Abstract
We used the psoralen gel retardation assay and Northern blot analysis in an in vivo yeast system to analyze effects of rDNA enhancer deletions on the chromatin structure and the transcription of tagged rDNA units. We found that upon deletion of a single enhancer element, transcription of the upstream and downstream rRNA gene was reduced by about 50%. Although removing both flanking enhancers of an rRNA gene led to a further reduction in transcription levels, a significant amount of transcriptional activity remained, either resulting from the influence of more distantly located enhancer elements or reflecting the basal activity of the polymerase I promoter within the nucleolus. Despite the reduction of transcriptional activity upon enhancer deletion, the activation frequency (proportion of nonnucleosomal to nucleosomal gene copies in a given cell culture) of the tagged rRNA genes was not significantly altered, as determined by the psoralen gel retardation assay. This is a strong indication that, within the nucleolus, the yeast rDNA enhancer functions by increasing transcription rates of active rRNA genes and not by activating silent transcription units.
Ribosome biosynthesis, the process entailing rRNA gene transcription, transcript processing, synthesis of ribosomal proteins, and assembly of riboprotein subunits, has been shown to be a tightly regulated process which adapts rapidly to changes in environmental conditions (12). A central process of this adaptation is the regulation of transcription of the rRNA genes. In most eukaryotic organisms, these are organized in clustered tandem arrays at one or several chromosomal sites (17). In the yeast Saccharomyces cerevisiae, about 100 to 200 copies of the rRNA gene are localized in one large cluster on chromosome XII (35). Transcription of the rRNA genes by RNA polymerase I (pol I) in the nucleolus yields the large 35S precursor rRNA, which is subsequently processed into the 17S, 5.8S, and 25S mature rRNAs. However, not all rRNA gene copies are active at a given time. Instead, only a subset is free of nucleosomes and actively transcribed, while the inactive fraction was found to be packaged into nucleosomes (5).
Adjacent rRNA transcription units in yeast are separated by an intergenic spacer region, which contains, in addition to the small pol III transcribed 5S gene (25), several regulatory elements, such as the rRNA gene promoter (23), an autonomously replicating sequence element (31), and a region that exhibits all attributes of a transcriptional pol I enhancer (7, 8). This pol I enhancer, located at the very 3′ end of the rRNA coding sequence, has been shown to increase the transcription level of a pol I gene in in vitro experiments as well as on episomal plasmids by 10- to 50-fold (7, 8, 10, 28). An in vivo study performed within the rDNA locus reported a lower stimulatory effect of about twofold (14). Although the various results presented so far are somewhat inconsistent as to which genes are affected by a single enhancer element, more recent data suggest that its enhancing effect is confined to its two upstream and downstream flanking rRNA genes and is rather equally distributed between them (14).
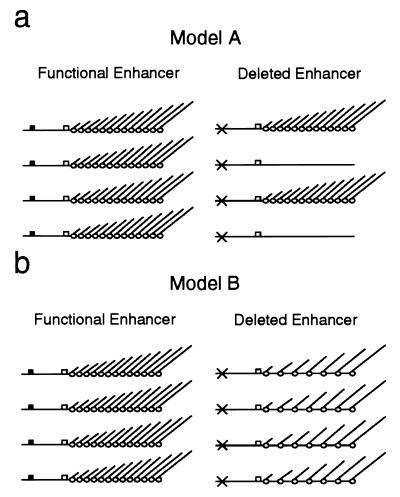
The mechanism by which the enhancer exerts its function is still controversal today. Principally, two models of enhancer action have been discussed in the literature (26) (Fig. 1). The first proposes a function of the enhancer in the first steps of transcription initiation, namely, in helping the formation of a stable promoter complex, and assumes that, once this complex has been formed, the enhancers are dispensable (28). Support for this model comes mainly from in vitro Sarkosyl experiments, in which only a single round of transcription initiation is believed to occur (11). The fact that enhancers still increased the amount of transcripts under these conditions was interpreted as meaning that a higher number of templates must have been transcribed. The recent report about an electron microscopic analysis of episomal rDNA genes injected into Xenopus oocytes indicates that metazoan rDNA enhancers may indeed function by such a mechanism (24). An alternative model proposes that the ribosomal enhancer acts by elevating the rate by which the rRNA genes are transcribed, resulting in an increased transcriptional activity of already active genes rather than in more genes being activated (21). Whereas some researchers favor this model for the yeast rDNA enhancer (10, 14), data against it have also been presented (3).
FIG. 1.
Models of rDNA enhancer action. (a) An enhancer could work by increasing the chance that an adjacent promoter is activated for transcription. In this case, deletion of the enhancer element would result in fewer genes being transcribed. (b) Alternatively, the enhancer might raise the polymerase initiation rate of active genes. Deletion of the enhancer would thus result in the same number of active genes transcribed by fewer polymerases. Black boxes denote enhancers, white boxes denote promoters, and crosses indicate deleted enhancer elements. Polymerases with nascent transcripts are depicted as empty circles.
In an earlier work, we developed a method to determine the chromatin structure of rRNA genes (4, 5). With this method, we were able to show that active rRNA gene copies are rather equally distributed within the yeast rDNA locus (6). We furthermore found that nucleosomefree (active) genes are always followed downstream by nucleosomefree enhancer elements and vice versa. While the open chromatin structure of the enhancers was interpreted as being the result of specific protein-DNA interactions, it remained unclear whether these interactions are involved in the activation process of the upstream gene.
In this study, we investigated whether the rDNA enhancer participates in the activation process of the rRNA genes. We therefore used an in vivo pol I system to analyze the effects of enhancer deletions on the nucleosomal packaging and the transcription level of the rRNA genes within their natural chromosomal context. We found no evidence that the enhancer is involved in altering the activation frequency of its adjacent gene promoters. The results of our study rather indicate that the pol I enhancer functions, at least in part, by increasing the rate of reinitiation on already-active promoter elements.
MATERIALS AND METHODS
Strains, media, and plasmids.
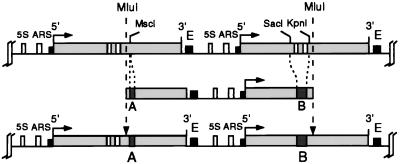
S. cerevisiae SC3 (MATa ura3-52 his3-1 trp1 gal2 gal10) (29) and derivatives constructed in this study (YMB1-1 to YMB3-2) were used for all analyses. All experiments were done with complex medium (yeast-peptone-dextrose [YPD]). In order to construct plasmids pMB119, pMB123, and pMB128, which were used to generate yeast strains with tagged rDNA genes, several intermediate constructs were generated (see simplified outline in Fig. 2). Plasmid pMB104r was produced by cutting S. cerevisiae partially purified rDNA (cesium chloride-actinomycin D) with MluI and cloning the 9.1-kb fragment into a pUC18 derivative whose polylinker (EcoRI-NdeI) had been replaced by an MluI linker. Tag sequences A and B were subcloned by inserting 227- and 479-bp fragments, respectively, from a StuI digest of simian virus 40 (SV40) into the SmaI site of pUC18, yielding plasmids pMB111I and pMB114I. Both tags were then excised from these plasmids by an EcoRI-BamHI (pMB111I) or an EcoRV-BamHI (pMB114I) double digest, and overhanging ends were filled in by the Klenow fragment. The resulting fragments were cloned into pMB104r, with tag sequence A cloned by insertion into the MscI-site within the 25S coding sequence and tag sequence B cloned by replacing the sequence between the KpnI and SacI sites encompassing internal transcribed sequences 1 and 2, the 5.8S sequence, and the corresponding processing sites, yielding plasmid pMB119. For generation of the enhancer deletion mutant, plasmid pMB123 was constructed by subcloning the rDNA intergenic spacer (region NarI-SmaI) from pMB119 into pUC18, removing a BfrI-HpaI fragment containing the whole 317-bp enhancer element, and reinserting the modified spacer sequence into the original plasmid. Plasmid pMB129, which carries the enhancer-promoter double deletion, was obtained by removing a 735-bp EcoRV fragment encompassing the pol I promoter and about 400 bp of the 5′ ETS of the downstream rRNA gene from plasmid pMB123.
FIG. 2.
Construction of yeast strains carrying tagged rRNA transcription units (YMB1-1 to YMB3-2). A 9.1-kb MluI-MluI fragment from the rDNA locus of S. cerevisiae SC3 was cloned into a pUC18-derived plasmid. Two short tag sequences were inserted at the sites indicated, and the resulting construct was reintegrated into the rDNA locus by homologous recombination (for details, see Materials and Methods).
Yeast cotransformation and selection of transformants.
Yeast transformation was done essentially as described by Becker and Guarente (1a), with S. cerevisiae SC3; 1 μg of MluI fragments from pMB119, pMB123, or pMB128, respectively; and 0.1 μg of pRS316 for complementation of Ura3 deficiency (30). Colonies grown on uracil-lacking medium were subjected to PCR and Southern blot analysis. For PCR analysis according to the method of Ling and coworkers (16), primers that anneal within the tag sequences and within endogenous ribosomal sequences outside the expected MluI integration sites were generated. Thus, only fragments that have integrated into the rDNA locus are amplified in this PCR. Furthermore, multiple integrations leading to double-tagged rRNA genes (gene 3 in YMB1-2, YMB2-2, and YMB3-2 [see Fig. 4a]) were identified by the use of primers annealing within both tag sequences. The number of fragments inserted was determined in Southern blot analysis by comparing the signals derived from the tags with one from the single-copy Trp1 gene by using a probe that consists of one of the tag sequences and a fragment of the Trp1 gene. The consistency of the introduced rRNA genes was confirmed by restriction fragment length analysis using various restriction enzymes (for further details, see reference 1).
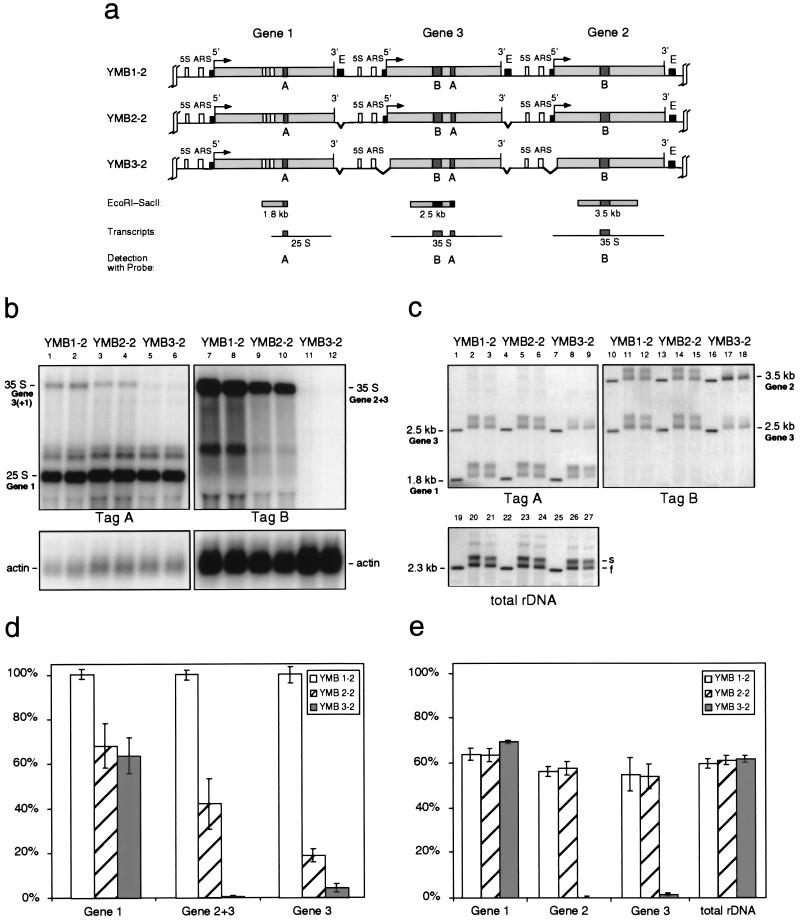
FIG. 4.
Analysis of double-integration clones. (a) Map showing the three tagged rRNA transcription units (gene 1 to 3) obtained by a double-integration event of the constructs into the rDNA locus, yielding yeast variants YMB1-2 to YMB3-2. See the legend to Fig. 3a for an explanation of abbreviations. (b) Northern blot analysis of the RNA levels of the tagged genes. Results from two independent clones of each yeast strain are shown. (c) Psoralen gel retardation analysis of the indicated restriction fragments. Two independent clones of each yeast strain are shown. (d) Statistical analysis of the data obtained from three Northern blot assays. All signals were corrected for loading by using the actin band as a standard. Signal strengths from strain YMB1-2 were defined as 100%. Note that tag B yields a composite signal derived from genes 2 and 3. (e) Statistical analysis of the percentage of slow-migrating bands (s bands, representing the nonnucleosomal, active gene fraction) from data obtained from three psoralen gel retardation assays. (d and e) Error bars, standard errors of the means.
Psoralen cross-linking.
Yeast cells were grown at 30°C in YPD to a density of 1 · 107 to 2 · 107 cells per ml. About 109 cells were spun down, washed twice with ice-cold water, resuspended in 1.6 ml of nuclear indicator buffer, broken as described by Wu and Gilbert (36), and then irradiated in the presence of 4,5′,8-trimethylpsoralen with a 366-nm UV lamp (model B-100A; Ultra Violet Products, Inc., San Gabriel, Calif.) at a distance of 6 cm essentially as described previously (5). A 0.05 volume of psoralen stock solution in ethanol (200 μg/ml) was added four times at intervals of 5 min, for a total irradiation time of 20 min. After washing the irradiated cells with 1 M sorbitol, DNA purification was continued as described previously (36).
DNA and RNA isolation, gel electrophoresis, transfer, hybridization, and quantification.
Genomic DNA was isolated according to the protocol of Wu and Gilbert (36), and total RNA was isolated according to the protocol of Kormanec and Farkasovsky (13). Electrophoretic separations of DNA fragments were done in 1.2% agarose gels at 1.9 V/cm for 18 h, and RNA separations were done in 0.8% formaldehyde gels at 1.7 V/cm for 20 h. Alkaline Southern blotting and hybridizations were performed as described elsewhere (18). RNA was blotted and hybridized according to standard protocols (27). Radioactive bands were detected by using Fuji X-ray films. All films were exposed without amplifier screens. Signals on Southern and Northern blots were quantitated by using a Molecular Dynamics PhosphorImager (176-μm pixel size). For quantification of rRNA transcripts, all signal intensities were corrected for loading by using the actin signal as a standard. Correction for transfer efficiency and hybridization, as necessary for comparing signals from different hybridizations, was achieved by using a DNA mix containing equimolar amounts of tagged fragments with a size similar to the rRNA transcripts as a standard. Signal distributions from psoralen gel retardation experiments were analyzed by peak deconvolution.
RESULTS
Generation of yeast strains carrying tagged rRNA transcription units.
In order to determine the effects of deletions of the rDNA enhancer element on the transcriptional activity and the chromatin structure of adjacent rRNA genes within their normal chromosomal context, we constructed yeast strains carrying tagged rRNA genes in the rDNA locus. For that purpose, we cloned a 9.1-kb rDNA repeat starting at a unique MluI site in the 25S sequence of a transcription unit and extending to the corresponding site in the next transcription unit (Fig. 2). Two small sequence tags (A and B) derived from SV40, which had been checked before to exclude cross-hybridization with endogenous S. cerevisiae sequences, were inserted at the MscI site and between the KpnI and SacI sites, respectively (for details, see Materials and Methods). Transformation of yeast strain SC3 with a linear MluI-MluI fragment excised from the plasmid resulted in reintegration of the fragment into the rDNA locus by homologous recombination and generation of two contiguous tagged rRNA transcription units. This allowed us to simultaneously monitor transcription and chromatin structure of two pol I transcription units flanking a defined enhancer element.
To determine the number of copies integrated and to exclude aberrant transcription units derived from abnormal integration events, we analyzed DNA restriction fragments from the transformants by using a hybrid probe visualizing simultaneously either of both tag sequences and the Trp1 gene (data not shown). Using the signal from the single copy Trp1 gene as a standard, we identified clones containing either two tagged rRNA transcription units (single integration; Fig. 2 and strain YMB1-1 in Fig. 3a) or three tagged units (double integration; strain YMB1-2 in Fig. 4a). All tagged units can be identified in Southern and Northern blot analyses according to their different restriction fragment and transcript sizes. Since recombination events in the chromosomal rDNA locus may occur occasionally, we verified the integrity of all constructs by restriction digest analysis in parallel to all experiments described below (data not shown).
FIG. 3.
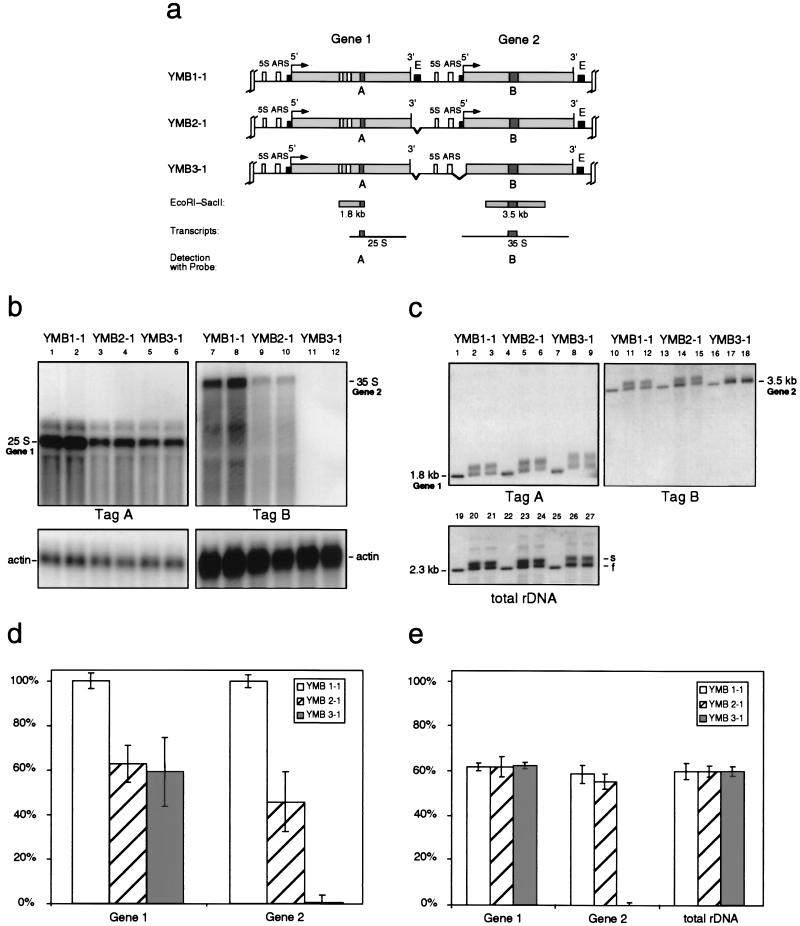
Analysis of single-integration clones. (a) Map showing the two tagged rRNA transcription units (genes 1 and 2) obtained by a single-integration event of the constructs into the rDNA locus, yielding yeast variants YMB1-1 to YMB3-1. The transcription initiation site (5′) and the 3′ end of the 35S genes are indicated. The small filled boxes near the 5′ and 3′ ends correspond to the promoter and enhancer (E) elements, respectively. The 5S genes and the autonomous replicating sequences (ARS) located in the intergenic spacers are also shown. Restriction fragments containing the SV40 sequence tags A and B are shown as well as the expected pol I transcripts. (b) Northern blot analysis of the RNA levels of the tagged genes. RNA was extracted, purified, and separated on a 0.8% formaldehyde gel. The gel was blotted and hybridized against the sequence tags A or B as indicated. The blots were then stripped and hybridized to an actin probe as a loading control. The results from two independent clones of each yeast strain are shown. (c) Psoralen gel retardation analysis of the indicated restriction fragments. Yeast cells growing exponentially in complex medium were photoreacted with psoralen. The purified DNA (extracted simultaneously with the RNA preparation for the Northern blot analysis) was digested with EcoRI and SacII, separated on a 1.2% agarose gel, and visualized after blotting by hybridization with the sequence tags A or B as indicated. The blots were then stripped and hybridized as a control to a probe complementary to the 25S gene (total rDNA) (for details, see the work of Dammann et al. [5]). The results from two independent clones of each yeast strain are shown. (d) Statistical analysis of the data obtained from three Northern blot assays. All signals were corrected for loading by using the actin band as a standard. Signal strengths from strain YMB1-1 were defined as 100%. (e) Statistical analysis of the percentage of low-migrating bands (s bands, representing the nonnucleosomal, active gene fraction) from data obtained from three psoralen gel retardation assays. (d and e) Error bars, standard errors of the means.
We first analyzed the transcripts arising from the tagged genes. As shown in Fig. 3a and 4a, only the first gene in the constructs (gene 1) was expected to yield a processed 25S rRNA transcript carrying tag sequence A. Transcription units whose internal processing sites have been replaced by tag sequence B will, after generation of the 35S precursor rRNA, not likely be processed. Therefore, we expected a 25S rRNA tagged with sequence A from gene 1, a 35S rRNA tagged with sequence B from gene 2, and, in the case of the double-integration strains, a second 35S rRNA arising from the middle gene (gene 3) and carrying both tag sequences (Fig. 4a). Total RNA was separated on a 0.8% formaldehyde gel, blotted, and hybridized to probes for the tag sequences. As the results in lanes 1, 2, 7, and 8 of Fig. 3b and 4b show, all expected transcripts can be visualized clearly. The signals above the 25S band originating from gene 1 most probably arise from processing intermediates, for instance the 27S intermediate (34). A comparison of the relative intensities of the transcripts from the tagged genes showed, after correction for loading, transfer efficiency, and hybridization (for details, see Materials and Methods), that the steady-state level of the 35S transcripts from genes 2 and 3 is approximately 5 to 10 times lower than that from the mature 25S rRNA from gene 1 (data not shown). This may be due to a shorter half-life of the 35S precursor, which would not be surprising given that the processing pathway of the rRNAs is believed to be tightly regulated (34). Abnormal 35S precursor rRNAs unable to be processed may well be removed rapidly. In order to allow a comparable quantification of the effects of deletions of regulatory elements in the experiments described below, all rRNA signals from YMB1-1 and YMB1-2 were defined as 100% (Fig. 3d and 4d). In the case of the double-integration strains, transcripts from the middle gene (gene 3) were quantified by using the 35S band from the tag A hybridization, as tag sequence B here gives a composite signal derived from genes 2 and 3.
Next, we addressed the frequency of activation of our tagged genes. In an earlier study from our laboratory, we developed a method which allowed us to determine the fraction of transcriptionally active rRNA gene copies present in a population of eukaryotic cells (4). This method allows the distinction between active and inactive gene copies due to their different chromatin structures and in vivo accessibility to the intercalating drug psoralen, which, upon irradiation, introduces cross-links into DNA sites not protected by nucleosomes (9, 32). We have been able to visualize these two distinct populations of rRNA genes by separating rDNA restriction fragments from psoralen cross-linked cells on native agarose gels: highly cross-linked DNA derived from nonnucleosomal genes migrates slower (s band) than only slightly cross-linked DNA originating from nucleosome-packaged genes (f band). Based on earlier work (4), the relative intensities of these two bands reflect the ratio of active to inactive gene copies in a given cell population and therefore, in the case of a single gene, the frequency of activation. For simplification, the s and f bands were explicitly labeled only in the total rDNA panels in Fig. 3c and 4c. Note that we cannot formally exclude the possible existence of a third fraction of rRNA genes which is nucleosome-free (and thus runs in parallel to the s band) but is not actively transcribed (i.e., is potentially active). The psoralen gel retardation assay would not allow the distinction between actively transcribed and potentially active genes. However, there is no evidence that such a potentially active rRNA gene fraction exists in a significant amount in exponentially growing yeast cells. We therefore consider the nonnucleosomal gene fraction as homogeneous and active.
We applied the psoralen gel retardation technique to the analysis of the chromatin structure of our tagged rRNA transcription units. Exponentially growing cells (parallel samples from the same cultures that were used in the previous experiment) were photoreacted in the presence of psoralen, and total DNA was purified and cut with EcoRI and SacII. The resulting fragments were separated on a native agarose gel, transferred to a nylon membrane, and hybridized to probes A and B, respectively (lanes 2, 3, 11, and 12 in Fig. 3c and 4c). As expected, the fragments from the tagged genes separate into two distinct bands (compare with non-cross-linked control DNA in lanes 1 and 10, respectively), representing the nonnucleosomal and nucleosomal gene copies, therefore mirroring the activation frequency of these genes. As a control experiment we used a probe complementary to the coding sequence of the ribosomal precursor, in order to determine the state of the remaining 100 to 200 untagged rRNA transcription units (lanes 19 to 21 in Fig. 3c and 4c). Quantification of the ratio of the intensities of the s and f bands revealed that about 60% of the tagged genes as well as the untagged rRNA genes were active (Fig. 3e and 4e), which is in good agreement with earlier data (5). We concluded from these experiments that the introduction of the tag sequences into the rRNA transcription units does not measurably alter the chromatin structure of the genes in terms of nucleosomal packaging.
Analysis of rRNA genes with deleted enhancer elements.
The first and foremost aim of this work was to more closely study the in vivo function of the pol I enhancer element, which is located within a 317-bp EcoRI-HpaI region approximately 100 bp downstream of the 3′ end of the mature 25S rRNA gene sequence (28). More specifically, we tried to answer the question of whether its transcription enhancing effect on its two neighboring rRNA genes (14) is based on increasing the chance that one or both flanking promoters will be activated (Fig. 1a) or on enhancing the expression level of already-active promoters (Fig. 1b). For that purpose we constructed an enhancer deletion mutant lacking the whole EcoRI-HpaI fragment. Following the cotransformation and selection procedure described above, we obtained single and double integration strains carrying either two tagged rRNA genes with one deleted enhancer in between (YMB2-1 in Fig. 3a) or three tagged genes with two deleted enhancer elements (YMB2-2 in Fig. 4a). The latter strain carries one tagged rRNA gene copy with both flanking enhancer elements deleted (gene 3).
We first analyzed RNA from the mutant strains by Northern blot hybridization as described above (lanes 3, 4, 9, and 10 in Fig. 3b and 4b). Quantification showed that all tagged genes were transcribed at a significantly lower level compared to the nondeletion strains (Fig. 3d and 4d). Steady-state levels of transcripts from genes with one enhancer deleted were reduced by about 40 to 60%, which is consistent with the results from a previous study which showed a reduction of approximately 50% (14). Our results with the double-integration strain YMB2-2, however, also clearly show that, although transcriptional activity of the enhancerless gene is further reduced compared to genes with only one missing enhancer element, a significant amount of transcription (∼20%) is retained (gene 3 in Fig. 4d).
We next wanted to correlate the observed reduction in transcription activity of the tagged genes with the activation frequency reflected in their chromatin structure. We therefore performed in vivo psoralen cross-linking experiments in order to determine changes in the nucleosomal packaging of the tagged transcription units. Separation of the digested, cross-linked DNA on a native agarose gel, blotting, and hybridization with the tag sequence probes as described above showed that the fragments of all tagged genes were resolved into the two characteristic s and f bands, composed of highly cross-linked fragments arising from the nonnucleosomal population and slightly cross-linked fragments from the nucleosomal gene copies (lanes 5, 6, 14, and 15 in Fig. 3c and 4c). We calculated from the ratio of these two bands that again about 60% of the cell population kept the tagged genes devoid of nucleosomes, resembling the active state (Fig. 3e and 4e). Even in the case of gene 3 of the double-integration strain YMB2-2, both of whose flanking enhancers had been deleted, no significant alteration in the ratio of nonnucleosomal to nucleosomal gene copies could be observed. Quantification of the two populations of the untagged rRNA genes confirmed that growth conditions were similar in all experiments (lanes 23 and 24 in Fig. 3c and 4c). Since no significant decrease in the activation frequency could be observed although rRNA levels are reduced to half their normal values (or even more in the case of gene 3), our results argue that the transcription-enhancing effect of the ribosomal pol I enhancer element is mainly based on increasing the amount of transcripts from a given active gene rather than increasing the number of active genes.
Is pol I able to traverse enhancer-deleted intergenic rDNA spacers?
Termination of transcribing pol I molecules in the ribosomal locus is believed to occur at a specific termination site located within the rDNA enhancer element (15) and to be mediated by Reb1 protein (15, 22). Although other termination sites further downstream from the enhancer element have also been identified (33), later studies failed to confirm this data (15). Since the principal termination site residing within the enhancer element had effectively been removed with the enhancer deletion in our experiments, nonterminated polymerases traversing the spacer region might thus be able to enter the next transcription unit. In order to ensure that such a phenomenon did not obscure our results, we constructed strains with enhancer-promoter double deletions. Note that the termination site upstream of transcription initiation (33) was removed in the promoter deletion. We reasoned that, if nonterminated polymerases do indeed traverse the spacer in our strains and account for a significant amount of detected transcripts from the downstream genes and also for their open chromatin structure, even a promoter deletion would not be able to shut down these genes completely.
After the cotransformation and selection procedure described above, we analyzed the transcriptional activity and chromatin structure of the genes in single- and double-integration strains (YMB3-1 in Fig. 3a and YMB3-2 in Fig. 4a). In these strains only the first tagged transcription unit (gene 1) possesses a functional promoter element, whereas the corresponding sequences of the rRNA genes tagged with tag sequence B are deleted (gene 2 and gene 3). The Northern blot analysis and the psoralen gel retardation experiment of these strains are shown in lanes 5, 6, 11, and 12 in Fig. 3b and 4b and in lanes 8, 9, 17, and 18 in Fig. 3c and 4c, respectively. As expected, the transcriptional activity of gene 1 was virtually unchanged compared to the enhancer deletion strains YMB2-1 and YMB2-2 (see the quantification in Fig. 3d and e and 4d and e). On the other hand, rRNA levels of the tag B-carrying genes, whose promoter regions had been deleted, practically reached zero. Nearly 100% of the cross-linked DNA fragments derived from these genes show a mobility consistent with nucleosomal, inactive transcription units. We note that, in lanes 5 and 6 of Fig. 4b, a faint signal, whose length corresponds to the expected transcript from gene 3, is visible (see also the quantification in Fig. 4d). Because there is no such signal detectable with tag sequence B (lanes 11 and 12), we believe that this weak band represents traces of the rapidly processed 35S precursor of the gene 1 transcript.
Taken together, the results show that nonterminated RNA polymerases originating from upstream promoters are not able in vivo to travel through the intergenic spacer region and to enter the next transcription unit and, therefore, demonstrate that the data from our enhancer deletion strains are not distorted by nonterminated upstream gene transcription.
DISCUSSION
We analyzed the effects of pol I enhancer deletions on transcriptional activity and chromatin structure of their flanking rRNA transcription units. In order to remain as close as possible to the normal chromosomal context, tagged full-size rRNA genes were introduced into the rDNA locus by homologous recombination. While Northern blot analysis showed that the deletion of an rDNA enhancer element reduces the rRNA levels of both flanking transcription units by a factor of approximately two, which is in good agreement with previous in vivo data (14), our experimental setup also allowed direct measurement of the effects of a deletion of both flanking enhancer elements on the enclosed gene (see gene 3 in YMB2-2 in Fig. 4). We found that, although a further decrease in transcriptional activity could be observed compared to genes with one deleted enhancer, the gene was not turned off completely as proposed previously (14). The fact that a significant amount of transcription remained indicates that control over rRNA gene transcription under natural conditions is not exerted by only its two closest enhancer elements. The residual activity observed may either be mediated by the influence of more distantly located enhancer elements or merely reflect the basal transcriptional activity of the pol I promoter within the nucleolus.
The effects of the pol I enhancer deletion on the transcriptional activity of the flanking genes were not reflected by corresponding changes in their chromatin structure as measured by psoralen accessibility. Instead, the ratio of nonnucleosomal (active [4, 5]) to nucleosomal (inactive [4, 5]) copies of our tagged genes remained virtually constant, independent of the presence or absence of one or both flanking enhancers. This clearly shows that the rDNA enhancer does not work by increasing the number of the nucleosome-free (active) rRNA genes (Fig. 1a). Instead, the reduced rRNA levels can only be explained by a lower average transcription rate of the same number of nonnucleosomal rRNA genes (Fig. 1b). We cannot formally exclude the possibility that this originates from a significant amount of transcriptionally silent but still nonnucleosomal gene copies in the absence of the flanking enhancers (Fig. 1a). However, we favor the alternative, namely, a more or less similar reduction in the transcription rate of all the nonnucleosomal tagged genes as shown in Fig. 1b, mainly because, as recent data from our group suggest, the open nonnucleosomal structure of actively transcribed rRNA genes is the result of RNA polymerase I molecules advancing through the rDNA template (6, 20). However, how many polymerase molecules are necessary for stable maintenance of an open chromatin structure, and whether the polymerase density on different transcribed rRNA gene copies is similar or may also be unequal to a certain extent is a question that remains extremely difficult to answer. Since only the passage of the replication machinery has been shown in yeast so far to be able to repackage active rRNA genes in nucleosomes (20), one might be tempted to speculate that, as soon as the nucleosomes have been displaced by a few transcribing polymerases, the chromatin structure of the gene may remain open until the next S phase (4). Therefore, we propose that the rDNA enhancer most probably does not determine the proportion of nonnucleosomal rRNA gene copies, as no obvious effects on the activation frequency of its flanking rRNA genes were observed. However, it appears that the enhancer plays a role in the average transcription initiation rate.
An intriguing observation is that in yeast the nonnucleosomal (active) rRNA genes are strictly followed downstream by nonnucleosomal enhancer elements and vice versa (6). The nonnucleosomal structure of the enhancer presumably reflects specific protein-DNA interactions at this element. Since advancing RNA polymerase I molecules are responsible for creating the nonnucleosomal chromatin state of the rRNA genes (6, 20), these protein-DNA interactions might be part of an activated promoter complex and therefore be rather the cause than the consequence of the activated upstream gene. This means that the information to activate an rRNA gene has to be transferred to the promoter and enhancer elements by a yet-unknown mechanism. How this may happen is difficult to speculate about, but we know at least that RNA polymerase I per se is not part of this process, since nucleosome-free enhancer elements are also present in pol I deletion mutants (6). Johnson and Warner (10) and Kulkens et al. (14) have hypothesized that the enhancer and the promoter elements are physically brought in contact with each other by a common protein, Reb1p, which binds to both elements. Principally, it could be imagined that Reb1p bound to promoter elements is one factor or can recruit those factors responsible for the observed open chromatin structure of the downstream enhancer element. Whereas there is today no real evidence for such a mechanism, some of the reasons for the recruitment of these factors to enhancers downstream of active rRNA genes have already become clear. For instance, two of the proteins that bind to the rDNA enhancer, Abf1p and Reb1p, are general transcription factors and may thus be involved in rDNA transcription enhancement (21, 28). Reb1p, furthermore, is essential for efficient transcription termination of active rRNA genes, leading to the already discussed hypothesis of polymerase recycling. Moreover, there are also some intriguing correlations between the activity of an rRNA gene as determined by its open chromatin structure and the process of replication termination. This process, in the form of a polar replication fork barrier located within the borders of the enhancer element (2), only seems to occur if the corresponding rRNA gene and therefore also its 3′ enhancer element are free of nucleosomes (19). It will require much more work to elucidate the significance of these findings, and it will be interesting, in this context, to extend our current enhancer deletion analysis to the effects on rDNA replication.
ACKNOWLEDGMENTS
We thank F. Thoma for correction of the manuscript, C. Weissmann for access to the PhosphorImager; R. Wellinger, B. Schweizer, V. Taylor, and M. Smerdon for helpful discussion; and U. Suter for continuous support. We also thank the anonymous referees for helpful suggestions.
This work was supported by grants from the Boehringer Ingelheim Fonds (to M.B.) and the Swiss National Science Foundation (to J.M.S.; grant 31-52246.97).
REFERENCES
- 1.Banditt M. Ph.D. thesis no. 12874. Zürich, Switzerland: Eidgenössische Technische Hochschule Zürich; 1998. [Google Scholar]
- 1a.Becker D M, Guarente L. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–187. doi: 10.1016/0076-6879(91)94015-5. [DOI] [PubMed] [Google Scholar]
- 2.Brewer B J, Fangman W L. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell. 1988;55:637–643. doi: 10.1016/0092-8674(88)90222-x. [DOI] [PubMed] [Google Scholar]
- 3.Butlin M, Quincey R. The yeast rRNA gene enhancer does not function by recycling RNA polymerase I and cannot act as a UAS. Curr Genet. 1991;20:9–16. doi: 10.1007/BF00312759. [DOI] [PubMed] [Google Scholar]
- 4.Conconi A, Widmer R M, Koller T, Sogo J M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 5.Dammann R, Lucchini R, Koller T, Sogo J M. Chromatin structures and transcription of rDNA in yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:2331–2338. doi: 10.1093/nar/21.10.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dammann R, Lucchini R, Koller T, Sogo J M. Transcription in the yeast rDNA locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol Cell Biol. 1995;15:5294–5303. doi: 10.1128/mcb.15.10.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elion E A, Warner J R. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell. 1984;39:663–673. doi: 10.1016/0092-8674(84)90473-2. [DOI] [PubMed] [Google Scholar]
- 8.Elion E A, Warner J R. An RNA polymerase I enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson C V, Shen K J, Hearst J E. Cross-linking of DNA in situ as a probe for chromatin structure. Science. 1976;193:62–64. doi: 10.1126/science.935855. [DOI] [PubMed] [Google Scholar]
- 10.Johnson S P, Warner J R. Unusual enhancer function in yeast rRNA transcription. Mol Cell Biol. 1989;9:4986–4993. doi: 10.1128/mcb.9.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato H, Nagamine M, Kominami R, Muramatsu M. Formation of the transcription initiation complex on mammalian rDNA. Mol Cell Biol. 1986;6:3418–3427. doi: 10.1128/mcb.6.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kief D R, Warner J R. Coordinated control of synthesis of ribosomal ribonucleic acid and ribosomal proteins during nutritional shift-up in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:1007–1015. doi: 10.1128/mcb.1.11.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kormanec J, Farkasovsky M. Isolation of total RNA from yeast and bacteria and detection of rRNA in Northern blots. BioTechniques. 1994;17:838–842. [PubMed] [Google Scholar]
- 14.Kulkens T, van der Sande C A F M, Dekker A F, van Heerikhuizen H, Planta R J. A system to study transcription by yeast RNA polymerase I within the chromosomal context: functional analysis of the ribosomal DNA enhancer and the RBP1/REB1 binding sites. EMBO J. 1992;11:4665–4674. doi: 10.1002/j.1460-2075.1992.tb05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang W H, Reeder R H. The REB1 site is an essential component of a terminator for RNA polymerase I in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:649–658. doi: 10.1128/mcb.13.1.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ling M, Merante F, Robinson B H. A rapid and reliable DNA preparation method for screening a large number of yeast clones by polymerase chain reaction. Nucleic Acids Res. 1995;23:4924–4925. doi: 10.1093/nar/23.23.4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long E O, Dawid I B. Repeated genes in eucaryotes. Annu Rev Biochem. 1980;49:727–764. doi: 10.1146/annurev.bi.49.070180.003455. [DOI] [PubMed] [Google Scholar]
- 18.Lucchini R, Sogo J M. Different chromatin structures along the spacers flanking active and inactive Xenopus rRNA genes. Mol Cell Biol. 1992;12:4288–4296. doi: 10.1128/mcb.12.10.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucchini R, Sogo J M. Chromatin structure and transcriptional activity around the replication forks arrested at the 3′ end of the yeast rRNA genes. Mol Cell Biol. 1994;14:318–326. doi: 10.1128/mcb.14.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucchini R, Sogo J M. Replication of transcriptionally active chromatin. Nature. 1995;374:276–280. doi: 10.1038/374276a0. [DOI] [PubMed] [Google Scholar]
- 21.Morrow B E, Johnson S P, Warner J R. The rDNA enhancer regulates rRNA transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1283–1289. doi: 10.1128/mcb.13.2.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrow B E, Ju Q, Warner J R. Purification and characterization of the yeast rDNA binding protein REB1. J Biol Chem. 1990;265:20778–20783. [PubMed] [Google Scholar]
- 23.Musters W, Knol J, Maas P, Dekker A F, van Heerikhuizen H, Planta R J. Linker scanning of the yeast RNA polymerase I promoter. Nucleic Acids Res. 1989;17:9661–9678. doi: 10.1093/nar/17.23.9661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osheim Y N, Mougey E B, Windle J, Anderson M, O’Reilly M, Miller O L, Jr, Beyer A, Sollner-Webb B. Metazoan rDNA enhancer acts by making more genes transcriptionally active. J Cell Biol. 1996;133:943–954. doi: 10.1083/jcb.133.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philippsen P, Thomas M, Kramer R A, Davis R W. Unique arrangement of coding sequences for 5S, 5.8S, 18S and 25S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loops and hybridisation analysis. J Mol Biol. 1978;123:387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- 26.Reeder R H. Regulatory elements of the generic ribosomal gene. Curr Opin Cell Biol. 1989;1:466–474. doi: 10.1016/0955-0674(89)90007-0. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Schultz M C, Choe S Y, Reeder R H. In vitro definition of the yeast RNA polymerase I enhancer. Mol Cell Biol. 1993;13:2644–2654. doi: 10.1128/mcb.13.5.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigurdson C D, Gaarder M E, Livingston D M. Characterization of the transmission during cytoductant formation of the 2μm DNA plasmid from Saccharomyces. Mol Gen Genet. 1981;183:59–65. doi: 10.1007/BF00270139. [DOI] [PubMed] [Google Scholar]
- 30.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skryabin K G, Eldarov M A, Larionov V L, Bayev A A, Klootwijk J, de Regt V C H F, Veldman G M, Planta R, Georgiev O I, Hadjiolov A A. Structure and function of the nontranscribed spacer region of yeast rDNA. Nucleic Acids Res. 1984;12:2955–2968. doi: 10.1093/nar/12.6.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sogo J M, Ness P J, Widmer R M, Parish R W, Koller T. Psoralen crosslinking as a probe for the structure of active nucleolar chromatin. J Mol Biol. 1984;178:897–928. doi: 10.1016/0022-2836(84)90318-8. [DOI] [PubMed] [Google Scholar]
- 33.van der Sande C A F M, Kulkens T, Kramer A B, de Wijs I J, van Heerikhuizen H, Klootwijk J, Planta R J. Termination of transcription by yeast RNA polymerase I. Nucleic Acids Res. 1989;17:9127–9146. doi: 10.1093/nar/17.22.9127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venema J, Tollervey D. Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast. 1995;11:1629–1650. doi: 10.1002/yea.320111607. [DOI] [PubMed] [Google Scholar]
- 35.Warner J R. Synthesis of ribosomes in Saccharomyces cerevisiae. Microbiol Rev. 1989;53:256–271. doi: 10.1128/mr.53.2.256-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J-R, Gilbert D M. Rapid DNA preparation for 2D gel analysis of replication intermediates. Nucleic Acids Res. 1995;23:3997–3998. doi: 10.1093/nar/23.19.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]