Abstract
The epithelial-to-mesenchymal transition (EMT), which conveys epithelial (E) carcinoma cells to quasi-mesenchymal (qM) states, enables them to metastasize and acquire resistance to certain treatments. Murine tumors composed of qM mammary carcinoma cells assemble an immunosuppressive tumor microenvironment (TME) and develop resistance to anti-CTLA4 immune checkpoint blockade therapy (ICB), unlike their E counterparts. Importantly, minority populations of qM cells within a tumor can cross-protect their more E neighbors from immune attack. The underlying mechanisms of immunosuppression and cross-protection have been unclear. We demonstrate that abrogation of qM carcinoma cell-derived factors (CD73, CSF1 or SPP1) prevents the assembly of an immunosuppressive TME and sensitizes otherwise refractory qM tumors partially or completely to anti-CTLA4 ICB. Most strikingly, mixed tumors in which minority populations of carcinoma cells no longer express CD73, are now sensitized to anti-CTLA4 ICB. Finally, loss of CD73 also enhances the efficacy of anti-CTLA4 ICB during the process of metastatic colonization.
Keywords: EMT, immune checkpoint blockade, immunosuppression, tumor microenvironment, breast carcinoma
Introduction
The development and use over the past decade of immunotherapy to treat various types of human tumors has attracted substantial attention, given its powers to create durable clinical responses in certain types of human tumors. However, the successes of these therapies, notably immune checkpoint blockade therapy (ICB), have been limited by the fact that responses remain heterogeneous between different groups of patients. Moreover, the factors that accurately predict these variable responses are elusive.
A number of studies have examined the utility of certain markers for predicting responses, prominent among them being the presence of T-cells, expression of PDL1, the presence of antigen presentation markers and neoantigen load(1–5). Nonetheless, not all PDL1-expressing tumors respond to anti-PDL1 therapy(6). In addition, somatic mutations affecting carcinoma cell-intrinsic pathways associated with antigen-presentation and/or IFNγ sensing induce resistance to ICB(5,7–11). While there have been numerous attempts at identifying markers that can actively predict successful clinical responses to ICB, none of these has explored systematically how the epithelial versus mesenchymal states of the carcinoma cells within tumors govern these outcomes.
As has been well described over the past two decades, the EMT is a cell-biological program that potentiates aggressive properties of carcinomas including their metastatic dissemination(12,13). Upon activation of this program, cells typically shed the expression of epithelial markers, such as E-Cadherin, and express instead mesenchymal markers, such as vimentin, fibronectin and certain master EMT-inducing transcription factors (EMT-TFs), notably Zeb1, Twist, Snail and Slug; once expressed, these EMT-TFs regulate the expression of genes associated with the more mesenchymal states of carcinoma cells. Among the acquired mesenchymal characteristics are invasiveness and motility, which empower carcinoma cells to disseminate to distant anatomical sites, to form tumor-initiating cancer stem-cells (CSCs), and to acquire an elevated resistance to various standard chemotherapeutic regimens(14–17). In fact, EMT programs do not operate as binary controls that switch cells between alternative epithelial and mesenchymal states, but instead usually convey cells from fully epithelial (E) states to cells of mixed epithelial/mesenchymal phenotype(18,19), which we refer to hereafter as quasi-mesenchymal (qM).
Several years ago, we and others demonstrated that tumors composed of more E or more qM carcinoma cells are differentially susceptible to immune attack and mount distinct responses to anti-CTLA4 ICB(20–23). In our own work, we used matched pairs of E and qM murine mammary adenocarcinoma cells that were derived from tumors arising in the transgenic MMTV-PyMT breast carcinoma mouse model, some of which bear reporter constructs that label cells expressing the endogenous Snail EMT-TF (20,24). This enabled us to fractionate tumors using the expression of Snail-YFP in conjunction with the epithelial marker, Epcam, yielding isogenic SnailHI (qM) or SnailLO (E) cell lines. These tumor cell lines offered multiple advantages, including the facts that they reside stably in either the E or qM phenotypic states and could be grown in syngeneic, immunocompetent hosts, spawning corresponding more E or more qM tumors in vivo.
As was reported, the more mesenchymal (qM) tumors contained (i) immunosuppressive cells such as M2-like macrophages and Tregs in the tumor microenvironment, (ii) excluded CD8+ T-cells to their periphery, and (iii) exhibited elevated resistance to anti-CTLA4 ICB therapy. This behavior contrasted sharply with that of their epithelial counterparts, which recruited functionally active cytotoxic CD8+ T-cells to the tumor core and were indeed sensitive to anti-CTLA4 therapy (20). Strikingly, in tumors arising from mixtures of E and qM carcinoma cells, we found that a small minority (10%) of qM cells could protect a large majority (90%) of E cells that were residing within the same tumor from immune attack(20). Thus, not only do qM carcinoma cells assemble an immunosuppressive TME and mount refractory responses to ICB, but they also cross-protect their nearby epithelial neighbors from elimination by anti-CTLA4 ICB.
This earlier work did not reveal the underlying mechanisms by which the EMT program contributes to immunosuppression, the resulting failure of a checkpoint blockade immunotherapy, and the mechanism of cross-protection. Understanding precisely how mesenchymal carcinoma cells resist anti-tumor immune attack is particularly important, as most human primary carcinomas are likely to contain subpopulations of qM cells or their derivatives residing within primary tumors (25). Furthermore, while there have been occasional reports in the literature that have correlated certain aspects of the EMT program with a lack of response to ICB(22,23), none of these has demonstrated whether the qM state plays a causal role in regulating refractory responses to ICB.
We present here our findings that delineate in a systematic fashion precisely how the EMT program can affect responses to ICB in a syngeneic, immunocompetent murine model of mammary adenocarcinoma and suggest that the described mechanisms are likely to operate as well in a diverse spectrum of other carcinomas that have activated components of the EMT program.
Results
Enrichment of immunosuppressive markers and pathways in quasi-mesenchymal breast carcinomas
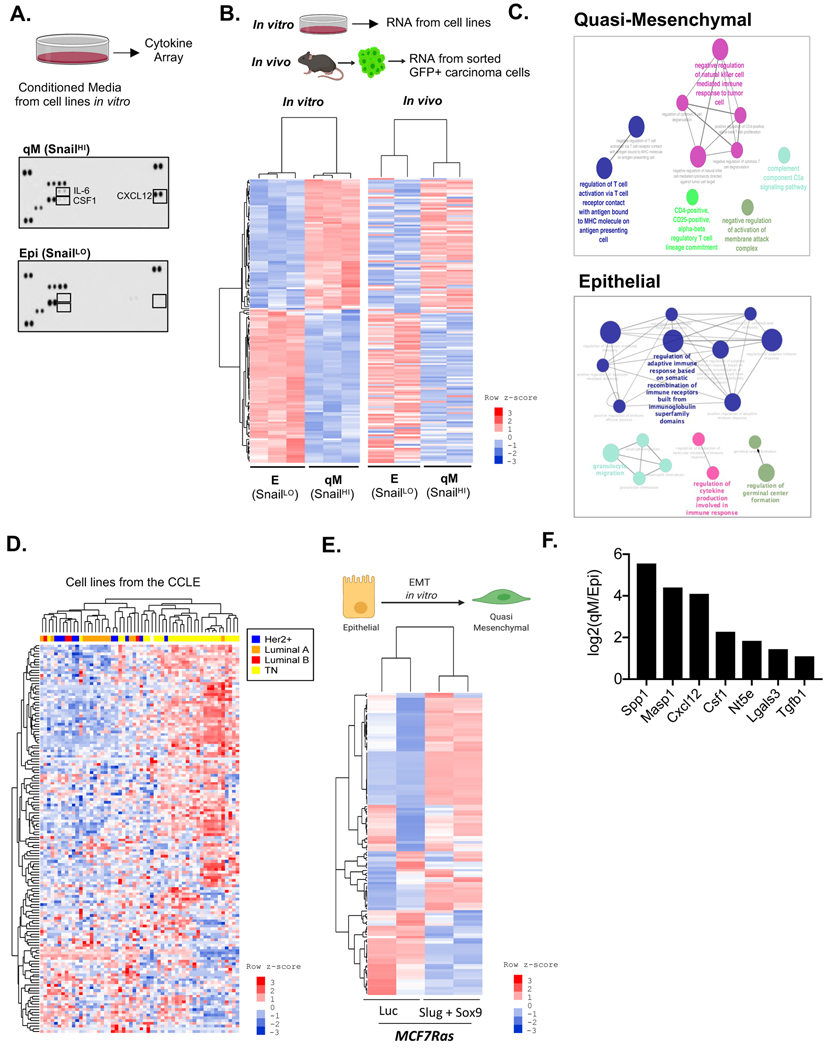
We began by determining whether E and qM carcinoma cells secrete distinct sets of cytokines and chemokines that could possibly operate via paracrine signaling to recruit different sets of immune cells to their corresponding tumors. Cytokine array analysis using conditioned media from various E and qM PyMT cell lines cultured in vitro revealed that qM cell lines secreted CSF1, CXCL12 and IL-6 (Fig 1A), which have been previously documented to play important roles in regulating the recruitment and function of macrophages and Tregs (26,27). In contrast, cells of the E lines lacked the expression of these cytokines (Fig 1A).
Figure 1: Immunosuppressive markers and pathways associated with qM breast carcinomas.
(A) Cytokine Array analysis for the indicated cell lines cultured in vitro. (B) Schema for isolation of RNA from the indicated cell lines cultured in vitro. RNA was also extracted from GFP-labelled E or qM carcinoma cells that were FACS-sorted from their corresponding tumors. Heat map of differentially expressed genes (nCounter PanCancer Immune Profiling Panel, Nanostring Technologies), between the indicated E and qM cell lines propagated in vitro or sorted from their corresponding tumors propagated in vivo. Differentially expressed genes were defined as a twofold change (up or down) and FDR adjusted p-value <0.056 and N=3 (in vitro); N=2 (in vivo). All differentially expressed genes were assayed using DESeq2, which uses the Wald test. For all heat maps, each row of the heatmap represents row-wise z-score from log-transformed normalized expression levels, where the colors red and blue represent up and down regulation, respectively. The left dendrogram in each heatmap indicates clustering of genes. (C) ClueGO pathway enrichment analysis for immune-modulatory pathways expressed by qM or E carcinoma cells. (D) Unsupervised hierarchical clustering of human breast cancer cell lines using the same list of differentially expressed genes in Fig 1B. (E) Schema showing that MCF7Ras cells can be induced to undergo an EMT in vitro by simultaneous expression of doxycycline-induced Slug and Sox9 relative to those expressing a Luciferase control. Heat map showing that these cells also differentially express the same list of immunomodulatory genes as (B) upon activation of the EMT program. FDR adjusted p-value <0.056 and N=2. (F) Transcript levels of genes expressed by qM carcinomas relative to E carcinomas.
We undertook to determine whether the observed differences in cytokine and chemokine production were also accompanied by changes in the expression of other immunomodulatory markers. To do so, we used the nCounter PanCancer Immune Profiling Panel (Nanostring Technologies)(28) to perform transcriptomic analyses of the E and qM mammary carcinoma cells; these cells were either cultured in vitro or prepared by FACS sorting of GFP-labelled carcinoma cells from their corresponding tumors (Fig 1B). Once again, we found marked differences between the E and qM carcinoma cell lines, in that they differentially expressed a number of immunomodulatory gene transcripts. (Heat Maps – Fig 1B, Supp Fig 1A,B; for Gene List see Supplementary Table #1). ClueGO pathway analysis, Gene Set enrichment, and DAVID pathway analysis(29–31) revealed that qM carcinomas express genes associated with immune-suppressive pathways, such as negative regulation of natural killer (NK) and effector T-cell function, induction of Tregs, and immune evasion and tolerogenesis. In sharp contrast, E carcinoma cells express immune genes associated with positive regulation of effector T-cell function, proliferation, antigen presentation and cytokine secretion (Fig 1C and Supp Fig 1C–E).
Of special interest, unsupervised hierarchical clustering of human breast cancer (BrCa) cell line expression profiles from the Cancer Cell Line Encyclopedia (CCLE) using the same list of immune genes that were differentially expressed by E and qM PyMT cell lines (Supplementary Table #1) revealed the existence of two distinct groups. The immunomodulatory genes expressed by qM murine PyMT cell lines were also found to be expressed by the triple-negative breast cancer (TNBC) subtype, whereas immune genes expressed by E PyMT cell lines were expressed by luminal A/B and HER2+ subtypes instead (Fig 1D). Taken together, these data demonstrate that E and qM breast carcinomas differ dramatically in their expression of immune modulatory genes, with an enrichment of immune-suppressive agents and pathways in the qM but not the E state. Moreover, the distinct phenotypes of the E and qM murine carcinoma cells echo the behavior of distinct subtypes of human breast cancers.
Finally, to demonstrate a direct association between activation of the EMT program and immunosuppression, we induced EMT programs in vitro in an array of cultured BrCa cell lines via the doxycycline-induced expression of various EMT-inducing transcription factors (EMT-TFs) (Supp Fig 2A). Unsupervised hierarchical clustering revealed that activation of the EMT program in vitro also resulted in increased expression of carcinoma cell-intrinsic, immunosuppressive factors and pathways (Fig 1E and Supp Fig 2B–F)(20,32–34). Hence, the acquisition of these immunosuppressive features is a direct consequence of the activation, in a cell-autonomous manner, of the EMT program operating within BrCa cells.
Abrogation of immunomodulatory factors specifically associated with the quasi-mesenchymal state.
In light of the strong enrichment of carcinoma cell-intrinsic immunosuppressive factors in the qM but not the E state, we asked whether any of these factors might play key functional roles in governing the assembly of an immunosuppressive TME. To address this question, we used multiple screening criteria, in particular (i) expression in at least two different qM PyMT cell lines, (ii) differential expression in luminal versus TNBC human BrCa cell lines, (iii) high levels of expression (normalized read counts >100) and (iv) association with immunosuppressive pathways (Supp Fig 2G). This screening strategy enabled us to converge on a list of seven immunomodulatory factors associated with paracrine signaling that were specifically associated with the qM but not the E cells (Fig 1F). These were Osteopontin (Spp1), MASP1 (Masp1), CXCL12 (Cxcl12), M-CSF (Csf1), CD73 (Nt5e), Galectin-3 (Lgals3) and TGFβ1 (Tgfb1).
The genes encoding these seven factors were knocked out (KO) individually via CRISPR/Cas9 in order to generate qM PyMT cells lines lacking the expression of the corresponding proteins (Supp Fig 3A–G). An important goal of these analyses was to focus on a set of genes whose abrogation preserved the continued residence of carcinoma cells in the qM state while at the same time, evoked significant changes in the associated TME. Immunofluorescence and western blot analysis of the various KO-cell lines (Fig 2A–C) as well as histopathological analyses of the corresponding tumor sections (Fig 2D) revealed that six of the seven KO-cell lines and derived tumors continued to exhibit a sarcomatoid-like morphology, expressed Zeb1 and Vimentin and lacked the expression of E-cadherin (Fig 2A–D). Hence, loss of any of these six factors did not destabilize continued residence of these cells in the qM phenotypic state.
Figure 2: Residence of carcinoma cells in a qM state.

(A) Phase contrast images and immunofluorescence analysis of qM control or KO cell lines for the indicated EMT markers. E-cadherin (white), Vimentin or Zeb1 (red), and DAPI (nuclear stain in blue). Scale bar = 20 μm (B) Western Blot analysis of qM control or KO cell lines for the indicated EMT markers. (C) Histogram showing surface levels of Epcam expression in qM control or KO cell lines as determined by flow cytometry. (D) Hemotoxylin and Eosin staining and immunofluorescence staining of tumor sections obtained from qM control or KO-tumor bearing mice stained for the indicated EMT markers. GFP (carcinoma cells in green). E-cadherin (white), Vimentin or Zeb1 (red), and DAPI (nuclear stain in blue) Scale bar = 20 μm. Data represent three independent experiments.
Of additional interest was the exception provided by the behavior of the TGF-β1 KO cells, which lapsed into a more epithelial state, as indicated by their acquisition of Epcam and loss of Vimentin. This observation, in concurrence with previously published work, suggested that the continued residence of these cells in the qM state depended on ongoing autocrine signaling driven by this cytokine(35,36). Hence, qM-derived TGF-β1 is poised to act in both a paracrine fashion on the nearby TME and in an autocrine fashion on the qM cells themselves. This shift to a more epithelial state confounded subsequent attempts to elucidate the immunological effects of cells residing in a true qM state while, at the same time, lacking the ability to produce this particular cytokine. Accordingly, we focused our further interest on the remaining six paracrine factors.
Ability of quasi-mesenchymal carcinoma-cell derived paracrine factors to regulate anti-tumor immunity
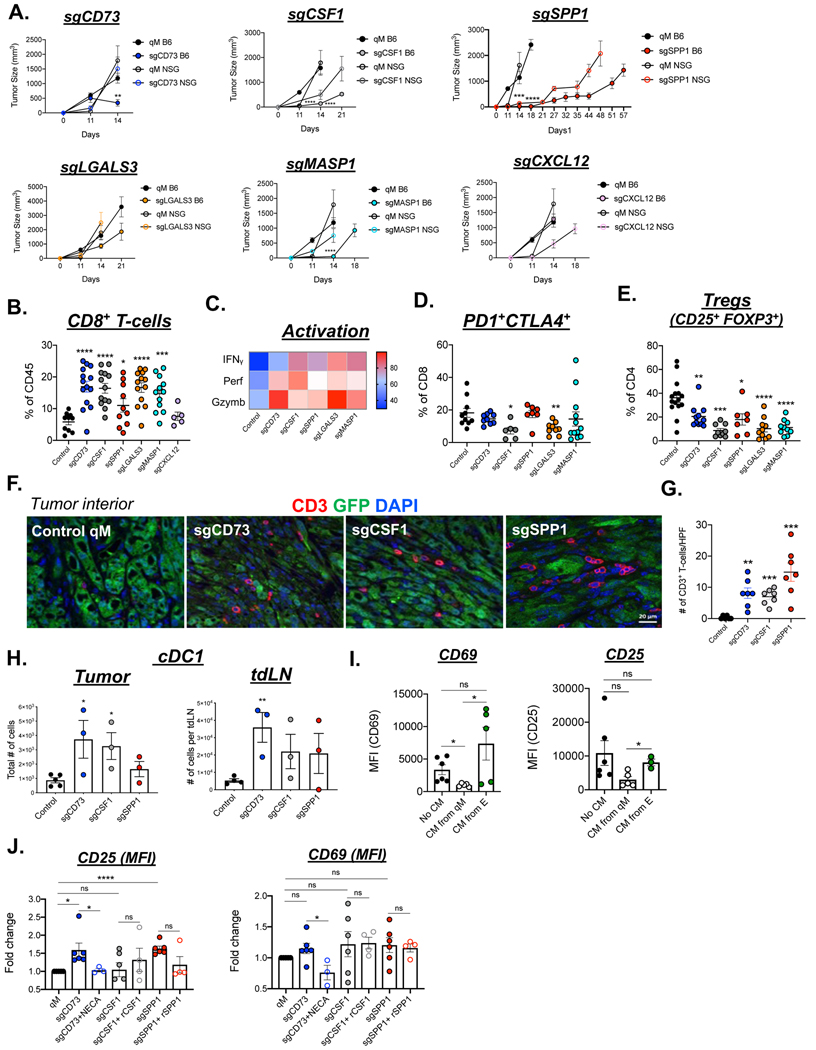
We proceeded to determine whether abrogating the expression of any of the remaining six paracrine factors could prevent the assembly of an immunosuppressive TME and thereby sensitize qM tumors to anti-tumor immune attack. Accordingly, we implanted the qM parental and various KO-PyMT cell lines orthotopically into syngeneic hosts. We observed that, with the exception of the CXCL12-KO, each of the other five KO-tumors demonstrated delayed kinetics of tumor growth relative to control qM tumors (Fig 3A). These differences were largely diminished when the various tumors were implanted in immune-compromised NSG hosts (Fig 3A). Additionally, all KO-cell lines proliferated just as efficiently as control qM cells in vitro (Supp Fig 4A). Taken together, these observations provided a strong indication that qM tumors lacking the expression of five out of the six immunomodulatory factors (CD73, CSF1, SPP1, LGALS3, or MASP1) were partially susceptible to anti-tumor immune attack even in the absence of an applied ICB (Fig 3A).
Figure 3: Ability of qM carcinoma-cell derived factors to regulate anti-tumor immunity.
(A) Kinetics of tumor growth of control qM cells or those knocked out for the indicated factors in C57BL6/J or NSG mice. N = 5–7 mice/group and represent pooled values from two independent experiments. (B) Bar graph representing the % of CD8+ T-cells after gating on the CD45+ population in the indicated tumors. Each data point represents a tumor from one mouse. N = 3–5 mice/group and represent pooled values from two-three independent experiments. (C) Heatmap indicating the % of CD8+ T-cells expressing the indicated cytokines as determined by flow cytometry. (D) % of CD8+ T-cells co-expressing PD1 and CTLA4 (E) % of CD4 cells co-expressing CD25 and FOXP3. (F) Immune-fluorescence analysis of tumor sections from control qM or KO tumors showing GFP (carcinoma cells in green) CD3 (T-cells in red) and DAPI (nuclear stain in blue). Scale bar = 20 μm. (G) Bar graph accompanying the immune-fluorescent image representing the numbers of CD3+ T-cells present per high power field (HPF) at 63X magnification. (H) Total numbers of conventional type 1 dendritic cells (cDC1) present in the tumor or tumor draining lymph nodes (tdlN) of the indicated tumors. (I) Mean Fluorescent Intensity for CD69 and CD25. CD8+ T-cells were activated in either no conditioned media, or conditioned media obtained from E or qM carcinoma cells. Surface expression of CD25 and CD69 was assessed after 48hrs by flow cytometry (J) Fold changes in CD25 and CD69 expression relative to levels expressed by qM cells. CD8+ T-cells were activated in the presence of conditioned media obtained from qM control or sgCD73, sgCSF1 or sgSPP1 cells. CD8+ T-cells were also activated in the conditioned media from the indicated cell lines that was reconstituted with either NECA, recombinant CSF1 or recombinant SPP1. Surface expression of CD25 and CD69 was assessed after 48hrs by flow cytometry. Data represent pooled values from three independent experiments. Error bars represent ± SEM. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001, ns = not significant
In addition to the delayed kinetics of tumor growth in immune-competent syngeneic hosts, qM tumors knocked out for Nt5e (CD73), Csf1, Spp1, Lgals3, or Masp1 all showed a significant increase in the numbers of CD8+ T-cells relative to control qM tumors or those knocked out for Cxcl12 (Fig 3B, Supp Fig 4B,C). Furthermore, the CD8+ T-cells present in each of these five KO-tumors expressed higher levels of the T-cell effector markers perforin, granzyme B and IFNγ and lower levels of exhaustion markers PD1 and CTLA4 relative to CD8+ T-cells present in control qM tumors (Fig 3C,D, Supp Fig 4D–H). Strikingly, the increase in the numbers of CD8+ T-cells was accompanied by a decrease in the numbers of immune-suppressive regulatory T-cells (Tregs) (Fig 3E, Supp Fig 4I–K). Hence, even in the absence of ICB therapy, five factors deployed by qM carcinoma cells can, to some extent, compromise attacks by CD8+ T-cells.
Mobilization of T-cells into the tumor core is considered to be a key factor that predicts a favorable response to ICB(37). Accordingly, we proceeded to determine whether the elevated numbers of T-cells in the aforementioned KO-tumors had indeed infiltrated into the tumor core. Strikingly, only qM tumors knocked out for Nt5e (CD73), Csf1 or Spp1 recruited T-cells into the tumor core. In sharp contrast, these T-cells were effectively excluded to the tumor periphery in control qM tumors and those knocked out for Lgals3, Masp1, Cxcl12 or Tgfb1, (Fig 3F, G and Supp Fig 4L).
In addition to the presence of tumor-infiltrating lymphocytes, another factor that is known to regulate the susceptibility of tumors to immune attack and ICB is the presence of conventional antigen-presenting dendritic cells (cDC1s). This is due, in part, to their ability to efficiently prime and subsequently activate T-cells(3). Strikingly, only the KO-tumors that demonstrated the presence of tumor-infiltrating lymphocytes also showed a concomitant increase in the numbers of cDC1s (Fig 3H, Supp Fig 5A–C). In sharp contrast, these cDC1s failed to be recruited to control qM tumors and those KO tumors that had excluded T-cells to the tumor periphery. (Supp Fig 5A–C). In fact, the co-existence of antigen-presenting dendritic cells and tumor-infiltrating T-cells has been found by others to be predictive of an active anti-tumor immune response and a favorable response to ICB(3). These various observations led us to conclude that qM carcinoma cell-derived CD73, CSF1 or SPP1 were most critically important in reducing the susceptibility of qM tumors to anti-tumor immune attack, causing us to focus on these three factors for the remainder of our analyses.
To determine whether any of these three factors could directly regulate the function of CD8+ T-cells in the absence of additional stromal components, we isolated CD8+ T-cells from the spleens of naïve mice and activated them in vitro with plate-bound anti-CD3ε and anti-CD28 antibodies in the presence of conditioned media (CM) obtained from either E, qM-control or qM-KO PyMT cell lines. Here, we found that CD8+ T-cells activated in the presence of CM from a control qM cell line failed to be optimally activated, as determined by low levels of cell-surface expression of the T-cell activation markers CD25 and CD69 relative to CD8+ T-cells activated in the presence of CM obtained from E carcinoma cells (Fig 3I). Strikingly, however, activation in the presence of CM derived from CD73-KO, CSF1-KO or SPP1-KO cell lines largely restored T-cell function as indicated by increased surface levels of CD25 and CD69 (Fig 3J). To further confirm the direct effects of each of these paracrine factors on T-cell activation, T-cells were also activated in KO-CM that was reconstituted with the specific paracrine factor (specifically, the low molecular weight adenosine receptor agonist NECA, recombinant CSF1 or recombinant SPP1). Activation of T-cells in sgCD73 CM that was reconstituted with NECA reduced T-cell activation as documented by reduced surface levels of CD25 and CD69, confirming the direct effects of CD73/Adenosine on T-cell activation. In contrast, neither rCSF1 nor rSPP1 sufficed to reduce T-cell activation (Fig 3J).
Taken together, these observations indicated that qM carcinoma cell-derived CD73, CSF1 and SPP1 can each function to inhibit the assembly of an immunosuppressive TME. Moreover, of these three, the CD73 pathway is far more effective at directly regulating the activity of CD8+ T-cells relative to CSF1 or SPP1. This provides the first indication that preventing the carcinoma cells from releasing any one of these carcinoma cell-derived factors partially sensitizes qM tumors to anti-tumor immune attack even in the absence of applied ICB. Moreover, this sensitization occurs without destabilizing the ongoing residence of these cells in the qM state.
Ability of quasi-mesenchymal carcinoma-cell derived immune factors to regulate macrophages
Tumor-associated macrophages (TAMs) play a pivotal role in influencing various immune cells in the TME, causing us to also focus on these cells in our further characterization of the TME. TAMs can exist in a pro-tumor M2 state or an anti-tumor M1 state (38,39). In addition, we and others have demonstrated that murine qM breast tumors attract large numbers of immunosuppressive, M2-like macrophages into the tumor core(20,40). Since some of the aforementioned secreted factors expressed by the qM cells are known to regulate chemoattraction and polarization of macrophages, we asked whether the representation and types of macrophages were altered in the various KO-tumors independent of any direct effects on T-cells. Of relevance here, in light of the known phenotypic plasticity of macrophages between these alternative states and the apparent involvement of a continuum of states arrayed along the M1-M2 polarization spectrum, we will refer to these TAMs as “M1-like” or “M2-like”(41,42).
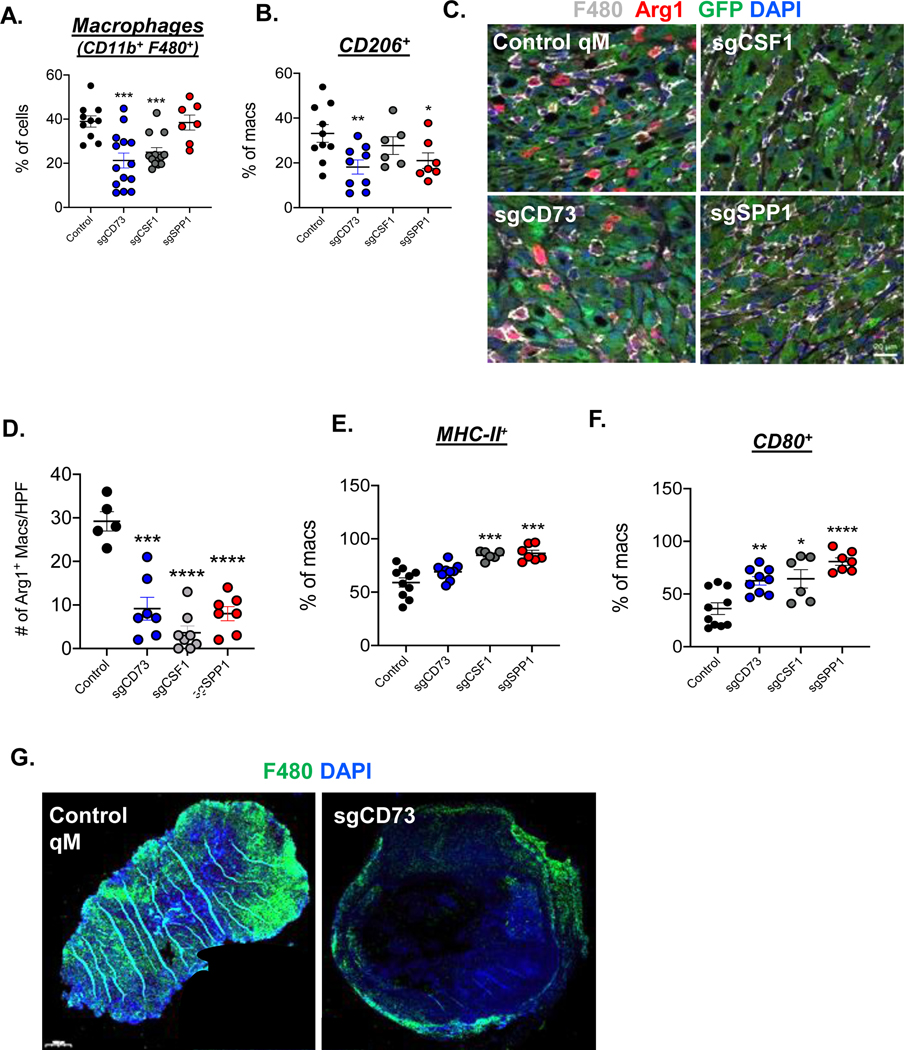
We found that qM tumors arising from cell lines knocked out for either CD73 or CSF1 had significantly fewer total macrophages relative to tumors arising from a control qM cell line (Fig 4A, Supp Fig 5D). Importantly, abrogation of either CD73, SPP1 or CSF1 led to decreased expression of the M2-associated macrophage markers CD206 and Arginase1 relative to macrophages present in control qM tumors which expressed these markers strongly (Fig 4B–D, Supp Fig 5E,F). Most strikingly, the decreased expression levels of the aforementioned M2-like markers were accompanied by significant increases in the expression of M1-like markers – MHC-II and CD80 in each of the three KO tumors (Fig 4E, F, Supp Fig 5G–I). Hence, abrogation of any one of these three secreted factors could alter the phenotypic state of macrophages recruited into qM tumors, favoring their entrance into an M1-like state that sharply contrasts with that of the M2-like, immunosuppressive macrophages present in control qM tumors.
Figure 4: Ability of qM carcinoma-cell derived factors to regulate tumor-associated macrophages.
(A) Bar graph representing % of CD11b+ F480+ total macrophages in the indicated tumors. Each data point represents a tumor from one mouse. N = 3–5 mice/group and represent pooled values from two-three independent experiments (B) % of CD11b+ F480+ total macrophages expressing CD206 (C) Immune-fluorescence analysis of tumor sections from control or KO tumors showing GFP (carcinoma cells in green), F480 (Macrophages in white), Arginase-1 (M2-like macrophages in red) and DAPI (nuclear stain in blue). Images represent fields of view from the tumor core. Scale bar = 20 μm. (D) Bar graph accompanying the immune-fluorescent image representing the numbers of Arginase positive macrophages present per high power field (HPF) at 63X magnification. (E) % of CD11b+ F480+ total macrophages expressing MHC-II (F) % of CD11b+ F480+ total macrophages expressing CD80. (G) Entire sections of the indicated tumors showing F480 (Macrophages in green) and DAPI (nuclear stain in blue). Scale bar = 1000 μm. Error bars represent ± SEM. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001
We have previously demonstrated that M2-like TAMs infiltrate into the core of qM tumors(20). Only qM tumors knocked out for CD73 excluded these TAMs to the tumor periphery (Fig 4G). Taken together, these observations demonstrate that CSF1 and SPP1 regulate the polarization state of TAMS, while CD73 regulates both, the polarization state, as well as the topological localization of macrophages within qM tumors.
Indirect regulation of immune-suppression by Macrophages
Several studies have demonstrated that TAMs, like those described above, can directly regulate the function of T-cells in the tumor microenvironment(26,43). Accordingly, we asked whether enrichment of functionally active T-cells in qM tumors was mediated directly by carcinoma cell- released factors or, alternatively, indirectly by the action of these factors on TAMs, enabling the latter in turn, to exert secondary effects on T-cells.
To address this question, we depleted macrophages in qM tumor-bearing mice via administration of an anti-CSF1 antibody, which results in neutralization of the macrophage chemoattractant CSF1. In a parallel set of experiments, we also repolarized macrophages by treating qM tumor-bearing mice either with a monocyte-specific pharmacological agent (IPI549) or by intratumoral delivery of a Toll-like receptor-9 (TLR9) ligand, CpG; both of these treatments have been reported to reprogram macrophages from an M2-like to an M1-like state(44,45) (Fig 5A). As anticipated, treatment of qM tumor-bearing mice with anti-CSF1 led to a significant decrease in the total number of macrophages. In addition, treatment with IPI549 led to a significant decrease in the numbers of Arginase+ M2-like macrophages while treatment with CpG led to significantly lower expression levels of both M2-markers - Arginase1 and CD206 relative to macrophages present in control qM tumors (Fig 5B–D, Supp Fig 5J). Importantly, the loss of M2 markers was associated with a gain of the M1 markers CD86 (in IPI549 and CpG treated mice) and MHC-II (in CpG treated mice) (Fig 5E, Supp Fig 5K), confirming that the reprogramming of macrophages in qM tumors from an M2-like to an M1-like state upon treatment with IPI549 or CpG had largely succeeded.
Figure 5: Regulation of immune-suppression in qM tumors by tumor-associated macrophages.

(A) Schema for macrophage depletion, repolarization or activation in qM tumor-bearing mice (B) Immune-fluorescent analysis of the indicated tumors for GFP (carcinoma cells in green), F480 (Macrophages in white), Arginase-1 (M2-like macrophages in red) and DAPI (nuclear stain in blue). Scale bar = 20 μm. (C) Bar graph accompanying the immune-fluorescent image representing the numbers of Arginase positive macrophages present per high power field (HPF) at 63X magnification. (D) Bar graph representing % of CD11b+ F480+ total macrophages expressing CD206. Each data point represents a tumor from one mouse. N = 3–5 mice/group and represent pooled values from two-three independent experiments. (E) % of CD11b+ F480+ total macrophages expressing CD86 or MHC-II. (F) % of CD8+ T-cells after gating on the CD45+ population (G) % of CD8+ T-cells expressing the indicated cytokines (H) % of CD4 cells co-expressing CD25 and FOXP3. (I) High (63X) and Low (20X) power images of Immune-fluorescence analysis of tumors receiving the indicated treatments stained for GFP (carcinoma cells in green) CD3 (T-cells in red) and DAPI (nuclear stain in blue). Scale bar = 20 μm for images at 63X magnification. Scale bar = 200 μm for images at 20X magnification. (J) Bar graph accompanying the immune-fluorescent image representing the numbers of CD3+ T-cells present per high power field (HPF) at 63X magnification. Error bars represent ± SEM. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001
As we found thereafter, both macrophage depletion (using Anti-Csf1) and reprogramming (using CpG), led to a significant increase in the numbers of CD8+ cytotoxic T-cells in qM tumors (Fig 5F, Supp Fig 5L). Furthermore, T-cells in tumors arising in IPI549 or CpG-treated mice stained positive for the expression of T-cell effector molecules (IFNγ, Perforin and Granzyme B) (Fig 5G, Supp Fig 5M). Importantly, this increase in the numbers and function of CD8+ T-cells was accompanied by a significant decrease in the numbers of immuosuppressive Tregs, especially in Anti-CSF1 and CpG treated cohorts (Fig 5H, Supp Fig 5N).
While depletion or reprogramming of macrophages, as described above, increased the numbers of T-cells, these T-cells were found to be largely excluded to the periphery of mesenchymal tumors (Fig 5I,J). These observations led us to conclude that the macrophages infiltrating qM tumors do in fact play a dominant role in the establishment of an immunosuppressive tumor microenvironment; however, moving these cells from an M2-like state to an M1-like state, does not, on its own, suffice to permit entrance of T-cells into the interior cores of qM tumors. Hence, while macrophages function as direct regulators of multiple aspects of immune suppression in qM tumors, the mobilization of T-cells relies on factors derived directly from qM carcinoma cells.
Regulation of responses of quasi-mesenchymal tumors to anti-CTLA4 checkpoint blockade immunotherapy
Having observed that abrogation of certain carcinoma cell-derived factors altered the immunosuppressive TME of qM tumors, we asked whether this reprogramming could sensitize previously refractory qM tumors to anti-CTLA4 ICB therapy (Fig 6A). To this end, we treated control qM and KO-tumor bearing mice with an anti-CTLA4 antibody. We found that control qM tumors as well as those knocked out for either Lgals3, Masp1, Tgfβ1 or Cxcl12 were resistant to anti-CTLA4 (Fig 6B and Supp Fig 6A). In contrast, tumors knocked out for Spp1 or Csf1 showed a delay in the kinetics of tumor growth and mounted heterogenous responses to anti-CTLA4, comprised of some responders and some non-responders (Fig 6C, D and Supp Fig 6B).
Figure 6: Abrogation of qM carcinoma cell-derived factors sensitizes qM tumors to anti-CTLA4.
(A) Schema for treatment of tumor-bearing mice with anti-CTLA4 (B-E) Tumor kinetics of the indicated control or treated tumors. N=5 mice/group and represent three independent experiments (F, G) Kinetics of tumor growth of sgCD73 tumor-bearing mice receiving the indicated antibodies. N=4 mice/group. Data represent pooled values from two independent experiments (H) % of sgCD73 tumor cells staining positive for dead cells after co-culture with different concentrations of CD8+ T-cells isolated from the spleens of responding mice. Data represent pooled values from three independent experiments (I) Splenocytes were isolated from naïve wild type or responding mice and co-cultured with sgCD73 cells for four days followed by flow cytometry analysis of the indicated markers after gating on CD8+ T-cells. The bar graph shows % of CD8+ T-cells expressing the indicated markers. Data represent pooled values from three independent experiments (J, K) Tumor kinetics of qM-tumor bearing mice receiving the indicated treatments. N=3–5 mice/group. Data represent pooled values from two independent experiments. Error bars represent ± SEM. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001
Most strikingly, however, qM tumors lacking CD73 were sensitized to anti-CTLA4 relative to control qM tumors which were resistant (Fig 6E). Out of the responding tumors, 65.7% showed complete tumor regression in response to anti-CTLA4 treatment, while 34.3% of responders had tumors that regressed but were not completely eliminated (Supp. Fig 6C). CD73-KO tumor-bearing mice that showed complete regression upon treatment with anti-CTLA4 remained tumor-free upon re-challenge with the same qM cell line knocked out for CD73, indicating the activation of immunological memory (Fig 6E). Furthermore, depletion of CD8+ T-cells or CD4+ T-cells from CD73-KO tumor-bearing mice abrogated their susceptibility to anti-CTLA4, demonstrating important functional roles of both, cytotoxic CD8+ T-cells and CD4+ T-cells in mediating tumor regression upon administration of ICB (Fig 6F, G and Supp Fig 6D).
To provide further evidence that the complete response of CD73-KO tumors to anti-CTLA4 therapy was mediated by the cytotoxic powers of antigen-specific T-cells, we isolated CD8+ T-cells from the spleens of responding mice and co-cultured them with CD73-KO tumors cells in cytotoxicity assays in vitro. CD8+ T-cells from responding mice were able to efficiently kill CD73-KO tumor cells in a dose-dependent manner (Fig 6H and Supp Fig 6E,F). Moreover, CD8+ T-cells from responding mice expressed significantly higher levels of markers associated with T-cell activation (CD25, CD69 and IFNγ) when exposed to CD73-KO cells in vitro, relative to those isolated from naïve mice, indicative of antigen-specific, effector function (Fig 6I and Supp Fig 6G,H). To summarize, these findings demonstrated that abrogation of CD73 strongly sensitizes qM tumors to anti-CTLA4 ICB in a T-cell dependent manner.
In more detail, CD73 is an ectoenzyme that dephosphorylates AMP, producing adenosine, a highly immunosuppressive molecule operating in the extracellular space (46). Once generated, adenosine binds to adenosine receptors (A1R, A2AR, A2BR and A3R), expressed on the surface of various immune cells and attenuates their cytolytic function (46,47). Given the striking sensitization of qM tumors lacking CD73 to anti-CTLA4, we asked whether perturbing the CD73-adenosine axis could potentiate the efficacy of ICB. To this end, we treated qM tumor-bearing mice with either anti-CD73 antibody or SCH-5861, a pharmacological antagonist of the adenosine receptors. Treatment of qM tumor-bearing mice with either anti-CD73 or SCH-5861 alone, led to an increase in the function of CD8+ T-cells as determined by increased expression of the effector molecules perforin and IFNγ and a decrease in the numbers of Tregs relative to control, untreated mice (Supp Fig 6I). Most importantly, combinations of either anti-CD73 or SCH-58261 with anti-CTLA4 led to reduced kinetics of tumor growth relative to control qM tumor-bearing mice (Fig 6J, K). Furthermore, combination treatment led to a dramatic increase in the numbers of T-cells infiltrating the tumor core relative to control qM tumors, in which these T-cells were excluded to the periphery (Supp Fig 6J). Hence, these observations demonstrate that perturbing the CD73/adenosine signaling axis could strongly potentiate the efficacy of anti-CTLA4 ICB.
Targeting CD73 enhances the efficacy of anti-CTLA4 ICB in mixed tumors
As mentioned above, we have previously demonstrated that in mixed tumors comprised of both E and qM carcinoma cells, a minority population (10%) of qM carcinoma cells can cross-protect the vast majority (90%) of their E neighbors from immune attack(20). These mixed tumors represent a model of human carcinomas, which are likely to harbor sub-populations of such qM cells. Accordingly, we asked whether abrogation of CD73 could sensitize previously refractory mixed tumors to anti-CTLA4 ICB. To this end, we mixed 10% of control qM cells or, alternatively, those qM cells lacking the expression of CD73 (qM-sgCD73) with 90% of E cells. As we have demonstrated previously, such mixes gave rise to heterogeneous tumors where E and qM cells were segregated topologically to distinct sectors within the same tumor (Fig 7A). Moreover, the 9:1 ratio of E:qM control or E:qM-sgCD73 cells was largely maintained during the outgrowth of the resulting primary tumors in vivo (Supp Fig 7A). We confirmed our previously reported observation that tumors arising from 9:1 mixtures of E:qM-control cells were resistant to anti-CTLA4 ICB (Fig 7B). Most striking, however, tumors arising from 9:1 mixtures where the minority subpopulations of qM cells lacked the expression of CD73 showed a significant reduction in tumor volume upon treatment with ICB (Fig 7B, Supp Fig 7B). This provided the first direct indication that the ability of the admixed, minority qM cells to cross-protect their E neighbors indeed depended in significant part on the ability of these qM cells to express CD73.
Figure 7: Targeting CD73 enhances the efficacy of anti-CTLA4 ICB in multiple models.
(A) Epi (pB2) PyMT cells were mixed with either qM (SnailHI) control or qM-sgCD73 PyMT cell lines at a 9:1 ratio and orthotopically implanted into C57BL6/J mice. Tumors were harvested when controls reached 2cm in size. N=3–5 mice/group. Data represent pooled values from two independent experiments. Immune-fluorescent analysis of the indicated tumors for GFP (qM carcinoma cells in green), E-cadherin (Epi cells in white), Vimentin or CD73 (red) and DAPI (nuclear stain in blue). Scale bar = 20 μm (B) Schema and kinetics of tumor growth for 9:1 Epi:qM or Epi:sgCD73 control tumors or those treated with anti-CTLA4. (C) Schema and kinetics of tumor growth of PyMT implant tumor-bearing mice receiving the indicated treatments. N=4 mice/group and data represent pooled values from two independent experiments. (D) Schema for intravenous injection of qM pB3-GFP/1.3g cells in mice receiving the indicated treatments. Representative images of lungs showing lung metastases (in green) from each treatment group. Bar graph represents numbers of lung metastases. N=3–5 mice/group and data represent pooled values from two independent experiments. (E) Venn diagram showing Snail-bound targets in pB3-GFP/1.3g cells that overlap with differentially expressed immunomodulatory genes obtained from the Nanostring Tumor Immunology panel (Supplementary Table #2). (F) Chip Seq signals (in cpm) for Snail binding at the Nt5e, Csf1 and Spp1 loci. Red arrows indicate transcription start sites. Error bars represent ± SEM. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001
To further demonstrate the importance of targeting CD73 in combination with anti-CTLA4 ICB in more-physiological settings of heterogeneous tumors, we isolated a late-stage tumor from the autochthonous MMTV-PyMT mouse and orthotopically transplanted 1cm sectors of this tumor into immunocompetent, syngeneic hosts. Since this approach did not involve isolation of E or qM carcinoma cells from the PyMT tumors, it represented an unbiased strategy for generating intra-tumoral phenotypic heterogeneity (Fig 7C-Schema). PyMT tumor-bearing mice were unresponsive to single-agent anti-CTLA4 or anti-CD73. In sharp contrast, PyMT tumor-bearing mice that received combination treatments (i.e., anti-CTLA4 plus anti-CD73 antibodies), showed a dramatic reduction in tumor size relative to control mice (Fig 7C, Supp Fig 7C). Taken together, these results underscore the importance of qM-derived CD73 in sensitizing not only qM tumors, but also heterogeneous tumors comprised of minority populations of qM carcinoma cells to anti-CTLA4 ICB.
Targeting CD73 in combination with anti-CTLA4 ICB reduces metastatic burden
The above-described results demonstrated the importance of CD73 in regulating the response of primary-qM tumors to anti-tumor immune attack. However, all most all breast cancer-related deaths can be attributed to the metastasis of carcinoma cells from the primary tumor to various distant organ sites(48). Given the importance of the EMT program in regulating the metastatic cascade(49), we proceeded to determine the functional role of CD73 in regulating metastatic colonization by qM cells and subsequent responses to ICB. To do so, we abrogated the expression of CD73 via CRISPR/Cas9 in a qM-PyMT cell line (pB3-GFP/1.3g), which is highly metastatic and forms abundant lung metastases upon intra-venous injection(24,50,51). As we found, qM cells lacking the expression of CD73 continued to reside in a qM state similar to that of the parental, unmodified cells and proliferated just as efficiently as the parental control qM cells in vitro (Supp Fig 7D–F).
To begin, we noted that while control qM cells formed lung metastases when introduced intra-venously, into syngeneic, immunocompetent hosts, 75% of the mice that had received sgCD73 cells showed a dramatic reduction in the total numbers of lung metastases, relative to those seen in the lungs of mice that had received control qM cells, this occurring even in the absence of ICB (Supp Fig 7G,H). Furthermore, treatment of sgCD73 recipient mice with anti-CTLA4 led to heterogeneous responses comprised of some responders (75 % of mice) and some non-responders (25% of mice) relative to control qM mice, which continued to harbor lung metastases even upon treatment with anti-CTLA4 ICB (Supp Figs 7G,H). Hence, we concluded that qM-derived CD73 is at least partially responsible for regulating metastatic colonization in response to ICB. Additionally, in contrast to primary tumors, no significant differences were observed in the numbers of various adaptive or innate immune cell types present in the lungs as a whole of mice injected intravenously with qM control or sgCD73 cells (Supp Fig 8A,B).
Several reports in the literature have underscored the functional importance of both – carcinoma cell-intrinsic as well as stromal cell associated CD73 expression in regulating metastatic colonization(52–54). For this reason, we treated qM recipient mice with an anti-CD73 antibody in order to target both carcinoma cells as well as stromal cells expressing CD73. This was also done in combination with an anti-CTLA4 antibody. Indeed, this combination led to a dramatic decrease in metastatic burden relative to control qM recipient mice or those treated with each antibody alone (Fig 7D). Taken together, these observations suggest that interrupting the CD73 signaling axis does indeed synergize with anti-CTLA4 ICB to reduce metastatic colonization. However, in contrast to primary tumors, where abrogation of carcinoma cell-intrinsic CD73, on its own, suffices to obtain complete sensitization to ICB, reduction in metastatic burden likely involves targeting CD73 expressed by both carcinoma cells as well as stromal/immune cells.
Our results indicate that activation of the EMT program leads to the expression of multiple immunosuppressive paracrine factors, three of which (CD73, CSF1 and SPP1) are critical for regulating the susceptibility of SnailHI qM tumors to anti-tumor immune attack and elimination by anti-CTLA4 ICB. To demonstrate a direct relationship between the EMT program and immunosuppression, we asked whether the Snail EMT-TF can directly regulate the expression of immune-modulatory genes by binding to their respective promoters. Thus, we overlapped Snail-Chip-Seq data from a metastatic, immunosuppressive qM cell line with the transcriptomic data generated using the Nanostring Tumor Immunology panel(24). We observed that Snail was found to bind to 89 immunomodulatory genes (Fig 7E and Supplementary Table #2). Most importantly, Snail binding was observed at the promoters of Nt5e (CD73), Csf1 and Spp1, suggesting a direct regulation of these targets (Fig 7F).
Discussion
A number of efforts over the past few years have been focused on identifying markers that can be used to accurately predict a patient’s response to ICB(55). While some of these markers have proven useful, their utility has been limited to a small subset of malignancies(1,3). We demonstrate that the pathophysiological state of the carcinoma cells themselves can be used as an important surrogate marker to predict future responses to at least on form of ICB. Understanding precisely how qM carcinoma cells exert immunosuppressive effects is particularly important, as most human breast carcinomas harbor minority populations of qM carcinoma cells that can cross-protect their E neighbors from anti-tumor immune attack.
Our findings suggest that these refractory responses of qM tumors to ICB are strongly dependent on carcinoma cell-derived immunomodulatory factors, as activation of the EMT program in vitro can directly alter the expression of several immunosuppressive factors in the absence of any collaborating stromal cell components. Indeed, the Snail-EMT-TF was found to bind to the promoters of multiple immunosuppressive genes. This implies that one way in which the EMT program contributes to immunosuppression is by direct regulation of immunosuppressive genes by EMT-TFs, in this case, the Snail-EMT-TF. This effect of carcinoma cells on the adjacent stroma contrasts with an induction of an EMT program in these cells, which is surely dependent on heterotypic interactions between them and cells of the adjacent TME.
A central goal of the present analyses was to focus on a set of genes expressed by qM carcinoma cells whose abrogation did not affect their continued residence in the qM phenotypic state but, at the same time, altered their susceptibility to anti-tumor immune attack. Indeed, loss of either CD73, CSF1, SPP1, Galectin-3, MASP1 or CXCL12 - did not dramatically perturb the expression of markers associated with the qM state orchestrated by the EMT. The only exception to this rule was created by qM cells that were deprived of TGFβ1 expression, which is known from other work to act in a paracrine manner to induce the formation of Tregs and suppress the activation of CD8 lymphocytes and NK cells(56). Importantly, TGF-β1 can also activate the expression of various EMT-TFs via multiple mechanisms and is itself a strong inducer of the EMT program(35,36). Indeed, the loss of this potent cytokine led to a discernible shift of carcinoma cell state into the epithelial direction undoubtedly due to the autocrine functions of this powerful cytokine(57). Accordingly, any shifts in the immune microenvironment observed following knockout of TGF-β1 in the genome of carcinoma cells might reflect the loss by these cells of a qM phenotype, the direct effects of this cytokine on cells of the immune microenvironment, or both.
While abrogation of five carcinoma cell-intrinsic immunosuppressive factors led to increased numbers and function of CD8+ T-cells, only the loss of CD73, CSF1 and SPP1, could additionally mobilize functionally active T-cells and cDC1s into the tumor core. The unique ability of these factors to regulate the topological localization of T-cells in addition to enhancing their function requires further investigation. Furthermore, in accordance with previous reports(47), we confirmed that the CD73 pathway is far more effective in directly regulating T-cell activity relative to CSF1 or SPP1. It remains to be established whether abrogation of carcinoma cell-intrinsic Csf1 or Spp1 has secondary effects on the expression of other immunosuppressive paracrine factors that could, in turn, affect T-cell activation. It also remains to be determined whether these pathways act independently or in collaboration with one another to modulate T-cell activity.
Of additional interest, while loss of CD73 entirely sensitized qM tumors to anti-CTLA4, loss of either CSF1 or SPP1 led to only partial sensitization. These observations suggest that the presence of tumor-infiltrating lymphocytes and cDC1 cells may not, on its own, be sufficient to predict robust responses to ICB. A growing body of recent studies has implicated the presence of a CD8+ T-cell subsets with enhanced stem-like, progenitor function in mounting efficient anti-tumor responses post ICB (58,59). Thus, whether the loss of CD73 from qM tumors also elicits the formation of stem-like, progenitor and effector-memory CD8+ T-cells remains to be determined.
We observed that both cytotoxic CD8+ T-cells as well as CD4+ helper T-cells were required for sensitizing tumors lacking CD73 to anti-CTLA4 ICB. Indeed, a very recent study demonstrated that both these populations of cells are required for mounting immunotherapy- induced anti-tumor responses (60). The precise dynamics of potential interactions between these two subsets of T-cells in the context of mesenchymal tumors lacking CD73 and their roles in sensitizing such tumors to ICB therapies remain to be established.
While abrogation of the aforementioned factors could sensitize qM tumors to anti-CTLA4, they did not affect their susceptibility to anti-PD1 (Supp Fig 8C). Given the different mechanisms of action of these two ICB agents, it is plausible that cell-intrinsic factors that regulate the susceptibility of qM tumors to anti-CTLA4 may be distinct from those that regulate responses to other forms of ICB(61).
Several studies have demonstrated the utility of perturbing the CD73/adenosine signaling axis to potentiate the efficacy of adoptive T-cell transfer therapy and ICB(46,47,62). More specifically, inhibition of CD73 dramatically abrogates the establishment of lung metastasis in the 4T1 mouse model of breast cancer(52). Indeed, we observed that targeting CD73 in combination with anti-CTLA4 significantly reduced the ability of metastatic qM carcinoma cells to colonize the lungs. These metastatic qM carcinoma cells were GFP-labelled and injected into C57BL/6 recipients, in which GFP is minimally immunogenic(50,51). Synergistic effects of anti-CD73 and anti-CTLA4 antibodies could also be extrapolated to more-physiological settings of mixed, heterogeneous tumors. Hence, targeting CD73 may provide a highly useful therapeutic strategy for potentiating the actions of ICB.
Taken together, our work indicates that carcinoma cell-intrinsic factors specifically associated with residence in the qM state can directly influence their response to ICB. As a result, this work brings to the forefront the possibility of using the epithelial-mesenchymal phenotypic states of carcinoma cells as an important surrogate marker that can be used to predict responses to ICB. In addition, our work provides clear indication of the clinical utility of inhibiting adenosine production in the TME of qM tumors and encouraging carcinoma cells to transit from more mesenchymal to more epithelial phenotypic states in order to render them more sensitive to subsequently applied checkpoint immunotherapies. Such induced changes may lead to significantly improved treatments for breast carcinomas and, we anticipate, for other types of carcinomas as well.
Methods
Mice.
C57BL/6 mice were obtained from the Jackson laboratory. A colony of NSG mice was generated and maintained in-house. All animals used for experiments were 6–8-week-old female mice. Animals were maintained in compliance with the guidelines and protocols approved by the Animal Care and Use Committees at the Massachusetts Institute of Technology
Cell lines and cell culture.
All PyMT cell lines - the SnailHI PyMT qM cell line, the pB3-GFP/1.3g qM cell line, the SnailLO EpcamHI PyMT epithelial cell line and the pB2 PyMT epithelial cell line were established and maintained as previously described(20,24). Control cells as well as those knocked out for target genes were cultured in DMEM/F12 medium containing 5% adult bovine serum supplemented with 1X penicillin-streptomycin and 1X non-essential amino acids for the duration of this study. MCF7 cells were established and maintained as previously described (32) in DMEM supplemented with 10% fetal bovine serum and 1X penicillin-streptomycin. HMLER cells were established and maintained in MEGM media as previously described (33,63). HCC1806 cells were obtained from ATCC and maintained in DMEM supplemented with 10% fetal bovine serum and 1X penicillin-streptomycin. Cells were stably transfected with GFP, RAS, the indicated EMT-TFs (HCC1806) or doxycycline-inducible EMT-TFs (MCF7Ras simultaneously expressing dox-inducible Slug and Sox9 and HMLERas expressing dox-inducible Zeb1) as previously described (32,64). All cell lines containing doxycycline-inducible expression vectors were treated with 1 μg/ml doxycycline hyclate every two days for a total period of 6 days (Sigma Aldrich). All cell lines were tested for mycoplasma using the MycoAlert Mycoplasma Detection Kit (Lonza). All cells tested negative for mycoplasma (2019) and have not been authenticated since first acquisition. All cell lines were obtained between 2009–2018 and used within 1–3 years of first acquisition.
CRIPSR/Cas9 mediated knockdown of target genes.
Target genes were knocked out from the control qM SnailHI or the pB3-GFP/1.3g cell lines via transient transfection using CRIPSR/Cas9 plasmids and protocols obtained from Santacruz Biotechnology (CD73: SC-423919; CSF1: SC-419838-KO-2; OPN: SC-423124; Galectin3: SC-421416-KO2; MASP1: SC-4211564; CXCL12/SDF1: SC-422854; TGFB1: SC-423364, Plasmid transfection medium (SC-108062), Ultracruz transfection reagent (SC-395739). Cell lines were seeded at 0.5 × 106 cells/well in each well of a 6-well plate for 12 hrs and transfected with the plasmid of interest according to the manufacturer’s protocol. After 48hrs of incubation, Cas9-GFP expressing cells were sorted as single cells into each well of a 96-well flat bottom plate. These single cell clones were then expanded and screened for the presence or absence of the protein of interest via western blotting (Anti-CSF1, Anti-Galectin-3 – Cell Signaling Technologies), ELISA (TGFB1-Life Technologies, OPN-Ray Biotech, CXCL12-Ray Biotech, MASP1-My Biosource) or flow cytometry (CD73-Thermo Fischer Scientific).
In vivo models, depletion of immune cells and treatment with immune checkpoint blockade antibodies.
For orthotopic tumor transplantations, 1 × 106 cells (SnailHI qM control or those knocked out for target genes) were resuspended in 20% Matrigel and implanted into the mammary fat pad of C57BL/6J mice or NSG mice. For mixed tumor experiments, PyMT E cells (pB2 for Fig 7 and SnailLOEpcamHI for Supp Fig 7) were mixed with SnailHI qM-control or qM-sgCD73 cells at a 9:1 ratio. A total of 1 × 106 cells were resuspended in 20% Matrigel and implanted into the mammary fat pad of C57BL/6J mice. Animals were sacrificed once control tumors reached 2cm in size. For metastatic colonization assays, 0.5 × 106 pB3-GFP (1.3g) control or sgCD73 cells were injected retro-orbitally into C57BL/6J mice. Lungs were harvested at Day 10. Tumors were digested and processed as previously described(20). For primary tumors, tumor volume was calculated using the modified ellipsoid formula tumor volume (mm3) = (L X W X W)/2, where L represents the largest tumor diameter and W represents the perpendicular measurement. For immune checkpoint blockade therapy experiments, primary tumor-bearing mice were treated with 200ug of anti-CTLA4 (9H10) every two days for a total of 6 days, with the first dose given two days after orthotopic implantation. For sgCD73, sgCSF1, sgSPP1 and mixed tumor-bearing mice, this treatment regimen was followed by 100ug of anti-CTLA4 once a week for two more weeks. sgCD73 tumor-bearing mice that mounted complete responses to anti-CTLA4 were re-challenged by orthotopic implantation of 1 × 106 sgCD73 cells that were resuspended in 20% Matrigel. For metastatic colonization assays, tumor-bearing mice were treated with anti-CTLA4 and anti-CD73 on Days 3 and 6 post injection. T-cells were depleted by administration of 200ug of anti-CD8 (53–6.7) or anti-CD4 (GK1.5) starting 3 days prior to tumor implantation followed by once every week for a total of four weeks. Macrophages were depleted by administration of 300ug of anti-CSF1 (5A1) starting three days prior to tumor implantation followed by two times a week for a total of four weeks. For macrophage repolarization, 200ug IPI549 (MedChemExpress) was administered daily by oral gavage starting one day post tumor implantation for a total of 20 days. 50ug of CpG (ODN M362-TLR9 ligand; Invivogen) was administered intra-tumor, two times a week, starting seven days post tumor implantation for a total of three weeks. 100ug of anti-CD73 (TY/23) was administered three days prior to tumor implantation followed by once a week for a total of four weeks. SCH-58261(Sigma-Aldrich) was administered daily, intra-peritoneally, at 1mg/kg starting one day after tumor implantation for a total of 20 days. All antibodies were purchased from Bioxcell unless otherwise noted.
T-cell isolation, in vitro activation and Cytotoxicity Assays.
Splenic CD8+ T-cells were isolated from the spleens of naïve C57BL6/J mice. Briefly, spleens were mashed in PBS using a 10ml syringe and filtered through a 70-micron filter. RBCs were lysed using ACK lysis buffer. CD8+ T-cells were isolated by magnetic separation using anti-mouse CD8 magnetic particles DM (BD Biosciences) and the BD Imag Cell Separation magnet (BD Biosciences) according to the manufacturer’s protocol. CD8+ T-cells were resuspended in T-cell media (50% DME, 50% RPMI, 25% FBS, 1X Pen/Strep, 1X L-glutamine). CD8+ T-cells were activated in vitro on plates pre-coated with anti-Hamster IgG (Sigma) o/n at 4C, followed by 1ug/ml of anti-CD3e (145–2C11, BD Biosceinces) and 1ug/ml of anti-CD28 (37.51, BD Biosciences) in each well of a 12-well plate. Conditioned media was obtained from E and SnailHI qM cell lines (control and KO) 48hrs after seeding cells at a density of 0.5×106 cells/ml in each well of a 12-well plate. Supernatant was collected after centrifugation to remove floating dead cells. 1ml of conditioned media from each cell line was then added to 1ml of T-cells that were resuspended in T-cell media. For re-constitution assays, conditioned media obtained from KO cell lines was reconstituted with 10ug/ml of NECA (Tocris), recombinant CSF1 (Biolegend #576406) or recombinanat SPP1 (Biolegend #763606). T-cells were harvested after 48hrs of incubation and analyzed for their surface expression of CD25 and CD69 (both from Thermo Fischer Scientific) by flow cytometry. For Cytotoxicity assays, sgCD73 cells were treated with 50uM of Mitomycin C (Sigma Aldrich) for 40 mins at 37C, washed two times in PBS, and plated at a density of 0.5×106 cells/ml in each well of a 12-well plate. Splenocytes were obtained from responding mice (sgCD73 tumor-bearing mice treated with anti-CTLA4) and added to the tumor cells at a density of 3×106 cells/ml. After 4–6 days of co-culture, splenocytes were harvested and CD8+ T-cells were isolated as described above. CD8+ T-cells were then added at the indicated ratios to 0.5×105 cells/ml of sgCD73 tumor cells in each well of a 12-well plate for 4hrs. The % of dead tumor cells was determined by flow cytometry using the LIVE/DEAD fixable near IR-dead cell stain kit 633nm (Life Technologies). T-cell activity in the co-culture system was determined by co-culturing splenocytes (3×106 cells/ml) obtained from responding mice or naïve C57BL/6 mice with sgCD73 tumor cells (0.5×105 cells/ml) in each well of a 12-well plate for 4 days. 1X Brefeldin A solution (Life Technologies) was added to each well for the last 4 hrs before harvesting. Cells were stained for the indicated T-cell activation markers and analyzed by flow cytometry.
Flow cytometry.
Dissociated tumors were resuspended in wash buffer (PBS containing 0.1% BSA) and stained for surface markers by incubating 1 × 106 cells with the respective antibodies for 30 mins on ice. All antibodies and staining kits were purchased from Thermo Fisher Scientific unless otherwise noted. CD45 PECy7 (30F-11), CD45 FITC (30F-11), CD4 PE (RM4–5), CD8a APC (53–6.7), CD25 PercpCy5.5 (pc61.5), CD69 E450 (H1.2F3), PD-1 FITC (J43), CTLA4 PE (UC10–4B9), CD11B PercpCy5.5 (M1/70), F480 PECY7 (BM8; BioLegend), MHCII PE (M5/114.15.2), CD80 E450 (16–10A1), CD86 APC (GL1), LY6C E450 (HK1.4), LY6G FITC (1A8), CD206 APC (C068C2; BioLegend), CD3 PercpCy5.5 (17A2), CD73 APC (AD2). The following antibodies were used for the cDC1 panel for primary tumors: CD19 eF450 (1D3; eBioscience), CD3e eF450 (17A2), CD45 BUV395 (30-F11; BD Horizon), MHCII AF700 (M5/114.15.2; eBioscience), Ly6C PE (HK1.4; BioLegend), F480 PE-Cy7 (BM8; BioLegend), CD11c APC-Cy7 (N418; BioLegend), CD11b PE-CF594 (M1/70; BD Horizon), CD8 BUV737 (53–6.7; BD Horizon), CD103 APC (2E7; eBioscience), SIRPa AF488 (P84; BioLegend), SiglecH BV605 (440c; BD Optibuild). The following antibodies were used for the cDC1 panel for the lungs: Live/Dead eFluor780, CD19 APC-Cy7 (6D5 BioLegend), CD3e APC-Cy7 (17A2 BioLegend), CD45 BUV395 (30-F11 BD Horizon), MHCII AF700 (M5/114.15.2), Ly6C PE (HK1.4 BioLegend), F4/80 PE-Cy7 (BM8 BioLegend), CD11c BV421 (N418 BioLegend), CD11b BUV737 (M1/70 BD Biosciences), CD8a BV711 (53–6.7 BioLegend), CD103 APC (2E7), SiglecH BV605 (440c BD Optibuild). For intra-cellular cytokine staining, CD8+ T-cells were sorted from digested tumor samples and co-cultured with the respective cell lines (1 × 106 cells/ ml) for 5 hrs in the presence of Monensin Golgi Stop (BD Biosciences). Intracellular cytokine staining was performed using the Intracellular Fixation and Permeabilization Buffer Set and the following antibodies- IFNγ PECY7 (XMG1.2), Granzyme B FITC (NGZB), Perforin PE (eBIOMAK-D) as per the manufacturer’s protocol. Intra-cellular staining for FOXP3 was performed using the FOXP3/Transcription Factor Staining Buffer Set using FOXP3 Alexa488 (FJK-16s) as per the manufacturer’s protocol. Flow cytometry data was acquired on a BD LSRFortessa and the data were analyzed using the FlowJo™ (V10) software.
Immunofluorescence Staining.
Tumors were fixed in 10% neutral buffered formalin for 12hrs and transferred to 70% ethanol, followed by embedding and sectioning. Tumor sections and cells were processed for immunofluorescence staining as described before (20). Primary antibodies used were E-cadherin (BD Biosciences), Vimentin (Cell Signaling Technology), Zeb1 (Santa Cruz Biotechnology), GFP (Rockland Immunochemicals), CD3 (AbCam), F480 (ThermoFisher), Arginase1 (Santa Cruz Biotechnology). Images were acquired using a Zeiss inverted microscope and analyzed using the ZEN imaging software.
Western Blot.
Whole cell lysates were made in Aqueous Lysis Buffer (50mM Tris pH 7.5, 150mM NaCl, 10mM EDTA pH8.0, 0.2% Sodium azide, 50mM NaF, 0.5%NP40) and resolved on a gradient gel as previously described(20). Primary antibodies used were E-cadherin, Vimentin, Zeb1, Snail, Slug, Sox9, GAPDH, CSF1, Galectin-3, MASP1/3, Fibronectin (BD Biosciences)
Cytokine Arrays.
E and qM PyMT cell lines were seeded at a density of 0.5 × 106 cells/ml in a 10cm plate and conditioned media was collected after 48hrs. This was then centrifuged to remove floating dead cells. Cytokine and chemokine expression was determined by using the mouse cytokine array kit, pa (Thermo Fisher Scientific) as per the manufacturer’s instructions.
Transcriptomic analysis.
RNA was extracted from the indicated cell lines cultured in vitro or from sorted GFP+ tumor cells using the RNeasy Mini Kit (Qiagen) and transcriptomic analysis was performed using the nCounter PanCancer Immune Profiling Panel (Nanostring Technologies). Normalization of Nanostring nCounter gene expression values and differential expression were assayed by DESeq2), which uses the Wald test [PMID: 25516281]. Differentially expressed genes were defined as a twofold change (up or down). CCLE bam files were obtained from the Genomic Data Commons (GDC) data portal, and normalized counts were obtained by HT-Seq Count [PMID: 25260700] and DESeq [PMID: 20979621]. Enrichment analysis were performed using the following software, ClueGO [PMID: 19237447], pre-ranked Gene Set Enrichment Analysis (GSEA) [PMID: 16199517], and DAVID [PMID: 12734009] where the uploaded background gene list were all the genes on the Nanostring panel. Hierarchical clustering, based on uncentered correlation and average linkage, was done using Cluster 3.0 [PMID: 14871861] and visualized in Java TreeView [PMID: 15180930]. ChIPSeq data peak calls were called by MACS2 [PMID: 18798982] and visualized in IGV as previously described(24). Data have been deposited in the GEO database under the accession code GSE161748.
Statistical Analysis.
All data are represented as mean ± SEM. Statistical Analysis was performed using the GraphPad Prism 8 software. P values were calculated using an unpaired two-tailed student’s t test. *, p<0.05, **, p<0.01, ***, p<0.001, ****, p<0.0001, ns - not significant.
Schematic Illustrations.
All illustrations and schematic diagrams were created using www.BioRender.com
Supplementary Material
Statement of Significance.
Minority populations of qM carcinoma cells, which likely reside in human breast carcinomas can cross-protect their E neighbors from immune attack. Understanding the mechanisms by which qM carcinoma cells resist anti-tumor immune attack can help identify signaling channels that can be interrupted to potentiate the efficacy of checkpoint blockade immunotherapies.
Acknowledgements
We thank Richard A. Goldsby, Barbara A. Osborne and George W. Bell for critical reading of the manuscript. We thank Whitney Henry for HCC1806 cell lines and all members of the Weinberg Lab for helpful discussion. We thank the Flow Cytometry Core facility at the Whitehead Institute for assistance with cell sorting and flow cytometry analysis; the Keck Imaging facility at the Whitehead Institute for assistance with microscopy and the Histology Facility at the Koch Institute/MIT for tissue sectioning. R.A.Weinberg received grants from the Breast Cancer Research Foundation, the Ludwig Center for Cancer Research, and NIH grants R35CA220487 and P01CA080111.
Footnotes
Conflict of Interest: Dr. Weinberg has a consulting agreement with Verastem Inc together with holding shares of this company. Dr Spranger reports personal fees from Arcus BioSciences, personal fees from Ribon, personal fees from TAKEDA, personal fees from Merck, personal fees from Dragonfly, and personal fees from Tango outside the submitted work. All other authors declare no conflict of interest.
References
- 1.Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell 2016;165(1):35–44 doi 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rashidian M, Ingram JR, Dougan M, Dongre A, Whang KA, LeGall C, et al. Predicting the response to CTLA-4 blockade by longitudinal noninvasive monitoring of CD8 T cells. J Exp Med 2017;214(8):2243–55 doi 10.1084/jem.20161950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature 2015;523(7559):231–5 doi 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 4.Hellmann MD, Callahan MK, Awad MM, Calvo E, Ascierto PA, Atmaca A, et al. Tumor Mutational Burden and Efficacy of Nivolumab Monotherapy and in Combination with Ipilimumab in Small-Cell Lung Cancer. Cancer Cell 2018;33(5):853–61 e4 doi 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sade-Feldman M, Jiao YJ, Chen JH, Rooney MS, Barzily-Rokni M, Eliane JP, et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat Commun 2017;8(1):1136 doi 10.1038/s41467-017-01062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: Mechanisms, response biomarkers, and combinations. Sci Transl Med 2016;8(328):328rv4 doi 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribas A. Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov 2015;5(9):915–9 doi 10.1158/2159-8290.CD-15-0563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-gamma Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016;167(2):397–404 e9 doi 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshihama S, Roszik J, Downs I, Meissner TB, Vijayan S, Chapuy B, et al. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proc Natl Acad Sci U S A 2016;113(21):5999–6004 doi 10.1073/pnas.1602069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375(9):819–29 doi 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalbasi A, Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat Rev Immunol 2020;20(1):25–39 doi 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019;20(2):69–84 doi 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 13.Nieto MA. Epithelial-Mesenchymal Transitions in development and disease: old views and new perspectives. Int J Dev Biol 2009;53(8–10):1541–7 doi 10.1387/ijdb.072410mn. [DOI] [PubMed] [Google Scholar]
- 14.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008;133(4):704–15 doi 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 2008;3(8):e2888 doi 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100(7):3983–8 doi 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene 2010;29(34):4741–51 doi 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Celia-Terrassa T, Meca-Cortes O, Mateo F, Martinez de Paz A, Rubio N, Arnal-Estape A, et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest 2012;122(5):1849–68 doi 10.1172/JCI59218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, et al. EMT Subtype Influences Epithelial Plasticity and Mode of Cell Migration. Dev Cell 2018;45(6):681–95 e4 doi 10.1016/j.devcel.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dongre A, Rashidian M, Reinhardt F, Bagnato A, Keckesova Z, Ploegh HL, et al. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res 2017;77(15):3982–9 doi 10.1158/0008-5472.CAN-16-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer Metastasis Is Accelerated through Immunosuppression during Snail-induced EMT of Cancer Cells. Cancer Cell 2009;15(3):195–206 doi 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Lou Y, Diao L, Cuentas ER, Denning WL, Chen L, Fan YH, et al. Epithelial-Mesenchymal Transition Is Associated with a Distinct Tumor Microenvironment Including Elevation of Inflammatory Signals and Multiple Immune Checkpoints in Lung Adenocarcinoma. Clin Cancer Res 2016;22(14):3630–42 doi 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Terry S, Savagner P, Ortiz-Cuaran S, Mahjoubi L, Saintigny P, Thiery JP, et al. New insights into the role of EMT in tumor immune escape. Mol Oncol 2017;11(7):824–46 doi 10.1002/1878-0261.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye X, Tam WL, Shibue T, Kaygusuz Y, Reinhardt F, Eaton EN, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015;525(7568):256-+ doi 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121(7):2750–67 doi 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141(1):39–51 doi 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol 2019;16(6):356–71 doi 10.1038/s41571-019-0175-7. [DOI] [PubMed] [Google Scholar]
- 28.Cesano A, Marincola FM, Thurin M. Status of Immune Oncology: Challenges and Opportunities. Methods Mol Biol 2020;2055:3–21 doi 10.1007/978-1-4939-9773-2_1. [DOI] [PubMed] [Google Scholar]
- 29.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009;25(8):1091–3 doi 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 2003;4(5):P3. [PubMed] [Google Scholar]
- 31.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell 2012;148(5):1015–28 doi 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frose J, Chen MB, Hebron KE, Reinhardt F, Hajal C, Zijlstra A, et al. Epithelial-Mesenchymal Transition Induces Podocalyxin to Promote Extravasation via Ezrin Signaling. Cell Rep 2018;24(4):962–72 doi 10.1016/j.celrep.2018.06.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroger C, Afeyan A, Mraz J, Eaton EN, Reinhardt F, Khodor YL, et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc Natl Acad Sci U S A 2019;116(15):7353–62 doi 10.1073/pnas.1812876116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res 2009;19(2):156–72 doi 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol 2014;15(3):178–96 doi 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keenan TE, Burke KP, Van Allen EM. Genomic correlates of response to immune checkpoint blockade. Nat Med 2019;25(3):389–402 doi 10.1038/s41591-019-0382-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 2004;25(12):677–86 doi 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 39.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 2014;41(1):14–20 doi 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim IS, Gao Y, Welte T, Wang H, Liu J, Janghorban M, et al. Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat Cell Biol 2019;21(9):1113–26 doi 10.1038/s41556-019-0373-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biswas SK, Sica A, Lewis CE. Plasticity of macrophage function during tumor progression: regulation by distinct molecular mechanisms. J Immunol 2008;180(4):2011–7 doi 10.4049/jimmunol.180.4.2011. [DOI] [PubMed] [Google Scholar]
- 42.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nat Immunol 2010;11(10):889–96 doi 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 43.DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19(6):369–82 doi 10.1038/s41577-019-0127-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 2016;539(7629):443–7 doi 10.1038/nature20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan R, Li S, Geng H, Wang X, Guan Q, Li X, et al. Reversing the polarization of tumor-associated macrophages inhibits tumor metastasis. Int Immunopharmacol 2017;49:30–7 doi 10.1016/j.intimp.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 46.Vigano S, Alatzoglou D, Irving M, Menetrier-Caux C, Caux C, Romero P, et al. Targeting Adenosine in Cancer Immunotherapy to Enhance T-Cell Function. Front Immunol 2019;10:925 doi 10.3389/fimmu.2019.00925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kjaergaard J, Hatfield S, Jones G, Ohta A, Sitkovsky M. A2A Adenosine Receptor Gene Deletion or Synthetic A2A Antagonist Liberate Tumor-Reactive CD8(+) T Cells from Tumor-Induced Immunosuppression. J Immunol 2018;201(2):782–91 doi 10.4049/jimmunol.1700850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steeg PS. Targeting metastasis. Nat Rev Cancer 2016;16(4):201–18 doi 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell 2017;168(4):670–91 doi 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skelton D, Satake N, Kohn DB. The enhanced green fluorescent protein (eGFP) is minimally immunogenic in C57BL/6 mice. Gene Ther 2001;8(23):1813–4 doi 10.1038/sj.gt.3301586. [DOI] [PubMed] [Google Scholar]
- 51.Ansari AM, Ahmed AK, Matsangos AE, Lay F, Born LJ, Marti G, et al. Cellular GFP Toxicity and Immunogenicity: Potential Confounders in in Vivo Cell Tracking Experiments. Stem Cell Rev Rep 2016;12(5):553–9 doi 10.1007/s12015-016-9670-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stagg J, Divisekera U, McLaughlin N, Sharkey J, Pommey S, Denoyer D, et al. Anti-CD73 antibody therapy inhibits breast tumor growth and metastasis. Proc Natl Acad Sci U S A 2010;107(4):1547–52 doi 10.1073/pnas.0908801107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Allard B, Turcotte M, Stagg J. Targeting CD73 and downstream adenosine receptor signaling in triple-negative breast cancer. Expert Opin Ther Targets 2014;18(8):863–81 doi 10.1517/14728222.2014.915315. [DOI] [PubMed] [Google Scholar]
- 54.Goswami S, Walle T, Cornish AE, Basu S, Anandhan S, Fernandez I, et al. Immune profiling of human tumors identifies CD73 as a combinatorial target in glioblastoma. Nat Med 2020;26(1):39–46 doi 10.1038/s41591-019-0694-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12(4):252–64 doi 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Viel S, Marcais A, Guimaraes FS, Loftus R, Rabilloud J, Grau M, et al. TGF-beta inhibits the activation and functions of NK cells by repressing the mTOR pathway. Sci Signal 2016;9(415):ra19 doi 10.1126/scisignal.aad1884. [DOI] [PubMed] [Google Scholar]
- 57.Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 2011;145(6):926–40 doi 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miller BC, Sen DR, Al Abosy R, Bi K, Virkud YV, LaFleur MW, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 2019;20(3):326–36 doi 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siddiqui I, Schaeuble K, Chennupati V, Fuertes Marraco SA, Calderon-Copete S, Pais Ferreira D, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 2019;50(1):195–211 e10 doi 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 60.Alspach E, Lussier DM, Miceli AP, Kizhvatov I, DuPage M, Luoma AM, et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019;574(7780):696–701 doi 10.1038/s41586-019-1671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, et al. Distinct Cellular Mechanisms Underlie Anti-CTLA-4 and Anti-PD-1 Checkpoint Blockade. Cell 2017;170(6):1120–33 e17 doi 10.1016/j.cell.2017.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang B. CD73: a novel target for cancer immunotherapy. Cancer Res 2010;70(16):6407–11 doi 10.1158/0008-5472.CAN-10-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev 2001;15(1):50–65 doi 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Cock JM, Shibue T, Dongre A, Keckesova Z, Reinhardt F, Weinberg RA. Inflammation Triggers Zeb1-Dependent Escape from Tumor Latency. Cancer Res 2016;76(23):6778–84 doi 10.1158/0008-5472.CAN-16-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.