Abstract
Type-specific serologic assays for herpes simplex virus (HSV) types 1 and 2 based on glycoprotein G-1 (gG-1) (HSV-1) and gG-2 (HSV-2) discriminate between antibodies against HSV-1 and HSV-2. We previously developed a Western blot assay using gG-1 and gG-2 expressed in baculovirus, performed extensive validation studies, and determined that it was both sensitive and specific for type-specific detection of HSV antibody. Here we report that, among a cohort of Thai military recruits, the serostatus of some individuals changed from positive to negative over time (6.6% among those ever positive for HSV-1, and 14.9% among those ever positive for HSV-2). We tested a subset of these specimens in three other gG-based assays: an enzyme-linked immunosorbent assay, an immunoblot strip assay, and a Western blot assay. Positive-to-negative shifts occurred in every assay; the frequency of the shifts ranged from 6.1% to 21.2% of the specimen sets tested. There was only limited agreement among the assays concerning which individuals lost reactivity. This inaccuracy, exhibited by all of the assay protocols, was not predicted by validation studies employing specimens from cross-sectional studies and was most pronounced in HSV-2 testing. This argues for the inclusion of serial blood specimens in serologic assay validation procedures.
Herpes simplex virus types 1 (HSV-1) and 2 (HSV-2) have approximately 83% nucleotide sequence similarity (7) and have as much as 85% amino acid sequence identity for some proteins (16). As a result, HSV-1 and HSV-2 show extensive serologic cross-reactivity (27). This has impeded seroepidemiologic studies of the two viruses. The discovery of glycoprotein G (gG) in the mid-1980s seemed to resolve this difficulty (19, 23), because it is antigenically distinct between HSV-1 and HSV-2. The only significant amino acid similarity between gG-1 (HSV-1) and gG-2 (HSV-2) occurs at the signal sequence and in the short membrane anchor (20). Since the signal sequence is removed during posttranslational processing (21) and the membrane anchor is sequestered from the immune system by the lipid bilayer, only the unrelated portions of the gG molecules are available as antigenic epitopes.
Several type-specific serologic assays based on gG-1 and gG-2 have been developed. These include the immunodot (17, 18), HSV-infected cell-based Western blotting (4), baculovirus-expressed gG immunoblotting (26), enzyme-linked immunosorbent assay (ELISA) (9), and immunoblot strip (1) methods. Western blotting of HSV-infected cell lysates (4) is regarded as the most reliable method, since it examines the antibody response to other immunogenic HSV proteins in addition to gG. Although there is cross-reactivity between HSV-1 and HSV-2 for these other proteins, differences in size result in distinct migration patterns in the Western blot format. This form of HSV-specific Western blotting detected HSV-2 antibodies in some sera that were not detected by a purified-gG-based ELISA (24).
Cross-sectional and clinic-based studies using the baculovirus-expressed gG-based immunoblot assays were consistent with other published findings (3, 5, 10–13, 15, 24, 29) in that comparable prevalences were observed in similar populations. However, when the assay was used in cohort studies, results for some participants shifted from seropositive to seronegative for either HSV-1 or HSV-2 over time.
Published HSV cohort studies using type-specific gG-based assays (3, 5, 10–13, 15, 24, 29) have been conducted largely in clinical settings (3, 11, 29). Although loss of antibody reactivity has not been observed in studies of clinic attendees, those results cannot be generalized to nonclinical populations, because they involve self-selected participants, usually with genital herpes disease. The aim of the present study was to characterize the underlying cause of shifts in HSV serostatus observed in this gG-based immunoblot assay and to determine whether similar shifts could be observed in other type-specific serologic assays for HSV.
MATERIALS AND METHODS
gG-based HSV assay protocols.
Four different gG-based assays were employed in this study. Each was performed blinded with regard to the results of other tests.
BIB.
Antigens used in the baculovirus-expressed gG-based immunoblot (BIB) assay were produced in Sf9 insect cells infected with baculoviruses expressing either gG-1 or gG-2 (25). The assay was previously validated against other type-specific HSV serologic methods (26). The published protocol was modified as follows. gG was partially purified by extraction in a buffer consisting of 50 mM Tris (pH 8.0), 500 mM NaCl, 1% Nonidet P-40, 100 μg of phenylmethylsulfonyl fluoride per ml, and 1 μg of aprotinin per ml. Infected Sf9 cells were scraped from confluent T150 flasks and pelleted, suspended in extraction buffer, sonicated three times for 30 s each on ice (output control, 4; duty cycle, 50%) in a cup horn sonicator (model W-375; Heat Systems-Ultrasonic, Inc., Farmingdale, N.Y.), and centrifuged at 14,000 × g for 15 min at 4°C, and the pellet was resuspended in standard buffer. A blocking step was also added. For this purpose, uninfected Sf9 cells (approximately 2 × 107) were suspended in 2 ml of phosphate-buffered saline, sonicated and centrifuged as described above, and stored at 4°C. Fifty microliters of this lysate in a total final volume of 100 μl was added to serum specimens to reduce antibody reactivity directed against insect proteins.
HWB.
Antigens used in the HSV-infected-cell-based Western blot (HWB) assay were prepared from HSV-1 (strain F)- and HSV-2 (strain G)-infected Hep-2 cells. The protocol for this assay was modified from previously published methods (2). The antigens are separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and electrophoretically transferred to nitrocellulose prior to incubation with test sera and colorimetric development.
Purified gG ELISA.
The purified gG ELISA is under development for commercial use by Biokit, S.A. (Barcelona, Spain). Reactions were performed in wells of a 96-well plate coated with purified gG-1 (baculovirus expressed) or gG-2 (lentil lectin-purified HSV-2-infected-cell lysate). Bound antibody was detected with rabbit anti-human immunoglobulin G (IgG) conjugated with peroxidase and developed with 3,3′,5,5′-tetramethylbenzidine. A450 was measured.
SIA.
The strip immunoblot assay (SIA) was developed by Chiron Vaccines Corporation (Emeryville, Calif.) and employs nitrocellulose strips impregnated with baculovirus-expressed gG-1, gG-2 (truncated), gD-2, and vp41, in discrete bands, along with anti-IgG antibody-level control bands. Test strips were prepared according to the manufacturer’s instructions, and diluted serum was added to each tube containing a test strip and incubated for 4.5 h. Strips were washed once in their reaction tubes, transferred to a common washing vessel, and washed three times with phosphate-buffered detergent solutions provided with the test kits. Bound antibody was detected by peroxidase-conjugated goat anti-human antibody and developed with 4-chloro-2-naphthol plus H2O2. After 15 min of color development, strips were washed twice with deionized water and dried on absorbent paper in the dark. Strips were read manually within 3 h. All test kits used were from the same lot.
Source of human serum specimens.
The specimens for this study were collected from 1991 to 1993 as a part of the HIV/AIDS Collaboration in Thailand (Thailand Ministry of Public Health and the Centers for Disease Control and Prevention). Blood was collected (and sera stored) from 1,158 participants at 6-month intervals (initial specimen plus up to three follow-ups) from Thai military recruits 21 to 27 years of age (28). A total of 3,116 specimens were available for this study.
Statistical analysis.
The χ2 test was used to assess the association between laboratory parameters and shifts from positive to negative results. The agreement between pairs of methods was measured with the kappa statistic; the relative strengths of agreement are 0.81 to 1.00 for almost perfect, 0.61 to 0.80 for substantial agreement, 0.41 to 0.60 for moderate agreement, 0.21 to 0.40 for fair agreement, and 0.00 to 0.21 for slight agreement (14).
Human subject approval.
The study protocol was approved by the Committee on Ethical Review of Research on Human Subjects of the Thai Ministry of Public Health and an Institutional Review Board of the Centers for Disease Control and Prevention.
RESULTS
With the BIB assay, 902 of 1,158 recruits (77.9%) had antibody specific for gG-1 at one or more time points, and 213 (18.4%) had at least one positive HSV-2 test. Since these prevalence values were similar to those for other published findings (19), suspicions were not raised about difficulties with the assay.
However, sequential sera from some individuals gave inconsistent results (Table 1). Among the 960 participants for whom two or more serum specimens were available, 91.4% of the specimen sets were consistent (always positive or always negative) for HSV-1, and 91.8% of the sets were consistent for HSV-2. Similar proportions of consistent and inconsistent result sets were observed regardless of the number of specimens in a set that were available for testing.
TABLE 1.
Frequency of occurrence of all possible BIB assay result patterns for the participants in the Thai military recruit study
| Pattern by participant group | No. with result (%)
|
|
|---|---|---|
| HSV-1 | HSV-2 | |
| 4 specimens | ||
| Consistently positive | 239 (68.5) | 37 (10.6) |
| Consistently negative | 72 (20.6) | 276 (79.1) |
| Converters | 4 (1.1) | 17 (4.9) |
| Inconsistent | 34 (9.7) | 19 (5.4) |
| 3 specimens | ||
| Consistently positive | 206 (68.7) | 29 (9.7) |
| Consistently negative | 71 (23.7) | 248 (82.7) |
| Converters | 8 (2.7) | 9 (3.0) |
| Inconsistent | 15 (5.0) | 14 (4.7) |
| 2 specimens | ||
| Consistently positive | 221 (71.1) | 36 (11.6) |
| Consistently negative | 68 (21.9) | 255 (82.0) |
| Converters | 7 (2.3) | 15 (4.8) |
| Inconsistent | 15 (4.8) | 5 (1.6) |
| 1 specimen | ||
| Consistently positive | 153 (77.3) | 32 (16.2) |
| Consistently negative | 45 (22.7) | 166 (83.8) |
We examined several features of the test results to identify laboratory-based causes for the positive-to-negative discordance. There was no association between inconsistent results and the date the assay was performed or the individual blot on which the result was produced. A trend between protein lot number and weak positives was suggested in the raw data, but did not achieve statistical significance for either HSV-1 or HSV-2.
We also investigated the association between inconsistent test results and test color intensity. All BIB data were rescored (blinded as to the original interpretation) according to the relative intensity of the virus-specific colorimetric bands (negative, weakly positive, or strongly positive). Relative intensity was assessed subjectively by comparing each test result for a given blot with an internal strongly positive control band. Results were scored strongly positive if they were more or nearly as intense as the control band. Results were scored as weakly positive if they were substantially weaker than the internal control band. Specimen sets from every individual with more than one time point available and who tested positive for at least one serum sample were used in this analysis. For HSV-1, among sets that underwent at least one positive-to-negative shift, 41 of 64 (64.1%) included one or more tests that were scored as weak with BIB compared with 341 of 685 (49.8%) pairs that underwent no shift (χ2 = 4.77; P = 0.029). For HSV-2, 32 of 38 (84.2%) sets including a positive-to-negative shift were associated with weak-intensity bands, compared with 94 of 143 (65.7%) among pairs that did not shift (χ2 = 4.85; P = 0.028).
To determine whether the observed positive-to-negative shifts could be accounted for by day-to-day fluctuations in the performance of the assay, we selected 100 Thai study specimens from collection period 2 in numerical order, beginning at an arbitrarily chosen point in the specimen list. Three coded panels consisting of an aliquot of each of these specimens were prepared for blind testing. Of the 100 specimens tested for HSV-1, 97 gave consistent results in the three test runs: 24 were consistently negative, and 73 were consistently positive. Ninety-three of the specimens gave consistent HSV-2 results; for 82, the results were negative, and 11 were consistently positive. Of the discrepant specimens, two of three inconsistent results for HSV-1 were negative in test 2 and positive for both others, as were five of the seven inconsistent HSV-2 results. One HSV-1 result and two HSV-2 results were negative in the first two tests and positive in the third. There were no other patterns. Side-by-side examination of the blots revealed a slight but noticeable reduction of the color intensity on blots from the second test run. In contrast, when 24 specimens (selected as described above) were divided into three aliquots each, coded, and tested in the same assay run, there was no variability between aliquots of the same specimen. Taken together, these results suggest that the positive-to-negative shifts in the day-to-day test may be accounted for by day-to-day variation in the sensitivity of the BIB assay.
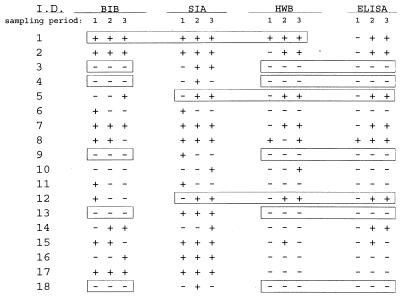
To compare the performance of the BIB assay with those of other type-specific HSV assays, we assembled a panel of 33 specimen sets that represented all four result patterns in the BIB assay (for HSV-2, consistently positive [n = 6], consistently negative [n = 15], positive to negative [n = 5], and negative to positive [n = 4]). The 99 specimens were coded and blindly tested with the ELISA, SIA, and HWB and BIB assays. All four assays gave positive-to-negative results with some serum sets. The results for HSV-2 (comparable to those obtained for HSV-1) are presented in Fig. 1. Since the HWB detects antibodies to all immunogenic HSV-infected-cell proteins, it was possible to detect differences in the response profiles among specimens from the same set. There was evidence that four of the test sets may have included serum specimens from more than one individual, and these have been excluded from the analysis. Elevan of 29 sets were consistent across all four assay methods (3 positive, 8 negative, 1 converter) and are not included in the table. Conversely, other sets with consistent profiles in the HWB assay produced positive-to-negative shifts in at least one assay (sets 4, 6, 8, 9, 11, 12, and 15). In 11 of 29 sets (31.0%), the results were identical for the four assays. Of the other 18 sets, 8 sets agreed between three of the assay methods, but these agreements did not occur consistently among the same three assays. Sequences of positive-to-negative results occurred in sets 4 (SIA), 6 (BIB and SIA), 8 (BIB and HWB), 9 (SIA), 11 (BIB and SIA), 12 (BIB), 15 (BIB, HWB, and ELISA), and 18 (SIA). The overall agreement among pairs of assays as measured by the kappa statistic was moderate. Immunoblotting showed 37.5, 45.2, and 50.1% agreement with SIA, HWB and ELISA, respectively. SIA showed 41.2 and 36.3% agreement with HWB and ELISA, respectively. The best agreement was between HWB and ELISA (77.2%).
FIG. 1.
Comparison of HSV-2 results from selected sequential serum sets among four gG-based serologic methods. Note that the sets were not consistent in all four assay methods.
There were rather marked differences in the total number of positive results detected by the four assays. SIA detected 46 of 87 samples as positive (52.9%), whereas, BIB detected 32 (36.8%), ELISA detected 26 (29.9%), and HWB detected 25 (28.7%) samples as positive.
Serum sets from 90 participants in the study (321 serum samples), representing individuals who were either singly seropositive by BIB for HSV-1 or HSV-2 or seronegative for both viruses, were also tested with a commercial assay that does not discriminate anti-HSV-1 from anti-HSV-2 antibody (data not shown). This assay always detected HSV antibody among those singly positive (45 serum sets), and it did not detect antibody in negative sera (45 serum sets). No inconsistent result patterns were observed with this assay.
DISCUSSION
The advent of gG-based serologic assays for type-specific detection of HSV infections provided a powerful epidemiological tool for understanding the biology of these closely related viruses in humans. Nevertheless, this cohort study of Thai military recruits revealed substantial inconsistency in results obtained from consecutive serum specimens. Although both the HSV-1 and HSV-2 BIB tests produced consistent results in over 90% of the specimen sets, among participants with more than one specimen and at least one positive result, a substantial number of serum sets included at least one positive-to-negative shift. We did not identify any single explanation for this; however, many of the positive-to-negative shifts in BIB were associated with weakly positive specimens. This association was also supported by the observation in the day-to-day reproducibility study of a concomitant increase in negative test results in BIBs with diminished color intensity. Nevertheless, a large number of weakly positive tests were not associated with positive-to-negative shifts, particularly for HSV-1. Variation in color intensity is thus likely to be a more important factor for interpreting HSV-2 results in this assay.
We tested a subset of specimens in four gG-based assays with different formats and found that similar inconsistencies occurred in all of them. The specimen sets used for comparison were selected on the basis of performance in the BIB assay and included consistently positive, consistently negative, and shifting sets (negative-to-positive and positive-to negative shifts).
Errors in specimen handling appear to account for some of the inconsistent results. Among the 33 specimen sets that were compared among four different HSV assays, HWB results suggested that three of the test sets may have included specimens from more than one individual. In a large study like this, such errors are bound to occur, and many of the test sets that were selected for comparison were chosen because of sequential positive-to-negative tests on BIB. However, the remaining inconsistent sets in the comparison study could not be explained on the basis of discrepant HWB profiles.
The inconsistencies observed in these studies were more prominent for HSV-2. Epidemiologic studies utilizing immunoassays based on gG-2 (6, 8, 10, 13, 22, 24, 30) have reported similar prevalence figures for the virus in comparable populations. Shifting of sequential test results has not been described. One reason could be that studies evaluating age-specific prevalence rates for HSV-2 have not routinely retested individuals who have seroconverted or who tested positive initially, on the supposition that they will always remain positive.
A limitation of this study is the absence of an undisputed “gold standard” test for detecting HSV antibodies. The data on day-to-day variability and color intensity and the observation that inconsistent test patterns did not occur in an assay that fails to discriminate HSV-1 from HSV-2 all suggest a a limitation in the sensitivity of gG-based assays. Nonetheless, these data do not rule out a contributory role for limits in assay specificity, or even the possibility of a biological phenomenon resulting in natural fluctuations in HSV antibody levels in vivo. All of the gG-based assays examined here are adaptable to semiquantitative formats (i.e., antibody titration) and could be used to assess individuals over time, including periods following known HSV reactivation. The question of sensitivity versus specificity could be addressed in humans by performing a reproducibility study examining sera from culture-confirmed HSV-1, HSV-2, and dually infected participants, both symptomatic and asymptomatic.
In any event, until these limitations in the reliability of gG-based serology are better understood and controlled, such tests should be employed with caution; they are best suited to performing cross-sectional studies of prevalence. Results of incidence studies would be difficult to interpret. In a broader context, the observation of limitations in the reliability of this test is likely to extend to many other serologic tests. Assays that measure immune responses to a single viral protein or to individual epitopes may be particularly prone to difficulties. Our results argue that testing of cohort specimens from population-based rather than clinical studies should be a part of any serologic assay validation scheme. On the basis of these observations, it is also recommended that serial testing be performed during studies or procedures for which determination of type-specific HSV seropositivity is necessary.
ACKNOWLEDGMENTS
We acknowledge the insight and assistance of the following individuals in the development of this article: James G. Dobbins, Michele Reyes, John A. Stewart, and Judith Graber.
REFERENCES
- 1.Alexander D, Kilpatrick J, Ashley R, Corey L, Dinello R, Polito A, Burke R L. Recombinant-truncated gG1 and gG2 as antigens for a type-specific serological assay to diagnose prior infection with HSV-1 or HSV-2, abstr. 283. Vancouver, Canada: International Herpesvirus Workshop; 1994. [Google Scholar]
- 2.Ashley R. Herpes simplex virus. In: Lennette E, editor. Diagnostic procedures for viral, rickettsial and chlamydial infections. 7th ed. Washington, D.C: American Public Health Association; 1995. pp. 375–395. [Google Scholar]
- 3.Benedetti J, Corey L, Ashley R. Recurrence rates in genital herpes after symptomatic first-episode infection. Ann Intern Med. 1994;121:847–854. doi: 10.7326/0003-4819-121-11-199412010-00004. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein D I, Lovett M A, Bryson Y J. Serologic analysis of first-episode nonprimary genital herpes simplex virus infection. Am J Med. 1984;77:1055–1060. doi: 10.1016/0002-9343(84)90188-8. [DOI] [PubMed] [Google Scholar]
- 5.Brown Z A, Benedetti J K, Watts D H, Selke S, Berry S, Ashley R L, Corey L. A comparison between detailed and simple histories in the diagnosis of genital herpes complicating pregnancy. Am J Obstet Gynecol. 1995;172:1299–1303. doi: 10.1016/0002-9378(95)91496-x. [DOI] [PubMed] [Google Scholar]
- 6.Davidson M, Schnitzer P G, Bulkow L R, Parkinson A J, Schloss M L, Fitzgerald M A, Knight J A, Murphy C M, Kiviat N B, Toomey K E, Reeves W C, Schmid D S, Stamm W E. The prevalence of cervical infection with human papillomaviruses and cervical dysplasia in Alaska native women. J Infect Dis. 1994;169:792–800. doi: 10.1093/infdis/169.4.792. [DOI] [PubMed] [Google Scholar]
- 7.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faruque S, Edlin B R, McCoy C B, Word C O, Larsen S A, Schmid D S, von Bargen J C, Serrano Y. Crack cocaine smoking and oral sores in three inner-city neighborhoods. J Acquired Immune Defic Syndr Hum Retroviruses. 1996;13:87–92. doi: 10.1097/00042560-199609000-00012. [DOI] [PubMed] [Google Scholar]
- 9.Ho D W T, Field P R, Sjögren-Jansson E, Jeansson S, Cunningham A L. Indirect ELISA for the detection of HSV-2 specific IgG and IgM antibodies with glycoprotein G (gG-2) J Virol Methods. 1992;36:249–264. doi: 10.1016/0166-0934(92)90056-j. [DOI] [PubMed] [Google Scholar]
- 10.Johnson R E, Nahmias A J, Magder L S, Lee F K, Brooke C A, Snowden C B. A seroepidemiologic survey of the prevalence of herpes simplex virus type 2 infection in the United States. N Engl J Med. 1989;321:7–12. doi: 10.1056/NEJM198907063210102. [DOI] [PubMed] [Google Scholar]
- 11.Kahlon J, Lakeman F D, Ackermann M, Whitley R J. Human antibody response to herpes simplex virus-specific polypeptides after primary and recurrent infection. J Clin Microbiol. 1986;23:725–730. doi: 10.1128/jcm.23.4.725-730.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koutsky L A, Ashley R L, Holmes K K, Stevens C E, Critchlow C W, Kiviat N, Lipinski C M, Wolner-Hanssen P, Corey L. The frequency of unrecognized type 2 herpes simplex virus infection among women. Implications for the control of genital herpes. Sex Transm Dis. 1990;17:90–94. doi: 10.1097/00007435-199004000-00009. [DOI] [PubMed] [Google Scholar]
- 13.Kulhanjian J A, Soroush V, Au D S, Bronzan R N, Yasukawa L L, Weylman L E, Arvin A M, Prober C G. Identification of women at unsuspected risk of primary infection with herpes simplex virus type 2 during pregnancy. N Engl J Med. 1992;326:916–920. doi: 10.1056/NEJM199204023261403. [DOI] [PubMed] [Google Scholar]
- 14.Landis J R, Koch G E. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 15.Langenberg A, Benedetti J, Jenkins J, Ashley R, Winter C, Corey L. Development of clinically recognizable genital lesions among women previously identified as having “asymptomatic” herpes simplex virus type 2 infection. Ann Intern Med. 1989;110:882–887. doi: 10.7326/0003-4819-110-11-882. [DOI] [PubMed] [Google Scholar]
- 16.Lasky L A, Dowbenko D J. DNA sequence analysis of the type-common glycoprotein D genes of herpes simplex types 1 and 2. DNA. 1984;3:23–29. doi: 10.1089/dna.1.1984.3.23. [DOI] [PubMed] [Google Scholar]
- 17.Lee F K, Coleman R M, Pereira L, Bailey P D, Tatsuno M, Nahmias A J. Detection of herpes simplex virus type-2 specific antibody with glycoprotein G. J Clin Microbiol. 1985;22:641–644. doi: 10.1128/jcm.22.4.641-644.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee F K, Pereira L, Griffen C, Reid E, Nahmias A J. A novel glycoprotein (gG-1) for detection of herpes simplex virus specific antibodies. J Virol Methods. 1986;14:111–118. doi: 10.1016/0166-0934(86)90041-8. [DOI] [PubMed] [Google Scholar]
- 19.Marsden H S, Buckmaster A, Palfreyman J W, Hope R G, Minson A C. Characterization of the 92,000-dalton glycoprotein induced by herpes simplex virus type 2. J Virol. 1984;50:547–554. doi: 10.1128/jvi.50.2.547-554.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch D J, Moss H W, McNab D, Frame M C. DNA sequence and genetic content of the HindIII region in the short unique component of herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J Gen Virol. 1987;68:19–38. doi: 10.1099/0022-1317-68-1-19. [DOI] [PubMed] [Google Scholar]
- 21.Rapoport T A. Protein translocation across and integration into membranes. Crit Rev Biochem. 1986;20:73–137. doi: 10.3109/10409238609115901. [DOI] [PubMed] [Google Scholar]
- 22.Reeves W C, Gary H E, Jr, Johnson P R, Icenogle J P, Brenes M M, deBritton R M, Dobbins J G, Schmid D S. Risk factors for genital papillomavirus infection in populations at high and low risk for cervical cancer. J Infect Dis. 1994;170:753–758. doi: 10.1093/infdis/170.4.753. [DOI] [PubMed] [Google Scholar]
- 23.Roizman B, Norrild B, Chan C, Pereira L. Identification and preliminary mapping with monoclonal antibodies of a herpes simplex virus 2 glycoprotein lacking a known type 1 counterpart. Virology. 1984;133:242–247. doi: 10.1016/0042-6822(84)90447-1. [DOI] [PubMed] [Google Scholar]
- 24.Safrin S, Arvin A, Mills J, Ashley R. Comparison of the Western immunoblot assay and a glycoprotein G enzyme immunoassay for detection of serum antibodies to herpes simplex virus type 2 in patients with AIDS. J Clin Microbiol. 1992;30:1312–1314. doi: 10.1128/jcm.30.5.1312-1314.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sánchez-Martínez D, Pellett P E. Expression of HSV-1 and HSV-2 glycoprotein G in insect cells using a novel baculovirus expression vector. Virology. 1991;182:229–238. doi: 10.1016/0042-6822(91)90666-y. [DOI] [PubMed] [Google Scholar]
- 26.Sánchez-Martínez D, Schmid D S, Whittington W, Brown D, Reeves W C, Chatterjee S, Whitley R J, Pellett P E. Evaluation of a test based on baculovirus-expressed glycoprotein G for detection of herpes simplex virus type-specific antibodies. J Infect Dis. 1991;164:1196–1199. doi: 10.1093/infdis/164.6.1196. [DOI] [PubMed] [Google Scholar]
- 27.Spear P G. Glycoproteins specified by herpes simplex viruses. In: Ruizman B, editor. The herpesviruses. Vol. 3. New York, N.Y: Plenum Press; 1985. pp. 315–356. [Google Scholar]
- 28.Sweat M D, Nopkesorn T, Mastro T D, Sangkharomya S, MacQueen K, Pokapanichwong W, Sawaengdee Y, Weniger B G. AIDS awareness among a cohort of young Thai men: exposure to information, level of knowledge, and perception of risk. AIDS Care. 1995;7:573–591. doi: 10.1080/09540129550126236. [DOI] [PubMed] [Google Scholar]
- 29.Tedder D G, Ashley R, Tyler K L, Levin M J. Herpes simplex virus infection as a cause of benign recurrent lymphocytic meningitis. Ann Intern Med. 1994;121:334–338. doi: 10.7326/0003-4819-121-5-199409010-00004. [DOI] [PubMed] [Google Scholar]
- 30.Wagner H U, Van Dyck E, Roggen E, Nunn A J, Kamali A, Schmid D S, Dobbins J G, Mulder D W. Seroprevalence and incidence of sexually transmitted diseases in a rural Ugandan population. Int J Sex Transm Dis AIDS. 1994;5:332–337. doi: 10.1177/095646249400500509. [DOI] [PubMed] [Google Scholar]