Abstract
The appearance of colonies on the chromogenic medium CHROMagar Candida combined with observation of morphology on corn meal–Tween 80 agar was used for the identification of 353 clinical yeast isolates. The results were compared with those obtained with API yeast identification kits. The accuracy of identification and the turnaround time were equivalent for each method, and our cultural method was less expensive.
Identification to the species level of yeasts isolated from clinical specimens is often problematic for diagnostic laboratories, but it has become increasingly necessary. Greater numbers of immunosuppressed patients, a widening range of recognized pathogens, and the discovery of resistance to antifungal drugs mean that the common practice of identification or exclusion of Candida albicans alone is no longer adequate.
Reference procedures that use biochemical, morphological, and temperature studies (4) are often not practicable for the clinical laboratory because they are labor-intensive and run over several weeks. Packaged kit systems are widely used, but they are expensive and are limited by the sizes of their databases, while automated systems have many of the same limitations.
With the favorable evaluation of CHROMagar Candida (CMA; CHROMagar Company, Paris, France) (5, 10), we attempted to devise a simple, rapid scheme for the routine identification of clinically important yeasts and also investigated whether it was possible to extend the range of usefulness of the medium. We used colony appearance on CMA in combination with morphology on corn meal–Tween 80 agar (CTA; Oxoid, Basingstoke, United Kingdom) and compared our identifications with the results obtained with the API 20C AUX or API 32C system.
MATERIALS AND METHODS
A total of 352 yeast isolates and 1 isolate of the achlorophyllous alga Prototheca wickerhamii, which has yeast-like morphology on routine isolation media, were collected from clinical specimens sent to our laboratory. Also included was an isolate of the newly described yeast Candida dubliniensis, which we had received as a specimen for identification in the Royal College of Pathologists of Australasia’s Quality Assurance Program.
Isolates were subcultured onto Sabouraud dextrose agar (Oxoid) and were incubated at 30°C for 48 h. Single colonies were suspended in sterile distilled water, and then the turbidity was adjusted to a McFarland no. standard 2 with a spectrophotometer (Densimat; Biomerieux) for inoculation of API 20C AUX strips. These were incubated at 30°C in air for 24 to 72 h. One loopful of the suspension was streaked onto a 65-mm-diameter CMA plate to give isolated colonies. A CTA plate was inoculated by the Dalmau method with yeasts from the same colony from which the suspension had been prepared for examination of morphology, including chlamydospore formation.
CMA plates were incubated at 37°C for 48 h in air, as recommended by the manufacturer. CTA plates were incubated at 30°C in air for 48 h prior to examination. The diameters of yeasts which did not produce hyphae or pseudohyphae were measured with a calibrated microscope. Colony color on CMA plates and morphology on CTA plates were noted, and a preliminary identification was made by using these features. The API strips were read, and the results were interpreted with the corresponding identification software. The identities determined by each method were compared. Isolates giving discordant results were reexamined by the same protocol, except that the API 32C system instead of the API 20C AUX system was used.
RESULTS
The results including the distinctive features on CMA and CTA plates are summarized in Table 1. The appearances on CMA plates and the microscopic morphologies on CTA plates are given in Fig. 1. Of the 11 species, none were incorrectly identified by the scheme with the CMA and CTA plates, although when only a few isolates were available for study, a confident identification could not be assumed. The API kits were not able to identify two species, C. dubliniensis and P. wickerhamii, because these species were not included in the database. The API kits incorrectly identified 1 of the 50 germ tube-positive strains of C. albicans as Candida parapsilosis and identified all strains of Candida krusei and Candida guilliermondii with only a low level of confidence.
TABLE 1.
Identification of clinical yeast isolates by API kits or the combination of CMA plus CTA, including growth characteristics on these media
| Species (no. of strains) | Colony characteristics on CMA | Morphologic features on CTA | Identification with CMA plus CTA | Identification by API 20C AUX or API 32C system |
|---|---|---|---|---|
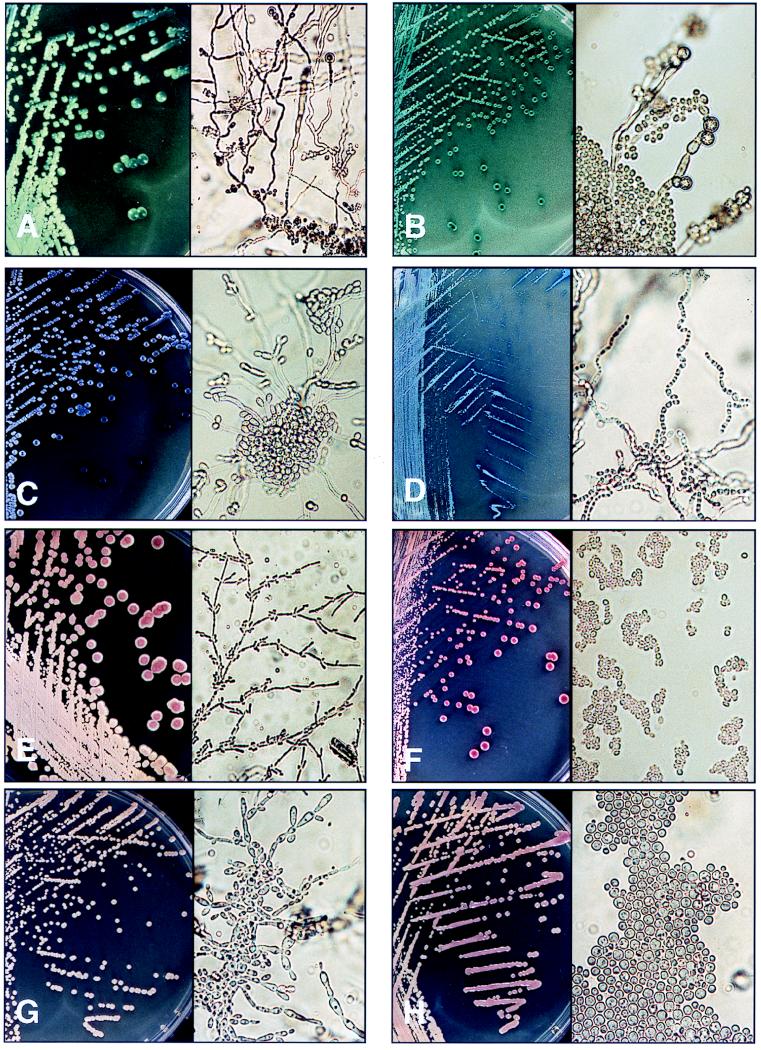
| C. albicans (50) | Apple green colonies; consistent distinctive appearance | Chlamydospores present, although >2 days required for some strains; abundant pseudohyphae, some true hyphae, clusters of blastospores along pseudohyphae; distinctive appearance | Accurate identification with both | Identified one strain as C. parapsilosis |
| C. glabrata (116) | Large pale pink to purple glossy colonies | No pseudohyphae or hyphae; yeast cells are ∼2.5 × 4 μm; consistent distinctive appearance | Accurate identification, morphology essential | Identified all strains as C. glabrata |
| C. tropicalis (87) | Dull blue, sometimes pink, colonies; all developed purple halo of pigment that diffused into surrounding agar; distinctive appearance | Abundant pseudohyphae often radiating with clusters of blastoconidia at the center; variable appearance | Accurate identification with CMA | Identified all strains as C. tropicalis |
| C. krusei (6) | Large, flat, spreading, pale pink colonies with matt surfaces; distinctive appearance | Extensive branched pseudomycelium with chains of elongate cells giving tree-like appearance; clusters and chains of blastospores along pseudohyphae; consistent distinctive appearance | Accurate identification with both | Strains identified as C. krusei with 50% confidence |
| C. parapsilosis (47) | Off-white to pale pink colonies; variable appearance | Branched chains of elongated cells with clusters of blastospores along them; occasional giant cells; variable appearance | Not always able to identify | Identified all strains as C. parapsilosis |
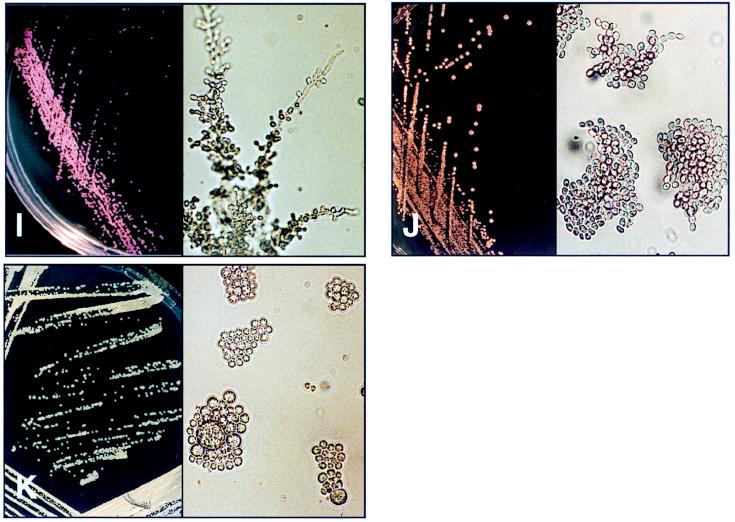
| C. guilliermondii (5) | Small pink to purple colonies; variable appearance | Pseudohyphae with clusters of blastospores; variable appearance | Not always able to identify | API 32C identified strains with 53.7% confidence |
| C. dubliniensis (1) | Dark green colonies | Abundant chlamydospores present; abundant pseudohyphae, some true hyphae, clusters of blastospores along pseudohyphae | Identification with both for the strain examined | Not in API database; identified as Candida sake with low confidence |
| Trichosporon spp. (37) | Tiny, rough, dry-looking, dirty blue-to-grey colonies, distinctive appearance | Pseudomycelium and abundant true mycelium breaking up into arthrospores; distinctive appearance | Accurate identification with both | Identified all strains as T. beigelii |
| C. neoformans (2) | Pink colonies, sometimes mucoid | No pseudohyphae or hyphae; Large spherical yeast cells ∼3–7.5 μm in diameter; distinctive appearance | Accurate identification, morphology essential | Identified as C. neoformans |
| S. cerevisiae (2) | Small purple colonies | Oval cells with very short multilateral budding | Accurate identification of strains examined, morphology essential | Identified one strain as Cryptococcus laurentii |
| P. wickerhamii (1) | Large, variably sized spherical cells, some containing sporangia; distinctive appearance | Large, variably sized spherical cells, 2–16 μm, some containing sporangia; distinctive appearance | Accurate identification, morphology essential | Not in API database; identified the strain as C. glabrata |
FIG. 1.
Appearances of colonies on CMA (left; magnification, ×1) and microscopic appearance on CTA (right; magnification, ×400) after 48 h of incubation. (A) C. albicans. (B) C. dubliniensis. (C) C. tropicalis. (D) T. beigelii. (E) C. krusei. (F) C. glabrata. (G) C. parapsilosis. (H) C. neoformans. (I) C. guilliermondii. (J) S. cerevisiae. (K) P. wickerhamii.
DISCUSSION
A number of researchers have found CMA to be effective for the presumptive identification of C. albicans, Candida tropicalis, and Trichosporon (5, 10). Pfaller et al. (7) also found it to be reliable for the presumptive identification of Candida glabrata, although others (5, 10) did not concur with this. We found that C. glabrata could not be distinguished by its appearance on CMA plates alone, having an appearance similar to those of C. parapsilosis, Saccharomyces cerevisiae, P. wickerhamii, Cryptococcus neoformans, and C. guilliermondii. However, the addition of information from CTA plates does allow identification, so that except for rare isolates of Candida famata, clinical yeasts that form pink glossy colonies on CMA but that have small yeast cells and no pseudomycelium on CTA can presumptively be identified as C. glabrata.
C. krusei can reliably be identified with the combination of CMA and CTA, having a distinctive morphology on both media, whereas kit systems do not cope well with this species. Pigment production by C. tropicalis on CMA allows discrimination of this species with >99% confidence (5). Trichosporon beigelii has a variable but distinctive appearance on CMA, with small dry-looking colonies, and the formation of arthroconidia on CTA gives a reliable confirmation of its identity. Other arthroconidium-forming species such as Coccidioides immitis, which takes much longer than 2 days to form arthroconidia, and Geotrichum candidum, which is a mold, will not be confused with Trichosporon on this medium.
C. neoformans colonies on CMA are a nondistinctive pink and may be mucoid. Dalmau plate morphology reveals large round yeast cells, often with the presence of capsules, suggesting the identity of C. neoformans. The clinical importance of C. neoformans requires confirmation of its identity by serological or biochemical methods.
The achlorophyllous alga P. wickerhamii, which may cause wound infections and meningitis (2, 13), has a yeast-like colony morphology on Sabouraud and blood agars and can easily be mistaken for a yeast. The API 20C AUX and API 32C systems identify it as C. glabrata because both species assimilate only glucose and trehalose among the sugars in the panel and P. wickerhamii is not included in the databases. Without observation of morphology, it might be reported as C. glabrata, but on CMA distinctive sporangia varying in size from 5 to 25 μm are readily observed.
Definitive identification of the newly described chlamydospore-positive, germ tube-positive species C. dubliniensis requires testing for β-glucosidase activity, an expensive and not widely available test (8), or DNA fingerprinting, which demonstrates the nonreactivity of its DNA with a C. albicans-specific oligonucleotide probe, Ca3 or 27A (6, 11, 12). Phenotypic tests would be more practical for clinical laboratory screening for this species. C. dubliniensis has much darker green colonies than C. albicans on CMA, and it usually produces abundant chlamydospores (1, 3, 11, 12). Chlamydospore formation in C. dubliniensis is radically different from that in C. albicans, with chlamydoconidia often attached in pairs, triplets, or larger clusters to the same suspensor cell rather than singly at the ends of hyphae or pseudohyphae as in C. albicans (1), although this characteristic may not be present in all strains (11). The formation of dark green colonies on CMA may be lost on repeated subculture or storage at −70°C (11). It does not fluoresce under Woods lamp illumination on methyl blue-Sabouraud agar, unlike C. albicans, although this property may also be lost on subculture (3, 11). Only one isolate of C. dubliniensis was available to the researchers, and this strain had been subcultured a number of times, yet it retained the ability to form dark green colonies on CMA. The growth of C. dubliniensis at 42°C is poor relative to that of C. albicans, and C. dubliniensis not grow at all at 45°C, although some strains of C. albicans also fail to grow at this temperature (8). Differential growth at one of these temperatures has been suggested as a useful method for differentiation between the two species (1, 3, 8, 9, 11, 12). Carbohydrate assimilation patterns have been reported to be unstable (1, 11), although the failure of C. dubliniensis to assimilate both xylose and α-methyl-d-glucoside compared with the utilization of one or both of these by C. albicans has been suggested as a useful test (9). It is important to differentiate between these species, because significant resistance to azoles has been reported in C. dubliniensis (12). We suggest initial screening with CMA and CTA and then confirmation with examination for growth at 42 or 45°C as a useful method for the presumptive identification of C. dubliniensis.
The turnaround time of 48 h for the CMA and CTA scheme is similar to that for commercial kit systems, which require 24 to 72 h. The average time taken by experienced technicians to set up and read CMA and CTA plates is 3 min per isolate, while the average time taken to set up and read an API strip is 10 to 15 min. The estimated cost of materials per isolate for the CMA and CTA plates is US$1 when 65-mm petri dishes are used for CMA and when four strains are tested on a 90-mm plate for CTA. We pay US$5.20 for each API strip.
It is important that isolated colonies be observed on CMA because the identifying descriptions are based on the form, color, and pigment production of single colonies. Recognition of yeast morphologies on Dalmau plates requires some experience, but once this is mastered, it has been our experience that laboratory staff find the scheme simple and reliable. We were able to use this scheme to identify more than 95% of yeast isolates in our laboratory.
Advances in medical technology have not fostered the development of traditional mycology skills among medical laboratory personnel, yet these skills are increasingly needed. Careful observation of yeast morphology can add confidence to the identification of the commonly encountered species and, more importantly, will alert the microbiologist to the presence of unusual isolates whose misidentification may have serious clinical implications.
REFERENCES
- 1.Coleman D C, Sullivan D J, Bennett D E, Moran G P, Barry H J, Shanley D B. Candidiasis: the emergence of a novel species, Candida dubliniensis. AIDS. 1997;11:557–567. doi: 10.1097/00002030-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Kaminski Z C, Kapila R, Sharer L R, Kloser P, Kaufman L. Meningitis due to Prototheca wickerhamii in a patient with AIDS. Clin Infect Dis. 1992;15:704–706. doi: 10.1093/clind/15.4.704. [DOI] [PubMed] [Google Scholar]
- 3.Kirkpatrick W R, Revankar S G, McAtee R K, Lopez-Ribot J L, Fothergill A W, McCarthy D I, Sanche S E, Cantu R A, Rinaldi M G, Patterson T F. Detection of Candida dubliniensis in oropharyngeal samples from human immunodeficiency virus-infected patients in North America by primary CHROMagar Candida screening and susceptibility testing of isolates. J Clin Microbiol. 1998;36:3007–3012. doi: 10.1128/jcm.36.10.3007-3012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kreger-van Rij N J W, editor. The yeasts: a taxonomic study. 3rd ed. Amsterdam, The Netherlands: Elsevier Science Publishers; 1984. [Google Scholar]
- 5.Odds F C, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odds F C, Van Nuffel L, Dams G. Prevalence of Candida dubliniensis isolates in a yeast stock collection. J Clin Microbiol. 1998;36:2869–2873. doi: 10.1128/jcm.36.10.2869-2873.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaller M A, Houston A, Coffmann S. Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. J Clin Microbiol. 1996;34:58–61. doi: 10.1128/jcm.34.1.58-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J Clin Microbiol. 1998;36:2093–2095. doi: 10.1128/jcm.36.7.2093-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salkin I F, Pruitt W R, Padhye A A, Sullivan D, Coleman D, Pincus D H. Distinctive carbohydrate assimilation profiles used to identify the first clinical isolates of Candida dubliniensis recovered in the United States. J Clin Microbiol. 1998;36:1467. doi: 10.1128/jcm.36.5.1467-1467.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.San-Millan R, Ribacoba L, Ponton J, Quindos G. Evaluation of a commercial medium for identification of Candida species. Eur J Clin Microbiol Infect Dis. 1996;15:153–158. doi: 10.1007/BF01591489. [DOI] [PubMed] [Google Scholar]
- 11.Schoofs A, Odds F C, Colebunders R, Ieven M, Goossens H. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur J Clin Microbiol Infect Dis. 1998;16:296–300. doi: 10.1007/BF01695634. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang W Y M, Lo K K, Lam W Y, Fung K S C, Koehler A, Cheng A F B. Cutaneous protothecosis: report of a case in Hong Kong. Br J Dermatol. 1995;133:479–482. doi: 10.1111/j.1365-2133.1995.tb02683.x. [DOI] [PubMed] [Google Scholar]