Abstract
Chlorhexidine gluconate (CHG) is an antiseptic that is widely used in healthcare due to its excellent safety profile and wide spectrum of activity. Daily bathing with CHG has proven to be effective in the prevention of healthcare-associated infections and multidrug-resistant pathogen decolonization. Despite the proven benefits of CHG use, there remain concerns and unanswered questions about the potential for unintended microbial consequences of routine CHG bathing. This review aims to explore some of these questions.
Keywords: chlorhexidine gluconate, healthcare-associated infections, decolonization, antibiotic resistance, antiseptic
Healthcare-associated infections (HAIs) due to multidrug-resistant organisms (MDROs) are associated with high attributable mortality, increased length of stay, and excess healthcare cost [1, 2]. Colonization with MDROs likely precedes clinical infection [3]. Clinical evidence has demonstrated the effectiveness of chlorhexidine gluconate (CHG) bathing for reduction in a patient’s MDRO bioburden and reduction of HAIs [4–11].
Chlorhexidine gluconate is a cationic biguanide that binds to negatively charged microbial cell membranes, thereby disrupting their function, leading to potential collapse and intracellular leakage [12]. It has been used widely in healthcare since its introduction in the 1950s. This is largely due to its excellent safety profile with only rare cases of hypersensitivity and anaphylaxis reported [13] and wide spectrum of activity, which includes gram-positive and gram-negative bacteria and fungi. Notably, it lacks activity against mycobacteria and bacterial spores [9, 14].
Chlorhexidine gluconate is used in a variety of applications for HAI prevention. These include routine handwashing, preprocedure skin preparation, indwelling catheter exit-site care, oral care for prevention of ventilator-associated pneumonia, and whole-body bathing [10]. A common CHG bathing regimen is to cleanse patients’ skin daily with a liquid or foam CHG preparation, which can be rinsed off or not, or with no-rinse CHG-impregnated washcloths. Chlorhexidine gluconate binds to the stratum corneum of the skin, providing an antimicrobial coating that is replenished with each bath [15]. Due to the above-mentioned evidence of CHG’s effectiveness in HAI reduction, routine CHG bathing has been recommended increasingly by professional societies [16]. A 2014 survey of over 4000 National Healthcare Safety Network participating centers found that CHG bathing was used routinely for MDRO control in approximately 49% of long-term care facilities and 63% of acute-care facilities [17].
As with many of our tools in the anti-infective armamentarium, widespread use may result in unintended consequences (Figure 1). The possibility of emergence of resistance to CHG is of particular concern. Assessment of phenotypic resistance to CHG has remained a challenge for a number of reasons. There is a lack of consensus regarding susceptibility definitions, with a variety of testing methods applied in the literature in attempt to explore this (ie, broth microdilution, agar dilution, time-kill, hard surface carrier tests) [18]. Minimum inhibitory concentrations (MICs) and minimum bactericidal concentrations (MBCs) have been the most frequently applied in this endeavor. However, MIC/MBC methods were developed to predict clinical response to systemically administrated antibiotics, and no standardized definitions of which MIC/MBC breakpoints represent phenotypic resistance to CHG exist. Moreover, antiseptics such as CHG are applied topically; thus, it is unclear whether there is clinical relevance to the detection of increased MICs/MBCs. Studies including more informative microbial comparative definitions that are generally accepted forms of antiseptic efficacy (eg, exposure-log reduction measures) are noticeably lacking [19, 20]. Despite the limitations of MIC/MBC methods for assessment of CHG resistance, many investigators have applied these methods. But in doing so they have used different quality-control organisms and different conditions for the same methods. Even when the same species of organisms are tested, MICs and MBCs have varied significantly and cannot be confidently assessed interchangeably across studies [21]. Given these uncertainties in testing and the uncertain clinical significance of MIC/MBC levels, we will avoid the use of the term “resistance“ when referring to phenotype. Rather, we will simply state the presence of increased MICs or MBCs when relevant.
Figure 1.
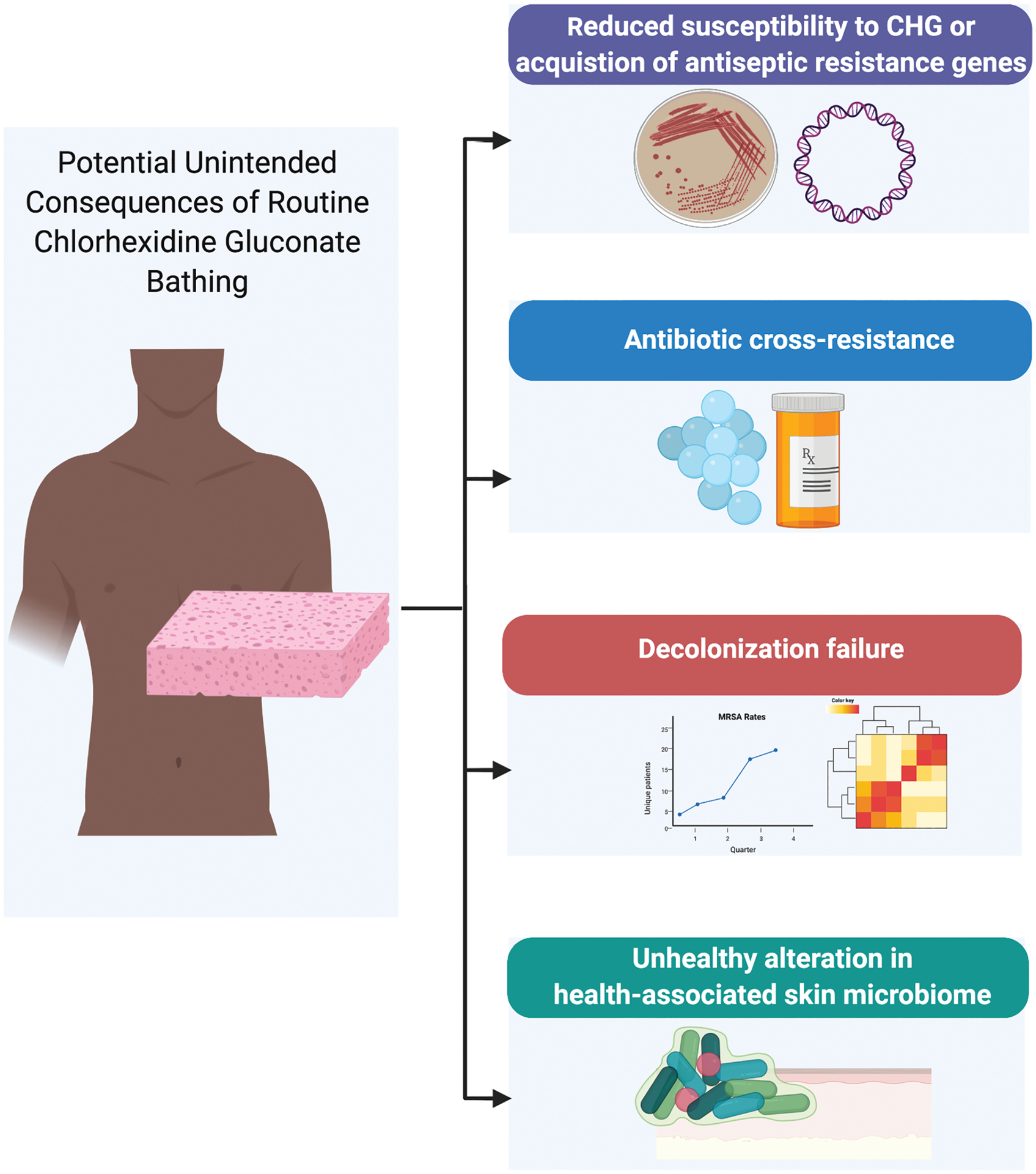
Potential unintended consequences of routine CHG bathing. Despite the proven benefits of CHG use, there remains concerns and unanswered questions on the potential for unintended microbial consequences of routine CHG bathing. These include 1) increase in MIC and/or AMR genes 2) emergence of antibiotic cross-resistance 3) decolonization failure 4) alteration of skin microbiome. To date there is minimal evidence for any of these adverse events. Figure created with Biorender.com. Abbreviations: CHG, chlorhexidine gluconate, MRSA, methicillin-resistant, Staphylococcus aureus.
Given the above challenges, many have attempted to characterize genotypic markers associated with increased CHG MICs as a surrogate for phenotype. Despite progress in the field of genomic characterization of resistance mechanisms and correlation with phenotypes [22], this has remained challenging for CHG. Among staphylococci, qacA/B are the genes most commonly associated with CHG MIC/MBC increases [23]. However, the presence of these genes does not uniformly correlate with increased MICs or MBCs [24–30]. This phenotype/genotype inconsistency may be due in part to the ability of the efflux pump encoded by qacA, but not that encoded by qacB, to utilize CHG as a substrate and the inability of polymerase chain reaction (PCR) to differentiate between the 2 due to their genetic sequence similarity [19, 31]. Other genes that have been less clearly associated with elevated CHG MICs/MBCs include smr genes [18] and norA/B genes, which also encode for efflux pumps [30]. However, these associations are weaker than those shown with qacA/B [32]. Among gram-negative bacteria, genes such as cepA, qacΔE, and qacE that encode for efflux pumps have been associated with an increase in CHG MICs [33]. However, as the majority of studies exploring genes associated with reduced susceptibility to CHG were performed in Staphylococcus aureus, in the remainder of the review we will refer to genotypic resistance as the presence of qacA/B unless otherwise stated.
DOES ROUTINE CHLORHEXIDINE GLUCONATE USE LEAD TO AN INCREASE IN MINIMUM INHIBITORY CONCENTRATIONS OR CHLORHEXIDINE GLUCONATE ANTISEPTIC–RESISTANCE GENES?
Despite increasing CHG use, methicillin-resistant S. aureus (MRSA) isolates with increased MICs and/or qacA/B genes have seldom been reported (<1%) from settings in the United States, where CHG decolonization strategies have been implemented [34–36]. Comparatively, global prevalence of the qacA/B genes in S. aureus isolates has ranged from 10% to 80% in countries where CHG use—but not necessarily routine CHG bathing—is standard practice [32, 37–41]. This wide range may stem from baseline difference in MDROs and differences in study methodology, including study periods.
Whether reduced susceptibility to CHG or the increased prevalence of antiseptic efflux genes is truly driven by routine use of CHG is yet to be ascertained. Secondary analyses of clinical trials assessing the efficacy of CHG for HAI reduction or MDRO decolonization have explored this. In the majority of these, examination of isolates has not demonstrated significant change in CHG MICs [4, 7, 11, 29] and/or qacA/B gene prevalence [24, 29, 35] during or after periods of CHG decolonization implementation [4, 11, 29, 35] and/or when compared with non-CHG control arms [7, 24, 29, 35, 42, 43]. This is exemplified by the analysis of 3173 isolates across 16 states from the REDUCE-MRSA trial [29]. Of the 3173 isolates, only 2 had elevated CHG MICs, and 5 out of 814 isolates were found to carry qacA/B, of which only one was identified in the CHG decolonization arm [29].
The abovementioned studies tend to be either multicenter or multiunit cluster-randomized or crossover studies with a large number of participants in study arms. While results point towards little or no significant reduction in susceptibility or increase in qacA/B genes among trial isolates, some limitations must be acknowledged. In some studies, the timing of isolate collection may have been at the time of patient enrollment and thus these patients would have had little exposure to CHG interventions. Furthermore, the periods of use and/or observation might have been too brief to capture the emergence of reduced susceptibility to CHG or increase in qacA/B genes.
Observational studies have been conducted to examine CHG susceptibility and/or prevalence of qacA/B or other efflux pump genes among S. aureus isolates after implementation of routine CHG bathing or during time periods with ongoing CHG bathing. In these studies, the time frames in which the authors examined for changes are variable (some going as far back as the 1920s [30]), as are results. Some did not find any change in CHG MICs after routine implementation of CHG [44], or identified nonlinear changes in qacA/B genes over time, despite increasing expansion of CHG bathing [45, 46]. Others found increases in MICs/MBCs and/or qacA/B gene presence over time or when comparing pre- and post–CHG era isolates [27, 28, 30, 47]. Zhang et al. [48] found a higher prevalence of qacA/B among isolates from healthcare workers versus non–healthcare workers (CHG-unexposed control group), despite similar rates of Staphylococcus spp. carriage. However, changes in MICs/MBCs did not consistently correlate with changes in qacA/B presence in some of the aforementioned studies [27, 28, 30, 45]. While MICs/MBCs are imperfect measures of CHG susceptibility profiling, the above results call into question the significance of the increasing rates of qacA/B demonstrated.
Among observational studies examining nonstaphylococcal bacteria, reports of CHG MIC increases among organisms causing intravenous catheter–associated bloodstream infections (mostly vancomycin-resistant Enterococcus [VRE]) [49] or antibiotic-resistant Acinetobacter baumannii have been reported with implementation of CHG decolonization protocols [50]. However, the above were single-center studies and included a limited number of isolates.
While observational data are inconclusive, in vitro experiments exposing bacteria to subinhibitory concentrations of CHG, hypothesized to simulate residual concentration on skin after use, have been found to induce increased CHG MICs. This has been demonstrated among MRSA [37], VRE [51], A. baumannii [52], Pseudomonas spp. [53, 54], and Klebsiella pneumoniae [55, 56]. Furthermore, exposure to subinhibitory concentrations of CHG has been found to induce qacA expression [23].
Despite this evidence, one must keep in mind that CHG is a topical antiseptic agent, applied directly to the skin, therefore achieving local skin concentrations that are magnitudes higher than MICs reported (0.25–16 μg/mL vs 10 000–40 000 μg/mL). Hence, the clinical significance of slightly raised MICs is unclear, especially when compared with the concentration that is achievable on the skin [57, 58]. Even among multidrug-resistant gram-negative bacteria that have significantly higher MICs than gram-positive organisms [21], a CHG skin concentration of 128 μg/mL has been reported to confer a protection against skin colonization [8].
Notably, achievable skin concentration may vary based on CHG-containing products used, application techniques, and body site [57–60]. Lower skin CHG concentrations have been found with use of 4% CHG soap versus 2% CHG-impregnated polyester clothes [57, 60] and among patients when rinsing with water after CHG solution bath (4% CHG in warm water) versus without rinsing and preimpregnated CHG cloths [59]. Similarly, lower concentrations of CHG and higher microbial colony counts were detected in the neck region compared with other sites [58]. Finally, using a colorimetric assay to determine the CHG skin concentration, investigators found suboptimal concentrations, which improved with healthcare staff feedback and re-education [15]. Another group of investigators found a low MDRO bioburden on skin irrespective of CHG skin concentration [61].
Evidence of isolates with CHG qacA/B genes being clonal as a result of CHG pressure is limited [62, 63]. A variety of studies from different settings and a plethora of countries show isolates with qacA/B genes to be of different lineages and of diverse sequence type [25, 27, 28, 32, 35, 37, 45, 46]. The majority of the studies used older typing methods and hence further genomic studies are needed for confirmation and further resolution [30].
DOES CHLORHEXIDINE GLUCONATE USE LEAD TO ANTIBIOTIC CROSS-RESISTANCE?
This question is of paramount clinical significance. If decreased CHG susceptibility is associated with increased antibiotic resistance, then could CHG decolonization strategies be leading to selection of or increasing rates of healthcare-associated MDROs? This is especially a concern in situations where suboptimal bathing methods lead to lower than desired skin concentrations.
It is hypothesized that antibiotic cross-resistance may occur in the presence of qacA/B genes either by the presence of co-resistance (acquisition of qacA/B plasmids carrying multiple antimicrobial resistance (AMR) genes [64]) or cross-resistance mechanisms (multi-substrate efflux pumps [65] or adaptions in membrane permeability) or co-selection of MDROs.
The correlation between CHG MICs and antibiotic resistance has been demonstrated in a variety of both gram-positive and gram-negative bacteria [21]. Among the gram-negative bacteria, the strongest correlation was seen with carbapenems and fluoroquinolones [21, 66]. Among gram-positive bacteria, this correlation was variable among the antibiotics tested [21]. However, higher CHG MBCs have been demonstrated in MRSA versus methicillin-susceptible S. aureus [23]. In a crossover trial evaluating the effect of CHG handwashing on AMR patterns of nurses’ hand flora, there was a significant increase in Staphylococcus epidermidis isolates resistant to oxacillin and gentamicin and Staphylococcus warneri isolates resistant to rifampin [67]. However, no accompanying analysis of CHG susceptibility or qacA/B gene presence was performed [67].
The correlation between qacA/B presence and antibiotic resistance has also been explored. Studies comparing isolates with and without qacA/B have found higher rates of antibiotic resistance in the presence of qacA/B [25, 45, 48, 62]—in particular, fluoroquinolone resistance [24, 47, 48]. However, these findings have not been universally reproduced [35] and assessment of other resistance determinants such as norA has not been accounted for [24].
Examination of in vitro subinhibitory exposure of CHG and the emergence of cross-resistance between CHG and antibiotics has similarly been explored [37]. Upregulation of liaXYZ and van operon genes was seen among VRE isolates with CHG exposure leading to daptomycin nonsusceptibility [51]. Similar exposures in Pseudomonas stutzeri led to variable increases in resistance to polymyxin B, gentamicin, nalidixic acid, erythromycin, and ampicillin [53]. Among K. pneumoniae, conflicting results of upregulation of efflux pumps (cepA, acrA, and kdeA) with exposure to CHG have been reported [33, 56]. However, cross-resistance to colistin with CHG exposure has been demonstrated [68]. Whole-genome sequencing of these strains revealed upregulation of pmrD and pmrK, which was hypothesized to be secondary to mutations in the regulator gene phoPQ [68]. Similar findings were observed on analysis of K. pneumoniae carbapenemase (KPC)-producing K. pneumoniae outbreak isolates that occurred in the setting of routine CHG bathing [69].
While these findings are not conclusive, they are suggestive of a link between genetic correlates of reduced CHG susceptibility genes and antibiotic resistance. However, the question of causation versus correlation is unanswered and requires further study. This unanswered question should not shift the focus from ongoing stewardship interventions that have tried and true impact on antibiotic-resistance rates.
DO INCREASED CHLORHEXIDINE GLUCONATE MINIMUM INHIBITORY CONCENTRATIONS LEAD TO DECOLONIZATION FAILURE OR ADVERSE CLINICAL OUTCOMES?
Reports of decolonization failure in the setting of increased reduced CHG susceptibility or qacA/B genes are limited. Only a few cases exist in the literature despite widespread CHG use in healthcare for over 50 years [38, 70, 71].
Investigators in Switzerland found qacA/B gene presence was significantly higher among patients who failed decolonization for MRSA (5 days of nasal mupirocin and 7 days of CHG bathing). On multivariable analysis, decolonization failure was associated with qacA/B in combination with low-level mupirocin resistance; qacA/B alone was not associated with decolonization failure [71]. Moreover, low-level mupirocin resistance has been previously linked to decolonization failure [10]. In a UK intensive care unit (ICU), adoption of a CHG-based antiseptic protocol (among other sequential interventions) led to continued transmission of qacA/B-containing isolates (some with increased CHG MBCs), while transmission of MRSA strains that did not carry qacA/B decreased. The authors deduced that the use of routine CHG may have led to the selection of this “outbreak” strain. However, this conclusion is in contrast to other studies in which typing of isolates from settings with high CHG use found no evidence of clonal selection [25, 27, 28, 32, 35, 37, 45, 46]. In an outbreak of 6 genetically related KPC-producing K. pneumoniae isolates in an ICU where patients had been routinely bathed with CHG, some of the isolates were found to have increased CHG MBCs. However, not all patients involved in the outbreak were washed regularly with CHG [69]. Moreover, CHG efficacy for decolonization of gram-negative bacteria remains controversial [8] and multidrug-resistant K. pneumoniae ST258 isolates may have de novo raised CHG MICs prior to exposure [56], illustrating this sequence type’s evolution to adapt to the hospital environment [72]. Furthermore, while selection of resistance may occur occasionally at a patient level, the likelihood of subsequent cross-transmission (across bacterial genera or patients) is what determines whether such resistance becomes prevalent and hence problematic [73].
Whether the presence of increased CHG MICs and/or qacA/B gene presence affects treatment outcomes is unknown. Data on this topic are limited to 3 studies from the same pediatric center that is the site of a long-term prospective S. aureus surveillance study. These studies comparing clinical characteristics and outcomes among patients infected with qacA/B-containing MRSA isolates compared with qacA/B-negative isolates have demonstrated increased central-line use and bacteremia in the qacA/B group. This is likely reflective of the use of CHG in central-line care. There was no difference in mortality between groups [45, 47, 74]. In a careful case study, Johnson et al. [70] reported on a recurrent MRSA infection in a soldier participating in a prospective cluster-randomized trial aimed at preventing skin and soft tissue infections (SSTIs). Despite the patient receiving weekly CHG bathing he developed a second MRSA skin and soft tissue infection. The patient’s first clinical isolate was qacA/B negative. The second clinical isolate was positive for qacA, which was carried on a large uncharacterized plasmid, and had a slight increase in CHG MIC (0.3–0.8 μg/mL) as well as fluoroquinolone resistance. While the authors concluded that selection of this second strain was secondary to CHG use, whole-genome sequencing revealed more than 140 single-nucleotide polymorphisms difference between the 2 isolates, suggesting that the 2 isolates were genetically distinct [70]. No information on underlying risk factors leading to reacquisition of MRSA was provided in the report.
DOES CHLORHEXIDINE GLUCONATE RESULT IN ALTERATION OF THE HEALTH-ASSOCIATED SKIN MICROBIOME?
Frequent washing/bathing with soap or antiseptic products can lead to skin drying and irritation and to subsequent microbial community shifts [75, 76]. This skin damage is mediated through denaturation of stratum corneum proteins, changes in intercellular lipids (either depletion or reorganization of lipid moieties), decreased corneocyte cohesion, and decreased stratum corneum water-binding capacity [76, 77]. In a crossover design study that compared daily bathing of ICU patients with soap and water versus 2% no-rinse CHG-impregnated cloths, more patients had deterioration in skin condition during the soap and water period than during the CHG period [11]. With daily CHG bathing, patients’ skin microbial richness and diversity have been found to decrease sequentially with repeated application of CHG [78]. Hospitalized patients bathed with CHG were found to have decreased richness and diversity when compared with controls and/or compared with pre-CHG bathing sampling [78, 79]. Another study suggested that this effect might be limited and that changes in relative abundance are site specific and hence likely dependent on CHG concentration [80]. These studies have been mostly conducted among hospitalized patients, who are likely to have aberrant skin microbiome compared with healthy community patients [79, 80]. In contrast, among community-dwelling patients, richness and diversity did not change significantly with repeated application of CHG [78]. The above studies were limited in sample size and design, and more research and monitoring are required to investigate the potential collateral impact to the health-associated microbiota caused by CHG bathing. However, these may be challenging to conduct. Other investigators have found that despite the proven antibacterial effects of CHG, bathing was associated with little change in bacterial community structures. Further investigation demonstrated that CHG may bind bacterial DNA released from lysed bacteria to the skin. This may render estimates of shifts in bacterial community by 16S sequencing difficult to evaluate [81].
Due to its wide spectrum of action, it is conceivable that CHG may disrupt the balance of health-associated microbiota on the skin, an important component of colonization resistance to MDROs [82]. While culture-based studies have shown an overall reduction in microbial density [58], metagenomics allows us to examine this with added power, sensitivity, and resolution.
CHLORHEXIDINE GLUCONATE AND CANDIDA AURIS
Candida auris is a recently emerged nosocomial multidrug-resistant yeast that can cause invasive infections, colonize human skin for long periods, and persist in the dry, inanimate hospital environment. The efficacy of CHG in reducing the bioburden of C. auris on patients’ skin remains to be seen. Chlorhexidine gluconate has activity against planktonic and sessile C. auris forms [83]. However, CHG activity seems to be significantly lower than that seen with C. albicans and the effect is dependent on concentration of CHG [84]. Also, CHG MICs are higher among C. auris than most Candida species [85].
Some have reported ongoing outbreaks despite the use of CHG for decolonization. In a neurosciences ICU outbreak linked to reusable equipment, the mean duration of C. auris colonization was 2–3 months. This was despite all patients in the unit undergoing routine CHG bathing with 2% CHG washcloths [86]. Likewise, residents of a skilled nursing facility in Chicago were found to have persistent C. auris colonization over extended time periods despite routine CHG bathing [87]. And in a reportedly ongoing outbreak of C. auris, patients underwent twice-daily CHG bathing and use of central-line CHG-impregnated protective disks, which did not abate the outbreak [88]. Re-colonization from the environment or other colonized patients has been postulated to explain these findings versus persistent skin colonization [89].
CONCLUSIONS
The preponderance of evidence suggests that the likelihood of clinically relevant reductions in bacterial susceptibility to CHG resulting from topical applications of CHG is very low, and hence should not be a barrier to its use on patients where evidence of effectiveness of CHG in preventing HAIs is great. This review highlighted gaps in knowledge about the consequences of routine CHG bathing that require continued investigation. Critically, a consensus on testing methodology and/or determination of clinically relevant measurement of CHG effect is needed, after which periodic phenotypic surveillance among different taxa can be performed; genome-wide association studies may be useful to identify genomic correlates of reduced susceptibility to CHG or to antibiotics. Although investigators should continue to monitor and explore the possible unintended clinical consequences, this should not impede the implementation of CHG bathing in settings where it has been demonstrated to confer benefit.
Financial support.
M.K.H is supported in part by Centers for Disease Control and Prevention Cooperative Agreement U54 CK000481.
Potential conflicts of interest.
M. K. H. reports grants from Clorox and is co-investigator on research projects for Sage/Stryker Inc, Molnycke, Medline, and Clorox. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Footnotes
Publisher's Disclaimer: Disclaimer. The findings and conclusions presented here are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
References
- 1.Cosgrove SE, Qi Y, Kaye KS, Harbarth S, Karchmer AW, Carmeli Y. The impact of methicillin resistance in Staphylococcus aureus bacteremia on patient outcomes: mortality, length of stay, and hospital charges. Infect Control Hosp Epidemiol 2005; 26:166–74. [DOI] [PubMed] [Google Scholar]
- 2.Roberts RR, Scott RD 2nd, Hota B, et al. Costs attributable to healthcare-acquired infection in hospitalized adults and a comparison of economic methods. Med Care 2010; 48:1026–35. [DOI] [PubMed] [Google Scholar]
- 3.Davis KA, Stewart JJ, Crouch HK, Florez CE, Hospenthal DR. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin Infect Dis 2004; 39:776–82. [DOI] [PubMed] [Google Scholar]
- 4.Climo MW, Yokoe DS, Warren DK, et al. Effect of daily chlorhexidine bathing on hospital-acquired infection. N Engl J Med 2013; 368:533–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang SS, Septimus E, Kleinman K, et al. ; CDC Prevention Epicenters Program; AHRQ DECIDE Network and Healthcare-Associated Infections Program. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 2013; 368:2255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang SS, Septimus E, Kleinman K, et al. ; ABATE Infection Trial Team. Chlorhexidine versus routine bathing to prevent multidrug-resistant organisms and all-cause bloodstream infections in general medical and surgical units (ABATE Infection trial): a cluster-randomised trial. Lancet 2019; 393:1205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bleasdale SC, Trick WE, Gonzalez IM, Lyles RD, Hayden MK, Weinstein RA. Effectiveness of chlorhexidine bathing to reduce catheter-associated bloodstream infections in medical intensive care unit patients. Arch Intern Med 2007; 167:2073–9. [DOI] [PubMed] [Google Scholar]
- 8.Lin MY, Lolans K, Blom DW, et al. ; Centers for Disease Control and Prevention Epicenter Program. The effectiveness of routine daily chlorhexidine gluconate bathing in reducing Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae skin burden among long-term acute care hospital patients. Infect Control Hosp Epidemiol 2014; 35:440–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milstone AM, Passaretti CL, Perl TM. Chlorhexidine: expanding the armamentarium for infection control and prevention. Clin Infect Dis 2008; 46:274–81. [DOI] [PubMed] [Google Scholar]
- 10.Septimus EJ, Schweizer ML. Decolonization in prevention of health care-associated infections. Clin Microbiol Rev 2016; 29:201–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vernon MO, Hayden MK, Trick WE, Hayes RA, Blom DW, Weinstein RA; Chicago Antimicrobial Resistance Project (CARP). Chlorhexidine gluconate to cleanse patients in a medical intensive care unit: the effectiveness of source control to reduce the bioburden of vancomycin-resistant enterococci. Arch Intern Med 2006; 166:306–12. [DOI] [PubMed] [Google Scholar]
- 12.Sheldon AT Jr. Antiseptic “resistance”: real or perceived threat? Clin Infect Dis 2005; 40:1650–6. [DOI] [PubMed] [Google Scholar]
- 13.Krautheim AB, Jermann TH, Bircher AJ. Chlorhexidine anaphylaxis: case report and review of the literature. Contact Dermatitis 2004; 50:113–6. [DOI] [PubMed] [Google Scholar]
- 14.McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev 1999; 12:147–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Supple L, Kumaraswami M, Kundrapu S, et al. Chlorhexidine only works if applied correctly: use of a simple colorimetric assay to provide monitoring and feedback on effectiveness of chlorhexidine application. Infect Control Hosp Epidemiol 2015; 36:1095–7. [DOI] [PubMed] [Google Scholar]
- 16.Marschall J, Mermel LA, Fakih M, et al. Strategies to prevent central line–associated bloodstream infections in acute care hospitals: 2014 update. Infect Control Hosp Epidemiol 2014; 35:753–71. [DOI] [PubMed] [Google Scholar]
- 17.Weiner LM, Webb AK, Walters MS, Dudeck MA, Kallen AJ. Policies for controlling multidrug-resistant organisms in US healthcare facilities reporting to the national healthcare safety network, 2014. Infect Control Hosp Epidemiol 2016; 37:1105–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horner C, Mawer D, Wilcox M. Reduced susceptibility to chlorhexidine in staphylococci: is it increasing and does it matter? J Antimicrob Chemother 2012; 67:2547–59. [DOI] [PubMed] [Google Scholar]
- 19.Kampf G Acquired resistance to chlorhexidine—is it time to establish an “antiseptic stewardship” initiative? J Hosp Infect 2016; 94:213–27. [DOI] [PubMed] [Google Scholar]
- 20.Russell AD. Bacterial resistance to disinfectants: present knowledge and future problems. J Hosp Infect 1999; 43Suppl:S57–68. [DOI] [PubMed] [Google Scholar]
- 21.Kõljalg S, Naaber P, Mikelsaar M. Antibiotic resistance as an indicator of bacterial chlorhexidine susceptibility. J Hosp Infect 2002; 51:106–13. [DOI] [PubMed] [Google Scholar]
- 22.Babiker A, Mustapha MM, Pacey MP, et al. Use of online tools for antimicrobial resistance prediction by whole-genome sequencing in methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). J Glob Antimicrob Resist 2019; 19:136–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith K, Gemmell CG, Hunter IS. The association between biocide tolerance and the presence or absence of qac genes among hospital-acquired and community-acquired MRSA isolates. J Antimicrob Chemother 2008; 61:78–84. [DOI] [PubMed] [Google Scholar]
- 24.Schlett CD, Millar EV, Crawford KB, et al. Prevalence of chlorhexidine-resistant methicillin-resistant Staphylococcus aureus following prolonged exposure. Antimicrob Agents Chemother 2014; 58:4404–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conceição T, Coelho C, de Lencastre H, Aires-de-Sousa M. High prevalence of biocide resistance determinants in Staphylococcus aureus isolates from three African Countries. Antimicrob Agents Chemother 2016; 60:678–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Longtin J, Seah C, Siebert K, et al. Distribution of antiseptic resistance genes qacA, qacB, and smr in methicillin-resistant Staphylococcus aureus isolated in Toronto, Canada, from 2005 to 2009. Antimicrob Agents Chemother 2011; 55:2999–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McGann P, Kwak YI, Summers A, Cummings JF, Waterman PE, Lesho EP. Detection of qacA/B in clinical isolates of methicillin-resistant Staphylococcus aureus from a regional healthcare network in the eastern United States. Infect Control Hosp Epidemiol 2011; 32:1116–9. [DOI] [PubMed] [Google Scholar]
- 28.Wang JT, Sheng WH, Wang JL, et al. Longitudinal analysis of chlorhexidine susceptibilities of nosocomial methicillin-resistant Staphylococcus aureus isolates at a teaching hospital in Taiwan. J Antimicrob Chemother 2008; 62:514–7. [DOI] [PubMed] [Google Scholar]
- 29.Hayden MK, Lolans K, Haffenreffer K, et al. Chlorhexidine and mupirocin susceptibility of methicillin-resistant Staphylococcus aureus isolates in the REDUCE-MRSA trial. J Clin Microbiol 2016; 54:2735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy K, Sunnucks K, Gil H, et al. Increased usage of antiseptics is associated with reduced susceptibility in clinical isolates of Staphylococcus aureus. mBio 2018; 9:e00894–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulsen IT, Brown MH, Littlejohn TG, Mitchell BA, Skurray RA. Multidrug resistance proteins QacA and QacB from Staphylococcus aureus: membrane topology and identification of residues involved in substrate specificity. Proc Natl Acad Sci USA 1996; 93:3630–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sheng WH, Wang JT, Lauderdale TL, Weng CM, Chen D, Chang SC. Epidemiology and susceptibilities of methicillin-resistant Staphylococcus aureus in Taiwan: emphasis on chlorhexidine susceptibility. Diagn Microbiol Infect Dis 2009; 63:309–13. [DOI] [PubMed] [Google Scholar]
- 33.Abuzaid A, Hamouda A, Amyes SG. Klebsiella pneumoniae susceptibility to biocides and its association with cepA, qacΔE and qacE efflux pump genes and antibiotic resistance. J Hosp Infect 2012; 81:87–91. [DOI] [PubMed] [Google Scholar]
- 34.McDanel JS, Murphy CR, Diekema DJ, et al. Chlorhexidine and mupirocin susceptibilities of methicillin-resistant Staphylococcus aureus from colonized nursing home residents. Antimicrob Agents Chemother 2013; 57:552–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fritz SA, Hogan PG, Camins BC, et al. Mupirocin and chlorhexidine resistance in Staphylococcus aureus in patients with community-onset skin and soft tissue infections. Antimicrob Agents Chemother 2013; 57:559–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mc Gann P, Milillo M, Kwak YI, Quintero R, Waterman PE, Lesho E. Rapid and simultaneous detection of the chlorhexidine and mupirocin resistance genes qacA/B and mupA in clinical isolates of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis 2013; 77:270–2. [DOI] [PubMed] [Google Scholar]
- 37.Vali L, Davies SE, Lai LL, Dave J, Amyes SG. Frequency of biocide resistance genes, antibiotic resistance and the effect of chlorhexidine exposure on clinical methicillin-resistant Staphylococcus aureus isolates. J Antimicrob Chemother 2008; 61:524–32. [DOI] [PubMed] [Google Scholar]
- 38.Batra R, Cooper BS, Whiteley C, Patel AK, Wyncoll D, Edgeworth JD. Efficacy and limitation of a chlorhexidine-based decolonization strategy in preventing transmission of methicillin-resistant Staphylococcus aureus in an intensive care unit. Clin Infect Dis 2010; 50:210–7. [DOI] [PubMed] [Google Scholar]
- 39.Miyazaki NH, Abreu AO, Marin VA, Rezende CA, Moraes MT, Villas Bôas MH. The presence of qacA/B gene in Brazilian methicillin-resistant Staphylococcus aureus. Mem Inst Oswaldo Cruz 2007; 102:539–40. [DOI] [PubMed] [Google Scholar]
- 40.Mayer S, Boos M, Beyer A, Fluit AC, Schmitz FJ. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J Antimicrob Chemother 2001; 47:896–7. [DOI] [PubMed] [Google Scholar]
- 41.Lee H, Lim H, Bae IK, et al. Coexistence of mupirocin and antiseptic resistance in methicillin-resistant Staphylococcus aureus isolates from Korea. Diagn Microbiol Infect Dis 2013; 75:308–12. [DOI] [PubMed] [Google Scholar]
- 42.Ridenour G, Lampen R, Federspiel J, Kritchevsky S, Wong E, Climo M. Selective use of intranasal mupirocin and chlorhexidine bathing and the incidence of methicillin-resistant Staphylococcus aureus colonization and infection among intensive care unit patients. Infect Control Hosp Epidemiol 2007; 28:1155–61. [DOI] [PubMed] [Google Scholar]
- 43.Johnson MD, Schlett CD, Grandits GA, et al. Chlorhexidine does not select for resistance in Staphylococcus aureus isolates in a community setting. Infect Control Hosp Epidemiol 2012; 33:1061–3. [DOI] [PubMed] [Google Scholar]
- 44.Musuuza JS, Sethi AK, Roberts TJ, Safdar N. Analysis of multidrug-resistant organism susceptibility to chlorhexidine under usual clinical care. Infect Control Hosp Epidemiol 2017; 38:729–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McNeil JC, Kok EY, Vallejo JG, et al. Clinical and molecular features of decreased chlorhexidine susceptibility among nosocomial Staphylococcus aureus isolates at Texas Children’s Hospital. Antimicrob Agents Chemother 2016; 60:1121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren DK, Prager M, Munigala S, et al. Prevalence of qacA/B genes and mupirocin resistance among methicillin-resistant Staphylococcus aureus (MRSA) isolates in the setting of chlorhexidine bathing without mupirocin. Infect Control Hosp Epidemiol 2016; 37:590–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNeil JC, Hulten KG, Kaplan SL, Mahoney DH, Mason EO. Staphylococcus aureus infections in pediatric oncology patients: high rates of antimicrobial resistance, antiseptic tolerance and complications. Pediatr Infect Dis J 2013; 32:124–8. [DOI] [PubMed] [Google Scholar]
- 48.Zhang M, O’Donoghue MM, Ito T, Hiramatsu K, Boost MV. Prevalence of antiseptic-resistance genes in Staphylococcus aureus and coagulase-negative staphylococci colonising nurses and the general population in Hong Kong. J Hosp Infect 2011; 78:113–7. [DOI] [PubMed] [Google Scholar]
- 49.Suwantarat N, Carroll KC, Tekle T, et al. High prevalence of reduced chlorhexidine susceptibility in organisms causing central line-associated bloodstream infections. Infect Control Hosp Epidemiol 2014; 35:1183–6. [DOI] [PubMed] [Google Scholar]
- 50.Apisarnthanarak A, Yang Hsu L, Lim TP, Mundy LM. Increase in chlorhexidine minimal inhibitory concentration of Acinetobacter baumannii clinical isolates after implementation of advanced source control. Infect Control Hosp Epidemiol 2014; 35:98–9. [DOI] [PubMed] [Google Scholar]
- 51.Bhardwaj P, Hans A, Ruikar K, Guan Z, Palmer KL. Reduced chlorhexidine and daptomycin susceptibility in vancomycin-resistant Enterococcus faecium after serial chlorhexidine exposure. Antimicrob Agents Chemother 2018; 62:e01235–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuangthong M, Julotok M, Chintana W, et al. Exposure of Acinetobacter baylyi ADP1 to the biocide chlorhexidine leads to acquired resistance to the biocide itself and to oxidants. J Antimicrob Chemother 2011; 66:319–22. [DOI] [PubMed] [Google Scholar]
- 53.Russell AD, Tattawasart U, Maillard JY, Furr JR. Possible link between bacterial resistance and use of antibiotics and biocides. Antimicrob Agents Chemother 1998; 42:2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thomas L, Maillard JY, Lambert RJ, Russell AD. Development of resistance to chlorhexidine diacetate in Pseudomonas aeruginosa and the effect of a “residual” concentration. J Hosp Infect 2000; 46:297–303. [DOI] [PubMed] [Google Scholar]
- 55.Bock LJ, Wand ME, Sutton JM. Varying activity of chlorhexidine-based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J Hosp Infect 2016; 93:42–8. [DOI] [PubMed] [Google Scholar]
- 56.Naparstek L, Carmeli Y, Chmelnitsky I, Banin E, Navon-Venezia S. Reduced susceptibility to chlorhexidine among extremely-drug-resistant strains of Klebsiella pneumoniae. J Hosp Infect 2012; 81:15–9. [DOI] [PubMed] [Google Scholar]
- 57.Edmiston CE Jr, Krepel CJ, Seabrook GR, Lewis BD, Brown KR, Towne JB. Preoperative shower revisited: can high topical antiseptic levels be achieved on the skin surface before surgical admission? J Am Coll Surg 2008; 207:233–9. [DOI] [PubMed] [Google Scholar]
- 58.Popovich KJ, Lyles R, Hayes R, et al. Relationship between chlorhexidine gluconate skin concentration and microbial density on the skin of critically ill patients bathed daily with chlorhexidine gluconate. Infect Control Hosp Epidemiol 2012; 33:889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alserehi H, Filippell M, Emerick M, et al. ; CDC Prevention Epicenter Program. Chlorhexidine gluconate bathing practices and skin concentrations in intensive care unit patients. Am J Infect Control 2018; 46:226–8. [DOI] [PubMed] [Google Scholar]
- 60.Rhee Y, Palmer LJ, Okamoto K, et al. ; Centers for Disease Control and Prevention Epicenter Program. Differential effects of chlorhexidine skin cleansing methods on residual chlorhexidine skin concentrations and bacterial recovery. Infect Control Hosp Epidemiol 2018; 39:405–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nadimpalli G, O’Hara LM, Leekha S, et al. ; CDC Prevention Epicenter Program. Association between chlorhexidine gluconate concentrations and resistant bacterial bioburden on skin. Infect Control Hosp Epidemiol 2019; 40:1430–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Otter JA, Patel A, Cliff PR, Halligan EP, Tosas O, Edgeworth JD. Selection for qacA carriage in CC22, but not CC30, methicillin-resistant Staphylococcus aureus bloodstream infection isolates during a successful institutional infection control programme. J Antimicrob Chemother 2013; 68:992–9. [DOI] [PubMed] [Google Scholar]
- 63.Alam MM, Kobayashi N, Uehara N, Watanabe N. Analysis on distribution and genomic diversity of high-level antiseptic resistance genes qacA and qacB in human clinical isolates of Staphylococcus aureus. Microb Drug Resist 2003; 9:109–21. [DOI] [PubMed] [Google Scholar]
- 64.Sidhu MS, Heir E, Leegaard T, Wiger K, Holck A. Frequency of disinfectant resistance genes and genetic linkage with beta-lactamase transposon Tn552 among clinical staphylococci. Antimicrob Agents Chemother 2002; 46:2797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.DeMarco CE, Cushing LA, Frempong-Manso E, Seo SM, Jaravaza TA, Kaatz GW. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob Agents Chemother 2007; 51:3235–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernández-Cuenca F, Tomás M, Caballero-Moyano FJ, et al. ; Spanish Group of Nosocomial Infections (GEIH) from the Spanish Society of Clinical Microbiology and Infectious Diseases (SEIMC) and the Spanish Network for Research in Infectious Diseases (REIPI). Reduced susceptibility to biocides in Acinetobacter baumannii: association with resistance to antimicrobials, epidemiological behaviour, biological cost and effect on the expression of genes encoding porins and efflux pumps. J Antimicrob Chemother 2015; 70:3222–9. [DOI] [PubMed] [Google Scholar]
- 67.Cook HA, Cimiotti JP, Della-Latta P, Saiman L, Larson EL. Antimicrobial resistance patterns of colonizing flora on nurses’ hands in the neonatal intensive care unit. Am J Infect Control 2007; 35:231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wand ME, Bock LJ, Bonney LC, Sutton JM. Mechanisms of increased resistance to chlorhexidine and cross-resistance to colistin following exposure of Klebsiella pneumoniae clinical isolates to chlorhexidine. Antimicrob Agents Chemother 2017; 61:AAC.01162–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stein C, Vincze S, Kipp F, Makarewicz O, Al Dahouk S, Pletz MW. Carbapenem-resistant Klebsiella pneumoniae with low chlorhexidine susceptibility. Lancet Infect Dis 2019; 19:932–3. [DOI] [PubMed] [Google Scholar]
- 70.Johnson RC, Schlett CD, Crawford K, Lanier JB, Merrell DS, Ellis MW. Recurrent methicillin-resistant Staphylococcus aureus cutaneous abscesses and selection of reduced chlorhexidine susceptibility during chlorhexidine use. J Clin Microbiol 2015; 53:3677–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee AS, Macedo-Vinas M, François P, et al. Impact of combined low-level mupirocin and genotypic chlorhexidine resistance on persistent methicillin-resistant Staphylococcus aureus carriage after decolonization therapy: a case-control study. Clin Infect Dis 2011; 52:1422–30. [DOI] [PubMed] [Google Scholar]
- 72.Sundermann AJ, Babiker A, Marsh JW, et al. Outbreak of vancomycin-resistant Enterococcus faecium in interventional radiology: detection through whole genome sequencing-based surveillance. Clin Infect Dis 2020; 70:2336–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hetem DJ, Bootsma MC, Bonten MJ. Prevention of surgical site infections: decontamination with mupirocin based on preoperative screening for Staphylococcus aureus carriers or universal decontamination? Clin Infect Dis 2016; 62:631–6. [DOI] [PubMed] [Google Scholar]
- 74.McNeil JC, Ligon JA, Hulten KG, et al. Staphylococcus aureus Infections in Children With Congenital Heart Disease. J Pediatr Infect Dis Soc 2013; 2: 337–44. [DOI] [PubMed] [Google Scholar]
- 75.Rocha LA, Ferreira de Almeida E Borges L, Gontijo Filho PP. Changes in hands microbiota associated with skin damage because of hand hygiene procedures on the health care workers. Am J Infect Control 2009; 37:155–9. [DOI] [PubMed] [Google Scholar]
- 76.Kownatzki E Hand hygiene and skin health. J Hosp Infect 2003; 55:239–45. [DOI] [PubMed] [Google Scholar]
- 77.Larson E, Girard R, Pessoa-Silva CL, Boyce J, Donaldson L, Pittet D. Skin reactions related to hand hygiene and selection of hand hygiene products. Am J Infect Control 2006; 34:627–35. [DOI] [PubMed] [Google Scholar]
- 78.Burnham CA, Hogan PG, Wallace MA, et al. Topical decolonization does not eradicate the skin microbiota of community-dwelling or hospitalized adults. Antimicrob Agents Chemother 2016; 60:7303–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cassir N, Papazian L, Fournier PE, Raoult D, La Scola B. Insights into bacterial colonization of intensive care patients’ skin: the effect of chlorhexidine daily bathing. Eur J Clin Microbiol Infect Dis 2015; 34:999–1004. [DOI] [PubMed] [Google Scholar]
- 80.Kates AE, Zimbric ML, Mitchell K, Skarlupka J, Safdar N. The impact of chlorhexidine gluconate on the skin microbiota of children and adults: a pilot study. Am J Infect Control 2019; 47:1014–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.SanMiguel AJ, Meisel JS, Horwinski J, Zheng Q, Bradley CW, Grice EA. Antiseptic agents elicit short-term, personalized, and body site-specific shifts in resident skin bacterial communities. J Invest Dermatol 2018; 138:2234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woodworth MH, Hayden MK, Young VB, Kwon JH. The role of fecal microbiota transplantation in reducing intestinal colonization with antibiotic-resistant organisms: the current landscape and future directions. Open Forum Infect Dis 2019; 6:ofz391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sherry L, Ramage G, Kean R, et al. Biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis 2017; 23:328–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moore G, Schelenz S, Borman AM, Johnson EM, Brown CS. Yeasticidal activity of chemical disinfectants and antiseptics against Candida auris. J Hosp Infect 2017; 97:371–5. [DOI] [PubMed] [Google Scholar]
- 85.Abdolrasouli A, Armstrong-James D, Ryan L, Schelenz S. In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses 2017; 60:758–63. [DOI] [PubMed] [Google Scholar]
- 86.Eyre DW, Sheppard AE, Madder H, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med 2018; 379:1322–31. [DOI] [PubMed] [Google Scholar]
- 87.Pacilli M, Kerins JL, Clegg WJ, et al. Regional emergence of Candida auris in Chicago and lessons learned from intensive follow-up at one ventilator-capable skilled nursing facility. Clin Infect Dis 2020; ciaa435. doi: 10.1093/cid/ciaa435.Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schelenz S, Hagen F, Rhodes JL, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 2016; 5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vallabhaneni S, Kallen A, Tsay S, et al. Investigation of the first seven reported Cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013-August 2016. Am J Transplant 2017; 17:296–9. [DOI] [PubMed] [Google Scholar]


