Abstract
Circadian rhythms, referring to 24-h daily oscillations in biological and physiological processes, can significantly regulate host immunity to pathogens, as well as commensals, resulting in altered susceptibility to disease development. Furthermore, vaccination responses to microbes have also shown time-of-day-dependent changes in the magnitude of protective immune responses elicited in the host. Thus, understanding host circadian rhythm effects on both gut bacteria and viruses during infection is important to minimize adverse effects on health and identify optimal times for therapeutic administration to maximize therapeutic success. In this review, we summarize the circadian modulations of gut bacteria, viruses and their interactions, both in health and during infection. We also discuss the importance of chronotherapy (i.e., time-specific therapy) as a plausible therapeutic administration strategy to enhance beneficial therapeutic responses.
Keywords: circadian, bacteria, virus, infection, immunity
Circadian Rhythms
Circadian rhythms refer to 24-h oscillations of biological or physiological activity, which are found in almost all life on earth (Loudon, 2012; Westwood et al., 2019). In mammals, the timing of these rhythms is determined by a central core located in the suprachiasmatic nucleus (SCN) of the mammalian brain, with additional inputs from peripheral oscillators (circadian modulators located in cells or tissues outside of the SCN). These rhythms are strongly induced by light signals via the retinohypothalamic tract (Meijer et al., 1986, 1992) within the SCN, stimulating neuronal firing and inducing rhythmic neurotransmitter secretion, e.g., glucocorticoids such as vasoactive intestinal polypeptide (Shinohara et al., 1994), which coordinate and regulate peripheral rhythms located in other tissues of the body. This coordination by the SCN enables synchronization of these light-controlled rhythms for maximal host benefit, that includes induction of metabolic pathways in anticipation of dietary intake. Peripheral rhythms themselves are also influenced by other factors including hormones and food intake/nutrient availability (Balsalobre et al., 2000; Damiola et al., 2000). Together, the regulation of these rhythms is vital to maintain good health, as disruptions to these rhythms alter susceptibility to infections, autoimmunity, metabolic diseases, and cancer (Turek et al., 2005; Lee et al., 2010; Gibbs et al., 2014; Edgar et al., 2016; Voigt et al., 2016; Ehlers et al., 2018; Hopwood et al., 2018; Krakowiak and Durrington, 2018).
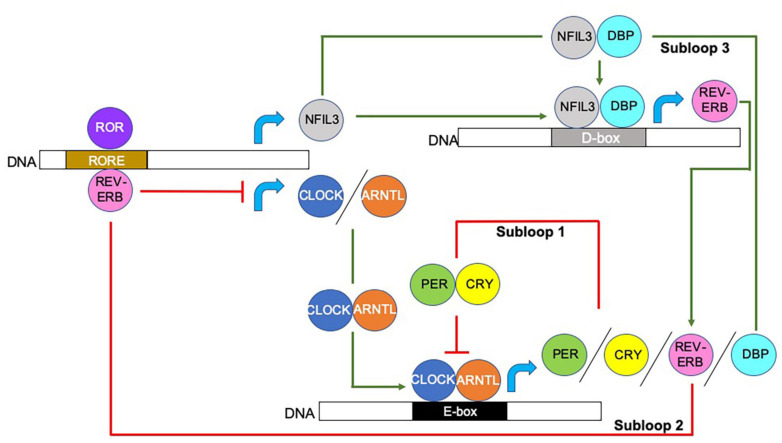
At the transcriptional level, circadian rhythms are controlled by integrated autoregulatory transcription-translation feedback loops, which direct proteins to bind to sequence-specific elements, resulting in the initiation or repression of circadian gene expression. The interlocking of the three transcriptional feedback loops, shown in Figure 1, generates ∼24-h cycles of transcription with varied phases of expression depending on the proteins available to bind to the promotors or enhancers of target clock-controlled genes (CCGs) (Takahashi, 2017). Together these loops enable a more responsive feedback mechanism that modulates the circadian rhythm in response to environmental changes or stimulations, and these include temperature, hormones, food intake, and microbes.
FIGURE 1.
Molecular modulation of circadian rhythm. Circadian rhythms are initiated and regulated by transcriptional/translational feedback loops. Circadian locomotor output cycles kaput (CLOCK) and Aryl hydrocarbon receptor nuclear translocator like (ARNTL; also known as BMAL1) proteins heterodimerize and bind to the E-box motifs of Period (Per) and Cryptochrome (Cry) genes, driving their transcription (Gekakis et al., 1998; Yoo et al., 2005; Xu et al., 2015). When PER and CRY protein concentrations increase, they too form heterodimers and counter-regulate the CLOCK/ARNTL complex, and thus, inhibit their own expression. CLOCK/ARNTL heterodimer proteins also drive the expression of the nuclear receptors REV-ERBα (encoded by Nr1d1; known as nuclear receptor subfamily group D member (Nr1d) 1) and REV-ERBβ (encoded by Nr1d2) (as shown by REV-ERB), which bind to retinoic acid-related orphan binding elements (ROREs) located in the CLOCK and ARNTL promoters, repressing Clock/Arntl transcription. A third subloop driven by the CLOCK/ARNTL complex, involves the proline and acidic amino acid-rich basic leucine zipper factors, D-box binding protein (DBP), thyrotroph embryonic factor and hepatic leukemia factor proteins (represented as DBP in the figure) (Takahashi, 2017). These proteins interact with the transcriptional repressor Nuclear Factor, Interleukin 3 Regulated (NFIL3; also known as E4BP4), the expression of which is driven by the REV-ERB/ROR loop, at sites containing D-boxes (Takahashi, 2017; Xie et al., 2019). Red lines indicate repression, green arrows indicate activation, blue curved arrows indicate gene transcription. Additional abbreviations used include deoxyribonucleic acid (DNA), Enhancer box (E-box) Destruction box (D-box), retinoic acid-related orphan receptor (ROR).
Microbes and Health
It has been estimated that ∼1,000 microbial species reside in the human intestines, encoding a vast metagenome with millions of unique genes, modulating host metabolism and regulation of immune responses (Whitman et al., 1998; Qin et al., 2010; Li et al., 2014). Microbial composition can be strongly influenced by many factors including genetics, age, lifestyle and diet (Ley et al., 2006; Turnbaugh et al., 2009; Wu et al., 2011; Goodrich et al., 2014; Bokulich et al., 2016; Odamaki et al., 2016; Sonnenburg and Bäckhed, 2016; Sonnenburg et al., 2016). Crosstalk between bacteria and the immune system is important for the education and development of a mature immune system, including the development of adaptive T-helper 17 cells (Th17) and regulatory T cells (Treg) (Ivanov et al., 2009; Atarashi et al., 2011, 2013), as well as the development of innate lymphoid cells (Gury-BenAri et al., 2016). Importantly, host bacterial composition influences disease susceptibility to obesity (Bäckhed et al., 2004; Turnbaugh et al., 2006; Vrieze et al., 2012), autoimmunity (Frank et al., 2007; Wen et al., 2008; Feng et al., 2010; Lee et al., 2011; de Goffau et al., 2013; Cekanaviciute et al., 2017; Pearson et al., 2019a), cancer (Arthur et al., 2012), and infectious diseases (Dubourg et al., 2017; Kaul et al., 2020; Yeoh et al., 2021). Furthermore, host bacterial composition can modulate responses to immunotherapy (Gopalakrishnan et al., 2018; Routy et al., 2018; Mager et al., 2020) and drugs (Wu et al., 2017; Zimmermann et al., 2019). Taken together, these reciprocal interactions highlight the important role bacteria play in shaping both our immune responses and therapeutic success against a variety of microbial infections, but also other conditions.
Similarly to bacteria, viruses, which are obligate parasites requiring infection of host cells in order to multiply, have co-adapted to their hosts and endogenous viral elements have been incorporated into host genomes millions of years ago (Simmonds et al., 2019). Alongside bacteria, viruses are often associated with many public health outbreaks, e.g., norovirus and rotavirus (Atmar and Estes, 2006; Harris et al., 2008; Lysén et al., 2009; Wikswo and Hall, 2012). Viruses have also been associated with global pandemics including the H1N1 influenza virus (Smith et al., 2009a, b) and SARS-CoV-2 (Wu Y. et al., 2020; Zhu et al., 2020), which have been responsible for the deaths of millions of people worldwide. Thus, there is great need to better understand how antiviral responses are stimulated and how they can be modulated to achieve protective antiviral responses, without promoting significant inflammation and cell damage. Therefore, better understanding of the mechanisms by which microbes can be modulated, including by circadian rhythms, are essential for identifying ways to improve the harnessing of microbes for promoting disease protection and therapeutic success.
Circadian Oscillations of Commensal Bacteria
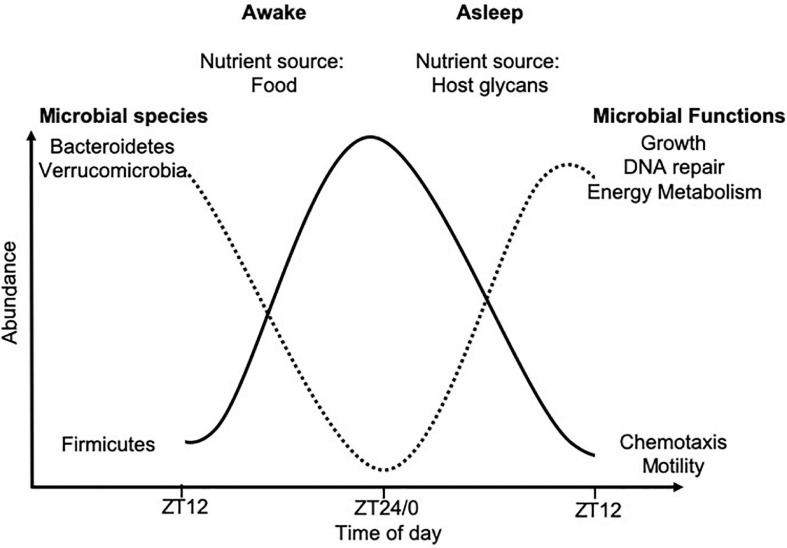
Within the intestine, circadian rhythms alter the regeneration of intestinal stem cells, production of gastric acid, gut motility, nutrient absorption and mucosal immunity (Larsen et al., 1991; Pan and Hussain, 2009; Hoogerwerf et al., 2010; Karpowicz et al., 2013; Mukherji et al., 2013; Yu et al., 2013; Wang et al., 2017). In addition, circadian rhythms alter the gut microbial composition and functions (Thaiss et al., 2014, 2016; Zarrinpar et al., 2014; Liang et al., 2015). This microbial rhythmicity is strongly linked to food intake and thus, nutrient availability. Mice housed in standard 12-h light/dark cycles with food ad libitum, were found to consume the majority of their food in the dark cycle, when they are most active, corresponding with cyclical changes in the cecal microbial composition, as studied by 16S rRNA sequencing (Hatori et al., 2012; Zarrinpar et al., 2014). Driving these microbial oscillations were time-of-day-dependent changes in the microbial abundances of species belonging to Firmicutes, Bacteroidetes, and Verrucomicrobia (Figure 2). Importantly, these time-of-day-dependent changes in microbial abundance were also associated with functional changes (Thaiss et al., 2014, 2016). Time-restricted feeding protocols, whereby mice are fed either in the light or dark cycles only, confirmed these microbial oscillations were regulated by dietary intake, rather than light exposure (Mukherji et al., 2015a, b).
FIGURE 2.
Time-of-day-dependent commensal microbial changes. Oscillations in microbial abundance and functions change at different times-of-day. The abundance of species belonging to the Firmicutes phylum (solid black line) increase in response to dietary intake, while species of the Bacteroidetes and Verrucomicrobia phyla (dotted black line) increase in abundance once all the nutrients from the food have been metabolized, due to their ability to metabolize host glycans. These data were compiled from animal studies with data shown over a 24-h period from ZT12 on 1 day, to ZT12 the following day. ZT12 refers to 12 h post light onset, while ZT24/0 refers to the end of the dark period and the start of the light cycle in standard 12-h light/dark housing conditions, with mice fed ad libitum.
Diet can also affect microbial rhythms, as mice given standard chow exhibited diurnal oscillations in microbial abundance, whereas, mice fed a high-fat diet showed significantly blunted microbial rhythms, which could be partially restored if given time-restricted feeding (Zarrinpar et al., 2014; Leone et al., 2015). A recent study in humans showed that arrhythmic bacteria (not changed in abundance at different times-of-day) could be linked to susceptibility to developing type 2 diabetes (Reitmeier et al., 2020), which confirmed the importance of these microbial rhythms in human health. It is not clear whether arrhythmic or rhythmic bacteria regulate susceptibility to other diseases. Obesity and obesity-associated health problems, e.g., hypertension and type 2 diabetes, are continuing to increase in prevalence worldwide (Blüher, 2019), and these individuals are more likely to develop severe complications and higher mortality from infections including influenza H1N1 (Huttunen and Syrjänen, 2010; Fezeu et al., 2011; Louie et al., 2011; Milner and Beck, 2012) and more recently SARS-CoV-2 (Sattar et al., 2020; Wu C. et al., 2020; Zhou et al., 2020). Thus, greater numbers of the people within the population are likely to be more susceptible to infectious disease in the future. Therefore, it is important to understand the contribution of the microbial rhythms in mediating susceptibility to infection and whether time-restricted feeding can reduce an individual’s susceptibility to infection.
It should also be noted that some bacteria have their own circadian rhythms. Cyanobacteria, e.g., Synechococcus elongatus, encodes three core clock genes (kaiA, kaiB, and kaiC), which regulate different bacterial biology including cell division, photosynthesis, and amino acid uptake, providing a survival advantage to the bacteria (Kondo et al., 1994). Klebsiella (previously designated Enterobacter) aerogenes, a commensal bacterium from the human gastrointestinal system, exhibits rhythms in swarming/motility in response to oscillating host melatonin levels (Paulose and Cassone, 2016; Paulose et al., 2016). This was postulated to be related to the bacteria expressing sequences similar to human melatonin binding sites, although the similarity is only ∼24–42%; thus, further studies are needed (Paulose et al., 2016). This bacterium also modulates circadian rhythms in response to temperature changes (Paulose et al., 2019), which affects host temperature changes after infection, and, in turn, it is likely to influence microbial commensal circadian rhythms. In addition, Escherichia coli and Pseudomonas aeruginosa encode receptors for blue and red light, respectively, enabling them to respond to light (Davis et al., 1999; Perlova et al., 2019), a dominant influencer of circadian rhythms; however, it is unknown if these receptors would induce bacterial oscillations in the intestine, given that there is no direct light source. Similarly, Legionella pneumophilia encodes homologues of the KaiBC proteins of cyanobacteria that regulate circadian gene expression; however, these gene homologues in Legionella pneumophilia appear to only enhance stress resistance (Loza-Correa et al., 2014). Given ∼1,000 microbial species live in the human intestine, it is likely that other bacteria may express genes or responses that can be controlled intrinsically in a circadian manner. More studies are needed to probe whether these intrinsic oscillations have any impact on other microbe rhythmicity or immune functions.
Host Influences on Bacterial Circadian Oscillations
Host circadian rhythms can also alter bacterial oscillations. Studies using mice deficient in either Bmal1 or Per1/2, or mice with a mutation in the Clock gene (altering the period, precision, and persistence of circadian rhythms) revealed that all these mice exhibit altered microbial rhythmicity and composition (Thaiss et al., 2014; Liang et al., 2015; Voigt et al., 2016). It is likely that this is a two-way interaction as germ-free (GF) mice, which lack microbiota, have different intestinal and hepatocyte circadian gene expression when compared to either specific pathogen-free (SPF; which have microbiota) mice or conventionalized mice (GF mice colonized with microbiota from SPF mice) (Leone et al., 2015; Weger et al., 2019). Leone et al. (2015) found that these diurnal oscillations modulated bacterial metabolite production, particularly butyrate, which had direct influence on circadian rhythm gene expression in the hepatocytes. Importantly, diet also modulated bacterial metabolites – butyrate oscillated in normal chow-fed mice, whereas hydrogen sulfide oscillated in high-fat diet fed mice. This further highlights the important link between diet, microbiota, and circadian rhythms.
The initial recognition of bacteria and virus by the host is through pathogen recognition receptors (PRR) including toll-like receptors (TLRs), nod-like receptors (NLRs), and inflammasomes, all of which oscillate in their expression at different times-of-day, in both hematopoietic and non-hematopoietic cells (Silver et al., 2012a, b, 2018; Mukherji et al., 2013; Wang et al., 2017, 2018; Pourcet et al., 2018). The presence of gut microbiota influences both their circadian clock and PRR expression (Mukherji et al., 2013; Wang et al., 2017). Interestingly, the circadian clock in intestinal epithelial cells relies on IL-23 secreted by TLR-stimulated dendritic cells, which activates type 3 innate lymphocyte (ILC3) cells to subsequently secrete IL-22 that in turn activates the circadian rhythms in epithelial cells (Wang et al., 2017). Inflammasomes are also regulated by circadian genes as Rev-erbα represses nlrp3 transcription by directly binding to the promoter region of the nlrp3 gene (Wang et al., 2018). While bacterial composition is regulated in a circadian manner, it is likely therefore, that microbial ligands will oscillate and thus responses to bacterial/viral ligands may vary in a time-dependent manner. This is likely to be important, as studies in macrophages have shown that the expression of different TLRs peak at different times (Silver et al., 2012a), suggesting that anti-microbe responses may also change at different times-of-day. Furthermore, these time-of-day-dependent PRR rhythms can also contribute to vaccine responses, as has been shown for B cell-secreted antibody responses (Silver et al., 2012b) (discussed in more detail later). To our knowledge, no studies to date have correlated the microbial oscillations with ligand abundance which could regulate PRR signaling at different times-of-day. This may partially explain the disparity between responses to different microbes at different times-of-day.
Hormones, e.g., glucocorticoids, are known to modulate circadian rhythms (Sollars et al., 2014), including influencing immune cell trafficking (Shimba et al., 2018), and immune responses (Gibbs et al., 2014). Interestingly, gut microbiota is also interconnected with glucocorticoids, as the microbiota can metabolize glucocorticoids, generating steroid metabolites (Morris and Brem, 2019), which affects the host’s immunity and behaviour (Wohlgemuth et al., 2011; Luo et al., 2018), whereas glucocorticoid treatment can alter the gut microbiota composition (Wohlgemuth et al., 2011; Huang et al., 2015; Guo et al., 2020). Thus far, little is known about the crosstalk between circadian rhythms, microbiota and glucocorticoid responses.
Sex has also been shown to influence microbial rhythmicity, with female mice exhibiting greater oscillations in microbial abundance (as determined by 16S copies/g feces), and a possible reduced fecal bacterial load compared to males (Liang et al., 2015). Given that sex can alter the microbial composition and disease susceptibility (Markle et al., 2013; Yurkovetskiy et al., 2013; Ding and Schloss, 2014; Singh and Manning, 2016; Sinha et al., 2019), it is possible that these alterations in microbial rhythmicity may play an important role in contributing to susceptibility to the diseases that affect more women than men, e.g., breast and thyroid cancers and autoimmune diseases such as systemic lupus erythematosus (Rees et al., 2016; Westergaard et al., 2019).
Importantly, changes in time-of-day or disruptions to the circadian rhythms can have large impacts on the host’s susceptibility to a number of health issues including obesity and susceptibility to infections. Bellet et al. (2013); Nguyen et al. (2013); Gibbs et al. (2014), viral pathogens (Edgar et al., 2016; Gagnidze et al., 2016; Ehlers et al., 2018) (discussed next), and parasites (Hopwood et al., 2018), indicating the importance of the host circadian rhythm in susceptibility to disease.
Circadian Rhythm Influences the Host’s Susceptibility to Pathogens
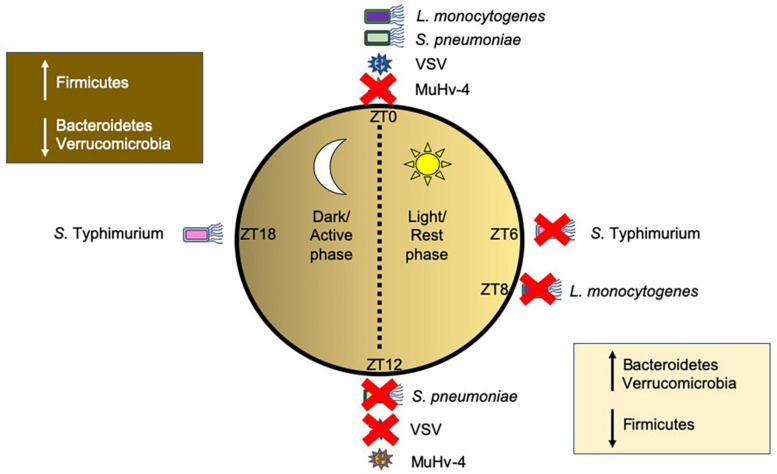
Both bacterial and viral pathogens exhibit time-of-day-dependent differences in their ability to replicate or induce immune responses (Figure 3). Time-of-day is often designated by Zeitgeber (ZT) measurements, whereby ZT0 represents the start of the circadian rhythm, i.e., when the light starts; any time after ZT0, for example ZT6, represents the number of hours (6 h) past the initiation of the circadian rhythm. In mice housed in standard 12 h light/dark cycles, ZT0-ZT12 represent the light phase when mice are resting, while ZT12–ZT24 (ZT0) represent the dark phase, when mice are most active. Mice infected with Salmonella Typhimurium (Bellet et al., 2013) or Streptococcus pneumoniae (Gibbs et al., 2014) showed enhanced ability of the host to reduce pathogen colonization and boost anti-bacterial immunity when infected during the active phase (ZT16 or ZT12, respectively), compared to the rest phase (ZT4 or ZT0, respectively). Similarly, mice infected with Listeria monocytogenes (Nguyen et al., 2013) exhibited better resolution of infection when infected later in the rest phase at ZT8, compared with infection at the start of the rest phase at ZT0. Clearly, host circadian rhythms modulate susceptibilities to bacterial pathogens at different times-of-day. Using myeloid cell-specific Bmal1-deficient mice (ArntlLoxP/LoxPLyz2Cre) to study diurnal rhythms of inflammatory Ly6Chi monocytes, Nguyen et al. (2013) found that the altered immune responses to Listeria monocytogenes infection of the mice correlated with different times-of-day. The ArntlLoxP/LoxPLyz2Cre mice developed enhanced bacterial-induced inflammation driven by increased recruitment of inflammatory monocytes and secretion of chemokines (CCL2, CCL8) as well as cytokines (IL-1β and IL-6). In addition, TNFα+iNOS+ DCs and IFNγ-secreting T cells were also increased. Thus, Bmal1is important in the control of Ly6Chi oscillations, limiting their recruitment at specific times-of-day to prevent inflammation and promote host survival. Similarly, in response to bacterial LPS and Salmonella Typhimurium infection, Clock-mutated bone marrow derived monocytes (BMDMs), causing a phase shift of 8 h, exhibited reduced proinflammatory cytokine responses compared to wild-type BMDMs (Bellet et al., 2013). While macrophages and monocytes had been implicated in the circadian responses to both Salmonella Typhimurium (Bellet et al., 2013) and Listeria monocytogenes (Nguyen et al., 2013), they were not essential in mediating host susceptibility to Streptococcus pneumoniae infection at different times-of-day (Gibbs et al., 2014). Instead, bronchial airway epithelial cells were found to modulate host susceptibility to infection mediated through rhythmic glucocorticoid signaling, which drove oscillations in Cxcl5 expression from the epithelium, and thus differences in recruitment of neutrophils to the site of infection. Both responses to Salmonella Typhimurium (Bellet et al., 2013) and Streptococcus pneumoniae (Gibbs et al., 2014) involve bacterial ligand (LPS/pneumolysin toxin) binding to TLR4 to trigger an immune response. As mentioned earlier, host TLRs can oscillate at different times of day both in the intestinal epithelium and in immune cell subsets, including macrophages and neutrophils (Silver et al., 2012a, b, 2018; Mukherji et al., 2013; Wang et al., 2017). Thus, it is likely that TLR oscillations are also involved in the regulation of the immune responses to these bacteria. While TLR4 oscillations were not observed in the mouse lung (Gibbs et al., 2014), this does not exclude the role of TLR4 rhythmicity, as a heterogeneous population of immune cells is present in the lung and each cell subset may have altered TLR rhythmicity. Thus, more studies are required to elucidate whether TLR signaling is vital in modulating these circadian differences.
FIGURE 3.
Time-of-day-dependent changes in modulating host susceptibility to pathogens. Host circadian rhythms influence the promotion of bacterial and viral disease depending on the time-of-day that the host was infected. Pathogens are shown around a circle representing different times of day with ZT0 representing the start of the circadian cycle (Bmal1 induction) and rest/light phase and ZT6, ZT8, ZT12, and ZT18 representing 6, 8, 12, and 18 h past light onset, where at ZT12 the dark/active phase starts. Red crosses indicate a time-of-day the host immune response is better prepared to prevent infection, while pathogens without a red cross indicate a time that is better suited for the pathogens to cause disease. This figure was generated from published murine studies. Other pathogens, including human pathogens, are likely to follow similar patterns. Changes in commensal gut microbial species at different times-of-day are also shown in boxes. Abbreviations: ZT, Zeitgeber, S. pneumoniae (Streptococcus pneumoniae), S. Typhimurium (Salmonella Typhimurium), L. monocytogenes (Listeria monocytogenes); VSV, Vesicular stomatitis virus; Mu-HV-4, Murid herpesvirus 4.
Crosstalk between viruses and host circadian rhythms influences host circadian rhythm modulation of viral infections and reciprocal viral modulation of host circadian rhythms. Unlike bacteria, viruses do not themselves have circadian rhythms of their own but viral infection of the host cell can influence and be influenced by host circadian rhythms. Mice infected with either murid herpesvirus 4 (MuHV-4) (Edgar et al., 2016) or herpes simplex virus 2 (HSV-2) (Matsuzawa et al., 2018) during the rest phase (ZT0–ZT12) showed greater protection from viral infection, compared to the mice infected during the active phase (ZT12–24). This protection correlated with high Bmal1 gene expression in infected fibroblasts/keratinocytes, which was confirmed by the studies using Bmal1-/- mice, whereby viral load was significantly higher in Bmal1-/- mice compared to wild-type mice. In addition, mice infected with the DNA virus, herpes simplex virus 1 (HSV-1) as well as the RNA viruses, Sendai virus and murine influenza A (IAV) also showed enhanced viral replication in vivo in Bmal1-/- hosts (Edgar et al., 2016; Ehlers et al., 2018). Moreover, viral replication of the RNA viruses, respiratory syncytial virus (RSV) and parainfluenza virus type 3 (PIV3) was also increased in Bmal1-deficient cells in vitro (Majumdar et al., 2017). It was intriguing that Sendai virus, IAV, RSV, PIV3 (all RNA viruses), which encode their own RNA-dependent RNA polymerases, and therefore do not utilize host transcriptional machinery for viral gene expression, unlike the herpes viruses (DNA viruses) which are more reliant on host transcription for gene expression, yet Bmal1-deficiency increases viral replication in all these viruses. Treatment of human A549 cells (an alveolar carcinoma line) with 100 nM siRNA targeting Bmal1 (72% knockdown) led to higher PIV3 and RSV infections, respectively, agreeing with studies conducted in Bmal1-/- mice (Majumdar et al., 2017). Seasonal variation in Bmal1 expression in humans, namely Bmal1 expression is lower in the winter months, coincides with higher disease risk marker expression. This seasonal variation in Bmal1 may explain the increased susceptibility to viral infections during winter months (Dowell, 2001; Dopico et al., 2015; Martinez, 2018). Protein analysis comparing primary fibroblasts from WT and Bmal1-/- mice indicated that circadian rhythms can control viral replication through many mechanisms, including alteration of proteins related to virus particle uncoating, genome trafficking, chromatin assembly, virus protein biosynthesis, viral assembly and egress (Edgar et al., 2016). In addition, expression of Nectin1, the receptor for HSV-2 (Taylor et al., 2007), oscillates in both mouse skin and human keratinocytes, peaking at ZT18, suggesting a mechanism for the increased infection at ZT18 vs. ZT6 (Matsuzawa et al., 2018). Altered immune responses to viruses have also been implicated in time-of-day-dependent susceptibilities to infection. For example, Sendai virus infections in Bmal1-/- mice exacerbated inflammation, by increasing CCL2, CXCL5, IFNs, and IL-6 production (Ehlers et al., 2018). Furthermore, in studies of RSV infection, human nasal wash samples showed virally-modulated impacts on circadian genes. In infants hospitalized with RSV bronchiolitis, reduced Bmal1 expression (Ehlers et al., 2018) was found in the nasal wash samples compared to uninfected controls, and this likely contributed to the higher levels of viral replication, and thus, worse clinical manifestation.
Interestingly, some viral infections exploit host susceptibility during the rest phase, suggesting different virus-specific requirements at different times-of-day. Mice intranasally infected with vesicular stomatitis virus (VSV) developed encephalitis, but interestingly, the mice were more protected from the disease if the infection was at ZT12 compared with those infected at ZT0 (40% vs. 95% mortality, respectively) (Gagnidze et al., 2016). Further, the protective effect was due to Rev-Erbα-mediated ccl2 repression at the start of the active phase, leading to reduced recruitment of inflammatory cells, and thus reduced disease severity. Similarly, Rev-Erbα agonists repressed the replication of hepatitis B and C viruses (HBV, Zhuang et al., 2021; HCV, Zhuang et al., 2019) and human immunodeficiency virus (HIV, Chang et al., 2018; Borrmann et al., 2020), whereas Bmal1 expression promoted viral replication. It is noteworthy that Bmal1 is negatively regulated by Rev-Erbα. It is clear Bmal1 has a pleiotropic role whereby Bmal1 promotes VSV, HBV, HCV, and HIV replication, while suppressing IAV, RSV, HSV-2, PIV3, HSV-1, and MuHV-4 replication. Therefore, there are likely to be additional virus-specific pathways, which modulate circadian rhythms that need to be further elucidated.
Other studies have shown that viral infections disrupt epigenetic mechanisms, affecting the functioning of the circadian clock. Using an in vitro HCV infection system, HCV core protein (genotype 1b) decreased PER2 and CRY2 protein levels in infected HuH-7 cells (human hepatoma cell line), whereas overexpression of Per2 in HuH-7 cells reciprocally decreased HCV RNA replication (Benegiamo et al., 2012). Further study suggested that the CDH1 (E-cadherin) promoter region of the HCV core protein(+) Huh-7 cells becomes substantially hypermethylated, and the reduced CDH1 protein expression in hepatocellular carcinoma patients was associated with poor prognosis (Chen et al., 2014). Hypermethylation of the CDH1 (E-cadherin) promoter region also increased SIRT1 protein levels in HCV core protein+ Huh-7 cells (Ripoli et al., 2011). Importantly, SIRT1 is known to regulate the circadian clock in hepatocytes, controlling regeneration, proliferation, and metabolism of hepatocytes (Bellet et al., 2016; Sato et al., 2017). Thus, HCV modulates circadian rhythms via manipulation of the epigenetic architecture. Further investigation into epigenetic changes induced by viruses and bacteria in modulating circadian rhythms is needed.
It should be noted that the effect of circadian rhythms on the same virus could have different effects, dependent on whether infection is active or latent. Acute herpes viral infections can be influenced by circadian rhythms, whereas there was no evidence suggesting that latent infections are similarly influenced (Edgar et al., 2016). However, whether particular stimuli that induce reactivation of latent viral infections can modify the influence on circadian rhythms remains to be studied. In addition, whether circadian dysregulation promotes viral reactivation also is yet to be studied.
Together, these studies, just discussed, indicate that the time-of-day can substantially alter the potency of the pathogenicity and the host immunity. Importantly, the time at which the host is most susceptible to pathogen infection varies and depends on the type of virus and bacteria, all of which may have important clinical implications. Thus far, circadian rhythms and timed viral co-infection have not been studied; however, this type of investigation may enable us to better understand how circadian rhythm modulated pathways and the pathogens interact. Studying bacterial and viral pathogens known to differentially modulate host susceptibility at different times-of-day may provide important insight into new pathways that could be therapeutically targeted to boost host protection. Alterations in the oscillations of gut commensal microbiota composition have yet to be studied in response to viral and bacterial pathogens, which likely play an important role in modulating host susceptibility to infection, as discussed next.
Bacterial-Viral Crosstalk in Infection
It is known that viral infections can alter the gut microbiota composition, including HIV, influenza virus, norovirus, rotavirus, HBV and HCV in humans, which can in turn influence disease severity (reviewed in Li et al., 2019; Yuan et al., 2020). Individuals with HIV infection had reduced microbial richness and depleted Bacteroidia members but their stool samples were enriched in species belonging to the Proteobacteria phyla (Vujkovic-Cvijin et al., 2013). Interestingly, the reduced alpha diversity (number of different microbial species present) (Noguera-Julian et al., 2016), was associated with the severity of immunodeficiency (Noguera-Julian et al., 2016). However, this could be restored following anti-retroviral therapy (Ji et al., 2018). Importantly, the alterations in microbial composition were also associated with altered microbial functions, whereby HIV infection repressed the generation of proline, phenylalanine, and lysine (all amino acids) (Serrano-Villar et al., 2016), while promoting tryptophan catabolism (Vujkovic-Cvijin et al., 2013) by the microbiota. Given Proteobacteria and Bacteroidetes (which includes the Bacteroidia members) oscillate, it is conceivable that circadian rhythms influence the microbiota composition in those individuals. Administration of antibiotics/probiotics/prebiotics in mice, as well as studies in germ-free (GF; which do not have any bacteria) or gnotobiotic mice (which have a defined microbial community), have confirmed that viral-bacterial crosstalk can modulate susceptibility to viral infection and disease, e.g., norovirus infection (Schaffer et al., 1963; Isaak et al., 1988; Cadwell et al., 2010; Ichinohe et al., 2011; Basic et al., 2014; Osborne et al., 2014; Robinson K. M. et al., 2014; Pearson et al., 2019b).
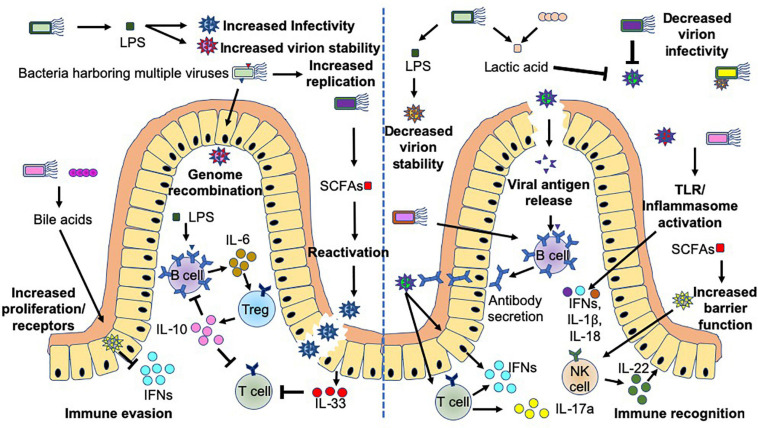
Commensal bacteria exhibit both stimulatory and suppressive functions in controlling viral infection, through direct and indirect mechanisms, which can lead to harmful or beneficial outcomes for the host (Figure 4). Commensal microbiota can promote viral infections directly through the production of microbial ligands (e.g., LPS) or metabolites (e.g., Bile acids or SCFAs), which can promote: (1) target cell proliferation and upregulation of virus receptors, e.g., CD300lf (a receptor for murine norovirus) on Tuft cells (von Moltke et al., 2016; Nelson et al., 2018; Wilen et al., 2018), (2) reactivation of viruses (Gorres et al., 2014), (3) increased virion stability (Robinson C. M. et al., 2014; Berger et al., 2017), and (4) increased viral replication (Kuss et al., 2011). Importantly, norovirus infection of B cells can be promoted or prevented depending on the presence of bacteria expressing H-type histo-blood group antigens (Jones et al., 2014; Karst, 2015). In addition, enteric bacteria have been reported to enhance enteric viral (poliovirus) co-infection, which consequently can facilitate genetic recombination and enhance viral fitness (Erickson et al., 2018). Importantly, commensal bacteria can also modulate the immune response, promoting viral infection (Jude et al., 2003; Kane et al., 2011; Young et al., 2012; Uchiyama et al., 2014; Baldridge et al., 2015; Wilks et al., 2015). Two examples with different mechanisms of virus-mediated immune suppression include mouse mammary tumor virus (MMTV), which primarily infects intestinal dendritic cells, B cells and T cells (Held et al., 1993; Beutner et al., 1994) and Norovirus, which primarily infects intestinal Tuft cells (Wilen et al., 2018). In MMTV infection, MMTV in conjunction with bacterial LPS (signaling through TLR4), strongly induces proinflammatory IL-6, which in turn promotes Tregs to produce IL-10, a potent immune-suppressing cytokine, promoting viral escape from the immune system (Jude et al., 2003; Kane et al., 2011; Wilks et al., 2015). However, in norovirus infection, the presence of gut bacteria downregulates antiviral IFNλ receptor expression in the intestine and subsequently reduces intracellular downstream signaling via Stat1 and Irf3 (Baldridge et al., 2015). This should limit antiviral immunity and the survival of the virus.
FIGURE 4.
Bacteria-virus crosstalk can promote virus infection or enhance host protection. Interactions between bacteria and viruses can promote infection (left hand-side), or protection of the host (right-hand side) by multiple different mechanisms. These mechanisms focus on alterations to viral stability, replication, genetic recombination, antigen release, mucosal barrier functions and host immunity including cytokine release and antibodies. Bacteria influence viral crosstalk both directly, e.g., Lipopolysaccharide (LPS) binding to the virus or bacterial stimulation of host Toll-like receptor (TLR) or inflammasomes, and indirectly, including through the production of short chain fatty acids (SCFAs e.g., butyrate) or metabolism of bile acids. Additional abbreviations include: IFN (interferons) and IL- (Interleukin-).
On the other hand, commensal microbiota can also suppress viral infections by multiple mechanisms including: (1) directly binding to the virus, whereby the virus may be destabilized, prevented from being internalized and/or viral replication is suppressed (Botić et al., 2007; Conti et al., 2009; Mastromarino et al., 2011; Chen et al., 2016; Bandoro and Runstadler, 2017), and (2) boosting antiviral immunity and mucosal barrier integrity. This antiviral immunity includes enhancing viral antigen presentation to antigen-specific T and B cells, leading to the activation of cytotoxic CD8+ T cells, generation of IFN-γ-producing Th1 cells and IL-17a-secreting Th17 cells (Hensley-McBain et al., 2016), as well as synthesis of virus-specific antibodies (Ichinohe et al., 2011; Oh et al., 2014) to control and limit the infection. Furthermore, bacterial and viral ligands can activate toll-like receptors and inflammasomes, both of which promote the secretion of IFNs, IL-1β and IL-18 (Ichinohe et al., 2011; Wang et al., 2012; Oh et al., 2014) from a range of cells, while reducing IL-33 secretion (Oh et al., 2016) and enhancing IL-22 (Hensley-McBain et al., 2016) and IFN responses (Steed et al., 2017). Together, they boost antiviral immune responses and mucosal barrier integrity.
Interestingly, microbial products, such as LPS, can also both promote and suppress viral infections, which may be due to different LPS structures that have different immunostimulatory effects (Vatanen et al., 2016). This has been reported in MMTV studies, where MMTV virions bound to E. coli LPS had a greater ability to stimulate IL-6 production from splenocytes than B. theta LPS (Wilks et al., 2015). However, further studies are required to determine whether the differences in LPS from the various bacterial sources, and/or their structures, differentially modulate virus stability. Interestingly, different serotypes of E. coli LPS can differentially alter body temperature in rats (Dogan et al., 2000). As the peripheral circadian oscillators sense temperature changes, it is also possible that the origin of the LPS may differentially induce temperature changes in vivo and alter circadian rhythmicity. Furthermore, as both bacteria and TLRs oscillate, including TLR4, it is highly likely this will have an effect on viral promotion or suppression.
Bacteriophages, viruses which infect and subsequently destroy bacteria, are highly abundant and can modulate bacterial composition of the host (Hsu et al., 2019; Khan Mirzaei et al., 2020) and can prevent infection of the host by pathogenic bacteria by adhering to the mucus layer (Dubos et al., 1943; Matsuzaki et al., 2003). Importantly, bacteriophages have also been shown to be influenced by circadian rhythms (Kao et al., 2005; Liu et al., 2019). Cyanophages, which infect cyanobacteria, have been shown to exhibit diurnal rhythms in their ability to infect bacteria and in gene expression, which relates to the ability of the cyanobacteria to undergo photosynthesis (Liu et al., 2019). While this provides the first evidence of circadian modulation of phages, future studies are needed to determine whether phages in humans or mice exhibit similar changes. It will also be important to develop new technologies to fully understand the potential of these as microbial modulators and as modulators of circadian rhythms (Khan Mirzaei and Deng, 2021).
Further studies evaluating crosstalk between viruses and bacteria in circadian rhythms are urgently needed. Studies involving GF and gnotobiotic mice would be greatly insightful, so would co-infection models. Better understanding of these interactions would undoubtedly help not only to identify novel pathways for therapeutic targeting but also to better understand how therapy may inadvertently alter susceptibility to other pathogens, including opportunistic pathogens.
Circadian Influences on Antibacterial and Antiviral Therapy
Therapies targeting the circadian clock have shown promise in controlling bacterial and viral infection. SR9009, a REV-ERB agonist, which suppresses Bmal1, has successfully inhibited viral entry and replication of HCV (Zhuang et al., 2019), HBV (Zhuang et al., 2021), and HIV-1 (Borrmann et al., 2020). Likewise, SR8278, a REV-ERB antagonist, which represses Rev-erbα and promotes Bmal1 expression, has rendered mice more susceptible to VSV infection when infected at ZT12, compared to the more protected mice infected at ZT12 without REV-ERB antagonist treatment (Gagnidze et al., 2016). It is unknown, thus far, if Rev-erb agonists change the gut bacterial composition and function. Thus, these studies need to be conducted, and combined with studies of viral infection. While the therapies targeting circadian proteins have shown promise with respect to infection, it is important to consider broader effects. For example, Rev-erbα modulates Th17 cell development (Yu et al., 2013), and thus Rev-erbα agonists may promote cell development, while antagonists may prevent Th17 expansion. It is possible that Th17-promoting Rev-erbα agonists may subsequently result in intestinal dysbiosis in the host, due to dysregulated Th17 immune responses to gut bacteria. In addition, given the plasticity of Th17, Treg, and Th1 responses (Lee et al., 2009; Wei et al., 2009; Mukasa et al., 2010), any Rev-erbα modulation needs to be considered in all these aspects for additional immune impacts that may alter both the efficacy of treatment and susceptibility to other health problems.
The specific timing of therapeutic intervention, referred to as chronotherapy, may also play a role in modulating bacterial and viral crosstalk. In murine HSV-2 infection, time-of-day influences the survival of the infected mice, with infections at ZT6 repressing viral replication and promoting survival of the host, compared to infections at ZT18 (Matsuzawa et al., 2018). Interestingly, when these mice were infected at ZT18, a fourfold higher dose of acyclovir (administered 30 min prior to infection) was required for mice infected at ZT18, compared to those infected at ZT6 to obtain similar survival and clinical scores (100 mg/kg vs. 25 mg/kg). Therefore, the time of infection can make a substantial difference to the drug dosage needed to control the infection, which for some therapies may not be possible due to the enhanced toxicity or side effects of the drug when higher doses are used. In addition, the use of higher doses may also promote host resistance to the effective therapy. It is unclear whether antibiotics may have a similar effect on bacterial pathogens at different times-of-day. If so, administering the antibiotics at a time synchronized with the timing of optimal anti-microbial host immune responses may prove beneficial and prevent suboptimal doses of antibiotics being used, and thus reduce antibiotic resistance.
Given that the microbiota have the ability to metabolize many drugs (Zimmermann et al., 2019), and that the abundance of bacteria can oscillate, it is likely that this change in the microbiota at different times-of-day may alter the therapeutic responses that the drugs elicit. For example, certain bacterial species can enhance the success of therapy with immune checkpoint blockade in mice and cancer patients (Sivan et al., 2015; Vétizou et al., 2015; Mager et al., 2020) and gut bacteria could also enhance the responses to chemotherapy via modulating the tumor microenvironment and immune cell function (Iida et al., 2013; Viaud et al., 2013). Importantly, not only does drug-metabolism by microbiota have a local effect, but systemic consequences can occur, particularly in the liver, where hepatic drug metabolism can also be influenced by circadian oscillations (Kim and Lee, 1998; Lin et al., 2019; Wang et al., 2019). Interestingly, in the liver, Bmal1 regulates oscillations in drug metabolizing genes, as Bmal1-deficient mice showed higher levels of toxicity from metabolizing xenobiotics (Lin et al., 2019). Given that Bmal1 promotes oscillations in drug-metabolizing genes and toxicity sensitivity, and in addition, Bmal1 promotes both HBV and HCV infection in hepatocytes, some potential adverse effects of circadian therapies (i.e., Rev-erb agonists) on drug metabolism, which may result in increasing toxicity, should also be considered.
Circadian oscillations also modulate vaccine responses to bacterial and viral pathogens. The magnitude of antibody response to the trivalent inactivated influenza vaccine in humans correlated with early TLR5 expression (Oh et al., 2014). Microbial flagellin is a ligand for TLR5 (Hayashi et al., 2001) and stimulation of TLR5 boosted plasma cell numbers and antibody titers in response to the influenza vaccine (Oh et al., 2014). TLR5 was also involved in boosting antibody responses to the inactivated polio vaccine but not to adjuvanted (Tetanus-Diphtheria-pertussis) vaccine or the live-attenuated yellow fever vaccine (YF-17D). Interestingly, antibody responses to the trivalent influenza vaccine are also altered at different times-of-day, with higher titers observed in elderly individuals vaccinated in the morning compared with the afternoon (Long et al., 2016). It is unclear whether circadian oscillations in gut bacteria contribute to the alteration seen in anti-influenza antibody response; however, it is certainly possible, as TLRs oscillate and are likely to be involved in bacterial and viral ligand sensing. Further studies need to be conducted for other vaccinations and in different age groups. Given that elderly people have reduced immune responses compared to younger individuals (Gustafson et al., 2020), timing of the vaccination to boost the protective response is clinically relevant.
Applicability of Animal Models for Human Circadian Studies
As discussed, it is clear animal models have a circadian rhythm and that both bacterial and viral organisms are able to modulate these rhythms to either promote better survival of the host or better advantage to the pathogen. While these studies have been enlightening and have some similarities with human circadian rhythm studies, it is important to consider the true implication of animal circadian studies and how these rhythms may be different to humans.
Both mice and humans have an evolutionarily preserved molecular clock and SCN, which controls the circadian rhythms in response to light; however, as mice are nocturnal and humans are diurnal their responses to light can be different. Mice can entrain to light exposure of 1 lux for a few minutes, while humans require both a high light intensity (>100 lux’s) and a longer duration (>30 min) to entrain (Daan and Pittendrigh, 1976; Khalsa et al., 2003). Thus, differences in study design related to light intensity and duration can have different effects between the species and thus the induction of circadian rhythms. Similarly, laboratory animals are generally housed in abrupt 12-h light/dark cycles, whereas humans can experience seasonal variations in light exposure (Jasser et al., 2006; Higuchi et al., 2007). Interestingly, nocturnal vs. diurnal animal species can also have a different time window and magnitude of response when serotonin is able to modulate circadian rhythms (Horikawa and Shibata, 2004; Novak and Albers, 2004; Cuesta et al., 2008). Thus, changes in response and timing need to be considered when considering translation and applicability of animal studies into human studies.
There are many other factors that may also be involved in modulating circadian differences between mice and humans. For example, changes in microbial composition between the species, as well as food type and time eaten, will also be important in determining microbial influences on circadian rhythms. Furthermore, the laboratory animals studied are often in-bred and thus, in comparison to humans, exhibit much less genetic diversity, which also may influence the applicability of murine studies to humans. However, as for other types of research that use animal models, murine studies provide a signpost for areas of investigation when considering circadian rhythms in humans. Thus, it is important for studies conducted in animal models to be confirmed in humans.
Summary
Circadian rhythms modulate the composition and function of commensal bacteria, and host susceptibility to bacterial and viral infection. While the interactions between circadian rhythms and viral or bacterial infection have been separately studied, there is a significant knowledge gap regarding the three-way crosstalk among viruses, bacteria and circadian rhythms. Thus, the studies with co-infection (bacteria and virus) may provide important insight into the crosstalk. It is clear that circadian clock manipulation and vaccination at particular times-of-day may have important implications for boosting host protection from infection and inflammation. Thus, there is a need to better understand these interactions. As discussed earlier, not all viruses have the same time-of-day virulence to infect the host; thus, we need to consider how to target individual viruses that may alter susceptibility to infection by other viruses, opportunistic bacteria or immune-mediated diseases. By better understanding these interactions we may be able to reduce antiviral/antibiotic resistance by using lower doses of drugs, which may also result in fewer toxic side effects. Although it is difficult to pinpoint the time when an infection occurs in humans, it is possible to boost the protective immune responses at the optimal time to enhance therapeutic success, whether by vaccination or immune-modulating therapies. Finally, given the impact that circadian rhythms have on microbes and immunity, it would be important to consider the role of circadian rhythms in the design and implementation of experiments in both animal and human studies.
Author Contributions
JAP wrote the manuscript. ACV and KB-B conducted research for the manuscript. FSW and LW edited the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding. This work was funded by a Medical Research Council Career Development Award (MR/T010525/1) and a JDRF UK grant (1-SGA-2021-002) to JAP; MRC research grant (MR/K021141/1) to FSW and National Institutes of Health (DK 045735, HD 097808), Diabetes Action Research and Education Foundation and Diabetes Research Connection to LW.
References
- Arthur J. C., Perez-Chanona E., Mühlbauer M., Tomkovich S., Uronis J. M., Fan T. J., et al. (2012). Intestinal inflammation targets cancer-inducing activity of the microbiota. Science. 338 120–123. 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500 232–236. 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., et al. (2011). Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331 337–341. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmar R. L., Estes M. K. (2006). The epidemiologic and clinical importance of norovirus infection. Gastroenterol. Clin. North Am. 35 275–290. 10.1016/j.gtc.2006.03.001 [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101 15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge M. T., Nice T. J., McCune B. T., Yokoyama C. C., Kambal A., Wheadon M., et al. (2015). Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 347 266–269. 10.1126/science.1258025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsalobre A., Brown S. A., Marcacci L., Tronche F., Kellendonk C., Reichardt H. M., et al. (2000). Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science 289 2344–2347. 10.1126/science.289.5488.2344 [DOI] [PubMed] [Google Scholar]
- Bandoro C., Runstadler J. A. (2017). Bacterial lipopolysaccharide destabilizes influenza viruses. mSphere 2:e267–17. 10.1128/mSphere.00267-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basic M., Keubler L. M., Buettner M., Achard M., Breves G., Schröder B., et al. (2014). Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis. 20 431–443. [DOI] [PubMed] [Google Scholar]
- Bellet M. M., Deriu E., Liu J. Z., Grimaldi B., Blaschitz C., Zeller M., et al. (2013). Circadian clock regulates the host response to Salmonella. Proc. Natl. Acad. Sci. U.S.A. 110 9897–9902. 10.1073/pnas.1120636110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellet M. M., Masri S., Astarita G., Sassone-Corsi P., Della Fazia M. A., Servillo G. (2016). Histone deacetylase SIRT1 controls proliferation, circadian rhythm, and lipid metabolism during liver regeneration in mice. J. Biol. Chem. 291 23318–23329. 10.1074/jbc.M116.737114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benegiamo G., Vinciguerra M., Mazzoccoli G., Piepoli A., Andriulli A., Pazienza V. (2012). DNA methyltransferases 1 and 3b expression in Huh-7 cells expressing HCV core protein of different genotypes. Dig. Dis. Sci. 57 1598–1603. 10.1007/s10620-012-2160-1 [DOI] [PubMed] [Google Scholar]
- Berger A. K., Yi H., Kearns D. B., Mainou B. A. (2017). Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog. 13:e1006768. 10.1371/journal.ppat.1006768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutner U., Kraus E., Kitamura D., Rajewsky K., Huber B. T. (1994). B cells are essential for murine mammary tumor virus transmission, but not for presentation of endogenous superantigens. J. Exp. Med. 179 1457–1466. 10.1084/jem.179.5.1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blüher M. (2019). Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 15 288–298. 10.1038/s41574-019-0176-8 [DOI] [PubMed] [Google Scholar]
- Bokulich N. A., Chung J., Battaglia T., Henderson N., Jay M., Li H., et al. (2016). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8:343ra82. 10.1126/scitranslmed.aad7121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrmann H., Davies R., Dickinson M., Pedroza-Pacheco I., Schilling M., Vaughan-Jackson A., et al. (2020). Pharmacological activation of the circadian component REV-ERB inhibits HIV-1 replication. Sci. Rep. 10:13271. 10.1038/s41598-020-70170-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botić T., Klingberg T. D., Weingartl H., Cencic A. (2007). A novel eukaryotic cell culture model to study antiviral activity of potential probiotic bacteria. Int. J. Food Microbiol. 115 227–234. 10.1016/j.ijfoodmicro.2006.10.044 [DOI] [PubMed] [Google Scholar]
- Cadwell K., Patel K. K., Maloney N. S., Liu T. C., Ng A. C., Storer C. E., et al. (2010). Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell. 141 1135–1145. 10.1016/j.cell.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cekanaviciute E., Yoo B. B., Runia T. F., Debelius J. W., Singh S., Nelson C. A., et al. (2017). Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc. Natl. Acad. Sci. U.S.A. 114 10713–10718. 10.1073/pnas.1711235114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. C., Naranbhai V., Stern J., Roche M., Dantanarayana A., Ke R., et al. (2018). Variation in cell-associated unspliced HIV RNA on antiretroviral therapy is associated with the circadian regulator brain-and-muscle-ARNT-like-1. AIDS 32 2119–2128. 10.1097/QAD.0000000000001937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. W., Liu P. F., Liu Y. T., Kuo S., Zhang X. Q., Schooley R. T., et al. (2016). Nasal commensal Staphylococcus epidermidis counteracts influenza virus. Sci. Rep. 6:27870. 10.1038/srep27870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhao J., Ma R., Lin H., Liang X., Cai X. (2014). Prognostic significance of E-cadherin expression in hepatocellular carcinoma: a meta-analysis. PLoS One. 9:e103952. 10.1371/journal.pone.0103952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C., Malacrino C., Mastromarino P. (2009). Inhibition of herpes simplex virus type 2 by vaginal lactobacilli. J. Physiol. Pharmacol. 60(Suppl. 6) 19–26. [PubMed] [Google Scholar]
- Cuesta M., Mendoza J., Clesse D., Pévet P., Challet E. (2008). Serotonergic activation potentiates light resetting of the main circadian clock and alters clock gene expression in a diurnal rodent. Exp. Neurol. 210 501–513. 10.1016/j.expneurol.2007.11.026 [DOI] [PubMed] [Google Scholar]
- Daan S., Pittendrigh C. S. A. (1976). Functional analysis of circadian pacemakers in nocturnal rodents. J. Comp. Physiol. 106 253–266. [Google Scholar]
- Damiola F., Le Minh N., Preitner N., Kornmann B., Fleury-Olela F., Schibler U. (2000). Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14 2950–2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis S. J., Vener A. V., Vierstra R. D. (1999). Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science. 286 2517–2520. 10.1126/science.286.5449.2517 [DOI] [PubMed] [Google Scholar]
- de Goffau M. C., Luopajärvi K., Knip M., Ilonen J., Ruohtula T., Härkönen T., et al. (2013). Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 62 1238–1244. 10.2337/db12-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding T., Schloss P. D. (2014). Dynamics and associations of microbial community types across the human body. Nature. 509 357–360. 10.1038/nature13178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogan M. D., Ataoglu H., Akarsu E. S. (2000). Effects of different serotypes of Escherichia coli lipopolysaccharides on body temperature in rats. Life Sci. 67 2319–2329. 10.1016/s0024-3205(00)00821-3 [DOI] [PubMed] [Google Scholar]
- Dopico X. C., Evangelou M., Ferreira R. C., Guo H., Pekalski M. L., Smyth D. J., et al. (2015). Widespread seasonal gene expression reveals annual differences in human immunity and physiology. Nat. Commun. 6:7000. 10.1038/ncomms8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell S. F. (2001). Seasonal variation in host susceptibility and cycles of certain infectious diseases. Emerg. Infect. Dis. 7 369–374. 10.3201/eid0703.010301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos R. J., Straus J. H., Pierce C. (1943). The multiplication of bacteriophage in vivo and its protective effects against an experimental infection with Shigella dysenteriae. J. Exp. Med. 78 161–168. 10.1084/jem.78.3.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourg G., Surenaud M., Lévy Y., Hüe S., Raoult D. (2017). Microbiome of HIV-infected people. Microb Pathog. 106 85–93. 10.1016/j.micpath.2016.05.015 [DOI] [PubMed] [Google Scholar]
- Edgar R. S., Stangherlin A., Nagy A. D., Nicoll M. P., Efstathiou S., O’Neill J. S., et al. (2016). Cell autonomous regulation of herpes and influenza virus infection by the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 113 10085–10090. 10.1073/pnas.1601895113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers A., Xie W., Agapov E., Brown S., Steinberg D., Tidwell R., et al. (2018). BMAL1 links the circadian clock to viral airway pathology and asthma phenotypes. Mucosal. Immunol. 11 97–111. 10.1038/mi.2017.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson A. K., Jesudhasan P. R., Mayer M. J., Narbad A., Winter S. E., Pfeiffer J. K. (2018). Bacteria facilitate enteric virus co-infection of mammalian cells and promote genetic recombination. Cell Host Microbe 23 77.e–88.e. 10.1016/j.chom.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng T., Wang L., Schoeb T. R., Elson C. O., Cong Y. (2010). Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. J. Exp. Med. 207 1321–1332. 10.1084/jem.20092253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fezeu L., Julia C., Henegar A., Bitu J., Hu F. B., Grobbee D. E., et al. (2011). Obesity is associated with higher risk of intensive care unit admission and death in influenza A (H1N1) patients: a systematic review and meta-analysis. Obes Rev. 12 653–659. 10.1111/j.1467-789X.2011.00864.x [DOI] [PubMed] [Google Scholar]
- Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104 13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnidze K., Hajdarovic K. H., Moskalenko M., Karatsoreos I. N., McEwen B. S., Bulloch K. (2016). Nuclear receptor REV-ERBα mediates circadian sensitivity to mortality in murine vesicular stomatitis virus-induced encephalitis. Proc. Natl. Acad. Sci. U.S.A. 113 5730–5735. 10.1073/pnas.1520489113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., et al. (1998). Role of the CLOCK protein in the mammalian circadian mechanism. Science. 280 1564–1569. [DOI] [PubMed] [Google Scholar]
- Gibbs J., Ince L., Matthews L., Mei J., Bell T., Yang N., et al. (2014). An epithelial circadian clock controls pulmonary inflammation and glucocorticoid action. Nat. Med. 20 919–926. 10.1038/nm.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J. K., Waters J. L., Poole A. C., Sutter J. L., Koren O., Blekhman R., et al. (2014). Human genetics shape the gut microbiome. Cell 159 789–799. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V., Spencer C. N., Nezi L., Reuben A., Andrews M. C., Karpinets T. V., et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 359 97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorres K. L., Daigle D., Mohanram S., Miller G. (2014). Activation and repression of Epstein-Barr Virus and Kaposi’s sarcoma-associated herpesvirus lytic cycles by short- and medium-chain fatty acids. J. Virol. 88 8028–8044. 10.1128/JVI.00722-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Wang H., Xu S., Zhuang Y., An J., Su C., et al. (2020). Alteration in gut microbiota is associated with dysregulation of cytokines and glucocorticoid therapy in systemic lupus erythematosus. Gut Microbes. 11 1758–1773. 10.1080/19490976.2020.1768644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gury-BenAri M., Thaiss C. A., Serafini N., Winter D. R., Giladi A., Lara-Astiaso D., et al. (2016). The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166 1231–1246.e13. 10.1016/j.cell.2016.07.043 [DOI] [PubMed] [Google Scholar]
- Gustafson C. E., Kim C., Weyand C. M., Goronzy J. J. (2020). Influence of immune aging on vaccine responses. J. Allergy Clin. Immunol. 145 1309–1321. 10.1016/j.jaci.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J. P., Edmunds W. J., Pebody R., Brown D. W., Lopman B. A. (2008). Deaths from norovirus among the elderly, England and Wales. Emerg. Infect. Dis. 14 1546–1552. 10.3201/eid1410.080188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M., Vollmers C., Zarrinpar A., DiTacchio L., Bushong E. A., Gill S., et al. (2012). Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell. Metab. 15 848–860. 10.1016/j.cmet.2012.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F., Smith K. D., Ozinsky A., Hawn T. R., Yi E. C., Goodlett D. R., et al. (2001). The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 410 1099–1103. 10.1038/35074106 [DOI] [PubMed] [Google Scholar]
- Held W., Shakhov A. N., Izui S., Waanders G. A., Scarpellino L., MacDonald H. R., et al. (1993). Superantigen-reactive CD4+ T cells are required to stimulate B cells after infection with mouse mammary tumor virus. J. Exp. Med. 177 359–366. 10.1084/jem.177.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley-McBain T., Zevin A. S., Manuzak J., Smith E., Gile J., Miller C., et al. (2016). Effects of fecal microbial transplantation on microbiome and immunity in simian immunodeficiency virus-infected macaques. J. Virol. 90 4981–4989. 10.1128/JVI.00099-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi S., Motohashi Y., Ishibashi K., Maeda T. (2007). Less exposure to daily ambient light in winter increases sensitivity of melatonin to light suppression. Chronobiol. Int. 24 31–43. 10.1080/07420520601139805 [DOI] [PubMed] [Google Scholar]
- Hoogerwerf W. A., Shahinian V. B., Cornélissen G., Halberg F., Bostwick J., Timm J., et al. (2010). Rhythmic changes in colonic motility are regulated by period genes. Am. J. Physiol. Gastrointest Liver Physiol. 298 G143–G150. 10.1152/ajpgi.00402.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood T. W., Hall S., Begley N., Forman R., Brown S., Vonslow R., et al. (2018). The circadian regulator BMAL1 programmes responses to parasitic worm infection via a dendritic cell clock. Sci. Rep. 8:3782. 10.1038/s41598-018-22021-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa K., Shibata S. (2004). Phase-resetting response to (+)8-OH-DPAT, a serotonin 1A/7 receptor agonist, in the mouse in vivo. Neurosci. Lett. 368 130–134. 10.1016/j.neulet.2004.06.072 [DOI] [PubMed] [Google Scholar]
- Hsu B. B., Gibson T. E., Yeliseyev V., Liu Q., Lyon L., Bry L., et al. (2019). Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe 25 803–814.e5. 10.1016/j.chom.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E. Y., Inoue T., Leone V. A., Dalal S., Touw K., Wang Y., et al. (2015). Using corticosteroids to reshape the gut microbiome: implications for inflammatory bowel diseases. Inflamm Bowel Dis. 21 963–972. 10.1097/MIB.0000000000000332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen R., Syrjänen J. (2010). Obesity and the outcome of infection. Lancet Infect Dis. 10 442–443. 10.1016/S1473-3099(10)70103-1 [DOI] [PubMed] [Google Scholar]
- Ichinohe T., Pang I. K., Kumamoto Y., Peaper D. R., Ho J. H., Murray T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. U.S.A. 108 5354–5359. 10.1073/pnas.1019378108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida N., Dzutsev A., Stewart C. A., Smith L., Bouladoux N., Weingarten R. A., et al. (2013). Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 342 967–970. 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaak D. D., Bartizal K. F., Caulfield M. J. (1988). Decreased pathogenicity of murine leukemia virus-Moloney in gnotobiotic mice. Leukemia. 2 540–544. [PubMed] [Google Scholar]
- Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139 485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasser S. A., Hanifin J. P., Rollag M. D., Brainard G. C. (2006). Dim light adaptation attenuates acute melatonin suppression in humans. J. Biol. Rhythms. 21 394–404. 10.1177/0748730406292391 [DOI] [PubMed] [Google Scholar]
- Ji Y., Zhang F., Zhang R., Shen Y., Liu L., Wang J., et al. (2018). Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: influence of CD4+ T cell count. Emerg. Microbes Infect. 7:113. 10.1038/s41426-018-0117-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. K., Watanabe M., Zhu S., Graves C. L., Keyes L. R., Grau K. R., et al. (2014). Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 346 755–759. 10.1126/science.1257147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jude B. A., Pobezinskaya Y., Bishop J., Parke S., Medzhitov R. M., Chervonsky A. V., et al. (2003). Subversion of the innate immune system by a retrovirus. Nat. Immunol. 4 573–578. 10.1038/ni926 [DOI] [PubMed] [Google Scholar]
- Kane M., Case L. K., Kopaskie K., Kozlova A., MacDearmid C., Chervonsky A. V., et al. (2011). Successful transmission of a retrovirus depends on the commensal microbiota. Science. 334 245–249. 10.1126/science.1210718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao C. C., Green S., Stein B., Golden S. S. (2005). Diel infection of a cyanobacterium by a contractile bacteriophage. Appl. Environ. Microbiol. 71 4276–4279. 10.1128/AEM.71.8.4276-4279.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpowicz P., Zhang Y., Hogenesch J. B., Emery P., Perrimon N. (2013). The circadian clock gates the intestinal stem cell regenerative state. Cell Rep. 3 996–1004. 10.1016/j.celrep.2013.03.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst S. M. (2015). Identification of a novel cellular target and a co-factor for norovirus infection - B cells & commensal bacteria. Gut Microbes. 6 266–271. 10.1080/19490976.2015.1052211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul D., Rathnasinghe R., Ferres M., Tan G. S., Barrera A., Pickett B. E., et al. (2020). Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Nat. Commun. 11:2537. 10.1038/s41467-020-16429-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalsa S. B., Jewett M. E., Cajochen C., Czeisler C. A. (2003). A phase response curve to single bright light pulses in human subjects. J. Physiol. 549(Pt 3) 945–952. 10.1113/jphysiol.2003.040477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Mirzaei M., Deng L. (2021). New technologies for developing phage-based tools to manipulate the human microbiome. Trends Microbiol. 10.1016/j.tim.2021.04.007 Epub ahead of print, [DOI] [PubMed] [Google Scholar]
- Khan Mirzaei M., Khan M. A. A., Ghosh P., Taranu Z. E., Taguer M., Ru J., et al. (2020). Bacteriophages isolated from stunted children can regulate gut bacterial communities in an age-specific manner. Cell Host Microbe 27 199–212.e5. 10.1016/j.chom.2020.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. C., Lee S. J. (1998). Temporal variation in hepatotoxicity and metabolism of acetaminophen in mice. Toxicology. 128 53–61. 10.1016/s0300-483x(98)00046-8 [DOI] [PubMed] [Google Scholar]
- Kondo T., Tsinoremas N. F., Golden S. S., Johnson C. H., Kutsuna S., Ishiura M. (1994). Circadian clock mutants of cyanobacteria. Science. 266 1233–1236. 10.1126/science.7973706 [DOI] [PubMed] [Google Scholar]
- Krakowiak K., Durrington H. J. (2018). The role of the body clock in asthma and COPD: implication for treatment. Pulm Ther. 4 29–43. 10.1007/s41030-018-0058-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss S. K., Best G. T., Etheredge C. A., Pruijssers A. J., Frierson J. M., Hooper L. V., et al. (2011). Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 334 249–252. 10.1126/science.1211057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen K. R., Moore J. G., Dayton M. T. (1991). Circadian rhythms of acid and bicarbonate efflux in fasting rat stomach. Am. J. Physiol. 260(4 Pt 1) G610–G614. 10.1152/ajpgi.1991.260.4.G610 [DOI] [PubMed] [Google Scholar]
- Lee S., Donehower L. A., Herron A. J., Moore D. D., Fu L. (2010). Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 5:e10995. 10.1371/journal.pone.0010995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Menezes J. S., Umesaki Y., Mazmanian S. K. (2011). Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1) 4615–4622. 10.1073/pnas.1000082107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y. K., Turner H., Maynard C. L., Oliver J. R., Chen D., Elson C. O., et al. (2009). Late developmental plasticity in the T helper 17 lineage. Immunity. 30 92–107. 10.1016/j.immuni.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone V., Gibbons S. M., Martinez K., Hutchison A. L., Huang E. Y., Cham C. M., et al. (2015). Effects of diurnal variation of gut microbes and high-fat feeding on host circadian clock function and metabolism. Cell Host Microbe. 17 681–689. 10.1016/j.chom.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S., et al. (2014). An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 32 834–841. 10.1038/nbt.2942 [DOI] [PubMed] [Google Scholar]
- Li N., Ma W. T., Pang M., Fan Q. L., Hua J. L. (2019). The commensal microbiota and viral infection: a comprehensive review. Front. Immunol. 10:1551. 10.3389/fimmu.2019.01551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X., Bushman F. D., FitzGerald G. A. (2015). Rhythmicity of the intestinal microbiota is regulated by gender and the host circadian clock. Proc. Natl. Acad. Sci. U.S.A. 112 10479–10484. 10.1073/pnas.1501305112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wang S., Zhou Z., Guo L., Yu F., Wu B. (2019). Bmal1 regulates circadian expression of cytochrome P450 3a11 and drug metabolism in mice. Commun. Biol. 2:378. 10.1038/s42003-019-0607-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Liu Y., Chen Y., Zhan Y., Zeng Q. (2019). Cyanobacterial viruses exhibit diurnal rhythms during infection. Proc. Natl. Acad. Sci. U.S.A. 116 14077–14082. 10.1073/pnas.1819689116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J. E., Drayson M. T., Taylor A. E., Toellner K. M., Lord J. M., Phillips A. C. (2016). Morning vaccination enhances antibody response over afternoon vaccination: a cluster-randomised trial. Vaccine. 34 2679–2685. 10.1016/j.vaccine.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loudon A. S. (2012). Circadian biology: a 2.5 billion year old clock. Curr. Biol. 22 R570–R571. 10.1016/j.cub.2012.06.023 [DOI] [PubMed] [Google Scholar]
- Louie J. K., Acosta M., Samuel M. C., Schechter R., Vugia D. J., Harriman K., et al. (2011). A novel risk factor for a novel virus: obesity and 2009 pandemic influenza A (H1N1). Clin. Infect. Dis. 52 301–312. 10.1093/cid/ciq152 [DOI] [PubMed] [Google Scholar]
- Loza-Correa M., Sahr T., Rolando M., Daniels C., Petit P., Skarina T., et al. (2014). The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments. Environ. Microbiol. 16 359–381. 10.1111/1462-2920.12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Zeng B., Zeng L., Du X., Li B., Huo R., et al. (2018). Gut microbiota regulates mouse behaviors through glucocorticoid receptor pathway genes in the hippocampus. Transl. Psychiatry. 8:187. 10.1038/s41398-018-0240-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysén M., Thorhagen M., Brytting M., Hjertqvist M., Andersson Y., Hedlund K. O. (2009). Genetic diversity among food-borne and waterborne norovirus strains causing outbreaks in Sweden. J. Clin. Microbiol. 47 2411–2418. 10.1128/JCM.02168-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager L. F., Burkhard R., Pett N., Cooke N. C. A., Brown K., Ramay H., et al. (2020). Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science. 369 1481–1489. 10.1126/science.abc3421 [DOI] [PubMed] [Google Scholar]
- Majumdar T., Dhar J., Patel S., Kondratov R., Barik S. (2017). Circadian transcription factor BMAL1 regulates innate immunity against select RNA viruses. Innate Immun. 23 147–154. 10.1177/1753425916681075 [DOI] [PubMed] [Google Scholar]
- Markle J. G., Frank D. N., Mortin-Toth S., Robertson C. E., Feazel L. M., Rolle-Kampczyk U., et al. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 339 1084–1088. 10.1126/science.1233521 [DOI] [PubMed] [Google Scholar]
- Martinez M. E. (2018). The calendar of epidemics: seasonal cycles of infectious diseases. PLoS Pathog. 14:e1007327. 10.1371/journal.ppat.1007327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastromarino P., Cacciotti F., Masci A., Mosca L. (2011). Antiviral activity of Lactobacillus brevis towards herpes simplex virus type 2: role of cell wall associated components. Anaerobe. 17 334–336. 10.1016/j.anaerobe.2011.04.022 [DOI] [PubMed] [Google Scholar]
- Matsuzaki S., Yasuda M., Nishikawa H., Kuroda M., Ujihara T., Shuin T., et al. (2003). Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage phi MR11. J. Infect. Dis. 187 613–624. 10.1086/374001 [DOI] [PubMed] [Google Scholar]
- Matsuzawa T., Nakamura Y., Ogawa Y., Ishimaru K., Goshima F., Shimada S., et al. (2018). Differential Day-Night Outcome to HSV-2 Cutaneous Infection. J. Invest. Dermatol. 138 233–236. 10.1016/j.jid.2017.07.838 [DOI] [PubMed] [Google Scholar]
- Meijer J. H., Groos G. A., Rusak B. (1986). Luminance coding in a circadian pacemaker: the suprachiasmatic nucleus of the rat and the hamster. Brain Res. 382 109–118. [DOI] [PubMed] [Google Scholar]
- Meijer J. H., Rusak B., Gänshirt G. (1992). The relation between light-induced discharge in the suprachiasmatic nucleus and phase shifts of hamster circadian rhythms. Brain Res. 598 257–263. [DOI] [PubMed] [Google Scholar]
- Milner J. J., Beck M. A. (2012). The impact of obesity on the immune response to infection. Proc. Nutr. Soc. 71 298–306. 10.1017/S0029665112000158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris D. J., Brem A. S. (2019). Role of gut metabolism of adrenal corticosteroids and hypertension: clues gut-cleansing antibiotics give us. Physiol. Genomics. 51 83–89. 10.1152/physiolgenomics.00115.2018 [DOI] [PubMed] [Google Scholar]
- Mukasa R., Balasubramani A., Lee Y. K., Whitley S. K., Weaver B. T., Shibata Y., et al. (2010). Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 32 616–627. 10.1016/j.immuni.2010.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A., Kobiita A., Chambon P. (2015a). Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc. Natl. Acad. Sci. U.S.A. 112 E6683–E6690. 10.1073/pnas.1519735112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A., Kobiita A., Damara M., Misra N., Meziane H., Champy M. F., et al. (2015b). Shifting eating to the circadian rest phase misaligns the peripheral clocks with the master SCN clock and leads to a metabolic syndrome. Proc. Natl. Acad. Sci. U.S.A. 112 E6691–E6698. 10.1073/pnas.1519807112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherji A., Kobiita A., Ye T., Chambon P. (2013). Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 153 812–827. 10.1016/j.cell.2013.04.020 [DOI] [PubMed] [Google Scholar]
- Nelson C. A., Wilen C. B., Dai Y. N., Orchard R. C., Kim A. S., Stegeman R. A., et al. (2018). Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc. Natl. Acad. Sci. U.S.A. 115 E9201–E9210. 10.1073/pnas.1805797115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen K. D., Fentress S. J., Qiu Y., Yun K., Cox J. S., Chawla A. (2013). Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science. 341 1483–1488. 10.1126/science.1240636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguera-Julian M., Rocafort M., Guillén Y., Rivera J., Casadellà M., Nowak P., et al. (2016). Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 5 135–146. 10.1016/j.ebiom.2016.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak C. M., Albers H. E. (2004). Novel phase-shifting effects of GABAA receptor activation in the suprachiasmatic nucleus of a diurnal rodent. Am. J. Physiol. Regul. Integr. Comp. Physiol. 286 R820–R825. 10.1152/ajpregu.00575.2003 [DOI] [PubMed] [Google Scholar]
- Odamaki T., Kato K., Sugahara H., Hashikura N., Takahashi S., Xiao J. Z., et al. (2016). Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 16:90. 10.1186/s12866-016-0708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. E., Kim B. C., Chang D. H., Kwon M., Lee S. Y., Kang D., et al. (2016). Dysbiosis-induced IL-33 contributes to impaired antiviral immunity in the genital mucosa. Proc. Natl. Acad. Sci. U.S.A. 113 E762–E771. 10.1073/pnas.1518589113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. Z., Ravindran R., Chassaing B., Carvalho F. A., Maddur M. S., Bower M., et al. (2014). TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity. 41 478–492. 10.1016/j.immuni.2014.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne L. C., Monticelli L. A., Nice T. J., Sutherland T. E., Siracusa M. C., Hepworth M. R., et al. (2014). Coinfection. Virus-helminth coinfection reveals a microbiota-independent mechanism of immunomodulation. Science 345 578–582. 10.1126/science.1256942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Hussain M. M. (2009). Clock is important for food and circadian regulation of macronutrient absorption in mice. J. Lipid Res. 50 1800–1813. 10.1194/jlr.M900085-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose J. K., Cassone C. V., Graniczkowska K. B., Cassone V. M. (2019). Entrainment of the circadian clock of the enteric bacterium klebsiella aerogenes by temperature cycles. iScience 19 1202–1213. 10.1016/j.isci.2019.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose J. K., Cassone V. M. (2016). The melatonin-sensitive circadian clock of the enteric bacterium Enterobacter aerogenes. Gut Microbes. 7 424–427. 10.1080/19490976.2016.1208892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulose J. K., Wright J. M., Patel A. G., Cassone V. M. (2016). Human gut bacteria are sensitive to melatonin and express endogenous circadian rhythmicity. PLoS One. 11:e0146643. 10.1371/journal.pone.0146643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. A., Kakabadse D., Davies J., Peng J., Warden-Smith J., Cuff S., et al. (2019a). Altered gut microbiota activate and expand insulin B15-23-reactive CD8+ T-Cells. Diabetes. 68 1002–1013. 10.2337/db18-0487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson J. A., Tai N., Ekanayake-Alper D. K., Peng J., Hu Y., Hager K., et al. (2019b). Norovirus changes susceptibility to type 1 diabetes by altering intestinal microbiota and immune cell functions. Front. Immunol. 10:2654. 10.3389/fimmu.2019.02654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlova T., Gruebele M., Chemla Y. R. (2019). Blue light is a universal signal for Escherichia coli chemoreceptors. J. Bacteriol. 201:e762–18. 10.1128/JB.00762-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcet B., Zecchin M., Ferri L., Beauchamp J., Sitaula S., Billon C., et al. (2018). Nuclear receptor subfamily 1 Group D Member 1 regulates circadian activity of NLRP3 inflammasome to reduce the severity of fulminant hepatitis in mice. Gastroenterology 154 1449–1464.e20. 10.1053/j.gastro.2017.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464 59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees F., Doherty M., Grainge M., Davenport G., Lanyon P., Zhang W. (2016). The incidence and prevalence of systemic lupus erythematosus in the UK, 1999-2012. Ann. Rheum. Dis. 75 136–141. 10.1136/annrheumdis-2014-206334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitmeier S., Kiessling S., Clavel T., List M., Almeida E. L., Ghosh T. S., et al. (2020). Arrhythmic gut microbiome signatures predict risk of type 2 diabetes. Cell Host Microbe 28 258–272.e6. 10.1016/j.chom.2020.06.004 [DOI] [PubMed] [Google Scholar]
- Ripoli M., Barbano R., Balsamo T., Piccoli C., Brunetti V., Coco M., et al. (2011). Hypermethylated levels of E-cadherin promoter in Huh-7 cells expressing the HCV core protein. Virus Res. 160 74–81. 10.1016/j.virusres.2011.05.014 [DOI] [PubMed] [Google Scholar]
- Robinson C. M., Jesudhasan P. R., Pfeiffer J. K. (2014). Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 15 36–46. 10.1016/j.chom.2013.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson K. M., McHugh K. J., Mandalapu S., Clay M. E., Lee B., Scheller E. V., et al. (2014). Influenza A virus exacerbates Staphylococcus aureus pneumonia in mice by attenuating antimicrobial peptide production. J. Infect. Dis. 209 865–875. 10.1093/infdis/jit527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B., Le Chatelier E., Derosa L., Duong C. P. M., Alou M. T., Daillère R., et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 359 91–97. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- Sato S., Solanas G., Peixoto F. O., Bee L., Symeonidi A., Schmidt M. S., et al. (2017). Circadian reprogramming in the liver identifies metabolic pathways of aging. Cell 170 664–677.e11. 10.1016/j.cell.2017.07.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar N., McInnes I. B., McMurray J. J. V. (2020). Obesity is a risk factor for severe COVID-19 infection: multiple potential mechanisms. Circulation. 142 4–6. 10.1161/CIRCULATIONAHA.120.047659 [DOI] [PubMed] [Google Scholar]
- Schaffer J., Beamer P. R., Trexler P. C., Breidenbach G., Walcher D. N. (1963). Response of germ-free animals to experimental virus monocontamination. I. Observation on Coxsackie B virus. Proc. Soc. Exp. Biol. Med. 112 561–564. 10.3181/00379727-112-28105 [DOI] [PubMed] [Google Scholar]
- Serrano-Villar S., Rojo D., Martínez-Martínez M., Deusch S., Vázquez-Castellanos J. F., Sainz T., et al. (2016). HIV infection results in metabolic alterations in the gut microbiota different from those induced by other diseases. Sci. Rep. 6:26192. 10.1038/srep26192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba A., Cui G., Tani-Ichi S., Ogawa M., Abe S., Okazaki F., et al. (2018). Glucocorticoids drive diurnal oscillations in T Cell distribution and responses by inducing interleukin-7 receptor and CXCR4. Immunity 48 286–298.e6. 10.1016/j.immuni.2018.01.004 [DOI] [PubMed] [Google Scholar]
- Shinohara K., Honma S., Katsuno Y., Abe H., Honma K. (1994). Circadian rhythms in the release of vasoactive intestinal polypeptide and arginine-vasopressin in organotypic slice culture of rat suprachiasmatic nucleus. Neurosci. Lett. 170 183–186. 10.1016/0304-3940(94)90269-0 [DOI] [PubMed] [Google Scholar]
- Silver A. C., Arjona A., Hughes M. E., Nitabach M. N., Fikrig E. (2012a). Circadian expression of clock genes in mouse macrophages, dendritic cells, and B cells. Brain Behav. Immun. 26 407–413. 10.1016/j.bbi.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver A. C., Arjona A., Walker W. E., Fikrig E. (2012b). The circadian clock controls toll-like receptor 9-mediated innate and adaptive immunity. Immunity. 36 251–261. 10.1016/j.immuni.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver A. C., Buckley S. M., Hughes M. E., Hastings A. K., Nitabach M. N., Fikrig E. (2018). Daily oscillations in expression and responsiveness of Toll-like receptors in splenic immune cells. Heliyon. 4:e00579. 10.1016/j.heliyon.2018.e00579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P., Aiewsakun P., Katzourakis A. (2019). Prisoners of war - host adaptation and its constraints on virus evolution. Nat. Rev. Microbiol. 17 321–328. 10.1038/s41579-018-0120-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P., Manning S. D. (2016). Impact of age and sex on the composition and abundance of the intestinal microbiota in individuals with and without enteric infections. Ann. Epidemiol. 26 380–385. 10.1016/j.annepidem.2016.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha T., Vich Vila A., Garmaeva S., Jankipersadsing S. A., Imhann F., Collij V., et al. (2019). Analysis of 1135 gut metagenomes identifies sex-specific resistome profiles. Gut Microbes. 10 358–366. 10.1080/19490976.2018.1528822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J. B., Aquino-Michaels K., Earley Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 350 1084–1089. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. J., Bahl J., Vijaykrishna D., Zhang J., Poon L. L., Chen H., et al. (2009a). Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 106 11709–11712. 10.1073/pnas.0904991106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. J., Vijaykrishna D., Bahl J., Lycett S. J., Worobey M., Pybus O. G., et al. (2009b). Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 459 1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- Sollars P. J., Weiser M. J., Kudwa A. E., Bramley J. R., Ogilvie M. D., Spencer R. L., et al. (2014). Altered entrainment to the day/night cycle attenuates the daily rise in circulating corticosterone in the mouse. PLoS One. 9:e111944. 10.1371/journal.pone.0111944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg E. D., Smits S. A., Tikhonov M., Higginbottom S. K., Wingreen N. S., Sonnenburg J. L. (2016). Diet-induced extinctions in the gut microbiota compound over generations. Nature 529 212–215. 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg J. L., Bäckhed F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535 56–64. 10.1038/nature18846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed A. L., Christophi G. P., Kaiko G. E., Sun L., Goodwin V. M., Jain U., et al. (2017). The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science. 357 498–502. 10.1126/science.aam5336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi J. S. (2017). Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18 164–179. 10.1038/nrg.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Lin E., Susmarski N., Yoon M., Zago A., Ware C. F., et al. (2007). Alternative entry receptors for herpes simplex virus and their roles in disease. Cell. Host Microbe. 2 19–28. 10.1016/j.chom.2007.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaiss C. A., Levy M., Korem T., Dohnalová L., Shapiro H., Jaitin D. A., et al. (2016). Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167 1495–1510.e12. 10.1016/j.cell.2016.11.003 [DOI] [PubMed] [Google Scholar]
- Thaiss C. A., Zeevi D., Levy M., Zilberman-Schapira G., Suez J., Tengeler A. C., et al. (2014). Transkingdom control of microbiota diurnal oscillations promotes metabolic homeostasis. Cell. 159 514–529. 10.1016/j.cell.2014.09.048 [DOI] [PubMed] [Google Scholar]
- Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., et al. (2005). Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308 1043–1045. 10.1126/science.1108750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457 480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Uchiyama R., Chassaing B., Zhang B., Gewirtz A. T. (2014). Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J. Infect. Dis. 210 171–182. 10.1093/infdis/jiu037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vatanen T., Kostic A. D., d’Hennezel E., Siljander H., Franzosa E. A., Yassour M., et al. (2016). Variation in Microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 165 842–853. 10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vétizou M., Pitt J. M., Daillère R., Lepage P., Waldschmitt N., Flament C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 350 1079–1084. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud S., Saccheri F., Mignot G., Yamazaki T., Daillère R., Hannani D., et al. (2013). The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 342 971–976. 10.1126/science.1240537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt R. M., Summa K. C., Forsyth C. B., Green S. J., Engen P., Naqib A., et al. (2016). The circadian clock mutation promotes intestinal Dysbiosis. Alcohol. Clin. Exp. Res. 40 335–347. 10.1111/acer.12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Ji M., Liang H. E., Locksley R. M. (2016). Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529 221–225. 10.1038/nature16161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A., Van Nood E., Holleman F., Salojärvi J., Kootte R. S., Bartelsman J. F., et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143 913–916.e7. 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I., Dunham R. M., Iwai S., Maher M. C., Albright R. G., Broadhurst M. J., et al. (2013). Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci. Transl. Med. 5:193ra91. 10.1126/scitranslmed.3006438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Lin Y., Yuan X., Li F., Guo L., Wu B. (2018). REV-ERBα integrates colon clock with experimental colitis through regulation of NF-κB/NLRP3 axis. Nat. Commun. 9:4246. 10.1038/s41467-018-06568-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Lin Y., Zhou Z., Gao L., Yang Z., Li F., et al. (2019). Circadian clock gene Bmal1 regulates bilirubin detoxification: a potential mechanism of feedback control of Hyperbilirubinemia. Theranostics. 9 5122–5133. 10.7150/thno.35773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Kuang Z., Yu X., Ruhn K. A., Kubo M., Hooper L. V. (2017). The intestinal microbiota regulates body composition through NFIL. Science. 357 912–916. 10.1126/science.aan0677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., MacLeod D. T., Di Nardo A. (2012). Commensal bacteria lipoteichoic acid increases skin mast cell antimicrobial activity against vaccinia viruses. J. Immunol. 189 1551–1558. 10.4049/jimmunol.1200471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weger B. D., Gobet C., Yeung J., Martin E., Jimenez S., Betrisey B., et al. (2019). The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell. Metab 29 362–382.e8. 10.1016/j.cmet.2018.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G., Wei L., Zhu J., Zang C., Hu-Li J., Yao Z., et al. (2009). Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 30 155–167. 10.1016/j.immuni.2008.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Ley R. E., Volchkov P. Y., Stranges P. B., Avanesyan L., Stonebraker A. C., et al. (2008). Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 455 1109–1113. 10.1038/nature07336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergaard D., Moseley P., Sørup F. K. H., Baldi P., Brunak S. (2019). Population-wide analysis of differences in disease progression patterns in men and women. Nat. Commun. 10:666. 10.1038/s41467-019-08475-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwood M. L., O’Donnell A. J., de Bekker C., Lively C. M., Zuk M., Reece S. E. (2019). The evolutionary ecology of circadian rhythms in infection. Nat. Ecol. Evol. 3 552–560. 10.1038/s41559-019-0831-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikswo M. E., Hall A. J. (2012). Prevention CfDCa. Outbreaks of acute gastroenteritis transmitted by person-to-person contact–United States, 2009-2010. MMWR Surveill Summ. 61 1–12. [PubMed] [Google Scholar]
- Wilen C. B., Lee S., Hsieh L. L., Orchard R. C., Desai C., Hykes B. L., et al. (2018). Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science. 360 204–208. 10.1126/science.aar3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks J., Lien E., Jacobson A. N., Fischbach M. A., Qureshi N., Chervonsky A. V., et al. (2015). Mammalian lipopolysaccharide receptors incorporated into the retroviral envelope augment virus transmission. Cell Host Microbe. 18 456–462. 10.1016/j.chom.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth S., Keller S., Kertscher R., Stadion M., Haller D., Kisling S., et al. (2011). Intestinal steroid profiles and microbiota composition in colitic mice. Gut Microbes. 2 159–166. 10.4161/gmic.2.3.16104 [DOI] [PubMed] [Google Scholar]
- Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. (2020). Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 Pneumonia in Wuhan, China. JAMA Intern. Med. 180 934–943. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G. D., Chen J., Hoffmann C., Bittinger K., Chen Y. Y., Keilbaugh S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334 105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Esteve E., Tremaroli V., Khan M. T., Caesar R., Mannerås-Holm L., et al. (2017). Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23 850–858. 10.1038/nm.4345 [DOI] [PubMed] [Google Scholar]
- Wu Y., Ho W., Huang Y., Jin D. Y., Li S., Liu S. L., et al. (2020). SARS-CoV-2 is an appropriate name for the new coronavirus. Lancet. 395 949–950. 10.1016/S0140-6736(20)30557-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Tang Q., Chen G., Xie M., Yu S., Zhao J., et al. (2019). New insights into the circadian rhythm and its related diseases. Front. Physiol. 10:682. 10.3389/fphys.2019.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Gustafson C. L., Sammons P. J., Khan S. K., Parsley N. C., Ramanathan C., et al. (2015). Cryptochrome 1 regulates the circadian clock through dynamic interactions with the BMAL1 C terminus. Nat. Struct. Mol. Biol. 22 476–484. 10.1038/nsmb.3018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh Y. K., Zuo T., Lui G. C., Zhang F., Liu Q., Li A. Y., et al. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 70 698–706. 10.1136/gutjnl-2020-323020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. H., Ko C. H., Lowrey P. L., Buhr E. D., Song E. J., Chang S., et al. (2005). A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. U.S.A. 102 2608–2613. 10.1073/pnas.0409763102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G. R., Eksmond U., Salcedo R., Alexopoulou L., Stoye J. P., Kassiotis G. (2012). Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 491 774–778. 10.1038/nature11599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Rollins D., Ruhn K. A., Stubblefield J. J., Green C. B., Kashiwada M., et al. (2013). TH17 cell differentiation is regulated by the circadian clock. Science. 342 727–730. 10.1126/science.1243884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Hensley C., Mahsoub H. M., Ramesh A. K., Zhou P. (2020). Microbiota in viral infection and disease in humans and farm animals. Prog. Mol. Biol. Transl. Sci. 171 15–60. 10.1016/bs.pmbts.2020.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Burrows M., Khan A. A., Graham L., Volchkov P., Becker L., et al. (2013). Gender bias in autoimmunity is influenced by microbiota. Immunity. 39 400–412. 10.1016/j.immuni.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrinpar A., Chaix A., Yooseph S., Panda S. (2014). Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell. Metab. 20 1006–1017. 10.1016/j.cmet.2014.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. (2020). Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 395 1054–1062. 10.1016/S0140-6736(20)30566-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. (2020). A novel coronavirus from patients with Pneumonia in China, 2019. N. Engl. J. Med. 382 727–733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Forde D., Tsukuda S., D’Arienzo V., Mailly L., Harris J. M., et al. (2021). Circadian control of hepatitis B virus replication. Nat. Commun. 12:1658. 10.1038/s41467-021-21821-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X., Magri A., Hill M., Lai A. G., Kumar A., Rambhatla S. B., et al. (2019). The circadian clock components BMAL1 and REV-ERBα regulate flavivirus replication. Nat. Commun. 10:377. 10.1038/s41467-019-08299-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M., Zimmermann-Kogadeeva M., Wegmann R., Goodman A. L. (2019). Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 570 462–467. 10.1038/s41586-019-1291-3 [DOI] [PMC free article] [PubMed] [Google Scholar]