Abstract
Nosocomial vancomycin-resistant Enterococcus (VRE) infections have been described in only small numbers of pediatric patients. In none of these studies were multivariate analyses performed to assess which factors were independent risk factors in these patients. In the present cohort study of patients admitted to our hematology/oncology unit, surveillance cultures revealed a colonization rate of 24% and all isolates were identified as Enterococcus faecium. Risk factors associated with colonization with VRE identified by multiple logistic regression analysis included young age and chemotherapy with antineoplastic agents, cefotaxime, vancomycin, and ceftazidime. A molecular epidemiological tool, pulsed-field gel electrophoresis, was used to determine the relatedness of the VRE isolates detected. DNA analysis by this method identified two major clusters of VRE isolates. Young children with gastrointestinal colonization with VRE, without evidence of clinical infection, can serve as a reservoir for the spread of VRE.
The evolution of antimicrobial resistance has become a global problem (2, 5, 20). Analysis from the National Nosocomial Infections Surveillance system at the Centers for Disease Control and Prevention has demonstrated a 20-fold increase in nosocomial infections due to vancomycin-resistant enterococci (VRE) (4). This development limits the therapeutic options for treating serious infections (15). Acquisition of antimicrobial resistance by enterococci can be facilitated by interstrain spread of conjugative transposons, pheromone-responsive plasmids, and broad-host-range plasmids (18).
Nosocomial VRE infections have been described frequently in adult intensive care patients (3, 7, 9, 14, 16, 23, 25). However, VRE infections have been reported in only small numbers of pediatric patients (1, 10, 21). In none of these studies were multivariate analyses performed to identify independent risk factors in these patients. Young children colonized gastrointestinally with VRE may transmit this organism by fecal-oral spread and by contamination of their environment. The purpose of the present study was to determine the risk factors associated with VRE colonization in hospitalized children by using univariate and multiple logistic regression analyses. In addition, a molecular epidemiological tool was employed to establish the relatedness of the VRE isolates detected.
On 21 June 1994, a blood culture collected from a bone marrow transplant recipient yielded the first VRE identified at Children’s National Medical Center (CNMC). This finding prompted an investigation of the prevalence of VRE colonization among high-risk patients at our institution—a multidisciplinary, regional referral center.
MATERIALS AND METHODS
Study design.
Surveillance was conducted between 1 August and 20 October 1994 in our hematology/oncology (H/O) unit during an investigation of the circumstances surrounding the index case described above. The goal of this cohort study was to determine the prevalence of VRE colonization in our H/O unit. The H/O unit at CNMC has 27 beds, including 6 in individual positive-pressure rooms (intended for bone marrow transplant patients) and 5 in individual rooms reserved for other patients. The remaining beds are located within rooms containing two beds. One hundred twenty-five patients were admitted to the H/O unit during the surveillance period. The purpose of the surveillance cultures was explained to the patients and their parents, and specimens were collected on admission or soon thereafter. The majority of patients had an underlying diagnosis of malignancy or sickle cell disease. This unit also served as an overflow unit for a small number of acute care patients. Data collected from each patient included age, gender, weight, and admitting or underlying diagnosis on the day of admission. Diagnoses were categorized by ICD9 code. The remaining variables (e.g., antimicrobial agent administration prior to specimen collection, use of invasive devices, operative status, number of hospitalizations within the prior year, immune system status, and length of hospitalization) were assessed prior to culture. Information from patient medical records was recorded on a standardized data form. History of antimicrobial agent administration prior to specimen collection was obtained from patients before admission and from patient medical records while at the hospital. Some antimicrobial agent administration histories prior to hospital encounter may have been incomplete due to poor recollection by patients. A history of use included agents given for infectious disease prophylaxis or agents administered in the emergency department prior to admission. The use of vancomycin, ceftazidime, cefotaxime, and other commonly prescribed antimicrobial agents were analyzed as independent variables.
Invasive device use was defined as placement of a central venous line, a Foley catheter, or mechanical ventilation prior to specimen collection. All patients had peripheral intravenous lines placed prior to culture collection.
Immunosuppressed status was defined by the following criteria: administration of antineoplastic therapy within 6 months of specimen collection, bone marrow transplantation prior to culture, or an absolute neutrophil count of <500 per mm3. Operative procedure status was considered relevant when documentation of an operating room procedure during the same admission prior to specimen collection was present in the medical record. These procedures included Broviac line placements and tumor debulking procedures.
Microbiological methods.
Colonization with VRE was determined from rectal swabs obtained on admission to the H/O unit or from weekly surveillance cultures of rectal swabs obtained from patients hospitalized on the unit. Rectal swabs from all active H/O patients were cultured at least once. Rectal swabs were inoculated onto Campy blood agar with 10 μg of vancomycin (Campy BAP; Becton Dickinson, Cockeysville, Md.) per ml (6). Any growth or haze on the medium surface after 24 to 48 h of incubation at 35°C in ambient air was considered an indication of resistance to vancomycin. Suspicious nonhemolytic or alpha-hemolytic colonies were Gram stained to rule out the presence of Lactobacillus spp. Gram-positive cocci were tentatively identified as Enterococcus spp. with a negative catalase test and a positive pyrrolidonyl arylamidase test. Species identification was determined with the MicroScan WalkAway instrument (Positive Breakpoint Combo Panel Type 6; Dade MicroScan, West Sacramento, Calif.). Only one isolate per patient was studied.
MICs of vancomycin and other antimicrobial agents were also determined with the MicroScan WalkAway instrument. Panels were inoculated with turbidity-standardized suspensions prepared from overnight cultures of isolates characterized as resistant when cultured on Campy BAP. Panels were incubated for 24 h at 35°C and read by the MicroScan WalkAway instrument. Isolates were categorized as susceptible, intermediate, or resistant to antimicrobial agents in accordance with criteria published by the National Committee for Clinical Laboratory Standards (19).
DNA analysis by pulsed-field gel electrophoresis (PFGE) was also performed. Genomic DNA from 30 VRE isolates was prepared for digestion and electrophoresis as described previously (7). In brief, after digestion with the restriction endonuclease SmaI, chromosomal DNA fragments were separated with a contour-clamped homogenous electric field unit (CHEF-DR II; Bio-Rad Laboratories, Hercules, Calif.) and applied to agarose gels. The gels were stained with ethidium bromide and photographed. Gel patterns were compared, and isolates were categorized as indistinguishable, closely related, possibly related, or different in accordance with the criteria for interpreting PFGE patterns of Tenover et al. (24).
Environmental cultures of swabs of the sink, bed rail, countertop, over-bed table, and room-exiting doorknob were performed in colonized patient rooms before and after discharge. Dacron-tipped swabs were moistened with sterile Trypticase soy broth and used to sample a 1-cm2 area of the appropriate surfaces.
Statistical methods.
The Kruskal-Wallis test for continuous variables and Fisher’s exact test for categorical variables were used for univariate comparisons of variables between patients who were VRE positive and those who were not (11, 12). A significance level of <0.05 was chosen. Odds ratios and 95% confidence intervals were determined for each of the variables. Multiple logistic regression analysis was performed on variables that were statistically significant during the univariate analysis. Selection of variables was done by first performing a Fisher’s exact test or t test for each binary variable or continuous variable. A significance level of P of <0.30 was chosen as a cutoff. The multiple logistic regression model was developed by using forward and backward subtraction, and the model was checked for multicollinearity. The SAS/STAT software (SAS Institute Inc., Cary, N.C.) was used for univariate and multivariate analysis (22).
RESULTS
One hundred twenty-five children were admitted to the H/O unit between 1 August and 20 October 1994. Two patients refused surveillance cultures. Of the 123 children who were cultured for VRE, 30 were positive in addition to the index case (colonization rate, 24%). Twenty-six of the cultured children had incomplete medical record information and were excluded from the analysis, leaving a study sample size of 97 patients. Of the 26 patients with incomplete records, only one was colonized with VRE. The patients with missing records did not differ significantly from those in the study sample with respect to their demographic characteristics. Of the 97 study patients (30 VRE positive and 67 VRE negative), 46 had an underlying diagnosis of malignancy, 40 had sickle cell disease, and 11 had other diagnoses. During the surveillance period, one patient colonized with VRE developed VRE bacteremia while undergoing total body irradiation and chemotherapy as a prelude to bone marrow transplantation. During the first 3 weeks of October, 43 patients had surveillance cultures performed, with only one positive culture resulting. By that time, all current H/O unit patients had had surveillance cultures performed at least once. No additional cases of VRE infection or intestinal colonization were recognized after 20 October 1994, and surveillance cultures were discontinued. All of the environmental cultures were negative.
Among the 30 VRE-positive and 67 VRE-negative children, the risk factors associated by univariate analysis with VRE colonization were young age, use of invasive devices, administration of antimicrobial therapy, immunosuppression, and an underlying diagnosis of malignancy or sickle cell disease (Table 1). Several of these risk factors are surrogate markers for frequent hospitalization. The multiple logistic regression analysis using variables from Table 1 is shown in Table 2. After controlling for other risk variables, our analysis showed that young patients who were given antineoplastic chemotherapy, cefotaxime, vancomycin, or ceftazidime prior to surveillance cultures had 10-, 38-, 50-, or 96-fold higher risks of VRE positivity, respectively (Table 2). The risk coefficients for patients receiving cefotaxime and vancomycin or ceftazidime and vancomycin were not additive.
TABLE 1.
Univariate analysis of Patient risk factors for VRE colonization
| Variable | Value for patients
|
Probabilitya | RRc | CId | |
|---|---|---|---|---|---|
| With VRE (N = 30) | Without VRE (N = 67) | ||||
| Mean age (SEM) (yr) | 4.60 (0.74) | 8.14 (0.70) | 0.008b | ||
| Mean wt (SEM) (kg) | 18.68 (2.59) | 30.48 (2.60) | 0.004b | ||
| No. of female patients (%) | 15 (50) | 29 (43.3) | 0.66 | 1.20 | 0.64–2.25 |
| No. (%) of patients with: | |||||
| Invasive device use | 22 (73.3) | 19 (28.4) | <0.001 | 3.76 | 1.91–7.37 |
| Antimicrobial therapy | 27 (90) | 40 (59.7) | 0.004 | 4.03 | 1.31–9.34 |
| Cefotaxime | 7 (23.3) | 5 (7.46) | 0.043 | 2.16 | 1.31–3.54 |
| Ceftazidime | 18 (60.) | 7 (10.5) | <0.001 | 4.32 | 2.42–7.72 |
| Vancomycin | 19 (63.3) | 9 (13.4) | <0.001 | 4.26 | 2.33–7.78 |
| Immunosuppression | 26 (86.7) | 16 (23.9) | <0.001 | 8.51 | 3.46–20.93 |
| Chemotherapy | 24 (80) | 14 (20.9) | <0.001 | 6.21 | 2.90–13.31 |
| Low absolute neutrophil count | 19 (63.3) | 3 (4.48) | <0.001 | 5.89 | 3.25–10.67 |
| Surgery | 5 (16.7) | 10 (14.9) | 1.0 | 1.09 | 0.59–2.04 |
| Previous hospitalization | 22 (73.3) | 56 (83.6) | 0.27 | 0.67 | 0.35–1.27 |
| Underlying diseasee | 26 (86.7) | 20 (29.9) | <0.001 | 7.21 | 2.99–17.35 |
Fisher’s exact test.
The Kruskal-Wallis test.
RR, Relative risk.
CI, 95% confidence interval.
Underlying disease signifies malignancy or sickle cell disease.
TABLE 2.
Maximum likelihood estimates of VRE colonization
| Variable | Regression coefficient | SE | Probabilitya | Adjusted odds ratio | 95% CIb |
|---|---|---|---|---|---|
| Intercept | −3.065 | 0.924 | 0.001 | ||
| Age | −0.202 | 0.078 | 0.009 | 0.82 | 0.70–0.95 |
| Vancomycin | 3.920 | 1.322 | 0.003 | 50.39 | 3.77–672.71 |
| Cefotaxime | 3.644 | 1.405 | 0.009 | 38.24 | 2.43–600.83 |
| Ceftazidime | 4.560 | 1.305 | 0.001 | 95.62 | 7.41–999.00 |
| Chemotherapy | 2.327 | 0.839 | 0.005 | 10.25 | 1.98–53.03 |
| Cefotaxime and vancomycin | −3.957 | 1.969 | 0.044 | ||
| Ceftazidime and vancomycin | −5.202 | 1.789 | 0.004 |
Wald’s chi-square test.
Confidence interval for odds ratio determined by the formula Az(SE) = 0.949 (0.029), where Az is the area under the receiver operating characteristic curve.
All of the isolates that appeared to be vancomycin resistant on Campy BAP were determined to be resistant with the MicroScan WalkAway instrument (MIC, >32 μg/ml). All VRE isolates were Enterococcus faecium and were resistant to several other antimicrobial agents. However, all were susceptible to chloramphenicol and the high-level gentamicin synergy test and all but one were susceptible to tetracycline. Susceptibility to teicoplanin was not determined.
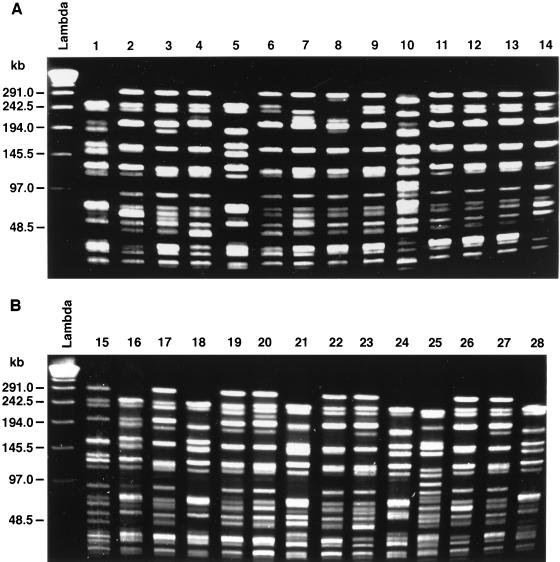
PFGE was performed on 30 of the VRE isolates. The PFGE patterns for 28 of the 30 isolates are shown in Fig. 1. Heterogeneity in these isolates was present. Fourteen different PFGE patterns were discerned. However, two pattern clusters encompassed 57% of the isolates tested: pattern cluster 1 encompassed 6 isolates (Fig. 1, lanes 1, 5, 16, 18, 24, and 28), pattern cluster 2 encompassed 10 isolates (lanes 4, 6, 9, 11, 12, 13, 17, 19, 22, and 27), pattern cluster 3 encompassed 2 isolates (lanes 2 and 26), and pattern cluster 4 encompassed 2 isolates (lanes 3 and 20). The remaining 8 isolates at best were possibly related. There was only a low-level correlation between the restriction endonuclease patterns and the antimicrobial agent resistance patterns.
FIG. 1.
PFGE of 28 of the 30 VRE isolates. There were two major clusters by SmaI restriction endonuclease patterns (clusters 1 and 2). Lanes 1, 5, 16, 18, 24, and 28, pattern cluster 1 (6 isolates); lanes 4, 6, 9, 11, 12, 13, 17, 19, 22, and 27, pattern cluster 2 (10 isolates); lanes 2 and 26, pattern cluster 3 (2 isolates); and lanes 3 and 20, pattern cluster 4 (2 isolates). The remaining 10 isolates could represent possibly related strains.
DISCUSSION
In this study, a molecular epidemiological technique revealed the existence of multiple clusters of genetically related VRE isolates in our patient population. PFGE yielded several different patterns for the 30 isolates that were tested. Two PFGE patterns predominated among the isolates, suggesting patient-to-patient spread of VRE within this cohort of patients. There was no correlation between the rooms to which patients were admitted during the current admission and the PFGE patterns. One can speculate that dissemination of VRE strains within these cohorts occurred during previous admissions, since recurrent hospitalization is frequent among high-risk H/O patients. In the United States, most acquisition of VRE occurs in the hospital. This is in contrast to the published experience in Europe, where community-acquired infections have been described (8). Little if any relation could be established between restriction endonuclease patterns and patterns of antimicrobial agent resistance.
We also identified a 24% carrier rate and risk factors associated with this carriage. Multivariate analysis indicated that VRE-colonized patients were young, tended to have received prior antimicrobial therapy, and were immunosuppressed. Our results are in keeping with those from a study of pediatric oncology patients in which neutropenia, exposure to broad-spectrum antimicrobial agents, and administration of vancomycin were important risk factors (10). Multiple logistic regression analysis of our data found, however, that ceftazidime therapy preceded VRE colonization more often than vancomycin therapy did (odds ratio, 95.6 versus 50.4). The same analysis revealed that administration of cancer chemotherapeutic agents within the previous 6 months also increased the risk of VRE colonization. During the surveillance period, one of our patients experienced VRE bacteremia while receiving chemotherapy in preparation for a bone marrow transplant. Intestinal tract colonization with VRE may put patients with malignancies at risk for VRE bacteremia, especially during neutropenic episodes (10).
The Hospital Infection Control Practices Advisory Committee guidelines on prevention and control of the spread of VRE were implemented in our H/O unit once patients were identified as colonized (13). Parents, family members, and staff were given VRE fact sheets, and the importance of observing infection prevention and control measures was emphasized. Because the gastrointestinal tract can remain colonized with VRE for prolonged periods without clinically apparent disease, early identification of infected patients is critical, especially when dealing with young children with poor hygiene who are prone to fecal-oral spread of microorganisms.
ACKNOWLEDGMENTS
We acknowledge the helpful suggestions of William Rodriguez. We thank Bruce Sprague of the CNMC Center for Health Services and Clinical Research for his help with data management; Eileen Cantwell, Judy Miles, and Dorleen Brown of the Hospital Epidemiology Department; and the staff of the H/O unit at the Children’s National Medical Center, Washington, D.C.
REFERENCES
- 1.Bingen E H, Denamur E, Lambert-Zechovsky N Y, Elion J. Evidence for the genetic unrelatedness of nosocomial vancomycin-resistant Enterococcus faecium strains in a pediatric hospital. J Clin Microbiol. 1991;29:1888–1892. doi: 10.1128/jcm.29.9.1888-1892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyce J M. Vancomycin-resistant enterococcus: detection, epidemiology, and control measures. Infect Dis Clin N Am. 1997;11:367–384. doi: 10.1016/s0891-5520(05)70361-5. [DOI] [PubMed] [Google Scholar]
- 3.Boyle J F, Soumakis S A, Rendo A, Herrington J A, Gianarkis D G, Thurberg B E, Painter B G. Epidemiologic analysis and genotypic characterization of a nosocomial outbreak of vancomycin-resistant enterococci. J Clin Microbiol. 1993;31:1280–1285. doi: 10.1128/jcm.31.5.1280-1285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention: Project ICARE—Intensive Care Antimicrobial Resistance Epidemiology. Unpublished data. National Nosocomial Infection System; 1998. [Google Scholar]
- 5.Chenworth C, Schaberg D. The epidemiology of enterococci. Eur J Clin Microbiol Infect Dis. 1990;9:80–89. doi: 10.1007/BF01963631. [DOI] [PubMed] [Google Scholar]
- 6.Edberg S C, Hardalo C J, Kontnick C, Campbell S. Rapid detection of vancomycin-resistant enterococci. J Clin Microbiol. 1994;32:2182–2184. doi: 10.1128/jcm.32.9.2182-2184.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmond M B, Ober J F, Weinbaum D L, Pfaller M A, Hwang T, Sanford M D, Wenzel R P. Vancomycin-resistant Enterococcus faecium bacteremia: risk factors for infection. Clin Infect Dis. 1995;20:1126–1133. doi: 10.1093/clinids/20.5.1126. [DOI] [PubMed] [Google Scholar]
- 8.Endtz H P, Van Den Braak N, Van Belkum A, Kluytmans J A J W, Koeleman J G M, Spanjaard L, Voss A, Weersink A J L, Vandenbroucke-Grauls C M J E, Buiting A G M, Van Duin A, Verbrugh H A. Fecal carriage of vancomycin-resistant enterococci in hospitalized patients and those living in the community in The Netherlands. J Clin Microbiol. 1997;35:3026–3031. doi: 10.1128/jcm.35.12.3026-3031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Handwerger S, Raucher B, Altarac D, Monka J, Marchione S, Singh K V, Murray B E, Wolf J, Walters B. Nosocomial outbreak due to Enterococcus faecium highly resistant to vancomycin, penicillin, and gentamicin. Clin Infect Dis. 1993;16:750–755. doi: 10.1093/clind/16.6.750. [DOI] [PubMed] [Google Scholar]
- 10.Henning K J, Delencastre H, Eagan J, Boone N, Brown A, Chung M, Wollner N, Armstrong D. Vancomycin-resistant Enterococcus faecium on a pediatric oncology ward: duration of stool shedding and incidence of clinical infection. Pediatr Infect Dis J. 1996;15:848–854. doi: 10.1097/00006454-199610000-00004. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch R P, Riegelman R K. Statistical operations: analysis of health care research data. 1996. pp. 270–274. , 326–330. Blackwell Science, Inc., Cambridge, Mass. [Google Scholar]
- 12.Hosmer D W, Lemeshow S. Applied logistic regression. New York, N.Y: John Wiley & Sons, Inc.; 1989. pp. 25–34. [Google Scholar]
- 13.Hospital Infection Control Practices Advisory Committee (HICPAC) Recommendation for preventing the spread of vancomycin resistance. Infect Control Hosp Epidemiol. 1995;16:105–113. doi: 10.1086/647066. [DOI] [PubMed] [Google Scholar]
- 14.Karanfil L V, Murphy M, Josephson A, Gaynes R, Mandel L, Hill B C, Swenson J M. A cluster of vancomycin-resistant Enterococcus faecium in an intensive care unit. Infect Control Hosp Epidemiol. 1992;13:195–200. doi: 10.1086/646509. [DOI] [PubMed] [Google Scholar]
- 15.Moellering R C. The enterococcus: a classic example of the impact of antimicrobial resistance on therapeutic options. J Antimicrob Chemother. 1991;28:1–12. doi: 10.1093/jac/28.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Morris J G, Shay D K, Hebden J N, McCarter R J, Perdue B E, Jarvis W, Johnson J A, Dowling T C, Polish L B, Schwalbe R S. Enterococci resistant to multiple antimicrobial agents, including vancomycin. Ann Intern Med. 1995;123:250–259. doi: 10.7326/0003-4819-123-4-199508150-00002. [DOI] [PubMed] [Google Scholar]
- 17.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray B E. Diversity among multidrug-resistant enterococci. Emerg Infect Dis. 1998;4:1–14. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing; eighth informational supplement. NCCLS document M100-S8. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 20.Rice L B, Shales D M. Vancomycin resistance in the enterococcus: relevance in pediatrics. Pediatr Clin N Am. 1995;42:601–617. doi: 10.1016/s0031-3955(16)38981-7. [DOI] [PubMed] [Google Scholar]
- 21.Rubin L G, Tucci E C, Cercenado E, Eliopoulos G, Isenberg H D. Vancomycin-resistant Enterococcus faecium in hospitalized children. Infect Control Hosp Epidemiol. 1992;13:700–705. doi: 10.1086/648342. [DOI] [PubMed] [Google Scholar]
- 22.SAS Institute. SAS/STAT user’s guide, version 6. 4th ed. Vol. 2. Cary, N.C: SAS Institute; 1990. pp. 1071–1126. [Google Scholar]
- 23.Stroud L, Edwards J, Danzig L, Culver D, Gaynes R. Risk factors for mortality associated with enterococcal bloodstream infections. Infect Control Hosp Epidemiol. 1996;17:576–580. doi: 10.1086/647386. [DOI] [PubMed] [Google Scholar]
- 24.Tenover F C, Arbeit R D, Goering G, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weinstein J W, Roe R, Towns M, Sanders L, Thorpe J J, Corey G R, Sexton D J. Resistant enterococci: a prospective study of prevalence, incidence, and factors associated with colonization in a university hospital. Infect Control Hosp Epidemiol. 1996;17:36–41. doi: 10.1086/647186. [DOI] [PubMed] [Google Scholar]