Abstract
Ubiquitin-mediated proteolysis controls diverse physiological processes in eukaryotes. However, few in vivo targets of the mammalian Cdc34 and Rad6 ubiquitin-conjugating enzymes are known. A yeast-based genetic assay to identify proteins that interact with human Cdc34 resulted in three cDNAs encoding bZIP DNA binding motifs. Two of these interactants are repressors of cyclic AMP (cAMP)-induced transcription: hICERIIγ, a product of the CREM gene, and hATF5, a novel ATF homolog. Transfection assays with mammalian cells demonstrate both hCdc34- and hRad6B-dependent ubiquitin-mediated proteolysis of hICERIIγ and hATF5. This degradation requires an active ubiquitin-conjugating enzyme and results in abrogation of ICERIIγ- and ATF5-mediated repression of cAMP-induced transcription. Consistent with these results, the endogenous ICER protein is elevated in cells which are null for murine Rad6B (mHR6B−/−) or transfected with dominant negative and antisense constructs of human CDC34. Based on the requirement for CREM/ICER and Rad6B proteins in spermatogenesis, we determined expression of Cdc34, Rad6B, CREM/ICER isoforms, and the Skp1–Cullin–F-box ubiquitin protein ligase subunits Cul-1 and Cul-2, which are associated with Cdc34 activity during murine testicular development. Cdc34, Rad6B, and the Cullin proteins are expressed in a developmentally regulated manner, with distinctly different patterns for Cdc34 and the Cullin proteins in germ cells. The Cdc34 and Rad6B proteins are significantly elevated in meiotic and postmeiotic haploid germ cells when chromatin modifications occur. Thus, the stability of specific mammalian transcription factors is the result of complex targeting by multiple ubiquitin-conjugating enzymes and may have an impact on cAMP-inducible gene regulation during both meiotic and mitotic cell cycles.
A fundamental mechanism for control of cellular processes is ubiquitin-mediated destruction of regulatory proteins. CDC34, a gene essential for the transition into S phase of the cell division cycle in Saccharomyces cerevisiae (6, 20), encodes a ubiquitin-conjugating (UBC) enzyme (named UBC3). Cdc34 is required for ubiquitination and proteolytic degradation of cyclins and cyclin-dependent kinase inhibitors (for reviews, see references 14 and 36). The Cdc34 protein consists of a highly conserved catalytic domain common to all UBC enzymes, including the DNA repair protein Rad6 (23, 30), and a unique carboxy-terminal extension or tail (20), which is essential for cell cycle function (34, 64). A chimera consisting of the Rad6 catalytic domain and the Cdc34 carboxy terminus can fulfill some of the in vivo function of both proteins (34, 64). Yeast RAD6 (UBC2) has been implicated in DNA repair, induced mutagenesis, retrotransposition, sporulation, and the degradation of proteins with destabilizing N-terminal amino acid residues (15, 25, 37, 67).
Highly related CDC34 genes in humans and mice have been cloned and characterized (26, 53, 55). Human CDC34 (hCDC34) can fully complement a cdc34-1 mutant strain for growth at the restrictive temperature (55). Similarly, both human RAD6 homologs (hRAD6A and hRAD6B) can complement the DNA repair and mutagenesis deficiencies of rad6Δ strains (33). Disruption of the mouse homolog of the hRAD6B gene results in partial arrest of gametes at the postmeiotic spermatid stage and alteration of postmeiotic chromatin remodeling (57). The role of hRAD6B in spermatogenesis appears to be indirect, since its deletion does not cause a complete and uniform block at a given point of spermatogenic differentiation.
Ubiquitination is a process in which ubiquitin, a small polypeptide, is covalently attached to a cellular protein and normally results in proteasome-dependent proteolysis (for reviews, see references 24 and 29). The UBC enzyme transfers the ubiquitin from a ubiquitin-activating enzyme (E1) to specific target proteins, in some cases requiring a third factor, the ubiquitin-protein ligase (E3), to mediate transfer (for reviews, see references 27 and 54). The yeast Cdc34 UBC enzyme is recruited to a large E3 complex called Skp1–Cullin–F-box protein (SCF) by interaction with the Cullin protein Cdc53 (32, 36, 54).
In S. cerevisiae, Cdc34 in conjunction with the SCF complex ubiquitinates cell cycle regulators (14) and other diverse substrates (for a review, see reference 61), including transcription factor Gcn4 (35). Phosphorylation of at least some Cdc34 targets (e.g., Sic1 and Cln2) is a prerequisite for its recognition and subsequent degradation by Cdc34-mediated ubiquitination (39, 63). The yeast Cdc34 and Rad6 enzymes appear to share certain targets, including Gcn4 (35). However, it has been difficult to identify specific targets of the mammalian enzymes because of the large number of UBC enzymes and the lack of cells containing conditional or null UBC alleles. We have used a yeast-based in vivo assay (two-hybrid cloning) to identify proteins that interact with mammalian Cdc34. As described here, two of the hCdc34 interactants obtained in this assay contain bZIP motifs and are repressors of cyclic AMP (cAMP)-induced transcription: inducible cAMP early repressor (ICER) and activating transcription factor 5 (ATF5).
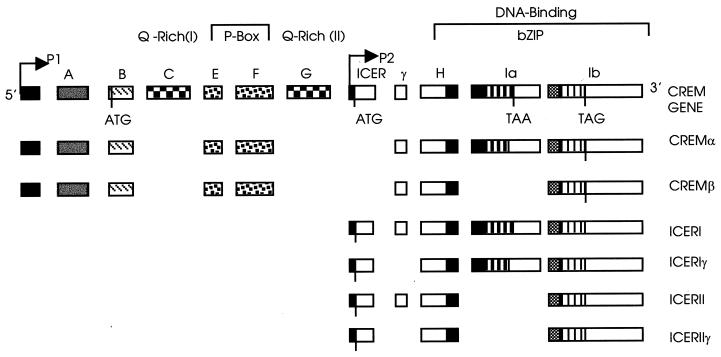
In eukaryotes, cAMP-mediated transcription regulates multiple physiological processes, including gametogenesis, circadian rhythm, and neuroendocrine functions (for a review, see reference 60). Stimulation of this pathway is mediated via phosphorylation by protein kinase A (PKA) of a single serine in the structurally similar transcription factors called cAMP-responsive element (CRE) binding proteins (CREB), CRE modulators (CREM), and ATFs. Transcription factors which regulate the response to cAMP belong to the bZIP family and bind as dimers to an 8-bp pallindromic DNA consensus sequence called the CRE (59). The CREM gene is controlled by two promoters and results in a large number of alternately spliced transcripts encoding activators and repressors of cAMP-dependent transcription that are expressed in a tissue- and developmentally regulated manner (Fig. 1). The upstream promoter (P1) directs the CREM τ, τ1, and τ2 activators and the CREM α, β, γ, and S repressors. The downstream intronic promoter (P2) directs the potent early repressors of cAMP-induced transcription (ICER) (18, 19, 38, 48). Four types of ICER transcripts (ICERI, -Iγ, -IIγ, and -II) are generated by alternate splicing of the DNA binding domain and γ domain exons (48). ICER proteins are small (estimated molecular mass, ∼13 kDa) and, unlike CREM, lack both the phosphorylation box (P box) and activation domain and escape from PKA-dependent phosphorylation. The principal determinant of ICER activity is not its degree of phosphorylation but its intracellular concentration (48), which depends on the transcription rate of the P2 promoter and the degradation rate of the ICER polypeptide (48, 49).
FIG. 1.
Schematic representation of the CREM/ICER gene structure. The CREM repressor isoforms, CREM α and CREM β, and ICER isoforms are indicated. The P1 promoter is GC rich and directs constitutive expression; the P2 promoter is strongly inducible by activation of the PKA-cAMP-dependent signaling pathway (modified from reference 38, printed with permission of the Royal Society).
CREM/ICER isoforms play a regulatory role in gene expression in haploid germ cells in mammals (for reviews, see references 11 and 12) and have recently been implicated in spermatogenesis in humans (40). CREM activates a number of testis-specific promoters of haploid cell-expressed genes. CREM gene products are highly abundant in adult testis, and their expression follows a developmental and quantitative switch (19); the activator forms are the dominant forms in postmeiotic germ cells, while the repressor forms are observed at only low levels before meiosis (12). Targeted disruption of the CREM gene in mice (5, 50) results in abrogation of spermatogenesis at the spermatid stage.
Here, we report that an hICER isoform (hICERIIγ) and a new ATF homolog (hATF5) are targeted by both hCdc34 and hRad6B UBC enzymes for degradation in vivo. Both Cdc34 and Rad6B proteins are expressed at high levels in pre- and postmeiotic germ cells, and targeting of CREM/ICER and ATF proteins by Cdc34 and Rad6B UBC enzymes may have a role in spermatogenesis. Thus, complex targeting of these repressors by multiple UBC enzymes may have a major impact on cAMP-dependent gene regulation during both meiotic and mitotic cell cycles.
MATERIALS AND METHODS
Two-hybrid reagents.
Reagents used in the two-hybrid screening include the Gal4 activation domain library, the Gal4 DNA binding domain vector (pPC97), the yeast host strain MV103 (MATa leu2 trp1 his3 gal1::HIS3 gal1::lacZ SPAL::URA3), and five constructs in MV103 for use as reference controls during screening (69, 70) and were kindly provided by M. Vidal (Massachusetts General Hospital Cancer Center, Charlestown).
Construction of the DNA binding domain-Cdc34 fusion (bait).
Human CDC34 cDNA (pKS-6110) (55) was digested with NotI followed by SmaI at the codon encoding the first methionine and inserted in frame into pPC97 (LEU2). The junction between GAL4 and hCDC34 has been verified by sequence analysis. The hCdc34C93S active-site mutant was generated from pKS-6110 by standard PCR mutagenesis methods (28), and the mutation was confirmed by sequencing. The mutant was then subcloned into pPC97 in a manner analogous to that for the hCDC34 bait construct described above. The cdc34-1 mutant yeast strain SJ1098-3d (MATa cdc34-2 leu2-3 ura3 trp1) was obtained from B. Byers, University of Washington, Seattle.
Activation domain cDNA library.
A human T-lymphocyte cDNA fusion library in the activation domain vector pPC86 (Trp+) was kindly provided by J. La Baer (Massachusetts General Hospital Cancer Center). The cDNAs were cloned into the EcoRI (5′) and SpeI (3′) sites. This library has approximately 2 × 106 clones, and the average insert size is 1 kb. The library was amplified once by electroporation with electrocompetent Escherichia coli JS4 cells (Bio-Rad, Hercules, Calif.) followed by replica plating onto Luria-Bertani agar–ampicillin plates. The DNA was prepared by using a Plasmid Maxi kit from Qiagen (Valencia, Calif.).
Selection of hCDC34-interacting genes.
The bait (LEU2) and the library plasmid (TRP1) were sequentially transformed into the yeast host strain MV103 by using the modified Li-acetate transformation protocol of Schiestl and Giets (62) with yeast total RNA and denatured salmon sperm DNA as the carrier to achieve a transformation efficiency of 300,000 colonies/μg of plasmid DNA. Three independent pools of library DNA were used to transform the MV103(pPC97-hCDC34) cells, and 500,000 transformants from each pool were obtained. The two-hybrid screen was performed by first selecting for growth of hCdc34 bait-library cotransformants on SC-His-Leu-Trp plus 25 mM 3-amino-1,2,4-triazole (3AT) (Sigma). Subsequently, activities of the additional reporter genes URA3 and lacZ in the 3AT-positive clones were determined. URA3 gene positive selection was on uracil-deficient medium and negative selection medium with 0.1% 5-fluoro-orotic acid. Induction of the lacZ gene was assayed qualitatively in the presence of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for blue colonies (58). The cDNA inserts of positive clones were PCR amplified and cloned into a pPCRII vector by using a TA cloning kit from Invitrogen (Torrey Pines, Calif.), and both strands were sequenced by using an automated sequencer (LI-COR, Inc., Lincoln, Nebr.).
Cell cultures and transfection.
NIH 3T3 and human choriocarcinoma JEG3 cells (both obtained from the American Type Culture Collection) were grown in Dulbecco’s modified Eagle’s medium (DMEM) and MEM, respectively, supplemented with 10% fetal bovine serum and were maintained at 37°C, 95% humidity, and 5% CO2. Cells used in the experiments were between 130 and 140 passages. Cells were transfected with appropriate plasmids in 100-mm-diameter dishes by the calcium phosphate method as previously described (58). A fixed amount of plasmid DNA was used in any given experiment. The total amount of expression vector DNA was equalized by adding blank vectors to control for the promoter competition effect. When necessary, transfection efficiency was monitored by use of 1 μg of cytomegalovirus β-galactosidase plasmid per transfection, and calorimetric β-galactosidase assays were performed with o-nitrophenyl β-d-galactopyranoside as a substrate (58). Fibroblast cell lines from wild-type mHR6B+/+ and knockout mHR6B−/− mice at passage 3 (kindly provided by H. P. Roest and J. Hoeijmaker) were grown in F10-DMEM supplemented with 10% fetal calf serum, and protein lysates were prepared as described below.
Plasmids.
The following plasmids were used for transfection. pCS2MT-hICERIIγ was constructed by ligation of the 500-bp EcoRV/PmacI fragment bearing the hICERIIγ cDNA, in frame at the end of the sixth Myc epitope in pCS2MT (B. Kelley, Fred Hutchinson Cancer Center, Seattle, Wash.), which had been digested with StuI. pFLAGCMV2-hCDC34 was generated by cloning the region of the hCDC34 gene corresponding to the N terminus of the product contained on a 1,298-bp NruI/KpnI fragment from pKS6110 (55) into pFLAGCMV2 (Kodak) that had been digested with EcoRV and KpnI. pFLAGCMV2-hCDC34C93S, which encodes the active-site C93S mutation, was also constructed analogously. pFLAGCMV2-hCDC34ΔCT was generated by cloning the hCDC34 cDNA corresponding to the amino-terminal end contained on a 629-bp NruI/ScaI fragment from pKS6110, encoding the first 189 amino acids (aa) from the putative first methionine of hCdc34 (55). Plasmid pCB6-hCDC34DN (kindly provided by M. Goebl, Indiana University, Indianapolis), containing the dominant negative human CDC34 mutant cDNA (66), was sequenced to verify the mutations C93S and L97S and subcloned into the pSG5 (Promega) (22) mammalian expression vector (pSG5-hCDC34DN). The hCDC34 antisense plasmid pSG5-hCDC34AS was constructed by subcloning the CDC34 fragment from pFLAGCMV2-hCDC34 (described above) in the reverse orientation relative to the promoter. pFLAGCMV2-hRAD6B was constructed by cloning a 766-bp HincII/SspI fragment from pRR518 (L. Prakash, University of Texas Medical Branch, Galveston) corresponding to the N terminus in frame into pFLGCMV2 digested with SmaI and EcoRV. Other plasmids used were pSomCAT and pSV-mouse CREM β (P. Sassone-Corsi, Institut de Genetique et de Biologie Moleculaire et Cellulaire, Strasbourg, France), pSG5 (Promega, Madison, Wis.), pCαEV (G. McKnight, University of Washington School of Medicine, Seattle), and the pMT133 and pMT107 vectors (hemagglutinin [HA] and His-tagged ubiquitin, respectively) (D. Bohman, EMBL, Heidelberg, Germany).
CAT assay.
hICERIIγ, hATF5, and hCDC34 sequences were cloned into the expression plasmid pSG5 for expression in mammalian cells. Forty-eight hours following transfection, protein extracts were made from freeze-thaw-lysed cells. β-d-Galactosidase activity was used for normalization of the amount of lysates to be used in the subsequent chloramphenicol acetyltransferase (CAT) assay. The CAT reaction was performed with cellular extracts, acetyl coenzyme A (Boehringer Mannheim, Indianapolis, Ind.), and [14C]chloramphenicol (NEN, Boston, Mass.), with incubation at 37°C for 3 to 4 h. Reaction products were extracted in ethyl acetate and separated on thin-layer chromatography plates (Whatman, Maidstone, England) in 95:5 chloroform-methanol. The CAT plates were visualized with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.), and the activity was quantified by measuring the percentage of chloramphenicol acetylated by using ImageQuant software (Molecular Dynamics).
Antisera.
Monoclonal antisera for human Cdc34 and Cul-2 (Transduction Lab, Lexington, Ky.), FLAG epitope and mouse β-actin (Sigma), c-Myc epitope and bacterial TrpE (Oncogene Research Products, Cambridge, Mass.), and HA epitope (BabCO, Richmond, Calif.) and polyclonal antisera for hRad6B (H. P. Roest and J. Hoeijmaker, Erasmus University, Rotterdam, The Netherlands), ICER (C. A. Molina, University of Medicine and Dentistry of New Jersey, Newark), CREM (P. Sassone-Corsi), pan-CREM (Upstate Biotechnology, Lake Placid, N.Y.), and Cul-1 (W. Krek, Friedrich Miescher Institut, Basel, Switzerland) were used.
Protein analysis and immunoprecipitation.
Cells were lysed directly on 100-mm-diameter tissue culture dishes in radioimmunoprecipitation assay buffer (1× phosphate-buffered saline [PBS], 1% Nonidet P-40, 0.1% sodium dodecyl sulfate [SDS], 0.5% sodium deoxycholate) or PBSTDS buffer (1× PBS, 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate) containing protease and phosphatase inhibitors (1 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 1 μg of pepstatin per ml, 30 μl of aprotinin per ml, 0.5 μg of leupeptin per ml, and 100 mM sodium orthovandate) (all from Sigma), followed by scraping and passing through a 21-gauge needle. After protein quantification (with Bio-Rad protein dye and bovine serum albumin as standards) and normalization, 10 to 40 μg of protein extract was electrophoresed on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride membrane (Millipore, Bedford, Mass.) with a Bio-Rad Mini Protein Blot apparatus according to the manufacturer’s protocol. The filters were initially blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween 20 for 1 to 2 h at room temperature and then probed with Myc (1.5 μg/ml), FLAG (2.5 μg/ml), β-actin (1:100,000), Cdc34 (1:2,000), Rad6B (1:250), Cul-1 (1:1,000), Cul-2 (1:500), ICER (1:1,000), and CREM (1:500) antibodies. The bound antibodies were visualized with the enhanced chemiluminescence detection system (Amersham, Buckinghamshire, England) in combination with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibodies as appropriate, and the intensities of the specific bands in the exposed films were quantified. Immunoprecipitation was performed as follows. Cell lysates (1.0 ml) were precleared by incubation with 10 μl of normal mouse immunoglobulin G and 20 μl of protein G plus agarose (Oncogene Research Products) at 4°C for 1 h on a rotator. The precleared lysate was collected after centrifugation at 750 × g for 15 min. Precleared lysates normalized for protein concentration (0.5 to 1.0 ml) were incubated at 4°C for 1 h with appropriate dilutions of the primary antibodies, and then 20 μl of protein G plus agarose was added. The mixture was then incubated at 4°C for another 12 to 16 h on a rotator. Precipitates were then washed four times with 1 ml of ice-cold PBS before electrophoresis and Western blot analysis.
Pulse-chase assay.
JEG3 cells at passages 138 and 140 transiently transfected with various plasmids were incubated for 2 h in methionine- and cysteine-free DMEM. Cells were incubated for 1 h with 125 μCi of [35S]methionine (NEN) per ml in the same medium. Cells were harvested (time zero) or washed four times in PBS and incubated for 3, 6, or 9 h in complete medium (MEM) supplemented with 4 mM methionine. At the end of each time period, cells were washed three times in ice-cold PBS and lysed in 2.5 ml of PBSTDS buffer. Following centrifugation at 1,250 × g for 15 min, the supernatant was collected and analyzed for protein content with a Bio-Rad protein assay kit and for incorporation of the [35S]methionine by trichloroacetic acid precipitation. The average incorporation among samples was found to be 40% ± 2.0%. An amount corresponding to 25 × 106 trichloroacetic acid-precipitable counts was immunoprecipitated with Myc antibody in accordance with the instructions of the manufacturer (Oncogene Research Products), resolved by SDS-polyacrylamide gel electrophoresis, fixed in acetic acid (10%)-methanol (40%), and analyzed in a Storm imager.
Proteasome inhibitors and detection of ubiquitin-ICERIIγ conjugates.
Peptide aldehydes MG115 and MG132 were obtained from Peptide Institute Inc. (Lexington, Ky.) and dissolved at 10 mM in dimethyl sulfoxide. Approximately 36 h after transfection and 5 h before harvest, cells were treated with 0.025 mM proteasome inhibitors. Cells were lysed as described above, with the addition of 5 mM N-ethylmaleimide (Sigma) to the lysis buffer as previously described (10). For the detection of HA-tagged ubiquitin-ICERIIγ conjugates, cells growing in 100-mm-diameter dishes were cotransfected with ICERIIγ and either HA (pMT133)- or His6 (pMT107)-tagged ubiquitin expression plasmid, followed by treatment with proteasome inhibitor, lysis, immunoprecipitation, and Western analysis as described above.
Testicular protein lysates and isolation of germ cells.
Testes were collected from a colony of wild-type C57 mice at various ages of development from birth until 40 days old at intervals of 5 days (kindly provided by S. Sharan and A. Bradley, Baylor College of Medicine, Houston, Tex.). Testes were dissected, washed several times with PBS, snap frozen in liquid nitrogen, and kept at −80°C until protein lysates were made. Germ cells were prepared from adult C57BL/6 × 129 (hybrid) mouse and Sprague-Dawley rat testes by trypsin digestion, followed by centrifugal elutriation as described previously (21, 45). Enriched populations of pachytene spermatocytes, round spermatids, and late spermatids were prepared by centrifugal elutriation, with a purity of ∼80% as shown by microscopic analysis. Protein lysates were made from these fractions by using radioimmunoprecipitation assay buffer. Testicular lysates from 12-week-old C3H × C57BL/6 jsd heterozygote (jsd/+) and mutant (jsd/jsd) mice (3) (kindly provided by G. Shetty) were made as described above. C57BL/6J mice (6 to 7 weeks old) (kindly provided by G. Wilson) treated with 16 Gy of radiation were sacrificed 4 weeks postirradiation, testes were collected, and lysates were made as described above (44). Testicular lysates from 7- to 8-week-old unirradiated littermates from the same colony were used as controls.
RNA extraction and Northern analysis.
Total RNA was extracted from transfected JEG3 cells by using a total RNA isolation kit from Qiagen. DNase-I treated RNAs were Northern blotted from 1% formaldehyde gels with Hybond N+ sheets (Amersham). The blots were hybridized overnight at 65°C with the nick-translated [γ-32P]dCTP-labeled hICERIIγ probe in 10% dextran sulfate–2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–1% SDS–250 μg of salmon sperm DNA per ml. The final wash was in 0.1× SSC–0.1% SDS at 65°C.
Nucleotide sequence accession number.
The hATF5 sequence reported in this paper has been deposited in the GenBank database under accession no. AF101388.
RESULTS
hCDC34 two-hybrid screen.
The two-hybrid cloning system used in this study has been described previously (69, 70). Reagents used in this screen include a human activated T-lymphocyte cDNA library fused to the Gal4 activation domain, the full-length hCDC34 cDNA (55) fused to the Gal4 DNA binding domain as bait (pPC97), and the yeast host strain MV103 with appropriate reporter constructs. The success of a two-hybrid screen depends on the production of a properly folded bait fusion protein in the host cells. The pPC97-hCDC34 bait construct encoded a functional hCdc34 protein as demonstrated by its ability to complement a cdc34-1 mutant yeast strain, SJ1098-3d, for growth at 37°C (data not shown). Complementation of cdc34-1 also suggests that the hCDC34 bait can form a functional complex with the SCF in yeast. Production of Gal4-hCdc34 bait and mutant Gal4-hCdc34C93S fusion proteins was also confirmed in Western blots with a monoclonal hCdc34 antiserum (result not shown). In a screen of 1.5 million transformants, 30 clones were found to be positive for all reporters tested (His/3AT+, Ura+, 5-fluoro-orotic acid negative, and X-Gal+). Of the 30 positive clones, 18 clones have been sequenced and 4 of them are novel genes. None of these 18 sequences represent human cDNAs encoding products with significant homology to members of the SCF complex, cyclins, or cyclin-dependent kinase inhibitors.
Three of the 18 clones, hICERIIγ, hATF5, and clone 30-17, encode proteins with bZIP motifs. hICERIIγ, an ICER isoform generated by alternate splicing, encodes a 108-aa protein and lacks the γ exon and one of the DNA binding and dimerization domains (Fig. 1). hICERIIγ has 97.2% homology with the mouse isoform mICERIIγ and 76.7% homology with hICERI, a 120-aa hICER protein. hATF5 is a novel clone; a partial sequence of a mouse homolog called ATFX has been reported (51). hATF5 encodes a 122-aa protein with extensive homology in the C-terminal domain with human (52.1%) and mouse (50.9%) ATF4. Mouse ATFX and human ATF5 are 96.6% homologous in their available sequences. In the leucine zipper domain, hATF5, like mouse ATFX, has only three leucines instead of the five that are present in hATF4 and hATF3, with the distal two leucines replaced by valines.
Expression of Cdc34/Rad6B and its targets in spermatogenesis.
Isolation of a particular interaction in a two-hybrid screen does not necessarily imply that these two proteins are coexpressed or normally interact in mammalian cells. Currently little is known about the expression pattern of the Cdc34 protein in normal mammalian tissues. Considering the requirement for ICER/CREM and Rad6B proteins during spermatogenesis, we determined whether human Cdc34 is also expressed during spermatogenesis.
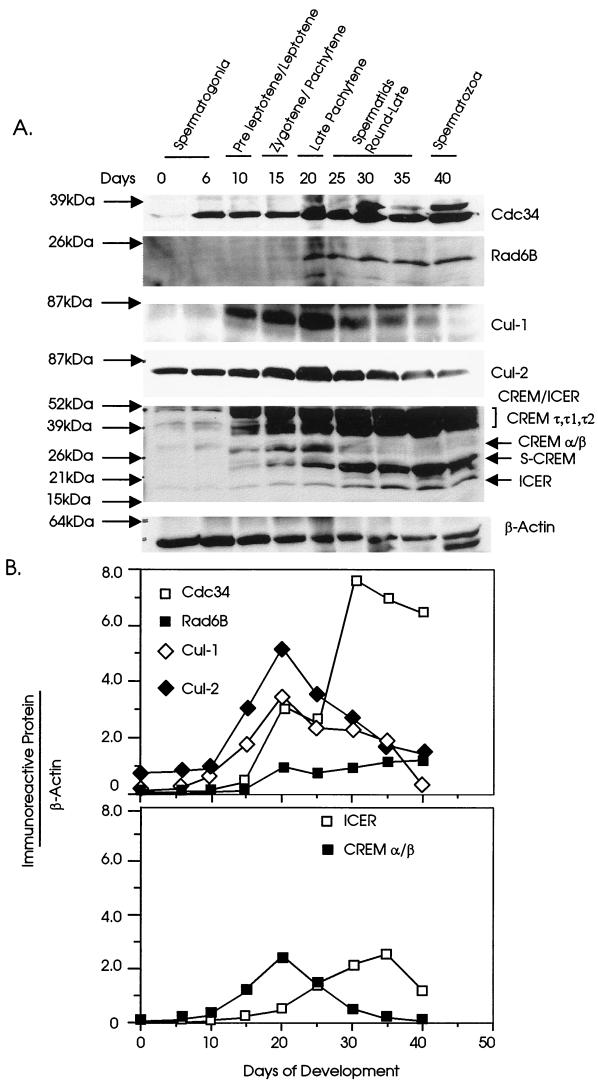
During spermatogenesis, the temporal appearance of the most advanced wave of spermatogenic cells has been well characterized (4). The ontogeny of germ cell development in mice is as follows: spermatogonia (types A and B), primary spermatocytes (preleptotene, leptotene, zygotene, and pachytene), secondary spermatocytes, spermatids, and sperm. By day 3 most of the germ cells are spermatogonia, by day 13 the most advanced stage of spermatogenesis is the pachytene spermatocyte stage, by day 22 the first wave of meiosis is completed and early spermatids first appear, and by day 35 mature spermatozoa are produced. To establish the time of appearance of Cdc34 and Rad6B proteins during development, testis extracts from mice at different ages were studied by Western analysis (Fig. 2A). Cdc34 protein is found at very low levels at birth. However, the level increases considerably at day 20, correlating with the time of an increased number of spermatocytes. The Cdc34 profile shows an initial peak at day 20, followed by a significant rise again by day 30, when spermatids become the predominant cell type. A similar pattern of expression is also observed for Rad6B. Rad6B protein appears at day 15, and the level increases considerably at day 20 and reaches a peak by day 35 (Fig. 2B). The levels of Cdc34 and Rad6B proteins remain high during postmeiotic development of round and then late spermatids. Given the requirement for the SCF complex in Cdc34-mediated degradation of proteins in budding yeast (1, 16, 65) and conservation of the SCF complex in higher eukaryotes (41), we studied the expression of the Cdc53 homologs Cul-1 and Cul-2 during testicular development. As shown in Fig. 2, both Cul-1 and Cul-2 follow a developmental profile which is distinct from that of Cdc34. Their levels, especially that of Cul-1, are extremely low in prepubertal testis (until day 6) and then increase sharply and reach a peak at day 20, coinciding with the initial increase in levels of Cdc34 and Rad6B proteins (Fig. 2B). However, the levels of both Cul-1 and Cul-2 proteins then decline gradually and are low in adult testis. The maximum expression of Cul-1 and Cul-2 along with elevated levels of Cdc34 at day 20, when pachytene spermatocytes are the predominant germ cell, indirectly suggests the formation of an active SCF complex during meiosis.
FIG. 2.
(A) Western blot analysis of the expression profiles of Cdc34, Rad6B, Cul-1, Cul-2, and CREM/ICER proteins during mouse testicular development. Testis extracts (40 μg/lane) from C57 mice at various ages of development as indicated were probed with monoclonal anti-hCdc34, anti-Cul-2, and anti-β-actin and polyclonal anti-Rad6B, anti-CREM/ICER, and anti-Cul-1 antisera as described in Materials and Methods. Molecular mass markers are on the left. The most advanced germ cell types during testicular development, as previously described (4), are noted on top. The profiles of the immunoreactive proteins shown are from four different Western blots, and each blot was probed with β-actin; one β-actin blot is shown as a representative to compare loading. (B) Densitometric analysis of the bands of the immunoreactive proteins.
Due to the presence of both germ cells and stromal cells in the analysis of whole testis, we also compared expression of Cdc34 and Rad6B proteins in testes from mice lacking differentiated germ cells, including the recessive mutant juvenile spermatogonial depletion (jsd) and irradiated testes. The adult testes from a jsd homozygous mouse are one-third of the normal size and lack cells undergoing spermatogenesis, compared to the jsd heterozygotes (jsd/+) (3). The majority of the tissue is composed of Sertoli’s, Leydig’s, and other interstitial cells, with rare type A spermatogonia. Figure 3A shows the result of Western blot analysis with antibodies to hCdc34 and hRad6B of lysates from whole testes of homozygous and heterozygous animals. These blots demonstrate a loss of Rad6B and a decrease in Cdc34 staining in testes from the homozygous jsd/jsd mice compared with heterozygous jsd/+ littermates. In addition, the higher-molecular-weight band recognized by the Cdc34 antibody which is seen in normal adult testis is absent. A similar change in the patterns of expression for Rad6B and Cdc34 is seen in the testes of irradiated mice (Fig. 3A), which also are deficient in germ cells secondary to radiation damage (44).
FIG. 3.
(A) Expression profiles of Cdc34 and Rad6B proteins in germ cell-deficient testis. Left panel, analysis of the testes of adult spermatocyte-deficient juvenile spermatogonial depletion (jsd/jsd) mutant mice and unaffected jsd heterozygote (jsd/+) mice. Right panel, pattern of expression in the testes from irradiated mice in comparison to nonirradiated testes. Testicular extracts were probed with monoclonal anti-hCdc34 and polyclonal anti-Rad6B antisera as described in Fig. 2. β-Actin is shown to compare loading. (B) Western blot analysis of the expression profiles of Cdc34, Rad6B, CREM α/β, and ICER proteins in elutriated germ cell fractions of C57BL/6 mice. Germ cell extracts were probed with monoclonal anti-hCdc34 and anti-β-actin and polyclonal anti-Rad6B and anti-CREM-ICER antisera as described in Materials and Methods. The profiles of the immunoreactive proteins shown are from three different Western blots, and each blot was probed with β-actin; one β-actin blot is shown as a representative to compare loading.
To further characterize the roles of Cdc34, Cul-1, Cul-2, and Rad6B in the germ cell component of the testis, we examined the expression of these proteins in separated germ cells from mouse and rat testes. Pachytene spermatocyte-, round spermatid-, and late spermatid-enriched fractions were obtained by centrifugal elutriation as described previously (21, 45). Western blot analysis of the protein lysates of these fractions demonstrated lower levels of Cdc34 and Rad6B proteins in mouse pachytene spermatocytes than in the round and late spermatids (Fig. 3B). Consistent with the developmental profile, the levels of these proteins are significantly higher in late spermatids, with a three- to fivefold increase over the protein levels in pachytene spermatocytes and round spermatids. Similar results were also obtained for Cdc34 protein in elutriated rat cells (results not shown). In both the developmental profile and the separated germ cell fractions there is the appearance of a higher-molecular-weight form of Cdc34 protein in the later stages of spermatogenesis. The patterns of expression of Cul-1 and Cul-2 are different and are distinct from that of Cdc34. There is maximal expression of Cul-2 in the spermatocyte fraction and of Cul-1 in the round spermatids, with both expressed at low levels in the late spermatid fractions.
With regard to potential targets of the Cdc34 and Rad6B UBC enzymes isolated in our screen, we investigated the developmental profile of CREM/ICER protein expression in the mouse testis. The polyclonal anti-CREM/ICER antiserum recognizes all CREM/ICER isoforms with the same efficiency, indicating that a common domain constitutes the antigenic epitope (12, 48). Five distinct immunoreactive bands were detected (Fig. 2A), with apparent molecular masses of ∼18 (ICER), 26 (S-CREM), ∼30 (CREM α/β), 39 to 40 (CREM τ1 and τ2), and ∼50 (CREM τ) kDa, respectively. ICER protein is found to be expressed at moderate levels in mouse testis after birth and reaches a maximum at around 35 days of age. The level of ICER is lower in prepubertal testis than in the postmeiotic testis in mice and remains higher through the stages of spermatids and spermatozoa. In contrast, the repressor CREM α/β protein (represented by the 30-kDa immunoreactive band) was found to be present at a very low level at birth, to increase significantly at day 10, and to reach a peak at day 20; thereafter its level declines sharply, and it remains undetectable after day 35 (Fig. 2B). The sharp decline and subsequent disappearance of CREM α/β coincide with the first wave of meiosis and the first appearance of spermatids.
The expression patterns of CREM/ICER isoforms were further examined in the separated germ cells of mouse (Fig. 3B) and rat (data not shown) testes as described above. ICER isoforms were expressed at low levels in pachytene spermatocytes but at increased levels in round and late spermatids of both mouse (Fig. 3B) and rat (data not shown). The 30-kDa CREM α/β, however, is found to be expressed at higher levels in pachytene spermatocytes and round spermatids, while it is not detected in late spermatids (Fig. 3B). Disappearance of the 30-kDa CREM α/β repressor isoform during the late puberty stage of testicular development and absence of the immunoreactive CREM α/β protein in the late spermatid fractions are correlated with high levels of Cdc34 and Rad6B proteins in late puberty and late spermatids (Fig. 2 and 3).
Ectopically expressed hICERIIγ and hATF5 proteins are degraded by the ubiquitin-proteasome machinery in a Cdc34- and Rad6B-dependent pathway.
The results of the previous experiments demonstrate that mammalian Cdc34 and the ICERIIγ proteins are both expressed in a complex pattern in germ cells during spermatogenesis. We therefore next determined if Cdc34 and the ICERIIγ proteins can directly interact in mammalian cells as suggested by the two-hybrid screen. We expressed epitope-tagged versions of hCdc34 and its targets by transient transfection in mammalian cells. The human choriocarcinoma cell line JEG3 has been used extensively to characterize the expression and function of ICER and other CREM gene products (18, 48) and therefore was chosen for use in the present study. Expression of Myc-tagged hICERIIγ (pCS2MT-hICERIIγ) resulted in production of a 24-kDa protein recognized by the Myc epitope antibody (Fig. 4A). Cotransfection of Myc-tagged hICERIIγ with FLAG-tagged hCDC34 (pFLAGCMV2-hCDC34) into JEG3 cells (Fig. 4A) and NIH 3T3 cells (data not shown) resulted in considerable and in some cases complete loss of the hICERIIγ fusion protein in multiple experiments. In contrast to the loss of protein, Northern analysis of the mRNA isolated from pCS2MT-hICERIIγ-transfected cells demonstrated no significant difference in the steady-state hICERIIγ transcripts compared to the cells cotransfected with pFLAGCMV2-hCDC34 (Fig. 4A).
FIG. 4.
Cotransfection assay for assessing the expression of ectopic hICERIIγ (IIγ) transcripts and protein in JEG3 cells. (A) Upper panel, Northern blot analysis of the mRNA isolated from the cells cotransfected with pCS2MT-hICERIIγ and the blank vector (pFLAGCMV2) or pFLAGCMV2-hCDC34 as described in Materials and Methods. The RNA was probed with the full-length hICERIIγ cDNA. Lane 1, untransfected control; lanes 2 to 5, results of duplicate experiments. Middle panel, Western blot analysis of the protein collected from the identical sets of experiments described above. Protein lysates were prepared as described in Materials and Methods, electrophoresed on an SDS–5 to 15% polyacrylamide gel, transferred onto a polyvinylidene difluoride membrane, and probed with the Myc epitope antibody. Blots were reprobed with the mouse β-actin antibody to compare loading (bottom panel). The numbers on the right represent molecular mass markers. The data shown are representative of those from 12 independent experiments. (B) Effect of a proteasome inhibitor, MG115, on the hCdc34-induced destabilization of hICERIIγ protein. Transfected JEG3 cells were incubated for 5 to 6 h in the presence or absence of 0.025 mM peptide aldehyde MG115 before harvest. Protein was analyzed as described above. The data shown are representative of those from three independent experiments. (C) Effect of mutant hCdc34 enzymes (C93S and ΔCT) on the stability of hICERIIγ protein in cotransfection assays. Vectors expressing the wild-type or mutant enzymes were cotransfected with the hICERIIγ vector into JEG3 cells, and protein was analyzed as before. The data shown are representative of those from three independent experiments.
To explore whether loss of hICERIIγ protein was secondary to ubiquitin-mediated proteolysis, we repeated the experiments in the presence of the potent 26S proteasome inhibitors MG115 and MG132 (42). Incubating the transfected cells for 5 to 6 h with 0.025 mM MG115 (Fig. 4B) or MG132 (data not shown) prevented the loss of hICERIIγ protein in cells cotransfected with hCDC34. The requirement for an active hCdc34 UBC enzyme in the loss of the hICERIIγ protein was determined by cotransfecting cDNAs encoding two different mutant hCdc34 proteins. The first, hCdc34C93S, has the highly conserved active-site cysteine of the enzyme replaced with a serine. Expression of this mutant protein, unlike the Cdc34 bait, was unable to complement the growth of cdc34-1 yeast at the restrictive temperature (data not shown). The second mutant, hCdc34ΔCT, is a truncated protein which has the carboxy terminus distal to amino acid 189 deleted. In S. cerevisiae, the comparable truncation has been shown to remove the binding site required for association with Cdc53 in yeast (43) and results in a protein unable to complement a cdc34 mutation (34, 64). Comparable expression of the wild-type, hCdc34C93S, and hCdc34ΔCT proteins in transfected JEG3 cells was verified by Western blot analysis (result not shown). In cotransfection assays with pCS2MT-hICERIIγ, the C93S and ΔCT mutant proteins had no effect on the steady-state level of hICERIIγ protein, compared to the loss of protein seen upon cotransfection of the wild-type hCdc34 (Fig. 4C), indicating that the loss of hICERIIγ protein by coexpression of hCdc34 required a fully functional UBC enzyme.
Given the homology between hCdc34 and hRad6B and the requirement for both murine Rad6B and CREM/ICER in spermatogenesis, we performed cotransfection assays with hRAD6B and the targets obtained in the hCDC34 screen. Coexpression of FLAG-tagged hRad6B (pFLAGCMV2-hRAD6B) with Myc-tagged hICERIIγ can mimic the loss of hICERIIγ protein to a similar but lesser degree as hCdc34 (Fig. 5A). This loss of hICERIIγ can also be reversed in the presence of proteasome inhibitors. Similar to the case for hICERIIγ, cotransfection of a Myc-tagged hATF5 fusion with either hCDC34 (Fig. 5B) or hRAD6B cDNA resulted in a significant loss of hATF5 protein, which can be reversed by incubation with MG115. However, once again, in comparison to hCdc34, the loss of hATF5 protein in the presence of hRad6B is lower (data not shown).
FIG. 5.
(A) Effect of pFLAGCMV2-hRAD6B on the stability of pCS2MT-hCERIIγ in JEG3 cells in the presence and absence of a proteasome inhibitor, MG115. (B) Effect of pFLAGCMV2-hCDC34 on the expression of ectopic pCS2MT-hATF5 in cotransfected JEG3 cells in the presence and absence of a proteasome inhibitor, MG115. β-Actin is shown to compare loading. The treatment and Western analysis of the protein were the same as for Fig. 4. The data shown are representative results from three independent experiments.
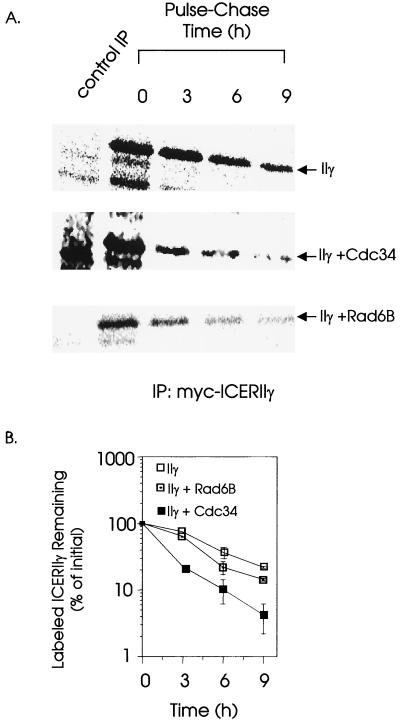
To determine whether the change in steady-state levels of hICERIIγ protein upon cotransfection with hCDC34 and hRAD6B is due to a change in protein half-life, we performed pulse-chase experiments with JEG3 cells (Fig. 6). The cells were transfected with pCS2MT-hICERIIγ in combination with pFLAGCMV2 vector, pFLAGCMV2-hCDC34, or pFLAGCMV2-hRAD6B. Forty hours following transfection, cells were metabolically labeled with [35S]methionine in a pulse-chase protocol. The Myc-ICERIIγ protein was then immunoprecipitated from the samples collected at various time points during the chase by using a monoclonal antibody against the Myc epitope. Immunoprecipitates were resolved on an SDS–15% polyacrylamide gel, and labeled protein was quantitated. The half-life of the transfected ICERIIγ protein during the chase was calculated to be 3.82 h. In the presence of hCdc34 or hRad6B, hICERIIγ protein degraded more rapidly, with estimated half-lives of 2.3 and 2.9 h, respectively (Fig. 6B). In duplicate experiments the kinetics of hICERIIγ protein degradation in the presence of hRAD6B suggests a biphasic pattern of instability with a decreased half-life later in the pulse (Fig. 6B).
FIG. 6.
Pulse-chase analysis of the stability of hICERIIγ protein in JEG3 cells alone or cotransfected with either pFLAGCMV2-hCDC34 or pCS2MT-hRAD6B. 35S pulse-chase labeling for the indicated periods of time was performed as described in Materials and Methods. Cell lysates were immunoprecipitated (IP) with monoclonal anti-Myc-tagged antibody, and the immunoprecipitates were analyzed on an SDS–15% polyacrylamide gel and quantitated. The control immunoprecipitation was performed with a mouse monoclonal antibody raised against the bacterial TrpE protein. (B) Quantitation of the bands corresponding to labeled hICERIIγ protein from panel A. Data are the averages and standard errors of the means from two experiments. In some cases the standard error of the mean is smaller than the symbol.
Coexpression of hCdc34 relieves the repression of cAMP-induced transcription mediated by hICERIIγ and hATF5 in JEG3 cells.
An important question to address is whether Cdc34-mediated degradation affects the biologically active nuclear repressor protein and not just excess (potentially misfolded) hICERIIγ and hATF5 protein. As previously reported by Molina et al. (48), we have found that hICERIIγ is a powerful repressor of PKA (cAMP)-induced transcription (Fig. 7). We used a pSomCAT reporter plasmid that contains a canonical rat somatostatin CRE sequence inserted upstream from the herpesvirus thymidine kinase promoter and the bacterial CAT gene (19). Activation of pSomCAT transcription was obtained by cotransfection of the mouse PKA subunit expression vector (pCαEV) (46) or treatment with 10 mM forskolin for 2 to 3 h before harvest (data not shown). JEG3 cells were transiently transfected with pSomCAT, pCαEV, pSG5-hICERIIγ, or pSG5-hATF5 in combination with pSG5-hCDC34. Both hICERIIγ and hATF5 show strong repression of PKA-induced CAT activity (Fig. 7). However, coexpression of hICERIIγ or hATF5 with hCDC34 completely abrogated the repression of cAMP-induced CAT activity. These results demonstrate that Cdc34 targets for degradation the biologically active transcriptional repressor proteins in mammalian cells.
FIG. 7.
(A) Assay for the effect of hCdc34 on the cAMP-induced transcriptional repression activities of hICERIIγ and hATF5. pSG5-ICERIIγ or pSG5-hATF5 with or without pSG5-hCDC34 was cotransfected into JEG3 cells along with β-d-galactosidase plasmid, pSomCAT, and pCαEV. Reporter pSomCAT was used to measure PKA-mediated activation of the somatostatin CRE element upstream of the CAT gene. pCαEV encodes murine PKA and activates the CRE element. β-d-Galactosidase plasmid was used to assess transfection efficiency. All experiments had equal amounts of plasmid DNA transfected into the JEG-3 cells by using the appropriate pSG5 vector control. (B) Average CAT activity (percent conversion of nonacetylated to acetylated chloramphenicol) from three different experiments as shown in panel A performed in duplicate. Each value represents the mean ± standard error of the mean for six observations.
The ICER-specific domain is not sufficient for degradation.
All four isoforms of human ICER proteins possess the same 9-amino-acid N-terminal domain. The isoforms differ based on the presence or absence of two exons, exon γ and exon Ia (Fig. 1). To analyze the potential role of the ICER-specific domain in the stability of hICERIIγ in vivo, we made a Myc epitope-tagged construct (pCS2MT-hICERIIγ 1-33) bearing the N-terminal 33 amino acids. Cotransfection of the mini-ICER construct with pFLAGCMV2-hCDC34 into either JEG3 cells (Fig. 8A) or NIH 3T3 cells (data not shown) did not result in significant loss of the truncated protein compared to the full-length hICERIIγ, indicating that the ICER-specific domain and exon γ are not sufficient for hCdc34 targeting and subsequent degradation. In addition, this result also demonstrates that specific sequences in the full-length hICERIIγ protein are required for targeting by hCdc34.
FIG. 8.
(A) Role of the ICER-specific domain in the hCdc34-mediated destabilization of the ICERIIγ protein. A Myc epitope-tagged construct (pCS2MT-ICERIIγ 1-33) bearing the N-terminal 33 amino acids of the ICER-specific domain and exon γ of the hICERIIγ protein (mini-IIγ) was made as described in Materials and Methods. Cotransfection of full-length hICERIIγ (IIγ) or pCS2MT-ICERIIγ 1-33 (mini-IIγ) with pFLAGCMV2-hCDC34 into JEG3 cells and Western analysis were performed as described for Fig. 4. Lanes 1 and 2, expression of the full-length Myc epitope-tagged hICERIIγ protein; lanes 3 and 4, expression of the mini-ICERIIγ protein; lanes 2 and 4, presence of FLAG-hCDC34; lanes 1 and 3, absence of FLAG-hCDC34. The data shown are representative results from three independent experiments. (B) Effect of pSG5-hCDC34 on the expression of ectopic pSG5-mCREMβ in cotransfected JEG3 cells in the presence and absence of a proteasome inhibitor, MG115. The treatment and Western analysis of the protein were the same as for Fig. 4. The data shown are representative results from three independent experiments.
Coexpression of hCDC34 destabilizes CREM β in JEG3 cells.
Given these results and an inverse correlation between Cdc34 and CREM α/β during spermatogenesis, we investigated the targeting of the CREM α/β repressor isoforms by hCdc34. Mammalian expression vectors encoding both mouse CREM β (mCREM β), which does not contain the ICER-specific exon, and hCdc34 were cotransfected into JEG3 cells (Fig. 8B) and NIH 3T3 cells (result not shown). The protein lysates from the transfected cells were analyzed in a Western blot with CREM antiserum as a probe. Coexpression of mCREMβ and hCDC34 resulted in a significant loss of the CREM β protein, which was reversed by addition of the proteasome inhibitor MG115.
The Myc-ICERIIγ protein is ubiquitinated by endogenous hCdc34.
As ubiquitination is not a prerequisite for the degradation of proteins via 26S proteasomes (8), we examined the polyubiquitination of hICERIIγ protein. Polyubiquitination has been detected for a number of substrates, including c-Jun (68), cyclin E (10), and p27 (52). To demonstrate the formation of the polyubiquitin-hICERIIγ conjugates, an immunoprecipitation-Western assay in which an HA-tagged ubiquitin construct was cotransfected along with the Myc-tagged hICERIIγ in the presence and absence of pFLAGCMV2-hCDC34 into JEG3 cells was used. Five hours before harvest, cells were treated with MG115. Extracts were made in the presence of N-ethylmaleimide and immunoprecipitated with either the anti-Myc or anti-HA monoclonal antiserum, followed by Western blotting with either anti-Myc or anti-HA antiserum. High-molecular-weight species of hICERIIγ protein were detected with either antibody and in cells transfected with both HA-ubiquitin and Myc-hICERIIγ or in cells transfected with FLAG-hCDC34, Myc-ICERIIγ, and HA-ubiquitin (Fig. 9A). However, no ubiquitin conjugates were observed in the presence of His6-tagged ubiquitin as opposed to HA-ubiquitin. Thus, the slower-migrating species contain both Myc-hICERIIγ protein and HA-ubiquitin and represent multiubiquitinated forms of hICERIIγ protein. A ladder of bands was found in the presence and absence of transfected hCDC34, which indicates that ICERIIγ can be targeted by endogenous UBC enzymes, confirming an earlier report (17).
FIG. 9.
(A) Identification of hICERIIγ-ubiquitin conjugates by an immunoprecipitation (IP)-Western blot analysis. JEG3 cells were transfected with the indicated plasmids and then treated with 0.025 mM MG115 5 h before harvest. Left panel, cell lysates were immunoprecipitated with HA antibody, followed by Western blotting with the Myc antibody (which recognizes the Myc epitope on hICERIIγ). Right panel, cell lysates were immunoprecipitated with the Myc antibody, and the Western blot was probed with anti-HA. Ubiquitin-hICERIIγ conjugates are detected only in cells transfected with both Myc- and HA epitope-tagged constructs and in the presence or absence of exogenous hCDC34. A His6-tagged ubiquitin construct (His-Ub) was used as a negative control. Immunoglobulin G (IgG) heavy chain is visualized in this analysis and is indicated. (B) Effect of the expression of Cdc34-inhibitory constructs hCDC34DN and hCDC34AS on the stability of ectopically expressed myc-ICERIIγ protein. JEG3 cells were transiently transfected with pCS2MT-ICERIIγ and the pSG5 empty vector, pSG5-hCDC34DN, or pSG5-hCDC34AS. Cell lysates were prepared and subjected to Western analysis as described for Fig. 4, using anti-Myc and anti-Cdc34 antibodies. β-Actin is shown to compare loading. The data shown are representative of those from two independent experiments.
To determine whether endogenous hCdc34 is one of the endogenous UBC enzymes responsible for the ubiquitination of ICERIIγ, we transfected cells with the Myc-tagged hICERIIγ construct and two Cdc34-inhibitory constructs. The first is a mammalian expression vector containing the corresponding double mutation in the human sequence (hCDC34DN-C93S and -L97S) of a dominant negative allele of yeast CDC34 (2). The hCDC34DN mutation has been previously reported to inhibit the ubiquitination of other Cdc34 substrates, including MyoD in mammalian cells (66). Transfection of the cDNA containing the dominant negative mutation subcloned into the pSG5 vector (pSG5-hCDC34DN) resulted in high-level expression of the mutant hCdc34 protein (Fig. 9B). A second inhibitory construct, containing the wild-type human cDNA subcloned into pSG5 in an antisense orientation (pSG5-hCDC34AS), was also constructed. Transfection of this construct resulted in a decrease in the steady-state level of endogenous Cdc34 protein (Fig. 9B). Cotransfection of JEG-3 cells with the Myc-tagged hICERIIγ and either pSG5 empty vector or dominant negative or antisense hCDC34 constructs was carried out in parallel. Expression of both the dominant negative and antisense hCDC34 constructs results in increased levels of Myc-tagged hICERIIγ protein compared to that with the empty vector control. These results suggest that the ubiquitination of the Myc-tagged hICERIIγ demonstrated in Fig. 9A is at least partially due to ubiquitination by the endogenous hCdc34 UBC enzyme.
Endogenous ICER protein expression is increased by mutation of Rad6B or inhibition of Cdc34 activity.
The data presented here are consistent with both the human Cdc34 and Rad6B conjugating enzymes targeting transfected hICERIIγ and ATF5 proteins for ubiquitination. In order to determine the impact of these enzymes on the endogenous ICER target proteins, we obtained mouse embryo fibroblasts (MEF) mutant for the orthologous gene mHR6B, which have been previously described (57). Whole protein lysates of MEF from mHR6B homozygous null mice (mHR6B−/−) or wild-type littermate controls (mHR6B+/+) were hybridized with an antibody to ICER proteins as described above. A distinct increase in expression of the ∼18-kDa band which comigrates with ICERIIγ in the null cells is seen compared with the wild-type cells (Fig. 10A). A number of other proteins recognized by this antibody are increased in the mHR6B null cells and represent other CREM isoforms.
FIG. 10.
(A) Western blot analysis of the expression profiles of endogenous ICER and other CREM isoforms in mHR6B wild-type (mHR6B+/+) and mutant (mHR6B−/−) MEF. Total cell lysates from exponentially growing cells were prepared and subjected to Western analysis as described for Fig. 4, using anti-ICER antibody. Molecular mass markers are indicated on the right. β-Actin is shown to compare loading. The data shown are representative of those from two independent experiments. (B) Effect of the expression of an hCDC34AS or hCDC34DN mutant construct on the stability of endogenous ICER protein. JEG3 cells were transiently transfected with vector alone (pSG5), pSG5-CDC34DN, or pSG5-CDC34AS. Cell lysates were prepared and subjected to Western analysis as described for Fig. 4, using anti-ICER and anti-Cdc34 antibodies. β-Actin is shown to compare loading. The data shown are representative of those from two independent experiments.
Although cdc34 null cells are not available, our results suggest that we can inhibit the endogenous Cdc34 enzyme by expression of either the dominant negative or antisense constructs. We therefore transfected JEG-3 cells with either the empty vector or dominant negative or antisense Cdc34 constructs and isolated whole-cell lysates 36 to 40 h after transfection. The transient-transfection technique used here results in a transfection efficiency of approximately 30% (data not shown). We therefore expect that the subpopulation of cells that are transfected with the antisense or dominant negative construct will have inhibition of Cdc34 activity, while the remainder of the cells will have normal Cdc34 activity. Despite this limitation, Western blot analysis of these lysates (Fig. 10B) with an ICER antibody demonstrates that the cells transfected with either the antisense or dominant negative construct have a significantly increased steady-state level of the endogenous ICER protein compared with the empty vector control.
DISCUSSION
In this study, we have found that the human Cdc34 and Rad6B UBC enzymes can target repressors of cAMP-inducible transcription for ubiquitination, including hICERIIγ (a previously described ICER isoform), CREM β, and hATF5 (a novel protein isolated in the present study). Most substrates of the yeast UBC enzymes have been identified by changes in stability of candidate target proteins in UBC mutant strains. The target proteins obtained in our study were initially identified as the result of a two-hybrid screen in yeast with hCDC34 cDNA as the bait, which demonstrates that this method is capable of detecting the potentially transient interaction between Cdc34 and its substrates. Our ability to detect this interaction was likely dependent on the finding that the hCDC34 bait encoded a functional protein in yeast.
In addition to the interactions detected in the yeast two-hybrid assay, several other biochemical experiments directly support the model in which both hICERIIγ and hATF5 are targeted by hCdc34 for ubiquitination and subsequent degradation via the 26S proteasome. First, transfection of hICERIIγ or hATF5 with hCDC34 or hRAD6B expression constructs in mammalian cells results in significant and sometimes complete loss of the target proteins; this loss is reversed in the presence of proteasome inhibitors. Second, coexpression of hCdc34 mutant proteins fails to destabilize the targets. Third, the half-life of the target protein is decreased in the presence of hCdc34 and hRad6B. Fourth, coexpression with hCDC34 in mammalian cells completely abrogates the repression of cAMP-induced transcription by hICERIIγ and hATF5. These findings taken together demonstrate that Cdc34 coexpression results in the loss of the biologically relevant repressor protein.
The involvement of the ubiquitin-proteasome pathway in the degradation of endogenous ICER protein in primary cardiocytes and myogenic cell lines has been previously reported (17). We also detect ubiquitin conjugates of hICERIIγ-Myc fusion protein in the presence of ectopic ubiquitin and peptide-aldehyde without addition of exogenous hCDC34 or hRAD6B. Further evidence for specific targeting of the endogenous ICER protein by endogenous Cdc34 and Rad6B enzymes was obtained by analysis of the mHR6B−/− MEF and JEG-3 cells transfected with Cdc34 inhibitory constructs. These experiments confirm the results seen with transfected ICERIIγ protein. Loss of either Rad6B or Cdc34 activity results in increased levels of endogenous ICER protein.
We have demonstrated that both the Cdc34 and Rad6B UBC enzymes are capable of targeting for degradation CREM, ICER, and ATF proteins. Although S. cerevisiae CDC34 (6) was originally identified due to a cell cycle phenotype and RAD6 (25) was identified through a DNA repair phenotype, the yeast enzymes also have common targets. For example, degradation of Gcn4, which contains a bZIP motif, also requires both Rad6 (UBC2) and Cdc34 (UBC3) (35). There are other examples of shared targeting, with Matα2 being targeted by UBC4, UBC5, UBC6, and UBC7 enzymes (7). In our experiments the targeting by multiple UBC enzymes (E2s) has different characteristics; e.g., hCdc34 has a more significant effect on the half-life of hICERIIγ than hRad6B does. The degradation kinetics with hRad6B was biphasic, with an initial lower rate. Similar differences in the kinetics have also been reported for the degradation of yeast Gcn4 by Cdc34 and Rad6 (35). Also consistent with the differential effect of Cdc34 and Rad6B, we detected an interaction in a mammalian two-hybrid assay with hRAD6B but not hCDC34 cDNA as bait and hICERIIγ as the target construct (data not shown), presumably because the degradation of the target is too rapid in the presence of the transfected hCdc34.
It was suggested by Kornitzer et al. (35) that targeting of Cdc34 and Rad6 may be specific to proteins containing PEST sequences. Our analysis of hCdc34 interactants does not support that hypothesis. In ICER proteins, the unique ICER-specific domain and the γ exon are rich in PEST-containing sequences. ICERIIγ lacks the characteristic γ exon, and a construct containing only the ICER-specific domain was not destabilized by hCdc34. hICERIIγ also lacks the amino-terminal phosphorylation domain (P box or kinase-inducible domain) unique to CREM and CREB, which indicates that this specific phosphorylation is not required for hCdc34-mediated targeting. Further studies to identify specific domains, if any, in these bZIP transcription factors that are targeted by hCdc34 are in progress. However, extensive deletion studies of Gcn4 by Kornitzer et al. (35) were not able to demonstrate a single region of Gcn4 sufficient for efficient ubiquitination by both Cdc34 and Gcn4. Three targets (hICERIIγ, hATF5, and clone 30-17) isolated from the two-hybrid screen and CREM β and Gcn4 (35) all have a characteristic bZIP domain. In these targets, the basic domain, flanking the characteristic leucine zipper, is rich in lysine and arginine residues, and this lysine-rich region may form the target for ubiquitination.
Currently, there is uncertainty as to whether Cdc34-mediated ubiquitination requires an SCF complex for all targets. Michel and Xiong (47) have indicated that the SCF pathway, although similarly used by the mammalian Cullin-1, is not shared by other Cullin members, which may use a Skp1–F-box-independent pathway. From our studies, we cannot be conclusive about the involvement of the SCF complex in the Cdc34-mediated targeting of hICERIIγ and hATF5. hCdc34 apparently can complex with the SCF complex in yeast, based on complementation of a cdc34-1 strain, and a carboxy-truncated mutant which cannot complex with the Cullin proteins is not capable of targeting hICERIIγ. However, we find that addition of exogenous hCdc34 or hRad6B protein alone in mammalian cells is sufficient to increase turnover of the ectopic hICERIIγ. Therefore, if the SCF complex is required for this degradation, it is found in excess and hCdc34 is limiting. At present it is also not known whether Rad6- and Cdc34-mediated ubiquitination of common targets requires shared proteins. Our development of a Cdc34-dependent in vivo assay for ubiquitination of these targets in mammalian cells should allow us to dissect these pathways and determine which components of the SCF complex are required for ubiquitination.
The expression pattern of mammalian Cdc34 and the SCF complex components has not been well described. We examined the expression profiles of both Cdc34 and Rad6B proteins along with the SCF subunit Cullin proteins (Cul-1 and Cul-2) during murine testicular development. In budding yeast, the Cdc34 protein level has been reported to remain constant throughout the cell cycle (20). We find that both UBC enzymes Cdc34 and Rad6B are expressed in a developmentally regulated manner in mouse testis and germ cells. The ratio of Cdc34 to the Cullin proteins does not remain constant during development, with maximal expression of Cul-1 and Cul-2 at day 20 in late pachytene and maximal expression of Cdc34 at day 30. Thus, at different points during spermatogenesis Cdc34 may form different complexes with unique targeting specificities. Cdc34 is maximally expressed late during spermatid differentiation when most intracellular proteins are being degraded and a complex series of chromatin modifications, including the ubiquitination of histone, takes place (for a review, see reference 9). We also reproducibly detect a higher-molecular-weight form of Cdc34 specifically in germ cells late in spermatogenesis, which may represent an additional form of regulation of the enzyme. Therefore, despite the evidence for Cdc34 function in mitotically growing cells, Cdc34 expression in the testis is highly regulated and is maximal in the postmeiotic phase of spermatogenesis. In addition, a second E1 protein, Ube1y, expressed in mouse testis is encoded by a gene on the Y chromosome (31). Therefore, a ubiquitin-activating (E1) enzyme plus multiple UBC (E2) enzymes are present in both X- and Y-containing gametes late in spermatogenesis when histone ubiquitination and massive protein degradation is occurring.
The transcriptional activity of ICER, including the repression of its own promoter, has been reported to be determined by the intracellular concentration of the ICER proteins (48, 49). Thus, turnover of ICER and its degradation via the ubiquitin-proteasome pathway may act as a regulatory mechanism to relieve transcriptional repression and to control the negative effect of ICER on the cAMP-inducible transcriptional response. Although we were unable to show a simple inverse correlation between the expression of ICER and Cdc34 and Rad6B proteins in germ cells, this may be for several reasons: (i) ICER protein expression is a sum of transcriptional regulation, alternative splicing, and protein stability; (ii) there may be a requirement for other regulatory proteins, including subunits of the SCF complex, to mediate ubiquitination of ICER isoforms; and (iii) ubiquitin-mediated proteolysis of ICER proteins may occur in only a specific subset of cells in the testis. On the other hand, a clear negative correlation was observed between the repressor CREM isoform CREM β and Cdc34 and Rad6B proteins during testicular development and in the haploid germ cells, indicating a potential targeting of CREM β by Cdc34 in late spermatids. This finding was further supported by the destabilization of CREM β by Cdc34 in transfection assays and warrants further investigation.
ICER isoforms regulate a number of biological processes, including spermatogenesis, circadian rhythm of transcription in the pineal gland, and cell proliferation (for a review, see reference 60). For example, ICER isoforms negatively regulate the expression of cyclin A mRNA, potentially resulting in specific induction of cyclin A from mid-G1 to early S, when the level of ICER is low (13) and the Cdc34-SCF complex is active (36). Therefore, Cdc34 may regulate cyclin A both at the transcriptional level and by the previously documented association of the cyclin A-CDK2 complex with the Cdc34-SCF complex (41). Conversely, a recent report has demonstrated that overexpression of ICERIIγ inhibits tumor cell growth and results in G2 arrest at a point in the cycle when the Cdc34-SCF complex may be less active (56). The cyclin A promoter is also suppressed by ATF4, which has considerable sequence homology to hATF5, a protein identified as a target of Cdc34. Thus, knowledge gained about mammalian Cdc34 function will be important in understanding how selective destabilization of proteins through ubiquitination regulates diverse processes, including both meiotic and mitotic cell cycles.
ACKNOWLEDGMENTS
We are grateful to a large number of investigators who provided reagents for this study. We thank M. Vidal for yeast two-hybrid reagents; L. Prakash, P. Sassone-Corsi, G. McKnight, B. Kelly, D. Bohmann, M. Goebl, and B. Clurman for strains and plasmids; J. La Baer for the cDNA library; C. Molina, H. Roest, J. Hoeijmaker, P. Sassone-Corsi, and W. Krek for antisera; S. Sharan and A. Bradley for mouse tissues; J. Hoeijmaker and H. Roest for mHR6B recombinant cells; G. Wilson for irradiated testes; G. Shetty for testes from jsd mice; Y. Zhang for elutriated germ cells; and S. Luo for assistance with the Northern analysis. We thank S. Elledge and V. Lundblad for comments on the manuscript and an anonymous reviewer for suggesting the experiments with the results shown in Fig. 10.
This study was supported by the grants from the U.S. Army Medical Research and Material Command (DAMD-17-96-1-6087 to D.P.), the American Cancer Society (JFRA-559 to S.E.P.), and the National Institutes of Health (HD16843 to M.L.M.) and by Baylor Child Health Research Center grant P30-HD27832.
REFERENCES
- 1.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee A, Deshaies R J, Chau V. Characterization of a dominant negative mutant of the cell cycle ubiquitin-conjugating enzyme Cdc34. J Biol Chem. 1995;270:26209–26215. doi: 10.1074/jbc.270.44.26209. [DOI] [PubMed] [Google Scholar]
- 3.Beamer W G, Cunliffe-Beamer T L, Shultz K L, Langley S H, Roderick T H. Juvenile spermatogonial depletion (jsd): a genetic defect of germ cell proliferation of male mice. Biol Reprod. 1988;38:899–908. doi: 10.1095/biolreprod38.4.899. [DOI] [PubMed] [Google Scholar]
- 4.Bellve A R, Cavicchia J C, Millette C F, O’Brien D A, Bhatnagar Y M, Dym M. Spermatogenic cells of the prepubertal mouse, isolation and morphological characterization. J Cell Biol. 1977;74:68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blendy J A, Kaestner K H, Weinbauer G F, Nieschlag E, Schutz G. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature. 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 6.Byers B, Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- 7.Chen P, Johnson P, Sommer T, Jentsch S, Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993;74:357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- 8.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 9.Clermont Y, Oko R, Hermo L. Cell biology of mammalian spermatogenesis. In: Desjardins C, Ewing L L, editors. The cell and molecular biology of the testis. Oxford, United Kingdom: Oxford University Press; 1993. pp. 332–376. [Google Scholar]
- 10.Clurman B E, Sheaff R J, Thress K, Groudine M, Roberts J M. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdck2 binding and cyclin phosphorylation. Genes Dev. 1996;10:1979–1990. doi: 10.1101/gad.10.16.1979. [DOI] [PubMed] [Google Scholar]
- 11.Delmas V, van der Hoorn F, Mellstrom B, Jegou B, Sassone-Corsi P. Induction of CREM activator proteins in spermatids: down-stream targets and implications for haploid germ cell differentiation. Mol Endocrinol. 1993;7:1502–1514. doi: 10.1210/mend.7.11.8114765. [DOI] [PubMed] [Google Scholar]
- 12.Delmas V, Sassone-Corsi P. The key role of CREM in the cAMP signaling pathway in the testis. Mol Cell Endocrinol. 1994;100:121–124. doi: 10.1016/0303-7207(94)90290-9. [DOI] [PubMed] [Google Scholar]
- 13.Desdouets C, Matesic G, Molina C A, Foulkes N S, Sassone-Corsi P, Brechot C, Sobczak-Thepot J. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol Cell Biol. 1995;15:3301–3309. doi: 10.1128/mcb.15.6.3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deshaies R J. Phosphorylation and proteolysis: partners in regulation of cell division in budding yeast. Curr Opin Genet Dev. 1997;7:7–16. doi: 10.1016/s0959-437x(97)80103-7. [DOI] [PubMed] [Google Scholar]
- 15.Dohmen R J, Madura K, Bartel B, Varshavsky A. The N-end rule is mediated by the UBC2 (RAD6) ubiquitin-conjugating enzyme. Proc Natl Acad Sci USA. 1991;88:7351–7355. doi: 10.1073/pnas.88.16.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feldman R M R, Correll C C, Kaplan K B, Deshaies R J. A complex of Cdc4p and cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- 17.Folco E J, Koren G. Degradation of the inducible cAMP early repressor (ICER) by the ubiquitin-proteasome pathway. Biochem J. 1997;328:37–43. doi: 10.1042/bj3280037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foulkes N S, Borrelli E, Sassone-Corsi P. CREM gene: use of alternative DNA binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 1991;64:739–749. doi: 10.1016/0092-8674(91)90503-q. [DOI] [PubMed] [Google Scholar]
- 19.Foulkes N S, Mellstorm B, Benusiglio E, Sassone-Corsi P. Developmental switch of CREM function during spermatogenesis: from antagonist to transcriptional activator. Nature. 1992;355:80–84. doi: 10.1038/355080a0. [DOI] [PubMed] [Google Scholar]
- 20.Goebl M, Yochem J, Jentsch S, McGrath J P, Varshavsky A, Byers B. The yeast cell cycle gene CDC34 encodes a ubiquitin-conjugating enzyme. Science. 1988;241:1331–1335. doi: 10.1126/science.2842867. [DOI] [PubMed] [Google Scholar]
- 21.Grabske R J, Lake S, Gledhill B L, Meistrich M L. Centrifugal elutriation: separation of spermatogenic cells on the basis of sedimentation velocity. J Cell Physiol. 1975;86:177–189. doi: 10.1002/jcp.1040860119. [DOI] [PubMed] [Google Scholar]
- 22.Green S, Issemann I, Sheer E. A versatile in vivo and in vitro eukaryotic vector for protein engineering. Nucleic Acids Res. 1988;16:369. doi: 10.1093/nar/16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas A L, Reback P B, Chau V. Ubiquitin conjugation by the yeast RAD6 and CDC34 gene products. Comparison to their putative rabbit homologs, E2(20K) and E2(32K) J Biol Chem. 1991;266:5104–5112. [PubMed] [Google Scholar]
- 24.Haas A L, Siepmann T J. Pathways of ubiquitin conjugation. FASEB J. 1997;11:1257–1268. doi: 10.1096/fasebj.11.14.9409544. [DOI] [PubMed] [Google Scholar]
- 25.Haynes R H, Kunz B A. DNA repair and mutagenesis in yeast. In: Strathern J, Jones E, Broach J, editors. The molecular biology of the yeast Saccharomyces cerevisiae: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1981. pp. 371–414. [Google Scholar]
- 26.Hemzawi Z, Mooney E E, Plon S E. Characterization of the mammalian CDC34 cell cycle gene. Am Assoc Cancer Res. 1995;36:A191. [Google Scholar]
- 27.Hershko A. Role of ubiquitin-mediated proteolysis in cell cycle control. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 28.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Hochstrasser M. Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol. 1995;7:215–223. doi: 10.1016/0955-0674(95)80031-x. [DOI] [PubMed] [Google Scholar]
- 30.Jentsch S, McGrath J P, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987;329:131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- 31.Kay G F, Ashworth A, Penny G D, Dunlop M, Swift S, Brockdroff N, Rostan S. A candidate spermatogenesis gene on mouse Y chromosome is homologous to ubiquitin-activating enzyme E1. Nature. 1991;345:486–489. doi: 10.1038/354486a0. [DOI] [PubMed] [Google Scholar]
- 32.Kishi T, Seno T, Yamao F. Grr1 functions in the ubiquitin pathway in Saccharomyces cerevisiae through association with Skp1. Mol Gen Genet. 1998;257:143–148. doi: 10.1007/s004380050633. [DOI] [PubMed] [Google Scholar]
- 33.Koken M H, Reynolds P, Jaspers-Dekker I, Prakash L, Prakash S, Bootsma D, Hoeijmakers J H. Structural and functional conservation of two human homologs of the yeast DNA repair gene RAD6. Proc Natl Acad Sci USA. 1991;88:8865–8869. doi: 10.1073/pnas.88.20.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolman C J, Toth J, Gonda D K. Identification of a portable determinant of cell cycle function with the carboxyl-terminal domain of the yeast CDC34 (UBC3) ubiquitin conjugating (E2) enzyme. EMBO J. 1992;11:3081–3090. doi: 10.1002/j.1460-2075.1992.tb05380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor GCN4. EMBO J. 1994;13:303–312. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krek W. Proteolysis and the G1-S transition: the SCF connection. Curr Opin Genet Dev. 1998;8:36–42. doi: 10.1016/s0959-437x(98)80059-2. [DOI] [PubMed] [Google Scholar]
- 37.Kupiec M, Simchen G. Regulation of the RAD6 gene of Saccharomyces cerevisiae in the mitotic cell cycle and in meiosis. Mol Gen Genet. 1986;203:538–543. doi: 10.1007/BF00422083. [DOI] [PubMed] [Google Scholar]
- 38.Lalli E, Lee J S, Lamas M, Tamai K, Zazopoulos E, Nantel F, Penna L, Foulkes N S, Sassone-Corsi P. The nuclear response to cAMP: role of transcription factor CREM. Philos Trans R Soc Lond B. 1996;351:201–209. doi: 10.1098/rstb.1996.0017. [DOI] [PubMed] [Google Scholar]
- 39.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 40.Lin W W, Lamb D J, Lipshultz L I, Kim E D. Absence of cyclic adenosine 3′:5′ monophosphate responsive element modulator expression at the spermatocyte arrest stage. Fertil Steril. 1998;69:533–538. doi: 10.1016/s0015-0282(97)00535-5. [DOI] [PubMed] [Google Scholar]
- 41.Lisztwan J, Marti A, Sutterluty H, Gstaiger M, Wirbelauer C, Krek W. Association of human CUL-1 and ubiquitin-conjugating enzyme CDC34 with the F-box protein p45SKP2: evidence for evolutionary conservation in the subunit composition of the CDC34-SCF pathway. EMBO J. 1998;17:368–383. doi: 10.1093/emboj/17.2.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maki C G, Howley P. Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol. 1997;17:355–363. doi: 10.1128/mcb.17.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathias N, Steussy C N, Goebl M G. An essential domain within Cdc34p is required for binding to a complex containing Cdc4p and Cdc53p in Saccharomyces cerevisiae. J Biol Chem. 1998;273:4040–4045. doi: 10.1074/jbc.273.7.4040. [DOI] [PubMed] [Google Scholar]
- 44.Meistrich M L, Hunter N R, Suzuki N, Trostle P K, Withers H R. Gradual regeneration of mouse testicular stem cells after exposure to ionizing radiation. Radiat Res. 1978;74:349–362. [PubMed] [Google Scholar]
- 45.Meistrich M L, Longtin J, Brock W A, Grimes S R, Jr, Mace M L. Purification of rat spermatogenic cells and preliminary biochemical analysis of these cells. Biol Reprod. 1981;25:1065–1077. doi: 10.1095/biolreprod25.5.1065. [DOI] [PubMed] [Google Scholar]
- 46.Mellon P L, Clegg C H, Correll L A, McKnight G S. Regulation of transcription by cyclic AMP-dependent protein kinase. Proc Natl Acad Sci USA. 1989;86:4887–4891. doi: 10.1073/pnas.86.13.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michel J J, Xiong Y. Human CUL-1, but not other cullin family members, selectively interacts with SKP1 to form a complex with SKP2 and cyclin A. Cell Growth Differ. 1998;9:435–449. [PubMed] [Google Scholar]
- 48.Molina C A, Foulkes N S, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 49.Monaco L, Foulkes N S, Sassone-Corsi P. Pituitary follicle-stimulating hormone (FSH) induces CREM gene expression in Sertoli cells: involvement in long-term desensitization of the FSH receptor. Proc Natl Acad Sci USA. 1995;92:10673–10677. doi: 10.1073/pnas.92.23.10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nantel F, Monaco L, Foulkes N S, Masquilier D, LeMeur M, Henriksen K, Dierich A, Parvinen M, Sassone-Corsi P. Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature. 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 51.Nishizawa M, Nagata S. cDNA clones encoding leucine-zipper proteins which interact with G-CSF gene promoter element 1-binding protein. FASEB Lett. 1992;299:36–38. doi: 10.1016/0014-5793(92)80094-w. [DOI] [PubMed] [Google Scholar]
- 52.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Del Sal G, Chau V, Yew P R, Draetta G F, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 53.Pati, D., and S. E. Plon. Unpublished data.
- 54.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 55.Plon S E, Leppig K A, Do N H, Groudine M. Cloning of the human homolog of the CDC34 cell cycle gene by complementation in yeast. Proc Natl Acad Sci USA. 1993;90:10484–10488. doi: 10.1073/pnas.90.22.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Razavi R, Ramos J C, Yehia G, Schlotter F, Molina C A. ICER-IIγ is a tumor suppressor that mediates the antiproliferative activity of cAMP. Oncogene. 1998;17:3015–3019. doi: 10.1038/sj.onc.1202225. [DOI] [PubMed] [Google Scholar]
- 57.Roset H P, Klaveren J V, de Wit J, vanGurp C G, Koken M H M, Vermey M, van Roijen J H, Hoogerbrugge J W, Vreeburg J T M, Baarends W M, Bootsma D, Grootegoed J A, Hoeijmakers J H J. Inactivation of the HR6B ubiquitin-conjugating DNA repair enzyme in mice causes male sterility associated with chromatin modification. Cell. 1996;86:799–810. doi: 10.1016/s0092-8674(00)80154-3. [DOI] [PubMed] [Google Scholar]
- 58.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 59.Sassone-Corsi P. Transcription factors responsive to cAMP. Annu Rev Cell Dev Biol. 1995;11:355–377. doi: 10.1146/annurev.cb.11.110195.002035. [DOI] [PubMed] [Google Scholar]
- 60.Sassone-Corsi P. Coupling gene expression to cAMP signaling: role of CREB and CREM. Int J Biochem Cell Biol. 1998;30:27–38. doi: 10.1016/s1357-2725(97)00093-9. [DOI] [PubMed] [Google Scholar]
- 61.Scheffner M, Smith S, Jentsch S. The ubiquitin-conjugating system. In: Peters J-M, Harris J R, Finley D, editors. Ubiquitin and the biology of cell. New York, N.Y: Plenum Publishing Corporation; 1998. pp. 65–98. [Google Scholar]
- 62.Schiestl R H, Giets R D. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr Genet. 1989;16:339–346. doi: 10.1007/BF00340712. [DOI] [PubMed] [Google Scholar]
- 63.Schneider B L, Yang Q H, Futcher A B. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–562. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 64.Silver E T, Gwozd T J, Ptak C, Goebl M, Ellison M J. A chimeric ubiquitin conjugating enzyme that combines the cell cycle properties of CDC34 (UBC3) and the DNA repair properties of RAD6 (UBC2): implications for the structure, function and evolution of the E2s. EMBO J. 1992;11:3091–3098. doi: 10.1002/j.1460-2075.1992.tb05381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Skowyra D, Craig K L, Tyres M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 66.Song A, Wang Q, Goebl M G, Harrington M A. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol Cell Biol. 1998;18:4994–4999. doi: 10.1128/mcb.18.9.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sung P, Berleth E, Pickart C, Prakash S, Prakash L. Yeast RAD6 encoded ubiquitin conjugating enzyme mediates protein degradation dependent on the N-end-recognizing E3 enzyme. EMBO J. 1991;10:2187–2193. doi: 10.1002/j.1460-2075.1991.tb07754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Treier M, Staszewski L, Bohann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 69.Vidal M, Brachmann R, Fattaey A, Harlow E, Boeke J D. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vidal M, Braun P, Chen E, Boeke J D, Harlow E. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc Natl Acad Sci USA. 1996;93:10321–10326. doi: 10.1073/pnas.93.19.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]