Abstract
Gestational diabetes mellitus (GDM) is the most common metabolic complication in pregnancy, which affects the future health of both the mother and the newborn. Its pathophysiology involves nutritional, hormonal, immunological, genetic and epigenetic factors. Among the latter, it has been observed that alterations in DNA (deoxyribonucleic acid) methylation patterns and in the levels of certain micro RNAs, whether in placenta or adipose tissue, are related to well-known characteristics of the disease, such as hyperglycemia, insulin resistance, inflammation and excessive placental growth. Furthermore, epigenetic alterations of gestational diabetes mellitus are observable in maternal blood, although their pathophysiological roles are completely unknown. Despite this, it has not been possible to determine the causes of the epigenetic characteristics of GDM, highlighting the need for integral and longitudinal studies. Based on this, this article summarizes the most relevant and recent studies on epigenetic alterations in placenta, adipose tissue and maternal blood associated with GDM in order to provide the reader with a general overview of the subject and indicate future research topics.
Keywords: gestational diabetes, epigenetics, DNA methylation, miRNAs, adipose tissue, placenta
1. Introduction
GDM is defined as any degree of glucose intolerance first identified during pregnancy [1]. The prevalence of this disorder varies globally, depending on the diagnostic criteria applied and on the ethnic group studied. It varies from 1% to more than 30% [2]. The population with GDM has been increasing worldwide, mainly as a result of the rising proportion of women with pre-pregnancy obesity, a sedentary lifestyle and advanced maternal age at birth. Additional risk factors include family history of diabetes, parity, previous GDM, prior macrosomia and excessive weight gain in pregnancy. GDM is associated with short-term and long-term adverse consequences in the mother and in the offspring. In the mother, it increases the risk of cesarean section and favors the development of gestational hypertension or pre-eclampsia. Furthermore, it has been estimated that women with GDM have a more than sevenfold increased risk of type 2 diabetes mellitus (T2DM) compared to normoglycemic pregnancies. In the fetus, the risk of macrosomia or a larger size for gestational age increases, which leads to complications such as shoulder dystocia and Erb’s palsy. Another important neonatal complication is immediate hypoglycemia at birth caused by fetal hyperinsulinemia [3,4]. In adulthood, the child is more likely to develop obesity, T2DM and cardiovascular disease [5].
In addition to lifestyle factors, genetic heritability is implicated in the etiology of GDM, and emerging data indicate contributions of environmental and dietary factors to the risk of developing GDM, through epigenetic changes. Epigenetics refers to changes in gene expression that occur without any alterations in the nucleotide sequence of DNA. Epigenetic changes include DNA methylation, chemical histone modifications (methylation, acetylation, etc.) and non-coding RNA molecules such as micro RNAs (miRNAs) and long non-coding RNAs [6,7]. The current evidence of epigenetic factors related to GDM is substantial and allows us to see that there are particular epigenetic signatures before the onset of hyperglycemia; during GDM and postpartum; and before the development of T2DM. Thus, the objectives of this review are to present an organized and updated summary of epigenetic alterations related to pathogenesis of GDM and indicate future topics for research.
2. The Pathophysiology of GDM
To understand the pathophysiology of GDM, it is necessary to know the metabolic changes that occur in healthy pregnancy. The first trimester of pregnancy is characterized by a progressive increase in insulin secretion, perhaps as a result of an increase in pancreatic β-cell mass, and increased sensitivity to this hormone. From the second trimester, a progressive decrease in insulin sensitivity, called insulin resistance, begins, mainly in the adipose tissue, skeletal muscle and liver [8,9]. Historically, this insulin resistance has been thought to be the result of increased concentrations of placental hormones such as human placental lactogen (hPL) and placental growth hormone, as their effects can interfere with insulin receptor signaling and cause a marked decrease in glucose utilization [10]. To compensate for this insulin resistance, the pancreatic β cells increase insulin secretion, thereby preventing the blood glucose concentration from exceeding normal values [11]. In addition to these hormonal changes, some studies have suggested the participation of adipose tissue in insulin resistance in pregnancy. During pregnancy, there is an increase in fat mass that can vary from 2 to 5 kg [12]. Adipose tissue functions as an endocrine organ, in which increase in fat is related to changes in the production of cytokines and adipokines, such as interleukin 1 beta (IL-1β), interleukin 6 (IL-6), tumoral necrosis factor alpha (TNF-α), leptin and adiponectin, whose effects intervene in insulin sensitivity in pregnancy [13].
The pathophysiology of GDM is still under investigation; however, it is characterized by two main mechanisms which result in hyperglycemia: dysfunction of pancreatic β cells and exacerbated insulin resistance. In women who develop GDM, there is a defect in the response of β cells that compensates for insulin resistance, which becomes evident in late pregnancy, when insulin resistance is greatest [14]. There is evidence that this defect in β cell function exists before pregnancy, but only becomes clinically apparent with the increased insulin resistance of pregnancy. For this reason, screening for GDM is usually done between weeks 24 and 28 of pregnancy, and there are various criteria for GDM diagnosis using the oral glucose tolerance test (OGTT) [15].
In addition, insulin resistance in women with GDM may be more pronounced due to the down-regulation of adiponectin and up-regulation of leptin and pro-inflammatory cytokines [16]. Interestingly, in many cases, insulin resistance already exists before pregnancy, especially in populations with high rates of obesity [17].
There is emerging evidence that epigenetic changes contribute to the pathophysiology of GDM. Recently, several studies have reported placental and adipose tissue epigenetic changes associated with GDM. In addition, GDM has been associated with epigenetic modifications that are detectable in maternal blood. A summary of the methodological characteristics of studies that compared epigenetic changes in placental tissue, adipose tissue and peripheral blood between GDM patients and controls is presented in Table 1.
Table 1.
The methodological characteristics of studies that compared epigenetics changes between GDM patients and controls.
| Authors | Study Design | n | Method Used for Epigenetic Analysis | Biological Sample Used for Epigenetic Analysis | Genes or Proteins Affected | GDM Criteria |
|---|---|---|---|---|---|---|
| Lesseur, et al. [18] | Cohort | 47 GDM and 488 non GDM women | BSP | Placental tissue (fetal side) | LEP | Not reported |
| Gagné-Ouellet, et al. [19] | Cohort | 12 GDM and 250 non GDM women | Infinium MethylationEPIC BeadChip |
Placental tissue (fetal side) | LEP | Canadian Diabetes Association (year not reported) |
| Bouchart, et al. [20] | Cohort | 31 IGT women and 67 NGT women | BSP | Placental tissue from both maternal and fetal sides | ADIPOQ | Only 2 women from IGT group fulfilled the ADA 2009 criteria for GDM. Women were classified as having IGT with a 2-h post-OGTT glucose level ≥7.8 mmol/L |
| Blazevic, et al. [21] | Case-control | 18 GDM and 32 NGT | DBS | Placental tissue (fetal side) | SLC6A4 | IADPSG 2010 |
| Côté, et al. [22] | Cohort | 33 GDM and 100 NGT | BSP | Placental tissue (fetal side) | PRDM16, BMP7, CTBP2 and PPARGC1α | WHO 2013 |
| Wang, et al. [23] | Case-control | 18 GDM and 32 control | DBS | Placental tissue (fetal side) | PPARGC1α and PDX1 | Two step: Step 1: 50 g GCT Cutoff value (mmol/L): ≥7.8 Step 2: 100 g OGTT Cutoff value (mmol/L) Fasting: >5.6 1 h: >10.3 2 h: >8.6 3 h: >6.7 Diagnosed: 4 abnormalities |
| Houde, et al. [24] | Cohort | 27 GDM and 99 NGT | Illumina Human Methylation 450 DNA Analysis Beadchip and BSP | Placental tissue (fetal side) | LPL | WHO 2013 |
| Qiu, et al. [25] | Case-control | 60 GDM and 60 controls | qRT-PCR | Plasma from maternal peripheral blood and placenta | miR-518d, NF-κB, COX-2, TNF-α, IL-1β, IL-6 and PPARα | Not reported |
| Sun, et al. [26] | Case-control | 204 GDM and 202 control | qRT-PCR | Placental tissue | miR-29b and HIF3A | WHO 2013 |
| Cao, et al. [27] | Case-control | 193 GDM and 202 control | qRT-PCR | Placental tissue | miR-98, MECP2 and TRPC3 | Not reported |
| Ding, et al. [28] | Case-control | Discovery stage: 8 GDM and 8 controls Validation stage: 28 GDM and 26 control |
RNA-seq and qRT-PCR | Placental tissue | miR-202-5p, miR-138-5p, miR-210-5p, miR-3158-5p, miR-4732-3p, TBL1X, NOTUM, FRMD4A, SLC16A2, CLDN19, CCL18, HTRA1 and SLC39A6 | IADPSG 2010 |
| Hepp, et al. [29] | Case-control | 40 GDM and 40 controls | Immunohistochemistry and double immunofluorescence | Placental tissue (maternal side) | Histones H3 | GSDB 2011 |
| Rancourt, et al. [30] | Nested case-control | 19 GDM and 22 controls matched for maternal age, socio-economic status, ethnic origin, parity and pre-pregnancy BMI | BSP | Subcutaneous and visceral adipose tissues | TNF-α and SOCS3 | GSGO 2018 |
| Ott, et al. [31] | Nested case-control | 25 GDM and 30 controls matched for maternal age, ethnic origin, socio-economic status, parity and pre-pregnancy BMI | BSP | DNA from blood cells, subcutaneous and visceral adipose tissue | ADIPOQ | GSGO 2018 |
| Deng, et al. [32] | Case-control | Discovery stage: 3 GDM and 3 controls Validation stage: 26 GDM and 24 controls |
Illumina Human Methylation 450 k DNA Analysis Beadchip and BSP | Visceral omental adipose tissue | HLA-DMB, MSLN, and HSPA6 | WHO 2013 |
| Shi, et al. [33] | Case-control | Discovery stage: 3 GDM and 3 controls. Validation stage: 13 GDM and 13 controls. |
AFFX miRNA expression chips and qRT-PCR | Omental adipose tissue | miR-222, ERα and GLUT4 | ADA 2006 |
| Dias, et al. [34] | Case-control | 12 GDM and 12 controls matched for age, gestational age and BMI. | Illumina’s Infinium HumanMethylationEPIC Bead Chip | DNA from maternal peripheral blood | SLC9A3, MEA1;KLHDC3, CAMTA1 and RASA3 | IADPSG 2010 |
| Wu, et al. [35] | Cohort | 11 GDM and 11 controls matched for age, BMI, ethnicity, smoking, treatment, and folate supplementation | Illumina HumanMethylation450 BeadChip and BSP | DNA from maternal peripheral blood | COPS8, PIK3R5, HAAO, C5orf34 and CCDC124 | Not reported |
| Michalczy, et al. [36] | Cohort | 27 pregnant women classified as non-diabetic (7 women), GDM who did not develop T2DM (8 women), GDM who developed T2DM (6 women), and women with pre-existing T2DM (6 women) | Western blot | White blood cells from maternal peripheral blood | Histones H3 | IADPSG 2010 |
| Zhao, et al. [37] | Nested case-control | Discovery stage: 24 GDM and 24 controls. Internal validation: 36 GDM and 36 controls. Two different external validations: 32 GDM and 32 controls in total. In all stages, the groups were matched for age, BMI, gestational age, and gravidity |
TaqMan low density arrays Chips and qRT-PCR | Serum from maternal peripheral blood | miR-29a, miR-222, INSIG1 and PCK2 | ADA 2004 |
| Zhu, et al. [38] | Case-control | 10 GDM and 10 controls matched for maternal age and gestational age | Ion Torrent high-throughput sequencing technology and qRT-PCR | Plasma from maternal peripheral blood | hsa-miR-16-5p, hsa-miR-17-5p, hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-20a-5p, MAPK-1, IRS-1, IRS-2, SOS-1, SMAD5, SMAD4 and AKT3 | ADA 2011 |
| Pheiffer, et al. [39] | Case-control | 28 GDM and 53 controls matched for age and BMI | qRT-PCR | Serum from maternal peripheral blood | hsa-miR-20a-5p | IADPSG 2010 |
BMI, body mass index; NGT, normal glucose tolerance; BSP, bisulfite pyrosequencing; DBS, direct bisulfite sequencing; qRT-PCR, quantitative reverse transcription—polymerase chain reaction; LEP, leptin; ADIPOQ, adiponectin; SLC6A4, solute carrier family 6 member 4; PRDM16, PR domain-containing protein 16; BMP7, bone morphogenetic protein 7; CTBP2, C-terminal binding protein 2; PPARGC1α, peroxisome proliferator-activated receptor-gamma, co-activator 1, alpha; PDX1, pancreatic and duodenal homeobox l; LPL, lipoprotein lipase; NF-κB, nuclear factor-kappa B; COX-2, cytochrome C oxidase subunit II; TNF-α, tumoral necrosis factor alpha; IL-1β, interleukin 1 beta; IL-6, interleukin 6; PPARα, peroxisome proliferator-activated receptor alpha; HIF3A, hypoxia inducible factors 3A; MECP2, methyl CpG binding protein 2; TRPC3, transient receptor potential 3; TBL1X, transducing beta like 1 X-linked; NOTUM, notum, palmitoleoyl-protein carboxylesterase; FRMD4A, FERM domain containing 4A; SLC39A6, solute carrier family 39 member 6; SLC16A2, solute carrier family 16 member 2; CLDN19, claudin 19; CCL18, C-C motif chemokine ligand 18; HTRA1, HtrA serine peptidase 1; SOCS3, suppressor of cytokine signaling 3; HLA-DMB, major histocompatibility complex, class II, DM beta; MSLN, mesothelin; HSPA6, heat shock protein family A (Hsp70) member 6; ERα, estrogen receptor alpha; GLUT4, glucose transporter 4; SLC9A3, solute carrier family 9 member A3; MEA1;KLHDC3, male-enhanced antigen 1; kelch domain-containing protein 3; CAMTA1, calmodulin binding transcription activator 1; RASA3, RAS P21 protein activator 3; COPS8, constitutive photomorphogenic homolog subunit 8; PIK3R5, phosphoinositide-3-kinase, regulatory subunit 5; HAAO, 3-hydroxyanthranilate 3,4-dioxygenase; C5orf34, chromosome 5 open reading frame 34; CCDC124, coiled-coil domain containing 124; INSIG1, insulin-induced gene 1; PCK2, phosphoenolpyruvate carboxy kinase 2; MAPK-1, mitogen-activated protein kinase 1; IRS-1, insulin receptor substrate 1; IRS-2, insulin receptor substrate 2; SOS-1, SOS Ras/Rac guanine nucleotide exchange factor 1; SMAD5, SMAD family member 5; SMAD4, SMAD family member 4; AKT3, AKT serine/threonine kinase 3; GSDB, German Society for Diabetes Mellitus; GSGO 2018, German Society for Gynecology and Obstetrics 2018; GCT, glucose challenge test; ADA, American Diabetes Association; WHO, World Health Organization; IADPSG, International Association of Diabetes and Pregnancy Study Group; IGT, impaired glucose tolerance.
3. Epigenetic Alterations Determined in the Placenta
The placenta secretes steroids, hormones and cytokines that are necessary for the correct course of pregnancy [40,41]. In GDM, the size and weight of the placenta are significantly greater than in normal pregnancy [42]. This increase in placental size is accompanied by higher concentrations of hPL, estradiol, and progesterone in the maternal circulation, which can promote the hyperglycemic state [43,44,45]. Moreover, GDM has a placental inflammatory component characterized by increased production of interleukin-8 (IL-8), TNF-α and leptin [46].
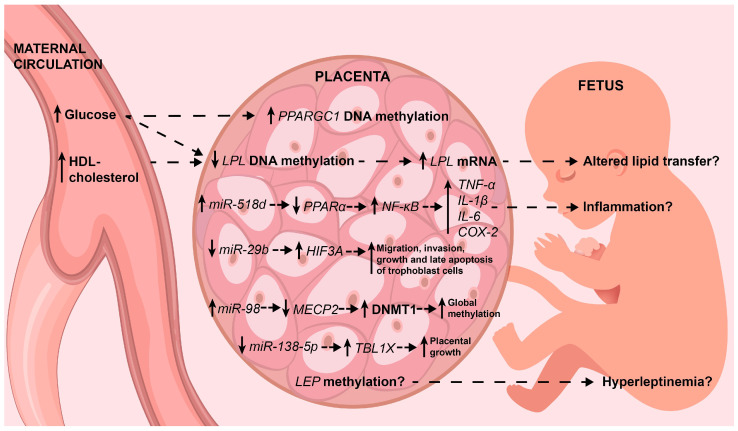
Interestingly, the diabetic placenta shows alterations in the proliferation, apoptosis and control of the trophoblast cell cycle [47]. Next, placental epigenetic profiles are described, some of which explain, in part, some of these associations between the placenta and GDM. The main findings are summarized in Figure 1.
Figure 1.
The main placental epigenetic alterations associated with GDM. Epigenetic alterations and their effects on target genes are shown (see the text for deeper details). Interestingly, some explain, in part, the well-known pro-inflammatory status and excessive growth of the placenta. On tissue, solid arrows indicate up-regulation or down-regulation in maternal circulation and the fetus indicate low or high circulating levels. Dotted line arrows indicate association. HDL-cholesterol: high-density lipoprotein cholesterol; mRNA: messenger ribonucleic acid; FFA: free fatty acids; PPARα: proliferator-activated receptor alpha; NF-κB: nuclear factor-kappa B; TNF-α: tumoral necrosis factor alpha; IL-1β: interleukin 1 beta; IL-6: interleukin 6; COX-2: cytochrome C oxidase subunit II; HIF3A: hypoxia inducible factors 3A; MECP2: methyl CpG binding protein 2; DNMT1, DNA methyltransferase 1; TBL1X: transducin β–like protein 1; LEP: leptin.
3.1. DNA Methylation
DNA methylation occurs when a methyl group is covalently added to 5-carbon of the cytosine in DNA by enzymes called DNA methyltransferases. Generally, this modification occurs when cytosine is followed by guanine, which is known as a CpG dinucleotide (the “p” refers to the phosphate group). Approximately 70–80% of the CpGs in the human genome are methylated, and the majority of the remainder are in gene promoter regions in clusters of CpGs called CpG islands that can be hundreds to tens of thousands of base pairs in length [48]. Generally, the level of DNA methylation in the promoter region of the gene inversely correlates with gene transcription, since methylation provides steric hindrance and favors compaction of chromatin, physically preventing DNA-binding proteins from recognizing their target sequences. CpG dinucleotides and CpG islands can also be located within genes and influence transcriptional activity; for example, when CpGs are close to or in splice sequences, methylation can influence the splice site and lead to alternative isoforms of mature mRNA [49,50,51]. Methylation can also occur in enhancer elements, which promotes the binding of proteins such as methyl-binding domain proteins, favoring transcription [52,53], or in repressor elements, which prevents the binding of methylation-sensitive proteins and leads to improved transcription, a rare case in which methylation positively correlates with transcription [54,55].
Lesseur et al. [18] evaluated the methylation levels of leptin (LEP) promoter in placental tissues from women with GDM in comparison to non-GDM women. The results showed that the average methylation of 23 CpGs sites analyzed in the LEP promoter was 2.5% higher in women with GDM. Interestingly, maternal pre-pregnancy obesity was not a significant predictor of LEP methylation, but it was strongly associated with GDM. Thus, the authors concluded that GDM mediates pre-pregnancy obesity’s effects on placental LEP methylation. In contrast, in the study by Gagné-Ouellet et al. [19] it was observed that higher maternal glycemia during pregnancy decreased methylation levels of LEP in the GDM placenta. The authors identified one CpG site (cg15758240) with more convincing results about the impact of maternal glucose levels on placental LEP DNA methylation. This CpG site has been characterized as an LEP interaction region located upstream of the LEP gene, with important roles for transcriptional regulation. In these studies, the placental expression of leptin was not measured, so the possible effect of differential methylation on mRNA and umbilical cord levels remains uncertain.
Bouchart et al. [20] analyzed the placental DNA methylation levels of adiponectin (ADIPOQ) in pregnant women classified according to glucose tolerance status in impaired glucose tolerance women (IGT) and normal glucose tolerant women (NGT). Although the authors did not observe significant differences in DNA methylation levels between the groups, they found that lower DNA methylation levels in the promoter of ADIPOQ on the fetal side of the placenta were correlated with higher maternal glucose levels during the second trimester of pregnancy. Furthermore, lower DNA methylation levels on the maternal side of the placenta were associated with higher insulin resistance during the second and third trimesters of pregnancy. Finally, on the maternal side of placenta, higher DNA methylation levels were associated with lower maternal circulating adiponectin levels throughout pregnancy and postdelivery.
A recent body of evidence suggests that serotonin, a multifunctional signaling molecule, contributes to obesity and related metabolic disorders, although the mechanisms are not well understood. The serotonin transporter, an integral membrane protein, mediates uptake of serotonin into cells, regulating the serotonin homeostasis [56,57]. Blazevic et al. [21] analyzed the methylation and mRNA levels of the placental serotonin transporter gene, also known as the solute carrier family 6 member 4 (SLC6A4), in placentas from GDM and NGT women. They observed that the DNA methylation across the seven analyzed loci was decreased in the GDM group in comparison to the control group and inversely correlated with placental SLC6A6 mRNA levels. Interestingly, maternal plasma glucose levels in the 24th to 28th weeks of gestation were negatively correlated with average DNA methylation.
Brown adipose tissue (BAT) is suspected to protect against obesity. Côté et al. [22] assessed DNA methylation variations in genes involved in BAT genesis and activation. They evaluated the DNA methylation levels at PR domain-containing protein 16 (PRDM16), bone morphogenetic protein 7 (BMP7), C-terminal binding protein 2 (CTBP2) and peroxisome proliferator-activated receptor-gamma co-activator 1 alpha (PPARGC1α) gene loci in the fetal side of placentas from 133 women (33 with GDM) followed during pregnancy (one visit at the end of each trimester), and observed that only BMP7 DNA methylation levels were lower in GDM compared to NGT. Interestingly, when analyzing all the women, higher PPARGC1α DNA methylation levels were associated with higher second trimester fasting glucose levels and 2 h post-OGTT glycemia; lower PRDM16 DNA methylation levels were associated with higher fasting glucose levels in the second and third trimesters, and lower BMP7 DNA methylation levels were associated with higher 2 h post-OGTT glycemia. In addition, Wang et al. [23] analyzed the methylation status and expression levels of the PPARGC1α and the pancreatic and duodenal homeobox l (PDX1) in placental tissue. They observed that the methylation frequency of PPARGC1α was higher and its expression was lower in GDM group compared to controls. There were no significant differences in relation to PDX1. Interestingly, PPARGC1α is a transcriptional co-activator that participates in lipid and carbohydrate metabolism in many tissues, including adipose tissue, muscle and liver.
Houde et al. [24] analyzed the placental methylation levels of the lipoprotein lipase (LPL) enzyme, which contributes to the transfer of free fatty acids from maternal lipoproteins to the fetus, and observed that the women with GDM had lower levels of methylation in each of the 3 CpG sites evaluated in comparison to NTG women. In addition, methylation levels of CpG2 and CpG3 sites (both located within the intronic CpG island) were negatively correlated with placental LPL mRNA levels, which were higher in GDM compared to NGT women. Interestingly, after adjusting for all potential confounding variables, it was observed that methylation levels at CpG1 (located within the promoter region) were negatively correlated with 2-h post-OGTT glucose concentrations, and CpG3 methylation levels negatively correlated with maternal concentrations of HDL-cholesterol during the third trimester of pregnancy.
3.2. miRNAs
miRNAs are small non-coding RNA sequences about 22 nucleotides in length that are capable of regulating gene expression. Most miRNAs are transcribed from DNA into primary sequences that are processed to give rise to mature miRNAs [58]. In most cases, miRNAs are loaded onto the Argonaute protein to form the minimal miRNA-induced silencing complex (miRISC). By base complementarity, miRNA leads to the interaction of miRISC with the 3′ untranslated region (UTR) of its target mRNA. If the complementarity is complete, the miRNA–mRNA interaction induces the endonuclease activity of Argonaute and the mRNA is degraded. In the case that the complementarity is lower, the miR–SC complex only inhibits the translation of the mRNA [59].
In the study of Qiu et al. [25] up-regulated expression of miR-518d in placenta was observed from women with GDM in comparison to the normal pregnancy group. Moreover, the authors identified increased mRNA levels of nuclear factor-kappa B (NF-κB), cytochrome C oxidase subunit II (COX-2), TNF-α, IL-1β and IL-6; and decreased mRNA levels of peroxisome proliferator-activated receptor α (PPARα) in placentas, in women with GDM. Using cultures of the human placental trophoblast cell line HTR8/SVneo transfected with miR-518d mimetics or inhibitors, the authors demonstrated that miR-518d promotes the mRNA expression of COX-2, TNF-α, IL-1β and IL-6. Furthermore, through luciferase assays, PPARα was validated as the target gene of miR-518d, and the decreased mRNA and protein levels of PPARα and its downstream genes in cultures transfected with miR-518d mimetics, suggesting that PPARα is negatively regulated by miR-518d. Inhibition of PPARα mRNA using PPARα-specific antagonist and PPARα siRNA interference resulted in increased mRNA and protein expression of NF-κB, COX-2, TNF-α, IL-1β and IL-6. Another series of experiments showed that the level of NF-κB in the nucleus was elevated with a high glucose concentration. It was reduced with the application of miR-518d inhibitors, and was increased with the knockdown of PPARα. This suggests that the development of GDM might be associated with an inflammatory response in the placenta which is regulated by miR-518d through the inhibition of PPARα and the activation of NF-κB.
Sun et al. [26] observed that the placental expression levels of miR-29b were lower in a GDM group compared to controls. Moreover, by using the trophoblast cell line HTR8/SVneo with an miR-29b mimetic or inhibitor, the authors observed that miR-29b over-expression inhibited cell growth, late apoptosis, migration and invasion; and miR-29b knockdown promoted cell migration and invasion. This suggests that, in women with GDM, the proliferation and infiltration abilities of trophoblast cells are strengthened. Using luciferase assays, the authors demonstrated that the binding site in the three prime untranslated region (3′-UTR) of hypoxia inducible factor 3A (HIF3A) is specific for miR-29b, and that miR-29b inversely regulates HIF3A expression in the trophoblast cell. In summary, miR-29b regulates trophoblast cells activities by modulating HIF3A expression.
Cao et al. [27] showed that miR-98 levels were up-regulated in placentas from women with GDM compared with normal placentas. By using JEG-3 cells transfected by miR-98 mimetics or inhibitors and measuring the global DNA methylation level, it was observed that miR-98 can positively regulate the global DNA methylation level. Further investigations revealed that methyl CpG binding protein 2 (MECP2) was the target gene of miR-98. In JEG-3 cells, the level of MECP2 protein was significantly down-regulated by an miR-98 mimetic and up-regulated by an miR-98 inhibitor, suggesting that MECP2 is negatively regulated by miR-98. These findings were supported by decreased expression of MCEP2 in placentas from women with GDM. To investigate the relationships of miR-98 and MCEP2 with the global DNA methylation level, the effects of the former two on the expression of DNA methyltransferase was evaluated. miR-98 mimetic and MECP2 siRNA increased the protein levels of DNA methyltransferase 1 (DNMT1), and MECP2 expression vector decreased DNMT1 protein level. Further investigations showed that miR-98 mimetic and MECP2 siRNA reduced the mRNA and protein levels of transient receptor potential 3 (TRPC3), one of the target genes of MECP2 involved in vasoconstriction and the regulation of blood pressure in metabolic syndrome [60], and the miR-98 inhibitor and MECP2 expression vector increased TRPC3 expression. These findings were supported by decreased mRNA and protein levels of TRPC3 in placentas from women with GDM, but only in certain age ranges. Thus, the authors demonstrated that miR-98 is involved in the occurrence of GDM through the MECP2-TRPC3 pathway.
Ding et al. [28] conducted an integrative analysis of the gene expression profiles of both mRNAs and miRNAs in placental tissues from eight women with GDM and eight controls: 281 mRNAs and 32 miRNAs with differential expression were observed. Among them, eight mRNAs (TBL1X, NOTUM, FRMD4A, SLC16A2, CLDN19, CCL18, HTRA1 and SLC39A6) and five miRNAs (miR-202-5p, miR-138-5p, miR-210-5p, miR-3158-5p and miR-4732-3p) were validated by quantitative reverse transcription polymerase chain reaction (qRT-PCR) in the entire study sample. The functional analysis based on the results of the two massive analyses identified a significant up-regulation of two biological functions: cellular development and cellular movement. Furthermore, the paired mRNA–miRNA analysis with 91 negatively expressed mRNA targets for 13 miRNAs allowed the construction of molecular interactions networks. The most enriched networks were related to cellular development and functions, organ morphology and organismal development. Notably, miR-138-5p was a central node in this network. The overexpression of miR-138-5p reduced both the migratory ability and the proliferation of HTR-8 cells, whereas the inhibition of miR-138-5p increased both the migratory ability and the proliferation of HTR-8 cells. Using luciferase assays, it was shown that miR-138-5p targets TBL1X and suppresses TBL1X expression in HTR-8 cells. Further investigations with HTR-8 cells co-transfected with miR-138-5p inhibitor and TBL1X siRNA indicated that miR-138-5p could inhibit the migration and proliferation of HTR-8 trophoblasts by targeting TBL1X. Both mRNA and protein levels of TBL1X were significantly up-regulated in GDM placentas and a significant negative correlation between the expression of miR-138-5p and TBL1X mRNA was observed. Interestingly, the placentas in women with GDM were heavier than in controls. The weight was negatively correlated with the expression of miR-138-5p and positively correlated with the mRNA levels of TBL1X. Thus, reduced expression of miR-138-5p contributed to the excessive growth of the placenta in GDM by enhancing the proliferation of trophoblasts by targeting TBL1X.
3.3. Histone Modifications
Genomic DNA is condensed into chromatin, which consists of said DNA bound to histone proteins. There are four types of histones that form an octamer in which DNA is wrapped, all of which constitutes a nucleosome. Histone tails can have chemical modifications, including acetylation, methylation and phosphorylation, among others, depending on the activity of different enzymes that add or remove such modifications [61,62]. These modifications alter the interactions of DNA with histones, which in turn can increase or decrease access to genes, thereby affecting gene expression. The most studied chemical modifications are acetylation and methylation (which can occur in the form of mono, di and tri-methylation), which occur mainly on lysine residues [63].
Hepp et al. [29] analyzed the levels of lysine acetylation of histones H3 at the H3K9 site (lysine at position 9 of histone H3) and the levels of lysine tri-methylation at the H3K4 site in nuclei of villous syncytiothrophoblast cells (SCT) and extra villous trophoblast cells (EVT) from term placentas from GDM women and controls. In both SCT and EVT cells, the levels of lysine acetylation at the H3K9 site were lower in GDM women in comparison to controls. The levels of lysine tri-methylation at H3K4 site were similar between the groups. Since an association between vitamin D deficiency and GDM has been documented [64], the authors, using cell cultures, evaluated the effects of different concentrations of vitamin D on acetylation levels of lysine at the H3K9 site, and observed that acetylation levels were not affected by low doses of calcitriol and decreased slightly at the highest concentration.
4. Epigenetic Alterations Determined in Adipose Tissue
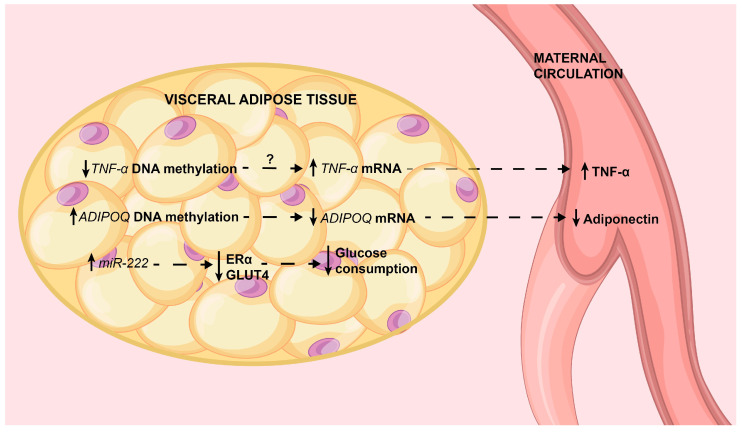
The main epigenetic alterations in adipose tissue are summarized in Figure 2.
Figure 2.
The main epigenetic alterations in visceral adipose tissue associated with GDM. Epigenetic alterations and their effects on target genes are shown (see the text for deeper details). These findings explain in part the well-known pro-inflammatory status, decreased production of adiponectin and insulin resistance in visceral adipose tissue. In tissue, solid line arrows indicate or up-regulation or down-regulation; in maternal circulation they indicate low or high circulating levels. Dotted arrows indicate associations. TNF-α: tumoral necrosis factor alpha; ADIPOQ: adiponectin; ERα: estrogen receptor alpha; GLUT4: glucose transporter 4.
4.1. DNA Methylation
Rancourt et al. [30] evaluated the mRNA expression of TNF-α and suppressor of cytokine signaling 3 (SOCS3), two important components in the inflammation and apoptosis processes in adipose tissue [65,66], and the methylation in their promoters in subcutaneous and visceral adipose tissues from women with GDM in comparison with women with NGT. It was observed that the mRNA expression levels of TNF-α and SOCS3 were significantly higher only in visceral adipose tissue from women with GDM compared to controls. Curiously, circulating maternal TNF-α levels correlated with both visceral adipose tissue TNF-α and SOC3 mRNA levels. In visceral adipose tissue, the methylation analysis showed decreased methylation percentages for three CpGs sites of the TNF-α promoter in the GDM group, and the methylation on the SOCS3 promoter was not different between the groups. No significant correlation was observed between methylation and mRNA levels of TNF-α. Assuming that TNF-α mRNA levels are not regulated by promotor methylation, there are other mechanisms that can regulate this transcript, such as the repressive effect of miR-19a-3p or the transcript stabilization by Hu antigen R [67].
Hypoadiponectinemia has been observed in people with insulin resistance, T2DM, GDM and obesity [68,69]. Adipose tissue is the main source of adiponectin, and recently, it has been suggested that the production of this hormone is greater in subcutaneous adipose tissue than in visceral adipose tissue [70]. This motivated Ott et al. [31] to evaluate adiponectin plasma concentrations and mRNA and DNA methylation levels in blood cells, subcutaneous adipose tissue and visceral adipose tissue from women with GDM compared to controls. Women with GDM showed hypoadiponectinemia accompanied by decreased mRNA levels in both adipose tissue types. There were greater differences between visceral adipose tissues than subcutaneous adipose tissues. Maternal adiponectin levels positively correlated with mRNA levels from both visceral and subcutaneous adipose tissues, but the correlation was stronger with subcutaneous adipose tissue mRNA levels. Four CpGs sites were at the ADIPOQ locus had different methylation levels (two in visceral adipose tissue and two in maternal blood) in the groups, but only the R3 CpG1 site in visceral adipose tissue, which showed greater methylation in GDM, inversely correlated with mRNA levels of adiponectin. This study highlights the importance of the two types of adipose tissue in the pathophysiology of GDM.
Deng et al. [32] while analyzing the global DNA methylation and whole genome expression in visceral omental adipose tissue from women with GDM and controls, observed 485 down-regulated and 485 up-regulated genes, and 1298 hypomethylated genes and 1568 hypermethylated genes in the GDM group. After integrating the data from the two massive techniques and validating the results, only three genes (HLA-DMB, MSLN and HSPA6) showed significantly different methylation and expression levels, and only MSLN showed a strong negative correlation between expression and methylation levels. Based on the integration of Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Genomes database pathway analysis of expression and methylation profiles, the authors found that the antigen processing and presentation pathway and immune-related genes were closely associated with GDM in visceral adipose tissue.
4.2. MiRNAs
Shi et al. [33] while analyzing the global expression of miRNAs in omental adipose tissue in women with GDM against controls, observed differential expression of 17 miRNAs. With qRT-PCR, only the increased expression of miR-222 in GDM was validated. Interestingly, the authors observed that the expression of miR-222 negatively correlated with the protein expression of estrogen receptor alpha (ERα) and glucose transporter 4 (GLUT4) in this same tissue. When observing that estradiol levels were higher in the GDM group, the authors used 3T3-L1 murine adipocyte cultures and observed that the increase in the concentration of 17β-estradiol in the medium increased the gene expression of miR-222 and decreased the protein expression of ERα and GLUT4. Through silencing assays in this same line cell, the authors observed that the silencing of miR-222 resulted in increased protein expression of ERα (later, by luciferase assays, it was shown that miR-222 was capable of binding to the 3′-UTR of ERα mRNA) and augmented glucose consumption after insulin stimulation—about 40% higher than in non-silenced cells. This was due to higher synthesis and translocation of GLUT-4. Thus, the authors propose that miR-222 plays an important role in insulin resistance through the regulation of ERα and GLUT-4.
To our knowledge, there are no studies on histone chemical modifications in adipose tissue associated with GDM.
5. Epigenetic Alterations Determined in Blood
It is important to point out that in most studies that have analyzed epigenetic profiles in blood, whether by analyzing DNA, histones or miRNAs, a relationship between the findings and the pathophysiology of GDM could not be established. Therefore, the studies are limited to describing epigenetic characteristics associated with GDM.
5.1. DNA Methylation
Dias et al. [34] evaluated the peripheral blood DNA methylation patterns in women with GDM and in NGT sampled around week 19 of gestation, and identified 1046 differentially methylated CpG sites (148 were hypermethylated and 898 were hypomethylated) in those women who developed GDM. The authors selected the top five significantly differentially methylated CpG sites for further analysis. These five CpG sites were associated with four unique genes, including solute carrier family 9 member A3 (SLC9A3), male-enhanced antigen 1 and kelch domain-containing protein 3 (MEA1 and KLHDC3), calmodulin binding transcription activator 1 (CAMTA1), RAS P21 protein activator 3 (RASA3) and one unknown gene. The KEGG analysis identified canonical pathways related to signal transduction, cell growth, proliferation, differentiation and apoptosis, insulin resistance, glucose metabolism, inflammation, neurological signaling and oncogenesis. The GO analysis identified biological processes associated with structural organization and development, and molecular functions associated with regulatory or binding activities. In this study, pyrosequencing validation was not performed, so these potential epigenetic markers need to be validated longitudinally in a larger population.
Wu et al. [35] in gestational weeks 12–16, analyzed the global maternal blood DNA methylation patterns in a cohort of 22 women (11 who developed GDM, and their respective matched controls) and identified 100 CpG sites (comprising 66 genes) differentially methylated between women who developed GDM and healthy pregnant women. After using rigorous criteria for the absolute β-value differences across all 11 matched pairs and validating by bisulfite pyrosequencing (BSP), the constitutive photomorphogenic homolog subunit 8 (COPS8), phosphoinositide-3-kinase, regulatory subunit 5 (PIK3R5), 3-hydroxyanthranilate 3,4-dioxygenase (HAAO), chromosome 5 open reading frame 34 (C5orf34) and coiled-coil domain containing 124 (CCDC124) genes, each with varied functions, were the only ones that maintained differential methylation between the groups. Thus, the authors proposed that changes in the methylation of these genes before the diagnosis of GDM have potential use as predictive biomarkers of this disorder, although they did not perform any type of analysis in this regard.
5.2. Histone Chemical Modifications
Michalczyk et al. [36] analyzed the levels of lysine di-methylation of histones H3 at sites H3K27 (lysine at position 27 of histone H3), H3K4, H3K79, H3K36 and H3K9 in DNA from white blood cells from pregnant women evaluated at week 30 of gestation and at weeks 8–10 and 20 postpartum. The women were classified as non-diabetic, GDM who did not develop T2DM, GDM who developed T2DM and women with pre-existing T2DM. Just focusing on the results on GDM, the authors observed that di-methylation in H3K4 was approximately 80% lower in women with GDM who developed T2DM compared to women with GDM who did not develop T2DM by week 10 (roughly) postpartum, and that di-methylation in H3K27 was approximately 50% lower in women with GDM who developed T2DM compared to women with GDM who did not develop T2DM by week 20 postpartum. Thus, the authors propose that these changes in histone methylation have potential utility as predictive biomarkers for the development of T2DM in women with GDM, but they did not carry out analyses to support this proposal.
5.3. MiRNAs
Zhao et al. [37] analyzed miRNAs in maternal serum from women who developed GDM and their respective controls. Both groups were sampled in one gestational week from 16 to 19. They identified significantly lower expression of miR-132, miR-29a and miR-222 in women who developed GDM, but only miR-29a and miR-222 were externally validated. Further investigations in HepG2 cells showed that insulin-induced gene 1 (INSIG1) was the target gene of miR-29a and that the knockdown of miR-29a increases the protein levels of INSIG1. Since INSIG1 has become related to sterol regulatory element-binding protein (SREBP)-mediated regulation of phosphoenolpyruvate carboxy kinase 2 (PCK2), a key enzyme in hepatic gluconeogenesis, the authors evaluated the expression of PCK2 in this same knockdown model and observed increased mRNA levels of PCK2. This suggests that miR-29a is a negative regulator of serum glucose. A recent study supports the observation that GDM is characterized by lower circulating levels of miR-29a [71].
Zhu et al. [38] constructed two small circulating RNA libraries from pooled plasma, one from women who subsequently developed GDM and another from controls, all sampled in gestational a week from 16 to 19. The authors identified 32 miRNAs with differential expression; however, only five miRNAs (hsa-miR-16-5p, hsa-miR-17-5p, hsa-miR-19a-3p, hsa-miR-19b-3p and hsa-miR-20a-5p) with increased expression levels in GDM were validated by qRT-PCR. The target prediction, GO analysis and pathway identification of these five miRNAs allowed the construction of a regulatory network with seven target genes (MAPK-1, IRS-1, IRS-2, SOS-1, SMAD5, SMAD4 and AKT3) and five signaling pathways (mitogen-activated protein kinase, insulin, type 2 diabetes mellitus, transforming growth factor β and mammalian target of rapamycin) for GDM. The results of Zhu et al. [38] regarding hsa-miR-20a-5p differ from those of Pheiffer et al. [39] who observed lower circulating levels of hsa-miR-20a-5p in women with GDM compared with controls; moreover, this miRNA and one or more risk factor were significant predictors of GDM.
Qiu et al. [25] while analyzing the expression levels of miR-518d in peripheral plasma and placentas from women with normal pregnancies and women with GDM, sampled between the 36th and 42th week of pregnancy, observed up-regulated expression of miR-518d in plasma from women with GDM in comparison to controls.
6. Conclusions
Several studies have investigated epigenetic changes in the placentas, adipose tissue and peripheral blood of GDM patients. Some authors have found alterations in the methylation of genes that modulate insulin resistance during pregnancy. Others have found alterations in miRNAs involved in inflammation, insulin resistance and metabolism. Notably, other epigenetic mechanisms, such as chemical histone modification, have been reported as predictive biomarkers for the development of T2DM at postpartum. These findings suggest that epigenetic modifications are related to the etiology of GDM and to its progression to T2DM. However, causality was not established in the majority of the studies, and it is therefore not clear whether epigenetic changes predate GDM. Interestingly, some epigenetic changes correlate with glucose levels and may be indicators of metabolic dysfunction. On the other hand, most studies did not explore whether epigenetic modifications change gene expression, and did not validate their findings. Therefore, additional studies seeking causality are necessary to elucidate the underlying mechanisms for the epigenetic changes and to find out how epigenetic changes regulate pathways crucial for the pathophysiology of GDM.
Acknowledgments
J.V.-O., R.S., M.A.S.-R. and E.G.R.M. hold fellowship positions in the National System of Investigators, CONACYT.
Author Contributions
All authors drafted the manuscript and provided significant intellectual contributions in the critical review of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.American Diabetes Association 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes—2020. Diabetes Care. 2020;43:S14–S31. doi: 10.2337/dc20-S002. [DOI] [PubMed] [Google Scholar]
- 2.McIntyre H.D., Catalano P., Zhang C., Desoye G., Mathiesen E.R., Damm P. Gestational diabetes mellitus. Nat. Rev. Dis. Primers. 2019;5:47. doi: 10.1038/s41572-019-0098-8. [DOI] [PubMed] [Google Scholar]
- 3.Bellamy L., Casas J.-P., Hingorani A., Williams D. Type 2 diabetes mellitus after gestational diabetes: A systematic review and meta-analysis. Lancet. 2009;373:1773–1779. doi: 10.1016/S0140-6736(09)60731-5. [DOI] [PubMed] [Google Scholar]
- 4.Catalano P.M. The impact of gestational diabetes and maternal obesity on the mother and her offspring. J. Dev. Orig. Heal. Dis. 2010;1:208–215. doi: 10.1017/S2040174410000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catalano P.M., Kirwan J.P., Haugel-de Mouzon S., King J. Gestational Diabetes and Insulin Resistance: Role in Short- and Long-Term Implications for Mother and Fetus. J. Nutr. 2003;133:1674S–1683S. doi: 10.1093/jn/133.5.1674S. [DOI] [PubMed] [Google Scholar]
- 6.Feil R., Fraga M. Epigenetics and the environment: Emerging patterns and implications. Nat. Rev. Genet. 2012;13:97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 7.Lacal I., Ventura R. Epigenetic Inheritance: Concepts, Mechanisms and Perspectives. Front. Mol. Neurosci. 2018;11:292. doi: 10.3389/fnmol.2018.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phelps R.L., Bergenstal R., Freinkel N., Rubenstein A.H., Metzger B.E., Mako M. Carbohydrate Metabolism in Pregnancy: XIII. Relationships Between Plasma Insulin and Proinsulin During Late Pregnancy in Normal and Diabetic Subjects. J. Clin. Endocrinol. Metab. 1975;41:1085–1091. doi: 10.1210/jcem-41-6-1085. [DOI] [PubMed] [Google Scholar]
- 9.Ryan E.A., O’Sullivan M.J., Skyler J.S. Insulin Action During Pregnancy: Studies with the Euglycemic Glucose Clamp Technique. Diabetes. 1985;34:380–389. doi: 10.2337/diab.34.4.380. [DOI] [PubMed] [Google Scholar]
- 10.McIntyre H.D. Discovery, Knowledge, and Action—Diabetes in Pregnancy Across the Translational Spectrum: The 2016 Norbert Freinkel Award Lecture. Diabetes Care. 2018;41:227–232. doi: 10.2337/dci17-0056. [DOI] [PubMed] [Google Scholar]
- 11.Catalano P.M., Tyzbir E.D., Roman N.M., Amini S.B., Sims E.A. Longitudinal changes in insulin release and insulin resistance in nonobese pregnant women. Am. J. Obstet. Gynecol. 1991;165:1667–1672. doi: 10.1016/0002-9378(91)90012-G. [DOI] [PubMed] [Google Scholar]
- 12.Institute of Medicine (US) Committee on Nutritional Status During Pregnancy and Lactation . Nutrition During Pregnancy. National Academies Press; Washington, DC, USA: 1990. pp. 121–134. [Google Scholar]
- 13.Kwon H.J., Pessin J.E. Adipokines Mediate Inflammation and Insulin Resistance. Front. Endocrinol. 2013;4:71. doi: 10.3389/fendo.2013.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plows J.F., Stanley J.L., Baker P.N., Reynolds C.M., Vickers M.H. The Pathophysiology of Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2018;19:3342. doi: 10.3390/ijms19113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchanan T.A. Pancreatic B-Cell Defects in Gestational Diabetes: Implications for the Pathogenesis and Prevention of Type 2 Diabetes. J. Clin. Endocrinol. Metab. 2001;86:989–993. doi: 10.1210/jcem.86.3.7339. [DOI] [PubMed] [Google Scholar]
- 16.Lekva T., Norwitz E.R., Aukrust P., Ueland T. Impact of Systemic Inflammation on the Progression of Gestational Diabetes Mellitus. Curr. Diabetes Rep. 2016;16:1–11. doi: 10.1007/s11892-016-0715-9. [DOI] [PubMed] [Google Scholar]
- 17.Catalano P.M., Huston L., Amini S.B., Kalhan S.C. Longitudinal changes in glucose metabolism during pregnancy in obese women with normal glucose tolerance and gestational diabetes mellitus. Am. J. Obstet. Gynecol. 1999;180:903–916. doi: 10.1016/S0002-9378(99)70662-9. [DOI] [PubMed] [Google Scholar]
- 18.Lesseur C., Armstrong D.A., Paquette A.G., Li Z., Padbury J.F., Marsit C.J. Maternal obesity and gestational diabetes are associated with placental leptin DNA methylation. Am. J. Obstet. Gynecol. 2014;211:654.e1–654.e9. doi: 10.1016/j.ajog.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagné-Ouellet V., Breton E., Thibeault K., Fortin C.-A., Cardenas A., Guérin R., Perron P., Hivert M.-F., Bouchard L. Mediation Analysis Supports a Causal Relationship between Maternal Hyperglycemia and Placental DNA Methylation Variations at the Leptin Gene Locus and Cord Blood Leptin Levels. Int. J. Mol. Sci. 2020;21:329. doi: 10.3390/ijms21010329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchard L., Hivert M.-F., Guay S.-P., St-Pierre J., Perron P., Brisson D. Placental Adiponectin Gene DNA Methylation Levels Are Associated With Mothers’ Blood Glucose Concentration. Diabetes. 2012;61:1272–1280. doi: 10.2337/db11-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blazevic S., Horvaticek M., Kesic M., Zill P., Hranilovic D., Ivanisevic M., Desoye G., Stefulj J. Epigenetic adaptation of the placental serotonin transporter gene (SLC6A4) to gestational diabetes mellitus. PLoS ONE. 2017;12:e0179934. doi: 10.1371/journal.pone.0179934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Côté S., Gagné-Ouellet V., Guay S.-P., Allard C., Houde A.-A., Perron P., Baillargeon J.-P., Gaudet D., Guérin R., Brisson D., et al. PPARGC1α gene DNA methylation variations in human placenta mediate the link between maternal hyperglycemia and leptin levels in newborns. Clin. Epigenetics. 2016;8:1–13. doi: 10.1186/s13148-016-0239-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L., Fan H., Zhou L., Wu Y., Lu H., Luo J. Altered expression of PGC-1 α and PDX1 and their methylation status are associated with fetal glucose metabolism in gestational diabetes mellitus. Biochem. Biophys. Res. Commun. 2018;501:300–306. doi: 10.1016/j.bbrc.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Houde A.A., St-Pierre J., Hivert M.F., Baillargeon J.P., Perron P., Gaudet D., Brisson D., Bouchard L. Placental lipoprotein lipase DNA methylation levels are associated with gestational diabetes mellitus and maternal and cord blood lipid profiles. J. Dev. Orig. Heal. Dis. 2014;5:132–141. doi: 10.1017/S2040174414000038. [DOI] [PubMed] [Google Scholar]
- 25.Qiu H., Liu X., Yao S., Zhou J., Zhang X., Du J. Regulation and Mechanism of miR-518d through the PPARα-Mediated NF-κB Pathway in the Development of Gestational Diabetes Mellitus. J. Diabetes Res. 2020;2020:7019597. doi: 10.1155/2020/7019597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun D.-G., Tian S., Zhang L., Hu Y., Guan C.-Y., Ma X., Xia H.-F. The miRNA-29b Is Downregulated in Placenta During Gestational Diabetes Mellitus and May Alter Placenta Development by Regulating Trophoblast Migration and Invasion Through a HIF3A-Dependent Mechanism. Front. Endocrinol. 2020;11:169. doi: 10.3389/fendo.2020.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao J.-L., Zhang L., Li J., Tian S., Lv X.-D., Wang X.-Q., Su X., Li Y., Hu Y., Ma X., et al. Up-regulation of miR-98 and unraveling regulatory mechanisms in gestational diabetes mellitus. Sci. Rep. 2016;6:32268. doi: 10.1038/srep32268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding R., Guo F., Zhang Y., Liu X.-M., Xiang Y.-Q., Zhang C., Liu Z.-W., Sheng J.-Z., Huang H.-F., Zhang J.Y., et al. Integrated Transcriptome Sequencing Analysis Reveals Role of miR-138-5p/ TBL1X in Placenta from Gestational Diabetes Mellitus. Cell. Physiol. Biochem. 2018;51:630–646. doi: 10.1159/000495319. [DOI] [PubMed] [Google Scholar]
- 29.Hepp P., Hutter S., Knabl J., Hofmann S., Kuhn C., Mahner S., Jeschke U. Histone H3 lysine 9 acetylation is downregulated in GDM Placentas and Calcitriol supplementation enhanced this effect. Int. J. Mol. Sci. 2018;19:4061. doi: 10.3390/ijms19124061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rancourt R.C., Ott R., Ziska T., Schellong K., Melchior K., Henrich W., Plagemann A. Visceral Adipose Tissue Inflammatory Factors (TNF-Alpha, SOCS3) in Gestational Diabetes (GDM): Epigenetics as a Clue in GDM Pathophysiology. Int. J. Mol. Sci. 2020;21:479. doi: 10.3390/ijms21020479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott R., Stupin J.H., Melchior K., Schellong K., Ziska T., Dudenhausen J.W., Henrich W., Rancourt R.C., Plagemann A. Alterations of adiponectin gene expression and DNA methylation in adipose tissues and blood cells are associated with gestational diabetes and neonatal outcome. Clin. Epigenetics. 2018;10:1–12. doi: 10.1186/s13148-018-0567-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng X., Yang Y., Sun H., Qi W., Duan Y., Qian Y. Analysis of whole genome-wide methylation and gene expression profiles in visceral omental adipose tissue of pregnancies with gestational diabetes mellitus. J. Chin. Med. Assoc. 2018;81:623–630. doi: 10.1016/j.jcma.2017.06.027. [DOI] [PubMed] [Google Scholar]
- 33.Shi Z., Zhao C., Guo X., Ding H., Cui Y., Shen R., Liu J. Differential Expression of MicroRNAs in Omental Adipose Tissue From Gestational Diabetes Mellitus Subjects Reveals miR-222 as a Regulator of ERα Expression in Estrogen-Induced Insulin Resistance. Endocrinology. 2014;155:1982–1990. doi: 10.1210/en.2013-2046. [DOI] [PubMed] [Google Scholar]
- 34.Dias S., Adam S., Rheeder P., Louw J., Pheiffer C. Altered Genome-Wide DNA Methylation in Peripheral Blood of South African Women with Gestational Diabetes Mellitus. Int. J. Mol. Sci. 2019;20:5828. doi: 10.3390/ijms20235828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu P., Farrell W.E., Haworth K.E., Emes R.D., Kitchen M.O., Glossop J.R., Hanna F.W., Fryer A.A. Maternal genome-wide DNA methylation profiling in gestational diabetes shows distinctive disease-associated changes relative to matched healthy pregnancies. Epigenetics. 2018;13:122–128. doi: 10.1080/15592294.2016.1166321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michalczyk A.A., Dunbar J.A., Janus E.D., Best J.D., Ebeling P.R., Ackland M.J., Asproloupos D., Ackland M.L. Epigenetic markers to predict conversion from gestational diabetes to type 2 diabetes. J. Clin. Endocrinol. Metab. 2016;101:2396–2404. doi: 10.1210/jc.2015-4206. [DOI] [PubMed] [Google Scholar]
- 37.Zhao C., Dong J., Jiang T., Shi Z., Yu B., Zhu Y., Chen D., Xu J., Huo R., Dai J., et al. Early Second-Trimester Serum MiRNA Profiling Predicts Gestational Diabetes Mellitus. PLoS ONE. 2011;6:e23925. doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y., Tian F., Li H., Zhou Y., Lu J., Ge Q. Profiling maternal plasma microRNA expression in early pregnancy to predict gestational diabetes mellitus. Int. J. Gynecol. Obstet. 2015;130:49–53. doi: 10.1016/j.ijgo.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Pheiffer C., Dias S., Rheeder P., Adam S. Decreased Expression of Circulating miR-20a-5p in South African Women with Gestational Diabetes Mellitus. Mol. Diagn. Ther. 2018;22:345–352. doi: 10.1007/s40291-018-0325-0. [DOI] [PubMed] [Google Scholar]
- 40.Newbern D., Freemark M. Placental hormones and the control of maternal metabolism and fetal growth. Curr. Opin. Endocrinol. Diabetes Obes. 2011;18:409–416. doi: 10.1097/MED.0b013e32834c800d. [DOI] [PubMed] [Google Scholar]
- 41.Szukiewicz D. Cytokines in Placental Physiology and Disease. Mediat. Inflamm. 2012;2012:1–2. doi: 10.1155/2012/640823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallace J.M., Horgan G.W., Bhattacharya S. Placental weight and efficiency in relation to maternal body mass index and the risk of pregnancy complications in women delivering singleton babies. Placenta. 2012;33:611–618. doi: 10.1016/j.placenta.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Espinoza I., Smith R.F., Gillmer M., Schidlmeir A., Hockaday T.D. High levels of growth hormone and human placental lactogen in pregnancy complicated by diabetes. Diabetes Res. 1986;3:119–125. [PubMed] [Google Scholar]
- 44.Lassance L., Haghiac M., Minium J., Catalano P., Hauguel-de Mouzon S.H.-D. Obesity-Induced Down-Regulation of the Mitochondrial Translocator Protein (TSPO) Impairs Placental Steroid Production. J. Clin. Endocrinol. Metab. 2015;100:E11–E18. doi: 10.1210/jc.2014-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S.R., Choi W.-Y., Heo J.H., Huh J., Kim G., Lee K.-P., Kwun H.-J., Shin H.-J., Baek I.-J., Hong E.-J. Progesterone increases blood glucose via hepatic progesterone receptor membrane component 1 under limited or impaired action of insulin. Sci. Rep. 2020;10:1–11. doi: 10.1038/s41598-020-73330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pantham P., Aye I., Powell T. Inflammation in maternal obesity and gestational diabetes mellitus. Placenta. 2015;36:709–715. doi: 10.1016/j.placenta.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aires M.B., Dos Santos A.C.V. Effects of maternal diabetes on trophoblast cells. World J. Diabetes. 2015;6:338–344. doi: 10.4239/wjd.v6.i2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziller M.J., Gu H., Müller F., Donaghey J., Tsai L.T.Y., Kohlbacher O., De Jager P.L., Rosen E.D., Bennett D.A., Bernstein B.E., et al. Charting a dynamic DNA methylation landscape of the human genome. Nat. Cell Biol. 2013;500:477–481. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin B., Ernst J., Tiedemann R.L., Xu H., Sureshchandra S., Kellis M., Dalton S., Liu C., Choi J.-H., Robertson K.D. Linking DNA Methyltransferases to Epigenetic Marks and Nucleosome Structure Genome-wide in Human Tumor Cells. Cell Rep. 2012;2:1411–1424. doi: 10.1016/j.celrep.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lev Maor G.L., Yearim A., Ast G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015;31:274–280. doi: 10.1016/j.tig.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Gutierrez-Arcelus M., Ongen H., Lappalainen T., Montgomery S.B., Buil A., Yurovsky A., Bryois J., Padioleau I., Romano L., Planchon A., et al. Tissue-Specific Effects of Genetic and Epigenetic Variation on Gene Regulation and Splicing. PLoS Genet. 2015;11:e1004958. doi: 10.1371/journal.pgen.1004958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Defossez P.-A., Stancheva I. Biological Functions of Methyl-CpG-Binding Proteins. Prog. Mol. Biol. Transl. Sci. 2011;101:377–398. doi: 10.1016/b978-0-12-387685-0.00012-3. [DOI] [PubMed] [Google Scholar]
- 53.Hite K.C., Adams V.H., Hansen J.C. Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 2009;87:219–227. doi: 10.1139/O08-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bahar Halpern K., Vana T., Walker M.D. Paradoxical Role of DNA Methylation in Activation of FoxA2 Gene Expression during Endoderm Development. J. Biol. Chem. 2014;289:23882–23892. doi: 10.1074/jbc.M114.573469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gellersen B., Kempf R. Human Prolactin Gene Expression: Positive Correlation between Site-Specific Methylation and Gene Activity in a Set of Human Lymphoid Cell Lines. Mol. Endocrinol. 1990;4:1874–1886. doi: 10.1210/mend-4-12-1874. [DOI] [PubMed] [Google Scholar]
- 56.Crane J.D., Palanivel R., Mottillo E.P., Bujak A.L., Wang H., Ford R.J., Collins A., Blümer R.M., Fullerton M.D., Yabut J.M., et al. Inhibiting peripheral serotonin synthesis reduces obesity and metabolic dysfunction by promoting brown adipose tissue thermogenesis. Nat. Med. 2015;21:166–172. doi: 10.1038/nm.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe H., Nakano T., Saito R., Akasaka D., Saito K., Ogasawara H., Minashima T., Miyazawa K., Kanaya T., Takakura I., et al. Serotonin Improves High Fat Diet Induced Obesity in Mice. PLoS ONE. 2016;11:e0147143. doi: 10.1371/journal.pone.0147143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jo M.H., Shin S., Jung S.-R., Kim E., Song J.-J., Hohng S. Human Argonaute 2 Has Diverse Reaction Pathways on Target RNAs. Mol. Cell. 2015;59:117–124. doi: 10.1016/j.molcel.2015.04.027. [DOI] [PubMed] [Google Scholar]
- 60.Liu D., Zhu Z., Tepel M. The Role of Transient Receptor Potential Channels in Metabolic Syndrome. Hypertens. Res. 2008;31:1989–1995. doi: 10.1291/hypres.31.1989. [DOI] [PubMed] [Google Scholar]
- 61.Vaquero A., Loyola A., Reinberg D. The constantly changing face of chromatin. Sci. Aging Knowl. Environ. 2003;2003:4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 62.Strahl B.D., Allis C.D. The language of covalent histone modifications. Nat. Cell Biol. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 63.Lee H.-T., Oh S., Ro D.H., Yoo H., Kwon Y.-W. The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis. J. Lipid Atheroscler. 2020;9:419–434. doi: 10.12997/jla.2020.9.3.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soheilykhah S., Mojibian M., Rashidi M., Rahimi-Saghand S., Jafari F. Maternal Vitamin D Status in Gestational Diabetes Mellitus. Nutr. Clin. Pract. 2010;25:524–527. doi: 10.1177/0884533610379851. [DOI] [PubMed] [Google Scholar]
- 65.Liu Z., Gan L., Zhou Z., Jin W., Sun C. SOCS3 promotes inflammation and apoptosis via inhibiting JAK2/STAT3 signaling pathway in 3T3-L1 adipocyte. Immunobiology. 2015;220:947–953. doi: 10.1016/j.imbio.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 66.Hasan A., Akhter N., Al-Roub A., Thomas R., Kochumon S., Wilson A., Koshy M., Al-Ozairi E., Al-Mulla F., Ahmad R. TNF-α in Combination with Palmitate Enhances IL-8 Production via The MyD88- Independent TLR4 Signaling Pathway: Potential Relevance to Metabolic Inflammation. Int. J. Mol. Sci. 2019;20:4112. doi: 10.3390/ijms20174112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q., Roy B., Turecki G., Shelton R.C., Dwivedi Y. Role of Complex Epigenetic Switching in Tumor Necrosis Factor-α Upregulation in the Prefrontal Cortex of Suicide Subjects. Am. J. Psychiatry. 2018;175:262–274. doi: 10.1176/appi.ajp.2017.16070759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weyer C., Funahashi T., Tanaka S., Hotta K., Matsuzawa Y., Pratley R.E., Tataranni P.A. Hypoadiponectinemia in Obesity and Type 2 Diabetes: Close Association with Insulin Resistance and Hyperinsulinemia. J. Clin. Endocrinol. Metab. 2001;86:1930–1935. doi: 10.1210/jcem.86.5.7463. [DOI] [PubMed] [Google Scholar]
- 69.Fasshauer M., Blüher M., Stumvoll M. Adipokines in gestational diabetes. Lancet Diabetes Endocrinol. 2014;2:488–499. doi: 10.1016/S2213-8587(13)70176-1. [DOI] [PubMed] [Google Scholar]
- 70.Meyer L.K., Ciaraldi T.P., Henry R.R., Wittgrove A.C., Phillips S.A. Adipose tissue depot and cell size dependency of adiponectin synthesis and secretion in human obesity. Adipocyte. 2013;2:217–226. doi: 10.4161/adip.24953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng L., Huang Y., Li L., Chen H., Su J. Serum miR-29a/b expression in gestational diabetes mellitus and its influence on prognosis evaluation. J. Int. Med. Res. 2020;48:300060520954763. doi: 10.1177/0300060520954763. [DOI] [PMC free article] [PubMed] [Google Scholar]