Abstract
Objective: To investigate the role and functions of FAM225A in gastric cancer. Methods: The expressions of FAM225A, miR-206, ADAM12 and epithelial-mesenchymal transition (EMT)-related genes were detected by quantitative real-time PCR and Western blot. Functional experiments including cell counting kit-8, colony formation, wound-healing, and Transwell assays were conducted to analyze the biological characteristics of gastric cancer cells in different groups. Bioinformatics, dual-luciferase reporter assay and Pearson correlation coefficients were performed for determining the regulatory relationship of lncRNA-miRNA-mRNA. In vivo nude mouse xenografts and immunohistochemistry were used to verify the results in vitro. Results: In gastric cancer, FAM225A and ADAM12 expressions were up-regulated, while miR-206 expression was down-regulated. Opposite to the regulatory effects of overexpressed FAM225A, blocking FAM225A expression reduced cell viability, migration, invasion and number of cell clones, increased E-Cadherin expression, inhibited N-Cadherin and Vimentin expressions, and ultimately promoted tumor growth. MiR-206 inhibitor partially offset the effects of siFAM225A. Moreover, FAM225A competitively bound to miR-206 to up-regulate ADAM12 expression. Overexpressed ADAM12 partially reversed the effect of miR-206 mimic on the biological characteristics of gastric cancer cells and EMT-related proteins. Conclusion: Our research revealed that FAM225A-miR-206-ADAM12 axis may be a potential pathway for regulating gastric cancer.
Keywords: Gastric cancer, lncRNA FAM225A, miR-206, ADAM12, proliferation
Introduction
The World Cancer Report 2014 released by the World Health Organization shows that the incidence and mortality of gastric cancer in China was the highest in 2012 [1]. Gastric cancer has the characteristics of hidden onset and unclear early symptoms, which impose great difficulty on the prevention and treatment of the cancer [2].
Increasing evidence supports that long non-coding RNAs (lncRNAs) are involved in the regulation of gastric cancer [3-5]. It has been reported that the high expressions of LINC00152 and lncRNA MALAT1 (MALAT1) are associated with an increased risk of developing gastric cancer [6,7]. LncRNAs could act as potential molecular markers for the diagnosis of gastric cancer [8]. It was found in a study that lncRNA FAM225A promoted the occurrence and metastasis of nasopharyngeal carcinoma [9], but its effects on gastric cancer have not been determined.
High expression of FAM225A was detected in stomach adenocarcinoma (STAD) using bioinformatics analysis. In recent years, it has been found in related studies that lncRNAs have a variety of biological functions by regulating miRNAs in cancer [10]. Competitive endogenous RNAs (ceRNAs) are widely studied among multiple interactions between lncRNAs and miRNAs [11,12]. Liu et al. showed that the up-regulated expression of lncRNA HOTAIR in patients with gastric cancer could regulate the inhibition of human epidermal growth factor receptor through competitively binding to miR-331-3p, thereby promoting the invasion of gastric cancer cells [13]. Therefore, we speculated that FAM225A could also act as a ceRNA in gastric cancer.
The role of miRNAs in gastric cancer has been previously investigated [14]. For example, Xie et al. found that up-regulating miR-374b-5p expression promoted the metastasis and invasion of gastric cancer cells by inhibiting the expression of RECK [15]. Based on the research above, we applied bioinformatics and predicted that FAM225A could share its binding site with miR-206. It was also demonstrated in multiple studies that the complex biological regulatory network involving both miR-206 and its target genes plays important roles in tumorigenesis and development [16,17]. Zhang et al. found that overexpression of miR-206 inhibited the growth of gastric cancer cells and the ability to form clones. Moreover, further research also showed that miR-206 could inhibit the proliferation of gastric cancer cells by targeting cyclin 2 [18].
However, whether FAM225A had a critical function in gastric cancer through regulating miR-206 remained to be confirmed. This study, therefore, explored the molecular regulatory pathway of FAM225A through the regulation of miR-206 on proliferation and metastasis of gastric cancer cells.
Materials and methods
Ethics statement
The acquisition of clinical samples was approved by the Ethics Committees of Nanjing First Hospital (NF202001019). The experiments with animals were conducted in accordance with the guideline of Institutional Animal Care and Use Committee (NF 202003017) at Nanjing First Hospital. All the patients signed a written informed consent.
Tissue, cell and culture
The clinical specimens were obtained from 40 patients with gastric cancer from February 2020 to May 2020 at Nanjing First Hospital. The tissues were frozen at -80°C after the surgery.
GES-1 cell lines (CL-0563) were cultured in RPMI-1640 medium (PM150110) containing 10% FBS (164210-500) and 1% P/S (PB180120). Gastric cancer cell lines were incubated in a cell incubator at 37°C in 5% CO2. Specifically, AGS (CL-0022) was cultured in Ham’s F-12 medium (CM-0022), KATO III (CL-0372) and SNU-5 (CL-0444) were cultured in IMDM medium (PM150510), and HGC27 (CL-0107) and NCI-N87 (CL-0169) were cultured in RPMI-1640 medium. All the cells and reagents were purchased from the Procell company (China).
Cell transfection
FAM225A or ADAM12 overexpression plasmid (FAM225A, ADAM12), siRNA interference plasmid for FAM225A (siFAM225A) and the corresponding control plasmids were ordered from GenePharma Co., Ltd. (China). MiR-206 mimic (miR10000462-1-5, UGGAAUGUAAGGAAGUGUGUGG), miR-206 inhibitor (miR20000462-1-5, CCACACACUUCCUUACAUUCCA) and mimic or inhibitor control (miR1N0000002-1-5, miR2N0000002-1-5) were obtained from Guangzhou RIBOBIO Co., Ltd. (China). AGS and NCI-N87 cells (1 × 105/well) were transfected with different plasmids strictly according to the instructions of Lipofectamine 3000 reagent (L3000015, ThermoFisher, USA).
Quantitative real-time PCR
Total RNA was isolated by TRIzol reagent (15596018, Invitrogen, CA), and RNAiso for Small RNA (9753Q, Takara, China) was used for the isolation of miRNA. PrimeScrip RT Master Mix (RR036Q, China) and External Standard Kit (3789, Takara) were applied to perform the reverse transcription reaction. The cDNA was served as a template for RT-PCR detection in the QuantStudio 5 PCR instrument (ABI, USA), according to the manual of the RT-PCR kit (A46113, Applied Biosystems, USA). The conditions for the reaction were set as follows: a pre-denaturation (at 95°C for 10 min), followed by a total of 40 cycles of denaturation (at 95°C for 30 s), and annealing (60°C, 1 min). For determining the relative expressions of genes, GAPDH and U6 were used as internal parameters. The gene values were calculated using 2-ΔΔCt method [19]. All the primers are listed below (5’-3’): FAM225A: (CCGATGGAGGTGTTGATGGATGT, CTGTGGCCCTTGGGCAAGTAAGT); GAPDH: (CGTAGTCAGTACGTAGGTA, CGTACGTAGCTACGTGGGC); miR-206: (GTAAGGAAGTGTGTGGGTCGT, TATCCAGTGCGTGTCGTGG); U6: (CTCGCTTCGGCAGCACA, AACGCTTCACGAATTTGCGT).
Cell counting kit (CCK)-8
The trypsin-digested AGS and NCI-N87 cells (1 × 104 cells/mL) were transferred to a cell incubator for the cultivation. After culture for 24, 48, or 72 h, the AGS or NCI-N87 cells were incubated with 10 μL CCK-8 reagent (41003, Dokdo, Japan) for 2 h. Finally, the absorbance at 450 nm was measured using a Stat Fax 4200 microplate reader (Awareness, USA).
Colony formation assay
After the transfection for 48 h, AGS and NCI-N87 cells (1 × 102 cells) were respectively cultured in a petri dish for 2 weeks. The colonies were fixed with 4% paraformaldehyde (158127, Sigma-Aldrich, Germany) for 15 min, and stained by Giemsa Stain (48900, Sigma-Aldrich) at room temperature for 30 min. Zeiss L LSM800 confocal microscope (Germany) was used for cell counting, photography, and analysis.
Wound-healing assay
A horizontal line was drawn on the bottom of the 6-well plate. The suspension of AGS or NCI-N87 cells (1 × 105 cell/well) was transferred to a 6-well plate for an overnight routine culture. A sterile pipette tip was applied to create a gap perpendicular to the bottom line of the 6-well plate containing the cells. Then, the fresh medium was added to the 6-well plate. After 48 h of further incubation, the cells were observed with a microscope (Zeiss L LSM800, Germany) at 100 × magnification.
Transwell invasion assay
Matrigel (354230, BD, USA) was diluted in serum-free RPMI-1640 culture medium for later use. Each group of pre-transfected AGS or NCI-N87 cells was prepared into a single cell suspension (1 × 105/well). Next, the basement membrane of the Transwell chamber (CLS3398, Sigma, Germany) was coated with 50 μL of diluted Matrigel and inserted into a 24-well plate. 50 μL of cell suspension was inoculated into the upper chamber, whereas 500 μL of RPMI-1640 medium containing 10% FBS was added to the lower chamber. The culture plate was then incubated in a cell incubator for 48 h. After the incubation, the medium was removed from the upper chamber. The cells were fixed with 4% paraformaldehyde, washed with PBS solution twice, and stained with 0.1% crystal violet at room temperature for 30 min. After rinsing, the cells on the surface of the upper chamber were gently wiped off by a sterile cotton ball. Cell invasion was randomly observed and calculated from 5 view fields of the microscope.
Western blot analysis
Western blot analysis was used to detect the relative protein expressions of genes as previously reported [20]. The total protein was extracted by RIPA lysate (P0013, Beyotime, China) and the protein concentration was determined by BCA method with BCA kit (A53227, ThermoFisher, USA). Each group of 20 µg protein was separated on 10% SDS-PAGE and transferred to PVDF membrane (IPVH00010, Millipore, USA). The PVDF membrane was incubated with the following specific primary antibodies (at 4°C overnight): E-cadherin (ab231303, 97 kDa, 1 µg/ml, Abcam, UK), N-cadherin (ab18203, 100 kDa, 1 µg/ml), Vimentin (ab20346, 54 kDa, 1 µg/ml), ADAM12 (ab28747, 95 kDa, 0.3 µg/ml), followed by incubation with the corresponding secondary antibodies (at room temperature for 2 h) as follows: Goat Anti-Rabbit (ab205718), 1/5000; Goat Anti-Mouse (ab205719, ab9167), 1/5000; Donkey Anti-Goat (ab6885), 1/2000. Finally, ECL luminescent substrate (SL1350-100 ml, Coolaber, China) and Quantity One image analysis software (Bio-Rad Lab, USA) were used for the analysis on the result. GAPDH served as an internal reference.
Prediction and verification of targeted relationship
The target miRNA and expressions of FAM225A in STAD were predicted using StarBase website, while the target gene of miR-206 was predicted using TargetScan v7.2 website. The relationship between target genes was verified by conducting dual-luciferase reporter assay (FR201-01, TransGen Biotech, China). Recombinant fluorescent reporter vectors were constructed using pmirGLO (CL414-01, Biomed, China). The wild-type or mutant gene 3’UTR recombinant reporter plasmids (FAM225A or ADAM12) were co-transfected with miR-206 mimic into AGS or NCI-N87 cells. After the transfection for 48 h, the supernatants of cells were mixed with the firefly luciferase substrate to measure the value of luciferase activity. Afterwards, the stop solution was added and Renilla luciferase substrates were used for determining the value of Renilla luciferase activity.
Tumor formation assay
BALB/c athymic nude mice (4 weeks old, male) were purchased from Guangdong Medical Laboratory Animal Center (China), kept in the SPF experimental animal center, and provided with free access to food and water. Pre-suspended AGS cells (1 × 106/mL) transfected with siNC or siFAM225A were used for the establishment of the xenograft nude mouse model through subcutaneous injection. The mice in the siNC or siFAM225A group received supplemental injections once a week. Vernier calipers were used to measure the longest and shortest diameters of xenograft tumors every 3 days to record the size of the tumor. After 28 days, the mice were sacrificed under anesthesia by intraperitoneal injection of excessive pentobarbital sodium (B5646-50 mg, ApexBio, USA). After complete resection of tumor and photographing, the tumor tissues were collected for further experiments.
Immunohistochemistry (IHC)
After routine dewaxing and hydration, the paraffin sections of xenograft tumor were repaired with high-temperature and high-pressure antigen repair methods, followed by treatment with hydrogen peroxide reagent (3% for 10 min) and goat serum (30 min, C0265, Beyotime, China). After washing the sections, anti-Ki67 antibody (SP6) (ab16667, Abcam, UK) was added and the sections were incubated at 4°C overnight. Then, the corresponding secondary antibody goat anti-rabbit IgG H&L (HRP) (ab205718) was added for another 30 min. The sections were then stained with SABC-POD immunohistochemistry kit (I001-2-1, Nanjing Jiancheng). After dehydration, the sections were transparentized, sealed, and the immunostained images were analyzed with an Olympus FluoView FV1000 microscope (Tokyo, Japan).
Statistical analysis
SPSS 20.0 or Graphpad prism 8.0 was used for statistical analysis. The measurement data were expressed as mean ± standard deviation, and the comparison was conducted using paired sample t test; correlation was analyzed using Pearson correlation analysis. In the tumor model experiment, the independent sample t test was used for two-group comparison, while the one-way ANOVA and Tukey’s test were used for comparison among multiple groups. P<0.05 was considered as statistically significant.
Results
LncRNA FAM225A was highly expressed in gastric cancer and regulated the biological characteristics of gastric cancer cells
It was found in bioinformatics analysis that FAM225A was abnormally highly expressed in stomach adenocarcinoma (STAD), suggesting that it plays a certain role in gastric cancer (P = 2.4e-11, Figure 1A). Moreover, we found that the expression of FAM225A was also higher in cancer tissues than in adjacent tissues (P<0.001, Figure 1B). In addition, whether a similar result could be detected in gastric cancer cell lines was examined. As expected, the expression of FAM225A in AGS, KATO III, HGC27, NCI-N87, and SNU-5 cells was significantly higher than that in GES-1 cells (P<0.001, Figure 1C). Noticeably, FAM225A showed the highest expression in AGS cells, but the lowest expression in NCI-N87 cells. Thus, the two cells were selected as the research cells.
Figure 1.
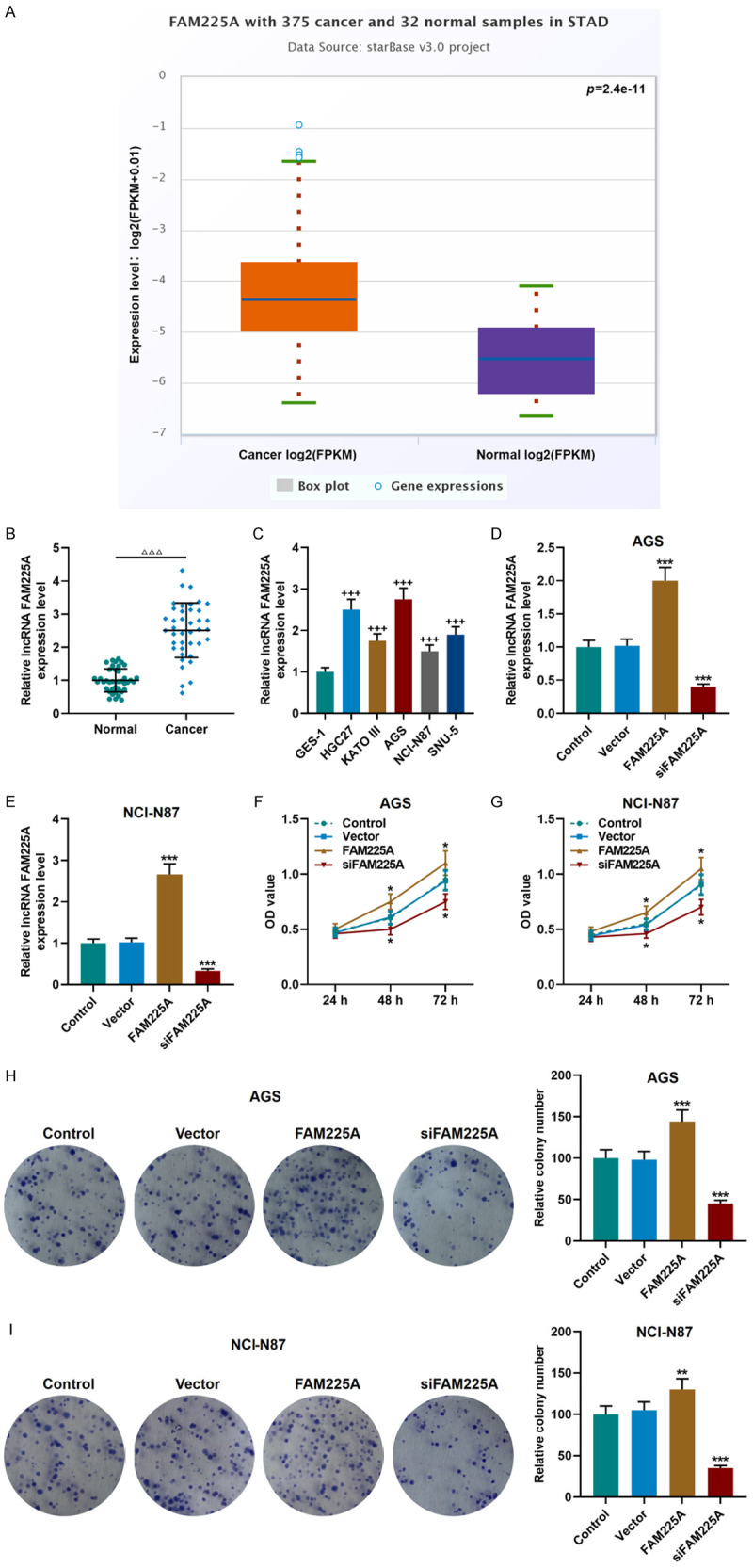
High expression of lncRNA FAM225A affected cell viability and proliferation in gastric cancer. A. LncRNA FAM225A was high-expressed in stomach adenocarcinoma (STAD), as detected by StarBase website (cancer = 375, normal = 32). B. LncRNA FAM225A was high-expressed in gastric cancer tissues, as detected by qRT-PCR (n = 40). C. The expression of lncRNA FAM225A in gastric cancer cell lines (HGC27, KATO III, AGS, NCI-N87 and SNU-5) and normal GES-1 cells was detected by qRT-PCR. D, E. The transfection efficiency of lncRNA FAM225A overexpression or silencing plasmid into AGS and NCI-N87 cells was detected by qRT-PCR. F, G. Overexpression of FAM225A promoted cell viability, while silencing FAM225A had the opposite effect in AGS and NCI-N87 cells, as detected by CCK-8 assay. H, I. The effect of FAM225A on the proliferation of AGS and NCI-N87 cells was tested by colony formation experiment. Each experiment was repeated three times independently. GAPDH was used as a control. qRT-PCR: quantitative reverse transcription real time polymerase chain reaction. CCK-8: Cell Counting Kit-8. ΔΔΔP<0.001 vs. Normal; +++P<0.001 vs. GES-1; *P<0.05, **P<0.01, ***P<0.001 vs. Vector.
Next, the FAM225A overexpression plasmid and siRNA silencing FAM225A plasmid were transfected into the above two groups of cells, and the transfection of plasmids was successful (P<0.001, Figure 1D, 1E). We further performed functional experiments to examine the effects of FAM225A on the biological characteristics of gastric cancer cells. We observed that the FAM225A overexpression group showed promoted viability, migration and invasion of gastric cancer cell, and increased number of cell clones, while the siFAM225A group showed the opposite changes (P<0.05, Figures 1F-I, 2A-D).
Figure 2.
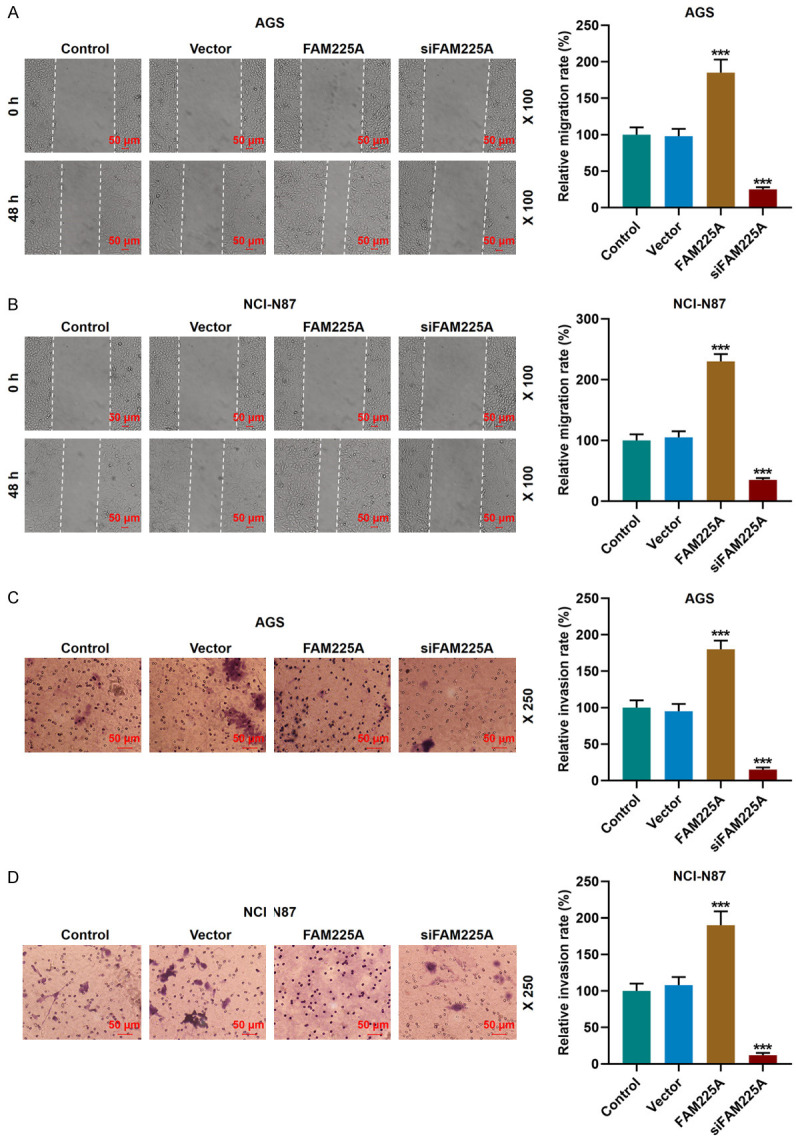
Overexpression of lncRNA FAM225A promoted the migration and invasion of gastric cancer cells, while silencing FAM225A produced opposite results. A, B. The effect of overexpression or silence of FAM225A on the migration of AGS and NCI-N87 cells was determined by the wound-healing assay. Scale: 50 µm; magnification: × 100. C, D. Overexpression of FAM225A promoted the invasion of AGS and NCI-N87 cells, while silencing FAM225A caused the opposite effect. The results were tested by Transwell assay. Scale: 50 µm; magnification: × 250. ***P<0.001 vs. Vector.
Competitive binding of FAM225A to miR-206 interfered biological function and affected the expressions of EMT-related proteins in gastric cancer cells
It was demonstrated from bioinformatics analysis that FAM225A and miR-206 shared mutual binding sites (Figure 3A), which was also verified by the dual-luciferase reporter assay, as the binding of miR-206 mimic to FAM225A-WT reduced the luciferase activity (P<0.001, Figure 3B, 3C), which suggested that FAM225A may regulate gastric cancer cells through miR-206. Our data revealed that miR-206 was abnormally low-expressed in the tissue samples of gastric cancer (P<0.001, Figure 3D), and results from correlation coefficient analysis showed that FAM225A was negatively correlated with miR-206 (r = -0.4109, P = 0.0084, Figure 3E).
Figure 3.
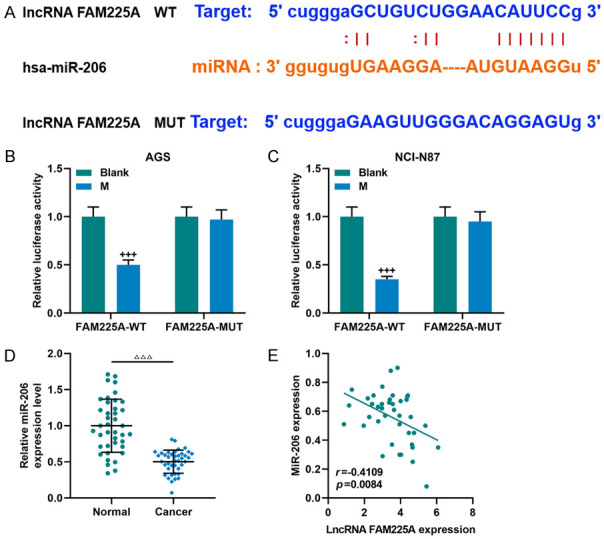
LncRNA FAM225A was proved to target and negatively regulate miR-206. A. The StarBase website was used to predict the binding site of lncRNA FAM225A and miR-206. B, C. Dual-luciferase reporter assay was used to verify the targeting relationship between lncRNA FAM225A and miR-206. D. MiR-206 showed a low expression in gastric cancer tissues (n = 40), as detected by qRT-PCR. Each experiment was repeated three times independently. E. The Pearson correlation coefficient was used to detect the linear relationship between LncRNA FAM225A and miR-206. qRT-PCR: quantitative reverse transcription real time polymerase chain reaction; M: miR-206 mimic. ΔΔΔP<0.001 vs. Normal; +++P<0.001 vs. Blank.
Next, we examined the effects of miR-206 inhibitor on AGS and NCI-N87 cells, and observed that miR-206 inhibitor was successfully transfected (Figure 4A, 4B). It was also found that siFAM225A could up-regulate miR-206 expression, and miR-206 inhibitor reversed the effect of siFAM225A (P<0.001, Figure 4C, 4D).
Figure 4.
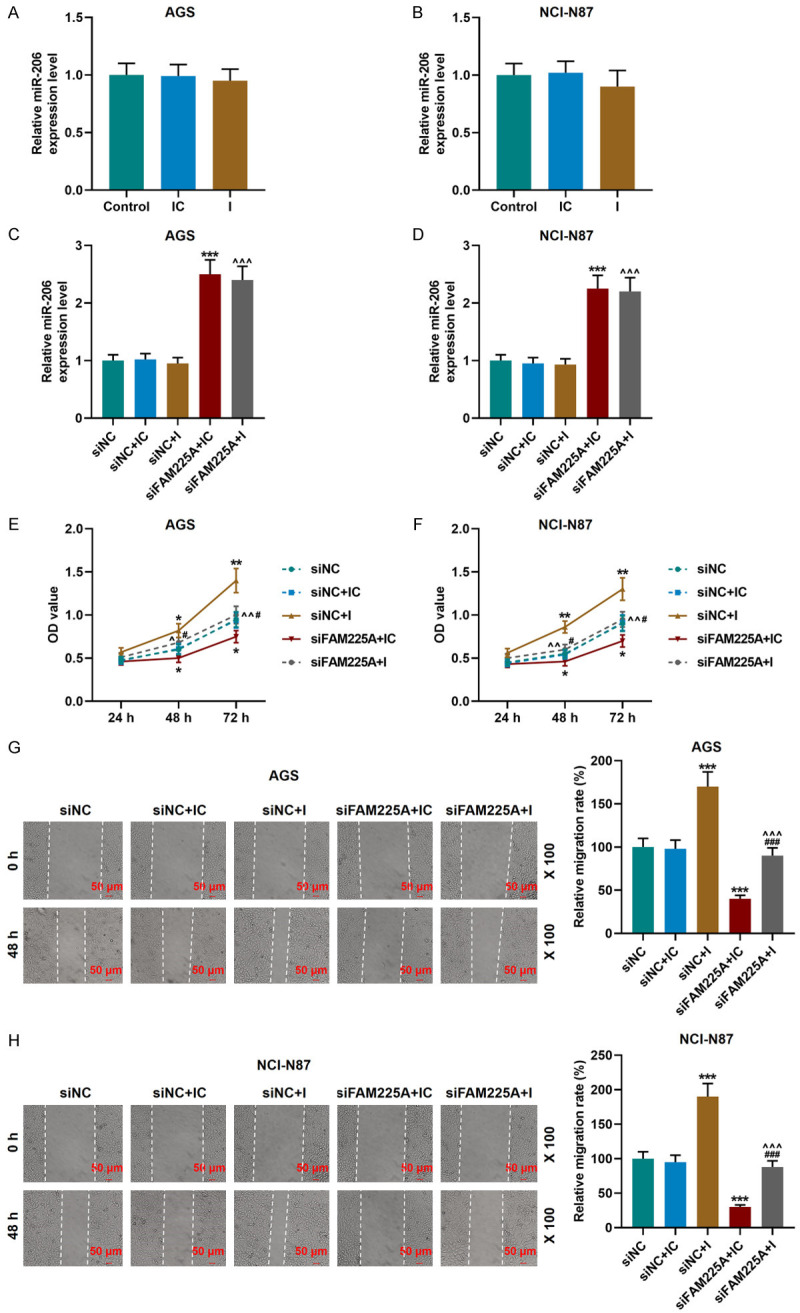
MiR-206 binding to LncRNA FAM225A affected the viability and migration of gastric cancer cells. A, B. qRT-PCR was used to detect the transfection efficiency of gastric cancer cells transfected with miR-206 inhibitor. C, D. Silencing FAM225A expression promoted the expression of miR-206. The results were determined by qRT-PCR. Each experiment was repeated three times independently. U6 was used as a control. E, F. miR-206 inhibitor promoted cell viability and reversed the regulatory effect of silencing FAM225A on the viability of AGS and NCI-N87 cells, as detected by CCK-8 assay. G, H. miR-206 inhibitor promoted cell migration and reversed the regulatory effect of silencing FAM225A on the migration of AGS and NCI-N87 cells, as measured by wound-healing assay. Scale: 50 µm; magnification: × 100. qRT-PCR: quantitative reverse transcription real time polymerase chain reaction; CCK-8: Cell Counting Kit-8; IC: inhibitor control; I: miR-206 inhibitor; siNC: small interfering RNA of negative control. *P<0.05, **P<0.01, ***P<0.001 vs. siNC+IC; ^P<0.05, ^^P<0.01, ^^^P<0.001 vs. siNC+I; #P<0.05, ###P<0.001 vs. siFAM225A+IC.
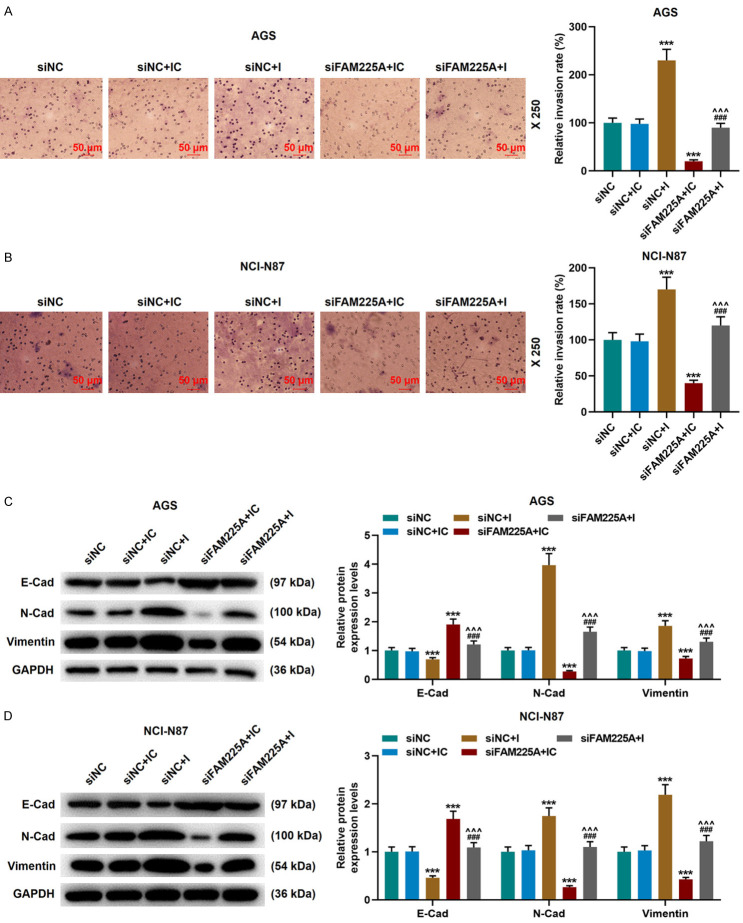
Subsequently, we explored the specific effects of siFAM225A and miR-206 inhibitor on gastric cancer cells. MiR-206 inhibitor significantly promoted the cell viability, migration and invasion, and partially offset the inhibitory effect of siFAM225A on the biological functions of gastric cancer cells (P<0.05, Figures 4E-H, 5A, 5B). Moreover, siFAM225A inhibited the expressions of N-Cadherin and Vimentin and promoted that of E-Cadherin, while miR-206 inhibitor partially reversed the regulation of siFAM225A on EMT-related proteins (P<0.001, Figure 5C, 5D).
Figure 5.
LncRNA FAM225A affected the invasion and EMT-related protein expressions in gastric cancer by regulating miR-206. A, B. miR-206 inhibitor reversed the effect of silencing FAM225A on inhibiting the invasion of AGS and NCI-N87 cells. The results were detected by Transwell assay. Scale: 50 µm; magnification: × 250. C, D. The expressions of N-Cadherin, Vimentin, E-Cadherin in siNC, siNC+IC, siNC+I, siFAM225A+IC, siFAM225A+I groups were determined by western blot. GAPDH was used as a control. IC: inhibitor control; I: miR-206 inhibitor; siNC: small interfering RNA of negative control. ***P<0.001 vs. siNC+IC; ^^^P<0.001 vs. siNC+I; ###P<0.001 vs. siFAM225A+IC.
The regulatory relationship and functions of FAM225A-miR-206-ADAM12 in gastric cancer
Bioinformatics analysis and dual-luciferase reporter assay proved that miR-206 and ADAM12 shared a targeted binding relationship (Figure 6A-C). Thus, the functions of ADAM12 were further detected. As shown in Figure 6D, ADAM12 expression was abnormally elevated in gastric cancer tissue samples (P<0.001). Further exploration of the linear relationship of FAM225A-miR-206-ADAM12 revealed that ADAM12 was positively correlated with FAM225A (r = 0.3617, P = 0.0218, Figure 6E) and negatively correlated with miR-206 (r = -0.3704, P = 0.0186, Figure 6F).
Figure 6.
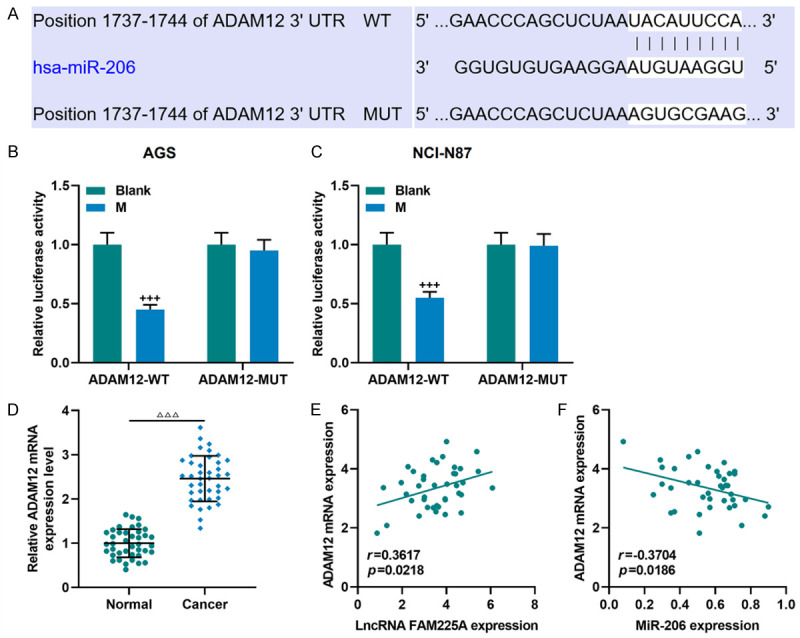
LncRNA FAM225A-miR-206-ADAM12 axis in gastric cancer. A. The binding site of miR-206 and ADAM12 was predicted by TargetScan7.2 website. B, C. The targeting relationship between miR-206 and ADAM12 was verified by dual-luciferase reporter assay. D. The expression of ADAM12 in gastric cancer and adjacent tissues (n = 40) was detected by qRT-PCR. Each experiment was repeated three times independently. GAPDH was used as a control. E, F. Pearson correlation coefficient was used to analyze the relationship between lncRNA FAM225A and ADAM12, ADAM12 and miR-206 in gastric cancer tissues. qRT-PCR: quantitative reverse transcription real time polymerase chain reaction; M: miR-206 mimic. ΔΔΔP<0.001 vs. Normal; +++P<0.001 vs. Blank.
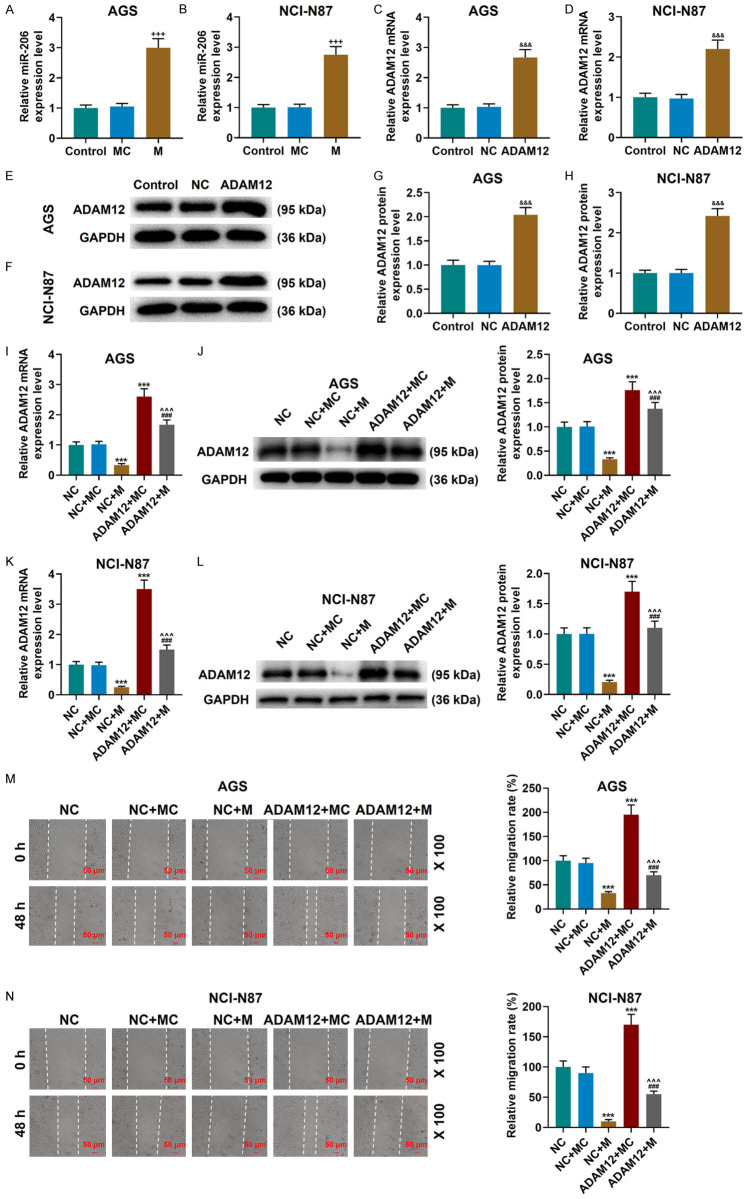
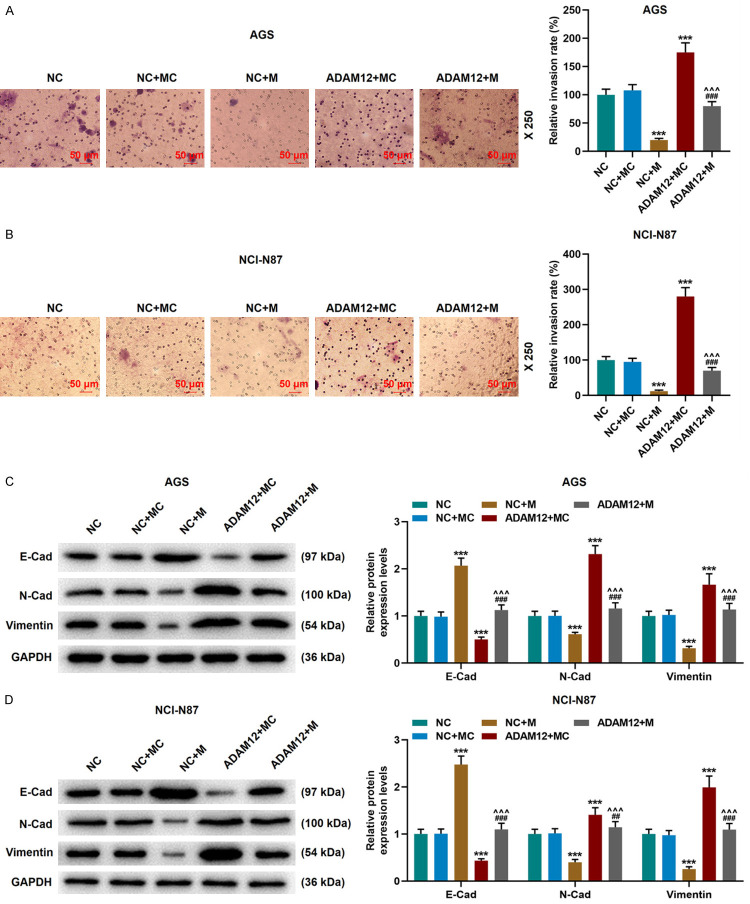
Subsequently, we verified the transfection efficiency of the cells transfected with miR-206 mimic and ADAM12 overexpression plasmids, and the expression of miR-206 was increased in the M group compared with the MC group, while the mRNA and protein expressions of ADAM12 were increased in ADAM12 group compared with the NC group (Figure 7A-H). The data also demonstrated that compared with the NC group, the expression of ADAM12 was down-regulated in the NC+M group but was up-regulated in the ADAM12+MC group, and that the expression of ADAM12 in the ADAM12+M group was lower than that in the ADAM12+MC group (P<0.001, Figure 7I-L). In addition, ADAM12 overexpression promoted the migration and invasion of gastric cancer cells, up-regulated the expressions of N-Cadherin and Vimentin, but down-regulated that of E-Cadherin. Interestingly, ADAM12 overexpression partially reversed the effects of miR-206 mimic on gastric cancer cells (P<0.001, Figures 7M-N, 8A-D).
Figure 7.
MiR-206 regulated gastric cancer migration via ADAM12. A, B. miR-206 mimic was transfected into AGS and NCI-N87 cells, and the transfection efficiency was detected by qRT-PCR. C-H. ADAM12 overexpression plasmid was transfected into AGS and NCI-N87 cells, and the transfection efficiency was detected by qRT-PCR and western blot. I-L. miR-206 suppressed ADAM12 expression, while the effect was partially reversed by overexpressing ADAM12. The results were determined by qRT-PCR and western blot. M, N. ADAM12 reversed the inhibitory effect of miR-206 mimic on cell migration. Scale: 50 µm; magnification: × 100. Each experiment was repeated three times independently. U6 and GAPDH were used as controls. qRT-PCR: quantitative reverse transcription real time polymerase chain reaction; NC, negative control; MC, mimic control; M, miR-206 mimic. +++P<0.001 vs. MC; &&&P<0.001 vs. NC; ***P<0.001 vs. NC+MC; ^^^P<0.001 vs. NC+M; ###P<0.001 vs. ADAM12+MC.
Figure 8.
MiR-206 targeted ADAM12 to regulate gastric cancer invasion and EMT-related protein expressions. A, B. The Transwell assay was used to detect the invasion of gastric cancer cells in the NC, NC+MC, NC+M, ADAM12+MC, and ADAM12+M groups. Scale: 50 µm; magnification: × 250. C, D. The expressions of N-Cadherin, Vimentin and E-Cadherin in gastric cancer cells of each group were determined by Western blot. NC, negative control; MC, mimic control; M, miR-206 mimic. ***P<0.001 vs. NC+MC; ^^^P<0.001 vs. NC+M; ###P<0.001 vs. ADAM12+MC.
Silent FAM225A inhibited tumor growth via miR-206/ADAM12 axis
The nude mouse xenograft model was constructed to further examine the effects of FAM225A on the cell proliferation of gastric cancer. As shown in Figure 9A, 9B, though the tumor size of mice in different groups was increased to some extent with time, the tumor size and volume of the siFAM225A group were significantly smaller than those of the siNC group (P<0.05). Ki-67 is a marker of cell proliferation [21]. It was found in our study that the Ki-67 level (brown part) in the siFAM225A group was greatly reduced (Figure 9C). Since the regulatory relationship of FAM225A-miR-206-ADAM12 network has been detected in the experiments in vitro, we further verified the expressions of FAM225A, miR-206, and ADAM12 in the mouse tissues in vivo. Compared with the siNC group, the expressions of FAM225A and ADAM12 were down-regulated, while that of miR-206 was up-regulated in the siFAM225A group (P<0.001, Figure 9D-F).
Figure 9.
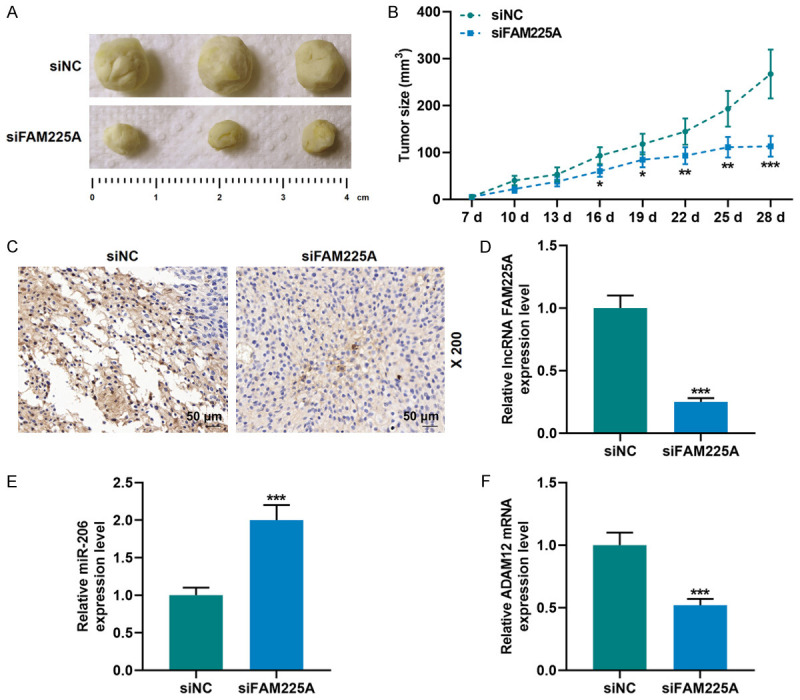
Silent lncRNA FAM225A inhibited gastric cancer tumor growth through miR-206/ADAM12. A, B. AGS cells transfected with siNC and siFAM225A were used to establish xenograft models, and tumor size was observed and tumor volume was determined. C. SiFAM225A inhibited Ki-67 expression, and the results were detected by immunohistochemistry. D-F. The expressions of lncRNA FAM225A, miR-206 and ADAM12 in mice injected with cells transfected with siNC and siFAM225A were detected by qRT-PCR. U6 and GAPDH were used as controls. Each experiment was repeated three times independently. qRT-PCR: quantitative reverse transcription real time polymerase chain reaction; siNC: small interfering RNA of negative control. ***P<0.001 vs. siNC.
Discussion
Increasing evidence has proved the participation of LncRNAs in a variety of biological functions, including the migration, proliferation, and metastasis of cancers by regulating miRNAs [22]. The latest research showed that FAM225A can act as a ceRNA to bind to miR-613 and regulate NOTCH3 expression, thus promoting the progression of colon cancer [23]. FAM225A is a relatively novel lncRNA, and its functions have been reported in two studies only [9,23]. FAM225A was found to competitively bind with miRNA to promote cancer growth [9,23]. Similarly, it was also found in our study that FAM225A expression was up-regulated in tissue samples of gastric cancer. Furthermore, silencing FAM225A inhibited the biological characteristics of gastric cancer cells in vitro and tumor growth in vivo. Our results indicated that FAM225A may be used as a predictor for the diagnosis of gastric cancer.
It was also found in this study that FAM225A can competitively bind with miR-206. The specific mechanism of action of gastric cancer-related lncRNAs in the cancer occurrence has been reported by many researchers. For example, Qi et al. found that lncRNA TUSC7 may serve as a ceRNA to inhibit the effect of miR-23b on target genes, which thus can be used to predict the overall survival of patients with gastric cancer [24]. Some scholars found that the combination of lncRNA HOTAIR and polycomb repressive complex 2 inhibits the expression of miR-34a and targets the growth factor-hepatocyte growth factor receptor pathway, thereby accelerating the EMT, tumor invasion and metastasis of gastric cancer cells [25]. However, this study is the first to reveal the role and mechanism of FAM225A in gastric cancer.
The role of miR-206 as a tumor suppressor gene has been previously reported [17,26]. Wang et al. found that miR-206 inhibited the proliferation, invasion and migration of thyroid cancer cells by inhibiting the expression of RAP1B [16]. Zhang et al. confirmed that miR-206 expression was down-regulated in esophageal cancer, and its overexpression inhibited the proliferation of esophageal cancer cells and induced apoptosis via the c-Met/AKT/mTOR signaling pathway [27]. Similarly, it has also been shown that miR-206 can inhibit the biological functions of gastric cancer cells through regulating the expression of MUC1 gene [28]. Our research indicated that miR-206 inhibitor promoted the metastasis of gastric cancer cells and could partially reverse the regulatory effects of siFAM225A. In addition, it has also been shown in previous studies that miRNAs act on the downstream target genes or signaling pathways to affect the malignant phenotype of tumors [29]. In this study, ADAM12 was confirmed as a downstream target gene of miR-206, suggesting that ADAM12 may regulate many biological behaviors of gastric cancer cells.
The results from current study suggested that ADAM12 expression was up-regulated in gastric cancer samples, and overexpression of ADAM12 promoted cell migration and invasion, accelerated the EMT process of gastric cancer cells, and partially reversed the tumor-suppressive effect of miR-206. The role of ADAM12 has also been studied and reported in other cancers [30]. For example, Wang et al. revealed that ADAM12 induced the EMT in pituitary adenoma cells through the EGFR/ERK signaling pathway to enhance the migration of cancer cells [31]. Huang et al. demonstrated that ADAM12 promoted the metastasis of breast cancer through sponging miR-34a [32]. The reports above further support the results obtained in the current research. In addition, by performing the experiments in vivo, we found that FAM225A interfered with tumor growth through the miR-206/ADAM12 axis, which is a new lncRNA-miRNA-mRNA regulatory network in gastric cancer.
To conclude, our research indicates that in gastric cancer, the expressions of FAM225A and ADAM12 are up-regulated and that of miR-206 is down-regulated. FAM225A competitively binds to miR-206 to regulate ADAM12 and promote the development of gastric cancer. The results of our research provide a better understanding of the clinical significance of FAM225A in the diagnosis and treatment of gastric cancer.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (Grant number: 2017YFC 1308900).
Disclosure of conflict of interest
None.
References
- 1.McGuire S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv Nutr. 2016;7:418–419. doi: 10.3945/an.116.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guggenheim DE, Shah MA. Gastric cancer epidemiology and risk factors. J Surg Oncol. 2013;107:230–236. doi: 10.1002/jso.23262. [DOI] [PubMed] [Google Scholar]
- 3.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–3165. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 5.Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437–445. doi: 10.1007/s00432-012-1324-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao Y, Tie Y, Du J, He J. LINC00152 promotes the proliferation of gastric cancer cells by regulating B-cell lymphoma-2. J Cell Biochem. 2019;120:3747–3756. doi: 10.1002/jcb.27655. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Y, Pan J, Geng Q, Wang G. LncRNA MALAT1 increases the stemness of gastric cancer cells via enhancing SOX2 mRNA stability. FEBS Open Bio. 2019;9:1212–1222. doi: 10.1002/2211-5463.12649. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Li T, Mo X, Fu L, Xiao B, Guo J. Molecular mechanisms of long noncoding RNAs on gastric cancer. Oncotarget. 2016;7:8601–8612. doi: 10.18632/oncotarget.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng ZQ, Li ZX, Zhou GQ, Lin L, Zhang LL, Lv JW, Huang XD, Liu RQ, Chen F, He XJ, Kou J, Zhang J, Wen X, Li YQ, Ma J, Liu N, Sun Y. Long noncoding RNA FAM225A promotes nasopharyngeal carcinoma tumorigenesis and metastasis by acting as ceRNA to sponge miR-590-3p/miR-1275 and upregulate ITGB3. Cancer Res. 2019;79:4612–4626. doi: 10.1158/0008-5472.CAN-19-0799. [DOI] [PubMed] [Google Scholar]
- 10.Zhang G, Pian C, Chen Z, Zhang J, Xu M, Zhang L, Chen Y. Identification of cancer-related miRNA-lncRNA biomarkers using a basic miRNA-lncRNA network. PLoS One. 2018;13:e0196681. doi: 10.1371/journal.pone.0196681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G, Zhao Q, Wu D, Gong W, Du M, Chu H, Wang M, Zhang A, Zhang Z. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol Cancer. 2018;17:87. doi: 10.1186/s12943-018-0829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J, Tang Z, Liao QX, Zhang H, Zeng LS, Cui SZ. LINC01133 as ceRNA inhibits gastric cancer progression by sponging miR-106a-3p to regulate APC expression and the Wnt/beta-catenin pathway. Mol Cancer. 2018;17:126. doi: 10.1186/s12943-018-0874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, Wang KM, Wang ZX. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shin VY, Chu KM. MiRNA as potential biomarkers and therapeutic targets for gastric cancer. World J Gastroenterol. 2014;20:10432–10439. doi: 10.3748/wjg.v20.i30.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie J, Tan ZH, Tang X, Mo MS, Liu YP, Gan RL, Li Y, Zhang L, Li GQ. MiR-374b-5p suppresses RECK expression and promotes gastric cancer cell invasion and metastasis. World J Gastroenterol. 2014;20:17439–17447. doi: 10.3748/wjg.v20.i46.17439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang P, Gu J, Wang K, Shang J, Wang W. miR-206 inhibits thyroid cancer proliferation and invasion by targeting RAP1B. J Cell Biochem. 2019;120:18927–18936. doi: 10.1002/jcb.29213. [DOI] [PubMed] [Google Scholar]
- 17.Samaeekia R, Adorno-Cruz V, Bockhorn J, Chang YF, Huang S, Prat A, Ha N, Kibria G, Huo D, Zheng H, Dalton R, Wang Y, Moskalenko GY, Liu H. miR-206 inhibits stemness and metastasis of breast cancer by targeting MKL1/IL11 pathway. Clin Cancer Res. 2017;23:1091–1103. doi: 10.1158/1078-0432.CCR-16-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Liu X, Jin H, Guo X, Xia L, Chen Z, Bai M, Liu J, Shang X, Wu K, Pan Y, Fan D. miR-206 inhibits gastric cancer proliferation in part by repressing cyclinD2. Cancer Lett. 2013;332:94–101. doi: 10.1016/j.canlet.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 20.Kim B. Western blot techniques. Methods Mol Biol. 2017;1606:133–139. doi: 10.1007/978-1-4939-6990-6_9. [DOI] [PubMed] [Google Scholar]
- 21.Menon SS, Guruvayoorappan C, Sakthivel KM, Rasmi RR. Ki-67 protein as a tumour proliferation marker. Clin Chim Acta. 2019;491:39–45. doi: 10.1016/j.cca.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 22.Tam C, Wong JH, Tsui SKW, Zuo T, Chan TF, Ng TB. LncRNAs with miRNAs in regulation of gastric, liver, and colorectal cancers: updates in recent years. Appl Microbiol Biotechnol. 2019;103:4649–4677. doi: 10.1007/s00253-019-09837-5. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, Shi H, Yao J, Li Y, Gao B, Zhang Y, Wang C, Zhou H, Zhang L. FAM225A facilitates colorectal cancer progression by sponging miR-613 to regulate NOTCH3. Cancer Med. 2020;9:4339–4349. doi: 10.1002/cam4.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi P, Xu MD, Shen XH, Ni SJ, Huang D, Tan C, Weng WW, Sheng WQ, Zhou XY, Du X. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer. 2015;137:1269–1278. doi: 10.1002/ijc.29516. [DOI] [PubMed] [Google Scholar]
- 25.Liu YW, Sun M, Xia R, Zhang EB, Liu XH, Zhang ZH, Xu TP, De W, Liu BR, Wang ZX. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. doi: 10.1038/cddis.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao H, Xiao W, Cao J, Li H, Guan W, Guo X, Chen K, Zheng T, Ye Z, Wang J, Xu H. miR-206 functions as a novel cell cycle regulator and tumor suppressor in clear-cell renal cell carcinoma. Cancer Lett. 2016;374:107–116. doi: 10.1016/j.canlet.2016.01.032. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Fa X, Zhang Q. MicroRNA206 exerts antioncogenic functions in esophageal squamous cell carcinoma by suppressing the cMet/AKT/mTOR pathway. Mol Med Rep. 2019;19:1491–1500. doi: 10.3892/mmr.2018.9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng M, Qin Y, Chen X, Wang Q, Wang J. MiR-206 inhibits proliferation, migration, and invasion of gastric cancer cells by targeting the MUC1 gene. Onco Targets Ther. 2019;12:849–859. doi: 10.2147/OTT.S180021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roy R, Dagher A, Butterfield C, Moses MA. ADAM12 is a novel regulator of tumor angiogenesis via STAT3 signaling. Mol Cancer Res. 2017;15:1608–1622. doi: 10.1158/1541-7786.MCR-17-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J, Zhang Z, Li R, Mao F, Sun W, Chen J, Zhang H, Bartsch JW, Shu K, Lei T. ADAM12 induces EMT and promotes cell migration, invasion and proliferation in pituitary adenomas via EGFR/ERK signaling pathway. Biomed Pharmacother. 2018;97:1066–1077. doi: 10.1016/j.biopha.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 32.Liu Z, Ren Z, Zhang J, Chuang CC, Kandaswamy E, Zhou T, Zuo L. Role of ROS and nutritional antioxidants in human diseases. Front Physiol. 2018;9:477. doi: 10.3389/fphys.2018.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]