. These results implied that circUBXN7 could improve the function of cardiomyocyte by suppressing cell apoptosis and inflammation after AMI.
As downstream targets of circRNAs, more and more studies have demonstrated that miRNAs also exert important functions in H/R injury. Ren et al. found that miR-320 aggravated cardiomyocyte death and apoptosis in I/R injury via targeting and suppressing HSP20 expression [37]. Overexpression of miR-451 alleviated myocardial I/R injury through inhibiting HMGB1 expression [38]. Accordingly, miR-622 was predicted and identified as a novel downstream miRNA target of circUBXN7 in cardiomyocytes for the first time in this study. Although miR-622 has not been reported to be involved in myocardial H/R injury, it could be sponged by circANRIL to aggravate oxygen-glucose deprivation and reperfusion-induced human brain microvascular endothelial cell apoptosis, which provided a clue that miR-622 might be associated with myocardial H/R injury [39]. Importantly, we observed that miR-622 was highly expressed in mice with AMI and H/R-treated cells, and overexpression of miR-622 reversed circUBXN7-mediated effects on cell apoptosis and inflammation. We first revealed the function of miR-622 in myocardial H/R damage and demonstrated that miR-622 was targeted by circUBXN7 to regulate H/R injury.
Several downstream targets of miR-622 have been identified, such as NUAK1, CCL18 and K-Ras [40-42]. We identified MCL1 as a novel target of circUBXN7/miR-622. MCL1 serves as a key anti-apoptotic regulator in various cells. Consistently, knockdown of MCL1 abrogated circUBXN7-mediated suppression of cell apoptosis and inflammation. However, further studies are needed to clarify the downstream signaling pathway of MCL1 in regulating myocardial H/R injury.
In summary, we demonstrated that circUBXN7 mitigated cardiomyocyte apoptosis and inflammation in cardiac H/R injury through targeting miR-622 and enhancing MCL1 expression (Figure 7). Our study not only identifies novel roles of the circUBXN7-miR-622-MCL1 axis in H/R injury, but also provides potential therapeutic targets for management of H/R injury. However, to achieve this, further investigations are required to elucidate the nature of the regulation in detail. In addition, more in vivo assays could be performed in samples from patients with myocardial H/R injury and AMI mouse models to further evaluate the roles of the circUBXN7-miR-622-MCL1 axis in H/R injury. Moreover, only miR-622 and MCL1 were identified in this study. However, as circRNA can have many target miRNAs and miRNA also has many target genes, whether other miRNAs and genes are implicated in circUBXN7-mediated regulation in cardiac H/R injury is unclear. We plan to seek other potential downstream miRNAs and genes of circUBXN7 in future studies.
Figure 7.
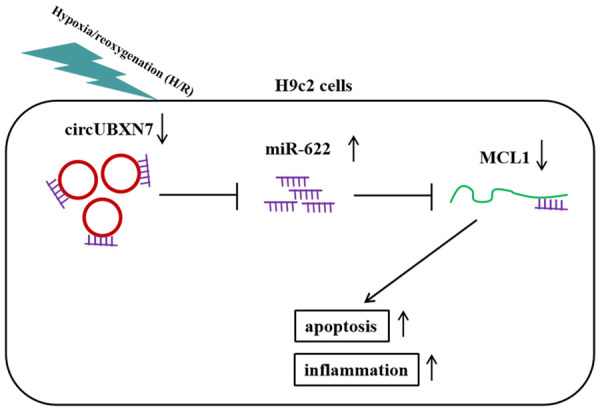
The schematic diagram of this study.
Disclosure of conflict of interest
None.
References
- 1.Boateng S, Sanborn T. Acute myocardial infarction. Dis Mon. 2013;59:83–96. doi: 10.1016/j.disamonth.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 2.Gong FF, Vaitenas I, Malaisrie SC, Maganti K. Mechanical complications of acute myocardial infarction: a review. JAMA Cardiol. 2021;6:341–349. doi: 10.1001/jamacardio.2020.3690. [DOI] [PubMed] [Google Scholar]
- 3.Anderson JL, Morrow DA. Acute myocardial infarction. N Engl J Med. 2017;376:2053–2064. doi: 10.1056/NEJMra1606915. [DOI] [PubMed] [Google Scholar]
- 4.Burke AP, Virmani R. Pathophysiology of acute myocardial infarction. Med Clin North Am. 2007;91:553–572. doi: 10.1016/j.mcna.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Bagai A, Dangas GD, Stone GW, Granger CB. Reperfusion strategies in acute coronary syndromes. Circ Res. 2014;114:1918–1928. doi: 10.1161/CIRCRESAHA.114.302744. [DOI] [PubMed] [Google Scholar]
- 6.Cowled P, Fitridge R. Pathophysiology of Reperfusion Injury. In: Fitridge R, Thompson M, editors. Mechanisms of vascular disease: a reference book for vascular specialists. South Australia: University of Adelaide Press; 2011. [PubMed] [Google Scholar]
- 7.Samokhvalov V, Jamieson KL, Fedotov I, Endo T, Seubert JM. SIRT is required for EDP-mediated protective responses toward hypoxia-reoxygenation injury in cardiac cells. Front Pharmacol. 2016;7:124. doi: 10.3389/fphar.2016.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neri M, Riezzo I, Pascale N, Pomara C, Turillazzi E. Ischemia/reperfusion injury following acute myocardial infarction: a critical issue for clinicians and forensic pathologists. Mediators Inflamm. 2017;2017:7018393. doi: 10.1155/2017/7018393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghafouri-Fard S, Shoorei H, Taheri M. Non-coding RNAs participate in the ischemia-reperfusion injury. Biomed Pharmacother. 2020;129:110419. doi: 10.1016/j.biopha.2020.110419. [DOI] [PubMed] [Google Scholar]
- 10.Smani T, Mayoral-Gonzalez I, Galeano-Otero I, Gallardo-Castillo I, Rosado JA, Ordonez A, Hmadcha A. Non-coding RNAs and ischemic cardiovascular diseases. Adv Exp Med Biol. 2020;1229:259–271. doi: 10.1007/978-981-15-1671-9_15. [DOI] [PubMed] [Google Scholar]
- 11.Zhang B, Zhou M, Li C, Zhou J, Li H, Zhu D, Wang Z, Chen A, Zhao Q. MicroRNA-92a inhibition attenuates hypoxia/reoxygenation-induced myocardiocyte apoptosis by targeting Smad7. PLoS One. 2014;9:e100298. doi: 10.1371/journal.pone.0100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79. doi: 10.1007/978-981-13-1426-1_6. [DOI] [PubMed] [Google Scholar]
- 13.Kristensen LS, Ebbesen KK, Sokol M, Jakobsen T, Korsgaard U, Eriksen AC, Hansen TB, Kjems J, Hager H. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat Commun. 2020;11:4551. doi: 10.1038/s41467-020-18355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang L, Meng X, Li G, Zhou Q, Xiao J. Circular RNAs in cardiovascular diseases. Adv Exp Med Biol. 2018;1087:191–204. doi: 10.1007/978-981-13-1426-1_15. [DOI] [PubMed] [Google Scholar]
- 15.Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The circular RNA cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B, Li G, Peng J, Ren L, Lei L, Ye H, Wang Z, Zhao S. CircMACF1 attenuates acute myocardial infarction through miR-500b-5p-EMP1 axis. J Cardiovasc Transl Res. 2020;14:161–172. doi: 10.1007/s12265-020-09976-5. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Chen D, Bi J, Han J, Yang M, Dong W, Lin T, Huang J. Circular RNA circUBXN7 represses cell growth and invasion by sponging miR-1247-3p to enhance B4GALT3 expression in bladder cancer. Aging (Albany NY) 2018;10:2606–2623. doi: 10.18632/aging.101573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DC. Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramaniam D, Natarajan G, Ramalingam S, Ramachandran I, May R, Queimado L, Houchen CW, Anant S. Translation inhibition during cell cycle arrest and apoptosis: Mcl-1 is a novel target for RNA binding protein CUGBP2. Am J Physiol Gastrointest Liver Physiol. 2008;294:G1025–1032. doi: 10.1152/ajpgi.00602.2007. [DOI] [PubMed] [Google Scholar]
- 20.Thomas RL, Roberts DJ, Kubli DA, Lee Y, Quinsay MN, Owens JB, Fischer KM, Sussman MA, Miyamoto S, Gustafsson AB. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev. 2013;27:1365–1377. doi: 10.1101/gad.215871.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao C, Liu J, Ge W, Li Z, Lv M, Feng Y, Liu X, Liu B, Zhang Y. Identification of regulatory circRNAs involved in the pathogenesis of acute myocardial infarction. Front Genet. 2020;11:626492. doi: 10.3389/fgene.2020.626492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu H, Wu J, Cao C, Ma L. Exosomes derived from regulatory T cells ameliorate acute myocardial infarction by promoting macrophage M2 polarization. IUBMB Life. 2020;72:2409–2419. doi: 10.1002/iub.2364. [DOI] [PubMed] [Google Scholar]
- 23.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rong D, Sun H, Li Z, Liu S, Dong C, Fu K, Tang W, Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siqueira CADS, de Souza DLB. Reduction of mortality and predictions for acute myocardial infarction, stroke, and heart failure in Brazil until 2030. Sci Rep. 2020;10:17856. doi: 10.1038/s41598-020-73070-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ioacara S, Popescu AC, Tenenbaum J, Dimulescu DR, Popescu MR, Sirbu A, Fica S. Acute myocardial infarction mortality rates and trends in romania between 1994 and 2017. Int J Environ Res Public Health. 2019;17:285. doi: 10.3390/ijerph17010285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Huo Y. Early reperfusion strategy for acute myocardial infarction: a need for clinical implementation. J Zhejiang Univ Sci B. 2011;12:629–632. doi: 10.1631/jzus.B1101010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grech ED, Jackson MJ, Ramsdale DR. Reperfusion injury after acute myocardial infarction. BMJ. 1995;310:477–478. doi: 10.1136/bmj.310.6978.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N Engl J Med. 2007;357:1121–1135. doi: 10.1056/NEJMra071667. [DOI] [PubMed] [Google Scholar]
- 30.Yao L, Chen H, Wu Q, Xie K. Hydrogen-rich saline alleviates inflammation and apoptosis in myocardial I/R injury via PINK-mediated autophagy. Int J Mol Med. 2019;44:1048–1062. doi: 10.3892/ijmm.2019.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalogeris T, Baines CP, Krenz M, Korthuis RJ. Cell biology of ischemia/reperfusion injury. Int Rev Cell Mol Biol. 2012;298:229–317. doi: 10.1016/B978-0-12-394309-5.00006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Cheng R, Wan H. Overexpression of TGR5 alleviates myocardial ischemia/reperfusion injury via AKT/GSK-3beta mediated inflammation and mitochondrial pathway. Biosci Rep. 2020;40:BSR20193482. doi: 10.1042/BSR20193482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu W, Zhang L, Zhang Y, Zhang K, Wu Y, Jin D. TRAF1 exacerbates myocardial ischemia reperfusion injury via ASK1-JNK/p38 signaling. J Am Heart Assoc. 2019;8:e012575. doi: 10.1161/JAHA.119.012575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26:29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Z, Yu T, Jiao Y, He D, Wu J, Duan W, Sun Z. Expression profiles and ontology analysis of circular RNAs in a mouse model of myocardial ischemia/reperfusion injury. Biomed Res Int. 2020;2020:2346369. doi: 10.1155/2020/2346369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Y, Zhu C, Wang B, Zheng H, McAlister V, Lacefield JC, Quan D, Mele T, Greasley A, Liu K, Zheng X. Foxo3 in cardiac ischemia reperfusion injury in heart transplantation: a new regulator and target. Am J Transplant. 2020 doi: 10.1111/ajt.16475. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 37.Ren XP, Wu J, Wang X, Sartor MA, Jones K, Qian J, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119:2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao J, Da Y, Li H, Peng Y, Hu X. Upregulation of microRNA-451 attenuates myocardial I/R injury by suppressing HMGB1. PLoS One. 2020;15:e0235614. doi: 10.1371/journal.pone.0235614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang S, Zhao G, Lu J, Jiang M, Wu Z, Huang Y, Huang J, Shi J, Jin J, Xu X, Pu X. Silencing of circular RNA ANRIL attenuates oxygen-glucose deprivation and reoxygenation-induced injury in human brain microvascular endothelial cells by sponging miR-622. Biol Res. 2020;53:27. doi: 10.1186/s40659-020-00295-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orlandella FM, Mariniello RM, Mirabelli P, De Stefano AE, Iervolino PLC, Lasorsa VA, Capasso M, Giannatiempo R, Rongo M, Incoronato M, Messina F, Salvatore M, Soricelli A, Salvatore G. miR-622 is a novel potential biomarker of breast carcinoma and impairs motility of breast cancer cells through targeting NUAK1 kinase. Br J Cancer. 2020;123:426–437. doi: 10.1038/s41416-020-0884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T, Sun X, Xu K. The suppressing role of miR-622 in renal cell carcinoma progression by down-regulation of CCL18/MAPK signal pathway. Cell Biosci. 2018;8:17. doi: 10.1186/s13578-018-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Fang Y, Sun B, Li Z, Chen Z, Xiang J. MiR-622 inhibited colorectal cancer occurrence and metastasis by suppressing K-Ras. Mol Carcinog. 2016;55:1369–1377. doi: 10.1002/mc.22380. [DOI] [PubMed] [Google Scholar]