ABSTRACT
Permafrost microbes may be metabolically active in microscopic layers of liquid brines, even in ancient soil. Metagenomics can help discern whether permafrost microbes show adaptations to this environment. Thirty-three metagenome-assembled genomes (MAGs) were obtained from six depths (3.5 m to 20 m) of freshly cored permafrost from the Siberian Kolyma-Indigirka Lowland region. These soils have been continuously frozen for ∼20,000 to 1,000,000 years. Eight of these MAGs were ≥80% complete with <10% contamination and were taxonomically identified as Aminicenantes, Atribacteria, Chloroflexi, and Actinobacteria within bacteria and Thermoprofundales within archaea. MAGs from these taxa have been obtained previously from nonpermafrost environments and have been suggested to show adaptations to long-term energy starvation, but they have never been explored in ancient permafrost. The permafrost MAGs had greater proportions in the Clusters of Orthologous Groups (COGs) categories of energy production and conversion and carbohydrate transport and metabolism than did their nonpermafrost counterparts. They also contained genes for trehalose synthesis, thymine metabolism, mevalonate biosynthesis, and cellulose degradation, which were less prevalent in nonpermafrost genomes. Many of these genes are involved in membrane stabilization and osmotic stress responses, consistent with adaptation to the anoxic, high-ionic-strength, cold environments of permafrost brine films. Our results suggest that this ancient permafrost contains DNA of high enough quality to assemble MAGs from microorganisms with adaptations to survive long-term freezing in this extreme environment.
IMPORTANCE Permafrost around the world is thawing rapidly. Many scientists from a variety of disciplines have shown the importance of understanding what will happen to our ecosystem, commerce, and climate when permafrost thaws. The fate of permafrost microorganisms is connected to these predicted rapid environmental changes. Studying ancient permafrost with culture-independent techniques can give a glimpse into how these microorganisms function under these extreme low-temperature and low-energy conditions. This will facilitate understanding how they will change with the environment. This study presents genomic data from this unique environment ∼20,000 to 1,000,000 years of age.
KEYWORDS: permafrost, MAGs, exobiology, environmental, bioinformatics
INTRODUCTION
Arctic soils are often studied by sampling the upper, seasonally thawed, active layer (1). Climate change is rapidly increasing the depths of permafrost active layers worldwide (2), making it necessary to learn more about the microbes living in the deeply buried permafrost before it thaws completely. Evidence for microbes persisting in ancient permafrost has come in the form of revivable cultured isolates, microscopic visualization of intact cells, live/dead cell staining, low d/l-amino acid ratios in cellular biomass, high-quality metagenomic reconstructions of microbial genomes, and the presence of high-quality RNA molecules (3–8). Evidence from other studies of 33,000-year-old permafrost in Alaska suggests that microbial communities are equipped to survive in this type of environment by using unique energy acquisition mechanisms and stress responses such as scavenging detrital biomass and environmental sensing (6). Variations in metagenomic functions of the in situ microbial populations in 30,000-year-old permafrost in the Kolyma-Indigirka Lowland region in northeastern Siberia corresponded to local geochemistry, suggesting that the populations were alive (8). Additionally, a study of >500,000-year-old permafrost samples from northeastern Russia contained microbial DNA with likely DNA repair mechanisms (9). Thus, some microbes in ancient permafrost may not be dormant (7, 10, 11) but instead maintain a continuous torpor-like state that allows them to subsist in the low-energy environment.
The Yedoma geological suite in the Kolyma-Indigirka Lowland region in northeastern Siberia originates from the late Pleistocene Era (12). Yedoma, or Ice Complex, permafrost deposits were frozen syngenetically (deposited and frozen simultaneously) ∼20,000 to ∼60,000 years ago (13). Yedoma spans approximately 1 million km2 (14) and is found, on average, up to 10 m below the surface (13, 15). The next layer down, called Olyor, is unlike Yedoma since sediments were frozen epigenetically (deposited and then frozen) after being deposited ∼0.6 million to 1.6 million years ago (15, 16) and they contain few ice formations (17). Some of the perennially frozen soils of this region have been dated as up to 3 million years old (18, 19). Although the permafrost of the Kolyma area is of freshwater origin (12), other studies showed that, during freezing, small solutes were occluded from the surrounding mineral material, creating small brine films (5).
To live in the permanently frozen soils underneath the active layer without seasonal nutrient replenishment, a microbe must be able to sustain cellular integrity with a limited source of liquid water (18). Liquid water exists in permafrost in the form of cryopegs or small brine films (5), which are hypersaline liquid water films and lenses that can occur throughout the permafrost at temperatures of −10°C and salt contents of 140 to 300 g/liter (18). These saline fluids can be in the shape of flattened films along sediment grains or in small pockets, depending on depositional characteristics (20).
While it is thought that the presence of liquid water in these brine films is sufficient to maintain live microbes within the thermostable deep permafrost, the interactions between microbial cells and the brine film have not been visualized at a microscopic level (21). However, plump cells with unique membrane adaptations have been visualized with electron microscopy in 200,000-year-old permafrost samples (22). Additionally, cells visualized from ice core and permafrost samples are notably very small (<1-μm diameter) (23, 24) but not small enough to be completely immersed in the brine vein (18). Since these brine films are so small, ∼1 nm wide (25), the cell membrane would have to make contact with the brine in order to act as a conduit for small molecule transport. Necessary microbial metabolic processes could occur in permafrost as long as physical mass transfer can occur in the environment (26). Since the cell’s internal environment remains unfrozen (27), the cell could transfer small metabolites (26) between the intracellular matrix and the brine films.
Many organisms have been able to be cultured from Kolyma-Indigirka Lowland permafrost, including representatives from the Carnobacterium genus (28), Gammaproteobacteria, Actinobacteria, and Firmicutes (29). Similar groups have been detected from 40,000-to 50,000-year-old permafrost (30). Microbes have also been isolated from Siberian permafrost in high salt concentrations (0.5% to 15% NaCl) (18, 31, 32) and with various medium types (33), suggesting that the microbes could survive in brines. Culture work, however, does not assess the entire microbial community, since many organisms have not been cultured, despite many attempts (34).
To supplement culturing studies, we used metagenomes to study the microbial populations in ∼20,000- to 1,000,000-year-old permafrost formed near the freshwater Alazeya River in the Kolyma-Indigirka Lowland region in northeastern Siberia. We determined the geological age of the upper Yedoma suite by 14C dating. The deeper soil age was determined to be 650,000 to 1,000,000 years in previous studies (12, 14, 16, 18, 35). Microbial communities, largely Firmicutes and Proteobacteria, have been identified in permafrost in this location by 16S rRNA gene analysis and are likely alive since they have substantial amounts of intracellular DNA (4). In this study, we used metagenomes to determine whether microbes in these ancient sediments (i) produce metagenome-assembled genomes (MAGs), which suggest the presence of high-quality nucleic matter, and (ii) contain genes that suggest adaptation to long-term subsistence under permafrost environmental conditions. We identified 8 MAGs with ≥80% genome completeness and ≤10% contamination from five taxonomic groups. A unique set of functional genes were present in permafrost MAGs but absent in publicly available genomes of the same taxonomic groups in nonpermafrost environments (referred to as nonpermafrost genomes). These ancient permafrost MAGs give preliminary insights into the survival capabilities of these microbes in 20,000- to 1,000,000-year-old permafrost.
RESULTS
Site characteristics.
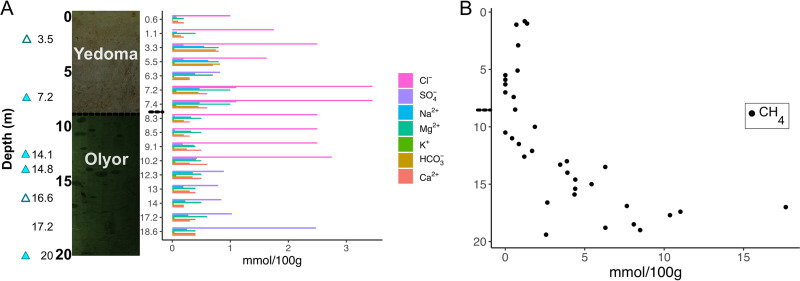
A 20-m-deep permafrost core (AL3-15) from the Kolyma-Indigirka Lowland region (69.339600, 154.996450) was subsectioned for microbiology and geology immediately upon retrieval in the field (Fig. 1A). The permafrost geology enters a transition zone at about 6.2 m, where it changes from the Yedoma geological suite to Olyor at 7.7 m. The temperature inside the borehole decreased from −2°C 1 m below the surface to −8.7°C 5 m below the surface and stayed in a constant range of −8°C to −8.6°C in the lower 10 m of the borehole (see Fig. S1 in the supplemental material). The δ13C of the inorganic carbon fraction (−3.4 to −6%) was more 13C enriched than the organic carbon (−28.8 to −24.3%) at 2.9, 3.5, and 5.6 m (see Table S1). The 14C ages of the inorganic carbon increased from 21,760 ± 120 years to 33,900 ± 550 years with stratigraphic depth over the upper 5.6 m of the Yedoma suite. However, the 14C ages of the two organic carbon pools showed no stratigraphic trend, with great variability at different depths (see Table S1). The large difference between 2.9 m (38,590 ± 980 to 41,700 ± 1,400 years) and 3.5 m (18,228 ± 78 to 20,158 ± 99 years) suggested that the ice-rich layer at 3.5 m might be an ice wedge that was much younger than the surrounding strata. The underlying sediments of the Olyor suite were too old for radiocarbon dating. With other methods such as fossil and palynology records, however, these soils were previously dated as between 650,000 and 1,000,000 years old (12, 14, 16, 18, 35). The porewater extracted from thawed subsamples of permafrost showed fluctuations in dissolved ions downcore but they generally increased with depth (Fig. 1A). Chloride had the greatest increase down the core, ranging from 1 mmol/100 g of soil at 3.5 m to 2.7 mmol/100 g of soil at 10.2 m. No chloride measurements were made below this depth. Methane increased from 1.2 to 17.6 mmol/100 g of soil and sulfate increased from 0.019 to 2.5 mmol/100 g of soil at depths of 3 m to 17.2 m, respectively (Fig. 1B). Methane levels were high only in the older Olyor deposits below about 13 m. The pH stayed in a constant range of 8 to 8.5 throughout the core (see Fig. S1). Cells were visualized with 4′,6-diamidino-2-phenylindole (DAPI) and SYBR gold staining (see Fig. S2A) but were below the quantification limit (36) for direct microscopic cell counts. Further evidence of intact cells comes from flow cytometry, which showed a population of similarly sized particles that stained with SYBR gold (see Fig. S2B and C). None of the 10 sequenced single-cell amplified genomes (SAGs) had high enough quality for use in this study (0% to 45% completeness). The only Genome Taxonomy Database-Toolkit (GTDB-Tk)-classifiable SAG was a Firmicutes organism (45% complete, with 5% contamination) from 7.2 m that matched the taxonomy of MAG_07_7.2m (which was not analyzed in this study due to its poor quality but can be found under BioProject accession number PRJNA596250).
FIG 1.
Downcore permafrost borehole characteristics. (A) Photos of Yedoma and Olyor sediments and their relative depths, with triangles indicating depths that produced MAGs and filled triangles indicating depths with MAGs that were >80% complete and <10% contaminated. Concentrations of major ions are shown for each depth. Note that the depth intervals are discrete in panel B to spread the data points evenly across the depths. (B) Concentrations of methane for each depth. Other geochemical measurements, including temperature, pH, and total carbon, are available in Fig. S1 in the supplemental material. The small dashed line at 7.7 m shows the depth of the transition between the Yedoma and Olyor layers.
Metagenomes.
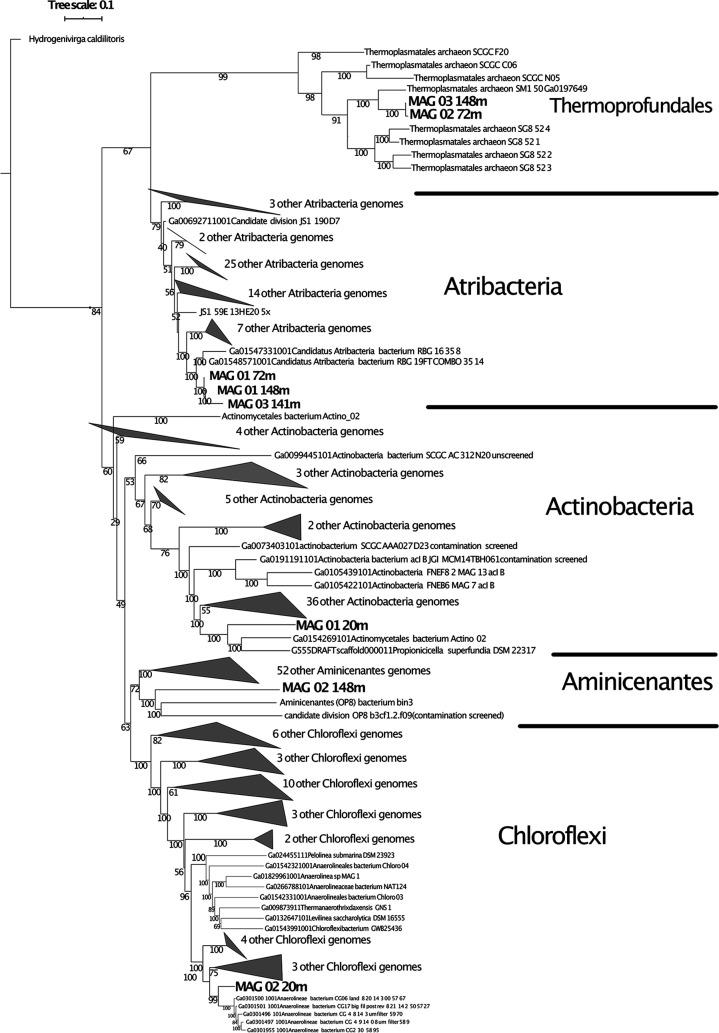
Of the seven depths from which DNA was extracted (3.5, 7.2, 14.1, 14.8, 16.6, 17.2, and 20 m), six samples produced metagenomic assemblies (Table 1). The metagenome from 17.2 m produced only ∼1 MB of data that did not assemble; therefore, it was not included in the analysis. The numbers of reads, numbers of assembled contigs, mean read lengths, N50 values, and total numbers of MAGs did not trend with the sample depth or the total amount of retrieved data (Table 1). Every assembled metagenome had at least 91% of the contigs incorporated into the MAGs, suggesting that the MAGs represented most of the microbial community that was assembled (Table 1). The 6 metagenomes produced a total of 33 MAGs (see Table S2), 8 of which were ≥80% complete with <10% contamination (see Fig. S2). GTDB-Tk and a maximum likelihood tree with 139 conserved proteins placed these MAGs into the groups Atribacteria (n = 3), Chloroflexi (n = 1), Aminicenantes (n = 1), Actinobacteria (n = 1), and Thermoprofundales (n = 2) among a total of 230 publicly available reference genomes (Fig. 2). A search through the National Center for Biotechnology Information (NCBI) and Joint Genome Institute (JGI) Integrated Microbial Genomes (IMG) public databases resulted in no MAGs from these groups from other ancient permafrost studies. The remaining 25 Siberian MAGs did not meet the quality standards for inclusion in our analysis; these were classified as Acidobacteriota (n = 1), Actinobacteriota (n = 9), Aminicenantes (n = 3), Atribacterota (n = 2), Chloroflexota (n = 2), Crenarchaeota (n = 1), Bathyarchaeota (n = 1), Firmicutes (n = 2), Planctomycetota (n = 3), and Thermoplasmatota (n = 1) (see Table S2).
TABLE 1.
Metagenome details for the seven sampling depths
| Sample depth (m) | Total no. of unassembled reads | Total no. of assembled reads | Total no. of assembled contigs | N50 (bp) | Total no. of MAGs | % of contigs binned |
|---|---|---|---|---|---|---|
| 3.5 | 14,187,214 | 13,195,548 | 115 | 2,842 | 1 | 91 |
| 7.2 | 9,621,340 | 9,045,678 | 7,556 | 4,323 | 8 | 94 |
| 14.1 | 11,513,548 | 10,956,796 | 4,513 | 4,527 | 7 | 93 |
| 14.8 | 13,544,854 | 12,728,740 | 7,098 | 4,625 | 11 | 93 |
| 16.6 | 5,698,300 | 5,115,016 | 900 | 2,871 | 2 | 99 |
| 17.2a | 420 | NA | NA | NA | 0 | 0 |
| 20 | 13,338,036 | 11,783,820 | 3,058 | 4,353 | 4 | 96 |
The sample depth of 17.2 m did not yield enough data to curate a metagenome. NA, not applicable.
FIG 2.
Phylogenetic tree of Siberian MAGs. A maximum likelihood phylogenetic tree of 139 concatenated conserved genes for 8 Siberian MAGs and 230 total reference MAGs and genomes that were used in the comparative analysis is shown. No reference MAGs or genomes from these groups were available from permafrost. The tree was visualized with iTOL. A full list of shared genes can be found in Supplemental Data File S6 in the supplemental material.
Description of major taxonomic groups. (i) Thermoprofundales.
Two MAGs, 1 from 7.2 m (96% complete, with 6% contamination) and 1 from 14.8 m (94% complete, with 2% contamination) grouped with Thermoprofundales genomes (Fig. 2; also see Fig. S2). Thermoprofundales is an order (37) of archaea within the phylum Euryarchaeota and the class Thermoplasmata (see Table S2). Thermoplasmata has been redesignated as the phylum Thermoplasmatota, based on GTDB taxonomy (38), and the class Izemarchaea (39). It was previously called DHVEG, which stands for deep-sea hydrothermal vent euryarchaeotal group (40), and marine benthic group D (41). Thermoprofundales are uncultured archaea that are predominantly found in marine subseafloor seeps worldwide (37, 41–43) and were recently detected in anoxic freshwater sediments (44). All 12,042 of the Thermoprofundales 16S rRNA gene sequences in the SILVA r138 database were from anoxic marine sediments. To our knowledge, only 9 Thermoprofundales genomes are publicly available (see Supplemental Data File S1), originating from sediment from the White Oak River estuary in North Carolina (5 MAGs) and the Aarhus Bay in Denmark (4 SAGs).
(ii) Atribacteria.
Atribacteria MAGs were retrieved from 7.2 m (90% complete, with 0% contamination), 14.1 m (81% complete, with 3% contamination), and 14.8 m (89% complete, with 0% contamination) (Fig. 2; also see Fig. S2). The phylum Atribacteria was previously called JS1/OP10 (45) and has been proposed to be renamed through GTDB taxonomy as Caldatribacteriota (38). The 66 nonpermafrost Atribacteria MAGs (see Supplemental Data File S2) originated across a variety of locations, including marine sediment from Aarhus Bay, Denmark; Great Boiling Springs, Nevada; soil from an acetate-fed aquifer in Rifle, Colorado; Green River sediment in Utah; Sakinaw Lake in Canada; a terephthalate-degrading reactor biofilm in Mexico; Etoliko Lagoon in Greece; and sediment from Lake Baikal in Russia. The 455 Atribacteria 16S rRNA genes in the SILVA r138 database are typically from temperate soil, deep marine sediment, and bioreactors. To our knowledge, neither 16S rRNA genes nor MAGs from this group from permafrost environments have been reported previously.
(iii) Chloroflexi.
One MAG from 20 m (81% complete, with 10% contamination) grouped within the Chloroflexi order Anaerolineales (Fig. 2; also see Fig. S2). Anaerolineales is abundant in many different types of environments; a meta-analysis of 1,504 metagenomes showed that Chloroflexi DNA sequences were in the top 10 most abundant groups in metagenomes from marine sediment, host-associated environments, hypersaline environments, freshwater, hot springs, and terrestrial subsurface (34). Forty-two nonpermafrost Chloroflexi genomes (see Supplemental Data File S3) were chosen to represent the phylogenetic and environmental diversity available among the nonpermafrost genomes, from deep Pacific Ocean basalt-hosted subsurface hydrothermal fluid; White Oak River estuary sediment; Utah Grand County groundwater; a biological phosphorous bioreactor; marine samples from the TARA04 Ocean Project; groundwater from an acetate-fed aquifer in Rifle, Colorado; freshwater from Green River, Utah; sediment from Australia; and marine aquatic samples from unreported locations.
(iv) Actinobacteria.
One MAG (MAG_01_20m; 96% complete, with 8% contamination) was in the Actinobacteria phylum, with the family Dermatophilaceae and genus Cutibacterium, which is commonly cultured from human skin (46). A complete nuclease gene from this MAG had 100% similarity to Cutibacterium acnes in a BLASTN search, and the entire MAG had 99.72% average nucleotide identity (ANI) (47) to Propionibacterium, Cutibacterium sp. strain KPL2009, from the Human Genome Project (46). The other Siberian permafrost Actinobacteria MAGs were not closely related to the skin microbe (see Table S2) but, since they had lower completeness and higher contamination levels, their phylogenetic relatedness could not be accurately determined.
(v) Aminicenantes.
One MAG (MAG_02_14.8m) with 94% completeness and 3% contamination came from the 14.8-m sample and grouped with Aminicenantes genomes on a maximum likelihood tree based on concatenated conserved genes (Fig. 2; also see Fig. S2). Aminicenantes was previously called OP8 and has been suggested to be in the Acidobacteriota phylum based on the GTDB reclassification (45). The Aminicenantes MAG was compared to 54 nonpermafrost Aminicenantes genomes (see Supplemental Data File S5), chosen so that each available study site was represented, i.e., Etoliko Lagoon in Greece; Sakinaw Lake in Canada; soil in Rifle, Colorado; hydrothermal fluid from the Juan de Fuca Ridge flank in the Pacific Ocean; and marine sediment from Baltimore, Maryland.
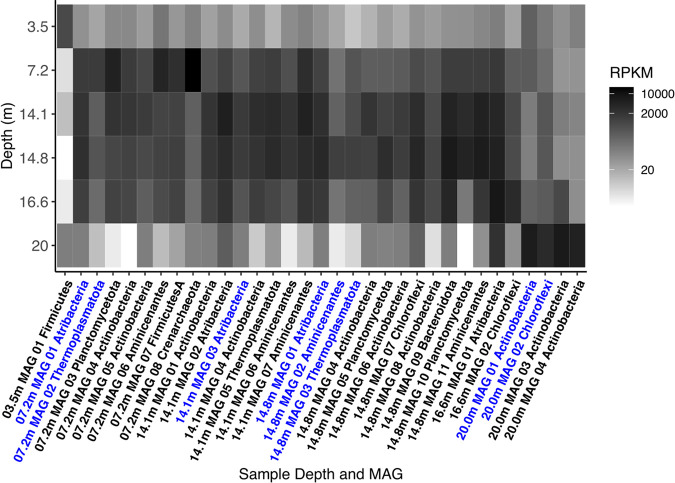
Aminicenantes has been found in a variety of marine and terrestrial environments but only up to 10.2% maximum relative abundance (48). Aminicenantes represented a slightly higher percentage of mapped reads in most of our permafrost metagenomes (3.5 m, 12.77%; 7.2 m, 15.77%; 14.1 m, 16.81%; 14.8 m, 14.7%; 16.6 m, 5.6%; 20 m, 0.14%) (see Fig. 5; also see Supplemental Data File S7). This suggests that Aminicenantes may be at higher relative abundance in ancient permafrost, or the discrepancy may be due to primer bias (49) during PCR amplification in the published studies.
FIG 5.
Metagenomic read coverage for each MAG at each sample depth. Abundance is reported in RPKM. MAGs in blue text are MAGs analyzed in this study (≥80% completeness and <10% contamination); MAGs in black text came from these samples but were not analyzed extensively in this study (although they are available under BioProject accession number PRJNA596250). Calculations can be found in Supplemental Data File S7 in the supplemental material.
Comparison of Siberian MAGs and nonpermafrost genomes.
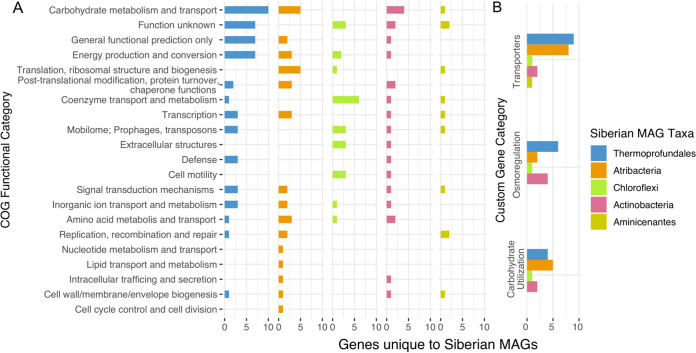
Genes that were present in Siberian MAGs that were rare or absent in the nonpermafrost MAGs/SAGs included 21 major Clusters of Orthologous Groups (COG) categories (Fig. 3A), excluding COG categories chromatin structure and dynamics, RNA processing, secondary metabolites, and cytoskeleton. The COG categories with the largest numbers of genes unique to the Siberian MAGs were carbohydrate metabolism and transport, energy production and conversion, function unknown, and general functional prediction only.
FIG 3.
Genes unique to the Siberian MAGs. (A) Numbers of genes in each COG category that are present in the Siberian MAG groups but absent in the reference genomes of the same groups (except for Atribacteria, for which the genes were in less than one-third of the reference genomes). Exact names of the genes can be found in the main text and in Supplemental Data Files S1 to S5 in the supplemental material. (B) Functional categories encompassing genes from various other COG categories that are known to have multiple functions. The full list of genes can be found in Supplemental Data File S6.
The Siberian Thermoprofundales MAGs had the most genes that were absent in nonpermafrost genomes (47 genes) (see Supplemental Data File S6). The greatest numbers were carbohydrate metabolism and transport, with four components of an ABC-type sugar transport system, ABC-type glycerol phosphate systems, cellobiose phosphorylase, and pyruvate kinase, energy production and conversion, with FoF1-type ATP synthase and isocitrate dehydrogenase, and inorganic ion transport and metabolism, with two components of a Ca2+/H+ antiporter and a Na+/H+ antiporter-related arsenite permease (Fig. 3A). The Siberian Thermoprofundales MAGs also had more genes per genome related to the carbohydrate metabolism and transport (2.5×) and defense (3×) COG categories than the nonpermafrost genomes (see Fig. S3).
There were no genes that were present in all Siberian Atribacteria MAGs that were also absent from all of the nonpermafrost MAGs. Therefore, we investigated the genes in all Siberian Atribacteria MAGs found in only 10% to 33% of the nonpermafrost Atribacteria MAGs (see Supplemental Data File S6). Of those 32 genes, 5 were in the carbohydrate metabolism and transport COG category (Fig. 3; also see Supplemental Data File S2). The 7 genes with the lowest representation (11% to 23%) in nonpermafrost Atribacteria were pyruvate/oxaloacetate carboxyltransferase, trehalose-6-phosphate synthase/hydroxylamine reductase (hybrid-cluster protein), cellobiose phosphorylase, nucleoside diphosphate kinase, cation transport ATPase, and ribosomal protein L32 (see Supplemental Data Files S2 and S6).
Thirty-three genes were present in at least 1 of 3 Siberian Atribacteria MAGs but were absent in all nonpermafrost Atribacteria. Six of these were from the energy production and conversion COG category, i.e., four subunits of NADH:ubiquinone oxidoreductase and two subunits of a heme/copper type cytochrome/quinol oxidase. Three Siberian Atribacteria MAG genes were in the carbohydrate metabolism and transport COG category, including mevalonate-3-phosphate 5-kinase, aryl-phospho-β-d-glucosidase, and a phosphoglycerate mutase. The three permafrost Atribacteria MAGs had twice the number of genes per genome related to the energy production and conversion, inorganic ion transport and metabolism, replication, recombination, and repair, cell wall/membrane/envelope biogenesis, and lipid transport and metabolism COG categories as did nonpermafrost genomes (see Fig. S3).
The Siberian Chloroflexi MAG had 20 genes that were absent in all nonpermafrost genomes (Fig. 3; also see Supplemental Data File S6), including a Na+/alanine symporter, fumarate hydratase class II, and nitric oxide reductase large subunit. A complete methyl-coenzyme M reductase contig (subunits α, β, and γ and operon protein D; 3,483 amino acids long) binned in the Chloroflexi MAG, but this contig had no other genes on it (see Supplemental Data File S3). The COG categories with the most genes that were only found in the Siberian Chloroflexi MAG, relative to nonpermafrost MAGs, were coenzyme transport and metabolism, extracellular structures, and cell motility (Fig. 3; also see Supplemental Data File S3). The Siberian Chloroflexi MAG generally had 1 to 1.5× genes per genome in each COG category, compared with the 42 nonpermafrost Chloroflexi (see Fig. S3).
Eleven genes were unique to the Siberian Aminicenantes MAG, relative to nonpermafrost Aminicenantes MAGs, including 2 genes in the replication, recombination, and repair COG category and 2 in the function unknown category (see Supplemental Data File S6). The Aminicenantes MAG had only 1 gene (endo-1,4-β-mannosidase) unique to the carbohydrate metabolism and transport COG category, but it had 2.7× more genes per genome than in the nonpermafrost genomes (see Fig. S3). Even though there were few genes that were unique to the Aminicenantes Siberian MAG, it did have more genes per genome in the COG categories than did the nonpermafrost genomes (see Fig. S3).
Since many of the genes that were unique to the Siberian MAGs were involved in transport, osmoregulation, and carbohydrate utilization but spanned multiple COG categories, we made custom groups for these three functions to examine them together (Fig. 3B; also see Supplemental Data File S6 and Table S3). The transporters group had the most genes that were unique to the Siberian MAGs, led by Thermoprofundales and Atribacteria, with 9 and 8 genes, respectively. Atribacteria and Thermoprofundales also had the most unique genes in the carbohydrate utilization group, with 5 and 4 genes, respectively. Carbohydrate metabolism and transport had the greatest numbers of genes, 4 of which were ABC-type sugar/transport system genes (Fig. 3B). The average numbers of genes within each COG category in the Actinobacteria MAG were less than or equal to the number of genes per genome in the nonpermafrost genomes (see Fig. S3). Thermoprofundales had the most unique genes in the osmoregulation group too, and all taxa except Aminicenantes had a Na+ symporter for osmoregulation. The only gene from Aminicenantes that fell within these three groups was a succinate-acetate transporter.
Comparison of Siberian MAGs to each other.
While no genes had identical annotations among all 8 of the Siberian MAGs (Fig. 4), each MAG had some version of three Na+/H+ antiporters, NhaD, MnhC, and MnhG, as well as a biotin transporter. Also, many genes had similar annotations in different unions (see Supplemental Data File S6). Forty genes were present in 7 Siberian permafrost MAGs that were not in the Actinobacteria MAG (Fig. 4; also see Supplemental Data File S6); these included pyruvate-formate lyase-activating enzyme, formate hydrogenlyase, carbamoyltransferase, Na+/H+ antiporter, M28 family peptidase, predicted nucleotidyltransferase component of viral defense system, protein-l-isoaspartate O-methyltransferase, and cellobiose phosphorylase.
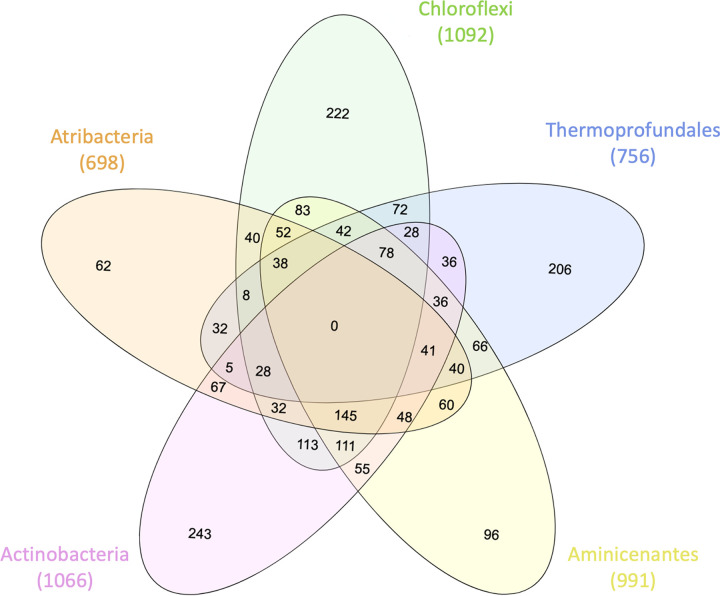
FIG 4.
Numbers of unique genes shared by different Siberian MAG groups. The numbers in parentheses are total numbers of genes present in all of the MAGs for that group. The complete list of genes in each MAG group and each union can be found in Supplemental Data File S6 in the supplemental material.
The 6 bacterial Siberian MAGs shared 145 gene annotations with each other, which was more than they did with the Thermoprofundales, the only archaeal group (Fig. 4). The Siberian Actinobacteria had the largest number of genes (243 genes) that were uniquely annotated, compared with the other 7 Siberian MAGs (Fig. 4). Some of these genes included H+/Cl−, Na+/H+, and K+/H+ antiporters with C-terminal TrkAC and CorC domains (see Supplemental Data File S6). The Siberian Thermoprofundales MAGs contained 206 genes that were uniquely annotated, relative to the other Siberian MAGs (Fig. 4; also see Supplemental Data File S6); these included acetate kinase, isocitrate dehydrogenase, Ca2+/H+ antiporter, HSP90 ATPase, and a Na+/proline symporter (see Supplemental Data File S4). Other genes specific to the Siberian Thermoprofundales MAGs included 21 different archaea-type regulatory proteins and an uncharacterized conserved protein related to pyruvate-formate lyase-activating enzyme, many archaea-type synthetases, and many genes with general or predicted functions (see Fig. S6A to C and Supplemental Data File S3).
Membrane stability genes were shared between two or more groups. Multiple genes and enzyme kinases for the mevalonate pathway were found in the 2 Thermoprofundales MAGs, 1 Chloroflexi MAG, and 1 Atribacteria MAG (see Supplemental Data File S6). Apart from Thermoprofundales, the other Siberian permafrost MAGs shared membrane-associated genes, such as peptidoglycan biosynthesis protein MviN/MurJ, bacterial cell division protein FtsW, and energy-coupling factor transporter ATP-binding protein EcfA2 (Fig. 4; also see Supplemental Data File S6). The Aminicenantes MAG had 96 unique genes that were not present in any other Siberian MAG (Fig. 4). This MAG also contained 111 genes similar to genes annotated in Actinobacteria and Chloroflexi MAGs, which are two other widely found soil and permafrost microbe groups (50, 51).
DISCUSSION
Comparison of the Siberian MAG communities to those found in other permafrost studies.
Apart from the 3.5-m depth, which was an ice wedge with few sediment inclusions, and the 17.2-m depth, which did not produce a metagenome for unknown reasons, DNA was present in sufficient quantities to produce MAGs throughout the 20,000- to 1,000,000-year-old permafrost. The identities of the MAGs from ancient Siberian permafrost differed greatly from those found in active layer permafrost environments. In the active layer of the Stordalen Mire, Woodcroft et al. found 1,434 MAGs (>70% completeness and <10% contamination), including 27% Actinobacteria, 27% Aminicenantes, 14% Proteobacteria, 4% Chloroflexi, and 6% Euryarchaeota (52). Our ancient Siberian permafrost contained Chloroflexi, Actinobacteria, and Aminicenantes members but also had MAGs from groups that were absent in the Stordalen Mire active layer, such as Atribacteria and Thermoprofundales. This suggests that there could be large differences in microbial community compositions between seasonally thawed active layers and ancient permafrost. Atribacteria and Thermoprofundales were absent in Alaskan ancient permafrost from 19,000 to 33,000 years ago (6, 7). The ancient Alaskan permafrost study showed an increase of Firmicutes 16S rRNA genes and a decrease of Actinobacteria with permafrost age (6). A similar trend was observed in the borehole adjacent to this study, AL3-15, called AL1-15 (4), where Firmicutes and Proteobacteria dominated 16S rRNA gene amplicon libraries. These taxa, Firmicutes, Planctomycetota, Crenarchaeota, and Bacteroidota, were found in Siberian MAGs but were not analyzed due to insufficient quality. This discrepancy may be due to the differential biases of amplicon-based libraries and metagenome-based libraries in low-biomass samples (34, 49). The presence of groups such as Atribacteria, Thermoprofundales, and Aminicenantes, which are not commonly found in ancient permafrost or freshwater environments but are more commonly found in marine environments, suggest that these ancient freshwater Siberian deposits may contain organisms adapted to saline conditions.
Genes common in Siberian permafrost MAGs indicate adaptation to a cold, saline, low-energy environment. (i) Saline regulatory genes.
Many of the genes shared among two or more groups of Siberian MAGs were involved in the transport of small molecules like sodium and carbohydrates. Having a variety of genes for ion and salt transportation could mean that these MAGs resemble organisms that are adapted to function in high-ionic-strength environments, like brines. All groups shared a similar annotation for three Na+/H+ antiporters and osmoregulators (NhaD, MnhC, and MnhG), as well as a biotin transporter. All Siberian MAGs had unique genes in the transporter category, compared to the nonpermafrost genomes (Fig. 3B).
(ii) Trehalose.
Many of the genes that were specific to Siberian MAGs and were not present in nonpermafrost outgroup genomes were involved in transport, osmoregulation, and carbohydrate utilization. The prominence of these categories suggests the Siberian MAGs had unique adaptations to interact with their environment, including dealing with osmotic stress and using carbohydrates as energy sources. Trehalose-6-phosphate synthase was found in the Siberian MAGs and lacked homologues in nonpermafrost genomes. It was in both Thermoprofundales MAGs (22% of the nonpermafrost outgroup), all 3 Atribacteria MAGs (13% of the nonpermafrost outgroup), the Aminicenantes MAG (2% of the nonpermafrost outgroup), and the Actinobacteria MAG (59% of the nonpermafrost outgroup). The Chloroflexi MAG had a gene annotated as trehalose and maltose hydrolase (possible phosphorylase). Trehalose-6-phosphate synthase has been suggested to help deep subsurface inhabitants maintain a low-energy state in marine sediments by producing trehalose, a disaccharide that prevents aggregation of degraded proteins, protects against osmotic stress, and increases cellular longevity (53, 54). Trehalose is a cryoprotectant and stabilizes cellular membranes and DNA at low temperatures and high osmolarity to increase cell longevity and to slow replication rates (55–57). Trehalose synthase has also been found in metagenomes from Antarctic soil and sediment samples (58).
(iii) Mevalonate pathway.
Multiple genes and enzyme kinases for the mevalonate pathway were found in both Thermoprofundales MAGs, the Chloroflexi MAG, and the Atribacteria MAG. The mevalonate pathway uses acetic acid for the biosynthesis of isoprenoids, which have been shown to stabilize membranes, increasing the organism’s survival at low temperatures (59–61), and have been found in bacteria (62, 63). A functional mevalonate pathway has also been identified in a Methanosarcina MAG from a deep Antarctic permafrost enrichment (64). The acetate ions for the mevalonate pathway could be from fermentation of the surrounding organic matter from the cellobiose, arabinoxylans, and proteinaceous peptides. Genes to ferment these substrates were also identified in the Siberian MAGs. Additionally, Psychrobacter arcticus 273-4, cultured from Siberian permafrost, has phenotypic evidence of an acetate-based metabolism (65, 66), and all 8 MAGs from this study had genes indicating mixed acid-acetate metabolism (see Fig. S6 in the supplemental material).
(iv) Cellobiose phosphorylase and carbamate kinase.
Metagenomes of thawed permafrost have shown increases in degradation of cellulose (67). Genes for cellobiose phosphorylase, which is an enzyme that aids in the degradation of cellulose as a carbon source (68–70), were identified in all of our Siberian MAGs except the Actinobacteria MAG. Cellobiose phosphorylase converts cellobiose into glucose and glucose-1-phosphate, offering an energetic advantage in anoxic environments. Cellobiose phosphorylase suggests that permafrost organisms may be able to slowly degrade available carbon substrates (43, 44). The Siberian Atribacteria MAGs had other genes indicative of adaptation to low-energy environments. Further, two Siberian Atribacteria MAGs contained aconitase A, α-l-arabinofuranosidase, and α-amylase/α-mannosidase, which were present in only 6%, 15%, and 19% of nonpermafrost genomes, respectively. These are important in cellulose degradation and carbohydrate metabolism (45). Carbon starvation protein (CstA) was in two Siberian Atribacteria MAGs and 30% of nonpermafrost Atribacteria genomes. This protein enhances peptide catabolism during carbon starvation (44). Carbamate kinase was in two Siberian Atribacteria MAGs and 25% of nonpermafrost Atribacteria genomes, suggesting that the permafrost Atribacteria can conserve energy by creating ADP and carbamoyl phosphate from the combination of ATP, CO2, and NH3 (46). This would benefit organisms in a low-energy environment, because this enzyme has roles in purine, glutamate, proline, and nitrogen metabolism (47).
(v) Biotic methane.
Through their genome-centric analysis of functional genes in active layer soils, Woodcroft et al. found genetic evidence for organic matter decomposition into CO2 and CH4 (52). Studies of 33,000-year-old permafrost (6) and recently thawed permafrost (67) from Alaska also generated a variety of methanogens. A gene for methyl coenzyme M reductase (mcr) was identified in another study’s metagenome from a 30,696-year-old Siberian permafrost sample from where methane was measured (8). Additionally, biogenic methane was detected in one of two boreholes in another study of Kolyma-Indigirka Lowland permafrost, while methanogens were distributed throughout both boreholes (71). Other permafrosts up to 33,000 years old, where methane was absent, did not yield 16S rRNA genes from known methanogenic groups (7) or any mcr genes (8). There was no evidence of methane metabolism in any of the Siberian Thermoprofundales MAGs, unlike their nonpermafrost counterparts. We found four subunits of the mcr gene on a single contig from the 20-m metagenome sample in the Chloroflexi MAG, suggesting that the methane observed there could have been biotically produced (Fig. 1B). The gene’s top BLASTP hit was an mcr gene found in the “Candidatus Methanoperedens ferrireducens” archeaon from an Australian marine sediment incubation (72). Since this MAG did not contain a full methanogenic pathway and no bacteria were previously shown to contain mcr, it is likely that this mcr gene was part of the 10% contamination.
(vi) DNA scavenging.
Ureidoglycolate dehydrogenase was present in 2 Siberian Atribacteria MAGs and 48% of nonpermafrost Atribacteria genomes. This is involved in the degradation of allantoin, a DNA decomposition product, and allows the use of DNA as a nitrogen source. Other Atribacteria strains have been suggested to use an allantoin degradation pathway to access detrital DNA as an energy source under extreme starvation conditions in deep subsurface marine sediment (38). The presence of DNA-foraging enzymes like those involved in allantoin degradation (54) and cellular debris recycling (30) may be another key to survival in low-energy environments. The Siberian Aminicenantes MAG had a protein-degrading metabolic pathway similar to that inferred for other Aminicenantes (54), since they all had the same types of extracellular peptidases, such as dipeptidase, dipeptidyl aminopeptidase, acylaminoacyl peptidase, RecA-mediated SOS response autopeptidase, d-alanyl-d-alanine carboxypeptidase, and cyanophycinase exopeptidases (see Fig. S6 and Supplemental Data File S5). Atribacteria, Thermoprofundales, and Aminicenantes from subseafloor sediments also have been found to have these extracellular enzymes (73). Additionally, Thermoprofundales have been suggested to ferment proteinaceous organic matter, since they have a high genomic content of extracellular peptidases whose activity could be measured in the bulk sediment (42). Therefore, the presence of genes for osmotic, cold, and energetic stress tolerance in our ancient Siberian permafrost MAGs suggests that they may be adapted to this environment.
Conclusions.
The Siberian permafrost MAGs belong to taxonomic groups that are commonly found in low-energy, anoxic, saline habitats. Finding MAGs related to marine-associated microbes like Thermoprofundales, Aminicenantes, and Atribacteria further supports that these MAGs are adapted to increased salinity (74) due to freezing of freshwater sediments, since the Yedoma and Olyor deposits in this area of Siberia have never been inundated by seawater (12, 13, 16). The concentrations of chloride increased with depth (Fig. 1) up to 879.8 ppm (248 mM), which is hypersaline, compared to accepted baseline tested agriculture soils, where 50 ppm is considered excessive (75). The high prevalence of Na+/H+ and Ca2+/alanine antiporters, mechanosensitive channels, osmoprotectants, and other ion transporters (Fig. 3B), relative to the nonpermafrost genomes, may suggest adaptation to the saline conditions observed in our geochemistry (Fig. 1), since these genes are common in halotolerant organisms (18, 76). Additionally, microbial cultures from Siberian permafrost have osmosis-specific adaptations and activity in −15°C permafrost (77, 78). Genes for osmotic stress tolerance have also been found within Antarctic (30) and Canadian (77) permafrost metagenomes.
We speculate that, upon burial and freezing, these permafrost microbes became the dominant organisms because those not adapted to the high-salinity liquid water films in permafrost died. If these organisms are adapted to surviving in saline brine films, then it is unlikely that they will retain their dominance in these microbial communities upon permafrost thawing due to climate change. As the permafrost thaws, these native brine-adapted microbial communities will likely be replaced by freshwater-adapted organisms when surface waters penetrate the newly thawed permafrost.
Further evidence to support extant life in this ancient Siberian permafrost is that the DNA was intact enough to produce reads that assembled into MAGs, with less than 9% of the assemblies remaining unbinned (Table 1) and high read recruitment in the MAGs (Fig. 5). Each MAG had some DNA read recruitment from other sample depths, which suggests that, with even greater sequencing depth, a more diverse population of MAGs might have been observed.
Finding evidence for these organisms in ancient 1,000,000-year-old permafrost samples furthers the idea of a microbial community being able to persist in ancient permafrost within the high-salinity brine films. The Siberian permafrost MAGs analyzed in this study demonstrate how individual organisms can be adapted to their environment, relative to members of the same taxonomic groups in nonpermafrost environments. Although it is impossible to have absolute certainty that these organisms were alive at the time of sampling, their genome functions and unique functionality, compared to nonpermafrost genomes (Fig. 3 and 4), coupled with the geochemistry of the environment (Fig. 1), suggest adaptations to the liquid brine films in ancient permafrost.
MATERIALS AND METHODS
Field sampling details.
Permafrost cores were taken from the northeastern Kolyma-Indigirka Lowland region in Siberia, Russia (69°20.376N, 154°59.787E), at the end of July 2015. A 20-m-long core (AL3-15) was retrieved in a similar fashion as its sibling core (AL1-15), which was taken at the foothill in the same location, as described previously (4). The temperature was measured inside the borehole at the end of each sampling day, to allow the borehole to calibrate back to the in situ temperature, using an Onset HOBO data logger. The geological suites changed from Yedoma to Olyor depositional characteristics near 7.7 m below the surface.
Environmental characteristics.
Permafrost samples were analyzed for pH, methane levels, and total carbon concentrations. pH was measured with a benchtop рН meter (SevenEasy pH meter; Mettler Toledo). Conductivity was measured using a conductometer (Ekspert-002; Soil Services, Russia). To measure ions in the permafrost, pore water was extracted from 100 g of permafrost following procedures described by Van Reeuwijk (79). Porewater analyses were performed following the standards of the Russian Federation GOST-R Certificate. Methane was liberated from the frozen permafrost soon after sample retrieval in the field, as described previously (11), using a headspace method (80). Concentrations of the gas were analyzed on a GC-mini-2 gas chromatograph (Shimadzu, Tokyo, Japan) with a hydrogen flame ionization detector and argon as a carrier gas. The 14C age of inorganic and organic carbon from three subsamples of the upper layers (2.9 to 3, 3.5, and 5.6 m) was determined at the Arizona Accelerator Mass Spectrometry Laboratory according to the following procedures. The inorganic carbon was released by acidifying the bulk material using phosphoric acid (under vacuum) and collecting the CO2. The residue was dried and combusted at 400°C to release a relatively “younger” organic carbon fraction (81) (i.e., microbial cell material) from the permafrost sediment. The resulting residue was further combusted at 800°C to oxidize any tightly bound carbon fraction that was potentially associated with clay particles in the sediments. This presumptive “older” carbon fraction could be inherited photosynthate or detrital carbon through geological time. An additional feature of this gas collection is the ability to determine the δ13C of the sample, compared to Pee Dee Belemnite.
Metagenomic processing and analysis.
Seven subsampled depths (3.5, 7.2, 14.1, 14.8, 16.6, 17.2, and 20 m below the surface), spanning ∼20,000 to 1,000,000 years of age, were used for metagenome sequencing. DNA was extracted from approximately 5 g of permafrost using 10 technical replicate extractions with the DNeasy PowerSoil kit (Qiagen, Germantown, MD, USA) with 0.5 g of sample in each extraction. The 10 extractions per sample were pooled, and DNA was precipitated with ethanol and resuspended in 50 μl molecular-grade water. DNA concentrations ranged from 2.9 to 29.2 ng/μl. Metagenomic libraries were prepared using the Nextera XT DNA library preparation kit (Illumina, USA), following the manufacturer’s instructions, and sequenced at the University of Tennessee Genomics Core on an Illumina MiSeq system with the 2 × 250-bp protocol.
A diagram describing the following bioinformatic methods can be found in Fig. S7 in the supplemental material. The raw metagenomic files were analyzed using KBase.us (82), and all links are provided in “Data availability” (registration with https://www.kbase.us is free and is required to access the narrative). The sample depths were kept separate for single-sample assembling and metagenomic binning, in order to avoid combining contigs from different sample depths into the same MAG. Trimmomatic v0.36 (83) was used for poor-quality data trimming, removal of barcodes, and creation of a paired-end assembly with the following parameters: leading_min_quality, 3; min_length, 36; sliding_window, 28; sliding_window_size, 10; trailing_min_quality, 3. MetaSPAdes v3.11.1 (84) was used for metagenomic assembly with minimum contig length of 2,000 bp. MaxBin2 v2.2.4 (85) with default settings was used to place contigs into metagenomic bins with the following parameters: prob_threshold, 0.8; min_contig_length, 2000. Prodigal (86) was used for gene finding and DIAMOND (87) for gene translation within anvi’o v6 esther (88) using the parameter anvi-run-ncbi-cogs in the anvi-gen-contigs-database command. GTDB-Tk v1.4.0 was used for initial taxonomic classification of the created bins (38) with the classify_wf command with a minimum alignment percent (min_perc_aa) of 10. A maximum likelihood phylogenetic tree was made in CLC Workbench v6 using 139 shared conserved genes gathered by using the anvi-get-sequences-for-hmm-hits command with -return-best-hit, -get-aa-sequences, and -concatenate in anvi’o (see Supplemental Data File S6). MAGs were quality assessed with CheckM v1.0.18 (89) for completeness and contamination percentages. Bowtie2 v2.3.3 (90) with default settings was used to map the paired metagenomes against each of the MAGs to determine the presence of MAGs at other depths. This mapping also helped to find similarities and difference of the MAGs in the metagenomic samples. Mapping results were standardized by calculating reads per kilobase per million mapped reads (RPKM), which accounts for the gene length and each sample’s library size (see Supplemental Data File S7). Nonpermafrost genomes were downloaded from the JGI IMG database for comparison of genetic content (91). The five taxonomic groups to which the MAGs belonged were Actinobacteria (n = 1, o = 57), Aminicenantes (n = 1, o = 54), Thermoprofundales (n = 2, o = 9), Atribacteria (n = 3, o = 66), and Chloroflexi (n = 1, o =42), where n is the number of permafrost MAGs and o is the number of nonpermafrost genomes downloaded from the JGI IMG database.
The program anvi’o (88) was used to compare the annotated genes for similarity between genomes using DIAMOND (87) and the NCBI database with an MCL inflation score of 2 within the groups. Comparisons were made between two experimental groups: (i) MAGs from this study against downloaded genomes (MAGs and SAGs) from the same phylogenetic groups and (ii) all of the Siberian MAG groups against each other. These comparisons were made on the basis of gene annotations and, more broadly, COG categories (92). To investigate the KEGG pathways and metabolic pathways of the annotated Siberian MAGs, the KEGG Orthology (KO) numbers from protein fasta files were compiled in GhostKoala and analyzed with KEGG-Decoder (93).
Cell visualization.
Cells were visualized by filtering from sonicated bulk soil with Nanopore 0.2-μm filters and staining with SYBR gold and DAPI (94) and were imaged under a Zeiss Imager M2 with ×100 magnification (see Fig. S4). A Guava easyCyte 12HT benchtop cytometer was also used with SYBR green cell dye to examine cells. Only cells frozen at −80°C were available, without cell preservation in the field; therefore, many of the cells might have lysed. Cell counting and live/dead staining methods did not yield quantifiable results. Additionally, SAGs were made from sample depths of 5.6 m and 7.2 m at the Bigelow Single Cell Genomics Center (East Boothbay, ME, USA) (95). Ten single cells with the best whole-genome amplification (see Fig. S5) with Bigelow’s whole-genome amplification and multiple displacement amplification methods were sequenced using an Illumina MiSeq 2 × 250-bp protocol at the University of Tennessee (Knoxville, TN).
Data availability.
The 6 assembled metagenome files and resulting MAGs are available in the NCBI database under BioProject accession number PRJNA596250. Sample methods are available to view, as follows: 3.5 m, https://narrative.kbase.us/narrative/27695; 7.2 m, https://narrative.kbase.us/narrative/27731; 14.1 m, https://narrative.kbase.us/narrative/27724; 14.8 m, https://narrative.kbase.us/narrative/27725; 16.6 m, https://narrative.kbase.us/narrative/37690; 17.2 m, https://narrative.kbase.us/narrative/37878; 20 m, https://narrative.kbase.us/narrative/37504.
ACKNOWLEDGMENTS
We thank Andrey Abramov, Nikita Demidov, Denis Shmelev, and Victor Sorokovikov from the Institute of Physicochemical and Biological Problems of Soil Science (Pushchino, Russia) for cooperation in the collection of permafrost samples. Peibo Li and Mackenzie Thorton helped extensively with microscopy. Nicholas T. Sipes helped with Python scripts; all in-house scripts are available at https://github.com/sipesk/SiberianMAGsPaper.
This study was supported by the National Science Foundation (grants DEB-1442262 and DEB-1460058), the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomic Science Program (grant DE-SC0020369), the Russian Government (assignment AAAA-A18-118013190181-6), and the Russian Foundation for Basic Research (grant 19-29-05003-mk).
We declare no conflicts of interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Karen G. Lloyd, Email: klloyd@utk.edu.
Robert M. Kelly, North Carolina State University
REFERENCES
- 1.Morgalev YN, Lushchaeva IV, Morgaleva TG, Kolesnichenko LG, Loiko SV, Krickov IV, Lim A, Raudina TV, Volkova II, Shirokova LS, Morgalev SY, Vorobyev SN, Kirpotin SN, Pokrovsky OS. 2017. Bacteria primarily metabolize at the active layer/permafrost border in the peat core from a permafrost region in western Siberia. Polar Biol 40:1645–1659. 10.1007/s00300-017-2088-1. [DOI] [Google Scholar]
- 2.Park H, Kim Y, Kimball JS. 2016. Widespread permafrost vulnerability and soil active layer increases over the high northern latitudes inferred from satellite remote sensing and process model assessments. Remote Sens Environ 175:349–358. 10.1016/j.rse.2015.12.046. [DOI] [Google Scholar]
- 3.Abramov A, Vishnivetskaya T, Rivkina E. 2021. Are permafrost microorganisms as old as permafrost? FEMS Microbiol Ecol 97:fiaa260. 10.1093/femsec/fiaa260. [DOI] [PubMed] [Google Scholar]
- 4.Liang R, Lau M, Vishnivetskaya T, Lloyd KG, Wang W, Wiggins J, Miller J, Pfiffner S, Rivkina EM, Onstott TC. 2019. Predominance of anaerobic, spore-forming bacteria in metabolically active microbial communities from ancient Siberian permafrost. Appl Environ Microbiol 85:e00560-19. 10.1128/AEM.00560-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilichinsky D, Rivkina E, Bakermans C, Shcherbakova V, Petrovskaya L, Ozerskaya S, Ivanushkina N, Kochkina G, Laurinavichuis K, Pecheritsina S, Fattakhova R, Tiedje JM. 2005. Biodiversity of cryopegs in permafrost. FEMS Microbiol Ecol 53:117–128. 10.1016/j.femsec.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Mackelprang R, Burkert A, Haw M, Mahendrarajah T, Conaway CH, Douglas TA, Waldrop MP. 2017. Microbial survival strategies in ancient permafrost: insights from metagenomics. ISME J 11:2305–2318. 10.1038/ismej.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burkert A, Douglas TA, Waldrop MP, Mackelprang R. 2019. Changes in the active, dead, and dormant microbial community structure across a pleistocene permafrost chronosequence. Appl Environ Microbiol 85:e02646-18. 10.1128/AEM.02646-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivkina E, Petrovskaya L, Vishnivetskaya T, Krivushin K, Shmakova L, Tutukina M, Meyers A, Kondrashov F. 2016. Metagenomic analyses of the late Pleistocene permafrost: additional tools for reconstruction of environmental conditions. Biogeosciences 13:2207–2219. 10.5194/bg-13-2207-2016. [DOI] [Google Scholar]
- 9.Johnson SS, Hebsgaard MB, Christensen TR, Mastepanov M, Nielsen R, Munch K, Brand T, Gilbert MTP, Zuber MT, Bunce M, Rønn R, Gilichinsky D, Froese D, Willerslev E. 2007. Ancient bacteria show evidence of DNA repair. Proc Natl Acad Sci U S A 104:14401–14405. 10.1073/pnas.0706787104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Willerslev E, Hansen AJ, Poinar HN. 2004. Isolation of nucleic acids and cultures from fossil ice and permafrost. Trends Ecol Evol 19:141–147. 10.1016/j.tree.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Gilichinsky DAA, Wilson GSS, Friedmann EII, Mckay CPP, Sletten RSS, Rivkina EMM, Vishnivetskaya TAA, Erokhina LGG, Ivanushkina NEE, Kochkina GAA, Shcherbakova VA, Soina VSS, Spirina EVV, Vorobyova EAA, Fyodorov-Davydov DG, Hallet B, Ozerskaya SMM, Sorokovikov VAA, Laurinavichyus KSS, Shatilovich AVV, Chanton JPP, Ostroumov VEE, Tiedje JMM, Scherbakova VA, Soina VSS, Spirina EVV, Vorobyova EAA, Frodorov-Davydov DG, Hallet B, Ozerskaya SMM, Sorokovikov VAA, Laurinavichyus KSS, Shatilovich AVV, Chanton JPP, Ostroumov VEE, Tiedje JMM. 2007. Microbial populations in Antarctic permafrost: biodiversity, state, age, and implication for astrobiology. Astrobiology 7:275–311. 10.1089/ast.2006.0012. [DOI] [PubMed] [Google Scholar]
- 12.Veremeeva A, Gubin S. 2009. Modern tundra landscapes of the Kolyma Lowland and their evolution in the Holocene. Permafrost Periglac Process 20:399–406. 10.1002/ppp.674. [DOI] [Google Scholar]
- 13.Strauss J, Schirrmeister L, Grosse G, Fortier D, Hugelius G, Knoblauch C, Romanovsky V, Schädel C, Schneider von Deimling T, Schuur EAG, Shmelev D, Ulrich M, Veremeeva A. 2017. Deep Yedoma permafrost: a synthesis of depositional characteristics and carbon vulnerability. Earth-Sci Rev 172:75–86. 10.1016/j.earscirev.2017.07.007. [DOI] [Google Scholar]
- 14.Grosse G, Robinson JE, Bryant R, Taylor MD, Harper W, DeMasi A, Kyker-Snowman E, Veremeeva A, Schirrmeister L, Harden J. 2013. Distribution of late Pleistocene ice-rich syngenetic permafrost of the Yedoma Suite in east and central Siberia, Russia. Open-file report 2013-1078. U.S. Geological Survey, Reston, VA. 10.3133/ofr20131078. [DOI] [Google Scholar]
- 15.Soina VS, Vorobiova EA, Zvyagintsev DG, Gilichinsky DA. 1995. Preservation of cell structures in permafrost: a model for exobiology. Adv Space Res 15:237–242. 10.1016/S0273-1177(99)80090-8. [DOI] [PubMed] [Google Scholar]
- 16.Schirrmeister L, Kunitsky V, Grosse G, Wetterich S, Meyer H, Schwamborn G, Babiy O, Derevyagin A, Siegert C. 2011. Sedimentary characteristics and origin of the Late Pleistocene Ice Complex on north-east Siberian Arctic coastal lowlands and islands: a review. Quat Int 241:3–25. 10.1016/j.quaint.2010.04.004. [DOI] [Google Scholar]
- 17.Lupachev AV, Gubin SV, Gerasimova MI. 2019. Problems of the cryogenic soils’ diagnostics in the recent Russian Soil Classification System. Eurasian Soil Sci 52:1170–1174. 10.1134/S1064229319080106. [DOI] [Google Scholar]
- 18.Gilichinsky D, Rivkina E, Shcherbakova V, Laurinavichuis K, Tiedje J. 2003. Supercooled water brines within permafrost: an unknown ecological niche for microorganisms: a model for astrobiology. Astrobiology 3:331–341. 10.1089/153110703769016424. [DOI] [PubMed] [Google Scholar]
- 19.Knelman JE, Legg TM, O’Neill SP, Washenberger CL, González A, Cleveland CC, Nemergut DR. 2012. Bacterial community structure and function change in association with colonizer plants during early primary succession in a glacier forefield. Soil Biol Biochem 46:172–180. 10.1016/j.soilbio.2011.12.001. [DOI] [Google Scholar]
- 20.Tolstikhin NI, Tolstikhin ON. 1974. Groundwater and surface water in the permafrost region. USSR Academy of Sciences, Siberian Branch, Novosibirsk, Russia. [Google Scholar]
- 21.Price PB, Sowers T. 2004. Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci U S A 101:4631–4636. 10.1073/pnas.0400522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soina VS, Mulyukin AL, Demkina EV, Vorobyova EA, El-Registan GI. 2004. The structure of resting bacterial populations in soil and subsoil permafrost. Astrobiology 4:345–358. 10.1089/ast.2004.4.345. [DOI] [PubMed] [Google Scholar]
- 23.Sheridan PP, Miteva VI, Brenchley JE. 2003. Phylogenetic analysis of anaerobic psychrophilic enrichment cultures obtained from a Greenland glacier ice core. Appl Environ Microbiol 69:2153–2160. 10.1128/AEM.69.4.2153-2160.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilichinsky DA, Soina VS, Petrova MA. 1993. Cryoprotective properties of water in the Earth cryolithosphere and its role in exobiology. Orig Life Evol Biosph 23:65–75. 10.1007/BF01581991. [DOI] [PubMed] [Google Scholar]
- 25.Anderson DM. 1967. Ice nucleation and the substrate-ice interface. Nature 216:563–566. 10.1038/216563a0. [DOI] [Google Scholar]
- 26.Ostroumov VE, Siegert C. 1996. Exobiological aspects of mass transfer in microzones of permafrost deposits. Adv Space Res 18:79–86. 10.1016/0273-1177(96)00002-6. [DOI] [Google Scholar]
- 27.McGrath J, Wagener SGD, Gilchinsky D. 1994. Cryobiological studies of ancient microorganisms isolated from the Siberian permafrost, p 48–67. In Gilchinsky D (ed), Viable microorganisms in permafrost. Russian Academy of Sciences, Pushchino, Russia. [Google Scholar]
- 28.Nicholson WL, Krivushin K, Gilichinsky D, Schuerger AC. 2013. Growth of Carnobacterium spp. from permafrost under low pressure, temperature, and anoxic atmosphere has implications for Earth microbes on Mars. Proc Natl Acad Sci U S A 110:666–671. 10.1073/pnas.1209793110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vishnivetskaya TA, Petrova MA, Urbance J, Ponder M, Moyer CL, Gilichinsky DA, Tiedje JM. 2006. Bacterial community in ancient Siberian permafrost as characterized by culture and culture-independent methods. Astrobiology 6:400–414. 10.1089/ast.2006.6.400. [DOI] [PubMed] [Google Scholar]
- 30.Goordial J, Davila A, Greer CW, Cannam R, DiRuggiero J, McKay CP, Whyte LG. 2017. Comparative activity and functional ecology of permafrost soils and lithic niches in a hyper-arid polar desert. Environ Microbiol 19:443–458. 10.1111/1462-2920.13353. [DOI] [PubMed] [Google Scholar]
- 31.Wilhelm RC, Niederberger TD, Greer C, Whyte LG. 2011. Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian High Arctic. Can J Microbiol 57:303–315. 10.1139/w11-004. [DOI] [PubMed] [Google Scholar]
- 32.Mitzscherling J, Winkel M, Winterfeld M, Horn F, Yang S, Grigoriev MN, Wagner D, Overduin PP, Liebner S. 2017. The development of permafrost bacterial communities under submarine conditions. J Geophys Res Biogeosci 122:1689–1704. 10.1002/2017JG003859. [DOI] [Google Scholar]
- 33.Vishnivetskaya T, Kathariou S, McGrath J, Gilichinsky D, Tiedje JM. 2000. Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments. Extremophiles 4:165–173. 10.1007/s007920070031. [DOI] [PubMed] [Google Scholar]
- 34.Lloyd KG, Steen AD, Ladau J, Yin J, Crosby L. 2018. Phylogenetically novel uncultured microbial cells dominate earth microbiomes. mSystems 3:e00055-18. 10.1128/mSystems.00055-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kraev G, Rivkina E, Vishnivetskaya T, Belonosov A, van Huissteden J, Kholodov A, Smirnov A, Kudryavtsev A, Teshebaeva K, Zamolodchikov D. 2019. Methane in gas shows from boreholes in epigenetic permafrost of Siberian Arctic. Geosciences 9:67. 10.3390/geosciences9020067. [DOI] [Google Scholar]
- 36.Buongiorno J, Turner S, Webster G, Asai M, Shumaker AK, Roy T, Weightman A, Schippers A, Lloyd KG. 2017. Interlaboratory quantification of bacteria and archaea in deeply buried sediments of the Baltic Sea (IODP Expedition 347). FEMS Microbiol Ecol 93:fix007. 10.1093/femsec/fix007. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Z, Liu Y, Lloyd KG, Pan J, Yang Y, Gu J-D, Li M. 2019. Genomic and transcriptomic insights into the ecology and metabolism of benthic archaeal cosmopolitan, Thermoprofundales (MBG-D archaea). ISME J 13:885–901. 10.1038/s41396-018-0321-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil PA, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 39.Adam PS, Borrel G, Brochier-Armanet C, Gribaldo S. 2017. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J 11:2407–2425. 10.1038/ismej.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takai K, Gamo T, Tsunogai U, Nakayama N, Hirayama H, Nealson KH, Horikoshi K. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269–282. 10.1007/s00792-004-0386-3. [DOI] [PubMed] [Google Scholar]
- 41.Vetriani C, Reysenbach AL, Doré J. 1998. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol Lett 161:83–88. 10.1111/j.1574-6968.1998.tb12932.x. [DOI] [PubMed] [Google Scholar]
- 42.Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD, Stepanauskas R, Richter M, Kleindienst S, Lenk S, Schramm A, Jørgensen BB, Jorgensen BB. 2013. Predominant archaea in marine sediments degrade detrital proteins. Nature 496:215–218. 10.1038/nature12033. [DOI] [PubMed] [Google Scholar]
- 43.Lauer A, Sørensen K, Teske A. 2016. Phylogenetic characterization of marine benthic archaea in organic-poor sediments of the Eastern Equatorial Pacific Ocean (ODP site 1225). Microorganisms 4:32. 10.3390/microorganisms4030032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borrel G, Lehours A-C, Crouzet O, Jézéquel D, Rockne K, Kulczak A, Duffaud E, Joblin K, Fonty G. 2012. Stratification of archaea in the deep sediments of a freshwater meromictic lake: vertical shift from methanogenic to uncultured archaeal lineages. PLoS One 7:e43346. 10.1371/journal.pone.0043346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 46.Corvec S. 2018. Clinical and biological features of Cutibacterium (formerly Propionibacterium) avidum, an underrecognized microorganism. Clin Microbiol Rev 31:e00064-17. 10.1128/CMR.00064-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon S-H, Ha S, Lim J, Kwon S, Chun J. 2017. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286. 10.1007/s10482-017-0844-4. [DOI] [PubMed] [Google Scholar]
- 48.Farag IF, Davis JP, Youssef NH, Elshahed MS. 2014. Global patterns of abundance, diversity and community structure of the Aminicenantes (candidate phylum OP8). PLoS One 9:e92139. 10.1371/journal.pone.0092139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eloe-Fadrosh EA, Ivanova NN, Woyke T, Kyrpides NC. 2016. Metagenomics uncovers gaps in amplicon-based detection of microbial diversity. Nat Microbiol 1:15032. 10.1038/nmicrobiol.2015.32. [DOI] [PubMed] [Google Scholar]
- 50.Jansson JK, Taş N. 2014. The microbial ecology of permafrost. Nat Rev Microbiol 12:414–425. 10.1038/nrmicro3262. [DOI] [PubMed] [Google Scholar]
- 51.Xue Y, Jonassen I, Øvreås L, Taş N. 2020. Metagenome-assembled genome distribution and key functionality highlight importance of aerobic metabolism in Svalbard permafrost. FEMS Microbiol Ecol 96:fiaa057. 10.1093/femsec/fiaa057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodcroft BJ, Singleton CM, Boyd JA, Evans PN, Emerson JB, Zayed AAFF, Hoelzle RD, Lamberton TO, McCalley CK, Hodgkins SB, Wilson RM, Purvine SO, Nicora CD, Li C, Frolking S, Chanton JP, Crill PM, Saleska SR, Rich VI, Tyson GW. 2018. Genome-centric view of carbon processing in thawing permafrost. Nature 560:49–54. 10.1038/s41586-018-0338-1. [DOI] [PubMed] [Google Scholar]
- 53.Argüelles JC. 2000. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch Microbiol 174:217–224. 10.1007/s002030000192. [DOI] [PubMed] [Google Scholar]
- 54.Bird JT, Tague ED, Zinke L, Schmidt JM, Steen AD, Reese B, Marshall IPG, Webster G, Weightman A, Castro HF, Campagna SR, Lloyd KG. 2019. Uncultured microbial phyla suggest mechanisms for multi-thousand-year subsistence in Baltic Sea sediments. mBio 10:e02376-18. 10.1128/mBio.02376-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Diniz-Mendes L, Bernardes E, de Araujo PS, Panek AD, Paschoalin VMF. 1999. Preservation of frozen yeast cells by trehalose. Biotechnol Bioeng 65:572–578. . [DOI] [PubMed] [Google Scholar]
- 56.Brauer MJ, Yuan J, Bennett BD, Lu W, Kimball E, Botstein D, Rabinowitz JD. 2006. Conservation of the metabolomic response to starvation across two divergent microbes. Proc Natl Acad Sci U S A 103:19302–19307. 10.1073/pnas.0609508103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kyryakov P, Beach A, Richard VR, Burstein MT, Leonov A, Levy S, Titorenko VI. 2012. Caloric restriction extends yeast chronological lifespan by altering a pattern of age-related changes in trehalose concentration. Front Physiol 3:256. 10.3389/fphys.2012.00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koo H, Hakim J, Morrow C, Crowley M, Andersen D, Bej A. 2018. Metagenomic analysis of microbial community compositions and cold-responsive stress genes in selected Antarctic lacustrine and soil ecosystems. Life 8:29. 10.3390/life8030029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee M, Gräwert T, Quitterer F, Rohdich F, Eppinger J, Eisenreich W, Bacher A, Groll M. 2010. Biosynthesis of isoprenoids: crystal structure of the [4Fe-4S] cluster protein IspG. J Mol Biol 404:600–610. 10.1016/j.jmb.2010.09.050. [DOI] [PubMed] [Google Scholar]
- 60.Smit A, Mushegian A. 2000. Biosynthesis of isoprenoids via mevalonate in archaea: the lost pathway. Genome Res 10:1468–1484. 10.1101/gr.145600. [DOI] [PubMed] [Google Scholar]
- 61.Dibrova DV, Galperin MY, Mulkidjanian AY. 2014. Phylogenomic reconstruction of archaeal fatty acid metabolism. Environ Microbiol 16:907–918. 10.1111/1462-2920.12359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voynova NE, Rios SE, Miziorko HM. 2004. Staphylococcus aureus mevalonate kinase: isolation and characterization of an enzyme of the isoprenoid biosynthetic pathway. J Bacteriol 186:61–67. 10.1128/JB.186.1.61-67.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilding EI, Brown JR, Bryant AP, Chalker AF, Holmes DJ, Ingraham KA, Iordanescu S, So CY, Rosenberg M, Gwynn MN. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in Gram-positive cocci. J Bacteriol 182:4319–4327. 10.1128/JB.182.15.4319-4327.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vishnivetskaya TA, Buongiorno J, Bird J, Krivushin K, Spirina EV, Oshurkova V, Shcherbakova VA, Wilson G, Lloyd KG, Rivkina EM. 2018. Methanogens in the Antarctic Dry Valley permafrost. FEMS Microbiol Ecol 94:fiy109. 10.1093/femsec/fiy109. [DOI] [PubMed] [Google Scholar]
- 65.Ayala-Del-Río HL, Chain PS, Grzymski JJ, Ponder MA, Ivanova N, Bergholz PW, Bartolo G, Di Hauser L, Land M, Bakermans C, Rodrigues D, Klappenbach J, Zarka D, Larimer F, Richardson P, Murray A, Thomashow M, Tiedje JM. 2010. The genome sequence of Psychrobacter arcticus 273-4, a psychroactive Siberian permafrost bacterium, reveals mechanisms for adaptation to low-temperature growth. Appl Environ Microbiol 76:2304–2312. 10.1128/AEM.02101-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tribelli PM, López NI. 2018. Reporting key features in cold-adapted bacteria. Life (Basel) 8:8. 10.3390/life8010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, Rubin EM, Jansson JK. 2011. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 480:368–371. 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- 68.Chomvong K, Kordić V, Li X, Bauer S, Gillespie AE, Ha S-J, Oh E, Galazka JM, Jin Y-S, Cate JHD. 2014. Overcoming inefficient cellobiose fermentation by cellobiose phosphorylase in the presence of xylose. Biotechnol Biofuels 7:85. 10.1186/1754-6834-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schimz KL, Broll B, John B. 1983. Cellobiose phosphorylase (EC 2.4.1.20) of Cellulomonas: occurrence, induction, and its role in cellobiose metabolism. Arch Microbiol 135:241–249. 10.1007/BF00413475. [DOI] [Google Scholar]
- 70.Haft RJF, Gardner JG, Keating DH. 2012. Quantitative colorimetric measurement of cellulose degradation under microbial culture conditions. Appl Microbiol Biotechnol 94:223–229. 10.1007/s00253-012-3968-5. [DOI] [PubMed] [Google Scholar]
- 71.Krivushin K, Kondrashov F, Shmakova L, Tutukina M, Petrovskaya L, Rivkina E. 2015. Two metagenomes from late Pleistocene northeast Siberian permafrost. Genome Announc 3:e01380-14. 10.1128/genomeA.01380-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai C, Leu AO, Xie GJ, Guo J, Feng Y, Zhao JX, Tyson GW, Yuan Z, Hu S. 2018. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J 12:1929–1939. 10.1038/s41396-018-0109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chakraborty A, Ruff SE, Dong X, Ellefson ED, Li C, Brooks JM, McBee J, Bernard BB, Hubert CRJ. 2020. Hydrocarbon seepage in the deep seabed links subsurface and seafloor biospheres. Proc Natl Acad Sci U S A 117:11029–11037. 10.1073/pnas.2002289117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zheng S, Ponder MA, Shih JYJ, Tiedje JM, Thomashow MF, Lubman DM. 2007. A proteomic analysis of Psychrobacter articus 273-4 adaptation to low temperature and salinity using a 2-D liquid mapping approach. Electrophoresis 28:467–488. 10.1002/elps.200600173. [DOI] [PubMed] [Google Scholar]
- 75.Horneck DA, Sullivan DM, Owen JS, Hart JM. 2011. Soil test interpretation guide. Oregon State University, Corvallis, OR. [Google Scholar]
- 76.Perreault NN, Andersen DT, Pollard WH, Greer CW, Whyte LG. 2007. Characterization of the prokaryotic diversity in cold saline perennial springs of the Canadian high Arctic. Appl Environ Microbiol 73:1532–1543. 10.1128/AEM.01729-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mykytczuk NCS, Foote SJ, Omelon CR, Southam G, Greer CW, Whyte LG. 2013. Bacterial growth at −15°C: molecular insights from the permafrost bacterium Planococcus halocryophilus Or1. ISME J 7:1211–1226. 10.1038/ismej.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tuorto SJ, Darias P, McGuinness LR, Panikov N, Zhang T, Häggblom MM, Kerkhof LJ. 2014. Bacterial genome replication at subzero temperatures in permafrost. ISME J 8:139–149. 10.1038/ismej.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Reeuwijk LP. 2002. Procedures for soil analysis, 6th ed. International Soil Reference and Information Center, Wageningen, The Netherlands. [Google Scholar]
- 80.Alperin MJ, Reeburgh WS. 1985. Inhibition experiments on anaerobic methane oxidation. Appl Environ Microbiol 50:940–945. 10.1128/aem.50.4.940-945.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Donahue DJ, Linick TW, Jull AJT. 1990. Isotope-ratio and background corrections for accelerator mass spectrometry radiocarbon measurements. Radiocarbon 32:135–142. 10.1017/S0033822200040121. [DOI] [Google Scholar]
- 82.Arkin AP, Cottingham RW, Henry CS, Harris NL, Stevens RL, Maslov S, Dehal P, Ware D, Perez F, Canon S, Sneddon MW, Henderson ML, Riehl WJ, Murphy-Olson D, Chan SY, Kamimura RT, Kumari S, Drake MM, Brettin TS, Glass EM, Chivian D, Gunter D, Weston DJ, Allen BH, Baumohl J, Best AA, Bowen B, Brenner SE, Bun CC, Chandonia J-M, Chia J-M, Colasanti R, Conrad N, Davis JJ, Davison BH, DeJongh M, Devoid S, Dietrich E, Dubchak I, Edirisinghe JN, Fang G, Faria JP, Frybarger PM, Gerlach W, Gerstein M, Greiner A, Gurtowski J, Haun HL, He F, Jain R, Joachimiak MP, Keegan KP, Kondo S, Kumar V, Land ML, Meyer F, Mills M, Novichkov PS, Oh T, Olsen GJ, Olson R, Parrello B, Pasternak S, Pearson E, Poon SS, Price GA, Ramakrishnan S, Ranjan P, Ronald PC, Schatz MC, Seaver SMD, Shukla M, Sutormin RA, Syed MH, Thomason J, Tintle NL, Wang D, Xia F, Yoo H, Yoo S, Yu D. 2018. KBase: the United States Department of Energy Systems Biology Knowledgebase. Nat Biotechnol 36:566–569. 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res 27:824–834. 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wu Y-W, Simmons BA, Singer SW. 2016. MaxBin 2.0: an automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 32:605–607. 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 86.Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 88.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, Delmont TO. 2015. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ 3:e1319. 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Markowitz VM, Chen IMA, Chu K, Szeto E, Palaniappan K, Grechkin Y, Ratner A, Jacob B, Pati A, Huntemann M, Liolios K, Pagani I, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res 40:D123–D129. 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Graham ED, Heidelberg JF, Tully BJ. 2018. Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J 12:1861–1866. 10.1038/s41396-018-0091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hobbie JE, Daley RJ, Jasper S. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol 33:1225–1228. 10.1128/aem.33.5.1225-1228.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stepanauskas R, Fergusson EA, Brown J, Poulton NJ, Tupper B, Labonté JM, Becraft ED, Brown JM, Pachiadaki MG, Povilaitis T, Thompson BP, Mascena CJ, Bellows WK, Lubys A. 2017. Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat Commun 8:84. 10.1038/s41467-017-00128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data File S1 (<i>Thermoprofundales</i> information). Download AEM.00972-21-s0001.xls, XLS file, 0.3 MB (316.5KB, xls)
Supplemental Data File S2 (<i>Atribacteria</i> information). Download AEM.00972-21-s0002.xls, XLS file, 0.6 MB (593KB, xls)
Supplemental Data File S3 (<i>Chloroflexi</i> information). Download AEM.00972-21-s0003.xls, XLS file, 2.4 MB (2.4MB, xls)
Supplemental Data File S4 (<i>Actinobacteria</i> information). Download AEM.00972-21-s0004.xls, XLS file, 1.0 MB (985KB, xls)
Supplemental Data File S5 (<i>Aminicenantes</i> information). Download AEM.00972-21-s0005.xls, XLS file, 0.5 MB (543.5KB, xls)
Supplemental Data File S6 (data used in analysis). Download AEM.00972-21-s0006.xlsx, XLSX file, 0.05 MB (53.1KB, xlsx)
Supplemental Data File S7 (data used for Fig. 5). Download AEM.00972-21-s0007.xlsx, XLSX file, 5.8 MB (5.8MB, xlsx)
Fig. S1 to S7, legends to Supplemental Data Files S1 to S7, Tables S1 to S3. Download AEM.00972-21-s0008.pdf, PDF file, 8.0 MB (8MB, pdf)
Data Availability Statement
The 6 assembled metagenome files and resulting MAGs are available in the NCBI database under BioProject accession number PRJNA596250. Sample methods are available to view, as follows: 3.5 m, https://narrative.kbase.us/narrative/27695; 7.2 m, https://narrative.kbase.us/narrative/27731; 14.1 m, https://narrative.kbase.us/narrative/27724; 14.8 m, https://narrative.kbase.us/narrative/27725; 16.6 m, https://narrative.kbase.us/narrative/37690; 17.2 m, https://narrative.kbase.us/narrative/37878; 20 m, https://narrative.kbase.us/narrative/37504.