Abstract
Polyadenylation (PA) is the process by which the 3′ ends of most mammalian mRNAs are formed. In nature, PA is highly coordinated, or coupled, with splicing. In mammalian systems, the most compelling mechanistic model for coupling arises from data supporting exon definition (2, 34, 37). We have examined the roles of individual functional components of splicing and PA signals in the coupling process by using an in vitro splicing and PA reaction with a synthetic pre-mRNA substrate containing an adenovirus splicing cassette and the simian virus 40 late PA signal. The effects of individually mutating splicing elements and PA elements in this substrate were determined. We found that mutation of the polypyrimidine tract and the 3′ splice site significantly reduced PA efficiency and that mutation of the AAUAAA and the downstream elements of the PA signal decreased splicing efficiency, suggesting that these elements are the most significant for the coupling of splicing and PA. Although mutation of the upstream elements (USEs) of the PA signal dramatically decreased PA, splicing was only modestly affected, suggesting that USEs modestly affect coupling. Mutation of the 5′ splice site in the presence of a viable polypyrimidine tract and the 3′ splice site had no effect on PA, suggesting no effect of this element on coupling. However, our data also suggest that a site for U1 snRNP binding (e.g., a 5′ splice site) within the last exon can negatively effect both PA and splicing; hence, a 5′ splice site-like sequence in this position appears to be a modulator of coupling. In addition, we show that the RNA-protein complex formed to define an exon may inhibit processing if the definition of an adjacent exon fails. This finding indicates a mechanism for monitoring the appropriate definition of exons and for allowing only pre-mRNAs with successfully defined exons to be processed.
Polyadenylation (PA) is the process by which the 3′ ends of most mammalian mRNAs are formed. In a tightly coupled set of nuclear reactions, the precursor RNA is endonucleotypically cleaved at a specific site; then, approximately 250 adenosine residues are polymerized to the cleaved end, forming the poly(A) tail of the final mRNA. Nearly all mammalian cellular and many viral mRNAs are processed in this manner. It has been clearly established that a poly(A) tail is essential for the survival, transport, stability, and translation of most mRNAs (8, 41, 49, 50, 52).
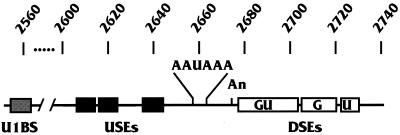
The PA signal in the precursor RNA defines the site of PA through specific binding with a complex of proteins which orchestrate the cleavage and PA of the precursor RNA; binding specificity is provided by elements in the RNA. The PA signal used in our studies is the simian virus 40 (SV40) late polyadenylation (SVLPA) signal shown diagrammatically in Fig. 1. The central, nearly invariant, feature of mammalian PA signals is the hexanucleotide consensus element AAUAAA. This is the binding site for the 160-kDa component of the cleavage and PA specificity factor (CPSF) (17, 30). The AAUAAA sequence is located between 11 and 25 nucleotides upstream of the actual cleavage and PA site. AAUAAA sequences are also found within the coding regions of many genes; thus, additional elements are needed to select the correct AAUAAA. These additional elements are found within sequences located 14 to 70 nucleotides downstream of an AAUAAA. These downstream elements (DSEs) greatly increase the efficiency of utilization of the AAUAAA in directing PA (3, 9, 15, 16, 25–27, 40, 42, 43, 53–55). A comparison of DSEs of many PA signals provides no clear consensus sequence other than variable lengths (6 to 20 nucleotides) of GU- or U-rich sequences. These DSEs are the binding sites for the 64-kDa component of the cleavage stimulatory factor (CStF), a complex of three proteins which interacts with the CPSF. Coordinately, the CPSF and the CStF stably bind the PA signal RNA (8, 46).
FIG. 1.
Linear diagram of the SVLPA signal. Above the diagram are the SV40 nucleotide numbers corresponding to the SVLPA signal. Elements shown are as follows: AAUAAA, the consensus hexonucleotide; An, the site for cleavage and PA; USEs, indicated by three black boxes representing the homologous AUUUGURA core elements; DSEs, indicated by three white boxes representing the GU-rich, G-rich, and U-rich elements; and U1BS, the U1 snRNP binding site. All elements are described in the text.
While most mammalian PA signals are composed of an AAUAAA and a GU-rich DSE, some are more complex. The SVLPA signal contains both a more complex downstream region and additional efficiency elements upstream of the AAUAAA (see below). Three DSEs have been identified by mutagenesis. A GU-rich element (GU in Fig. 1) located 14 to 40 nucleotides downstream of the AAUAAA (9) appears to be the binding site for the 64-kDa component of the CStF. A second, G-rich DSE (G in Fig. 1) located 45 to 58 nucleotides downstream of the AAUAAA binds a 50-kDa protein designated DSE factor 1 (DSEF-1) (1, 36); DSEF-1 binding may enhance the efficiency of PA (1, 36). Finally, a U-rich DSE (U in Fig. 1) was identified 59 to 67 nucleotides downstream of the AAUAAA by deletion analysis (42, 43). It can function as an hnRNP C protein binding site and can function alone as a DSE in heterologous constructions with an AAUAAA (53, 54).
Efficiency elements located upstream of the AAUAAA have been characterized for a number of PA signals, including the SVLPA signal (4–6, 11, 12, 29, 38, 39, 44, 45, 47, 48). Our studies of the SVLPA signal suggest that upstream elements (USEs) impart characteristics which provide efficiency or special levels of control to the PA signal. Few generalizations can be made about USEs, and no consensus sequence or similarity has been seen. However, we have characterized three functional elements in the upstream region of the SVLPA signal (Fig. 1) which share the sequence AUUUGURA (45).
An additional unique element of the SVLPA signal was characterized by Wassarman and Steitz (51), who showed that the U1 snRNP can cross-link in vitro to the SV40 late RNA through RNA-RNA base pairing between the U1 RNA and the SV40 RNA. This U1 snRNP binding site (U1BS in Fig. 1) lies 103 nucleotides upstream of the AAUAAA. It has been proposed that this interaction facilitates PA in vitro (51). However, several studies have indicated that U1 snRNP binding sites (e.g., 5′ splice sites) in similar locations near PA elements are inhibitory to PA (14, 18). Our findings, described below, agree with an inhibitory role for the U1 snRNP binding site.
PA is not an isolated mRNA processing event; in nature it is highly coordinated, or coupled, with splicing. In mammals this coordination is believed to be involved with the proper definition of the last exon of the mRNA. Much work has been done to determine how introns and exons are defined. The most compelling model for mammalian systems is the exon definition mechanism proposed by Berget and colleagues (2, 34, 37). In general, interior exons (those flanked by 3′ and 5′ splice sites, where splice sites are defined by intronic polarity) of mammalian pre-mRNAs tend to be small, on the order of 150 nucleotides, whereas introns can be very large (tens of thousands of nucleotides). The Berget model suggests that the units to be spliced are established by a mechanism which defines the smaller of these. In mammals this is usually the exon; intron definition has been indicated in some cases, such as Drosophila (21). Exon definition is proposed to occur when a complex of splicing factors believed to contain the U2 and U1 snRNPs recognizes the 3′ splice site of an interior exon, followed by U1 snRNP recognition of the downstream 5′ splice site. This process defines the limits of the exon, and the positioned snRNPs then proceed to form the spliceosome across the intron.
This model does not account for the definition of the first and last exons, since first exons contain an m7GpppG 5′ cap structure instead of a 3′ splice site and last exons contain a PA signal instead of a 5′ splice site. However, the most compelling data in support of the exon definition model come from studies of first- and last-exon definition. The 5′ cap structure of a mammalian pre-mRNA is necessary for the efficient utilization of the adjacent 5′ splice site for definition of the first exon and removal of the first intron (20). Efficient recognition of the cap-proximal 5′ splice site by the U1 snRNP is facilitated by the nuclear cap binding complex (CBC) (22). This CBC-directed definition of the first exon is believed to be one of the earliest steps in pre-mRNA recognition and appears to involve an interaction between the CBC (bound to the cap) and the U1 snRNP (which binds at the 5′ splice site).
Processing of the last exon and removal of the last intron involve interactions between splicing components at the 3′ splice site of the last exon and components of the PA complex at the PA signal (23, 33, 35). Niwa and Berget used a coupled in vitro splicing-PA system to show that mutations in the PA signal which eliminated PA also caused decreased splicing, i.e., decreased the removal of the last intron (33). Likewise, mutations in the 3′ splice site of the last exon, which eliminated splicing, caused an inhibition of PA (35). Our laboratory has confirmed this observation in vivo by using RNA substrates containing a splice site and the SVLPA signal (7). In addition, data obtained by others (31, 32) have further established that the removal of the last intron and PA influence each other in vivo.
Interestingly, the mechanisms for defining the first and last exons have unexpected similarities. We have shown that the presence of a wild-type m7GpppG cap but not a cap analog can positively affect the efficiency of PA of a PA-only substrate (i.e., the SVLPA signal with no intron present) (10). Cap analogs do not stimulate PA because they fail to bind a titratable cap binding factor (10). This factor was shown to be the CBC, which mediates a physical association between the CBC-cap complex and factors of the PA complex at the AAUAAA (13). This interaction is strikingly similar to the proposed interaction between splicing factors at the 3′ splice site and the PA complex, which is believed to influence last-exon definition and PA. The failure of cap analogs to stimulate PA could be overcome when a 3′ splice site was inserted upstream of the PA signal, indicating that factors bound at either the cap or the 3′ splice site function similarly to affect PA efficiency and complete exon definition.
In the present work, we have used an in vitro coupled splicing-PA reaction with a substrate containing an adenovirus splicing cassette and the SVLPA signal, MXSVL (35), to examine how the mutation of individual splicing elements affects PA and vice versa. We present evidence that mutation of the polypyrimidine tract and the 3′ splice site not only affects splicing but also significantly reduces PA efficiency. Similarly, mutation of the AAUAAA and specific DSEs not only affects PA but also reduces splicing efficiency. The results suggest that these are the elements most significant for the coupling of splicing and PA. The USEs of the SVLPA signal appear to modestly affect coupling, whereas the 5′ splice site, under some conditions (10), does not appear to affect coupling. Our data also suggest that the binding site for U1 snRNP upstream of the AAUAAA in the SVLPA signal can negatively affect both PA and splicing; hence, it appears to be a modulator of coupling. Finally, we show that the RNA-protein complex formed to define an exon may inhibit processing if the definition of an adjacent exon fails. This finding indicates a checkpoint mechanism for monitoring the appropriate definition of exons and for allowing only pre-mRNAs with successfully defined exons to be processed.
MATERIALS AND METHODS
Plasmids encoding precursor RNA substrates.
Substrate RNAs for in vitro splicing and PA reactions were based on the MXSVL substrate (35), containing an adenovirus splicing cassette upstream of the SVLPA signal (see description in Results). Substrate RNAs were prepared by in vitro transcription from linearized plasmid templates; the parent plasmid for the wild-type MXSVL substrate was pSP64-MXSVL (35). Numerous variants of plasmid pSP64-MXSVL containing mutations in the splicing elements or the PA signal elements were prepared. Plasmids pSP64-MXSVL (−USE), pSP64-MXSVL (DSE LSMs), and pSP64-MXSVL (−AUA) contain PA signal element mutations. These were prepared by use of plasmid pGem2-UPAS, which contains only the wild-type SVLPA signal (SV40 nucleotides 2531 to 2729). Linker substitution (LS) mutagenesis was used to produce the desired mutation in the pGem2-UPAS background. LS mutagenesis methods used here were similar to those previously described for the construction of LS mutations in the USEs of the SVLPA signal (45). The resultant mutant SVLPA signal was then excised and used to replace the BamHI-SalI fragment of pSP64-MXSVL, resulting in replacement of the entire SVLPA signal. Plasmid pGem2-UM123 has been previously described (45); it was used as the source for replacing the wild-type SVLPA sequences in MXSVL to construct pSP64-MXSVL(−USE). To produce pSP64-MXSVL(DSE LSMs), the following LSs were made: DM1 (5′...GGTACC...3′), DM2 (5′...CGCGGGAGGTACC...3′), DM3 (5′...GGTACC...3′), DM4 (5′...ATAGGTACC...3′), and DM5 (5′...CATGGTACC...3′). Similarly, by use of linker scanning mutagenesis, the AAUAAA present in pGem2-MXSVL (see below) was changed from 5′...AATAAACAAGTTAAC...3′ to 5′...AAGAAACAAGTTAAC...3′ (change is underlined) to create the U-to-G mutation in the poly(A) signal and to add a KpnI linker immediately downstream (for cloning purposes). To produce the pGem2-U1BS Sp/PA substrate plasmid, the EcoRI-SalI fragment encoding MXSVL was isolated from pSP64-MXSVL and inserted into a pGem2 vector which was better suited for PCR-mediated LS mutagenesis. The resultant plasmid, pGem2-MXSVL, was then used as a template for PCR-mediated LS of the U1 binding site (51) (5′...ACATGATAAGATA...3′), replacing it with 5′...GAGCGGTAACCGCG...3′), which contains an NcoI site. Substrate plasmid pGem2-PPT Sp/PA, containing a mutation of the polypyrimidine tract, was constructed in a manner similar to that used for pGem2-U1BS Sp/PA; the polypyrimidine tract (5′...TCCCTTTTTTTTCC...3′) was replaced with a BglII site and additional changes from pyrimidines to purines (5′...TCGAGATCTAAACC...3′).
Preparation of precursor RNA substrates.
Templates for in vitro transcription were linearized with DraI, with the exception of the DM5 mutant substrate, which was linearized with KpnI (KpnI was used in this case because the DraI site had been mutated by LS mutagenesis, which substituted a KpnI site). In vitro transcription reactions were performed with SP6 polymerase (Promega Biotec) under conditions described by the manufacturer plus 50 μCi of [32P]UTP (Amersham), 250 ng of linearized template DNA, and 0.5 mM m7GpppG cap substrate (Pharmacia). 32P-labeled RNAs were extracted with phenol-chloroform-isoamyl alcohol (50:49:1), ethanol precipitated, and purified by electrophoresis through a 5% polyacrylamide–7 M urea gel. RNAs were eluted from the gel slices in 20 mM Tris-HCl (pH 7.5)–400 mM NaCl–0.01% sodium dodecyl sulfate at room temperature overnight. After extraction with phenol-chloroform-isoamyl alcohol (50:49:1) and chloroform-isoamyl alcohol (49:1), the RNAs were ethanol precipitated. Incorporated [32P]UTP was quantitated by liquid scintillation counting.
Nuclear extracts and in vitro processing reactions.
HeLa-S3 cell nuclear extracts were prepared as described by Moore and Sharp (28) with cells obtained as pellets from the National Cell Culture Center. In vitro reaction mixtures contained (final concentrations) 32% (vol/vol) nuclear extract, 250 μM ATP (Pharmacia), 1 mM cordycepin 5′-triphosphate (Sigma), 20 mM phosphocreatine (Sigma), 2.6% polyvinyl alcohol, 40 U of RNasin (Promega), and approximately 5 fmol of 32P-labeled precursor RNA. The magnesium concentrations in the reactions were varied from 0.5 to 6.0 mM with MgCl2.
Optimal conditions for processing varies somewhat between different nuclear extract preparations. Therefore, each extract must be thoroughly characterized for specific optima. The length of incubation at 30°C needed to attain an optimal extent of processing (within the linear range of the reaction) was determined by evaluating nuclear extract activity over a time course with the wild-type splicing and polyadenylation substrate MXSVL (denoted WT Sp/PA). Extracts were also characterized for the magnesium concentration which provided optimal coupling; the optimal concentration has been noted to vary between 0.5 and 2.0 mM. Extracts with different optima provided similar coupling results in magnesium titration experiments, except that the curves were offset to correspond to the optimal magnesium concentration. The majority of experiments described in this report were conducted with the same nuclear extract, which had a 35-min incubation optimum and a 1.5 mM magnesium optimum. The experiments were repeated with other nuclear extract preparations, with no qualitative variations in the data trends indicated in magnesium titration experiments.
Processing reactions were terminated by the addition of 8 volumes of 20 mM Tris (pH 7.5)–400 mM NaCl–0.01% sodium dodecyl sulfate. Reaction products were extracted once with phenol-chloroform-isoamyl alcohol, ethanol precipitated, and analyzed on a 5% polyacrylamide–7 M urea gel. Substrate RNAs and processed species were quantitated with a Molecular Dynamics PhosphorImager. Percent processed RNA was calculated as described in Results.
RESULTS
Effects of Mg2+ concentration on the coupling of in vitro splicing and PA.
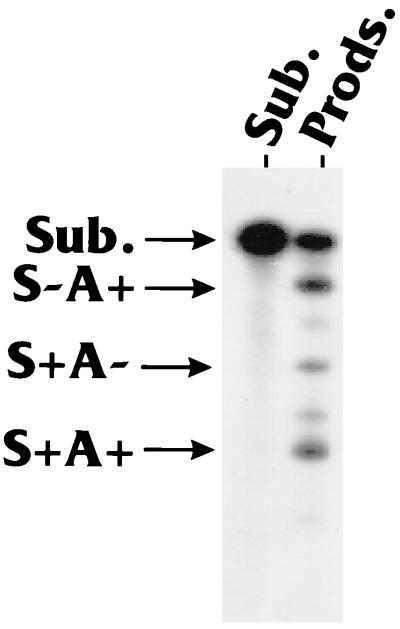
We determined the effects of splicing and PA signal elements on coupling by using an in vitro splicing-PA system (HeLa cell nuclear extracts; see Materials and Methods) (35) with the MXSVL RNA substrate (Fig. 2), which was constructed and studied by Niwa et al. (35). This MXSVL RNA substrate consists of a splicing cassette derived from the adenovirus major late region coupled to the SVLPA signal containing the elements described above (Fig. 1). The 32P-labeled MXSVL substrate was made by in vitro transcription and gel purification (see Materials and Methods). The purified substrate RNA was added to a HeLa cell nuclear extract under coupling conditions (see below), and the products were analyzed by gel electrophoresis. Figure 3 shows a typical gel analysis of the processing products. The reactions were carried out in the presence of the ATP analog cordycepin, a chain terminator that stops poly(A) tail elongation; thus, the products seen are essentially cleavage products. Cordycepin is used to inhibit poly(A) tail elongation because fully polyadenylated products migrate as smears [due to variable lengths of the poly(A) tails] and are difficult to quantitate.
FIG. 2.
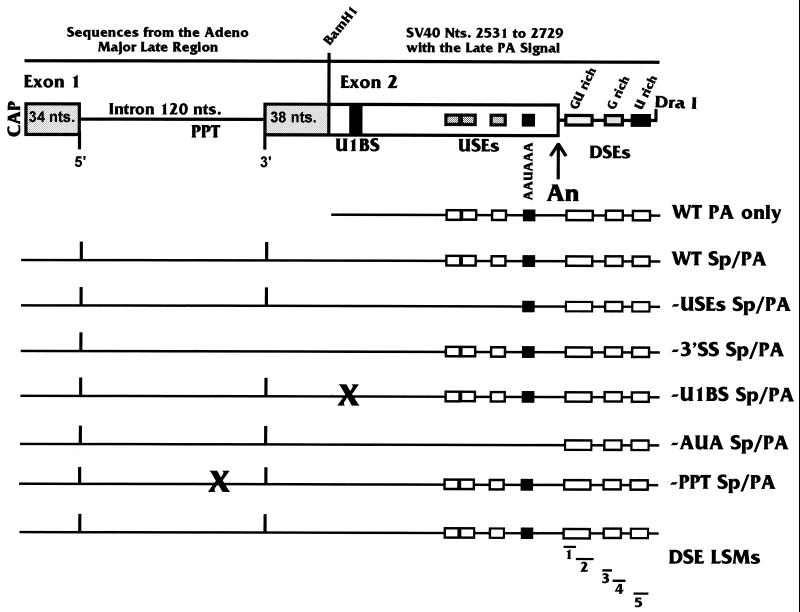
MXSVL splicing and PA substrate. At the top of the figure is a detailed map of the MXSVL substrate described in the text. Specific elements of the splice cassette are the 5′ splice site (5′), the polypyrimidine tract (PPT), and the 3′ splice site (3′). Specific elements of the SVLPA signal are the same as those shown in Fig. 1. Below the top diagram are simpler diagrams showing the WT Sp/PA substrate and a series of similar substrates with mutations in the various splicing and PA elements. Each is described in the text. In addition, LSMs were made across the DSEs of the PA signal; the positions of mutations DM1 to DM5 (1 to 5) are shown. Also shown is the WT PA only substrate, which we have used in our previous studies of the SVLPA signal. Adeno, adenovirus; nts., nucleotides.
FIG. 3.
Products of the splicing and PA reaction carried out with the MXSVL (WT Sp/PA) substrate. Lanes: Sub., migration position of the intact MXSVL (WT Sp/PA) substrate (Sub.); Prods., products (Prods.) of the splicing and PA reaction carried out under coupling conditions (see Materials and Methods). The major products are S−A+, S+A−, and S+A+.
The products observed are (i) polyadenylated and not spliced (S−A+), (ii) spliced and not polyadenylated (S+A−), and (iii) fully processed, spliced, and polyadenylated (S+A+). The percentage of each of these products can be determined by PhosphorImager analysis of the gels. In some cases, we show quantitation representing total splicing or total PA as a percentage of the total substrate added. For example, the total polyadenylated fraction would equal (S−A+ + S+A+)/(total of all products + remaining substrate); the result is converted to a percentage for presentation. Reaction results were quite reproducible, since substrate and product RNAs were stable in the reactions; thus, losses due to degradation were not a problem.
A potential complication of the in vitro coupled system is that the PA and splicing reactions, when carried out individually, have different Mg2+ optima. Thus, the coupled reaction must be performed with an Mg2+ concentration at which both processes occur relatively efficiently (35). It has been established with the MXSVL substrate that coupling occurs at low Mg2+ concentrations (e.g., 0.5 to 2 mM) and that the two reactions are uncoupled both in the absence of Mg2+ and at higher Mg2+ concentrations (35) (see below). The optimal Mg2+ concentration for coupling can vary moderately between extracts; therefore, the properties of each new extract must be characterized (see Materials and Methods for details of extract preparation and characterization).
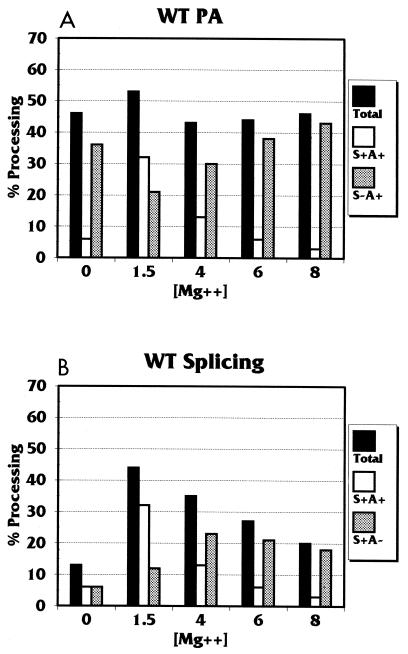
In order to demonstrate coupling and uncoupling, we titrated Mg2+ over a range of concentrations and quantitated the products. Figure 4 shows a bar graph of the PA (Fig. 4A) and splicing (Fig. 4B) results from an Mg2+ titration with the wild-type MXSVL substrate (percentage of total PA or total splicing is shown by black bars). For PA, note that there is no great difference in the total level of polyadenylated RNA over the entire range of Mg2+ concentrations. However, examination of the individual polyadenylated products (S+A+ [white bars] and S−A+ [gray bars]) indicates coupling. The fully processed product (S+A+) predominated at 1.5 mM Mg2+, which was the optimal level for the coupling of PA and splicing in the extract used. However, as the Mg2+ concentration was increased, the reactions were uncoupled, causing the level of the S+A+ product to decrease and to be replaced by increasing amounts of the S−A+ product. Thus, when the Mg2+ concentration is increased, the reactions are uncoupled, resulting in a shift in the polyadenylated fraction from predominantly fully processed product to predominantly partially processed product.
FIG. 4.
Quantitation of the splicing and PA products from the MXSVL (WT Sp/PA) substrate in the presence of increasing Mg2+ concentrations. The amounts of each splicing and PA product were quantitated from data similar to those shown in Fig. 3. These values were converted to the percentage of input substrate and shown as a bar graph. (A) Level of total PA (Total) as well as levels of the PA products S+A+ and S−A+. (B) Level of total splicing (Total) as well as levels of the splicing products S+A+ and S+A−. The error of the data shown was ±2 to 4%. WT, wild type.
It could be argued that the increase in the level of the S−A+ product at higher Mg2+ concentrations is due to the inhibition of splicing. However, examination of the analogous splicing data (Fig. 4B) shows only moderate inhibition of total splicing at higher Mg2+ concentrations. More importantly, at all Mg2+ concentrations above 1.5 mM, the partially processed S+A− product prevails. A comparison of the 1.5 and 4 mM Mg2+ reactions in Fig. 4A and B provides a particularly good illustration of the transition from coupled to uncoupled processing caused by higher Mg2+ concentrations. The total PA at each Mg2+ concentration is comparable and the total splicing at each concentration is comparable, but the level of the fully processed product (S+A+) is higher at 1.5 mM Mg2+ than at 4 mM Mg2+.
Effects of mutations in individual splicing and PA elements on the coupling of splicing and PA.
It could be argued that the above data only fortuitously indicate coupling and, in reality, result from the effects of varied Mg2+ concentrations on two separate reactions. However, the phenomenon of coupling is confirmed by the examination of substrates containing mutations in specific splicing and PA elements. The effects of these mutations, described below, define the role of each element in both of the individual processing reactions and in coupling.
Figure 2 shows mutations introduced into the WT Sp/PA substrate: (i) in −USEs Sp/PA, LS mutagenesis was used to replace the three homologous USEs (45); (ii) in −3′SS Sp/PA, a point mutation was introduced into the 3′ splice site; (iii) in −U1BS Sp/PA, an LS mutation (LSM) replaces the U1 snRNP binding site; (iv) in −AUA Sp/PA, a point mutation changes AAUAAA to AAGAAA and LSMs replace the USEs; (v) in −PPT Sp/PA, LS mutagenesis was used to replace the polypyrimidine tract; and (vi) DSE LSMs consist of five separate LSMs in the downstream region. A mutation of the 5′ splice site is not included since we have previously shown that mutation of the 5′ splice site in the presence of a wild-type polypyrimidine tract and a wild-type 3′ splice site has no significant effect on PA (10). Also shown in Fig. 2 is the wild-type PA-only (WT PA only) substrate which we have used in previous studies of the SVLPA signal (10, 23, 24, 45). Note that the entire SVLPA signal previously studied is contained in MXSVL.
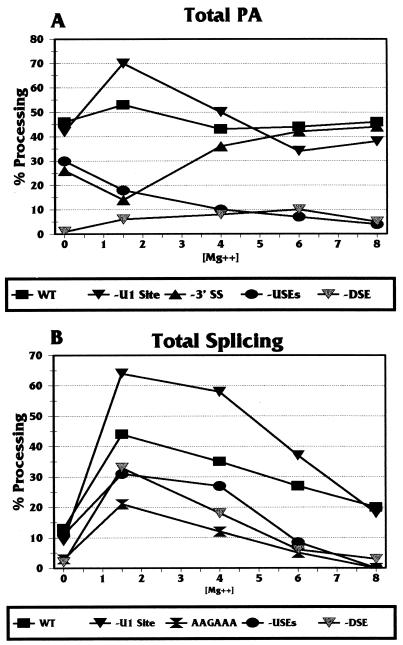
Each Sp/PA mutant substrate was tested for coupling in Mg2+ titration experiments similar to those shown in Fig. 4. In Fig. 5 we show only the total PA (Fig. 5A) and total splicing (Fig. 5B) results. The results obtained with the WT Sp/PA substrate for total PA and total splicing are similar to those shown in Fig. 4.
FIG. 5.
Analysis of total PA (A) and total splicing (B) of wild-type and mutant MXSVL substrates in the presence of increasing Mg2+ concentrations. The substrates analyzed are those shown in Fig. 2. Details are given in the text. The error of the data shown was ±2 to 4%.
(i) −3′SS Sp/PA substrate.
One of the first mutants tested was −3′SS Sp/PA (Fig. 2). The results are shown only in Fig. 5A (triangles), since this mutant has a point mutation in the 3′ splice site which completely eliminates splicing. This mutation has previously been shown to adversely affect PA; thus, it provided one of the first indications of coupling (35). Compared to the wild-type substrate (squares in Fig. 5), the −3′SS Sp/PA substrate showed that the mutation caused a significant loss of total PA at 1.5 mM Mg2+ (coupling conditions). Interestingly, at higher Mg2+ concentrations (uncoupling conditions), the total PA of the mutant substrate returned to a level similar to that of the wild-type substrate.
(ii) −PPT Sp/PA substrate.
The −PPT Sp/PA substrate (Fig. 2) had an Mg2+ titration profile similar to that of the −3′SS Sp/PA substrate, although the decrease in total PA at 1.5 mM Mg2+ was not as great. Total PA was consistently reduced from 26% to 21% (±1 to 2%) with −PPT Sp/PA, whereas it was reduced from 26% to 14% (±2 to 3%) with −3′SS Sp/PA. These results are not plotted in Fig. 5A due to their similarity to those obtained with the −3′SS Sp/PA substrate.
The above results indicate that mutations in the 3′ splice site or, to a lesser extent, the polypyrimidine tract which eliminate splicing result in the inhibition of PA specifically under coupling conditions. However, under uncoupling conditions, the mutations do not appear to affect PA. These findings strongly support the coupling of PA and splicing. A potential mechanism for the inhibitory effect of the −3′SS Sp/PA mutation under coupling conditions is proposed in the Discussion. The similarity of the effect when the polypyrimidine tract is mutated suggests that the coupling mechanism involves complexes interacting with both the 3′ splice site and the polypyrimidine tract.
(iii) −U1BS Sp/PA substrate.
Several reports have suggested that a 5′ splice site or potential U1 snRNP binding site located close to a PA signal (i.e., with no intervening 3′ splice site) inhibits PA (14, 18). This inhibition appears to be caused by U1 snRNP binding to the site, allowing the U1 snRNP 70K protein to interact with and inhibit the poly(A) polymerase in the PA complex (18). Considering this inhibition, we assessed the effect of mutating the U1 snRNP binding site in the SVLPA signal of the MXSVL substrate. The site (U1BS in Fig. 1 and 2) was mutated by LS (−U1BS Sp/PA in Fig. 2) and tested in an Mg2+ titration experiment (inverted solid triangles in Fig. 5). Mutation of the site increased total PA under coupling conditions (Fig. 5A), but total PA returned to a level similar to that of the wild-type substrate under uncoupling conditions. This result agrees with previous observations that such sites are inhibitory to PA (14, 18) and indicates further that inhibition is observed only when PA and splicing are coupled and is not effective under uncoupling conditions.
Total splicing (Fig. 5B) was also increased when the U1 snRNP binding site was mutated. Again, this effect was greatest under coupling conditions and declined under uncoupling conditions. Thus, it appears that the presence of the U1 snRNP binding site negatively modulates both splicing and PA under coupling conditions.
(iv) −USEs Sp/PA substrate.
LS mutagenesis of all three of the AUUUGURA upstream element motifs of the SVLPA signal (−USEs Sp/PA in Fig. 2) resulted in the inhibition of total PA. This inhibition increased with the Mg2+ concentration (circles in Fig. 5A). Conversely, these mutations had only a moderate effect on total splicing (circles in Fig. 5B), decreasing it to 75% the wild-type level under coupling conditions. Hence, the USEs appear to be primarily PA efficiency elements.
(v) −AUA Sp/PA substrate.
Mutation of AAUAAA to AAGAAA (−AUA Sp/PA in Fig. 2) completely eliminated PA, as expected, and also had the greatest effect of all of the mutations on total splicing (hourglasses in Fig. 5B). This mutation eliminates CPSF binding, suggesting that properly bound CPSF at AAUAAA is an important component in coupling, in agreement with previously proposed models (10, 23, 24, 33–35) (see Discussion).
(vi) DSE LSM substrates.
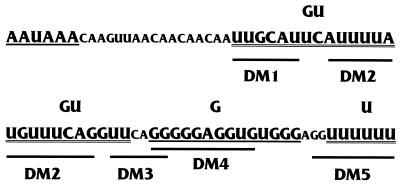
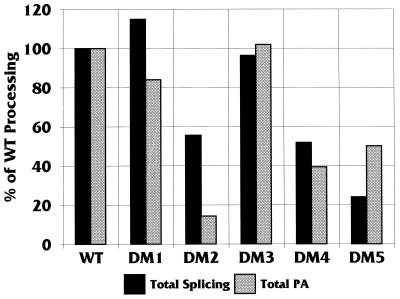
Figure 6 shows the sequence of the downstream region of the SVLPA signal, including the positions of the GU-, G-, and U-rich DSEs and the regions replaced by LS mutagenesis, DM1 through DM5 (the actual bases replaced are shown in Materials and Methods). Each mutation was inserted into the MXSVL substrate (Fig. 2) and tested for its effects on total splicing and total PA under coupling conditions (1.5 mM Mg2+). The data (Fig. 7) are presented as the percentage of wild-type total PA or the percentage of wild-type total splicing, where the wild-type (WT Sp/PA) levels of PA and splicing are set at 100%. The data suggest that three different DSE regions, defined by DM2, DM4, and DM5, significantly affected both PA and splicing. The other mutations, DM1 and DM3, had little effect on the coupling reaction. DM2, DM4, and DM5 each disrupted one of the three DSEs previously defined as being significant for PA (Fig. 1). Interestingly, in the WT PA only substrate (Fig. 2), the DM5 mutation had little effect on in vitro PA (10a). DM5 mutates the U-rich element, and the result suggests that this element has a unique function which is detected only under coupling conditions. In Fig. 5 (−DSE [inverted gray triangles]), the DM2 mutant substrate was subjected to the entire Mg2+ titration for comparison with the other mutant substrates.
FIG. 6.
Sequence of the downstream region of the SVLPA signal and positions of LSMs. The AAUAAA and the three DSEs (GU rich, G rich, and U rich) are shown in larger letters. The positions of LSMs DM1 to DM5 are indicated.
FIG. 7.
Total splicing and total PA of MXSVL substrates containing LSMs in the DSEs of the PA signal. Mutations DM1 to DM5 (Fig. 6) were introduced into the MXSVL substrate (see the DSM LSMs substrates in Fig. 2) and analyzed for in vitro splicing and PA under coupling conditions (1.5 mM Mg2+). Total splicing and total PA were quantitated as a percentage of processing obtained with the WT Sp/PA substrate (WT). The error of the data was ±2 to 4%.
DISCUSSION
This study represents a comprehensive examination of the effects of individual splicing elements and PA signal elements on the coupling of splicing and PA in an in vitro processing system. The results strongly support the coupling of PA and splicing. In addition, the data show that the 3′ splice site, the polypyrimidine tract, AAUAAA, and the individual DSEs each affect coupling. Further, the data suggest that coupling can be modulated by structures such as the U1 snRNP binding site (e.g., a 5′ splice site) in the last exon. The involvement of both the polypyrimidine tract and the 3′ splice site suggests that a pre-splicesosome or splicesosome structure is involved in coupling. Similarly, the involvement of the AAUAAA and the DSEs suggests that coupling requires the stable formation of at least the CPSF-CStF complex or, most likely, the entire PA complex, on the PA signal. Hence, it would appear that two relatively large complexes mediate coupling and last-exon definition.
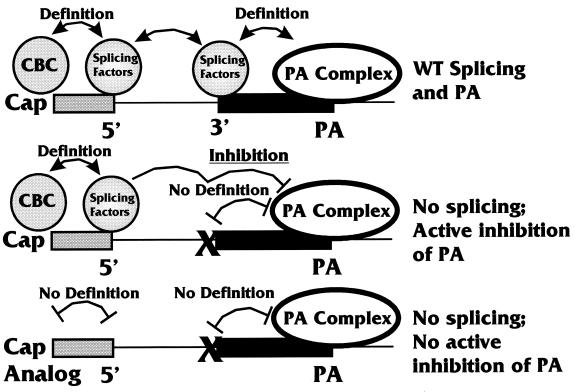
One of the most interesting manifestations of coupling was the inhibition of PA by mutations of the 3′ splice site, which was seen under conditions of coupling but not under uncoupling conditions. In order to postulate a mechanism for this finding, it is necessary to reconsider the suspected mechanisms of first-exon definition. As mentioned earlier, the 5′ cap structure of the pre-mRNA and the cap-proximal 5′ splice site are needed for first-exon definition (20). This process appears to involve an interaction between the CBC, bound at the cap, and U1 snRNP, bound at the 5′ splice site (22). It is also known that the CBC bound to the cap can affect PA (10). We showed that the WT PA only substrate (containing no splicing elements) (Fig. 2) was polyadenylated much less efficiently when it contained a cap analog to which the CBC could not bind (10). Mechanistically, the loss of PA efficiency results from the failure to complete a physical association between the CBC-cap complex and factors of the PA complex (13). Hence, the presence of a wild-type cap structure in the WT PA only substrate increases the efficiency of PA, and cap analogs decrease the efficiency. However, we noted that for the −3′SS Sp/PA substrate, a cap analog (to which the CBC could not bind) provided significantly higher levels of PA than did a wild-type cap on the same substrate (10). In other words, the inhibition of PA under coupling conditions was reduced when first-intron definition was debilitated by the use of a cap analog.
These data suggest the mechanism proposed in Fig. 8. The top diagram shows the processing of the wild-type substrate (wt Sp/PA), where wild-type splicing and PA would occur. The interactions between the CBC bound at a wild-type cap structure and the splicing factors bound at the 5′ splice site define the first exon. Similarly, the last exon is defined by interactions between splicing factors bound at the 3′ splice site (and polypyrimidine tract) and the PA complex bound at the PA site. These interactions may involve the U1 snRNP-specific U1A protein and the PA complex (19, 23, 24). With the exons properly defined, both splicing and PA can proceed. In the middle diagram in Fig. 8, the 3′ splice site has been mutated (indicated by the X), as in the −3′SS Sp/PA substrate. Here, the definition of the first exon can occur, but the subsequent exon cannot be defined due to the mutation. The data in Fig. 5A suggest that this situation leads to the inhibition of further processing. The suggestion that the complex defining the first exon mediates the inhibition is supported by our previous data (10) showing that the use of a cap analog (bottom diagram in Fig. 8) results not only in failure to form the complex which defines the first exon but also in decreased inhibition of PA. These data suggest a mechanism whereby the failure to properly define an exon causes the inhibition of subsequent processing of the RNA. This situation would provide a general processing checkpoint where a message would be completely processed only after the successful definition of all its exons.
FIG. 8.
Model for how the failure to properly define an exon may cause the definition complex on the previous exon to inhibit processing. WT, wild type.
Our data also suggest that the coupling of splicing and PA can be modulated by the U1 RNA binding site upstream of the SVLPA signal. It has been proposed that an interaction between this site and the U1 snRNP facilitates PA in vitro with a PA-only substrate (51). Using the MXSVL splicing and PA substrate, we found that mutation of the U1 binding site increased the efficiency of both PA and splicing. This result agrees with those of several studies indicating that U1 snRNP binding sites (e.g., 5′ splice sites) near PA signals inhibit PA (14, 18). Our findings extend this observation by suggesting that such sites may affect both PA and splicing; hence, they may modulate coupling.
In conclusion, our data suggest that specific splicing elements can affect PA and vice versa. This is an indication that they function in coupling. The effects of each element may be summarized as follows. (i) The 5′ splice site can be mutated with little effect on PA as long as the 3′ splice site end polypyrimidine tract is intact (10). (ii) Mutation of the polypyrimidine tract or the 3′ splice site significantly inhibits PA, suggesting that splicing complexes bound to these elements play a major role in coupling. Mutation of these elements, especially the 3′ splice site, results in the inhibition of PA under coupling conditions; this inhibition appears to be mediated by the complex which defines the previous exon. (iii) The USEs of the PA signal appear to be primarily PA elements; mutation of these elements has only a moderate effect on splicing. (iv) AAUAAA and the DSEs affect not only PA but also the efficiency of splicing. This result suggests that the stable formation of the PA complex on the PA signal affects the efficiency of splicing.
ACKNOWLEDGMENTS
We thank Susan Berget for providing extracts and reagents. We also thank the members of the Alwine laboratory for helpful discussion and support.
This work was supported by Public Health Service grant GM45773 provided to J.C.A. by the National Institutes of Health.
REFERENCES
- 1.Bagga P S, Ford L P, Chen F, Wilusz J. The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-factor. Nucleic Acids Res. 1995;23:1625–1631. doi: 10.1093/nar/23.9.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berget S M. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 3.Bhat B M, Wold W S M. ATTAAA as well as downstream sequences are required for RNA 3′-end formation in the E3 complex transcription unit of adenovirus. Mol Cell Biol. 1985;5:3183–3193. doi: 10.1128/mcb.5.11.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brackenridge S, Ashe H L, Giacca M, Proudfoot N J. Transcription and polyadenylation in a short human intergenic region. Nucleic Acids Res. 1997;25:2326–2335. doi: 10.1093/nar/25.12.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown P H, Tiley L S, Cullen B R. Efficient polyadenylation with the human immunodeficiency virus type I long terminal repeat requires flanking U3-specific sequences. J Virol. 1991;65:3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carswell S, Alwine J C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989;9:4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiou H, Dabrowski C, Alwine J C. Simian virus 40 late mRNA leader sequence invovled in augmenting mRNA accumulation via multiple mechanisms, including increased polyadenylation efficiency. J Virol. 1991;65:6677–6685. doi: 10.1128/jvi.65.12.6677-6685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 9.Conway L, Wickens M. A sequence downstream of AAUAAA is required for formation of simian virus 40 late mRNA 3′ termini in frog oocytes. Proc Natl Acad Sci USA. 1985;82:3949–3953. doi: 10.1073/pnas.82.12.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke C, Alwine J C. The cap and 3′-splice site have equivalent effects on polyadenylation efficiency. Mol Cell Biol. 1996;16:2579–2584. doi: 10.1128/mcb.16.6.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Cooke, C., and J. C. Alwine. Unpublished data.
- 11.DeZazzo J D, Imperiale M J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989;9:4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeZazzo J D, Kilpatrick J E, Imperiale M J. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type I mRNA 3′-end formation. Mol Cell Biol. 1991;11:1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaherty S M, Fortes P, Izaurralde E, Mattaj I W, Gilmartin G M. Participation of the nuclear cap binding complex in pre-mRNA 3′ processing. Proc Natl Acad Sci USA. 1997;94:11893–11898. doi: 10.1073/pnas.94.22.11893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furth P A, Choe W-T, Rex J H, Byrne J C, Baker C C. Sequences homologous to 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol Cell Biol. 1994;14:5278–5289. doi: 10.1128/mcb.14.8.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil A, Proudfoot N J. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit β-globin mRNA formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 16.Gil A, Proudfoot N J. A sequence downstream of AAUAAA is required for rabbit β-globin mTNS 3′-end formation. Nature. 1984;312:473–474. doi: 10.1038/312473a0. [DOI] [PubMed] [Google Scholar]
- 17.Gilmartin G M, Nevins J R. An ordered pathway of assembly of components required for polyadenylation site recognition and processing. Genes Dev. 1989;3:2180–2189. doi: 10.1101/gad.3.12b.2180. [DOI] [PubMed] [Google Scholar]
- 18.Gunderson S I, Polycarpou-Schwarz M, Mattaj I W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 19.Gunderson S I, Vagner S, Polycarpou-Schwarz M, Mattaj I W. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 20.Izaurralde E, Lewis J, McGuigan C, Jankowska M, Darzynklewicz E, Mattaj I W. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy C F, Kramer A, Berget S M. A role for SRp54 during intron bridging of small introns with pyrimidine tracts upstream of the branch point. Mol Cell Biol. 1998;18:5425–5434. doi: 10.1128/mcb.18.9.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis J D, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj I W. A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev. 1996;10:1683–1698. doi: 10.1101/gad.10.13.1683. [DOI] [PubMed] [Google Scholar]
- 23.Lutz C, Alwine J C. Direct interaction of the U1snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 24.Lutz C S, Murthy K G, Schek N, Manley J L, Alwine J C. Interaction between the U1snRNP-A protein and the 160 kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 25.McDevitt M A, Hart R P, Wong W W, Nevins J R. Sequences capable of restoring poly(A) site function define two distinct downstream elements. EMBO J. 1986;5:2907–2913. doi: 10.1002/j.1460-2075.1986.tb04586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDevitt M A, Imperiale M J, Ali H, Nevins J R. Requirement of a downstream sequence for generation of a poly(A) addition site. Cell. 1984;37:992–999. doi: 10.1016/0092-8674(84)90433-1. [DOI] [PubMed] [Google Scholar]
- 27.McLauchlan J, Gaffney D, Whitton J L, Clements J B. The consensus sequence YGTGTTYY located downstream from the AATAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 1985;13:1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore C L, Sharp P A. Site-specific polyadenylation in a cell-free reaction. Cell. 1984;36:581–591. doi: 10.1016/0092-8674(84)90337-4. [DOI] [PubMed] [Google Scholar]
- 29.Moreira A, Wollerton M, Monks J, Proudfoot N J. Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J. 1995;14:3809–3819. doi: 10.1002/j.1460-2075.1995.tb00050.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murthy K G K, Manley J L. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J Biol Chem. 1991;267:14804–14811. [PubMed] [Google Scholar]
- 31.Nesic D, Cheng J, Maquat L E. Sequences within the last intron function in RNA 3′-end formation in cultured cells. Mol Cell Biol. 1993;13:3359–3369. doi: 10.1128/mcb.13.6.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nesic D, Maquat L E. Upstream introns influence the efficiency of final intron removal and RNA 3′-end formation. Genes Dev. 1994;8:363–375. doi: 10.1101/gad.8.3.363. [DOI] [PubMed] [Google Scholar]
- 33.Niwa M, Berget S M. Mutation of the AAUAAA polyadenylation signal depresses in vitro splicing of proximal but not distal introns. Genes Dev. 1991;5:2086–2095. doi: 10.1101/gad.5.11.2086. [DOI] [PubMed] [Google Scholar]
- 34.Niwa M, MacDonald C C, Berget S M. Are vertebrate exons scanned during splice site selection? Nature (London) 1990;360:277–280. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- 35.Niwa M, Rose S D, Berget S M. In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev. 1990;4:1552–1559. doi: 10.1101/gad.4.9.1552. [DOI] [PubMed] [Google Scholar]
- 36.Qian Z, Wilusz J. An RNA-binding protein specifically interacts with a functionally important domain of the downstream element of the simian virus 40 late polyadenylation signal. Mol Cell Biol. 1991;11:5312–5320. doi: 10.1128/mcb.11.10.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robberson B L, Cote G J, Berget S M. Exon definition may facilitate splice site selection in RNAs with multiple exons. Mol Cell Biol. 1990;10:84–94. doi: 10.1128/mcb.10.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russnak R. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PS1 is orientation-dependent, distance-dependent, and additive. Nucleic Acids Res. 1991;19:6449–6456. doi: 10.1093/nar/19.23.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Russnak R, Ganem D. Sequences 5′ to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990;4:764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- 40.Ryner L C, Takagaki Y, Manley J L. Sequences downstream of AAUAAA signals affect pre-mRNA cleavage and polyadenylation in vitro both directly and indirectly. Mol Cell Biol. 1989;9:1759–1771. doi: 10.1128/mcb.9.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sachs A, Wahle E. Poly(A) tail metabolism and function in eucaryotes. J Biol Chem. 1993;268:22955–22958. [PubMed] [Google Scholar]
- 42.Sadofsky M, Alwine J C. Sequences on the 3′ side of hexanucleotide AAUAAA affect efficiency of cleavage at the polyadenylation site. Mol Cell Biol. 1984;4:1460–1468. doi: 10.1128/mcb.4.8.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sadofsky M, Connelly S, Manley J L, Alwine J C. Identification of a sequence element on the 3′ side of AAUAAA which is necessary for simian virus 40 late mRNA 3′-end processing. Mol Cell Biol. 1985;5:2713–2719. doi: 10.1128/mcb.5.10.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanfacon H, Brodmann P, Hohn T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 1991;5:141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- 45.Schek N, Cooke C, Alwine J C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal using in vitro cleavage and polyadenylation analyses. Mol Cell Biol. 1992;12:5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takagaki Y, Manley J L, MacDonald C C, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- 47.Valsamakis A, Schek N, Alwine J C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992;12:3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valsamakis A, Zeichner S, Carswell S, Alwine J C. The human immunodeficiency virus type 1 polyadenylation signal: a 3′-LTR element upstream of the AAUAAA necessary for efficient polyadenylation. Proc Natl Acad Sci USA. 1991;88:2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- 50.Wahle E, Keller W. The biochemistry of polyadenylation. Trends Biochem Sci. 1996;27:247–250. [PubMed] [Google Scholar]
- 51.Wassarman K M, Steitz J A. Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev. 1993;7:647–659. doi: 10.1101/gad.7.4.647. [DOI] [PubMed] [Google Scholar]
- 52.Wickens M. How the messenger got its tail: addition of poly(A) in the nucleus. Trends Biochem Sci. 1990;15:277–281. doi: 10.1016/0968-0004(90)90054-f. [DOI] [PubMed] [Google Scholar]
- 53.Wilusz J, Feig D I, Shenk T. The C proteins of heterogeneous nuclear ribonucleoprotein complexes interact with RNA sequences downstream of polyadenylation cleavage sites. Mol Cell Biol. 1988;8:4477–4483. doi: 10.1128/mcb.8.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilusz J, Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein-RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol Cell Biol. 1990;10:6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang F, Cole C N. Identification of a complex associated with processing and polyadenylation in vitro of herpes simplex virus type 1 thymidine kinase precursor RNA. Mol Cell Biol. 1987;7:3277–3286. doi: 10.1128/mcb.7.9.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]