Abstract
There have been increased reports of the isolation of unusual genotypic groups of Candida albicans (groups C and D) based on a well-defined genotypic method; this method uses cellular DNA digested with the EcoRI enzyme and the restriction fragment length polymorphisms (RFLPs) generated by agarose gel electrophoresis. The aim of the present study was to use additional molecular tools to characterize these unusual strains and to compare them with authentic strains of C. dubliniensis, a recently delineated species, and type I C. stellatoidea. The RFLPs of PCR products generated from the intergenic transcribed spacer (ITS) region did not differentiate among C. albicans genotypes A, B, and C and type I C. stellatoidea. However, this method did differentiate the C. albicans genotype D strains, which were identical to C. dubliniensis. The RFLPs generated by HaeIII digestion of the PCR products of the V3 region of the 25S rRNA gene (rDNA) could differentiate the same groups as RFLP analysis of the PCR amplicon of the ITS region. C. albicans genotype B isolates have been shown to have a transposable intron in the 25S rDNA, whereas genotype A isolates do not; C. dubliniensis strains also have an intron that is larger than that in genotype B C. albicans strains but that is in the same location. PCR designed to span this region resulted in a single product for C. albicans genotype A (450 bp), B (840 bp), type 1 C. stellatoidea (840 bp), and C. dubliniensis (1,080 bp), whereas the C. albicans genotype C isolates had two major products (450 and 840 bp). All C. albicans genotype D isolates gave a PCR product identical to that given by C. dubliniensis. These results indicate that those strains previously designated C. albicans genotype D are in fact C. dubliniensis, that no differences were found between type 1 C. stellatoidea and C. albicans genotype B strains, and that the C. albicans genotype C strains appear to have the transposable intron incompletely inserted throughout the ribosomal repeats in their genomes. The results of the antifungal susceptibility testing of 105 of these strains showed that, for fluconazole, strains of C. dubliniensis were significantly more susceptible than strains of each of the C. albicans genotypes (genotypes A, B, and C). The flucytosine susceptibility results indicated that strains of C. albicans genotype A were significantly less susceptible than either C. albicans genotype B or C. albicans genotype C strains. These results indicate that there is a correlation between the Candida groups and antifungal susceptibility.
There has been an increase in the occurrence of diseases caused by Candida over the recent past. The majority of these diseases are caused by Candida albicans (4, 11, 16, 17). Molecular typing methods have been used with increasing frequency for epidemiological investigations for the development of rational infection control measures (17). One of the earliest molecular methods for the differentiation of C. albicans strains used a simple technique of analyzing the restriction fragment length polymorphisms (RFLPs) of cellular DNA to divide isolates into two large groups on the basis of the position of a dimorphic band (group A strains have a band of 3.7 kb and group B strains have a band of 4.2 kb) and then subdivided them into types (19). The method was shown to be reliable and reproducible and was used for a large epidemiological study of strains isolated from multiple localities in the United States and the United Kingdom (21). In that study it was reported that there was an association of genotypic group A with increased resistance to the antifungal agent flucytosine (21). Recent studies by the same methodology have reported on the occurrence of C. albicans strains with unusual genotypes; these have been designated genotype C (strains with both the 3.7- and 4.2-kb bands) and genotype D (strains with neither band) (2, 9, 10). However, no association has been made between these newly described genotypes and antifungal susceptibility.
C. dubliniensis is a recently recognized species that is phenotypically very closely related to C. albicans. It has been isolated primarily from the oral cavities of patients infected with the human immunodeficiency virus (22). The characteristics and identification of this species have recently been reviewed (22). That review highlighted the difficulty in differentiating this species from C. albicans by routine laboratory methods (22). C. dubliniensis has been reported to be susceptible to the same range of antifungal agents as C. albicans (22).
Recently, a 379-nucleotide insert in the DNA encoding for the large-subunit rRNA (the 25S rRNA gene [rDNA]) has been shown to be responsible for the larger 4.2-kb band in the C. albicans genotype B isolates, whereas the smaller 3.7-kb band in the genotype A isolates lacks this insert of nucleotides (12). An homologous group 1 intron at the same insertion point was shown to be present in strains of C. dubliniensis; however, this intron was approximately 300 nucleotides larger than that in genotype B C. albicans (1).
The aim of the present study was to use molecular tools to characterize recently described new genotypic subgroups of C. albicans and to compare them with authentic strains of C. dubliniensis and type I C. stellatoidea. An additional aim was to assess a large, randomly chosen subset of these isolates for their comparative antifungal susceptibilities to fluconazole and flucytosine by the U.S. National Committee for Clinical Laboratory Standards (NCCLS) M27-A susceptibility assay (14).
MATERIALS AND METHODS
The isolates included in this study were made available from previous investigations (2, 3, 8, 9, 21). Authentic strains of C. dubliniensis were made available from D. Sullivan and have been submitted to the National Collection of Pathogenic Fungi (London, United Kingdom) as NCPF 3949 and to the Centraalbureau voor Schimmelcultures (Baarn, The Netherlands) as CBS 7987 and CBS 7988 (13, 24). Authentic strains of type I C. stellatoidea (B-4257 and B-4404) were provided by J. Kwon-Chung (National Institutes of Health, Bethesda, Md.) (5). Authentic strains of other Candida species were obtained from the American Type Culture Collection (ATCC). Finally, five strains identified as C. dubliniensis, including the ex-type strain, were provided by F. Odds; the other four strains were identified by atypical green color on CHROMagar Candida (CHROMagar Microbiology, Paris, France), repeated demonstration of the absence of intracellular beta-glucosidase activity, and weak hybridization by Southern blotting with the C. albicans-specific oligonucleotide sequence Ca3 (20).
All C. albicans isolates were identified by germ tube and chlamydospore formation, and the majority of genotype A, B, and C isolates were confirmed to be C. albicans with the API 20C system (bioMerieux, Marcy, l’Etoile, France).
Cellular DNA was isolated by previously described methods (19).
Primers for PCR were designed for three separate areas of the DNA encoding the rRNA. The first pair of primers, ITS1 (5′-TCC GTA GGT GAA CCT GCG G-3′) and ITS4 (5′-TCC TCC GCT TAT TGA TAT GC-3′), have been used in previous studies (15, 26). These give an expected PCR product extending from the 5′ end of the 18S rDNA to the 3′ end of the 25S rDNA and include both intergenic transcribed spacer (ITS) regions (ITS1 and ITS2) as well as the entire 5.8S rDNA.
The second pair of primers, CA25SV3-L (5′-TCT TAA CAG CTT ATC ACC CTG GAA TTG GTT-3′) and CA25SV3-R (5′-ATT GTG TCA ACA TCA CTT TCT GAC CAT CAC-3′), were designed from sequences submitted to GenBank (accession nos. X83718 and X83717). These give an expected PCR product that spans the V3 region of the 25S rDNA, a region previously used for the taxonomic differentiation of C. dubliniensis (24).
The final pair of primers results in an expected PCR product that spans the site of the transposable intron in the 25S rDNA, as published previously (12). The primers CA-INT-L (5′-ATA AGG GAA GTC GGC AAA ATA GAT CCG TAA-3′) and CA-INT-R (5′-CCT TGG CTG TGG TTT CGC TAG ATA GTA GAT-3′) were designed from sequences submitted to Genbank (accession nos. Z70663 and X74272).
For all PCRs, DNA was amplified in the buffer supplied by the Taq polymerase manufacturer (Gibco BRL, Grand Island, N.Y.) in a 50-μl volume containing 1 μM primers, 1.5 mM MgCl2, 2.5 U of Taq polymerase, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, and 200 μM dTTP. The reactions were performed with an automated thermal cycler (GeneMate; Lab-Line Instruments, Melrose Park, Ill.). DNA samples were denatured by incubation for 3 min at 94°C before 30 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 2.5 min. After the PCR, amplified DNA was purified with spin columns (Wizard PCR Preps; Promega, Madison, Wis.).
For RFLP analysis of the first (ITS region) and second (V3 25S rDNA region) PCR amplicons, the purified PCR products were digested with the restriction enzymes DdeI and HaeIII, respectively (Boehringer Mannheim, Indianapolis, Ind.). Endonuclease digestions were done by overnight incubation (to allow for complete digestion of the PCR amplimers) with 10 U of enzyme at the recommended temperature and with the corresponding buffer.
Ten microliters of the PCR products, with and without endonuclease digestion, were analyzed by electrophoresis through a 3% (wt/vol) agarose gel (2% Nusieve GTG, 1% SeaKem Gold; FMC BioProducts, Philadelphia, Pa.) in TAE buffer (40 mM Tris-acetate, 0.2 mM EDTA) for 2 h at 10 V/cm. Bands were visualized by Uv transillumination at 302 nm after ethidium bromide staining.
The testing of a representative sample of 105 strains for their susceptibilities to both fluconazole and flucytosine was done by the NCCLS M27-A method (14). The strains included in this analysis were randomly selected from a variety of geographically diverse sites and included 30 genotype A strains, 30 genotype B strains, 30 genotype C strains, and 15 genotype D strains (10). Statistical analysis of these antifungal susceptibility test results was done by transforming the resultant susceptibility level by log2. Initially, the transformed results were compared for all groups by a multiway analysis of variance (ANOVA). Where differences were observed at the level of a P value of <0.05, pairs of groups were compared by a two-sample t test with the assumption of unequal variances. Differences between groups were assumed to be significant when the probability (P) was ≤0.05.
RESULTS
Molecular analysis of Candida spp.
A total of 457 strains of Candida were analyzed by all three PCR methods. Four hundred thirty-nine of these strains derived from a large study of isolates identified as C. albicans (289 genotype A strains, 85 genotype B strains, 56 genotype C strains and 9 genotype D strains) obtained over a 20-year period from 15 geographically diverse areas) (10). Nine authentic strains of different species of Candida from ATCC, two authentic strains of C. dubliniensis, two authentic strains of type 1 C. stellatoidea, and a further five strains of C. dubliniensis were analyzed. A list of the authentic strains analyzed is given in Table 1.
TABLE 1.
Order, source, and designation of strains of the various Candida species and genotypes
| Lane no.a | Species/genotypeb | Strain designation |
|---|---|---|
| 1 | C. albicans/A | ATCC 64124 |
| 2 | C. albicans/A | ATCC 62342 |
| 3 | C. albicans/B | ATCC 38246 |
| 4 | C. albicans/B | ATCC 32354 |
| 5 | C. albicans/C | Singapore 7c |
| 6 | C. albicans/C | Singapore 8c |
| 7 | C. dubliniensis CD36 | NCPF 3949 and CBS 7987 |
| 8 | C. dubliniensis CM2 | CBS 7988 |
| 9 | C. stellatoidea | B-4257 |
| 10 | C. stellatoidea | B-4404 |
| 11 | C. parapsilosis | ATCC 22019 |
| 12 | C. tropicalis | ATCC 13803 |
| 13 | C. krusei | ATCC 6258 |
| 14 | C. lusitania | ATCC 4125 |
| 15 | C. glabrata | ATCC 66032 |
| 16 | Negative control |
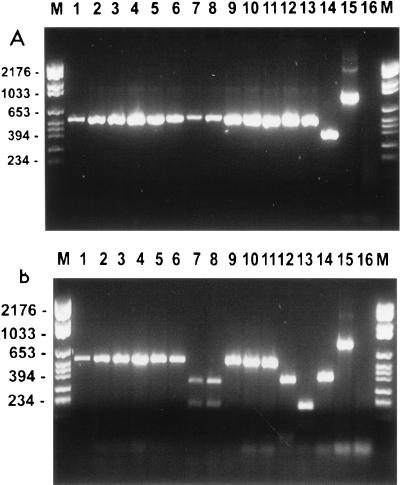
The RFLPs generated by DdeI digestion of the PCR products from the ITS region did not differentiate among C. albicans genotypes A, B, and C and type I C. stellatoidea (Fig. 1). However, this method did differentiate the C. albicans genotype D strains from C. albicans genotype A, B, and C strains (Fig. 1). The profiles of the C. albicans genotype D strains were identical to those of the C. dubliniensis strains.
FIG. 1.
Ethidium bromide-stained, UV-transilluminated PCR products (A) and the same PCR products obtained after digestion with DdeI (B) obtained by PCR with the primers for the ITS region. The photograph was obtained after electrophoresis in a 3% agarose gel. The DNA from the PCR (A) had first been purified with the Wizard PCR Preps purification system prior to overnight digestion with 10 U of the restriction endonuclease DdeI (B). Molecular size markers are in the lanes marked M, and their corresponding sizes (in base pairs) are given on the left. The isolate in each lane is specified in Table 1.
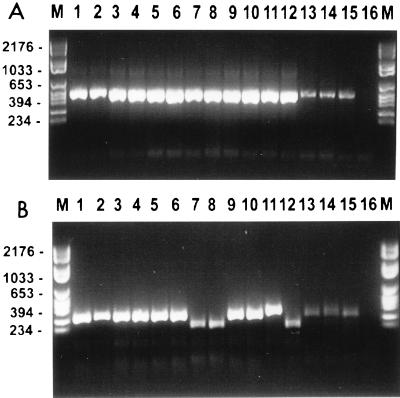
The RFLPs generated by HaeIII digestion of the PCR products of the V3 region of the 25S rDNA could differentiate the same groups as the RFLP analysis of the PCR amplicon of the ITS region (Fig. 2). The profiles of the C. albicans genotype D strains were identical to those of the C. dubliniensis strains.
FIG. 2.
Ethidium bromide-stained, UV-transilluminated PCR products (A) and the same PCR products obtained after digestion with HaeIII (B) obtained by PCR with the primers for the V3 region of the 25S rDNA. The photograph was obtained after electrophoresis in a 3% agarose gel. The DNA from the PCR (A) had first been purified with the Wizard PCR Preps purification system prior to overnight digestion with 10 U of the restriction endonuclease HaeIII (B). Molecular size markers are in the lanes marked M, and their corresponding sizes (in base pairs) are given on the left. The isolate in each lane is specified in Table 1.
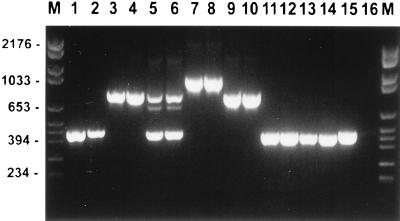
PCR designed to span the region that included the site of the transposable intron in genotype B C. albicans and C. dubliniensis strains (1, 12) resulted in a single product for C. albicans genotypes A (∼450 bp) and B (∼840 bp), C. stellatoidea (∼840 bp), and C. dubliniensis (∼1,080 bp) (Fig. 3). All of the C. albicans genotype C isolates had two PCR products (∼450 and ∼840 bp) that were identical in size to the respective products from C. albicans genotypes A and B (Fig. 3). All C. albicans genotype D isolates gave a PCR product identical in size to that of C. dubliniensis.
FIG. 3.
Ethidium bromide-stained, UV-transilluminated PCR products obtained by PCR with the primers that span the intron position in the 25S rDNA. The photograph was obtained after electrophoresis in a 3% agarose gel. Molecular size markers are in the lanes marked M, and their corresponding sizes (in base pairs) are given on the left. The isolate in each lane is specified in Table 1.
To assess if the C. albicans genotypic subgroup C strains were not a mixed culture of strains, two randomly chosen isolates were subcultured on yeast potato dextrose (YPD) agar for 48 h. Twenty-five individual colonies were chosen and grown in YPD broth for 24 h, and their DNAs were extracted and assessed for the presence of the intron in the 25S rDNA. All 25 colonies gave identical results, exhibiting DNA bands typical of those of genotype A and B strains; this result therefore indicated that the intron was partially present throughout their genomes.
Correlation of species, DNA type, and antifungal susceptibility.
Descriptive statistics of the MICs obtained by antifungal susceptibility testing of the randomly chosen 105 isolates (30 C. albicans genotype A strains, 30 C. albicans genotype B strains, 30 C. albicans genotype C strains, and 15 C. dubliniensis strains) are presented in Table 2.
TABLE 2.
Descriptive statistics of the MICs obtained by testing 105 strains their susceptibilities to both fluconazole and flucytosine
| Fluconazolea
|
Flucytosineb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| C. albicans genotype A | C. albicans genotype B | C. albicans genotype C | C. dubliniensis | C. albicans genotype A | C. albicans genotype B | C. albicans genotype C | C. dubliniensis | |
| Mean MIC (μg/ml) | 22.8 | 33.4 | 32.3 | 4.7 | 6.9 | 0.26 | 2.3 | 8.7 |
| SD MIC (μg/ml) | 29.8 | 29.1 | 30.5 | 9.3 | 3.5 | 0.07 | 2.1 | 5.8 |
| Geometric mean MIC (μg/ml) | 4.2 | 10.3 | 8.9 | 1.1 | 0.54 | 0.18 | 0.22 | 0.32 |
| No. of isolates | 30 | 30 | 30 | 15 | 30 | 30 | 30 | 15 |
| Confidence level (95.0%) | 11.1 | 10.8 | 11.4 | 5.2 | 7.2 | 0.14 | 4.3 | 12.4 |
ANOVA for the fluconazole susceptibility results of the four groups gave a P value of 0.003. Analysis of each pair of groups by a two-sample t test assuming unequal variance showed that the results for C. dubliniensis were significantly different from those for each of C. albicans genotypes A, B, and C (P = 0.018, 0.002, and 0.004, respectively). No other statistically significant differences were observed.
ANOVA for the flucytosine susceptibility results for the four groups gave a P value of 0.021. Analysis of each pair of groups by a two-sample t test assuming unequal variance showed that the results for the C. albicans genotype A strains were significantly different from those for each of C. albicans genotypes B and C (P = 0.004 and 0.003, respectively). No other statistically significant differences were observed.
Multiway ANOVA for the comparative fluconazole susceptibilities of the four groups gave a P value of 0.003. Analysis of each pair of groups by a two-sample t test assuming unequal variance showed that the C. dubliniensis strains were significantly more susceptible than each of the C. albicans genotype A, B, and C strains (P = 0.018, 0.002, and 0.004, respectively). No other statistically significant comparative differences were found for the results of fluconazole susceptibility testing.
Multiway ANOVA for the comparative flucytosine susceptibilities of the four groups gave a P value of 0.021. Analysis of each pair of groups by a two-sample t test assuming unequal variance showed that the C. albicans genotype A strains were significantly less flucytosine susceptible than either C. albicans genotype B or C. albicans genotype C (P = 0.004 and 0.003, respectively). No other significant differences were noted for the results of flucytosine susceptibility testing.
DISCUSSION
The results presented here indicate that those strains previously designated C. albicans genotype D (2, 9, 10) should be assigned to the species C. dubliniensis. All molecular methods for the differentiation of isolates to the species level described here used PCR techniques. The method that detects the presence and the size of the intron in the 25S rDNA is particularly easily adapted for use in reference laboratories for the rapid identification of large numbers of isolates. This simple PCR method has the additional advantage of differentiating strains of C. albicans into the described genotypic subgroups.
C. stellatoidea has previously been divided into two types, types I and II, with type II isolates shown to be sucrose-negative mutants of C. albicans (5). There has been extensive debate over the taxonomic relationship between type I C. stellatoidea and C. albicans. Genetic traits that have been used to differentiate these two species have relied on electrophoretic karyotyping (6), mitochondrial DNA restriction fragment patterns (6), cellular DNA restriction fragment length polymorphisms (7), or hybridization of cellular DNA with midrepeat probes (6). Recently, evidence that shows that it is not possible to support the differentiation of type I C. stellatoidea from C. albicans by a variety of molecular and nonmolecular methods has been accumulating (1, 18, 23). A recent study (24) which examined the homology of the rRNA V3 regions and also sequencing data concluded that C. stellatoidea types I and II are identical to each other and differ from C. albicans by only 1 or 2 bases. These results support the previous contention that type I C. stellatoidea does not merit species status (25). One of the earliest reports of the use of molecular methods for the differentiation of C. stellatoidea from C. albicans assessed the rDNA differences among 15 isolates of C. albicans and 2 isolates of C. stellatoidea (7). We note that all 15 of the C. albicans isolates assessed in this previous study were genotypic subgroup A C. albicans strains, because all strains had the 3.7-kb rDNA band, whereas the 2 C. stellatoidea isolates were genotypic subgroup B C. albicans strains (7). The strains of C. stellatoidea investigated in the present study were indistinguishable from the C. albicans genotype B strains by all methods by which the strains were assessed. Thus, our results indicate that type I C. stellatoidea is synonymous with C. albicans genotype B.
The results of the antifungal susceptibility testing for fluconazole showed that C. dubliniensis is more susceptible than C. albicans (genotypes A, B, and C). In early reports there were some differences in the antifungal susceptibilities of this species (8, 13, 24). However, the previous studies did not use the accepted NCCLS method for susceptibility testing. The recently reported development of stable fluconazole resistance at high frequency in vitro in C. dubliniensis (13) was not assessed in the present study, yet it may go a long way toward explaining the reported increasing occurrence of this species, particularly in patients with long-term exposure to prophylactic fluconazole (22).
The C. albicans genotype A strains studied in the present investigation showed increased levels of resistance to the antifungal agent flucytosine. This finding is consistent with previous data (12, 21), and it has been postulated that there is a direct causal relationship between the presence of the group 1 intron in the 25S rDNA (the presence of which determines that the strain should be classified as genotype B) and a decrease in the level of resistance to flucytosine (12). The results presented here indicate that this group 1 intron is only partially present throughout the rDNA repeats in the genomes of C. albicans genotype C strains. It may be postulated that we are observing these strains during a period when this intron is being lost and they are moving from a genotype B strain to a genotype A strain and concurrently developing an increased level of resistance to flucytosine. Such an hypothesis would partly explain the increase in the occurrence of genotype C strains worldwide (10), particularly in areas outside the United States where there may well be an increasing rate of use of flucytosine.
An alternative hypothesis for the present observations may be that the genotype C strains represent the genotype A isolates that have acquired the intron and that are becoming genotype B strains. The mechanisms by which this acquisition of genetic material occurs is as yet unknown, but an interesting possibility would be that genotype C strains represent the progeny of the sexual union between a genotype A strain with a genotype B (or type I C. stellatoidea) strain. Further research is required to elucidate whether or not the genotype C isolates have stable genotypes or whether these strains are in the process of losing or acquiring the group 1 intron in the 25S rDNA.
ACKNOWLEDGMENTS
We thank Lynda Treat-Clemons, diaDexus Corp., Santa Clara, Calif., for advice on the statistical analysis and PilSang Park for laboratory assistance.
This research was funded in part by a fellowship from the Commonwealth AIDS Research Grants Committee of the National Health and Medical Research Council of the Australian Federal Government.
REFERENCES
- 1.Boucher H, Mercure S, Montplaisir S, Lemay G. A novel group I intron in Candida dubliniensis is homologous to a Candida albicans intron. Gene. 1996;180:189–196. doi: 10.1016/s0378-1119(96)00453-2. [DOI] [PubMed] [Google Scholar]
- 2.Clemons K V, Feroze F, Holmberg K, Stevens D A. Comparative analysis of genetic variability among Candida albicans isolates from different geographic locales by three genotypic methods. J Clin Microbiol. 1997;35:1332–1336. doi: 10.1128/jcm.35.6.1332-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clemons K V, Shankland G S, Richardson M D, Stevens D A. Epidemiologic study by DNA typing of Candida albicans outbreak in heroin addicts. J Clin Microbiol. 1991;29:205–207. doi: 10.1128/jcm.29.1.205-207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon D M, McNeil M M, Cohen M L, Gellin B G, La Montagne J R. Fungal infections: a growing threat. Public Health Rep. 1996;111:226–235. [PMC free article] [PubMed] [Google Scholar]
- 5.Kwon-Chung K J, Hicks J B, Lipke P N. Evidence that Candida stellatoidea type II is a mutant of Candida albicans that does not express sucrose-inhibitable alpha-glucosidase. Infect Immun. 1990;58:2804–2808. doi: 10.1128/iai.58.9.2804-2808.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwon-Chung K J, Wickes B L, Merz W G. Association of electrophoretic karyotype of Candida stellatoidea with virulence for mice. Infect Immun. 1988;56:1814–1819. doi: 10.1128/iai.56.7.1814-1819.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magee B B, D’Souza T M, Magee P T. Strain and species identification by restriction fragment polymorphisms in the ribosomal DNA repeat of Candida species. J Bacteriol. 1987;169:1639–1643. doi: 10.1128/jb.169.4.1639-1643.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCullough M, Ross B, Reade P. Characterization of genetically distinct subgroup of Candida albicans strains isolated from oral cavities of patients infected with human immunodeficiency virus. J Clin Microbiol. 1995;33:696–700. doi: 10.1128/jcm.33.3.696-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCullough M J, Clemons K V, Del Palacio A, Stevens D A. Epidemiology of Candida albicans isolates from heroin addicts analysed by DNA typing. Med Mycol. 1998;36:213–217. [PubMed] [Google Scholar]
- 10.McCullough M J, Clemons K V, Stevens D A. Abstracts of the 98th General Meeting of the American Society for Microbiology 1998. Washington, D.C: American Society of Microbiology; 1998. Molecular epidemiology of the global and temporal diversity of C. albicans abstr. F-43; p. 260. [Google Scholar]
- 11.McCullough M J, Ross B C, Reade P C. Candida albicans: a review of its history, taxonomy, epidemiology, virulence attributes, and methods of strain differentiation. Int J Oral Maxillofac Surg. 1996;25:136–144. doi: 10.1016/s0901-5027(96)80060-9. [DOI] [PubMed] [Google Scholar]
- 12.Mercure S, Montplaisir S, Lemay G. Correlation between the presence of a self-splicing intron in the 25S rDNA of C. albicans and strain susceptibility to 5-fluorocytosine. Nucleic Acids Res. 1993;21:6020–6027. doi: 10.1093/nar/21.25.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.National Committee for Clinical Laboratory Standards. Reference method for broth dilution antifungal susceptibility testing of yeast. Approved standard. 1st. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 15.Nho S, Anderson M J, Moore C B, Denning D W. Species differentiation by internally transcribed spacer PCR and HhaI digestion of fluconazole-resistant Candida krusei, Candida inconspicua, and Candida norvegensis strains. J Clin Microbiol. 1997;35:1036–1039. doi: 10.1128/jcm.35.4.1036-1039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Odds F C. Ecology of Candida and epidemiology of candidiasis. In: Odds F C, editor. Candida and candidiasis. London, United Kingdom: Bailliere Tindall; 1988. pp. 85–86. [Google Scholar]
- 17.Pfaller M A. Epidemiology of fungal infections: the promise of molecular typing. Clin Infect Dis. 1995;20:1535–1539. doi: 10.1093/clinids/20.6.1535. [DOI] [PubMed] [Google Scholar]
- 18.Pujol C, Renaud F, Mallie M, de Meeus T, Bastide J M. Atypical strains of Candida albicans recovered from AIDS patients. J Med Vet Mycol. 1997;35:115–121. [PubMed] [Google Scholar]
- 19.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoofs A, Odds F C, Colebunders R, Ieven M, Goossens H. Recognition and identification of Candida dubliniensis isolates from HIV patients: use of specialized isolation media. Eur J Clin Microbiol Infect Dis. 1997;16:296–300. doi: 10.1007/BF01695634. [DOI] [PubMed] [Google Scholar]
- 21.Stevens D A, Odds F C, Scherer S. Application of DNA typing methods to Candida albicans epidemiology and correlation with phenotype. Rev Infect Dis. 1990;12:258–266. doi: 10.1093/clinids/12.2.258. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan D, Haynes K, Bille J, Boerlin P, Rodero L, Lloyd S, Henman M, Coleman D. Widespread geographic distribution of oral Candida dubliniensis strains in human immunodeficiency virus-infected individuals. J Clin Microbiol. 1997;35:960–964. doi: 10.1128/jcm.35.4.960-964.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 25.Wickes B L, Golin J E, Kwon-Chung K J. Chromosomal rearrangement in Candida stellatoidea results in a positive effect on phenotype. Infect Immun. 1991;59:1762–1771. doi: 10.1128/iai.59.5.1762-1771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williams D W, Wilson M J, Lewis M A, Potts A J. Identification of Candida species by PCR and restriction fragment length polymorphism analysis of intergenic spacer regions of ribosomal DNA. J Clin Microbiol. 1995;33:2476–2479. doi: 10.1128/jcm.33.9.2476-2479.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]