Abstract
The stability of Mycobacterium tuberculosis IS6110 fingerprint patterns and spoligotypes has been assessed by analyzing serial isolates from patients with drug-resistant tuberculosis. Altogether, 165 M. tuberculosis isolates obtained from 56 patients have been analyzed. The time spans between the first and the last or a changed isolate from one patient ranged from 1 to 772 days. Among the 56 patients, 5 (9%) were infected with isolates with changes in their IS6110 fingerprint patterns. According to the total number of strains analyzed, 5% of the subsequent isolates showed variations in their IS6110 restriction fragment length polymorphism patterns compared to the pattern of the first isolates. Up to 10 isolates from one patient sampled at time intervals of up to 772 days with no changes in their IS6110 patterns have been analyzed. A statistically significant correlation could be found between changes in insertion sequence (IS) patterns and the increased time intervals over which the isolates were obtained, whereas changes in IS patterns are not correlated to changes in the drug resistance of the isolates. In contrast to the observed variations in IS6110 fingerprint patterns, no changes in the spoligotypes of the isolates analyzed could be found. In conclusion, our results confirm that the IS6110 fingerprint patterns of M. tuberculosis isolates have high degrees of stability. Compared to IS6110, the direct repeat (DR) region, which is the basis for spoligotyping, has a lower rate of change. Partial deletions, e.g., deletions induced by homologous recombination between the repetitive DR elements, could not be detected in this study.
IS6110 DNA fingerprinting has become the standard method for analysis of the epidemiology of tuberculosis (TB) on a molecular level (1, 2, 14, 15). IS6110, a transposable sequence belonging to the IS3 family (7), is found with few exceptions in all members of the Mycobacterium tuberculosis complex and is apparently restricted to this group of organisms (2, 11). The epidemiological analysis of TB by IS6110 DNA fingerprinting is based on the observation that the polymorphism of the IS6110 restriction fragment length polymorphism (RFLP) patterns among unrelated clinical isolates is very high, whereas epidemiologically related M. tuberculosis strains show identical or similar (one band variation) fingerprint patterns (15). Hence, M. tuberculosis strains with identical fingerprint patterns represent strains that have possibly been recently transmitted, and these strains are likely to be members involved in a chain of transmission.
For the correct interpretation of IS6110 fingerprinting data, e.g., for determination of the percentage of reactivation versus the percentage of active transmission in a given population, it is mandatory that the stability of the genetic marker used for the analysis is known. Recently, the stability of IS6110 fingerprint patterns has been elucidated by analyzing serial isolates of 49 patients in the United States (16). In this study, a change in IS6110 banding pattern occurred in isolates from 12 of 49 patients (25%), indicating a relatively high degree of instability of IS6110. However, a comparable high degree of instability has not been observed in several other previously published studies, each comprising smaller numbers of M. tuberculosis strains (3, 9, 10, 14).
A new method for the characterization of M. tuberculosis complex isolates, spoligotyping, can be used for the diagnosis of M. tuberculosis infections and epidemiological studies and has recently been described by Kamerbeek et al. (6). It is based on the detection of various nonrepetitive spacer sequences located between small repetitive units (direct repeats [DRs]) in the DR locus of M. tuberculosis complex strains (6). Since the spoligotyping method is based on PCR, it is rapid and results can be obtained directly from an M. tuberculosis culture within 1 day. However, the discriminative power of spoligotyping compared to that of IS6110 fingerprinting is less (4, 6), and consequently, its use for epidemiological studies of M. tuberculosis is limited. Hence, a two-step strategy, initial spoligotyping analysis followed by subtyping of strains clustered by IS6110 fingerprinting to confirm the strains’ relationship, has been proposed (4). This strategy may allow fast and efficient analysis on large numbers of isolates (4). To apply this strategy, it is necessary to have information about the “molecular clock” of the DR region and the relative stability of spoligotypes compared to that of IS6110 fingerprint patterns. For example, one recombination event may result in the deletion of a large part of the DR region. As a consequence, the spoligotype of this strain will change dramatically, so that closely related strains may not be grouped together in the initial analysis and possible cases of transmission may be overlooked. So far, no data concerning the stability within the DR locus are available, and it is not yet clear in which way and how often changes in spoligotypes occur.
In this study we genotyped 165 isolates from 56 patients with drug-resistant TB to elucidate the stabilities of M. tuberculosis IS6110 fingerprint patterns and spoligotypes. Furthermore, from the data obtained in this study, the relative stability of spoligotypes compared to that of IS6110 fingerprint patterns and the feasibility of a two-step procedure that uses both methods should be clarified.
MATERIALS AND METHODS
Bacterial strains.
One hundred sixty-five M. tuberculosis isolates received by the National Reference Center for Mycobacteria between 1995 and 1997 were investigated by DNA fingerprinting and spoligotyping. The isolates were obtained from 56 patients living in Germany. Drug susceptibility testing of all isolates was done by the proportion method on Löwenstein-Jensen medium and/or the modified proportion method with BACTEC 460TB (Becton Dickinson Microbiology Systems, Cockeysville, Md.).
DNA genotyping techniques.
Extraction of DNA from mycobacterial strains and DNA fingerprinting with IS6110 as a probe were performed by standardized protocols (8, 12). The IS6110 fingerprint patterns of mycobacterial strains were analyzed with the GelCompar software (Windows 95, version 4.0; Applied Maths, Kortrijk, Belgium) as described previously (8, 12).
Spoligotyping of isolates was performed as described by Kamerbeek et al. (6). The spoligotyping analyses of serial isolates from one patient were done side by side to allow an accurate comparison of the spoligotypes.
Statistical analysis.
The chi-square test was performed to determine whether changes in IS6110 RFLP patterns were related to changes in drug resistance patterns or increased time intervals between the retrieval of isolates.
RESULTS
In this study 165 serial isolates from 56 patients were analyzed by the IS6110 DNA fingerprinting and spoligotyping methods. The isolates were selected from among approximately 600 drug-resistant M. tuberculosis isolates sent to the National Reference Center for Mycobacteria between 1995 and 1997. All available isolates from one patient up to the first isolate with a change in IS6110 fingerprint pattern have been analyzed. If no change in the IS6110 pattern occurred, the last isolate analyzed represents the last isolate from the patient that has so far been sent to the National Reference Center for Mycobacteria. Twenty-three additional isolates from 10 patients were excluded from this study because the dates of primary isolation were not known exactly. However, no changes in the IS6110 fingerprint patterns of these isolate have been observed.
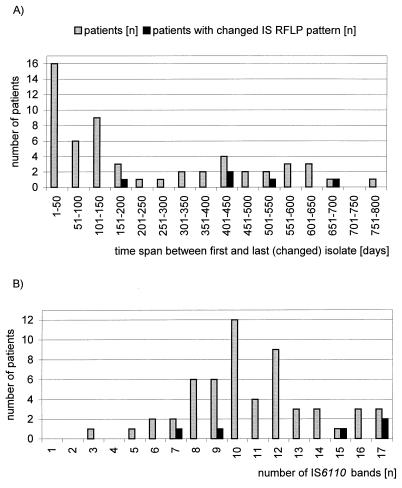
The time spans between the first and last or a changed isolate from one patient ranged from 1 to 772 days (Table 1; Fig. 1A). The number of IS6110 copies per isolate varied from 3 to 17, with a mean of 10 (Table 1, Fig. 1B). The drug resistance patterns of the isolates ranged from monodrug resistance, e.g., resistance to isoniazid, to multidrug resistance, e.g., resistance to isoniazid, ethambutol, rifampin, and pyrazinamide (data not shown).
TABLE 1.
Results of drug susceptibility tests, IS6110 DNA fingerprinting, and spoligotyping of serial isolates from 56 patients
| Patient no. | Age (yr) | Sexa | No. of isolates tested | No. of IS6110 bands | No. of spoligotype signals | Time spanb | Change in drug resistancec | Change in IS6110 patternd | Change in spoligotypee |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 33 | m | 2 | 11 | 35 | 200 | 0 | 0 | 0 |
| 2 | 38 | m | 2 | 7 | 39 | 150 | +1 | 0 | 0 |
| 3 | 34 | m | 3 | 16 | 29 | 195 | 0 | 0 | 0 |
| 4 | 51 | m | 4 | 7 | 34 | 447 | 0 | +1 | 0 |
| 5 | 26 | m | 2 | 10 | 29 | 9 | 0 | 0 | 0 |
| 6 | 40 | m | 2 | 10 | 38 | 38 | 0 | 0 | 0 |
| 7 | 55 | m | 2 | 8 | 34 | 11 | 0 | 0 | 0 |
| 8 | 34 | m | 3 | 12 | 32 | 28 | 0 | 0 | 0 |
| 9 | 26 | m | 2 | 13 | 32 | 3 | 0 | 0 | 0 |
| 10 | 33 | m | 2 | 12 | 36 | 1 | 0 | 0 | 0 |
| 11 | 30 | m | 2 | 10 | 35 | 120 | 0 | 0 | 0 |
| 12 | 23 | f | 6 | 10 | 35 | 448 | 0 | 0 | 0 |
| 13 | 32 | m | 3 | 3 | 37 | 42 | 0 | 0 | 0 |
| 14 | 79 | m | 3 | 15 | 29 | 193 | 0 | +1 | 0 |
| 15 | 42 | m | 3 | 9 | 9 | 42 | 0 | 0 | 0 |
| 16 | 73 | m | 4 | 9 | 32 | 609 | 0 | 0 | 0 |
| 17 | 63 | m | 3 | 12 | 34 | 302 | 0 | 0 | 0 |
| 18 | 44 | f | 2 | 11 | 38 | 5 | 0 | 0 | 0 |
| 19 | 49 | m | 4 | 11 | 38 | 131 | 0 | 0 | 0 |
| 20 | 46 | m | 2 | 13 | 31 | 140 | 0 | 0 | 0 |
| 21 | 37 | m | 3 | 10 | 38 | 634 | 0 | 0 | 0 |
| 22 | 25 | m | 2 | 8 | 39 | 81 | 0 | 0 | 0 |
| 23 | 26 | m | 3 | 14 | 9 | 459 | +2 | 0 | 0 |
| 24 | 22 | f | 2 | 10 | 9 | 52 | 0 | 0 | 0 |
| 25 | 39 | m | 3 | 10 | 16 | 57 | 0 | 0 | 0 |
| 26 | 75 | f | 2 | 11 | 37 | 132 | 0 | 0 | 0 |
| 27 | 67 | m | 3 | 10 | 39 | 548 | 0 | 0 | 0 |
| 28 | 27 | m | 4 | 16 | 9 | 616 | 0 | 0 | 0 |
| 29 | 48 | m | 2 | 12 | 35 | 90 | 0 | 0 | 0 |
| 30 | 40 | m | 3 | 9 | 27 | 397 | 0 | 0 | 0 |
| 31 | 28 | f | 2 | 12 | 27 | 68 | 0 | 0 | 0 |
| 32 | 71 | m | 2 | 8 | 39 | 9 | 0 | 0 | 0 |
| 33 | 31 | m | 6 | 17 | 9 | 448 | 0 | +1 | 0 |
| 34 | 49 | m | 10 | 10 | 29 | 772 | +2 | 0 | 0 |
| 35 | 60 | m | 5 | 10 | 35 | 106 | 0 | 0 | 0 |
| 36 | 35 | m | 2 | 17 | 9 | 6 | 0 | 0 | 0 |
| 37 | 57 | m | 2 | 10 | 39 | 47 | 0 | 0 | 0 |
| 38 | 68 | f | 2 | 8 | 39 | 3 | 0 | 0 | 0 |
| 39 | 36 | f | 3 | 9 | 11 | 262 | 0 | 0 | 0 |
| 40 | 43 | m | 2 | 8 | 33 | 106 | 0 | 0 | 0 |
| 41 | 98 | f | 2 | 12 | 39 | 218 | 0 | 0 | 0 |
| 42 | 91 | m | 2 | 6 | 29 | 72 | 0 | 0 | 0 |
| 43 | 47 | m | 2 | 12 | 32 | 38 | 0 | 0 | 0 |
| 44 | 44 | m | 8 | 9 | 37 | 683 | +1 | +1 | 0 |
| 45 | 64 | m | 2 | 5 | 23 | 35 | 0 | 0 | 0 |
| 46 | 36 | m | 5 | 17 | 9 | 512 | +1 | −1 | 0 |
| 47 | 38 | m | 3 | 14 | 9 | 578 | +2 | 0 | 0 |
| 48 | 32 | m | 4 | 12 | 36 | 600 | +1 | 0 | 0 |
| 49 | 58 | m | 2 | 12 | 38 | 307 | 0 | 0 | 0 |
| 50 | 34 | f | 3 | 13 | 30 | 471 | +2 | 0 | 0 |
| 51 | 63 | m | 2 | 8 | 32 | 487 | 0 | 0 | 0 |
| 52 | 45 | m | 2 | 14 | 9 | 560 | +1 | 0 | 0 |
| 53 | 47 | m | 2 | 6 | 39 | 378 | 0 | 0 | 0 |
| 54 | 28 | f | 3 | 16 | 9 | 133 | 0 | 0 | 0 |
| 55 | 61 | f | 2 | 10 | 34 | 39 | 0 | 0 | 0 |
| 56 | 62 | f | 2 | 9 | 33 | 132 | 0 | 0 | 0 |
| Total | 165 |
m, male; f, female.
Time span (days) between the first and the last or a changed isolate from one patient.
0, no change in drug resistance; +1, resistance to one additional drug; +2, resistance to two additional drugs.
0, no change in IS6110 RFLP pattern; +1, one additional IS6110 band; −1, one less IS6110 band.
0, no change in spoligotype.
FIG. 1.
Time spans between the first and the last or a changed isolate from the patients whose isolates were analyzed (A) and numbers of IS6110 fingerprint bands (B). Changes in IS6110 fingerprint patterns are shown as functions of the time interval between the times of retrieval of isolates (A) and numbers of fingerprint bands (B). IS, insertion sequence.
Among the 56 patients, 5 (9%) were infected with strains with changes in their IS6110 fingerprint patterns between the first and the last isolates that were obtained. All of these changes are based on differences in one band in the fingerprint patterns compared to the patterns for the first isolates (for four patients, one additional band; for one patient, one less band [Table 1]). In terms of the number of isolates analyzed, the proportion of IS6110 changes is 5% (if the numbers of first isolates from the patients were subtracted from the total number of isolates tested). Changes in IS6110 RFLP patterns have been observed over time intervals of from approximately 200 to 700 days in which the isolates were taken (Table 1; Fig. 1A). No changes could be observed for time intervals of less than approximately 190 days (Table 1). A significant correlation was found between changes in IS6110 patterns and increased time intervals between the time of retrieval of isolates (P < 0.05). Changes were found in isolates with an intermediate number (7 to 9) as well as with a large number (15 to 17) of bands (Fig. 1B). All changes occurred in isolates obtained from men. However, only 20% of the patients included in this study were women, and for half of them time intervals between the times of retrieval of the isolates were less than 200 days, which are likely to be too short for changes in IS6110 fingerprint patterns to occur (Table 1).
For isolates from nine patients, changes in the drug resistance patterns (resistance to up to two additional antituberculous drugs) of the isolates could be observed (Table 1). Although the serial isolates from two patients showed changes in their IS6110 fingerprints as well as in their drug resistance patterns, in total no significant correlation between these two events could be found.
The spoligotypes of the isolates analyzed had 9 to 39 hybridization signals (Table 1) and differed markedly among the isolates. In contrast to the observed changes in IS6110 fingerprint patterns, we found no variation in spoligotypes (Table 1). In addition, we saw no changes in the spoligotypes of the serial isolates from the 10 patients excluded from this study.
DISCUSSION
In this study, the stabilities of the M. tuberculosis IS6110 fingerprint patterns and spoligotypes have been analyzed. Compared to the study of Yeh et al. (16), we found a considerably higher degree of stability of IS6110 RFLP patterns (if changes were related to the total number of isolates analyzed). One possible reason for the higher degree of instability observed in the study of Yeh and coworkers (16) may be that only isolates whose times of retrieval were separated by at least 90 days have been analyzed. Nevertheless, limiting the number of patients in our study to this group of patients, the percentage of IS6110 changes remained less than that observed by Yeh et al. (16) (14% according to the number of patients and 6% according to the total number of isolates analyzed in the present study but 25% according to the total number of isolates analyzed in the study of Yeh et al. [16]). In any case, a study design that excluded patients whose isolates were retrieved over intervals of less than 90 days probably results in an overestimation of the real rate of change in IS6110 RFLP patterns, because isolates from patients in such studies are likely to have a higher rate of IS6110 fingerprint changes. In our study, only drug-resistant isolates have been analyzed, whereas Yeh et al. (16) tested drug-susceptible as well as drug-resistant strains. However, our results are in accordance with previously published studies that analyzed comparatively small numbers of regularly drug-susceptible isolates from patients repeatedly bearing positive cultures, and all of those studies found a high degree of stability of IS6110 patterns (3, 9, 10, 14). Hence, the rate of change of IS6110 fingerprint patterns in drug-resistant isolates seems not to differ from that in drug-susceptible isolates. Consequently, the high rate of IS6110 fingerprint changes found by Yeh et al. (16) may be a specific feature of the M. tuberculosis situation in San Francisco, Calif.
Our results confirm the high degree of stability of IS6110 RFLP patterns for M. tuberculosis bacteria that remain in the bodies of patients. Up to 10 isolates obtained from one patient at times separated by up to 772 days have been analyzed, and no change in the IS6110 RFLP patterns of the isolate were found. However, in contrast to Yeh et al. (16), we found a significant correlation between retrieval of isolates at increased time intervals and changes in IS6110 RFLP patterns. In conclusion, our results indicate that M. tuberculosis IS6110 patterns change at a relatively slow rate. In combination with the high degree of variability of IS6110 fingerprint patterns found for isolates from Germany (8) and other countries (5, 10, 13), our data stretch the hypothesis that patients whose M. tuberculosis isolates have identical IS6110 fingerprint patterns are likely to be part of a recent chain of transmission. On the basis of our data, clustering rates should further be based on identical fingerprint patterns, if other epidemiological data are not available.
In contrast to the observed changes in IS6110 fingerprint patterns, we found no changes in the spoligotypes of the isolates tested. Hence, the DR region seems to have a lower rate of change than IS6110. This fact is in accordance with the observed lower discriminative power of spoligotyping compared with that of IS6110 fingerprinting (4, 6). Partial deletions, e.g., via homologous recombination, do not seem to be frequent in the DR region. Hence, a two-step procedure for epidemiological studies of M. tuberculosis based on an initial typing of all isolates by spoligotyping, followed by subtyping of clustered isolates by IS6110 fingerprinting, as proposed by Gouguet de la Salmonière et al. (4), seems to be feasible. Since the initial spoligotyping analysis is fast and does not need a well-grown bacterial culture, this procedure should simplify and speed up epidemiological studies comprising large numbers of strains. Moreover, our results clearly demonstrate that isolates with different spoligotypes are most likely not related.
ACKNOWLEDGMENTS
We thank Andreas Zyzik, Borstel, Germany, for excellent technical assistance and, furthermore, Kristin Kremer, Dick van Soolingen, and Jan van Embden, Bilthoven, The Netherlands, for technical support.
Parts of this work were supported by the Robert Koch-Institut, Berlin, Germany.
REFERENCES
- 1.Bauer J, Yang Z, Poulsen S, Anderson A B. Results from 5 years of nationwide DNA fingerprinting of Mycobacterium tuberculosis complex isolates in a country with a low incidence of M. tuberculosis infection. J Clin Microbiol. 1998;36:305–308. doi: 10.1128/jcm.36.1.305-308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cave M D, Eisenach K D, McDermott P F, Bates J H, Crawford J T. IS6110: conservation of sequence in the Mycobacterium tuberculosis complex and its utilization in DNA fingerprinting. Mol Cell Probes. 1991;5:73–80. doi: 10.1016/0890-8508(91)90040-q. [DOI] [PubMed] [Google Scholar]
- 3.Cave M D, Eisenach K D, Templeton G, Salfinger M, Mazurek G, Bates J H, Crawford J T. Stability of DNA fingerprint pattern produced with IS6110 in strains of Mycobacterium tuberculosis. J Clin Microbiol. 1994;32:262–266. doi: 10.1128/jcm.32.1.262-266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goguet de la Salmonière Y-O, Li H M, Torrea G, Bunschoten A, van Embden J D A, Gicquel B. Evaluation of spoligotyping in a study of the transmission of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:2210–2214. doi: 10.1128/jcm.35.9.2210-2214.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hermans P W M, Messadi F, Guebrexabher H, van Soolingen D, de Haas P E W, Heersma H, de Neeling H, Ayoub A, Portaels F, Frommel D, Zribi M, Van Embden J D A. Analysis of the population structure of Mycobacterium tuberculosis in Ethiopia. Tunisia, and The Netherlands: usefulness of DNA typing for global tuberculosis epidemiology. J Infect Dis. 1995;171:1504–1513. doi: 10.1093/infdis/171.6.1504. [DOI] [PubMed] [Google Scholar]
- 6.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J D A. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McAdam R A, Hermans P W M, van Soolingen D, Zainuddin Z F, Catty D, van Embden J D A, Dale J W. Characterization of a M. tuberculosis insertion sequence belonging to the IS3 family. Mol Microbiol. 1990;4:1607–1613. doi: 10.1111/j.1365-2958.1990.tb02073.x. [DOI] [PubMed] [Google Scholar]
- 8.Niemann S, Rüsch-Gerdes S, Richter E. IS6110 fingerprinting of drug-resistant Mycobacterium tuberculosis strains isolated in Germany during 1995. J Clin Microbiol. 1997;35:3015–3020. doi: 10.1128/jcm.35.12.3015-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigouts L, Portaels F. DNA fingerprints of Mycobacterium tuberculosis do not change during the development of resistance to various antituberculous drugs. Tuberc Lung Dis. 1994;75:160. doi: 10.1016/0962-8479(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 10.Strässle A, Putnik J, Weber R, Fehr-Merhof A, Wüst J, Pfyffer G E. Molecular epidemiology of Mycobacterium tuberculosis strains isolated from patients in a human immunodeficiency virus cohort in Switzerland. J Clin Microbiol. 1997;35:374–378. doi: 10.1128/jcm.35.2.374-378.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thierry D, Brisson-Noël A, Vincent-Lévy-Frébault V, Nguyen S, Guesdon J-L, Gicquel B. Characterization of a Mycobacterium tuberculosis insertion sequence, IS6110, and its application in diagnosis. J Clin Microbiol. 1990;28:2668–2673. doi: 10.1128/jcm.28.12.2668-2673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnik T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Soolingen D, Hermans P W M, de Haas P E W, Soll D R, van Embden J D A. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Soolingen D, de Haas P E W, Hermans P W M, Groenen P M A, van Embden J D A. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Soolingen, D., and P. W. M. Hermans. 1995. Epidemiology of tuberculosis by DNA fingerprinting. Eur. Respir. J. 8(Suppl. 20):649s–656s. [PubMed]
- 16.Yeh R W, Ponce de Leon A, Agasino C B, Hahn J A, Daley C L, Hopewell P C, Small P M. Stability of Mycobacterium tuberculosis DNA genotypes. J Infect Dis. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]