Abstract
SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) is the causative agent of the COVID19 pandemic. The SARS-CoV-2 genome encodes for a small accessory protein termed Orf9b, which targets the mitochondrial outer membrane protein TOM70 in infected cells. TOM70 is involved in a signaling cascade that ultimately leads to the induction of type I interferons (IFN-I). This cascade depends on the recruitment of Hsp90-bound proteins to the N-terminal domain of TOM70. Binding of Orf9b to TOM70 decreases the expression of IFN-I; however, the underlying mechanism remains elusive. We show that the binding of Orf9b to TOM70 inhibits the recruitment of Hsp90 and chaperone-associated proteins. We characterized the binding site of Orf9b within the C-terminal domain of TOM70 and found that a serine in position 53 of Orf9b and a glutamate in position 477 of TOM70 are crucial for the association of both proteins. A phosphomimetic variant Orf9bS53E showed drastically reduced binding to TOM70 and did not inhibit Hsp90 recruitment, suggesting that Orf9b–TOM70 complex formation is regulated by phosphorylation. Eventually, we identified the N-terminal TPR domain of TOM70 as a second binding site for Orf9b, which indicates a so far unobserved contribution of chaperones in the mitochondrial targeting of the viral protein.
Keywords: SARS-CoV-2, COVID19, B.1.1.7, variant of concern, Orf9b, TOM70, mitochondria, Hsp90, interferon
1. Introduction
Since the beginning of the COVID19 pandemic, its causative agent, SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), has evolved into several lineages. The SARS-CoV-2 variant of concern (VoC) Alpha (B.1.1.7, VOC 202012/01) emerged in England in September 2020 and, within a few months, became the worldwide dominating lineage [1]. A hallmark of VoC Alpha is a higher transmissibility compared to preexisting virus variants, and recent studies indicate that it also poses a higher risk of hospitalization and ICU admission [2,3,4,5,6]. VoC Alpha is characterized by several genomic mutations, one of which is found within its N gene and seems to increase the expression of a sub-genomic open reading frame (ORF) encoding a small accessory protein termed Orf9b [7,8]. Recent unpublished data suggest that a sub-genomic RNA (sgRNA) encoding for Orf9b is expressed up to 16-fold in samples of patients infected with VoC Alpha, hinting at an important role of Orf9b for the pathogenicity of this variant [9].
Orf9b interacts with the mitochondria of infected cells by binding to TOM70, a versatile adaptor protein of the mitochondrial outer membrane involved in many cellular processes, including protein import, mediation of membrane contacts, and the innate immune response to viral infections [10,11]. Expression of Orf9b leads to a decreased induction of type I interferons (IFN-I) in infected cells by interfering with a signaling cascade involving the proteins RIG-I, MAVS, TOM70, TBK1, and IRF3 [12,13,14]. Upon viral RNA recognition, RIG-I binds to MAVS, which, in turn, interacts with TOM70 to allow association with Hsp90-bound TBK1 and IRF3. It has been speculated that inhibition of Hsp90 binding by Orf9b could be a possible reason for decreased IFN-I expression [13]. A study using isothermal titration calorimetry (ITC) indicated that the binding of an Orf9b-derived peptide inhibits the binding of an Hsp90-derived peptide to the N-terminal domain of TOM70, suggesting an allosteric inhibition [15]. However, the influence of Orf9b binding on the recognition of native Hsp90 was not investigated.
A high-resolution Cryo-EM structure of a core fragment of Orf9b in complex with human TOM70 depicts the central part of Orf9b (amino acids 39–76) buried within the C-terminal domain of TOM70 [16]. Additionally, a crystal structure of a similar fragment of Orf9b (amino acids 43–78) in complex with human TOM70 was published recently and confirmed the localization of the core part of Orf9b within TOM70 [15]. Within the interaction interface of the Orf9b–TOM70 complex, both structures show a prominent amino acid of Orf9b, a serine at position 53 (S53), to form a hydrogen bond with a glutamate at position 477 (E477) of TOM70 in an overall hydrophobic region. Interestingly, S53 was identified as an infection-driven phosphorylation site, suggesting a possible mechanism of regulation of the Orf9b–TOM70 interaction [17].
In this study, we characterized the binding site of Orf9b within the C-terminal domain of TOM70 and found that the interaction of Orf9b S53 with TOM70 E477 is crucial for complex formation. A phosphomimetic variant, Orf9bS53E, showed dramatically decreased incorporation into the C-terminal domain of TOM70, suggesting that the binding of Orf9b to the receptor can be regulated by the phosphorylation of Orf9b S53. We found that the binding of Orf9b inhibits the association of TOM70 with the cytosolic chaperone Hsp90, explaining the observed decrease in the IFN-I response in Orf9b-expressing cells. Eventually, we identified a second binding site of Orf9b within the N-terminal TPR domain of TOM70, defined by the arginine in position 192 (R192), which promotes the binding of Orf9b to the central part of TOM70.
2. Results
2.1. A Phosphomimetic Amino Acid Exchange in Orf9b Prevents Its Interaction with TOM70
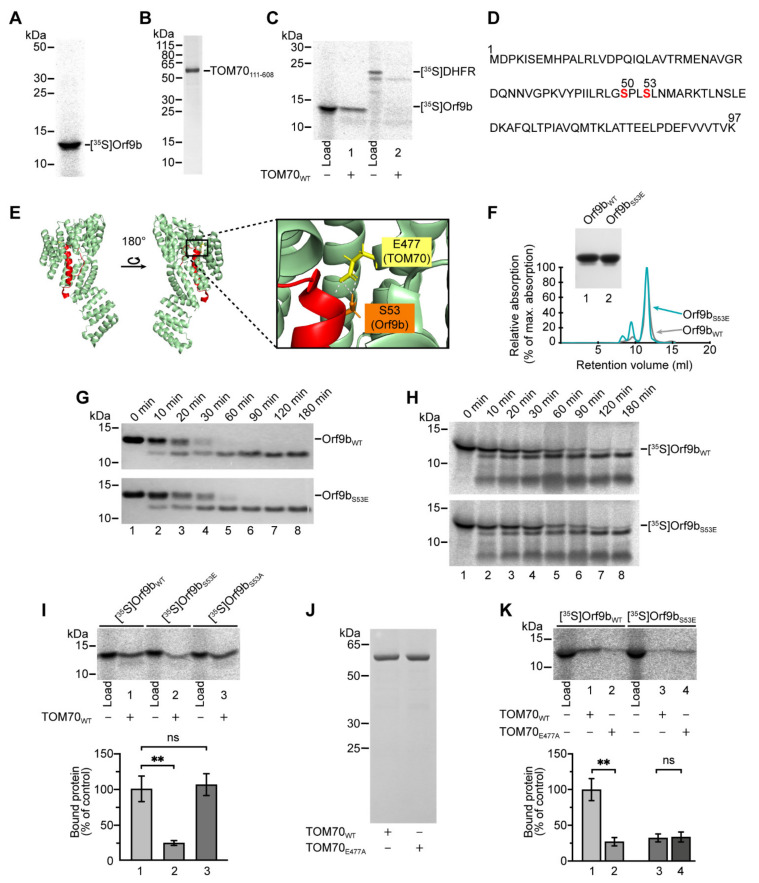
In previous studies, the complex formation of Orf9b and TOM70 was mainly characterized using purified proteins or synthetic peptides [15,16]. To investigate the interaction of both proteins under the conditions of a mammalian cytosol, we translated Orf9b in rabbit reticulocyte lysate (RL) in the presence of [35S]-methionine (Figure 1A). This system includes all cytosolic factors and molecular chaperones that may participate in the targeting and binding of proteins to the mitochondrial surface [18,19,20,21,22]. We then used samples of the reticulocyte lysate containing radiolabeled Orf9b for incubation with the cytosolic domain of human TOM70 (amino acids 111–608). For this purpose, TOM70 was bound to Ni-NTA agarose beads, following a basic protocol published by Brix et al. [23] (Figure 1B). The presence of equal amounts of TOM70 in the individual samples was confirmed by elution of the Ni-NTA-bound protein and Western blot analysis using an antibody against the N-terminal His tag of TOM70 (Figure S1). Using this system, we found that radiolabeled Orf9b associated with TOM70 (Figure 1C, lane 1). Under the same conditions, radiolabeled mouse dihydrofolate reductase (DHFR), a cytosolic protein of 21.6 kDa, often used as an established model protein [24,25,26], showed no binding to the receptor (Figure 1C, lane 2).
Figure 1.
Characterization of Orf9b binding to the C-terminal domain of TOM70. (A) Radiolabeled SARS-CoV-2 [35S]Orf9bWT was synthesized in rabbit reticulocyte lysate and was analyzed by SDS-PAGE. (B) Coomassie staining of TOM70111–608 purified using Ni-NTA agarose beads. (C) In vitro binding of [35S]Orf9bWT or [35S]DHFR (mouse dihydrofolate reductase) to 250 pmol Ni-NTA bound TOM70111–608. Signals obtained after SDS-PAGE and autoradiographic detection were quantified using 2% of the total added lysate as internal control (“Load”). (D) Primary structure of SARS-CoV-2 Orf9b highlighting the serine residues at position 50 and 53 of Orf9b. (E) Structure of the Orf9b–TOM70 complex highlighting the interaction of serine 53 of Orf9b with glutamate 477 of TOM70. Structural data were obtained from PDB file 7KDT [16]. (F) Size-exclusion chromatography of Orf9bWT and Orf9bS53E using a Superdex S75 increase 10/300 GL column depicting normalized UV absorption. DHFR-fused Orf9b variants were expressed in E. coli and purified from cell lysates by IMAC, IEC, removal of the DHFR tag by TEV digestion with subsequent affinity purification and SEC (Insert). (G) Trypsin treatment of purified Orf9bWT and Orf9bS53E yielded identical degradation products. For trypsin digestion, 5.5 µg/mL Orf9bWT or Orf9bS53E were incubated with 5 µg/mL trypsin on ice for the indicated times, and the reaction was stopped by incubation at 95 °C for 5 min. Samples were subsequently analyzed by SDS-PAGE and visualized by Coomassie staining. (H) Radiolabeled [35S]Orf9bWT and [35S]Orf9bS53E synthesized in reticulocyte lysate show similar trypsin degradation products. Trypsin digestion was performed as described in G, using 20% (v/v) reticulocyte lysate containing [35S]-labeled protein instead of purified proteins. (I) In vitro binding of radiolabeled Orf9b variants to Ni-NTA bound TOM70111-608. The experiment was carried out as described in C, and binding of [35S]Orf9bWT was set as control. Signals obtained after SDS-PAGE and autoradiographic detection were quantified using 2% of the total added lysate as internal control (“Load”). ** indicate p-values < 0.01, “ns” indicates no significance, error bars show SEM, n = 4. (J) Coomassie staining of TOM70WT and TOM70E477A purified using Ni-NTA agarose beads. (K) In vitro binding of radiolabeled [35S]Orf9bWT and [35S]Orf9bS53E to 250 pmol of either TOM70WT or TOM70E477A was carried out as described in (C). Orf9bWT binding to TOM70WT was set to 100% (control). ** indicate p-values < 0.01, “ns” indicates no significance, error bars show SEM, n = 4.
A recent study identified two phosphorylation sites on Orf9b: a serine at position 50 (S50) and a serine at position 53 (S53) (Figure 1D) [17]. Orf9b S53 forms a prominent hydrogen bond with a conserved glutamate at position 477 in the C-terminal domain of TOM70 (E477) within the overall hydrophobic interaction interface of both proteins (Figure 1E) [15,16]. We hypothesized that the phosphorylation of Orf9b S53 would destabilize the interaction with TOM70 E477, which, in turn, could weaken the association of both proteins. To test for the consequences of the phosphorylation of S53, we replaced serine 53 with a phosphomimetic glutamate residue, leading to the variant Orf9bS53E. Since Orf9b forms homodimers in the absence of TOM70 [15,16,27], we tested for a possible influence of the mutation of S53 on protein multimerization by purifying both variants Orf9bWT and Orf9bS53E from E. coli (Figure 1F, insert). The comparison with a standard curve of proteins of known molecular mass indicated that during size exclusion chromatography (SEC), both Orf9b variants were eluted as dimers of a molecular mass of approximately 28.7 kDa (Figure 1F). To exclude that the introduction of the phosphomimetic residue leads to structural changes within Orf9b, we incubated the purified variants in the presence of trypsin and subsequently compared the obtained digestion pattern of degradation products by SDS-PAGE. We observed a time-dependent appearance of a trypsin-resistant fragment of Orf9b for both variants, suggesting that the tertiary structures of the proteins were similar (Figure 1G). To exclude a negative effect of the S53E mutation on a possible association of Orf9b with cytosolic factors, we synthesized the two variants, Orf9bWT and Orf9bS53E, in the in vitro translation system and repeated the experiment. We found a similar appearance of the trypsin-dependent Orf9b fragment for both variants, suggesting that interaction with cytosolic factors is not mitigated for Orf9bS53E (Figure 1H).
We then used the binding assay to compare the interactions of radiolabeled Orf9bWT and Orf9bS53E with Ni-NTA-bound TOM70 (Figure 1I). We found that the binding of Orf9bS53E to isolated TOM70 was reduced by 76% compared to the wild-type protein (Figure 1I, lane 1 vs. lane 2). Conversely, the binding of a variant in which S53 was replaced by an alanine, Orf9bS53A, showed no reduction in TOM70 binding, demonstrating the specificity of this effect for the phosphomimetic variant (Figure 1I, lane 1 vs. lane 3).
To further characterize the effects of the different residues in position 53, we fused each variant of Orf9b to DHFR (to facilitate the resolution in SDS-PAGE), synthesized the hybrid proteins in reticulocyte lysate, and tested for the accessibility of trypsin cleavage sites in the presence or absence of purified soluble TOM70. In the absence of TOM70, trypsin treatment of radiolabeled Orf9bWT-DHFR in the reticulocyte lysate yielded a specific fragment corresponding to an N-terminally truncated product (Figure S2, fragment f1, lanes 2–4). In the presence of TOM70, we again observed an N-terminally shortened degradation product, albeit of higher molecular mass. This indicates that Orf9bWT-DHFR is efficiently bound to the C-terminal domain of TOM70, with most parts of the construct being shielded from the protease (Figure S2, fragment f2, lanes 6–8). The same pattern was obtained with Orf9bS53A-DHFR, indicating that the access of this construct to the binding site in the C-terminal domain of TOM70 was retained (Figure S2, fragment f2, lanes 6–8). In contrast, Orf9bS53E-DHFR was degraded to the same fragments, both in the presence and absence of TOM70, confirming that this variant is not incorporated into the receptor (Figure S2, fragment f1, lanes 6−7).
To further characterize the interaction between Orf9b S53 and TOM70 E477, we tested the binding of radiolabeled Orf9bWT and Orf9bS53E to isolated wild-type TOM70 (TOM70WT) and to a variant, in which the glutamate at position 477 was exchanged for an alanine (TOM70E477A; Figure 1J). The binding of in vitro-synthesized Orf9bWT to TOM70E477A was reduced by 73% in comparison to wild-type TOM70, thus to a similar extent as observed for the binding of Orf9bS53E to TOM70WT (Figure 1K, lane 2 vs. lane 3). This suggests that the interaction of the serine S53 in Orf9b with the glutamate E477 in TOM70 is crucial for the formation of the Orf9b–TOM70 complex since a mutation of either amino acid residue drastically reduces the association of the two proteins. Interestingly, the incubation of phosphomimetic Orf9bS53E with isolated TOM70E477A did not lead to a reduction in comparison to TOM70WT, indicating that the reduced association of Orf9bS53E with TOM70WT is not simply a result of the repulsion of two negative charges (Figure 1K, lane 3 vs. lane 4).
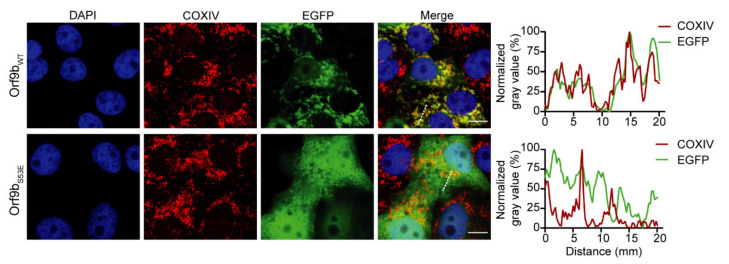
To test the relevance of our findings in mammalian cell culture, we expressed the variants Orf9bWT and Orf9bS53E C-terminally fused to EGFP in Vero E6 cells, and determined their cellular distribution by fluorescence microscopy (Figure 2). COXIV, a subunit of the cytochrome c oxidase (complex IV of the respiratory chain), was immunostained for the visualization of mitochondria. While Orf9bWT showed a strong co-localization with mitochondria, the phosphomimetic variant Orf9bS53E displayed a diffuse pattern, indicating a cytosolic location for the majority of molecules (Figure 2, profiles). Notably, we observed some co-localization of Orf9bS53E-EGFP with the DAPI-stained nuclei of several cells, which is likely a result of the lack of specific targeting of this construct.
Figure 2.
Subcellular localization of Orf9bWT and Orf9bS53E. Representative immunofluorescence images of Vero E6 cells expressing Orf9bWT-EGFP or Orf9bS53E-EGFP 24 h post-transfection (left panels). Gray value profiles of EGFP (green curves) and COXIV (red curves) (right) were measured with Image J (Fiji) software and normalized to the smallest and largest values in each data set. Gray value profiles were plotted along the white dotted lines. Cell nuclei are shown in blue (DAPI) and mitochondria in red. Scale bars, 10 µm.
The binding of Orf9b to TOM70 is obviously highly dependent on the interaction of serine 53 of Orf9b with glutamate 477 of TOM70. Our data further suggest that the association of both proteins is inhibited upon the phosphorylation of S53 of Orf9b.
2.2. Binding of Orf9b Inhibits the Interaction of TOM70 with Cytosolic Hsp90
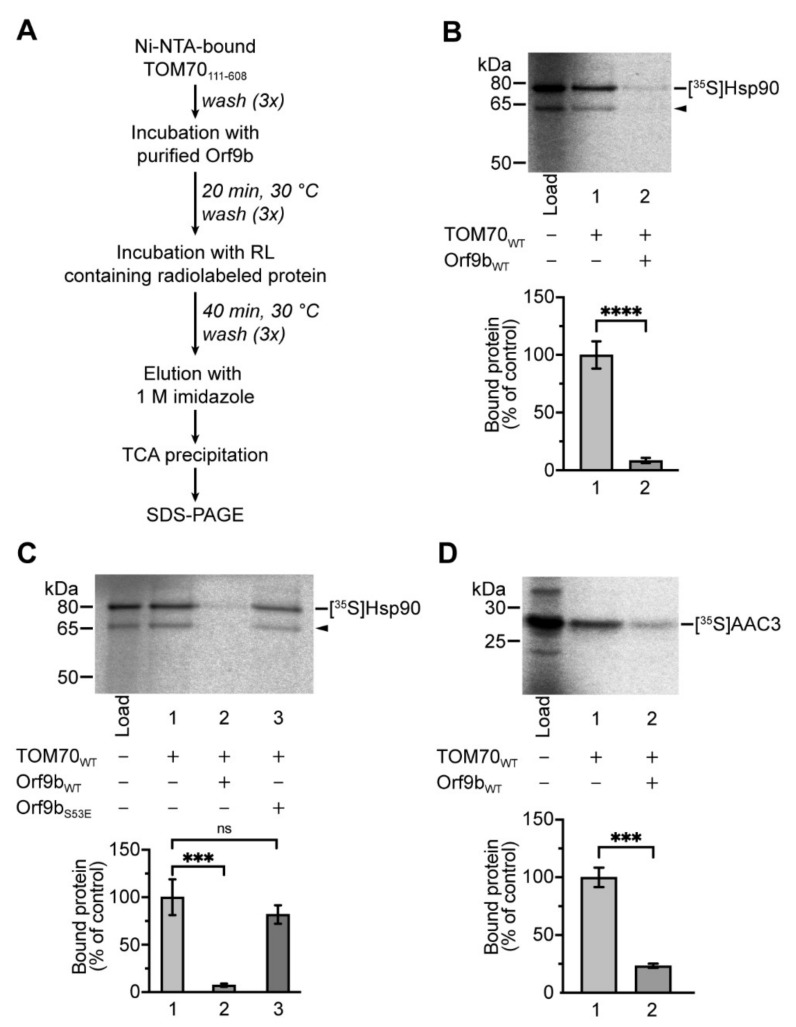
TOM70 is part of a signaling cascade that ultimately leads to the expression of IFN-I in response to RNA virus infections [11]. A crucial part of this cascade is the interaction of MAVS with TOM70 on the mitochondrial surface, which enables subsequent binding of Hsp90-bound TBK1 and IRF3 [12]. A previous study has shown that SARS-CoV-2 Orf9b targets TOM70 in mammalian cells and diminishes the expression of IFN-β, leading the authors to speculate that the binding of Orf9b to TOM70 might interfere with the recognition of Hsp90 [13]. A recent study determined the binding affinities of synthetic peptides to TOM70 by isothermal titration calorimetry (ITC) and found that an association of a short peptide containing the C-terminal EEVD motif of Hsp90 with the N-terminus of TOM70 was greatly decreased if the receptor was in complex with co-purified Orf9b [15]. Mammalian cytosol contains two isoforms of Hsp90, an inducible isoform, Hsp90-α, and the constitutively expressed isoform Hsp90-β [28]. To investigate if Orf9b binding to TOM70 affects its association with native full-length human Hsp90-β, we again used the procedure described by Brix et al. [23]. To this end, we pre-incubated isolated TOM70 with either purified Orf9bWT or Orf9bS53E and subsequently added reticulocyte lysate containing radiolabeled human Hsp90 (Figure 3A). The lysate contained a distinct second translation product of lower molecular mass, most likely caused by a second in-frame start codon within the open reading frame (Figure 3B, black arrow) [29]. The ability of this second product to associate with TOM70 is in agreement with the notion that complex formation is independent of the N-terminal part of Hsp90 and primarily mediated by its C-terminal EEVD motif, which binds to a tetratricopeptide repeat (TPR) domain within the N-terminus of TOM70 [30,31]. We found that the pre-incubation with Orf9bWT inhibited subsequent binding of Hsp90 almost completely (Figure 3B). No effect was observed when TOM70 was pre-incubated with the phosphomimetic variant Orf9bS53E, demonstrating that the interaction of S53 of Orf9b with E477 of TOM70 is essential not only for stable insertion into the Orf9b binding site but also for the inhibitory effect on the recruitment of Hsp90 (Figure 3C).
Figure 3.
Orf9b inhibits the binding of native Hsp90 to TOM70 in a phosphorylation-dependent manner. (A) Workflow of the binding assay used to investigate the effects of pre-incubation with Orf9b variants on the binding of native TOM70111–608 interactors. (B) Influence of Orf9b binding to TOM70 on the recognition of Hsp90. Ni-NTA-bound TOM70111–608 (50 pmol) was pretreated either with a 10-fold molar excess of isolated Orf9bWT or with buffer for 20 min at 30 °C and washed three times before addition of reticulocyte lysate (RL) containing in vitro-synthesized human [35S]Hsp90. Samples were incubated for 40 min at 30 °C and subjected to SDS-PAGE. The arrow indicates a second translation product of Hsp90. Binding of Hsp90 to TOM70111–608 without the addition of Orf9b was set to 100% (control). Signals obtained after SDS-PAGE and autoradiographic detection were quantified using 5% of the total added lysate as internal control (“Load”). **** indicate p-values < 0.0001, error bars show SEM, n = 6. (C) Binding of [35S]Hsp90 to Ni-NTA-bound TOM70111–608, pretreated with either buffer, isolated Orf9bWT, or isolated Orf9bS53E was performed as described in (B). Signals obtained after SDS-PAGE and autoradiographic detection were quantified using 5% of the total added lysate as internal control (“Load”). Binding of [35S]Hsp90 without Orf9b pre-incubation was set to 100% (control). *** indicate p-values < 0.001, “ns” indicates no significance, error bars show SEM, n = 4. (D) In vitro binding of [35S]AAC3 to Ni-NTA-bound TOM70111–608 pre-incubated with or without Orf9bWT. The experiment was carried out as described in (B). Signals obtained after SDS-PAGE and autoradiographic detection were quantified using 10% of the total added lysate as internal control (“Load”). Binding of [35S]AAC3 without Orf9bWT pre-incubation was set to 100% (control). *** indicate p-values < 0.001, error bars show SEM, n = 3.
TOM70 is known to act as a receptor for newly synthesized mitochondrial ADP/ATP carriers and related proteins of the same protein family [11]. In the cytosol, these preproteins are bound by chaperone proteins and, thereby, retained in a soluble state [18,19]. We recently found that in the mitochondrial import of carrier proteins, TOM70 functions essentially as a co-chaperone [24]. To test for possible effects of Orf9b on this system, we pre-incubated TOM70 with Orf9bWT and subsequently added reticulocyte lysate containing radiolabeled human ADP/ATP carrier 3 (AAC3), a model protein for chaperone-mediated interaction with TOM70. We found that Orf9b binding to TOM70 drastically reduced the binding of AAC3 to TOM70 by 77%, indicating a general inhibition of chaperone-bound TOM70 substrates (Figure 3D).
2.3. The Chaperone Binding Site of TOM70 Promotes Association with Orf9b
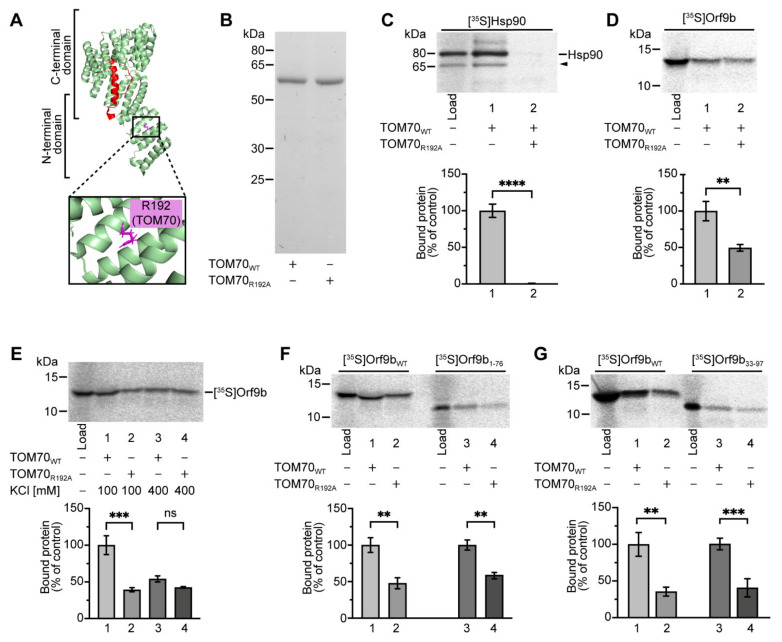
TOM70 contains a chaperone-binding site within its N-terminal part, including an important arginine residue in position 192 [31,32]. Considering the involvement of chaperones in the mitochondrial targeting of newly synthesized proteins, we reasoned that this chaperone-binding site may participate in the recognition of Orf9b. In a previous study, co-immunoprecipitation experiments were carried out with a TOM70 variant lacking the corresponding N-terminal TPR domain [13]. The results showed that Orf9b was still able to bind to this construct, suggesting a mechanism of direct access of Orf9b to the binding site in the C-terminal part of TOM70. However, using the cytosolic domain of TOM70 (residues 111–608), we found that upon disruption of this site (defined by residue E477) or by exchange of the corresponding residue within Orf9b (serine 53), a reduced but still significant association of both proteins was retained (Figure 1K).
To test if Orf9b may be a candidate for an attachment to the TOM70 chaperone binding site, we investigated the state of Orf9b in reticulocyte lysate. It is well established that mitochondrial precursor proteins translated in rabbit reticulocyte lysate form high molecular weight complexes of approximately 200–250 kDa that contain cellular chaperones [21,22]. We analyzed samples of reticulocyte lysate containing radiolabeled Orf9b by blue native polyacrylamide gel electrophoresis (BN-PAGE). Remarkably, two distinct states of the translation product were separated, with most of the Orf9b corresponding to a complex of approximately 220–240 kDa (Figure S3, labeled **) and smaller amounts being resolved at a lower molecular weight. Orf9bS53E showed slightly higher mobility in native gels compared to Orf9bWT, presumably because of the additional negative charge of Orf9bS53E (data not shown). A signal within the range of 200–250 kDa was not detected with the cytosolic protein DHFR, a small hydrophilic protein that is soluble independently of a permanent association with chaperone proteins (Figure S3, labeled **).
To investigate the possible participation of cytosolic chaperones in the formation of the Orf9b–TOM70 complex, we purified an established model protein, TOM70R192A, in which an arginine residue at position 192 within its N-terminal TPR domain is exchanged for an alanine to prevent the interaction of TOM70 with the C-terminal EEVD motif of Hsp70/Hsp90 chaperones (Figure 4A) [31]. We purified the variants TOM70WT and TOM70R192A using Ni-NTA beads (Figure 4B) and confirmed that the mutation R192A within TOM70 prevents the binding of Hsp90 in our assay (Figure 4C). In parallel, we also observed a reduction of binding, between 50% (Figure 4D) and 65% (Figure 4G), of Orf9b to TOM70R192A in comparison to TOM70WT. The results of these assays show that the chaperone binding site within the N-terminal domain of TOM70 is not essential for the recruitment of Orf9b; however, the binding of Orf9b is substantially facilitated.
Figure 4.
Orf9b has a second binding site in the N-terminal TPR domain of TOM70. (A) Overview of the N-terminal chaperone binding site of TOM70 defined by arginine R192. Structural data were obtained from PDB file 7KDT [16]. (B) Coomassie staining of TOM70WT and TOM70R192A purified using Ni-NTA agarose beads. (C) In vitro binding of [35S]Hsp90 to 50 pmol Ni-NTA-bound TOM70111–608 WT (TOM70WT) or TOM70111–608 R192A (TOM70R192A). Binding of [35S]Hsp90 to TOM70WT was set to 100% (control). **** indicate p-values < 0.0001, error bars show SEM, n = 7. (D) Binding of [35S]Orf9b to 250 pmol Ni-NTA-bound TOM70WT or TOM70R192A. Binding of [35S]Orf9b to TOM70WT was set to 100% (control). ** indicate p-value < 0.01, error bars show SEM, n = 5. (E) Binding of [35S]Orf9bWT to 250 pmol Ni-NTA-bound TOM70WT or TOM70R192A under high (400 mM KCl) or normal (100 mM KCl) salt conditions. Binding of [35S]Orf9bWT to TOM70WT in 100 mM KCl was set to 100% (control). *** indicate p-values < 0.001, “ns” indicates no significance, error bars show SEM, n = 3. (F) In vitro binding of [35S]Orf9bWT and [35S]Orf9b1–76 to 250 pmol Ni-NTA-bound TOM70WT or TOM70R192A. Binding of Orf9bWT or Orf9b1–76 to TOM70WT was set to 100% (control). ** indicate p-values < 0.01, error bars show SEM, n = 5. (G) In vitro binding of [35S]Orf9bWT and [35S]Orf9b33–97 to TOM70WT and TOM70R192A. Binding of Or9fbWT or Orf9b33–97 to TOM70WT was set to 100% (control). ** indicate p-values < 0.01; *** indicate p-values < 0.001, error bars show SEM, n = 9.
The binding of chaperones to the N-terminal domain of TOM70 is mediated by hydrophilic residues [31,32]. To verify the role of the chaperone binding site, we tested for the association of Orf9b with TOM70 under different salt concentrations. We found that a high salt concentration (400 mM KCl) reduced the binding of Orf9b by about 55% compared to normal salt levels (100 mM KCl), presumably by weakening the hydrophilic interactions in the N-terminus of TOM70 (Figure 4E, lane 1 vs. lane 2). In contrast, this effect was not observed using the variant TOM70R192A (lacking an intact chaperone binding site). Using this variant, the binding of Orf9b was not significantly reduced under high salt conditions (Figure 4E, lane 3 vs. lane 4). Additionally, we found that the core fragment of Orf9b (amino acids 33–76) was sufficient for chaperone-mediated binding as Orf9b variants lacking either the C-terminus (amino acids 77–97, Figure 4F) or the N-terminus (amino acids 1–32, Figure 4G) were still affected by the mutation of arginine 192 within the chaperone binding site of TOM70.
Our results indicate that interaction of Orf9b with TOM70 is mediated by two distinct binding sites: (i) a site within the C-terminal domain of TOM70 (defined by Orf9b S53 and TOM70 E477), and (ii) a second binding site within the N-terminal TPR domain of TOM70 (defined by TOM70 R192 and the EEVD motif of Orf9b-associated chaperones). While the C-terminal binding site is crucial for the interaction of Orf9b with TOM70, the N-terminal binding site appears to promote the binding of chaperone-associated Orf9b.
3. Discussion
3.1. The Mechanisms of Orf9b–TOM70 Interaction
Orf9b is a SARS-CoV-2-encoded accessory protein that can bind to a defined site within the bundle of helices that form the C-terminal part of the mitochondrial outer membrane protein TOM70 [15,16]. In this study, we investigated the process of complex formation and its functional implications.
We synthesized Orf9b in reticulocyte lysate and, thus, under conditions similar to the cytosol of a mammalian cell. Using BN-PAGE, we analyzed samples of the reticulocyte lysate and found that most of the Orf9b was a component of a high molecular weight complex of 220–240 kDa, corresponding to the size of newly synthesized chaperone-bound preproteins that are retained in a soluble state for subsequent import into mitochondria [21,22]. In mitochondrial protein import, TOM70 acts as a co-chaperone by binding chaperone-associated preproteins to facilitate the recognition of their targeting signals by the translocase of the outer membrane (TOM) complex [24]. In human TOM70, the interaction site for the recruitment of the chaperone-associated preproteins is defined by an arginine residue in position 192 within the N-terminal part of TOM70 [31]. In our experiments, we found that an exchange of this residue for alanine (R192A) reduced the efficiency of Orf9b binding to TOM70 by about 50%. Remarkably, this effect corresponds to similar values with regard to the contribution of TOM70 in the import of proteins into the inner mitochondrial compartments [24,33,34]. For efficient import of nuclear-encoded proteins into mitochondria, TOM70 is not essential; however, it substantially facilitates the targeting of its substrate proteins. Subsequent translocation across the outer membrane is mediated by the pore-forming protein TOM40 [11,18,19,35,36,37]. However, after release from the chaperone, Orf9b leaves the import pathway and is transferred to the binding site in the C-terminal part of TOM70. This final step can be inhibited by an exchange of the central glutamate E477 within this binding site. Under these conditions, the association of Orf9b with the modified TOM70 is drastically reduced by about 75%, but a substantial residual binding is still observed.
In summary, Orf9b initially follows a pattern that closely resembles the pathway of endogenous mitochondrial proteins that are imported from the cytosol. Orf9b diverges from this scheme after its initial interaction with the chaperone binding site at the arginine R192 of TOM70 to enter the binding cleft within its C-terminal domain. Orf9b is thus a protein with two binding sites within the same receptor protein.
The genome of the yeast Saccharomyces cerevisiae encodes two homologs of human TOM70 [11]. A comparison of the crystal structures of yeast Tom70 and its paralog Tom71 indicates that both receptors can exist in at least two distinct conformational states, an elongated “open” state and a “closed” state [11,38,39,40]. In yeast, the binding of the C-terminal EEVD motif of Hsp70 was suggested to induce the transition of Tom71 to its “open” state [39]. It is tempting to speculate that the binding of Orf9b to the N-terminal TPR domain of human TOM70, defined by the arginine at position 192, may trigger a switch to an “open” conformation of the receptor. This would allow for subsequent interaction of Orf9b with its binding site within the C-terminal domain of TOM70, defined by the glutamate E477. It is conceivable that Orf9b binding locks the receptor in its “open” state to prevent interaction with other Hsp90-bound proteins. In fact, we found that the binding of AAC3, a nuclear-encoded mitochondrial preprotein, to human TOM70 was drastically reduced under these conditions.
In parallel experiments, we also investigated the binding of isolated Orf9b to purified TOM70. Previous studies had suggested that Orf9b purified from E. coli is not able to associate with TOM70 in vitro [15,16]. Surprisingly, we found that the binding of isolated Orf9b to TOM70 is indeed possible when added in molar excess (1:10). Obviously, in the absence of chaperone proteins or additional factors, only a small fraction of the readily available Orf9b has the ability to bind to TOM70 in vitro. Notably, while purified dimeric Orf9b mainly forms β-strands, monomeric Orf9b displays an overall α-helical structure when in complex with TOM70 [15,16,27]. The ability of Orf9b to switch folds is also indicated by structure- and sequence-based predictions [41]. It is, therefore, tempting to speculate that a portion of Orf9b can undergo dynamic shifts between its dimeric state and its monomeric state.
3.2. Implications of Orf9b Binding to TOM70 for Health and Disease
SARS-CoV-2 VoC Alpha, also known as lineage B.1.1.7, is characterized by a higher transmissibility than preexisting virus variants [2,3,4]. Interestingly, recent data suggest a higher expression of Orf9b in cells of patients infected with the Alpha lineage and that this variant could lead to a higher number of hospitalizations and severe disease [5,6,9]. While it seems unlikely that an increased Orf9b expression plays a role in person-to-person transmission, it is conceivable that it represents a way to inhibit the innate immune response, which could provide a significant advantage over other virus variants. Previous studies have shown that expression of Orf9b in human cells leads to a reduced expression of IFN-I by the inhibition of a signaling cascade involving the proteins RIG-I, MAVS, TOM70, and Hsp90-bound TBK1 and IRF3 [13,14]. A compromised innate immune response has been suggested to be an important driver of COVID19, making the Orf9b–TOM70 interaction an attractive target for the development of therapeutic strategies [42]. In this regard, the mechanism of interference with the IFN-I inducing signaling cascade is of the utmost interest. Although it was concluded from experiments with short peptides derived from Hsp90 and Orf9b that the inhibition of Hsp90 binding to TOM70 could be a possible reason for the reduced IFN-I expression, the interplay of the three native proteins has not yet been investigated [15].
In this study, we found that the binding of Orf9b to TOM70 prevents the recognition of native Hsp90 and chaperone-associated TOM70 substrates. Due to technical limitations, we did not investigate the binding of a complex consisting of Hsp90/TBK1/IRF3; however, our data strongly suggest that inhibition of the interaction of Hsp90 with TOM70 represents the reason for the decreased IFN-I response in infected cells. Additionally, we could show that the substitution of the serine at position 53 by a phosphomimetic glutamate residue in Orf9b prevented its interaction with the C-terminal domain of TOM70 in vitro and in intact mammalian cells. During the preparation of this manuscript, a preprint that was published confirmed our finding that Orf9b binding is dependent on the state of its serine in position 53 [43]. In addition, the authors observed the suppression of host kinase activity in the early stages of infection, which likely represents a way of innate immune evasion for the virus.
Our data indicate that not only Hsp90 itself but also Hsp90-bound proteins are affected by the inhibitory effect of Orf9b, as suggested by the drastically reduced binding of in vitro-synthesized AAC3 to TOM70. The formation of the Orf9b–TOM70 complex may have additional implications for cellular processes beyond innate immunity, for example, in the recruitment of chaperone-bound preproteins [11,24,44]. In this context, it is interesting that peripheral blood mononuclear cells (PBMCs) from COVID19 patients seem to show metabolic alterations and reduced respiration that are compensated by a switch to glycolysis [45]. Therefore, the interaction of mitochondria with SARS-CoV-2-encoded proteins represents a promising avenue for further research with tremendous therapeutic potential.
The interaction of Orf9b with TOM70 will be an attractive target for the development of novel therapeutic strategies; however, it seems important to take into account the participation of two different binding sites in the formation of the Orf9b–TOM70 complex.
4. Materials and Methods
4.1. Plasmids
The plasmids used in this study are listed in Table 1. For the construction of the plasmids, DNA fragments were amplified by a standard PCR protocol or a protocol modified for FastCloning, as described by Li et al. (2011) [46], using Phusion DNA Polymerase (Thermo Scientific, Waltham, MA, USA). After restriction enzyme digestion, inserts were either ligated into vectors using T4 DNA Ligase (Thermo Scientific, Waltham, MA, USA) or mixed with DpnI-digested vector backbone prior to transformation in E. coli TOP10 (Thermo Scientific, Waltham, MA, USA).
Table 1.
Plasmids used in this study.
| Plasmid | Source |
|---|---|
| pProEx-HTA-TOM70(111–608) | Young et al., 2003; [31] |
| pProEx-HTA-TOM70(111–608)R192A | This study |
| pProEx-HTA-TOM70(111–608)E477A | This study |
| pCDNA3-Hsp90-HA | García-Cardeña et al., 1998; [47] |
| pYES2-Hsp90 | This study |
| pDONR334-SARS-CoV2 Orf9b | Kim et al., 2020; [48] |
| pProEx-His-DHFR-TEV-Orf9b | This study |
| pProEx-His-DHFR-TEV-Orf9bS53E | This study |
| pYES2-Orf9b | This study |
| pYES2-Orf9bS53E | This study |
| pYES2-Orf9bS53A | This study |
| pYES2-Orf9b1–76 | This study |
| pYES2-Orf9b33–97 | This study |
| pYES2-Orf9b-DHFR | This study |
| pYES2-Orf9bS53E-DHFR | This study |
| pYES2-Orf9bS53A-DHFR | This study |
| pYES2-AAC3 | This study |
| pYES2-DHFR | This study |
| pCAGGS-Orf9b-FLAG-EGFP | This study |
| pCAGGS-Orf9bS53E-FLAG-EGFP | This study |
4.2. Protein Purification
Orf9b was isolated from E. coli using the protocol published by Gordon et al. (2020) with minor modifications [16]. E. coli BL21 (DE3) cells were transformed with pProEx plasmids encoding for His-DHFR-TEV-Orf9b (His-DHFR fused to Orf9b via a TEV protease cleavage site) and tested for protein expression. Expression of His-DHFR-TEV-Orf9b was induced by the addition of 1 mM IPTG and carried out for 6 h at 37 °C. Cells were harvested at 6000× g for 5 min, frozen in appropriate aliquots, and stored at −70 °C until further use. After thawing, cells were resuspended in 10 mL/g lysis buffer (50 mM Tris/HCl; 500 mM NaCl; 10% (v/v) glycerol; 2 mM MgCl2; 2 mM PMSF; pH = 8.0) supplemented with 4.5 U/mL DNase I and protease inhibitors and incubated for 30 min at 4 °C. Complete lysis was obtained after four passages through an Emulsiflex C5 homogenizer (Avestin, Ottawa, ON, Canada). The lysate was cleared from cell debris by centrifugation at 47,500× g for 30 min. The supernatant was supplemented with 20 mM imidazole and loaded on two consecutive 1 mL Ni-NTA HisTrap High Performance columns (Sigma-Aldrich, St. Louis, MO, USA) equilibrated with lysis buffer using an ÄKTA start protein purification system (Cytiva, Marlborough, MA, USA). Columns were washed with 10 column volumes (cv) wash buffer (30 mM Tris/HCl; 150 mM KCl; 500 mM NaCl; 10% (v/v) glycerol; 0.2% Tween20; 2 mM ATP; 4 mM MgCl2; 40 mM imidazole; 1 mM DTT) followed by 5 cv buffer A (25 mM Tris/HCl, 50 mM KCl, 5% (v/v) glycerol; 1 mM DTT; pH = 8.5). Bound proteins were eluted with elution buffer (buffer A supplemented with 300 mM imidazole). The eluate was diluted with 4 volumes of buffer A, centrifuged for 10 min at 20,000× g and 4 °C. Proteins were further purified by ion-exchange chromatography (IEX) using a Resource Q 1 mL column (Cytiva, Marlborough, MA, USA) connected to an ÄKTA purifier system (Cytiva, Marlborough, MA, USA). Bound proteins were eluted with a gradient of buffer B (25 mM Tris/HCl; 1000 mM NaCl; 5% (v/v) glycerol; 1 mM DTT; pH = 8.5). Fractions containing the protein of interest were concentrated using an Amicon 10 kDa concentrator (Merck Millipore, Burlington, MA, USA) and washed three times with size exclusion chromatography (SEC) buffer (20 mM Tris/Cl, 150 mM NaCl, pH = 7.9). For removal of the His-DHFR tag, the solution was incubated with 0.6 mg/mL TEV protease overnight at 4 °C. The TEV protease was N-terminally fused to a His tag to allow for subsequent removal using Ni-NTA columns. After the addition of 10 mM imidazole, Orf9b was obtained from the TEV digestion mix as an unbound pass-through fraction using a 1 mL HisTrap High Performance column (Sigma-Aldrich, St. Louis, MO, USA) connected to an ÄKTA Start system, while the His-tagged TEV protease and the His-DHFR-TEV part of the fusion protein remained bound to the column bed. After concentration of the Orf9b fraction using an Amicon 3 kDa concentrator (Merck Millipore, Burlington, MA, USA), the protein solution was subjected to size exclusion chromatography (SEC) using a Superdex 75 increase 10/300 GL column (Cytiva, Marlborough, MA, USA). A typical retention volume for Orf9bWT and Orf9bS53E during SEC was 11.6 mL, which corresponds to an approximate molecular mass of 28.7 kDa by comparison with a protein standard curve. Eluted fractions containing the target protein were concentrated using an Amicon 3 kDa concentrator (Merck Millipore, Burlington, MA, USA), and the protein concentration was determined by Bradford assay using a bovine serum albumin standard (2 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) as a reference [49]. The solution containing purified Orf9b was divided into aliquots, frozen in liquid nitrogen, and stored at −70 °C until further use.
Human TOM70111–608 was purified as previously described, with minor modifications [24]. E. coli BL21 (DE3) cells were transformed with pProEx-HTA-TOM70111–608, and protein expression was induced at OD600 = 0.6 with 0.5 mM IPTG overnight at 16 °C. Cells were harvested and resuspended in lysis buffer (20 mM Tris/HCl; 500 mM NaCl; 10 mM imidazole; 2 mM PMSF; pH = 7.9) supplemented with 4.5 U/mL DNase I and protease inhibitors and incubated for 30 min at 4 °C. Cells were lysed by four passages through an Emulsiflex C5 homogenizer (Avestin, Ottawa, ON, Canada) and cleared from debris by centrifugation at 47,500× g for 30 min at 4 °C. After the addition of 10 mM imidazole, N-terminally His-tagged TOM70 was purified from the extract by affinity purification using Ni-NTA HisTrap High Performance columns (Sigma-Aldrich, St. Louis, MO, USA) and an ÄKTA Start protein purification system (Cytiva, Marlborough, MA, USA). Columns were washed with 10 cv wash buffer (20 mM Tris/HCl; 500 mM NaCl; 40 mM imidazole; pH = 7.9) and bound proteins were eluted with elution buffer (20 mM Tris/HCl; 500 mM NaCl; 300 mM imidazole; pH = 7.9). Protein-containing fractions were pooled and concentrated using an Amicon 30 kDa concentrator (Merck Millipore, Burlington, MA, USA) and washed three times with size exclusion chromatography (SEC) buffer (20 mM Tris/HCl; 150 mM NaCl; pH = 7.9). The protein was further purified using SEC with an ÄKTA purifier system equipped with a Superdex 200 increase 10/300 column (Cytiva, Marlborough, MA, USA). The protein concentration was determined by Bradford standard procedure using a bovine serum albumin standard (2 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) as reference. The purified protein was divided into aliquots, frozen in liquid nitrogen, and stored at −70 °C until further use.
For protein purification in small batches for in vitro binding experiments, we used a standard protocol published by Brix et al. with minor modifications [23]. Briefly, after protein expression, E. coli cells were harvested by centrifugation, resuspended in 10 mM MOPS buffer (pH = 7.2) and frozen in small aliquots until further use. For protein purification, bacteria were resuspended in binding buffer (20 mM Tris/HCl; 500 mM NaCl; 10 mM imidazole; 2 mM PMSF; pH = 7.9), and cells were disrupted by three rounds of 10 pulses using a Sonifier S-250A cell disruptor (Branson Ultrasonics, Brookfield, CT, USA) equipped with a micro tip. Insoluble fragments were sedimented by centrifugation at 20,000× g for 30 min at 4 °C. Protino Ni-NTA agarose beads (Macherey-Nagel, Düren, Germany) were added to the supernatant and incubated on a rotation mixer at 4 °C for one hour. Afterward, Ni-NTA beads were washed five times with a standard wash buffer (20 mM Tris/Cl; 150 mM NaCl; 40 mM imidazole; 2 mM PMSF; pH = 7.9). The amounts of protein bound to Ni-NTA were determined by SDS-PAGE and comparison to a BSA standard (2 mg/mL; Sigma-Aldrich, St. Louis, MO, USA), Coomassie stain, and densiometric analysis. Isolated proteins were kept on ice until further use.
4.3. In Vitro Translation of Radiolabeled Proteins
Radiolabeled proteins were synthesized by coupled transcription and translation in commercially available reticulocyte lysate (TNT Coupled Reticulocyte Lysate System, L4600; Promega, Madison, WI, USA) in the presence of [35S]-methionine (added from a stock solution of 10 μCi/μL, SCM01/37; Hartmann Analytic, Braunschweig, Germany), following the instructions of the manufacturer. The reaction was carried out for 120 min at 30 °C. Reticulocyte lysate containing the radiolabeled precursor protein was clarified by centrifugation (1 h; 20,000× g; 4 °C) before use. Small proteins (<15 kDa) were precipitated with a saturated ammonium sulfate solution by incubation on ice for 20 min, followed by centrifugation for 20 min at 20,000× g, and resuspended in untreated reticulocyte lysate (L4151; Promega, Madison, WI, USA).
4.4. In Vitro Binding Assay
To analyze the binding of proteins to the isolated cytosolic domain of human TOM70 (amino acids 111–608), we used an in vitro binding assay modified according to Brix et al. [23]. The purified cytosolic domain of TOM70 bound to Ni-NTA agarose beads was washed three times with assay buffer (10 mM MOPS/KOH; 20 mM imidazole; 100 mM KCl; 1% BSA (w/v); pH = 7.2) prior to use. A reaction mix containing 50–250 pmol of purified TOM70 per 100 µL assay buffer was added to Mobicol columns (M1002; MoBiTec, Göttingen, Germany), and the reaction was started by addition of 7% (v/v) reticulocyte lysate containing the [35S]-methionine-labeled protein. After incubation for 40 min at 30 °C while shaking (750 rpm), the column bed was washed three times with assay buffer without BSA (10 mM MOPS/KOH; 20 mM imidazole; 100 mM KCl; pH = 7.2). Proteins were eluted by the addition of 300 µL elution buffer (20 mM Tris/HCl; 1 M imidazole; 500 mM NaCl; pH = 7.9), incubation for 3 min at 30 °C, and subsequent centrifugation at 250× g for one minute. Eluates were precipitated and washed, following a standard trichloroacetic acid (TCA) precipitation protocol, and analyzed by SDS-PAGE. Individual binding experiments were carried out with separate samples in individual columns, and internal standard samples (“Load”) were included in each experiment. Quantification of bound protein was performed using the software Aida Image Analyzer V.4.19 (Elysia-raytest GmbH, Straubenhardt, Germany). Unspecific binding was analyzed by incubation of radiolabeled proteins with Ni-NTA beads that had been incubated with E. coli lysate prior to the assay, and unspecific binding values were subtracted from quantified binding values. The typical amounts of radiolabeled proteins bound to the immobilized TOM70 variants in these assays varied between 0.5% and 5%.
To investigate the influence of purified Orf9b on the binding of target proteins, Ni-NTA-bound TOM70 was washed three times with assay buffer containing BSA (10 mM MOPS/KOH; 20 mM imidazole; 100 mM KCl; 1% BSA (w/v); pH = 7.2) and was subsequently incubated with a 10-fold molar amount of Orf9bWT or Orf9bS53E. The beads were incubated at 30 °C while shaking (750 rpm) for 20 min and washed three times with assay buffer containing BSA (10 mM MOPS/KOH; 20 mM imidazole; 100 mM KCl; 1% BSA (w/v); pH = 7.2). Afterward, binding experiments using radiolabeled target proteins were performed as described above.
4.5. Trypsin Digestion
For trypsin digestion experiments, 5 µg/mL trypsin were added to either isolated proteins (5.5 µg/mL) or reticulocyte lysate containing radiolabeled proteins (20% (v/v) in a total volume of 100 µL). Samples were incubated in SEC buffer (20 mM Tris; 150 mM NaCl; pH = 7.9) and kept on ice during trypsin digestion. In the case of TOM70 pre-incubation, reticulocyte lysate containing radiolabeled proteins was incubated with isolated TOM70 at 25 °C for 10 min prior to the addition of the protease. The trypsin digestion was stopped by incubation at 95 °C for 5 min, and samples were subsequently prepared for SDS-PAGE.
4.6. Gel Electrophoresis
For SDS-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were denatured in SDS-sample buffer (50 mM Tris/HCl; 1% (w/v) SDS; 5% (v/v) 2-mercaptoehtanol; 0.01% (w/v) bromophenol blue; 10% (v/v) glycerol) by incubation for 5 min at 95 °C. Samples were loaded on polyacrylamide gels (12.5% (w/v) acrylamide; 0.33% (w/v) bis-acrylamide; 350 mM BisTris; pH = 6.5) and electrophoresis was carried out at 100 V for 10 min and 150 V for 30–50 min in either MOPS buffer (50 mM MOPS; 50 mM Tris/HCl; 1 mM EDTA; 0.1% (w/v) SDS; 5 mM Na2SO3; pH = 7.0) or MES buffer (50 mM MES; 50 mM Tris/HCl; 1 mM EDTA; 0.1% (w/v) SDS; 5 mM Na2SO3; pH = 7.0). Gels were either dried and incubated on image plates for read out by a Bas1800 II phosphorimager (Fujifilm Europe GmbH, Düsseldorf, Germany) and analyzed by Aida Image Analyzer V 4.19 (Elysia-raytest GmbH, Straubenhardt, Germany) or stained with colloidal Coomassie (5% (w/v) aluminum sulfate 14–18 hydrate; 5% phosphoric acid; 10% (v/v) ethanol; 0.02% (w/v) Coomassie Brilliant Blue G-250) for 1 h to overnight and destained for 2 h with destaining solution (2% (v/v) phosphoric acid; 10% (v/v) ethanol).
Blue native gel electrophoresis was performed essentially as described by Schägger and von Jagow (1991) [50], with minor modifications using freshly prepared 6–16.5% linear gradient gels. Briefly, a 6% gel solution (6% (w/v) acrylamide mixture; 50 mM BisTris; 500 mM 6-aminohexanoic acid) and a 16.5% gel solution (16.5% (w/v) acrylamide mixture; 50 mM BisTris; 500 mM 6-aminohexanoic acid; 20% (w/v) glycerol) were prepared from a 49.5% (w/v) acrylamide mixture (48% (w/v) acrylamide; 1.5% (w/v) bis-acrylamide) and a 3x gel buffer (150 mM BisTris; 1.5 M 6-aminohexanoic acid; pH = 7.0). Polymerization was initiated by addition of 0.5% (w/v) APS (ammonium persulfate) from a freshly prepared 10% (w/v) APS stock solution and 0.1% (v/v) TEMED (tetramethyl-etylenediamine). Using equal volumes of 6% and 16.5% gel solutions, gels were cast using a gradient mixer under constant flow. Freshly prepared reticulocyte lysate samples containing [35S]-labeled proteins were centrifuged at 20,000× g for 1 h and 4 °C. Afterward, samples were supplemented with digitonin buffer (20 mM Tris/Cl; 0.1 mM EDTA; 50 mM NaCl; 10% glycerol (w/v); 1% digitonin (w/v); pH = 7.0) and 1/10 vol. sample buffer (100 mM BisTris; 500 mM 6-aminohexanoic acid; 5% (w/v) Coomassie Brilliant Blue G-250; pH = 7.0) and loaded onto gels. Electrophoresis was performed at 100 V for 1 h and 200 V with the current limited to 15 mA for 2–3 h at 4 °C, using anode buffer (50 mM BisTris; pH = 7.0) and cathode buffer (50 mM Tricin; 15 mM BisTris; 0.02% (w/v) Coomassie Brilliant Blue G-250; pH = 7.0). Gels were dried and incubated with image plates for read out by a Bas1800 II phosphorimager (Fujifilm Europe GmbH, Düsseldorf, Germany) and analyzed by Aida Image Analyzer V 4.19 (Elysia-raytest GmbH, Straubenhardt, Germany).
4.7. Western Blot
Samples were analyzed after SDS-PAGE using Western blot following a standard protocol. After gel electrophoresis, the proteins were transferred to a nitrocellulose membrane (Amersham Protran 0.2 µM NC; GE Healthcare, Chicago, IL, USA) at 300 mA for 1 h in blotting buffer (10 mM NaHCO3; 3 mM Na2CO3; 20% (v/v) methanol; 0.01% (w/v) SDS; pH = 7.4). Membranes were dried, washed twice with TBS (10 mM Tris/Cl; 150 mM NaCl; pH = 7.5), and blocked with 5% (w/v) non-fat dry milk powder in TBS overnight at 4 °C under constant agitation. After two wash steps with TBS-TT (20 mM Tris/Cl; 500 mM NaCl; 0.05% (v/v) Tween-20; 0.2% (v/v) Triton X-100; pH = 7.5) and one subsequent wash step with TBS, membranes were incubated with a monoclonal mouse anti-His antibody (Invitrogen MA1-21315, 1:1000 in TBS; Thermo Scientific, Waltham, MA, USA) for one hour at room temperature and constant agitation. Wash steps were repeated, and the membrane was incubated with a fluorophore-conjugated goat anti-mouse antibody (IRDye 800CW, 1:15,000 in 10% (w/v) non-fat dry milk powder in TBS; LI-COR Biosciences, Lincoln, NE, USA) for one hour at room temperature and constant agitation. Membranes were scanned after four wash steps with TBS-TT, and signals were detected using an Odyssey Infrared Imaging System (LI-COR Bioscience, Lincoln, NE, USA).
4.8. Cell Culture
Vero E6 cells were maintained and cultured as a monolayer in complete Dulbecco’s modified Eagles medium (DMEM) at 37 °C, 95% humidity, and in a 5% CO2 atmosphere. Once cells reached 90% confluency, cells were detached from culture dishes with a 0.05% trypsin solution for 10 min at 37 °C, resuspended in appropriate media (prewarmed), and either used experimentally or re-seeded for further expansion in new culture dishes. Cells were exclusively seeded onto collagen-coated dishes.
4.9. Transfection and Immunofluorescence Microscopy
For transfection, Vero E6 cells were seeded at a density of 120,000 cells per well onto sterile coverslips embedded in a 24-well plate, incubated for 4 h, and subsequently transfected with 0.8 µg of pCAGGS vector containing the constructs of interest using Lipofectamine 2000 (100 µL Opti-MEM reduced serum media, 0.8 µg vector, 2.4 µg Lipofectamin 2000 per well). At 24 h post-transfection, cells were fixated (3% PFA), permeabilized (0.2% TritonX-100), blocked (5% horse serum), and stained with an anti-COXIV antibody (1:400; Cat. No. 4850S; Cell Signaling Technologies, Danvers, MA, USA). Cell nuclei were stained with DAPI. Before imaging, coverslips were mounted onto cover slides with 6 µL of Fluoromount. Images were acquired with a standard wide-field fluorescence microscope (BZ-X800E; Keyence, Osaka, Japan) in combination with a 60 × 1.40 numerical aperture (NA) oil-objective. The microscope is equipped with an 80 W metal halide lamp for the excitation of the DNA dye and a 2/3” monochrome CCD element (4080 × 3060 pixels per acquired image). DAPI (358 nm), green (488 nm), and red (568 nm) signals were acquired sequentially using BZ-X Filter DAPI, BZ-X Filter GFP, and BZ-X Filter TexasRed, respectively. Z-stack images were acquired with a 0.5 µm pitch, and full focus images were generated using BZ-X800 Analyzer.Ink (Keyence, Osaka, Japan) software.
4.10. Image Analysis
Gray value profiles in Figure 2 were analyzed using ImageJ software (Fiji) [51] and normalized to the smallest and largest values in each data set with GraphPad Prism version 9.1.2 for Windows (GraphPad Software, San Diego, CA, USA). Figures were generated using Illustrator CC 2021 (Adobe Inc., San Jose, CA, USA).
4.11. Statistical Analysis
For statistical analysis, the normality distribution of values was first tested by a Shapiro–Wilk test except for Figure 4F,G, where a Kolmogorov–Smirnov test was used. Normality distribution was confirmed for all tests. According to this result, two-tailed t-tests were performed for the comparison of two groups. Alternatively, one-way ANOVA was performed for statistical analysis of three or more groups. In Figure 1K and Figure 3C, one-way ANOVA was performed by Dunnett’s multiple comparisons test, with control group as indicated. In Figure 4E, one-way ANOVA was performed by Šídák’s multiple comparisons test to compare data sets in the two groups. All bar graphs contain error bars that display the standard error of means (SEMs). Statistical analysis was performed using GraphPad Prism 9 version 9.1.2 (GraphPad Software, San Diego, CA, USA).
Acknowledgments
We would like to thank Patricia Bartschies for her energetic support in construction of plasmids and isolation of proteins. We thank our colleagues of the department of Systems Biochemistry for support in protein isolation. We thank F. Ulrich Hartl (Max Planck Institute of Biochemistry, Martinsried) for kindly sharing the plasmid-encoding human TOM70. We thank Verian Bader (Ruhr University, Bochum) for the helpful discussion and support in statistical analysis. We would further like to thank Eike Steinmann (Ruhr University, Bochum) and Tim Rahmel (Universitätsklinikum Knappschaftskrankenhaus Bochum) for the helpful discussion. We acknowledge support by the Open Access Publication Funds of the Ruhr University Bochum.
Abbreviations
| [35S] | Radiolabeling by 35S-methionine residues |
| AAC3 | Human ADP/ATP carrier 3 |
| BN-PAGE | Blue native polyacrylamide gel electrophoresis |
| Co-IP | Co-immunoprecipitation |
| COVID19 | Coronavirus disease 2019 |
| COXIV | Subunit of the cytochrome c oxidase |
| Cryo-EM | Cryogenic electron microscopy |
| DAPI | 4′,6-diamidino-2-phenylindole |
| DHFR | Mouse dihydrofolate reductase |
| EGFP | Enhanced green fluorescent protein |
| Hsp90 | Human heat shock protein of 90 kDa |
| IEX | Ion exchange chromatography |
| IFN-I | Interferon type I |
| IMAC | Immobilized metal affinity chromatography |
| IRF3 | Interferon regulatory factor 3 |
| ITC | Isothermal titration calorimetry |
| kDa | Kilo Dalton |
| MAVS | Mitochondrial antiviral-signaling protein |
| Ni-NTA | Nickel-nitrilotriacetic acid |
| Orf9b | Protein encoded by the open reading frame 9b of SARS-CoV-2 |
| PBMC | Peripheral blood mononuclear cells |
| RIG-I | Retinoic acid-inducible gene I |
| RL | Rabbit reticulocyte lysate |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SDS-PAGE | Sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SEC | Size exclusion chromatography |
| SEM | Standard error of the means |
| TBK1 | TANK-binding kinase 1 |
| TEV | Tobacco etch virus nuclear-inclusion-a endopeptidase |
| TOM | Translocase of the outer mitochondrial membrane |
| TOM70 | Human TOM protein of 70 kDa |
| Tom70 | Yeast TOM Protein of 70 kDa |
| Tom71 | Yeast paralogue of Tom70 |
| TPR | Tetratricopeptide repeat |
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms22179233/s1.
Author Contributions
Designed the study, S.K. and J.R.; performed and analyzed experiments, L.B., A.M.K., and M.K.; analyzed data and provided significant intellectual contribution, Y.B. and S.P.; wrote the manuscript, S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a FoRUM grant from the Medical Faculty of the Ruhr University Bochum (F1014-2021).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available upon reasonable request from the corresponding author.
Conflicts of Interest
The authors of this study declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Toole Á.O., Hill V., Pybus O.G., Watts A., Bogoch I.I., Khan K., Messina J.P., The COVID-19 Genomics UK (COG-UK) Consortium. Network for Genomic Surveillance in South Africa (NGS-SA) Brazil-UK CADDE Genomic Network et al. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2 [version 1; peer review: 3 approved] Wellcome Open Res. 2021;6:1–14. doi: 10.12688/wellcomeopenres.16661.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372:3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davies N.G., Jarvis C.I., CMMID COVID-19 Working Group. Edmunds W.J., Jewell N.P., Diaz-Ordaz K., Keogh R.H. Increased mortality in community-tested cases of SARS-CoV-2 lineage B.1.1.7. Nature. 2021;593:270–274. doi: 10.1038/s41586-021-03426-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., Toole Á.O., et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature. 2021;593:266–269. doi: 10.1038/s41586-021-03470-x. [DOI] [PubMed] [Google Scholar]
- 5.Funk T., Pharris A., Spiteri G., Bundle N., Melidou A., Carr M., Gonzalez G., Garcia-Leon A., Crispie F., Connor L.O., et al. Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: Data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Eurosurveillance. 2021;26:1–10. doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Horby A.P., Bell I., Breuer J., Cevik M., Challen R., Davies N., Dabrera G., Edmunds J., Ferguson N., Funk S., et al. Update Note on B.1.1.7 Severity. [(accessed on 1 July 2021)];2021 Available online: https://www.gov.uk/government/publications/nervtag-update-note-on-b117-severity-11-february-2021.
- 7.Yang J., Zhang G., Yu D., Cao R., Wu X., Ling Y., Pan Y.-H., Yi C., Sun X., Sun B., et al. Evolutionary insights into a non-coding deletion of SARS-CoV-2 B.1.1.7. bioRxiv. 2021 doi: 10.1101/2021.04.30.442029. [DOI] [Google Scholar]
- 8.Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E., et al. Preliminary Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in the UK Defined by a Novel Set of Spike Mutations. [(accessed on 1 July 2021)];2020 Available online: https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563.
- 9.Parker M.D., Lindsey B.B., Shah D.R., Hsu S., Keeley A.J., Partridge D.G., Leary S., Cope A., State A., Johnson K., et al. Altered Subgenomic RNA Expression in SARS-CoV-2 B.1.1.7 Infections. bioRxiv. 2021 doi: 10.1101/2021.03.02.433156. [DOI] [Google Scholar]
- 10.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kreimendahl S., Rassow J. The mitochondrial outer membrane protein Tom70—Mediator in protein traffic, membrane contact sites and innate immunity. Int. J. Mol. Sci. 2020;21:7262. doi: 10.3390/ijms21197262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X.Y., Wei B., Shi H.X., Shan Y.F., Wang C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010;20:994–1011. doi: 10.1038/cr.2010.103. [DOI] [PubMed] [Google Scholar]
- 13.Jiang H., Zhang H., Meng Q., Xie J., Li Y., Chen H., Zheng Y., Wang X., Qi H., Zhang J., et al. SARS-CoV-2 Orf9b suppresses type I interferon responses by targeting TOM70. Cell. Mol. Immunol. 2020;17:998–1000. doi: 10.1038/s41423-020-0514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han L., Wei M., Jian Z., Yi D., Jing Z., Ling M., Xue N., Zhang J., Gao C., Wang P.H. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5–MAVS, TLR3–TRIF, and cGAS–STING signaling pathways. J. Med. Virol. 2021;93:1–14. doi: 10.1002/jmv.27050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X., Zhu K., Qin B., Olieric V., Wang M., Cui S. Crystal structure of SARS-CoV-2 Orf9b in complex with human TOM70 suggests unusual virus-host interactions. Nat. Commun. 2021;12:1–9. doi: 10.1038/s41467-021-23118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon D.E., Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;9403:1–38. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182:685–712. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bykov Y.S., Rapaport D., Herrmann J.M., Schuldiner M. Cytosolic Events in the Biogenesis of Mitochondrial Proteins. Trends Biochem. Sci. 2020;45:650–667. doi: 10.1016/j.tibs.2020.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Becker T., Song J., Pfanner N. Versatility of Preprotein Transfer from the Cytosol to Mitochondria. Trends Cell Biol. 2019;29:534–548. doi: 10.1016/j.tcb.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 20.Kim Y.E., Hipp M.S., Bracher A., Hayer-Hartl M., Hartl U.F. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 21.Sheffield W.P. Mitochondrial precursor protein. Effects of 70-kilodalton heat shock protein on polypeptide folding, aggregation, and import competence. J. Biol. Chem. 1990;265:11069–11076. doi: 10.1016/S0021-9258(19)38558-8. [DOI] [PubMed] [Google Scholar]
- 22.Dekker P.J.T., Müller H., Rassow J., Pfanner N. Characterization of the preprotein translocase of the outer mitochondrial membrane by blue native electrophoresis. Biol. Chem. 1996;377:535–538. doi: 10.1515/bchm3.1996.377.7-8.535. [DOI] [PubMed] [Google Scholar]
- 23.Brix J., Dietmeier K., Pfanner N. Differential Recognition of Preproteins by the Purified Cytosolic Domains of the Mitochondrial Import Receptors Tom20, Tom22, and Tom70. J. Biol. Chem. 1997;272:20730–20735. doi: 10.1074/jbc.272.33.20730. [DOI] [PubMed] [Google Scholar]
- 24.Kreimendahl S., Schwichtenberg J., Günnewig K., Brandherm L., Rassow J. The selectivity filter of the mitochondrial protein import machinery. BMC Biol. 2020;18:1–23. doi: 10.1186/s12915-020-00888-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiedemann N., Pfanner N., Ryan M.T. The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. EMBO J. 2001;20:951–960. doi: 10.1093/emboj/20.5.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eilers M., Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 1986;322:1–5. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- 27.Weeks S.D., De Graef S., Munawar A. X-ray Crystallographic Structure of Orf9b from SARS-CoV-2. RCSB PDB. 2020;12:1–9. doi: 10.2210/pdb6z4u/pdb. [DOI] [Google Scholar]
- 28.Schopf F.H., Biebl M.M., Buchner J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017;18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 29.Dasso M.C., Jackson R.J. On the fidelity of mRNA translation in the nuclease-treated rabbit reticulocyte lysate system. Nucleic Acids Res. 1989;17:3129–3144. doi: 10.1093/nar/17.8.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perez-Riba A., Itzhaki L.S. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr. Opin. Struct. Biol. 2019;54:43–49. doi: 10.1016/j.sbi.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Young J.C., Hoogenraad N.J., Hartl F.U. Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70. Cell. 2003;112:41–50. doi: 10.1016/S0092-8674(02)01250-3. [DOI] [PubMed] [Google Scholar]
- 32.Fan A.C.Y., Young J.C. Function of Cytosolic Chaperones in Tom70-Mediated Mitochondrial Import. Protein Pept. Lett. 2011;18:122–131. doi: 10.2174/092986611794475020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hines V., Brandt A., Griffiths G., Horstmann H., Brütsch H., Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto H., Fukui K., Takahashi H., Kitamura S., Shiota T., Terao K., Uchida M., Esaki M., Nishikawa S.I., Yoshihisa T., et al. Roles of Tom70 in import of presequence-containing mitochondrial proteins. J. Biol. Chem. 2009;284:31635–31646. doi: 10.1074/jbc.M109.041756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drwesh L., Rapaport D. Biogenesis pathways of α-helical mitochondrial outer membrane proteins. Biol. Chem. 2020;401:677–686. doi: 10.1515/hsz-2019-0440. [DOI] [PubMed] [Google Scholar]
- 36.Wiedemann N., Pfanner N. Mitochondrial Machineries for Protein Import and Assembly. Annu. Rev. Biochem. 2017;86:685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 37.Rampelt H., Sucec I., Bersch B., Horten P., Perschil I., Martinou J.C., Van Der Laan M., Wiedemann N., Schanda P., Pfanner N. The mitochondrial carrier pathway transports non-canonical substrates with an odd number of transmembrane segments. BMC Biol. 2020;18:1–14. doi: 10.1186/s12915-019-0733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu Y., Sha B. Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 2006;13:589–593. doi: 10.1038/nsmb1106. [DOI] [PubMed] [Google Scholar]
- 39.Li J., Qian X., Hu J., Sha B. Molecular Chaperone Hsp70/Hsp90 Prepares the Mitochondrial Outer Membrane Translocon Receptor Tom71 for Preprotein Loading. J. Biol. Chem. 2009;284:23852–23859. doi: 10.1074/jbc.M109.023986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J., Cui W., Sha B. The structural plasticity of Tom71 for mitochondrial precursor translocations. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:985–989. doi: 10.1107/S1744309110025522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter L.L. Predictable fold switching by the SARS-CoV-2 protein ORF9b. Protein Sci. 2021;2:1–7. doi: 10.1002/pro.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorne L.G., Bouhaddou M., Reuschl A.-K., Zuliani-Alvarez L., Polacco B., Pelin A., Batra J., Whelan M.V.X., Ummadi M., Rojc A., et al. Evolution of enhanced innate immune evasion by the SARS-CoV-2 B.1.1.7 UK variant. bioRxiv. 2021:1–36. doi: 10.1101/2021.06.06.446826. [DOI] [Google Scholar]
- 44.Backes S., Bykov Y.S., Flohr T., Räschle M., Zhou J., Lenhard S., Krämer L., Mühlhaus T., Bibi C., Jann C., et al. The chaperone-binding activity of the mitochondrial surface receptor Tom70 protects the cytosol against mitoprotein-induced stress. Cell Rep. 2021;35:108936. doi: 10.1016/j.celrep.2021.108936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ajaz S., McPhail M.J., Singh K.K., Mujib S., Trovato F.M., Napoli S., Agarwal K. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am. J. Physiol. Cell Physiol. 2021;320:C57–C65. doi: 10.1152/ajpcell.00426.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li C., Wen A., Shen B., Lu J., Huang Y., Chang Y. FastCloning: A highly simplified, purification-free, sequence- and ligation-independent PCR cloning method. BMC Biotechnol. 2011;11:1–10. doi: 10.1186/1472-6750-11-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.García-Cardeña G., Fan R., Shah V., Sorrentino R., Cirino G., Papapetropoulos A., Sessa W.C. Dynamic activation of endothelial nitric oxide synthase by Hsp90. Nature. 1998;392:821–824. doi: 10.1038/33934. [DOI] [PubMed] [Google Scholar]
- 48.Kim D.K., Knapp J.J., Kuang D., Chawla A., Cassonnet P., Lee H., Sheykhkarimli D., Samavarchi-Tehrani P., Abdouni H., Rayhan A., et al. A comprehensive, flexible collection of SARS-CoV-2 coding regions. G3 Genes Genomes Genet. 2020;10:3399–3402. doi: 10.1534/g3.120.401554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 50.Schägger H., von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-A. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study are available upon reasonable request from the corresponding author.