Abstract
Creatine pyruvate (CrPyr) is a new multifunctional nutrient that can provide both pyruvate and creatine. It has been shown to relieve the heat stress of beef cattle by improving antioxidant activity and rumen microbial protein synthesis, but the mechanism of CrPyr influencing rumen fermentation remains unclear. This study aimed to combine 16S rDNA sequencing and metaproteomics technologies to investigate the microbial composition and function in rumen fluid samples taken from heat-stressed beef cattle treated with or without 60 g/day CrPyr. 16S rDNA sequencing revealed that there were no significant differences in the α-diversity indices between the two groups. By analyzing the level profiles of 700 distinct proteins, we found that the CrPyr administration increased the expression of enzymes involved in specific metabolic pathways including (i) fatty acid β-oxidation; (ii) interconversion from pyruvate to phosphoenolpyruvate, oxaloacetate, acetyl-CoA, and malate; (iii) glycolysis/gluconeogenesis and citrate cycle metabolism; and (iv) biosynthesis of amino acids. These results indicated that the increased generation of adenosine triphosphate during fatty acid β-oxidation or citrate cycle and the up-regulation synthesis of microbial protein in rumen of beef cattle treated with CrPyr may help decrease oxidative stress, regulate energy metabolism, and further improve the rumen fermentation characteristic under heat stress.
Keywords: beef cattle, creatine pyruvate, heat stress, 16S rDNA sequencing, metaproteomics, rumen microbiota protein
Introduction
Ruminal microbiota is a large systematic microbial ecosystem that is composed of bacteria, archaea, protozoa, and fungi (Hobson and Stewart, 1997). Microbial fermentation of feedstuff in the rumen produces volatile fatty acids (VFA) and microbial protein to provide the bulk of the energy and protein required by the host animal (Thomas et al., 2017). Hence, rumen microbes are essential for ruminant growth and health. However, many studies show that heat stress leads to severe defects in rumen function and variation in rumen microbial abundance and diversity (Tajima et al., 2007; Salles et al., 2010; Uyeno et al., 2010). Uyeno et al. (2010) reported that high temperature and humidity increased the abundance of Streptococcus and decreased the abundance of Fibrobacter and Oscillospira. Sales et al. (2021) showed that beef cattle under heat stress presented lower prokaryote richness, genera Flavonifractor, Treponema, and Ruminococcus, higher genera Carnobacterium. Moreover, the ruminal pH usually decreases under heat stress conditions (Tajima et al., 2007). A drop in ruminal pH often depressed cellulolytic ruminal bacterial growth and fiber digestion, promoted the cytolysis of gram-negative bacteria, and released lipopolysaccharides, further leading to inflammatory response on the gastrointestinal tract (Dai et al., 2017). Especially in southern China, due to the hot weather and high humidity, feedlot beef cattle finished in the summer months often suffer from heat stress. Therefore, the possibility of controlling the rumen microbial metabolism to achieve more efficient nutrient utilization under heat stress conditions has become an appealing concept for nutritionists.
Manipulation of feed additives such as antibiotics and vitamins is an efficient tool for relieving the negative impact of heat stress. However, a possible linkage between antibiotics in animal feed and the transmission of antimicrobial-resistant bacteria to humans has prompted researchers to explore other alternative approaches. As a new multifunctional nutrient, creatine pyruvate (CrPyr, C7H13N3O5, 219.20) contains pyruvic acid and creatine at a ratio of 40:60 (Chen et al., 2012), which both are natural body intermediate metabolites. This means that CrPyr can be used safely in the feed. In heat-stressed ruminants, the reduction of energy supply for microbial growth results in the decreased generation of microbial crude protein (MCP; Hyder et al., 2017). Pyruvate is the intermediate product of carbohydrate fermentation by rumen microorganisms (Nagaraja, 2016). The adenosine triphosphate (ATP) yield in the net conversion of pyruvate to VFA can promote microbial growth. Creatine represents one of the most important nitrogen-containing compounds in protein and energy metabolism and can provide a source of nitrogen for microbial protein synthesis (Navrátil et al., 2009). Thus, we hypothesized that CrPyr may enhance the synchronization of energy and nitrogen supply and be beneficial for ruminal characteristics and microbial growth, and our previous study has shown that dietary supplementation with CrPyr improved rumen microbial protein synthesis. Creatine and pyruvate are not only essential substrates for energetics but also critical antioxidants (Navrátil et al., 2009; Nagaraja, 2016; Rieger et al., 2017). The results in our previous study showed that dietary supplementation with CrPyr improved stress resistance and increased the antioxidant status and rumen fermentation of heat-stressed beef cattle (Liu et al., 2020). It is implied that supplementing with CrPyr could be an effective option to mitigate heat stress suffered by beef cattle. However, to date, detailed information about the changes in rumen microbial communities and related metabolic enzymes associated with the effects of CrPyr on rumen fermentation has not yet been published. Herein, an intensive study of rumen microbial diversity and microbial protein synthesis helps clarify the regulatory mechanism of CrPyr on rumen fermentation.
With the rapid development of bioinformatics, more researchers are starting to use meta-omics technology to define the rumen ecosystem’s ecology and its responses to changes in diet and rearing conditions (Gruninger et al., 2019). Previous studies have shown that 16S rDNA sequencing was employed to characterize the composition and diversity of rumen microbes in Holstein heifers at elevated environmental temperatures and humidity, but with little information about their function (Uyeno et al., 2010; Sales et al., 2021), while metaproteomics can provide the means to study the role of key enzymes involved in ruminant feed utilization efficiency under a particular set of conditions that dictate function. Analysis of the metaproteomes of the rumen microbial community would reveal details about microbial community activity, structure, function, and metabolic pathway transformations that are presently lacking (Hart et al., 2018). Therefore, in this study, we combined 16S rDNA sequencing and metaproteomics technologies to understand the regulatory mechanism of CrPyr on rumen fermentation deeply.
Materials and Methods
This experiment was approved by the Committee for the Care and Use of Experimental Animals at Jiangxi Agricultural University (JXAULL-20190017).
Animal Treatments and Experimental Diets
Six ruminally cannulated Jinjiang steers (initial body weight = 405 ± 21 kg) were randomly allocated into two treatments. Jinjiang cattle are a breed of Chinese indigenous beef cattle bred in the northwest of the Jiangxi Province. The Jinjiang cattle received a CrPyr free basal diet (control group, CG) or a 60 g/day CrPyr supplemental basal diet (experiment group, EG) for 22 days (2019/08/02–2019/08/23). The purity of CrPyr is 99.9% (Hubei Ju Sheng Technology Co., Ltd., Wuhan, China). Our previous study results indicated that the Jinjiang cattle fed with 60 g/day CrPyr had a better resistance to heat stress (Liu et al., 2020). Therefore, the present experiment selected a 60-g/day dose to explore the regulatory mechanism of CrPyr on rumen fermentation. CrPyr was added to the concentrates, divided into two daily feeds (07:00 and 16:00). After the concentrate was fed, rice straw was given. All steers were housed in individual solid concrete floor pens in a closed cowshed, while clean, fresh water was available at all times. The composition and nutrient levels of the experimental diet were shown in Supplementary Table 1.
Measurement of Temperature and Humidity Index, Body Temperature, and Respiratory Rate
The temperature and humidity index (THI) was measured according to Song et al. (2014). The thermohygrometer was suspended on the wall around the cattle’s bodies on both sides of the experimental herd (approximately 1.5 m above the ground). The temperature and relative humidity at 08:00, 14:00, and 20:00 were recorded each day. Calculation of the average daily THI and the Livestock Weather Safety Index (LWSI) classifications for heat stress used the method of LCI (LCI, 1970). The feed volume and 2-h postprandial residual feed volume of each cattle per day were recorded accurately to calculate the average dry matter intake (ADMI = total dry matter intake per cattle/22 days). Body temperature was measured via the rectum using a veterinary thermometer at 08:00, and their respiratory rate was measured via manual count at 08:30 and 14:00 until day 21 during the experimental period.
Sample Collection
Samples of feed, feces, and rumen fluid were collected. All feces samples were collected from each steer on days 19 to 21. The feces were collected using a plastic bucket placed behind the cattle. The feces were thoroughly mixed, and 5% of the total feces were sampled once daily. After collection, the 3 days of fecal samples were mixed, and a subsample (300 g) was frozen at –20°C and used later to measure dry matter, ash, crude fat, neutral detergent fiber, and acid detergent fiber contents. The other subsample (100 g) was thoroughly mixed with 20 ml of 10% concentrated sulfuric acid and stored at –20°C until it could be analyzed for crude protein.
On day 21, rumen fluid samples were collected from the upper, middle, and lower sites in the rumen immediately post-feeding in the morning. Approximately 100 ml of rumen fluid was collected from each collection site. The rumen fluid was immediately measured for pH using a portable pH meter (HANNA Instruments, Cluj-Napoca, Romania). The rumen fluid samples were then filtered through four cheesecloth layers, and the samples were divided into three portions. The first 8 ml of rumen fluid was mixed with 2 ml of 25% (wt/vol) metaphosphoric acid and used for VFA analysis. One subsample (10 ml) of rumen fluid was mixed with 2 ml of H2SO4 (1% vol/vol) for determination of NH3-N, and another subsample (10 ml) of rumen fluid was used for MCP and 16S rDNA sequencing and meta-proteomics analysis. These samples were frozen at –20°C until analysis.
Chemical Analyses
The VFA concentrations in the rumen fluid samples were determined using gas chromatography (Shimadzu GC-2014, Japan) equipped with a capillary column (Stabilwax, Restek, Bellefonte, PA, United States). The NH3-N concentration was measured using a TU-1901 spectrophotometer (Beijing Purkinje General Instrument Co. Ltd., China) according to a method described by Broderick and Kang (Broderick and Kang, 1980). The MCP production was determined according to the method of Makkar and Becker (1999).
The feed and feces samples were dried at 60°C and ground through a 1-mm stainless steel screen. The dry matter, ash, and crude fat of the feed and fecal samples were determined according to methods 967.03, 924.05, and 920.39 of the AOAC (Cunniff, 1995), respectively. The neutral detergent fiber and acid detergent fiber were analyzed using an Ankom A200i Fiber Analyzer (ANKOM Technology Co., New York, NY, United States) according to the methods of Van Soest et al. (1991). The total nitrogen contents of the feed and feces samples were determined according to procedure 984.13 of the AOAC (Cunniff, 1995).
16S rDNA Sequencing Analysis
DNA Extraction and PCR Amplification
According to the manufacturer, microbial community genomic DNA was extracted from ruminal fluid samples using the E.Z.N.A.® soil DNA Kit (Omega Bio-Tek, Norcross, GA, United States). The DNA extract was checked on 1% agarose gel, and DNA concentration and purity were determined with NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, United States). The hypervariable region V3–V4 of the bacterial 16S rRNA gene was amplified with primer pairs 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by an ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, United States). The PCR amplification of the 16S rRNA gene was performed as follows: initial denaturation at 95°C for 3 min, followed by 27 cycles of denaturing at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, and single extension at 72°C for 10 min, and end at 4°C. The PCR mixtures contain 5 × TransStart FastPfu buffer 4 μl, 2.5 mM dNTPs 2 μl, forward primer (5 μM) 0.8 μl, reverse primer (5 μM) 0.8 μl, TransStart FastPfu DNA Polymerase 0.4 μl, template DNA 10 ng, and finally ddH2O up to 20 μl. PCR reactions were performed in triplicate. The PCR product was extracted from 2% agarose gel and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) according to the manufacturer’s instructions and quantified using QuantusTM Fluorometer (Promega, United States).
Illumina MiSeq Sequencing
Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, United States) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The raw reads were deposited into the NCBI BioProject database (Accession Number: PRJNA717402).
Processing of Sequencing Data
The raw 16S rDNA gene sequencing reads were demultiplexed, quality-filtered by Trimmomatic, and merged by FLASH with the following criteria. (i) The 300-bp reads were truncated at any site receiving an average quality score of <20 over a 50-bp sliding window, and the truncated reads shorter than 50 bp were discarded; reads containing ambiguous characters were also discarded. (ii) Only overlapping sequences longer than 10 bp were assembled according to their overlapped sequence. The maximum mismatch ratio of overlap region is 0.2. Reads that could not be assembled were discarded. (iii) Samples were distinguished according to the barcode and primers, and the sequence direction was adjusted, exact barcode matching, two-nucleotide mismatch in primer matching.
Operational taxonomic units (OTUs) with 97% similarity cutoff were clustered using UPARSE (version 7.1, http://drive5.com/uparse/), and chimeric sequences were identified and removed. The taxonomy of each OTU representative sequence was analyzed by RDP Classifier1 against the 16S rRNA database (e.g., Silva 132/16s_bacteria) using a confidence threshold of 0.7.
Meta-Proteomics Analysis
Protein Extraction
The processed rumen fluid sample was centrifuged at 4°C for 5 min at 800 g to collect the supernatant. The supernatant was centrifuged at 7,000 g for 20 min at 4°C, then borax/PVPP/phenol (BPP: 100 mM EDTA, 50 mM borax, 50 mM Vitamin C, 30% sucrose w/v, 100 mM Tris Base, 1% Triton X-100 v/v, and 5 mM DTT, pH 8.0) was added in the ratio of 1:10 to the resultant precipitate and ground with liquid nitrogen three times, and each grind lasted for 120 s. The solution was centrifuged at 12,000 g for 20 min at 4°C, and the supernatant was collected. The equal volume of Tris-saturated phenol was added and vortexed for 10 min at 4°C. The solution was centrifuged at 12,000 g for 20 min at 4°C, and the phenol phase was collected. The equal volume of BPP was added and vortexed for 10 min at 4°C. The solution was centrifuged at 12,000 g for 20 min at 4°C, and the phenol phase was collected. Five volumes of precooled 0.1 M ammonium acetate in methanol were added, and the protein was precipitated at –20°C overnight. The supernatant was discarded by centrifugation, and the precipitate was washed twice with 90% acetone. The supernatant by was discarded by centrifugation, and the precipitate was air-dried. The precipitate was resuspended with lysis buffer (1% SDS, 8 M urea, protease inhibitor cocktail), then sonicated for 2 min on ice. The lysates were centrifuged at 12,000 g for 20 min at 4°C, and supernatants were collected to test the protein concentration in all samples. Protein concentrations were determined by the bicinchoninic acid method.
Protein Digestion
Protein digestion was performed according to the standard procedure. Briefly, for each sample tube containing 100 μg protein, appropriate triethylammonium bicarbonate (TEAB) buffer was added to the final concentration of 100 mM. Then, Bond-BreakerTM TCEP solution (TCEP) was added to the final concentration of 10 mM, and the tubes were incubated at 37°C for 60 min. Appropriate iodoacetamide was added to the final concentration of 40 mM and reaction for 40 min in the dark. Six volumes of cold acetone were added to the sample tube, and the tube was incubated at –20°C for 4 h. The acetone was removed by centrifugation at 10,000 g for 20 min, and precipitated protein was resuspended with 100 μl 100 mM TEAB buffer. To each sample tube, according to the 1:50 proportion, the trypsin solution was added and the tubes were incubated at 37°C overnight.
Peptide Desalination and Quantification
The peptides were vacuum dried, then resuspended with 0.1% trifluoroacetic acid. Samples were desalted with an Oasis® HLB 96-well 300-μm plate and vacuum dried. Peptide concentrations were determined by the Thermo Fisher Scientific peptide quantification kit (Thermo, Cat. 23275). The loading buffer was added to each tube to prepare samples for mass spectrometry analysis, and the concentration of each sample was 0.25 μg/μl.
Mass Spectrometry Analysis
Experiments were performed on a Q Exactive HF-X mass spectrometer that was coupled with Easy-nLC 1200. Each peptide sample was injected for nanoLC-MS/MS analysis. The sample was loaded onto a C18 reversed-phase column (75 μm × 25 cm, Thermo, United States) in buffer A (2% acetonitrile and 0.1% formic acid) and separated with a linear gradient of buffer B (80% acetonitrile and 0.1% formic acid) at a flow rate of 300 nl/min. The electrospray voltage of 1.8 kV vs. the inlet of the mass spectrometer was used. A Q Exactive HF-X mass spectrometer was operated in data-dependent mode to switch automatically between MS and MS/MS acquisition. Survey full-scan MS spectra (m/z 350–1,300) were acquired with a mass resolution of 70 K, followed by 20 sequential high-energy collisional dissociation MS/MS scans with a resolution of 17.5 K. In all cases, one microscan was recorded using the dynamic exclusion of 30 s. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium2 via the iProX partner repository (Ma et al., 2019) with the dataset identifier PXD025118.
Taxonomic Analysis of Peptides and Identification of Proteins
MS/MS spectra were searched using Proteome DiscovererTM Software 2.2 software against the UniProt database (uniprot-taxonomy_3A171549_3A186802_3A191303_.fasta/LL_PEAKS_ exported_proteins.fasta) as the following parameters. The highest score for a given peptide mass (best match to that predicted in the database) was used to identify parent proteins. The parameters for protein searching were set as follows: tryptic digestion with up to two missed cleavages, carbamidomethylation of cysteines as fixed modification, and oxidation of methionines and protein N-terminal acetylation as variable modifications. Peptide spectral matches were validated based on q-values at a 1% false discovery rate.
Informatics Protocol
The protein quantification and calculation of statistical significance were carried out using the t-test in the R language. To perform functional analysis, the identified proteins were carried on the follow-up biological information function analysis using GO (Gene Ontology, http://www.geneontology.org/). Pathway mapping of identified proteins was performed using the KEGG database (Kyoto Encyclopedia of Genes and Genomes, http://www.genome.jp/kegg/).
Statistical Analysis
Statistical analysis of body temperature, respiratory rate, rumen fermentation, nutrient digestibility, and taxonomy was first tested for Gaussian distribution using the Shapiro–Wilk test and performed by independent sample T-test with SPSS (SPSS 2008) software. Mean values and standard error (SE) were reported, and differences were considered to be significant at P < 0.05.
Results
Temperature and Humidity Index
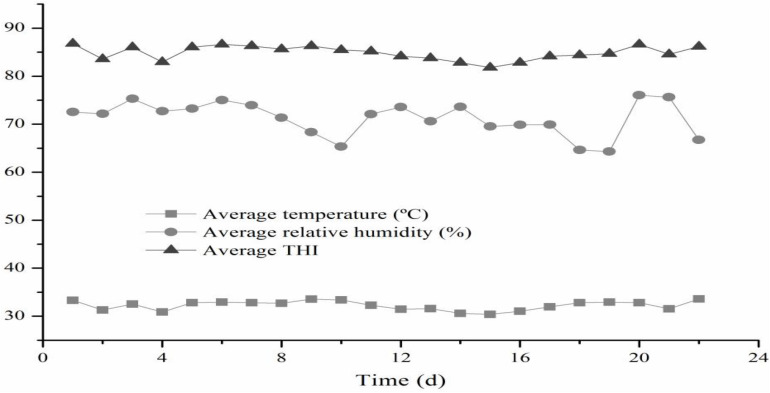
As shown in Figure 1, there were three times daily changes in THI values in the morning (08:00), noon (14:00), and evening (20:00) during the experimental period. The average daily THI values during the experimental period were higher than 79 for all 22 experimental days.
FIGURE 1.
Temperature-humidity index in the house of the beef cattle. THI, temperature and humidity index.
Average Dry Matter Intake, Body Temperature, Respiratory Rate, Rumen Fermentation, and Nutrient Digestibility
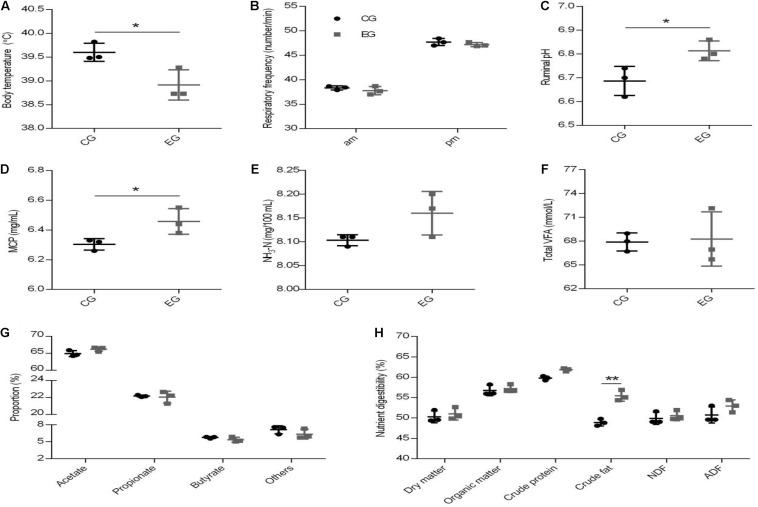
There was no difference in the ADMI between the EG and CG groups (4.89 kg vs. 4.89 kg, P = 0.081). Dietary supplementation with CrPyr decreased the body temperature of beef cattle (Figure 2A, P < 0.05), but did not affect the respiratory rate of beef cattle (Figure 2B). Diet supplemented with CrPyr increased the ruminal pH value (Figure 2C, P < 0.05) and MCP concentration (Figure 2D, P < 0.05). No effect was observed on the contents of NH3-N (Figure 2E), total VFA (Figure 2F), or individual VFA (Figure 2G). As shown in Figure 2H, CrPyr supplementation increased the crude fat digestibility (P < 0.05). No difference was found between groups for the digestibility of the dry matter, organic matter, crude protein, neutral detergent fiber, and acid detergent fiber.
FIGURE 2.
Effect of creatine pyruvate on body temperature (A), respiratory frequency (B), rumen fermentation parameter (C–G), and nutrient digestibility (H) of beef cattle. The rumen fermentation parameter including ruminal pH (C), the concentration of microbial crude protein [MCP; (D), NH3-N (E), and total VFA (F), the proportion of individual VFA (G). CG, control group; EG, experimental group, diet supplemented with 60 g/day CrPyr. n = 3 per group. *P < 0.05, **P < 0.01.
Diversity of Rumen Fluid Microbiota of Beef Cattle as Revealed by 16S rDNA High-Throughput Sequencing
The high-throughput sequencing technology was used to investigate the effects of CrPyr on microbial communities in the rumen of beef cattle. The sequences were clustered into 1,306 OTUs with a similarity of 97%. Among these, a total of 1,098 OTUs were common, 109 OTUs were specific for the EG, and the CG had 99 unique OTUs (Supplementary Figure 1). The rumen microbiota α-diversity of the two groups was evaluated by the Ace index, Chao1 index, Shannon index, and Simpson index (Supplementary Table 2). According to the result, there were no significant differences in the α-diversity index between the two groups.
Metaproteomics Analyses of Proteins in the Rumen Fluid Microbiota of Beef Cattle
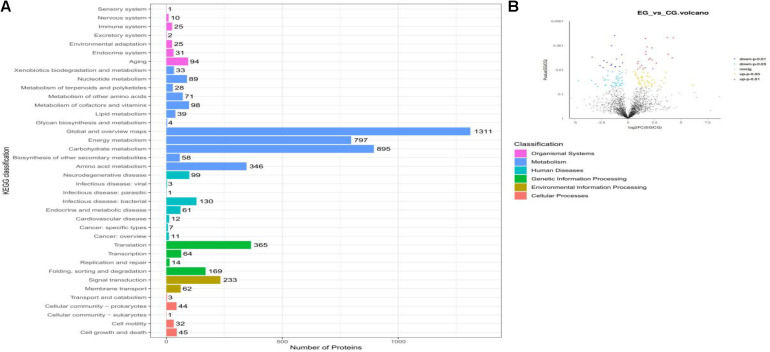
Label-free quantification proteomics was applied to investigate the rumen fluid microbiota proteins of beef cattle. A total of 716,150 MS/MS spectra were generated from the six rumen fluid samples. Of these, 11,361 peptides (16.2%) could be identified and assigned to 3,579 proteins (Supplementary Table 3). Of the proteins identified, most were related to cell metabolism (3,407). Others were related to organismal systems (188), human diseases (324), genetic information processing (612), environmental information processing (295), and cellular processes (125; Figure 3A). They included 2,997 proteins that were quantified in the rumen fluid microbiota of EG and CG. The number of overlapping proteins between the two groups was 2,153 (71.8%), while 373 and 206 community-specific proteins were unique to the rumen fluid microbiota in EG and CG, respectively, the number of quantified proteins without quantitative value (refers to a protein expressed in samples with less than two-thirds in both groups) was 265. Furthermore, in the 2,153 overlapping proteins, 121 differentially expressed proteins (DEPs) were identified in the EG compared with the CG, of which 67 proteins were up-regulated and 54 proteins were down-regulated (Figure 3B, P < 0.05).
FIGURE 3.
Metaproteomics analyses of proteins in the rumen fluid microbiota of beef cattle fed a CrPyr supplementation diet. (A) KEGG classification. (B) Volcano plots of differentially expressed proteins between EG and CG. EG, experimental group, diet supplemented with 60 g/day CrPyr; CG, control group. Each dot represents the mean expression level (n = 3) of a single protein. The yellow dots indicate the significantly up-regulated proteins at the P < 0.05 level, the red dots indicate the significantly up-regulated proteins at the P < 0.01 level, the light blue dot indicates the significantly down-regulated proteins at the P < 0.05 level, and the blue dot indicates the significantly down-regulated proteins at the P < 0.01 level.
Composition of Rumen Fluid Microbiota of Beef Cattle as Revealed by 16S rDNA Sequencing and Metaproteomics
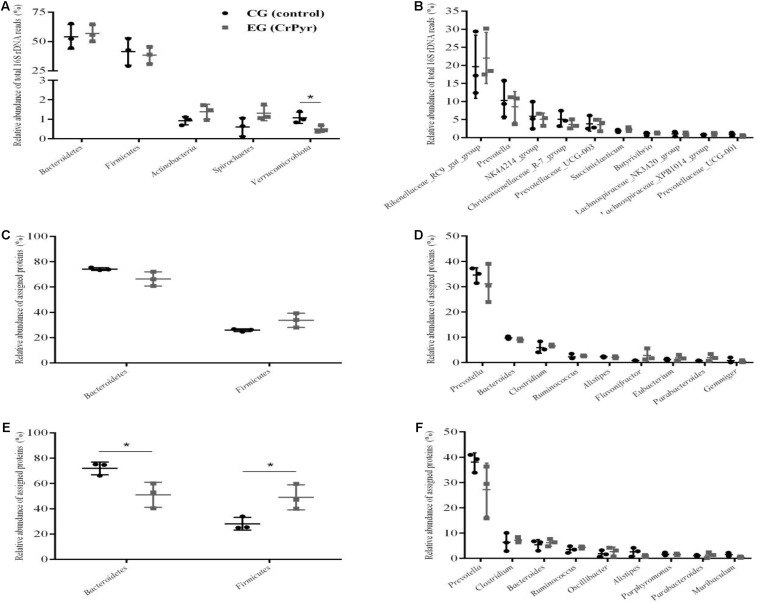
16S rDNA sequencing and metaproteomics were combined to investigate the effects of CrPyr on both the structure and function of rumen fluid microbiota of beef cattle. 16S rDNA sequencing revealed that bacteria belonging to the phyla Bacteroidetes and Firmicutes comprised most (the average coverage was ∼95.4%) of the total bacteria in the rumen fluid microbiota of the CG and EG. The remaining bacteria were mainly members of Actinobacteria, Spirochaetes, and Verrucomicrobiota (Figure 4A). The 16S rDNA relative abundances (abbreviated as 16SDA hereafter) of members of Bacteroidetes increased from 54.08% with CG to 56.89% with EG, the Firmicutes 16SDA decreased from 41.47% to 38.35%, and the Verrucomicrobiota 16SDA was significantly different between the two groups (P < 0.05; Figure 4A). At the genus level, the Rikenellaceae_RC9_gut_group was the dominant bacteria, and the other abundant genera were Prevotella, NK4A214_group, Christensenellaceae_R-7_group, Prevotellaceae_UCG-003, and Succiniclasticum (Figure 4B). At the species level, the unclassified_g_Rikenellaceae_RC9_gut_group 16SDA was significantly higher in the EG group than in the CG group (P < 0.05; Supplementary Figure 2).
FIGURE 4.
Phylogenetic classification and metaproteomics analyses of rumen fluid microbiota of beef cattle fed a CrPyr supplementation diet. (A,B) show phylum-level (A) and genus-level (B) rumen fluid community composition based on Illumina sequences of 16S rDNA amplicons (V3–V4 region). (C,D) show relative abundances of total identified proteins in rumen fluid assigned to bacterial phyla (C) and genera (D). (E,F) show relative abundances of differentially expressed proteins in rumen fluid assigned to bacterial phyla (E) and genera (F). CG, control group; EG, experimental group, diet supplemented with 60 g/day CrPyr. n = 3 per group. *P < 0.05.
Metaproteomics showed that the protein relative abundances (abbreviated as PRA hereafter) of members of Bacteroidetes in total quantified proteins decreased from 74.15% with CG to 66.30% with EG, and the Firmicutes PRA increased from 25.85% to 33.70% (Figure 4C). At the genus level, the proteomic data showed that members of Prevotella were the dominant proteins, and the other abundant proteins were members of Bacteroides, Clostridium, Ruminococcus, and Alistipes (Figure 4D).
Composition and Functional Classification of Differentially Expressed Proteins as Revealed by Metaproteomics
The metaproteomics data showed all 700 (373 unique in EG + 206 unique in CG + 121 differential overlapping, Supplementary Table 4). DEPs were members of Bacteroidetes and Firmicutes (Figure 4E). The Bacteroidetes PRA was increased, and the Firmicutes PRA was decreased significantly in the EG group than those in the CG group (P < 0.05). At the genus level, similar to the total quantified proteins (Figure 4D), the members of Prevotella were also the dominant proteins in DEPs (Figure 4F), and the other abundant proteins were Clostridium, Bacteroides, and Ruminococcus (Figure 4F).
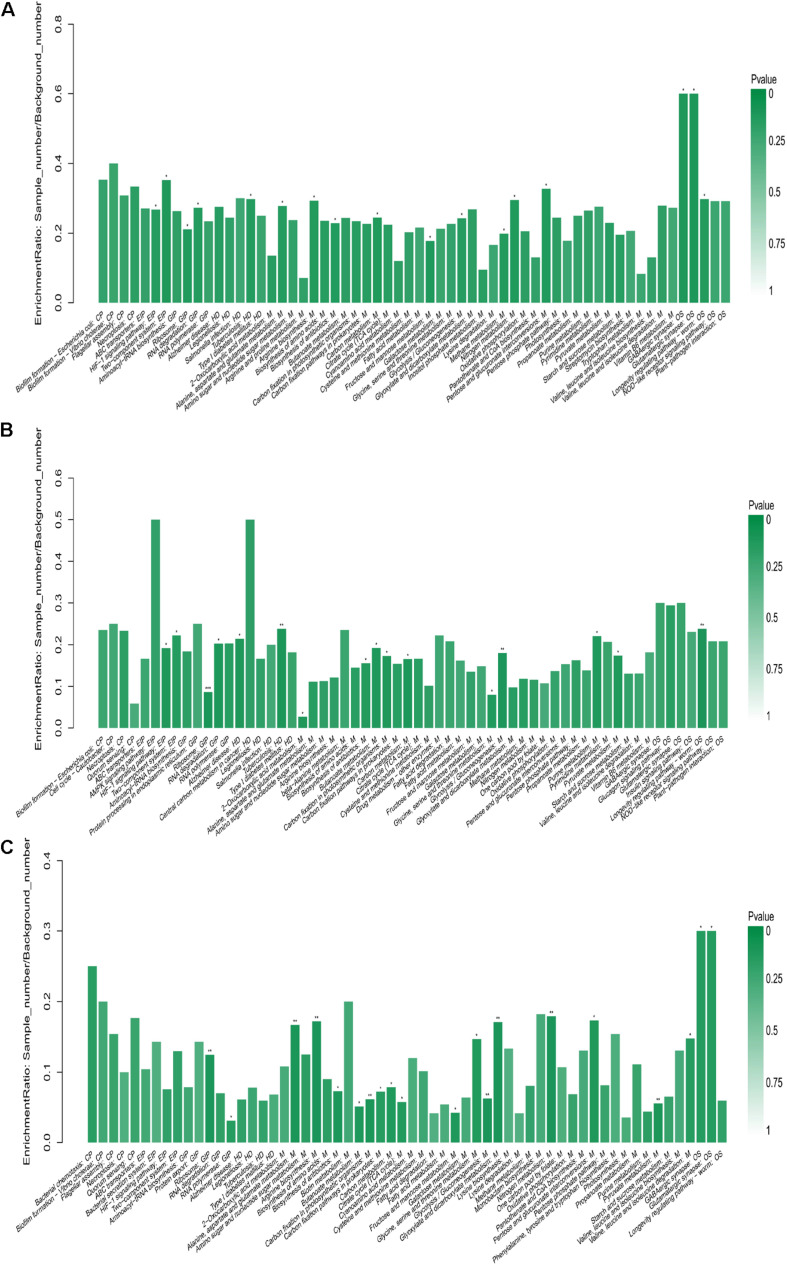
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was used to explore the biological pathways for DEPs between the rumen fluid microbiota of EG and CG. Figure 5A shows the KEGG pathway enrichment analysis of all DEPs. Figure 5B shows the KEGG pathway enrichment analysis of all up-regulated DEPs; diet supplemented with CrPyr significantly enriched proteins involved in environmental information processing (HIF-1 signaling pathway, two-component systems), genetic information processing (ribosome, RNA degradation), human diseases (Alzheimer’s disease, tuberculosis), metabolism (2-oxocarboxylic acid metabolism, biosynthesis of antibiotics, butanoate metabolism, carbon fixation in photosynthetic organisms, carbon metabolism, glycine, serine, and threonine metabolism, glycolysis/gluconeogenesis, purine metabolism, and pyruvate metabolism), organismal systems (longevity regulating pathway-worm). Figure 5C shows the KEGG pathway enrichment analysis of all down-regulated DEPs; diet supplemented with CrPyr significantly decreased proteins involved in genetic information processing (ribosome, RNA polymerase), metabolism (alanine, aspartate, and glutamate metabolism, arginine biosynthesis, biosynthesis of antibiotics, butanoate metabolism, carbon fixation in photosynthetic organisms, carbon fixation pathway in prokaryotes, carbon metabolism, citrate cycle, fructose and mannose metabolism, glycine, serine, and threonine metabolism, glycolysis/gluconeogenesis, glyoxylate and dicarboxylate metabolism, nitrogen metabolism, pentose and glucuronate interconversions, pyruvate metabolism, valine, leucine, and isoleucine degradation), and organismal systems (GABAergic synapse, glutamatergic synapse).
FIGURE 5.
KEGG enrichment of differentially expressed proteins. (A) KEGG enrichment analysis of all differentially expressed proteins. (B) KEGG enrichment analysis of all up-regulated differentially expressed proteins (experimental group vs. control group, EG vs. CG). (C) KEGG enrichment analysis of all down-regulated differentially expressed proteins (experimental group vs. control group, EG vs. CG). CP, cellular processes; EIP, environmental information processing; GIP, genetic information processing; HD, human diseases; M, metabolism; and OS, organismal systems.
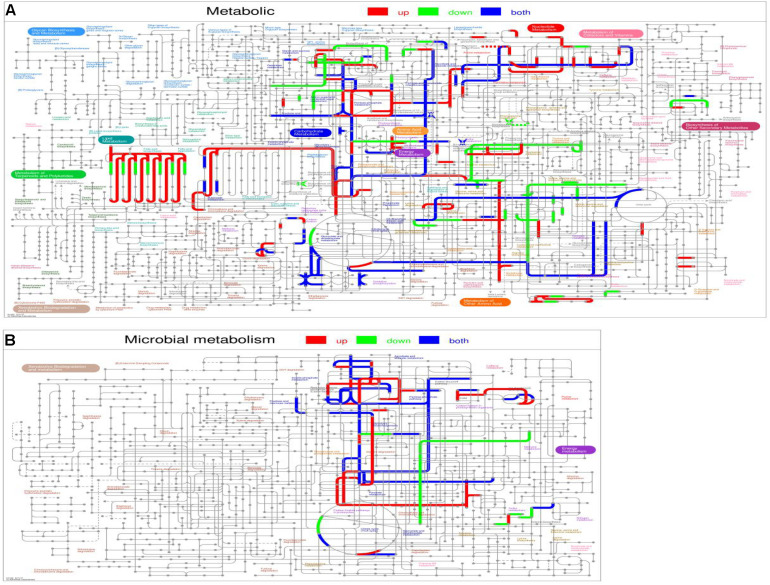
Interactive Pathways Explorer (IPath) analysis was also used to visualize the mutual relationship of DEPs in metabolic (Figure 6A) and microbial metabolism (Figure 6B), on which red lines show up-regulated pathways, green lines show down-regulated pathways, and blue lines show both up-regulated and down-regulated pathways. As shown in metabolic and microbial metabolism, the up-regulated pathways mainly including lipid metabolism, glycolysis/gluconeogenesis, and pyruvate metabolism.
FIGURE 6.
Interactive Pathways Explorer (iPath) analysis. (A) Metabolic. (B) Microbial metabolism. Red lines show up-regulated pathways, green lines show down-regulated pathways, and blue lines show both up-regulated and down-regulated pathways.
Administration of CrPyr Affected Core Enzymes Related to Key Metabolism
Fatty Acid Metabolism
As shown in Figure 7A and Supplementary Table 5, diet supplemented with CrPyr up-regulated the Acyl-CoA dehydrogenase (EC: 1.3.8.1) from Lachnoclostridium sp., Lachnospiraceae bacterium, and Oscillibacter sp. but down-regulated this enzyme from Muribaculum. Exposure to CrPyr up-regulated the acetyl-CoA C-acetyltransferase (ACAA; EC: 2.3.1.9) from Oscillibacter sp. Moreover, the 3-oxoacyl-[acyl-carrier-protein] synthase 2 (EC: 2.3.1.179) from Lentimicrobiaceae bacterium was up-regulated, and the 3-oxoacyl-[acyl-carrier-protein] reductase (EC: 1.1.1.100) from Prevotella sp. BP1-148 was down-regulated. The description of DEPs involved in fatty acid (FA) metabolism is shown in Supplementary Table 5.
FIGURE 7.
Protein identity and regulation involved in several important pathway in rumen fluid samples of beef cattle fed with a CrPyr supplementation diet (experimental group vs. control group, EG vs. CG). (A) Fatty acid metabolism. (B) Pyruvate metabolism. (C) Glycolysis/gluconeogenesis. (D) Citrate cycle (TCA cycle). (E) Nitrogen metabolism. (F) Biosynthesis of amino acids.
Pyruvate Metabolism
As shown in Figure 7B, for the pyruvate metabolism pathway, 62 DEPs were identified in the EG compared with the CG, of which 47 proteins were up-regulated and 15 proteins were down-regulated. Of the 47 up-regulated proteins, most were pyruvate, phosphate dikinase (PPDK; EC: 2.7.9.1; LCI, 1970). Others were pyruvate:ferredoxin (flavodoxin) oxidoreductase (Sales et al., 2021) and pyruvate-flavodoxin oxidoreductase (Thomas et al., 2017; EC: 1.2.7.1), phosphoenolpyruvate carboxykinase (ATP; Dai et al., 2017; EC: 4.1.1.49), and so on. The description of DEPs involved in pyruvate metabolism is shown in Supplementary Table 6.
Glycolysis/Gluconeogenesis
As shown in Figure 7C, for the glycolysis/gluconeogenesis, 101 DEPs were identified in the EG compared with the CG, of which 75 proteins were up-regulated and 26 proteins were down-regulated. Except for EC: 1.2.7.1, EC: 4.1.1.49, and EC: 1.2.7.11, which were also involved in pyruvate metabolism, the most up-regulated protein was glyceraldehyde-3-phosphate dehydrogenase (Van Soest et al., 1991), including 3 Type I glyceraldehyde-3-phosphate dehydrogenase (Fragment; EC: 1.2.1.12). Others were fructose-1,6-bisphosphate aldolase, class II and its isozyme (Dai et al., 2017; EC: 4.1.2.13), phosphoglycerate kinase (Uyeno et al., 2010; EC: 2.7.2.3), pyrophosphate-fructose 6-phosphate 1-phosphotransferase (Salles et al., 2010; EC: 2.7.1.90), enolase (Salles et al., 2010; EC: 4.2.1.11), and so on. The description of DEPs involved in glycolysis/gluconeogenesis is shown in Supplementary Table 7.
Citrate Cycle
As shown in Figure 7D, for the citrate cycle, 39 DEPs were identified in the EG compared with CG, of which 29 proteins were up-regulated and 10 proteins were down-regulated. Except for EC: 1.2.7.1, EC: 1.2.7.11, EC: 4.1.1.49, and EC: 6.4.1.1, which are also involved in pyruvate metabolism, the most up-regulated protein was succinate dehydrogenase/fumarate reductase and its isozyme (Salles et al., 2010; EC: 1.3.5.4; EC: 1.3.5.1). The description of DEPs involved in citrate cycle is shown in Supplementary Table 8.
Nitrogen Metabolism and Biosynthesis of Amino Acids
As shown in Figure 7E, for the nitrogen metabolism pathway, 28 DEPs were identified in EG compared with CG, of which 11 proteins were up-regulated and 17 proteins were down-regulated. Of the 11 up-regulated proteins, eight were glutamate dehydrogenase (GDH; EC: 1.4.1.4) and three glutamine synthetase (GS; EC: 6.3.1.2). As shown in Figure 7F, for the biosynthesis of amino acids, 94 DEPs were identified in the EG compared with the CG, of which 58 proteins were up-regulated and 36 proteins were down-regulated. The description of DEPs involved in nitrogen metabolism and biosynthesis of amino acids is shown in Supplementary Tables 9, 10.
Discussion
The LWSI classifications for heat stress are as follows: normal, ≤74; alert, 74 ≤ THI ≤ 79; danger, 79 ≤ THI ≤ 84; and emergency, THI ≥ 84 (LCI, 1970). The present study indicated that the experimental cattle were in a state of high heat stress due to the high temperature and humidity during the summer months. Consequently, to seek thermoregulation, the animal employs a series of physiological and metabolic changes, including an increased rectal temperature, respiration rate, panting and open-mouth breathing, water consumption, and a decrease in feed intake (Marai et al., 2007). Body temperature is an excellent indicator of an animal’s susceptibility to heat load, and the body core temperature of cattle in thermo-neutral conditions is maintained between 38 and 39.2°C (Ammer et al., 2016). In this study, in comparison to the control group, dietary supplementation with CrPyr significantly decreased the rectal temperature of cattle, which means CrPyr could relieve a heat stress-induced higher body temperature. Creatine is an osmotically active substance, and the osmotic effect of creatine might affect the thermoregulatory processes during exercise. Some research has suggested that, in humans, a potential mechanism involving increased extracellular fluid induced by added creatine may help the body deal with heat stress (Mendel et al., 2005).
Rumen pH value and rumen bacterial populations are decreased during heat stress (Tajima et al., 2007; Bernabucci et al., 2009). Rumen pH value is the result of the combined effect of factors such as the VFA in rumen fluid, buffer salt in saliva, organic acid produced by metabolism, NH3-N level, and emptying speed of rumen fluid. The production of NH3-N in the rumen can increase the rumen pH value. In the present study, CrPyr supplementation increased the rumen pH value. This might be attributed to the slow continuous release of ammonia from creatine. As previously reported, calcium pyruvate supplementation of goat diets could increase the content of VFA in the rumen (Chen et al., 2006; Ran et al., 2008). Moreover, creatine has been widely used as a nitrogen supplement in ruminant nutrition in the past few decades (McLaren, 1964). However, there was no significant difference in VFA or NH3-N concentration for beef cattle fed CrPyr in heat stress. This may be due to that the VFA production induced by pyruvate and the simultaneously slow continuous release of ammonia from creatine in the rumen were partly used by rumen microbes to synthesize proteins. Consistently, we found CrPyr increased the content of MCP in the present study.
In this study, CrPyr supplementation significantly increased the proportion of unclassified_g_Rikenellaceae_RC9_gut_group in the rumen of heat-stressed beef cattle (Supplementary Figure 2), which can produce VFA (mainly acetate and propionate) and promote cellulose digestion. Also, Cheng (2019) reported that rumen calcium pyruvate perfusion based on a high-concentrate diet (F:C, 6:4) increased the ratio of Rikenellaceae_RC9_gut_group in the rumen of nonpregnant dairy goats. This may illustrate that pyruvate is favorable for Rikenellaceae_RC9_gut_group reproduction and further contributes benefits to cellulose digestion. While our results showed that the neutral detergent fiber and acid detergent fiber digestibility between the EG and CG groups was not significantly different, this may be due to the main ruminal cellulolytic bacteria including Ruminococcus albus, Ruminococcus flavefaciens, and Fibrobacter succinogenes were not affected by CrPyr.
The results in the present study showed that supplementing with CrPyr significantly increased the crude fat digestibility. Simultaneously, the IPath analysis showed the administration of CrPyr-enriched proteins involved in lipid metabolism (Figure 6A), including those involved in FA degradation and FA biosynthesis, in the rumen fluid microbiota of beef cattle. The present study was the first metabolic evidence to demonstrate that CrPyr promotes the expression of enzymes involved in lipid metabolism of the rumen fluid microbiota of beef cattle. Indeed, in the rumen, bacteria are largely responsible for biohydrogenation of dietary unsaturated FA (Harfoot and Hazlewood, 1997), and few studies in the literature have reported rumen bacteria FA β-oxidation (Emmanuel, 1978). However, in the anaerobic digestion process long-chain FAs and short-chain FAs can be degraded via the β-oxidation pathway catalyzed by bacteria (Kim et al., 2004; Hao et al., 2020), which may suggest that FA β-oxidation may also occur under anaerobic conditions in the rumen. The metaproteomics screening in the present study detected that the expressions of short-chain acyl-CoA dehydrogenase (SCAD) and ACAA (Supplementary Figure 3 and Supplementary Table 5) were significantly up-regulated in the CrPyr-supplemented group compared with the control group. Acyl-CoA dehydrogenase catalyzes the initial rate-limiting step, and ACAA catalyzes the last step of the FA β-oxidation. They are all key enzymes of the FA oxidation (Andresen et al., 1999; Zhang et al., 2018). In the present study, the up-regulation of SCAD from Lachnoclostridium sp., L. bacterium, and Oscillibacter sp. catalyzes the dehydrogenation of hexanoyl-CoA and butanoyl-CoA. The up-regulation of ACAA from Oscillibacter sp. catalyzes the reaction of 3-oxo-hexanoyl-CoA and CoA to form acetyl-CoA and butanoyl-CoA, and the formation of two molecules of acetyl-CoA from acetoacetyl-CoA. Then, acetyl-CoA enters into the citrate cycle providing energy for metabolism. Moreover, Zhu et al. (2017) reported that the dominant species in the microbiota composition were L. bacterium 10_1 and Oscillibacter sp. 1_3 in stool samples of mice with resistance to high-fat diet-induced obesity. Ma et al. (2020) showed that L. bacterium DW67 was one of the key mediators in exerting antiobesity induced by cold-water brewed green tea. This may suggest that L. bacterium and Oscillibacter sp. have potential for FA oxidation in rumen. The above SCAD and ACAA catalyzed reactions could promote FA degradation, which may explain the increased digestibility of crude fat in the CrPyr-supplemented group. On the other hand, the ATP generation during FA β-oxidation may help decrease oxidative stress, regulate energy metabolism, and further improve the rumen fermentation characteristic under heat stress.
Pyruvate is the intermediate product of carbohydrate fermentation by rumen microorganisms (Nagaraja, 2016). Our metaproteomics analyses showed that the administration of CrPyr significantly increased the expression of enzymes involved in pyruvate metabolism, glycolysis/gluconeogenesis, and citrate cycle pathway. Phosphoenolpyruvate (PEP)/pyruvate interconversion is a major metabolic point in glycolysis/gluconeogenesis (Chen et al., 2019). In the case of the pyruvate-to-PEP conversion, the reaction proceeds through diversified metabolic reactions. For instance, in propionic acid bacteria, PPDK is the primary enzyme that directly converts pyruvate to PEP in one step (Evans and Wood, 1971). However, in most mammals, plants, and microorganisms, the reaction proceeds by two steps, catalyzed by pyruvate carboxylase (converts pyruvate to oxaloacetate) and PEP carboxykinase (interconverts oxaloacetate and PEP; Bräsen et al., 2014). In this study, the pyruvate-to-PEP conversion was promoted, proving that the PPDK was significantly enriched (including 17 up-regulated PPDK and 7 down-regulated PPDK from different bacteria). Besides, although the pyruvate carboxylase subunit B and the PEP carboxykinase (ATP) were up-regulated, this may not be the pyruvate-to-PEP conversion proceeded by two steps, since in some anaerobic bacteria, PEP carboxykinase carboxylates PEP to oxaloacetate with the conservation of energy as ATP (Zhang et al., 2009; Asanuma et al., 2010). The above up-expressed enzymes involved in pyruvate metabolism means that CrPyr supplementation promoted the conversions of pyruvate to PEP, pyruvate to PEP to oxaloacetate, and pyruvate to oxaloacetate in rumen bacteria.
Moreover, in pyruvate metabolism, we also found that the enzymes involved in the conversion of pyruvate to acetyl-CoA or (S)-malate were promoted in the CrPyr administration group. In several microorganisms, pyruvate-flavodoxin oxidoreductase is responsible for acetyl-CoA formation from pyruvate in a single step (Ferrer et al., 2007). Our metaproteomics analysis showed that the up-regulated enzymes involved in the pyruvate-to-acetyl-CoA conversion include 13 pyruvate-flavodoxin oxidoreductase and its isozymes from different bacteria, 2-oxoglutarate ferredoxin oxidoreductase subunit alpha from Prevotella sp. khp7, and 2-oxoglutarate ferredoxin oxidoreductase subunit beta from Prevotella sp. tf2-5. Generally, microbes have five metabolic pathways involved in malate biosynthesis. Two of these pathways take pyruvate as the metabolic starting point: (I) pyruvate is the first to convert to oxaloacetate via the pyruvate carboxylase, and then malate dehydrogenase converts oxaloacetate to malate; (II) pyruvate is directly converted to malate via malic enzyme in one step (Zhou et al., 2015). In the present study, CrPyr administration up-regulated the malate dehydrogenase from Bacteroides xylanolyticus, NAD-dependent malic enzyme from Clostridium homopropionicum DSM 5847, and allosteric NADP-dependent malic enzyme from Prevotella ruminicola. The above up-expressed enzymes involved in pyruvate metabolism means that CrPyr supplementation promoted the conversions of pyruvate to acetyl-CoA, pyruvate to malate, and pyruvate to oxaloacetate to malate in rumen bacteria.
In bacteria, external glucose is transported into the cells and phosphorylated by the PEP:sugar phosphotransferase system (PTS). PEP as the phosphoryl donor to phosphorylate sugars is the primary source of energy for glucose uptake and phosphorylation (Zhang et al., 2009; Erni, 2013). In the present study, CrPyr supplementation increased the expression of enzymes involved in the conversion of pyruvate to PEP, which may increase the pool of PEP available for PTS and to facilitate glucose phosphorylation transport in bacteria, thereby up-regulating glycolysis. Consistently, in this study, the enzymes involved in glycolysis/gluconeogenesis were up-regulated, including 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, phosphoglycerate kinase, glyceraldehyde-3-phosphate dehydrogenase, fructose-1,6-bisphosphate aldolase, pyrophosphate-fructose 6-phosphate 1-phosphotransferase, glucose-6-phosphate isomerase, phosphoglucomutase, and aldose 1-epimerase. Of those enzymes, the glyceraldehyde-3-phosphate dehydrogenase was most enriched, with 21 up-regulated and 6 down-regulated proteins from different bacteria.
Glycolysis and the citrate cycle provide sources of energy within the rumen (Czerkawski, 1986). Recently, one study using a metagenomics approach elucidated that the citrate cycle (Krebs) exists in rumen microbiota metabolism (Mu et al., 2021). In this study, the acetyl-CoA generated from FA β-oxidation and the oxaloacetate, malate, and acetyl-CoA generated from pyruvate metabolism could enter the citrate cycle, promote the citrate cycle metabolism, and provide energy. Consistent with this, we found that 29 proteins were up-regulated and 10 proteins were down-regulated in the citrate cycle pathway. The up-regulation of enzymes involved in citrate cycle metabolism might help to produce more α-ketoglutarate. When ammonia and α-ketoglutarate are in a certain ratio, the efficiency of MCP synthesis can be optimized without causing the accumulation of ammonia or α-ketoglutarate. This is rumen energy-nitrogen equilibrium (Feng and Lu, 2007). In rumen, ammonium is the preferred nitrogen source for microbial growth. Creatine, as an important nitrogen-containing compound in protein and energy metabolism, has been widely used as a nitrogen supplement in ruminant nutrition due to its slow continuous release of ammonia (McLaren, 1964; Navrátil et al., 2009). The results from nitrogen metabolism indicated that CrPyr administration led to 8 up-regulated bacterial reduced NADPH-specific GDH (EC: 1.4.1.4) and 13 down-regulated bacterial NADPH-specific GDH. We also found three up-regulated and three down-regulated bacterial GS (EC: 6.3.1.2) and one down-regulated bacterial glutamate synthase (GOGAT; EC: 1.4.1.13). The GDH and GS-GOGAT pathways are two classic routes for ammonia assimilation in bacteria (Wang and Tan, 2013). In the GDH pathway, the NADH-specific GDH serves to degrade glutamate:
| (1) |
while NADPH-specific GDH serves for glutamate synthesis:
| (2) |
Phibbs and Bernlohr (1971) in the GS-GOGAT pathway, GS is a ubiquitous enzyme that catalyzes the ATP-dependent amidation glutamate to generate glutamine; GOGAT catalyzes the reductive transfer of the amide group of glutamate. In this study, the GDH pathway appears to be the predominant route of ammonia assimilation. We further analyzed the DEPs involved in the biosynthesis of amino acids, of which 58 proteins were up-regulated and 36 proteins were down-regulated; these results showed that CrPyr was helpful to amino acid biosynthesis of bacteria in rumen fluid. Moreover, since this project assumed only the in vivo model, pure bacterial lines whose enzymatic abundance were changed between CrPyr-supplemented and -unsupplemented treatments were not isolated and assessed in vitro. These experiments need to further improve in the further study.
Conclusion
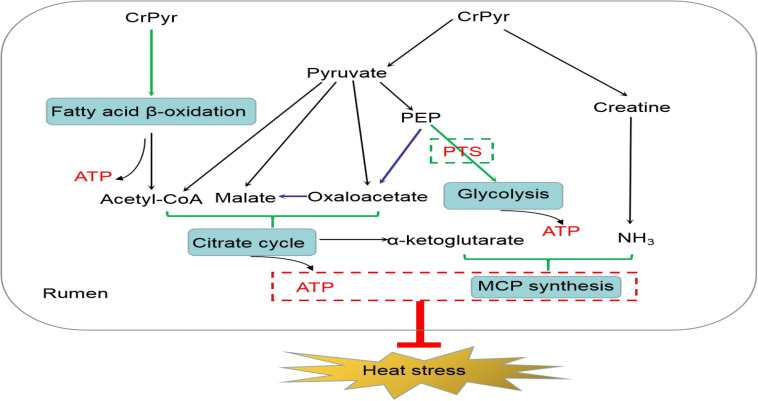
In conclusion, CrPyr administration increased the digestibility of crude fat through promoting the expression of enzymes involved in FA β-oxidation of rumen bacteria and up-regulated the expression of enzymes involved in glycolysis/gluconeogenesis through increasing the pool of PEP available for PTS and to facilitate glucose phosphorylation transport. The acetyl-CoA generated from FA β-oxidation and the oxaloacetate, malate, and acetyl-CoA generated from pyruvate metabolism promote the citrate cycle metabolism and might help to produce more α-ketoglutarate. α-Ketoglutarate, together with the NH3 released by creatine, promoted the synthesis of MCP in the rumen. The increased production of ATP and the up-regulation synthesis of MCP in rumen may help decrease oxidative stress, regulate energy metabolism, and further improve the rumen fermentation characteristic of beef cattle under heat stress (Figure 8).
FIGURE 8.
Proposed model of creatine pyruvate relieving heat stress in rumen of beef cattle.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The animal study was reviewed and approved by the Committee for the Care and Use of Experimental Animals at Jiangxi Agricultural University (JXAULL-20190017).
Author Contributions
YL and MQ designed the overall study. YL, YZ, XZ, LL, QQ, and KO performed the experiments. YL wrote the manuscript. All the authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors appreciate all the help from our colleagues and collaborators.
Funding. This work was supported by the China Agriculture Research System of MOF and MARA, the Educational Department of Jiangxi Province of China (GJJ180227), and the National Natural Science Foundation of China (No. 32060768).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.715088/full#supplementary-material
References
- Ammer S., Lambertz C., Gauly M. (2016). Comparison of different measuring methods for body temperature in lactating cows under different climatic conditions. J. Dairy Res. 83 165–172. 10.1017/S0022029916000182 [DOI] [PubMed] [Google Scholar]
- Andresen B. S., Olpin S., Poorthuis B. J., Scholte H. R., Vianey-Saban C., Wanders R., et al. (1999). Clear correlation of genotype with disease phenotype in very-long-chain acyl-CoA dehydrogenase deficiency. Am. J. Hum. Genet. 64 479–494. 10.1086/302261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asanuma N., Kanada K., Arai Y., Yoshizawa K., Ichikawa T., Hino T. (2010). Molecular characterization and significance of phosphoenolpyruvate carboxykinase in a ruminal bacterium, Streptococcus bovis. J. Gen. Appl. Microbiol. 56 121–127. 10.2323/jgam.56.121 [DOI] [PubMed] [Google Scholar]
- Bernabucci U., Lacetera N., Danieli P. P., Bani P., Nardone A., Ronchi B. (2009). Influence of different periods of exposure to hot environment on rumen function and diet digestibility in sheep. Int. J. Biometeorol. 53 387–395. 10.1007/s00484-009-0223-6 [DOI] [PubMed] [Google Scholar]
- Bräsen C., Esser D., Rauch B., Siebers B. (2014). Carbohydrate metabolism in Archaea: current insights into unusual enzymes and pathways and their regulation. Microbiol. Mol. Biol. Rev. 78 89–175. 10.1128/MMBR.00041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick G. A., Kang J. H. (1980). Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J. Dairy Sci. 63 64–75. 10.3168/jds.S0022-0302(80)82888-8 [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Mitra R., Zhang S. J., Zuo Z. Q., Lin L., Zhao D. H., et al. (2019). Unusual phosphoenolpyruvate (PEP) synthetase-Like protein crucial to enhancement of polyhydroxyalkanoate accumulation in Haloferax mediterranei revealed by dissection of PEP-Pyruvate interconversion mechanism. Appl. Environ. Microbiol. 85:e00984-19. 10.1128/AEM.00984-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Huang J., Deng J., Ma H., Zou S. (2012). Use of comparative proteomics to identify the effects of creatine pyruvate on lipid and protein metabolism in broiler chickens. Vet. J. 193 514–521. 10.1016/j.tvjl.2012.01.034 [DOI] [PubMed] [Google Scholar]
- Chen X., Wang X., Wang X., Yang J., Shen X., Jie C. (2006). Effect of calcium pyruvate on rumen metabolism of goats. Chin. Agric. Sci. Bull. 22 17–20. [Google Scholar]
- Cheng G. W. (2019). Study on the Mechanism of Dietary Carbohydrate Types Regulating C2/C3 Ratio in Rumen Fermentatino. Master’s thesis. Taian (SD): Shandong Agricultural University. [Google Scholar]
- Cunniff P. (1995). Official Methods of Analysis of AOAC International, 16th Edn, Vol. 1. Washington, DC: Association of Official Analytical Chemists. [Google Scholar]
- Czerkawski J. W. (1986). An Introduction to Rumen Studies. Great Britain: A. Wheaton and Co Ltd, Exeter. [Google Scholar]
- Dai H., Liu X., Yan J., Aabdin Z. U., Bilal M. S., Shen X. (2017). Sodium butyrate ameliorates high-concentrate diet-induced inflammation in the rumen epithelium of dairy goats. J. Agric. Food Chem. 65 596–604. 10.1021/acs.jafc.6b04447 [DOI] [PubMed] [Google Scholar]
- Emmanuel B. (1978). The relative contribution of propionate, and long-chain even-numbered fatty acids to the production of long-chain odd-numbered fatty acids in rumen bacteria. Biochim. Biophys. Acta 528 239–246. 10.1016/0005-2760(78)90198-4 [DOI] [PubMed] [Google Scholar]
- Erni B. (2013). The bacterial phosphoenolpyruvate:sugar phosphotransferase system (PTS): an interface between energy and signal transduction. J. Iran Chem. Soc. 10 593–630. 10.1007/s13738-012-0185-1 [DOI] [Google Scholar]
- Evans H. J., Wood H. G. (1971). Purifification and properties of pyruvate phosphate dikinase from propionic acid bacteria. Biochemistry 10 721–729. 10.1021/bi00781a001 [DOI] [PubMed] [Google Scholar]
- Feng Y. L., Lu Z. N. (2007). Nutritional Requirements and Feed Ingredients of Dairy Cows. Beijing: China Agricultural Press. [Google Scholar]
- Ferrer M., Golyshina O. V., Beloqui A., Golyshin P. N., Timmis K. N. (2007). The cellular machinery of Ferroplasma acidiphilum is iron-protein-dominated. Nature 445 91–94. 10.1038/nature05362 [DOI] [PubMed] [Google Scholar]
- Gruninger R. J., Ribeiro G. O., Cameron A., Mcallister T. A. (2019). Invited review: application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal 13 1843–1854. 10.1017/S1751731119000752 [DOI] [PubMed] [Google Scholar]
- Hao L., Michaelsen T. Y., Singleton C. M., Dottorini G., Kirkegaard R. H., Albertsen M., et al. (2020). Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics. ISME J. 14 906–918. 10.1038/s41396-019-0571-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfoot C. G., Hazlewood G. P. (1997). “Lipid metabolism in the rumen,” in The Rumen Microbial Ecosystem, 2nd Edn, eds Hobson P., Stewart C. S. (London: Chapman & Hall; ), 382–426. [Google Scholar]
- Hart E. H., Creevey C. J., Hitch T., Kingston-Smith A. H. (2018). Meta-proteomics of rumen microbiota indicates niche compartmentalisation and functional dominance in a limited number of metabolic pathways between abundant bacteria. Sci. Rep. 8:10504. 10.1038/s41598-018-28827-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson P. N., Stewart C. S. (1997). The Rumen Microbial Ecosystem. Cham: Springer. [Google Scholar]
- Hyder I., Reddy P. R. K., Raju J., Manjari P., Prasad C. S., Kumar K. A., et al. (2017). “Alteration in rumen functions and diet digestibility during heat stress in sheep,” in Sheep Production Adapting to Climate Change, eds Sejian V., Bhatta R., Gaughan J., Malik P., Naqvi S., Lal R. (Singapore: Springer International Publishing; ), 235–265. [Google Scholar]
- Kim S. H., Han S. K., Shin H. S. (2004). Two-phase anaerobic treatment system for fat-containing wastewater. J. Chem. Technol. Biot. 79 63–71. 10.1002/jctb.939 [DOI] [Google Scholar]
- LCI (1970). Patterns of Transit Losses. Omaha, NE: Livestock Conservation, Inc. [Google Scholar]
- Liu L., Zhang W., Yu H., Xu L., Qu M., Li Y. (2020). Improved antioxidant activity and rumen fermentation in beef cattle under heat stress by dietary supplementation with creatine pyruvate. Anim. Sci. J. 91:e13486. 10.1111/asj.13486 [DOI] [PubMed] [Google Scholar]
- Ma H., Zhang B., Hu Y., Li X., Wang S., Yang F., et al. (2020). The novel intervention effect of cold green tea beverage on high-fat diet induced obesity in mice. J. Funct. Foods. 75:104279. 10.1016/j.jff.2020.104279 [DOI] [Google Scholar]
- Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., et al. (2019). iProX: an integrated proteome resource. Nucleic Acids Res. 47 D1211–D1217. 10.1093/nar/gky869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makkar H. P., Becker K. (1999). Purine quantification in digesta from ruminants by spectrophotometric and HPLC methods. Brit. J. Nutr. 81 107–112. 10.1017/S0007114599000227 [DOI] [PubMed] [Google Scholar]
- Marai I. F. M., El-Darawany A. A., Fadiel A., Abdel-Hafez M. A. M. (2007). Physiological traits as affected by heat stress in sheep—A review. Small Ruminant. Res. 71 1–12. 10.1016/j.smallrumres.2006.10.003 [DOI] [Google Scholar]
- McLaren G. A. (1964). Symposium on microbial digestion in ruminants: nitrogen metabolism in the rumen. J. Anim. Sci. 23 577–590. 10.1007/978-1-62703-305-3_15 [DOI] [Google Scholar]
- Mendel R. W., Blegen M., Cheatham C. (2005). Effects of creatine on thermoregulatory responses while exercising in the heat. Nutrition 21 301–307. 10.1016/j.nut.2004.06.024 [DOI] [PubMed] [Google Scholar]
- Mu Y. Y., Qi W. P., Zhang T., Zhang J. Y., Mao S. Y. (2021). Gene function adjustment for carbohydrate metabolism and enrichment of rumen microbiota with antibiotic resistance genes during subacute rumen acidosis induced by a high-grain diet in lactating dairy cows. J. Dairy Sci. 104 2087–2105. 10.3168/jds.2020-19118 [DOI] [PubMed] [Google Scholar]
- Nagaraja T. G. (2016). “Microbiology of the rumen,” in Rumenology, eds Millen D. D., Arrigoni M. D. B., Pacheco R. D. L. (Cham: International Publishing; ), 39–61. [Google Scholar]
- Navrátil T., Kohlíková E., Petr M., Heyrovský M., Pelclová D., Přistoupilová K., et al. (2009). Contribution to explanation of the effect of supplemented creatine in human metabolism. Food Chem. 112 500–506. 10.1016/j.foodchem.2008.05.086 [DOI] [Google Scholar]
- Phibbs P. V., Bernlohr R. W. (1971). Purifification, properties, and regulation of glutamic dehydrogenase of Bacillus-licheniformis. J. Bacteriol. 106 375–385. 10.1128/JB.106.2.375-385.1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran L. W., Shen X. Z., Jie T. U., Lin P. F., Chen J. (2008). Study on the effect of calcium pyruvate on the dynamics of rumen VFA concentration in goats. Acta Ecol. Anim. Domastici. 29 40–44. [Google Scholar]
- Rieger E., de Franceschi I. D., Preissler T., Wannmacher C. (2017). Neuroprotective effect of creatine and pyruvate on enzyme activities of phosphoryl transfer network and dxidative stress alterations caused by leucine administration in wistar rats. Neurotox Res. 32 575–584. 10.1007/s12640-017-9762-5 [DOI] [PubMed] [Google Scholar]
- Sales G. F. C., Carvalho B. F., Schwan R. F., Vilela L. F., Meneses J. A. M., Gionbelli M. P., et al. (2021). Heat stress influence the microbiota and organic acids concentration in beef cattle rumen. J. Therm. Biol. 97:102897. 10.1016/j.jtherbio.2021.102897 [DOI] [PubMed] [Google Scholar]
- Salles M. S. V., Zanetti M. A., Salles F. A., Titto E. A. L., Conti R. M. C. (2010). Changes in ruminal fermentation and mineral serum level in animals kept in high temperature environments. Rev. Bras. Zootecn. 39 883–890. 10.1590/S1516-35982010000400025 [DOI] [Google Scholar]
- Song X., Luo J., Fu D., Zhao X., Bunlue K., Xu Z., et al. (2014). Traditional chinese medicine prescriptions enhance growth performance of heat stressed beef cattle by relieving heat stress responses and increasing apparent nutrient digestibility. Asian Austr. J. Anim. Sci. 27 1513–1520. 10.5713/ajas.2014.14058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima K., Nonaka I., Higuchi K., Takusari N., Kurihara M., Takenaka A., et al. (2007). Influence of high temperature and humidity on rumen bacterial diversity in Holstein heifers. Anaerobe 13 57–64. 10.1016/j.anaerobe.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Thomas M., Webb M., Ghimire S., Blair A., Olson K., Fenske G. J., et al. (2017). Metagenomic characterization of the effect of feed additives on the gut microbiome and antibiotic resistome of feedlot cattle. Sci. Rep. 7:12257. 10.1038/s41598-017-12481-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeno Y., Sekiguchi Y., Tajima K., Takenaka A., Kurihara M., Kamagata Y. (2010). An rRNA-based analysis for evaluating the effect of heat stress on the rumen microbial composition of Holstein heifers. Anaerobe 16 27–33. 10.1016/j.anaerobe.2009.04.006 [DOI] [PubMed] [Google Scholar]
- Van Soest P. J., Robertson J. B., Lewis B. A. (1991). Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74 3583–3597. 10.3168/jds.S0022-0302(91)78551-2 [DOI] [PubMed] [Google Scholar]
- Wang P. P., Tan Z. L. (2013). Ammonia assimilation in rumen bacteria: a review. Anim. Biotechnol. 24 107–128. 10.1080/10495398.2012.756402 [DOI] [PubMed] [Google Scholar]
- Zhang X., Jantama K., Moore J. C., Jarboe L. R., Shanmugam K. T., Ingram L. O. (2009). Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 106 20180–20185. 10.1073/pnas.0905396106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang Y., Wang X., Ji Y., Cheng S., Wang M., et al. (2018). Acetyl-coenzyme A acyltransferase 2 promote the differentiation of sheep precursor adipocytes into adipocytes. J. Cell Biochem. 120, 8021–8031. 10.1002/jcb.28080 [DOI] [PubMed] [Google Scholar]
- Zhou L., Cui W. J., Liu Z. M., Zhou Z. M. (2015). Advance in L-malate production based on metabolic engineering strategies. Sci. Technol. Food Ind. 36 383–394. [Google Scholar]
- Zhu H., Wei S., Jing W., Lu S., Yan Z., Shuai L., et al. (2017). Correlation of gut microbiota with susceptibility to high-fat diet-induced obesity in mice:a metagenomic study. J. Third Mil. Med. Univ. 39 773–780. 10.16016/j.1000-5404.201611223 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.