Abstract
Human transcription factor IIIC (hTFIIIC) is a multisubunit complex that mediates transcription of class III genes through direct recognition of promoters (for tRNA and virus-associated RNA genes) or promoter-TFIIIA complexes (for the 5S RNA gene) and subsequent recruitment of TFIIIB and RNA polymerase III. We describe the cognate cDNA cloning and characterization of two subunits (hTFIIIC63 and hTFIIIC102) that are present within a DNA-binding subcomplex (TFIIIC2) of TFIIIC and are related in structure and function to two yeast TFIIIC subunits (yTFIIIC95 and yTFIIIC131) previously shown to interact, respectively, with the promoter (A box) and with a subunit of yeast TFIIIB. hTFIIIC63 and hTFIIIC102 show parallel in vitro interactions with the homologous human TFIIIB and RNA polymerase III components, as well as additional interactions that may facilitate both TFIIIB and RNA polymerase III recruitment. These include novel interactions of hTFIIIC63 with hTFIIIC102, with hTFIIIB90, and with hRPC62, in addition to the hTFIIIC102–hTFIIIB90 and hTFIIIB90–hRPC39 interactions that parallel the previously described interactions in yeast. As reported for yTFIIIC131, hTFIIIC102 contains acidic and basic regions, tetratricopeptide repeats (TPRs), and a helix-loop-helix domain, and mutagenesis studies have implicated the TPRs in interactions both with hTFIIIC63 and with hTFIIIB90. These observations further document conservation from yeast to human of the structure and function of the RNA polymerase III transcription machinery, but in addition, they provide new insights into the function of hTFIIIC and suggest direct involvement in recruitment of both TFIIIB and RNA polymerase III.
A number of genes encoding small structural RNAs are transcribed by RNA polymerase III in conjunction with various accessory factors that, in the simplest cases (tRNA genes) applicable to both yeast and metazoans, include the multisubunit TFIIIB and TFIIIC complexes (reviewed in references 12, 18, 44, and 45). For virus-associated (VA) RNA or mammalian tRNA genes, preinitiation complex (PIC) assembly involves promoter recognition (A and B boxes) by TFIIIC, followed by sequential recruitment of TFIIIB and RNA polymerase III (reviewed in reference 42). In yeast, TFIIIC induces the formation of a stable TFIIIB-promoter complex that is sufficient (even after TFIIIC dissociation) for RNA polymerase III recruitment and function (16), although a similar phenomenon has not been reported for metazoans.
The structural and functional analysis of RNA polymerase III accessory factors is most advanced in yeast. Yeast TFIIIC contains six polypeptides of 138, 131, 95, 91, 60, and 55 kDa (reviewed in references 1 and 27); yeast TFIIIB contains TATA-binding protein (TBP), a 70-kDa TFIIB-related factor (TFIIIB70/BRF), and a 90-kDa subunit (TFIIIB90/B") (reviewed in reference 19); and yeast RNA polymerase III contains 16 subunits ranging from 10 to 160 kDa (11). Photocross-linking studies have localized various of these components to specific regions of the tRNA gene, including localization of the yeast TFIIIC138 (yTFIIIC138) subunit to the B box region, the yTFIIIC95 subunit to the A box region, the yTFIIIC131 subunit to regions both upstream of the start site and between the A and B boxes, the yTFIIIB70 and yTFIIIB90 subunits to a region upstream of the start site, and the yRPC34, yRPC31, and yRPC82 subunits of RNA polymerase III to a region surrounding the transcription start site (2–4). Consistent with these results and a simple sequential recruitment model, yTFIIIC131 has been shown to interact directly with both yTFIIIB70 and yTFIIIB90, albeit not yet with yTFIIIC95 as predicted from the cross-linking analyses, and yTFIIIB70 has been shown to interact with yRPC34 (8, 20, 32, 43).
Corresponding studies of the human RNA polymerase III machinery have revealed a comparable complexity, although not all essential factors have been purified to homogeneity (reviewed in references 40 and 41). Human TFIIIC can be resolved chromatographically into a subcomplex (TFIIIC2) that has five subunits (of 220, 110, 102, 90, and 63 kDa) and exhibits limited (namely, B box) DNA-binding activity (21, 47) and a partially purified subcomplex (TFIIIC1) that enhances the binding of TFIIIC2 (39, 46). The more recent purification to near-homogeneity of a TFIIIC complex exhibiting the combined properties of TFIIIC2 and TFIIIC1 has revealed several candidate TFIIIC1 subunits and suggested that TFIIIC functions as a single, stable complex (42). Human TFIIIB has been shown to contain TBP and a subunit (human TFIIIB90 [hTFIIIB90]) related to yTFIIIB70 (29, 38), although more recent studies have identified additional candidate subunits related to yTFIIIB90 (14a, 36a). Consistent with a potentially greater complexity of the metazoan RNA polymerase III machinery, other factors affecting various aspects of transcription have also been reported (reviewed in references 40 and 42). Consistent with a phylogenetic conservation of the RNA polymerase III recruitment mechanism, hTFIIIB90 has been shown to interact with a human RNA polymerase III subunit (hRPC39) that is related to yRPC34, is present in a subcomplex with homologues (hRPC32 and hRPC62) of yRPC31 and yRPC82, and is specifically required for transcription initiation (41).
Despite the conservation of RNA polymerase III and TFIIIB subunits, as well as common core promoter elements (A and B boxes) for recognition by TFIIIC, the cognate cDNA clones for the two largest subunits of TFIIIC2 (including a B box contact subunit) failed to reveal any sequence relationship with the reported yeast TFIIIC subunits (23, 26, 34). To further understand the structure and function of human TFIIIC, including both similarities and dissimilarities with yeast TFIIIC, we have cloned cognate cDNAs and characterized two human TFIIIC2 subunits that have counterparts in yeast TFIIIC and that function through novel interactions in TFIIIB and RNA polymerase III recruitment.
MATERIALS AND METHODS
Purification and cloning of hTFIIIC102 and hTFIIIC63.
TFIIIC2 was purified as described previously (21), and component subunits were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Microsequencing analysis was performed on the 102 (hTFIIIC102)- and 63 (hTFIIIC63)-kDa polypeptides as described elsewhere (23). cDNA clones encoding hTFIIIC102 and hTFIIIC63 were obtained by screening human cDNA libraries (Namalwa, BJAB, and HeLa cell libraries) with several degenerate oligonucleotide probes corresponding to the internal peptide sequences of hTFIIIC102 and hTFIIIC63. DNA sequences were obtained by the dideoxynucleotide chain termination method (United States Biochemical).
Antigen and antiserum preparation.
To produce His10-tagged recombinant proteins for use as antigens, cDNAs encoding hTFIIIC102 (amino acids 207 to 711) and hTFIIIC63 (amino acids 1 to 519) were subcloned into pET-19b vectors. The resulting plasmids were introduced into Escherichia coli BL21(DE3)pLysS (35). After induction with isopropyl-β-d-thiogalactopyranoside (IPTG), recombinant proteins were purified by Ni2+–nitrilotriacetic acid (NTA)–agarose affinity chromatography followed by SDS-PAGE. For the preparation of antisera, New Zealand White rabbits were injected and boosted subsequently every 3 weeks with excised gel slices emulsified with Freund’s adjuvant. Blood was collected 10 to 15 days after each boost. The antigens were cross-linked to CNBr-activated Sepharose 4B (Pharmacia) according to the manufacturer’s instructions and were then used to purify the corresponding antisera as described previously (13).
Immunodepletion and immunoprecipitation of HeLa nuclear extracts with anti-hTFIIIC102 and anti-hTFIIIC63 antibodies.
Antigen-purified anti-hTFIIIC102 and anti-hTFIIIC63 antibodies were first bound to protein A agarose (Oncogene Sciences) and then covalently cross-linked to the beads with dimethyl pimelimidate as described previously (13). As a control, the individual preimmune sera were processed the same way as immune sera. Antibody-coupled beads were incubated for 2 to 3 h at 4°C with HeLa nuclear extracts in buffer BC (20 mM Tris-HCl [pH 8], 20% glycerol, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 1 mM dithiothreitol [DTT]) containing 500 mM KCl (BC500) and 0.1% Nonidet P-40 (NP-40). After centrifugation, the supernatant was dialyzed against BC100 and aliquots were frozen. The beads were washed extensively with the same buffer. The bound proteins were eluted with glycine (pH 2.5) and then neutralized with Tris-HCl (pH 8).
In vitro transcription assays.
Transcription reactions for human tRNA and 5S genes and for adenovirus VAI genes were performed in a 25-μl reaction mixture with 250 ng of each supercoiled DNA template, 60 mM KCl, 6 mM MgCl2, 2 mM DTT, 8% glycerol, 10 mM HEPES (pH 7.9), 0.6 mM (each) ATP, CTP and UTP, 0.025 mM GTP, and 2.5 μCi of [α-32P]GTP. Reactions were allowed to proceed for 1 h at 30°C before termination by the addition of 50 μl of stop solution (100 mM sodium acetate, 20 mM EDTA, 1% SDS, and 1 mg of yeast tRNA/ml). The mixtures were extracted with phenol-chloroform, precipitated with ethanol, and resolved on 8% polyacrylamide–7 M urea gels.
Protein-protein interaction assays.
Purified recombinant proteins (0.5 to 2 μg) were bound to glutathione-Sepharose or M2 agarose beads and were incubated with 1 to 100 ng of the other purified recombinant protein in BC400–0.1% NP-40. The beads were washed extensively with BC400–0.1% NP-40. Residual proteins were eluted by boiling the beads in SDS gel sample buffer and were analyzed by Western blot analysis.
Assembly of the hTFIIIC102–hTFIIIC63–hTFIIIB90–TBP subcomplex.
Sf9 cell lysates containing independently expressed FLAG-hTFIIIB90, hemagglutinin (HA)-hTFIIIC102, and His10–hTFIIIC63 proteins and bacterial extracts containing glutathione S-transferase (GST)-TBP were mixed and incubated at 4°C in BC300–0.1% NP-40 for 2 to 3 h. The preassembled subcomplexes were first bound by Ni2+–NTA–agarose, then washed, and eluted in 1 M imidazole–BC300–0.1% NP-40. The eluted subcomplexes were immobilized on protein G-Sepharose coated with HA antibodies, washed, and eluted with BC300–0.1% NP-40–1 mg of HA peptides/ml. The HA peptide-eluted subcomplexes were then bound to glutathione-Sepharose, washed, and eluted with 100 mM glutathione–BC300–0.1% NP-40. The glutathione-eluted subcomplexes were finally immobilized on M2 agarose, washed, eluted by boiling in SDS gel sample buffer, and visualized by Western blot analysis.
Nucleotide sequence accession number.
The GenBank accession no. of the hTFIIIC102 sequence is AF133123. The GenBank accession no. of the hTFIIIC63 sequence is AF133124.
RESULTS
Cloning and sequence analysis of hTFIIIC63 and hTFIIIC102.
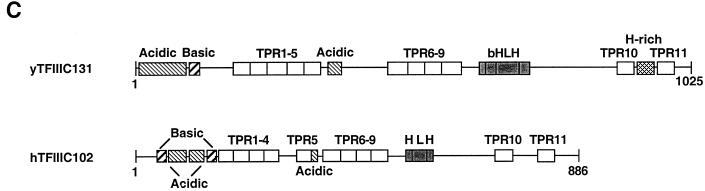
The purification of hTFIIIC2 has been described previously (21). The protein sequence information for four internal peptides of the 63-kDa subunit, designated hTFIIIC63, and four internal peptides of the 102-kDa subunit, designated hTFIIIC102, were obtained and used to clone cDNAs encoding all corresponding peptides. The deduced open reading frame of hTFIIIC63 cDNA encodes a polypeptide of 519 amino acids with a calculated molecular mass of 60 kDa (Fig. 1A). The deduced open reading frame of hTFIIIC102 cDNA encodes a protein of 886 amino acids with a predicted molecular mass of 101 kDa (Fig. 1B). A search of the National Center for Biotechnology Information nonredundant databases with the BLAST network service and the Genetics Computer Group GAP program servicer was performed to determine sequence similarities between hTFIIIC102 or hTFIIIC63 and other proteins. hTFIIIC63 shows significant sequence relationships to the TFIIIC95 (τ95) subunit of yeast TFIIIC (22% identity and 45% similarity) and to a hypothetical 50-kDa Caenorhabditis elegans protein (GenBank accession no. Z35603) (31% identity and 51% similarity). hTFIIIC102 shows 31% identity and 46% similarity to the TFIIIC131 (τ131) subunit of yeast TFIIIC, as well as 33% identity and 46% similarity to a hypothetical C. elegans protein (GenBank accession no. Z70783). In addition to the overall sequence similarity with yTFIIIC95, hTFIIIC63 shows conservation of a highly acidic region at the C terminus and a central helix-turn-helix motif (Fig. 1A). Like yTFIIIC131, hTFIIIC102 contains N-terminal acidic and basic regions, a central acidic region, a C-terminal helix-loop-helix (HLH) region, and, most notably, 11 tetratricopeptide repeats (TPRs) (Fig. 1C and D).
FIG. 1.
Predicted amino acid sequences and sequence alignments with yeast counterparts of the hTFIIIC102 and hTFIIIC63 subunits of the TFIIIC2 complex. The peptide sequences obtained by microsequence analyses are underlined. (A) Predicted amino acid sequence of hTFIIIC63 and alignment with yTFIIIC95 (31, 36). A highly acidic region at the C terminus and a central helix-turn-helix motif are double underlined. (B) Predicted amino acid sequence of hTFIIIC102 and alignment with yTFIIIC131 (28). (C) Schematic representation of hTFIIIC102 and yTFIIIC131 (28). Two basic regions are found at amino acid positions 23 to 34 and 130 to 147, and three acidic regions are found at amino acid positions 45 to 52, 92 to 114, and 363 to 374. The HLH motif is found at amino acid positions 552 to 600. (D) Consensus sequence of the TPR unit as defined by Sikorski et al. (33) and sequences of the 11 TPR units of hTFIIIC102. The four conserved residues that form “helix A” (residues 4, 7, 8, and 11), the three residues that form “helix B” (residues 20, 24, and 27), and the unique proline residue often found at position 32 (10, 14) are shown in boldfaced capital letters. Other residues that fit the TPR consensus sequence are shown in boldfaced lowercase letters. The following equivalences, based on the work of Marck et al. (28), are used: A, G, S, and V; E and D; K and R; I, L, M, and V; and L, F, Y, H, and W. Aspartate (D) residues, often found at positions 33 and 34 of TPR units, are underlined at those positions.
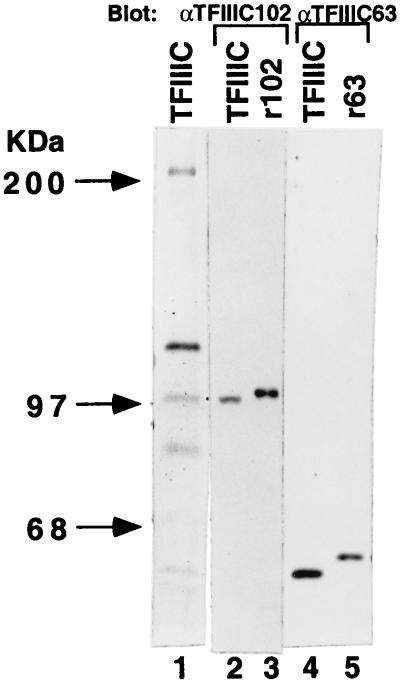
To verify that the cloned cDNAs indeed encode bona fide subunits of human TFIIIC, antibodies were raised against purified bacterially expressed recombinant proteins corresponding to the central portion (residues 207 to 711) of the hTFIIIC102 cDNA open reading frame and to the complete hTFIIIC63 cDNA open reading frame. Anti-hTFIIIC102 and anti-hTFIIIC63 antibodies reacted strongly and specifically only with protein bands of 102 and 63 kDa, respectively, in a highly purified TFIIIC2 fraction (Fig. 2, lanes 2 and 4) in immunoblot assays. To determine whether the cloned cDNAs contained complete coding sequences, the corresponding open reading frames were expressed either from a baculovirus vector in Sf9 cells as a recombinant His10-tagged protein (hTFIIIC63) or in a reticulocyte lysate system as a recombinant FLAG-tagged protein (hTFIIIC102). Immunoblot analyses indicated that the purified recombinant His10-tagged hTFIIIC63 (Fig. 2; compare lane 5 with lane 4) and the purified recombinant FLAG-tagged hTFIIIC102 (Fig. 2; compare lane 3 with lane 2) were slightly larger (as expected from the N-terminal tags) than their natural counterparts, indicating that the cDNA clones encode full-length proteins. In further support of this notion, polyadenylation signals followed by poly(A) tails were detected downstream of termination codons in both cDNAs.
FIG. 2.
Identification of the cDNA-encoded proteins as the 102- and 63-kDa hTFIIIC2 subunits. The polypeptide components of a highly purified TFIIIC2 fraction were analyzed in a silver-stained gel (lane 1). Anti-hTFIIIC102 and anti-hTFIIIC63 antibodies were used to detect hTFIIIC102 and hTFIIIC63 in a highly purified TFIIIC2 fraction (lanes 2 and 4) and in purified reticulocyte lysate-expressed FLAG-hTFIIIC102 and baculovirus-expressed His10–hTFIIIC63 (lanes 3 and 5).
The cDNA-encoded 102- and 63-kDa proteins are both bona fide subunits of the TFIIIC2 complex and are involved in RNA polymerase III-mediated transcription.
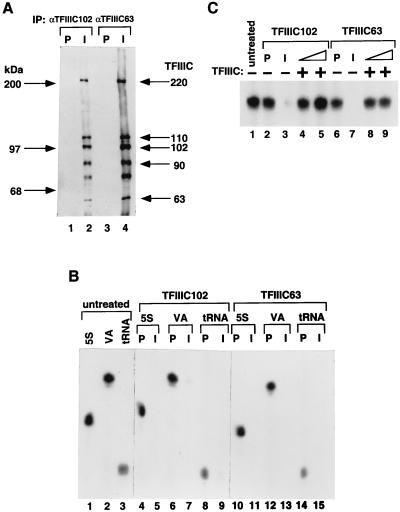
It has been established that human TFIIIC is required for the formation of stable PICs on 5S RNA, tRNA, and VA RNA genes (25). To determine whether the hTFIIIC102 and hTFIIIC63 cDNA-encoded proteins are components of the TFIIIC complex, antigen-purified anti-hTFIIIC102 and anti-hTFIIIC63 antisera, as well as control preimmune sera, were used to immunoprecipitate TFIIIC from HeLa nuclear extracts under stringent (0.5 M KCl–0.1% NP-40) conditions. As revealed by immunoblot analysis with antibodies to corresponding recombinant proteins (Fig. 3A), each immunoprecipitate, but neither preimmune serum precipitate, contained the 220-, 110-, 102-, 90-, and 63-kDa polypeptides that had been shown to copurify and correlate with the TFIIIC2 activity (21, 47). These results indicate that hTFIIIC102 and hTFIIIC63 are integral, tightly associated components of the TFIIIC2 complex. To determine the requirement for hTFIIIC102 and hTFIIIC63 in the transcription of 5S RNA, tRNA, and VA RNA genes, nuclear extract was depleted with either anti-hTFIIIC102 or anti-hTFIIIC63 antibodies and tested in a transcription assay. As shown in Fig. 3B, the high levels of transcription from 5S RNA, tRNA, and VAI RNA genes were reduced to undetectable levels after immunodepletion of either hTFIIIC102 (compare lane 5 with lane 1, lane 7 with lane 2, and lane 9 with lane 3) or hTFIIIC63 (compare lane 11 with lane 1, lane 13 with lane 2, and lane 15 with lane 3) but were unaffected by treatment with preimmune sera (lanes 4, 6, 8, 10, 12, and 14). Significantly, addition of an immunopurified TFIIIC complex containing both TFIIIC1 and TFIIIC2 (42), which were both required to restore transcription to anti-hTFIIIC110 immune serum-depleted extracts (34), restored to the depleted extracts transcription from the VAI template at levels similar to those observed with untreated extracts or with extracts treated with preimmune sera (Fig. 3C; compare lanes 4 and 5 with lanes 1 and 2, and lanes 8 and 9 with lanes 1 and 6). Thus, we conclude that hTFIIIC102 and hTFIIIC63, and associated polypeptides within TFIIIC, are necessary for RNA polymerase III-mediated transcription.
FIG. 3.
Immunoprecipitation of the TFIIIC2 complex and immunodepletion of TFIIIC transcription activity by anti-hTFIIIC102 and anti-hTFIIIC63 antibodies. (A) Immunoprecipitates from HeLa nuclear extracts treated with preimmune (P) and immune (I) anti-hTFIIIC102 (lanes 1 and 2) and anti-hTFIIIC63 (lanes 3 and 4) antibodies in BC500–0.1% NP-40 were subjected to Western blot analysis. The polypeptides were detected by a mixture of antibodies against hTFIIIC220, -110, -102, -90, and -63. The amounts of individual antibodies were adjusted so that the intensities of the hTFIIIC220, -110, -102, -90, and -63 immunoreactive bands were similar. The extra band between the 63- and 90-kDa polypeptides in lanes 2 and 4 reflects cross-reactivity with the anti-hTFIIIC110 antibodies and appears to represent a proteolytic breakdown product of hTFIIIC110 that was not consistently observed. (B) Nuclear extracts treated with preimmune (P) and immune (I) anti-hTFIIIC102 (lanes 4 to 9) and anti-hTFIIIC63 (lanes 10 to 15) antibodies were used for in vitro transcription assays with 5S RNA (lanes 1, 4, 5, 10, and 11), VAI RNA (lanes 2, 6, 7, 12, and 13), and tRNA (lanes 1, 8, 9, 14, and 15) templates. (C) Either 1 μl (lanes 4 and 8) or 2 μl (lanes 5 and 9) of an immunopurified TFIIIC was added to nuclear extracts depleted with immune anti-hTFIIIC102 (lanes 4 and 5) or with anti-hTFIIIC63 (lanes 8 and 9) and tested for transcription with the VAI RNA template.
Interaction of hTFIIIC63 and hTFIIIC102 with each other and with hTFIIIB90 and TBP.
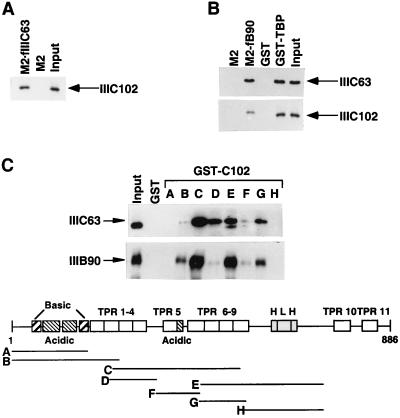
In yeast, it has been shown that PIC assembly on tRNA genes involves promoter recognition by TFIIIC followed by TFIIIB recruitment through protein-protein interactions. yTFIIIC95 and yTFIIIC131 are positioned, respectively, at and on both sides of the A box (2, 3), and yTFIIIC131 recruits TFIIIB through interactions with the TBP-interacting yTFIIIB70 subunit (8, 20) and with yTFIIIB90 (32). Since hTFIIIC102, hTFIIIC63, and hTFIIIB90 are homologues of yTFIIIC131, yTFIIIC95, and yTFIIIB70, respectively, we used purified recombinant proteins to test the potential interactions among hTFIIIC102, hTFIIIC63, hTFIIIB90, and TBP under stringent (0.4 M KCl–0.1% NP-40) conditions. As shown in Fig. 4A, purified baculovirus-expressed hTFIIIC102 (detected by anti-hTFIIIC102 antibodies) was bound to M2 agarose-immobilized FLAG-tagged hTFIIIC63 but not to M2 agarose alone. Similarly, purified baculovirus-expressed hTFIIIC102 and hTFIIIC63 (detected by anti-hTFIIIC102 and anti-hTFIIIC63 antibodies) were bound to M2 agarose-immobilized FLAG-TFIIIB90 but not to M2 agarose (Fig. 4B). Purified hTFIIIC102 and hTFIIIC63 showed similar interactions with glutathione-Sepharose-immobilized GST-TBP, but not with glutathione-Sepharose-immobilized GST alone (Fig. 4B). These results indicate that hTFIIIC102 and hTFIIIC63 associate tightly with each other and with hTFIIIB90 and TBP in the absence of other associated subunits.
FIG. 4.
Interactions of hTFIIIC102 with hTFIIIC63, hTFIIIB90, and TBP, and interactions of hTFIIIC63 with hTFIIIB90 and TBP. Input samples contained 10% of the amounts used for the interactions. (A) Purified baculovirus-expressed HA-hTFIIIC102 was incubated with M2 agarose or M2 agarose containing bound FLAG-hTFIIIC63 (M2-fIIIC63), and samples were washed and eluted as described in Materials and Methods. HA-hTFIIIC102 in input and eluted fractions was detected by immunoblotting with antisera against hTFIIIC102. (B) Purified baculovirus-expressed His10–hTFIIIC63 (top panel) or HA-hTFIIIC102 (bottom panel) was incubated with M2 agarose or M2 agarose-immobilized FLAG-hTFIIIB90 (M2-fB90) and with glutathione-Sepharose-immobilized GST or GST-TBP, and samples were washed and eluted as described in Materials and Methods. His10-hTFIIIC63 and HA-hTFIIIC102 were detected in input and eluted fractions by immunoblotting with anti-hTFIIIC63 and anti-hTFIIIC102 antibodies, respectively. (C) Sf9 cell extracts containing expressed His10–hTFIIIC63 (upper panel) or FLAG-hTFIIIB90 (lower panel) were incubated with glutathione-Sepharose-immobilized GST or with GST–truncated-hTFIIIC102 proteins (A, amino acids 1 to 148; B, amino acids 1 to 214; C, amino acids 207 to 507; D, amino acids 204 to 325; E, amino acids 419 to 711; F, amino acids 326 to 420; G, amino acids 419 to 531; and H, amino acids 527 to 724) in BC400–0.1% NP-40. After extensive washing of the beads with the same buffer, bound proteins were eluted by boiling in SDS sample buffer and analyzed by immunoblotting with antisera against hTFIIIC63 and hTFIIIB90. The amounts of GST–truncated-hTFIIIC102 proteins A through H and GST proteins were normalized by SDS-PAGE with Coomassie blue staining.
To elucidate the domains of hTFIIIC102 that mediate interactions with hTFIIIB90 and hTFIIIC63, we examined the binding of recombinant hTFIIIB90 and hTFIIIC63 to GST fusion proteins bearing different fragments of the N-terminal three-fourths of hTFIIIC102. As shown in Fig. 4C, fragments (A and H) lacking any TPRs showed no interaction with either hTFIIIC63 or hTFIIIB90. A fragment (F) containing almost exactly two complete TPRs (TPR5 and TPR6) and the central acidic region showed only barely detectable interactions with hTFIIIB90 or hTFIIIC63. A fragment (D) containing two complete TPRs (TPR3 and TPR4) but lacking the N-terminal acidic and basic regions showed barely detectable interactions with hTFIIIB90 but moderate interactions with hTFIIIC63, whereas a fragment (B) containing the N-terminal region and the two N-terminal TPRs showed moderate interactions with hTFIIIB90 but only very weak interactions with hTFIIIC63. In contrast, fragments containing six complete TPRs (C) or three complete TPRs plus the HLH region (E) showed strong interactions with both hTFIIIC63 and hTFIIIB90. These results are generally consistent with yeast studies implicating either a specific TPR (8) or regions containing TPRs (20) in interactions of yTFIIIC131 with yTFIIIB70. However, in the former case (8) there was a more stringent requirement for the acidic region in conjunction with TPRs.
hTFIIIC102, hTFIIIC63, hTFIIIB90, and TBP form a stable subcomplex.
The demonstrated interaction of hTFIIIC102 with hTFIIIB90 is similar to that reported for the yeast counterparts (8, 20), whereas the interactions of hTFIIIC63 with hTFIIIB90 and TBP and the interactions of hTFIIIC102 with hTFIIIC63 and with TBP represent novel observations that further strengthen the PIC assembly model. To test the prediction from these studies that hTFIIIC63, hTFIIIC102, hTFIIIB90, and TBP may form a subcomplex, Sf9 cell extracts containing independently expressed His10–hTFIIIC63, FLAG-hTFIIIB90, and HA-hTFIIIC102 proteins and bacterial extracts containing GST-TBP were mixed to form a presumptive subcomplex of four subunits. The mixture was then subjected to four successive affinity purification steps (with Ni2+–NTA–agarose, HA antibodies covalently linked to protein G-Sepharose, glutathione-Sepharose, and M2 agarose), each specific for a distinct recombinant protein, and the resulting preparation was analyzed by immunoblot analysis. The results in Fig. 5 show that the final immunopurified preparation contains hTFIIIC102, hTFIIIC63, hTFIIIB90, and TBP, indicating that these components can be assembled into a subcomplex that is sufficiently stable to survive four independent affinity purification methods at 300 mM KCl–0.1% NP-40. The results of gel filtration analyses (data not shown) are also consistent with the formation of a stable subcomplex.
FIG. 5.
hTFIIIC102, hTFIIIC63, hTFIIIB90, and TBP form a subcomplex in vitro. The subcomplex formed from the baculovirus-expressed His10–hTFIIIC63, HA-hTFIIIC102, and FLAG-hTFIIIB90 and bacterially expressed GST-TBP was purified by successive affinity chromatography on Ni2+–NTA–agarose, HA antibodies linked to protein G-Sepharose, glutathione-Sepharose, and then M2 agarose. The preparative subcomplex was analyzed by immunoblotting with a mixture of antibodies against hTFIIIC102, hTFIIIC63, hTFIIIB90, and TBP. The relative amounts of the individual antibodies were different from the amounts used in Fig. 3A and 6B, such that the different intensities of the hTFIIIC102 and hTFIIIC63 immunoreactive bands do not necessarily reflect different stoichiometric amounts of these components.
Interactions between TFIIIC, TFIIIB, and RNA polymerase III.
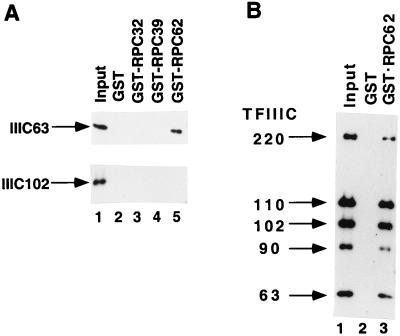
Yeast RNA polymerase III is recruited to the TFIIIB-TFIIIC promoter complex through interactions with TFIIIB (16), and this involves interactions of yTFIIIB70 with an RNA polymerase III subunit (yRPC34) that is part of a dissociable three-subunit complex (6, 43). A similar mechanism appears to operate in the human system, since binding of TFIIIB is dependent on prior binding of TFIIIC (5, 25) and since hTFIIIB90 and human TBP interact with an RNA polymerase III subunit (hRPC39) that is homologous to yRPC34 and part of an initiation-specific subcomplex containing hRPC62 and hRPC32 in addition to hRPC39 (41). Given the novel human TFIIIC and TFIIIB subunit interactions described above, as well as the availability of recombinant human RNA polymerase III subunits RPC62, RPC39, and RPC32, we tested whether any of the latter subunits could interact with hTFIIIC63 or hTFIIIC102. The GST fusion protein binding assays in Fig. 6A show that purified hTFIIIC63 interacts with GST-hRPC62 (lane 5) but not with equivalent amounts of GST-hRPC39 (lane 4), GST-hRPC32 (lane 3), or GST alone (lane 2). None of these RNA polymerase III subunits showed stable interactions with purified hTFIIIC102 (Fig. 6A). We further tested whether these interactions occur in the context of the TFIIIC complex. In support of this idea, an immunopurified TFIIIC complex, including all five TFIIIC2 subunits, bound to GST-hRPC62 but not to GST alone (Fig. 6B).
FIG. 6.
Interactions of hTFIIIC102 and hTFIIIC63 with RNA polymerase III subunits hRPC32, hRPC39, and hRPC62. Input samples contained 10% of the amounts used for the interactions. (A) Purified baculovirus-expressed His10–hTFIIIC63 (top panel) or HA-hTFIIIC102 (bottom panel) was incubated with glutathione-Sepharose-immobilized GST-hRPC32 (lane 3), GST-hRPC39 (lane 4), GST-hRPC62 (lane 5), and GST (lane 2) proteins in BC150–0.1% NP-40. After extensive washes with the same buffer, the beads were boiled in SDS sample buffer. His10–hTFIIIC63 and HA-hTFIIIC102 in input and eluted fractions were detected on immunoblots by antisera against hTFIIIC63 and hTFIIIC102, respectively. The amounts of GST-hRPC32, GST-hRPC39, GST-hRPC62, and GST proteins in the inputs were normalized by SDS-PAGE with Coomassie blue staining. (B) Beads containing glutathione-Sepharose-immobilized GST-hRPC62 or GST were incubated with an immunopurified TFIIIC in BC150–0.1% NP-40. After extensive washes with the same buffer, the beads were boiled in SDS sample buffer. hTFIIIC220, -110, -102, -90, and -63 in the SDS eluates were detected by immunoblotting with a mixture of antisera against hTFIIIC220, -110, -102, -90, and -63. The amounts of individual antibodies were adjusted so that the intensities of the hTFIIIC220, -110, -102, -90, and -63 immunoreactive bands were similar.
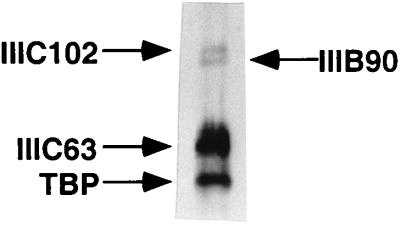
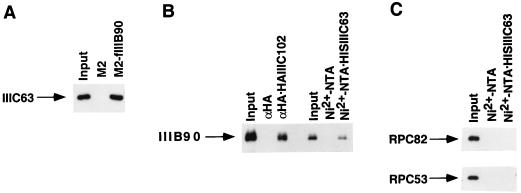
To further investigate the role of specific subunit interactions between TFIIIC and TFIIIB and between TFIIIC and RNA polymerase III, we analyzed binding of intact factors to individual immobilized polypeptides. The analysis in Fig. 7A shows that M2 agarose-immobilized FLAG-hTFIIIB90, but not M2 agarose alone, could bind immunopurified TFIIIC (detected by anti-hTFIIIC63 antibodies). The analysis in Fig. 7B shows that immobilized hTFIIIC63 and immobilized hTFIIIC102 could specifically bind a purified core hTFIIIB90-TBP complex (detected by anti-hTFIIIB90 antibodies), whereas immobilized hTFIIIC63 failed to stably bind an immunopurified RNA polymerase III (Fig. 7C).
FIG. 7.
Interactions between TFIIIB, TFIIIC, and RNA polymerase III. Input samples contained 10% of the amounts used for the interactions. (A) M2 agarose or M2 agarose-immobilized FLAG-hTFIIIB90 (M2-fIIIB90) was incubated with an immunopurified TFIIIC and washed with BC400–0.1% NP-40. Bound proteins were eluted with SDS buffer and analyzed by immunoblotting with antisera against hTFIIIC63. (B) Ni2+–NTA–agarose or Ni2+–NTA–agarose-immobilized His10–hTFIIIC63 and anti-HA agarose or anti-HA agarose-immobilized hTFIIIC102 were incubated with a purified core FLAG–hTFIIIB90–GST–TBP subcomplex. The latter subcomplex was isolated by successive affinity chromatography of a mixture of baculovirus-expressed FLAG-hTFIIIB90 and bacterially expressed GST-TBP on glutathione-Sepharose and M2 agarose, with BC400–0.1% NP-40 washes prior to elution (38). Bound proteins were eluted in the SDS buffer and analyzed by immunoblotting with antisera against hTFIIIB90. (C) Ni2+–NTA–agarose or Ni2+–NTA–agarose-immobilized His10–hTFIIIC63 was incubated with an immunopurified RNA polymerase III and washed with BC150–0.1% NP-40. Bound proteins were eluted in the SDS buffer and analyzed by immunoblotting with antisera against hRPC82 and hRPC53.
DISCUSSION
TFIIIC plays a primary role in promoter recognition and formation of stable PICs on a subset of genes transcribed by RNA polymerase III. Toward a more detailed analysis of the central role of human TFIIIC in these processes, we report the cloning and characterization of cDNAs encoding two subunits (hTFIIIC102 and hTFIIIC63) of the TFIIIC2 subcomplex that directly binds to tRNA and VA RNA promoters. These cDNAs have been used to investigate the primary structure, phylogenetic conservation, and mechanism of action of TFIIIC. In contrast to previously reported TFIIIC2 subunits with no yeast counterparts, hTFIIIC102 and hTFIIIC63 have yeast homologues and show overlapping but more-extensive interactions than those reported for their yeast counterparts.
Structure and evolutionary conservation of hTFIIIC102 and hTFIIIC63.
Given that yeast TFIIIC and human TFIIIC are multisubunit complexes and recognize similar promoter sequences (A and B boxes) in various class III genes, it was surprising to find that human TFIIIC220 (TFIIICα) and yeast TFIIIC138 (τ138), which both make B box contacts, show no significant sequence similarity (23, 26) and, further, that human TFIIIC110 (TFIIICβ) also shows no significant sequence similarity to any of the five cloned subunits (138, 131, 95, 91, and 55 kDa) of yeast TFIIIC (34). However, since most or all of the TFIIIB (14a, 29, 36a, 38) and RNA polymerase III (reviewed in references 40, 41, and 42a) subunits are conserved from yeast to human, it was anticipated that human TFIIIC would contain at least some subunits with sequence relationships to yeast TFIIIC (notably those interacting with conserved TFIIIB subunits). Consistent with this notion, hTFIIIC63 and hTFIIIC102 have significant sequence and functional relationships, respectively, to the TFIIIC95 and TFIIIC131 subunits of yeast TFIIIC. hTFIIIC63 preserves, minimally, the highly acidic region and helix-turn-helix motif of yTFIIIC95, and hTFIIIC102 preserves the charged regions, TPRs, and HLH domain structures of yTFIIIC131.
Interactions of human TFIIIC, TFIIIB, and RNA polymerase III in relation to PIC assembly.
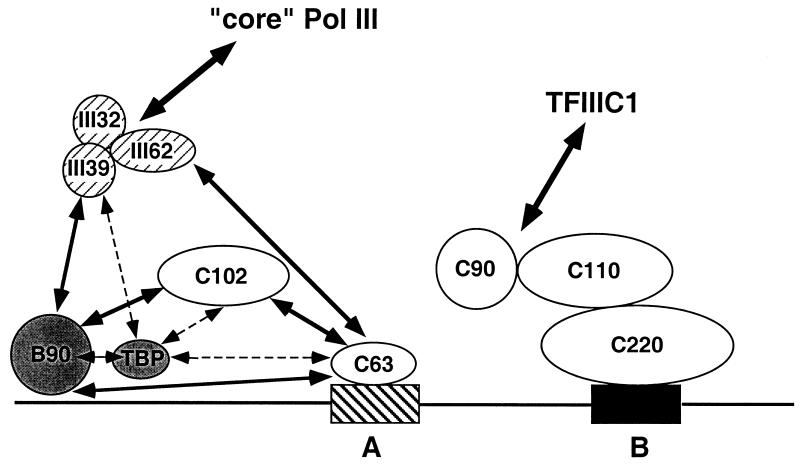
As detailed in the introduction, DNA binding and photoaffinity cross-linking studies revealed approximate topological positions of yeast TFIIIC, TFIIIB, and RNA polymerase subunits on the promoter and suggested interactions that were confirmed in part by direct protein-protein interactions and genetic interaction analyses. Consistent with the simple sequential recruitment model, the genetic interaction studies reported (i) interactions of yTFIIIC131 with yTFIIIB70 and yTFIIIB90, which are situated in the most distal occupied region of the promoter in the PIC (however, the expected physical interaction of yTFIIIC95, which is localized within the PIC in the A box region, with yTFIIIC131, part of which is situated upstream of the start site in the PIC, has not yet been demonstrated), and (ii) interaction of yTFIIIB70 with yRPC34, which also is localized to an upstream region of the A box in the PIC. In agreement with the sequence conservation of all these components from yeast to human, including the relationships of the newly described hTFIIIC63 and hTFIIIC102 to yTFIIIC95 and yTFIIIC131, respectively, we have demonstrated that hTFIIIC102 can interact with hTFIIIB90 (homologue of yTFIIIB70) and, in a previous report (41), that hTFIIIB90 can interact with hRPC39 (homologue of yRPC34). Significantly, however, we have also described novel interactions of human TFIIIC components not previously described for the homologous yeast components. These include the important interaction between hTFIIIC102 and hTFIIIC63, which documents a direct link between the TFIIIB-interacting and DNA-interacting subunits of TFIIIC, as well as interactions between hTFIIIB90 and hTFIIIC63, interactions between TBP and both hTFIIIC102 and hTFIIIC63, and interactions between hRPC62 and hTFIIIC63 (summarized in Fig. 8). The latter results suggest that additional TFIIIB-TFIIIC and TFIIIC-RNA polymerase III contacts may facililate, perhaps via concerted interactions, both TFIIIB and RNA polymerase III recruitment. Alternatively, these interactions may also be involved in the function of the PIC components during initiation, elongation, termination, or reinitiation steps. The hTFIIIC63–hRPC62 interaction is particularly interesting because the corresponding yeast components appear to be localized to a common position in the PIC (4, 9) and because hRPC62 is present, with hRPC32 and hRPC39, in a subcomplex required specifically for transcription initiation by human RNA polymerase III (41). These observations also suggest, contrary to the prevailing yeast model (18), that human TFIIIC may have functions in addition to TFIIIB recruitment, and they are reminiscent of recent indications that human TFIIIC has a (presumably distinct) role in transcription termination and reinitiation by RNA polymerase III (42).
FIG. 8.
An integrated view of the interactions detected between subunits of human TFIIIB, TFIIIC, and RNA polymerase III. The observed protein-protein interactions are indicated by double-headed arrows. The A box and B box of the VAI/tRNA gene promoter are shown as hatched and solid rectangles, respectively. Hatched symbols, subcomplex of initiation-specific RNA polymerase III subunits; shaded symbols, TFIIIB subunits; open symbols, TFIIIC2 subunits. Individual subunits involved in interactions between TFIIIC2 and TFIIIC1 or between core RNA polymerase III and the three-subunit subcomplex are unknown. The positioning of the hTFIIIC220 subunit is based on cross-linking studies (21), whereas the positioning of the hTFIIIC63 and hTFIIIC102 subunits, as well as that of the TFIIIB subunits, is based on studies of the cognate yeast components.
The interactions indicated in the working model presented in Fig. 8 are based largely on in vitro interaction data and remain to be substantiated by in vivo analysis. However, our belief that the in vitro interactions described between recombinant human TFIIIC, TFIIIB, and RNA polymerase III subunits are functionally relevant is supported by additional observations. First, hTFIIIC102, hTFIIIC63, hTFIIIB90, and TBP appear to form a stable subcomplex. Second, as described below, the hTFIIIC102 interactions depend on motifs implicated by both biochemical and genetic studies in yTFIIIC131 interactions. Third, the interactions appear in most cases to be maintained in the context of native complexes. Thus, hTFIIIB90 was found to interact with native TFIIIC; hTFIIIC102 and hTFIIIC63 were found to interact (individually) with core TFIIIB; and hRPC62 was found to interact with native TFIIIC. However, a reciprocal interaction of isolated hTFIIIC63 with native RNA polymerase III could not be demonstrated. One possibility is that TFIIIB interactions cause a conformational change in RNA polymerase III that exposes hRPC62 for subsequent interactions with hTFIIIC63. (This may provide a mechanism for cells to block the formation of nonproductive RNA polymerase III PICs that inhibit transcription.) Another possibility is that interactions between hTFIIIC63 and RNA polymerase III-associated hRPC62 may simply be too weak, and effective concentrations of interacting components too low, for stable binding in the absence of other interactions with RNA polymerase III.
Thus, based on past (5, 25) and present data, we propose that the overall mechanism for stepwise PIC formation in the human system is basically similar to that proposed for yeast, with the added complexity of supplementary interactions of TFIIIC with both TFIIIB and RNA polymerase III. Bearing in mind that there are some fundamental structural differences between human and yeast TFIIIC (discussed above) and that the human system may require other accessory factors not utilized in yeast (40, 42), these supplementary interactions could simply reflect a metazoan-specific extension of the fundamental mechanism—although it will be important to determine if similar interactions can be observed in yeast. In this regard it will also be important to determine other possible similarities or differences in yeast and mammalian TFIIIC and, specifically, how the TFIIIC1 subunits may relate to other yeast TFIIIC subunits. It also may be that additional TFIIIC interactions are required for target gene activation within chromatin; in this regard, recent studies have demonstrated a role for TFIIIC in chromatin antirepression mechanisms (7, 22) and the presence in human TFIIIC of several subunits with histone acetyltransferase activities (22). A final point concerns the possibility of novel TFIIIC interactions that might be important for the preassembly of RNA polymerase III and its accessory factors into a transcription-competent complex prior to PIC assembly. Although such a complex has been described (40), the extent to which transcription in vivo involves stepwise assembly of the PIC (from separate factors) versus preassembly of an RNA polymerase III holoenzyme remains to be determined.
Role of the TPRs in TFIIIC function.
Tandem arrays of the TPR have been found in a wide variety of eukaryotic proteins and are thought to mediate protein-protein interactions (reviewed in reference 24). In relation to the present study, yTFIIIC131 (homologue of hTFIIIC102) contains three clusters of TPR motifs (28) in N-terminal (TPR1 through -5), central (TPR6 through -9), and C-terminal (TPR10 through -11) regions. yTFIIIC131 fragments containing at least two repeats were shown to interact independently with yTFIIIB70 in vitro (20), whereas a fragment containing the N-terminal region and the adjoining TPR1 was sufficient for in vivo interactions with yTFIIIB70 (8). The failure of the in vivo (yeast two-hybrid) assays to detect yTFIIIB70 interactions with yTFIIIC131 lacking only the small N-terminal regions may have reflected autoinhibitory effects (within yTFIIIC131) that are normally reversed by conformational changes dependent on multiple interactions between full-length yTFIIIC131 and yTFIIIB70 (8).
Consistent with these results, the present study shows in vitro interactions between hTFIIIB90 (homologue of yTFIIIB70) and most tested fragments of hTFIIIC102 (homologue of yTFIIIC131) that contain at least two intact TPRs, as well as stronger interactions with fragments containing more than two repeats. Our analysis also shows similar interactions of hTFIIIC63 with hTFIIIC102 fragments containing TPR motifs. However, some specificity is evident in that a hTFIIIC102 fragment with the N-terminal amphiphilic and TPR1 to -2 regions interacts more strongly with hTFIIIB90 than with hTFIIIC63, whereas the reciprocal specificity is seen with a small hTFIIIC102 fragment containing TPR3 and -4. This apparent specificity is consistent with the demonstration that distinct TPR motifs within CYC8/SSN6 are involved in interactions with the corepressor TUP1 and with different DNA-binding repressors (37).
The specificity and relevance of TPRs within TFIIIC are further indicated by the identification of multiple mutations within yTFIIIC131 TPR2 that activate RNA polymerase III-mediated transcription through a mechanism that appears to involve both a conformational change in TFIIIC and enhanced TFIIIB recruitment (30). Other studies have shown conformational changes in yTFIIIC131 during TFIIIB binding to the PIC (17), as well as alternative TFIIIB and TFIIIC promoter arrangements that have been suggested to reflect TPR-mediated variations either in the folding of yTFIIIC131 or in its interactions with TFIIIB (15). The present results are consistent with these possibilities for the human homologues, as well as alternate interactions of hTFIIIC102 with hTFIIIC63. Clearly, it will be important to further document the role of individual TPRs in specific interactions with other polypeptides, and possible changes therein during PIC assembly and function both on a single promoter and on different promoters.
ACKNOWLEDGMENTS
We thank L. Bai for the TFIIIB-containing fraction free of RNA polymerase III and the RNA polymerase III-containing fraction free of TFIIIB, and M. Guermah for the His6-TBP baculoviruses. We also thank L. Bai, Y. Tao, and M. Teichmann for helpful discussions and data bank searching.
This work was supported by a grant (CA42567) from the National Institutes of Health to R.G.R.
REFERENCES
- 1.Arrebola R, Manaud N, Rozenfeld S, Marsolier M C, Lefebvre O, Carles C, Thuriaux P, Conesa C, Sentenac A. τ91, an essential subunit of yeast transcription factor IIIC, cooperates with τ138 in DNA binding. Mol Cell Biol. 1998;18:1–9. doi: 10.1128/mcb.18.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartholomew B, Kassavetis G A, Braun B R, Geiduschek E P. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking regent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartholomew B, Kassavetis G A, Geiduschek E P. Two components of Saccharomyces cerevisiae TFIIIB are stereospecifically located upstream of a tRNA gene and interact with the second largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartholomew B, Durkovich D, Kassavetis G A, Geiduschek E P. Orientation and topography of RNA polymerase III in transcription complexes. Mol Cell Biol. 1993;13:942–952. doi: 10.1128/mcb.13.2.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bieker J J, Martin P L, Roeder R G. Formation of a rate-limiting intermediate in 5S RNA gene transcription. Cell. 1985;40:119–127. doi: 10.1016/0092-8674(85)90315-0. [DOI] [PubMed] [Google Scholar]
- 6.Brun I, Sentenac A, Werner M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997;16:5730–5741. doi: 10.1093/emboj/16.18.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnol A F, Margottin F, Huet J, Almouzni G, Prioleau M N, Mechali M, Sentenac A. TFIIIC relieves repression of U6 snRNA transcription by chromatin. Nature. 1993;362:475–477. doi: 10.1038/362475a0. [DOI] [PubMed] [Google Scholar]
- 8.Chaussivert N, Conesa C, Shaaban S, Sentenac A. Complex interactions between yeast TFIIIB and TFIIIC. J Biol Chem. 1995;270:15353–15358. doi: 10.1074/jbc.270.25.15353. [DOI] [PubMed] [Google Scholar]
- 9.Chiannilkulchai N, Stalder R, Riva M, Carles C, Werner M, Sentenac A. RPC82 encodes the highly conserved, third-largest subunit of RNA polymerase C (III) from Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:4433–4440. doi: 10.1128/mcb.12.10.4433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das A K, Cohen P T W, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabrielsen O S, Sentenac A. RNA polymerase III(C) and its transcription factors. Trends Biochem Sci. 1991;16:412–416. doi: 10.1016/0968-0004(91)90166-s. [DOI] [PubMed] [Google Scholar]
- 12.Geiduschek E P, Kassavetis G A. RNA polymerase III transcription complexes. In: Yamamoto K, editor. Transcription regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 247–280. [Google Scholar]
- 13.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 14.Hirano T, Kinoshita N, Morikawa K, Yanagida M. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+ Cell. 1990;60:319–328. doi: 10.1016/0092-8674(90)90746-2. [DOI] [PubMed] [Google Scholar]
- 14a.Hsieh, Y., Z. Wang, and R. G. Roeder. Unpublished data.
- 15.Joazeiro C A P, Kassavetis G A, Geiduschek E P. Alternative outcomes in assembly of promoter complexes: the roles of TBP and a flexible linker in placing TFIIIB on tRNA genes. Genes Dev. 1996;10:725–739. doi: 10.1101/gad.10.6.725. [DOI] [PubMed] [Google Scholar]
- 16.Kassavetis G A, Braun B R, Nguyen L H, Geiduschek E P. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell. 1990;60:235–245. doi: 10.1016/0092-8674(90)90739-2. [DOI] [PubMed] [Google Scholar]
- 17.Kassavetis G A, Joazeiro C A P, Pisano M, Geiduschek E P, Colbert T, Hahn S, Blanco J A. The role of the TATA binding protein in the assembly and function of the multisubunit yeast RNA polymerase III, transcription factor TFIIIB. Cell. 1992;71:1055–1064. doi: 10.1016/0092-8674(92)90399-w. [DOI] [PubMed] [Google Scholar]
- 18.Kassavetis G A, Bardeleben C, Bartholomew B, Braun B R, Joazeiro C A P, Pisano M, Geiduschek E P. Transcription by RNA polymerase III. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 107–126. [Google Scholar]
- 19.Kassavetis G A, Kumar A, Ramirez E, Geiduschek E P. Functional and structural organization of Brf, the TFIIB-related component of the RNA polymerase III transcription initiation complex. Mol Cell Biol. 1998;18:5587–5599. doi: 10.1128/mcb.18.9.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoo B, Brophy B, Jackson S P. Conserved functional domains of the RNA polymerase III general transcription factor BRF. Genes Dev. 1994;8:2879–2890. doi: 10.1101/gad.8.23.2879. [DOI] [PubMed] [Google Scholar]
- 21.Kovelman R, Roeder R G. Purification and characterization of two forms of human transcription factor IIIC. J Biol Chem. 1992;267:24446–24456. [PubMed] [Google Scholar]
- 22.Kundu T K, Wang Z, Roeder R G. Human TFIIIC relieves chromatin-mediated repression of RNA polymerase III transcription and contains an intrinsic histone acetyltransferase activity. Mol Cell Biol. 1999;19:1605–1615. doi: 10.1128/mcb.19.2.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lagna G, Kovelman R, Sukegawa J, Roeder R G. Cloning and characterization of an evolutionarily divergent DNA-binding subunit of mammalian TFIIIC. Mol Cell Biol. 1994;14:3053–3064. doi: 10.1128/mcb.14.5.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb J R, Tugendreich S, Hieter P. Tetratricopeptide repeat interaction: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–258. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 25.Lassar A B, Martin P L, Roeder R G. Transcription of class III genes: formation of preinitiation complexes. Science. 1983;222:740–748. doi: 10.1126/science.6356356. [DOI] [PubMed] [Google Scholar]
- 26.L’Etoile M D, Fahnestock M L, Shen Y, Aebersold R, Berk A. Human transcription factor TFIIIC box B binding subunit. Proc Natl Acad Sci USA. 1994;91:1652–1656. doi: 10.1073/pnas.91.5.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manaud N, Arrebola R, Buffin-Meyer B, Lefebvre O, Voss H, Riva M, Conesa C, Sentenac A. A chimeric subunit of yeast transcription factor IIIC forms a subcomplex with τ95. Mol Cell Biol. 1998;18:3191–3200. doi: 10.1128/mcb.18.6.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marck C, Lefebvre O, Carles C, Riva M, Chaussivert N, Ruet A, Sentenac A. The TFIIIB-assembling subunit of yeast transcription factor TFIIIC has both tetratricopeptide repeats and basic helix-loop-helix motifs. Proc Natl Acad Sci USA. 1993;90:4027–4031. doi: 10.1073/pnas.90.9.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mital R, Kobayashi R, Hernandez N. RNA polymerase III transcription from the human U6 and adenovirus type 2 VAI promoters has different requirements for human BRF, a subunit of human TFIIIB. Mol Cell Biol. 1996;16:7031–7042. doi: 10.1128/mcb.16.12.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moir R A, Sethy-Coraci I, Puglia K, Librizzi M D, Willis I M. A tetratricopeptide repeat mutation in yeast transcription factor IIIC131 (TFIIIC131) facilitates recruitment of TFIIIB-related factor TFIIIB70. Mol Cell Biol. 1997;17:7119–7125. doi: 10.1128/mcb.17.12.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons M, Weil P A. Cloning of TFC1, the Saccharomyces cerevisiae gene encoding the 95-kDa subunit of transcription factor TFIIIC. J Biol Chem. 1992;267:2894–2901. [PubMed] [Google Scholar]
- 32.Ruth J, Conesa C, Dieci G, Lefebvre O, Dusterhoft A, Ottonello S, Sentenac A. A supressor of mutations in the class III transcription system encodes a component of yeast TFIIIB. EMBO J. 1996;15:1941–1949. [PMC free article] [PubMed] [Google Scholar]
- 33.Sikorski R S, Boguski M S, Goebl M, Hieter P. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell. 1990;60:307–317. doi: 10.1016/0092-8674(90)90745-z. [DOI] [PubMed] [Google Scholar]
- 34.Sinn E, Wang Z, Kovelman R, Roeder R G. Cloning and characterization of a TFIIIC2 subunit (TFIIICβ) whose presence correlates with activation of RNA polymerase III mediated transcription by adenovirus E1A expression and serum factors. Genes Dev. 1995;9:675–685. doi: 10.1101/gad.9.6.675. [DOI] [PubMed] [Google Scholar]
- 35.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 36.Swanson R N, Conesa C, Lefebvre O, Carles C, Ruet A, Quemenur E, Gagnon J, Sentenac A. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor τ (TFIIIC) Proc Natl Acad Sci USA. 1991;88:4887–4891. doi: 10.1073/pnas.88.11.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36a.Teichmann, M., Z. Wang, M. Ito, K. Seifart, and R. G. Roeder. Unpublished data.
- 37.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Roeder R G. Structure and function of a human transcription factor TFIIIB subunit that is evolutionarily conserved and contains both TFIIB- and high-mobility-group protein 2-related domains. Proc Natl Acad Sci USA. 1995;92:7026–7030. doi: 10.1073/pnas.92.15.7026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, Roeder R G. TFIIIC1 acts through a downstream region to stabilize TFIIIC2 binding to RNA polymerase III promoters. Mol Cell Biol. 1996;16:6841–6850. doi: 10.1128/mcb.16.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Z, Lou T, Roeder R G. Identification of an autonomously initiating RNA polymerase III holoenzyme containing a novel factor that is selectively inactivated during protein synthesis inhibition. Genes Dev. 1997;11:2371–2382. doi: 10.1101/gad.11.18.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Roeder R G. Three human RNA polymerase III specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 42.Wang Z, Roeder R G. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–751. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- 42a.Wang, Z., and R. G. Roeder. Unpublished data.
- 43.Werner M, Chaussivert N, Willis I M, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 44.White R J. RNA polymerase III transcription. R. G. Austin, Tex: Landes Co.; 1994. [Google Scholar]
- 45.Willis I M. RNA polymerase III. Genes, factors and transcriptional specificity. Eur J Biochem. 1993;212:1–11. doi: 10.1111/j.1432-1033.1993.tb17626.x. [DOI] [PubMed] [Google Scholar]
- 46.Yoshinaga S K, Boulanger P A, Berk A J. Resolution of human transcription factor TFIIIC into two functional components. Proc Natl Acad Sci USA. 1987;84:3585–3589. doi: 10.1073/pnas.84.11.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshinaga S K, L’Etoile N D, Berk A J. Purification and characterization of transcription factor IIIC2. J Biol Chem. 1989;264:10726–10731. [PubMed] [Google Scholar]