ABSTRACT
The human angiotensin-converting enzyme 2 acts as the host cell receptor for SARS-CoV-2 and the other members of the Coronaviridae family SARS-CoV-1 and HCoV-NL63. Here, we report the biophysical properties of the SARS-CoV-2 spike variants D614G, B.1.1.7, B.1.351, and P.1 with affinities to the ACE2 receptor and infectivity capacity, revealing weaknesses in the developed neutralizing antibody approaches. Furthermore, we report a preclinical characterization package for a soluble receptor decoy engineered to be catalytically inactive and immunologically inert, with broad neutralization capacity, that represents an attractive therapeutic alternative in light of the mutational landscape of COVID-19. This construct efficiently neutralized four SARS-CoV-2 variants of concern. The decoy also displays antibody-like biophysical properties and manufacturability, strengthening its suitability as a first-line treatment option in prophylaxis or therapeutic regimens for COVID-19 and related viral infections.
IMPORTANCE Mutational drift of SARS-CoV-2 risks rendering both therapeutics and vaccines less effective. Receptor decoy strategies utilizing soluble human ACE2 may overcome the risk of viral mutational escape since mutations disrupting viral interaction with the ACE2 decoy will by necessity decrease virulence, thereby preventing meaningful escape. The solution described here of a soluble ACE2 receptor decoy is significant for the following reasons: while previous ACE2-based therapeutics have been described, ours has novel features, including (i) mutations within ACE2 to remove catalytical activity and systemic interference with the renin/angiotensin system, (ii) abrogated FcγR engagement, reduced risk of antibody-dependent enhancement of infection, and reduced risk of hyperinflammation, and (iii) streamlined antibody-like purification process and scale-up manufacturability indicating that this receptor decoy could be produced quickly and easily at scale. Finally, we demonstrate that ACE2-based therapeutics confer a broad-spectrum neutralization potency for ACE2-tropic viruses, including SARS-CoV-2 variants of concern in contrast to therapeutic MAb.
KEYWORDS: ACE2-Fc, B.1.1.7, B.1.351, coronavirus, P.1, SARS-CoV-2, receptor decoy, spike affinity
INTRODUCTION
The emergence of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) at the end of 2019 (1) has caused a major coronavirus disease (COVID-19) worldwide pandemic outbreak, totaling over 100 million confirmed cases and over 2 million associated deaths as of January 2021 (https://covid19.who.int/). The rapid replication of SARS-CoV-2 has been shown in some patients to trigger an aggressive inflammatory response in the lung and acute respiratory disease syndrome (ARDS), leading to a cytokine release syndrome (CRS) due to the elevated expression of proinflammatory cytokines (2–4). Similar to SARS-CoV-1 (5), this enveloped virus belongs to the β-coronavirus genus with a positive-strand RNA genome and utilizes angiotensin-converting enzyme 2 (ACE2) as the receptor for host cell entry by binding to its spike (S) glycoprotein (1, 6). The S is arranged as a trimeric complex of heterodimers composed of S1, containing the receptor-binding domain (RBD), and S2, responsible for viral fusion and cell entry, which are generated from the proteolytical cleavage of the S precursor via furin in the host cell (6, 7).
Currently, more than 1,100 monoclonal antibodies (MAb) against SARS-CoV-2 have been reported in the literature, with over 20 currently in clinical evaluation (8, 9). The antibodies LY-CoV555 and LY-CoV016 developed by Eli Lilly and Company and the antibody cocktail REGN-COV2 (REGN10933 plus REGN10987) developed by Regeneron were granted emergency-use authorization (EUA) by the Food and Drug Administration (FDA). To maximize neutralization capacity, most of the antibodies in development are directed toward the RBD in order to disrupt interaction between the viral S protein and ACE2 (10). These recombinant antibodies block viral entry by binding various epitopes on the RBD in a manner that fundamentally differs from the binding of the glycoprotein to ACE2 and are therefore susceptible to viral mutational escape.
Several variants have emerged carrying mutations in S, including in the RBD. Of note is the identification of the D614G (clade 20A) that has rapidly become the dominant strain globally (11). Additional variants have also gained partial dominance in different regions of the globe. The variants A222V (clade 20A.EU1) and S477N (clade 20A.EU2) emerged in the summer of 2020 in Spain and have rapidly shown diffusion within Europe (12). Recently, two new variants, clade 20B/501Y.V1, B.1.1.7 and clade 20C/501Y.V2, B.1.351, characterized by multiple mutations in S, have been associated with a rapid surge in COVID-19 cases in the United Kingdom and South Africa, respectively, and have shown increased transmissibility and reduction of convalescent-phase serum neutralization capacity (13–15). Finally, two variants that emerged in Brazil (B.1.1.28 and P.1) contained mutational hallmarks of both the UK and South Africa variants, suggesting convergent evolution in SARS-CoV-2 due to similar selective pressures (16, 17). These variants have already been shown to affect MAb neutralization potency (18, 19).
Receptor-based decoy strategies have successfully been employed in the clinic (20–22); similarly, ACE2-based decoy strategies have been proposed for COVID-19. A key advantage is that mutations in S which disrupt viral interaction with the ACE2 decoy will by necessity decrease virulence, thereby preventing meaningful escape by mutation. Previously described ACE2-based decoys include the soluble human catalytically active ACE2, repurposed from its initial development for treatment of non-COVID-19 ARDS (23). Additionally, ACE2 mutants with enhanced affinity for the SARS-CoV-2 viral glycoprotein have also been described (24–26). However, limitations of these approaches include short circulating half-life, activity over the renin/angiotensin system which may prevent its use in prophylaxis, and viral mutational escape which may be enabled by engineering of the S protein-targeting domain of ACE2.
With a view to eliminate the risk of mutational escape, eliminate the physiological effects on the renin/angiotensin system, and increase circulating half-life, we generated a catalytically inactive ACE2 receptor decoy fused to a human Fc domain further engineered to bear minimal immunomodulatory activity. This molecule has shown complete lack of enzymatic activity and natural substrate sequestration, with no residual engagement to human FcγRs, adopting a set of Fc mutations reported to preserve long half-life and FcRn interaction (27). The construct showed broad neutralizing capacity with proven activity toward ACE2-tropic viruses, including the SARS-CoV-2 variants of concern B.1.1.7 and B.1.351, with improved consistency and resistance to viral mutational escape compared to those of leading monoclonal antibody therapeutics. Additionally, we report the biophysical characterization and ACE2 affinity measurements for the D614G, B.1.1.7, and B.1.351 SARS-CoV-2 S1 variants, with links to infective potency in a pseudotyped vector setting, with direct comparison to approved COVID-19 monoclonal antibodies.
RESULTS
Biophysical characterization of SARS-CoV-2 spike variants.
We first explored the binding kinetic between SARS-CoV-2 S1 and ACE2. Inhouse purified recombinant S1 domains from Wuhan, D614G, B.1.1.7, B.1.351, and P.1 variants demonstrated similar properties to commercially sourced S1 wild-type (WT) protein (Fig. 1A). Interestingly, the Wuhan and D614G variants displayed a similar thermal unfolding profile, with the first transition event (melting temperature; Tm) at 42.9 and 42.2°C, respectively, while the P.1, B.1.1.7, and B.1.351 resulted in a 4.1, 6.9, and 11.5°C increase in temperature compared to that of S1 Wuhan, respectively (Fig. 1B).
FIG 1.

Biophysical characterization of SARS-CoV-2 S1 variants. (A) Particle size distribution of recombinant purified SARS-CoV-2 S1 variants analyzed via MADLS. (B) Thermal stability analysis of purified SARS-CoV-2 variants showing increased Tm for B.1.1.7, B.1.351, and P.1 variants compared to that of WT S1. (C) Kinetic affinity of catalytically active ACE2-Fc with SARS-CoV-2 S1 WT, D614G, B1.1.7, B.1.351, and P.1. All sensograms fitted with 1:1 Langmuir binding model. Analyte starting concentration 250 nM with 2-fold serial dilutions. 3-fold and 2.3-fold higher affinity detected for S1 of B.1.1.7 and P.1 variants, respectively. Physical particle number determined via p24 ELISA (D) and infectious viral titer (E) comparison for SARS-CoV-1, SARS-CoV-2 Wuhan, SARS-CoV-2 D614G, SARS-CoV-2 B1.1.7, and SARS-CoV-2 B1.351 pseudotyped vectors, showing comparable particle concentration but diverse infectivity capacity (mean ± standard deviation [SD]). One-way analysis of variance (ANOVA) Dunnett’s multiple comparisons test (F = 105.2, df = 4, 10).
The binding affinity of the spike variants for the ACE2 receptor was assessed by surface plasmon resonance (SPR) using the recombinant S1 domains to allow for a monovalent binding interaction. The SARS-CoV-2 S1 WT, D614G, and B.1.351 displayed overall similar kinetic affinities, although the latter showed an off-rate (kd) 1.5-fold lower than that of WT S1, which was compensated by a slightly lower on-rate (ka). The B.1.1.7 and P.1 S1 variants, however, showed approximately 3-fold increase in affinity compared to that of Wuhan, mainly driven by a lower kd (Fig. 1C and Table 1).
TABLE 1.
Kinetic affinities
To assess the infectivity conferred by the SARS-CoV-2 spike variants, we engineered replication-deficient lentiviral vectors pseudotyped with the WT glycoprotein or carrying the D614G, B.1.1.7, and B.1.351 mutations, alongside SARS-CoV-1. Although all pseudotyped vectors showed equivalent physical particle concentrations, as measured by p24 enzyme-linked immunosorbent assay (ELISA), they exhibited vastly different infectivity capacity (Fig. 1D). SARS-CoV-1 resulted in the lowest viral titer, with a reduction of 3.2-fold in infectious units (IU)/ml compared to that of SARS-CoV-2 Wuhan. The SARS-CoV-2 D614G variant was instead the most efficient, with viral titer 2.6-fold higher than that of Wuhan. B.1.1.7 and B.1.351 showed viral titers 1.8- and 1.9-fold higher than that of SARS-CoV-2 Wuhan, respectively (Fig. 1E). All pseudotype titers were determined on permissive HEK-293T cell line stably transduced to express human ACE2 and TMPRSS2 enzymes.
Catalytically inactive ACE2-Fc fusion with streamlined purification.
The extracellular domain of human ACE2 (aa 18 to 740, UniProt Q9BYF1) was fused to the human IgG1 Fc via the human IgG1 hinge region to allow for homodimer stabilization (Fig. 2A). The ACE2 domain used included both the zinc metallopeptidase and the collectrin domains to allow full receptor representation. The Fc domain was included to improve circulating half-life and to capitalize on the streamlined antibody purification processes. In order to generate an inert receptor decoy, the catalytic site of the enzyme was mutated at residues 374 (H374N) and 378 (H378N), termed HH:NN, to inhibit enzymatic activity and prevent conversion of the angiotensin-(1–8) (Ang II) substrate to angiotensin-(1–7). This mutation is predicted to remove interaction with zinc ions (Zn+2) mediated by the two original Histidine (His) residues, with a spatially conservative mutation (Fig. 2B).
FIG 2.
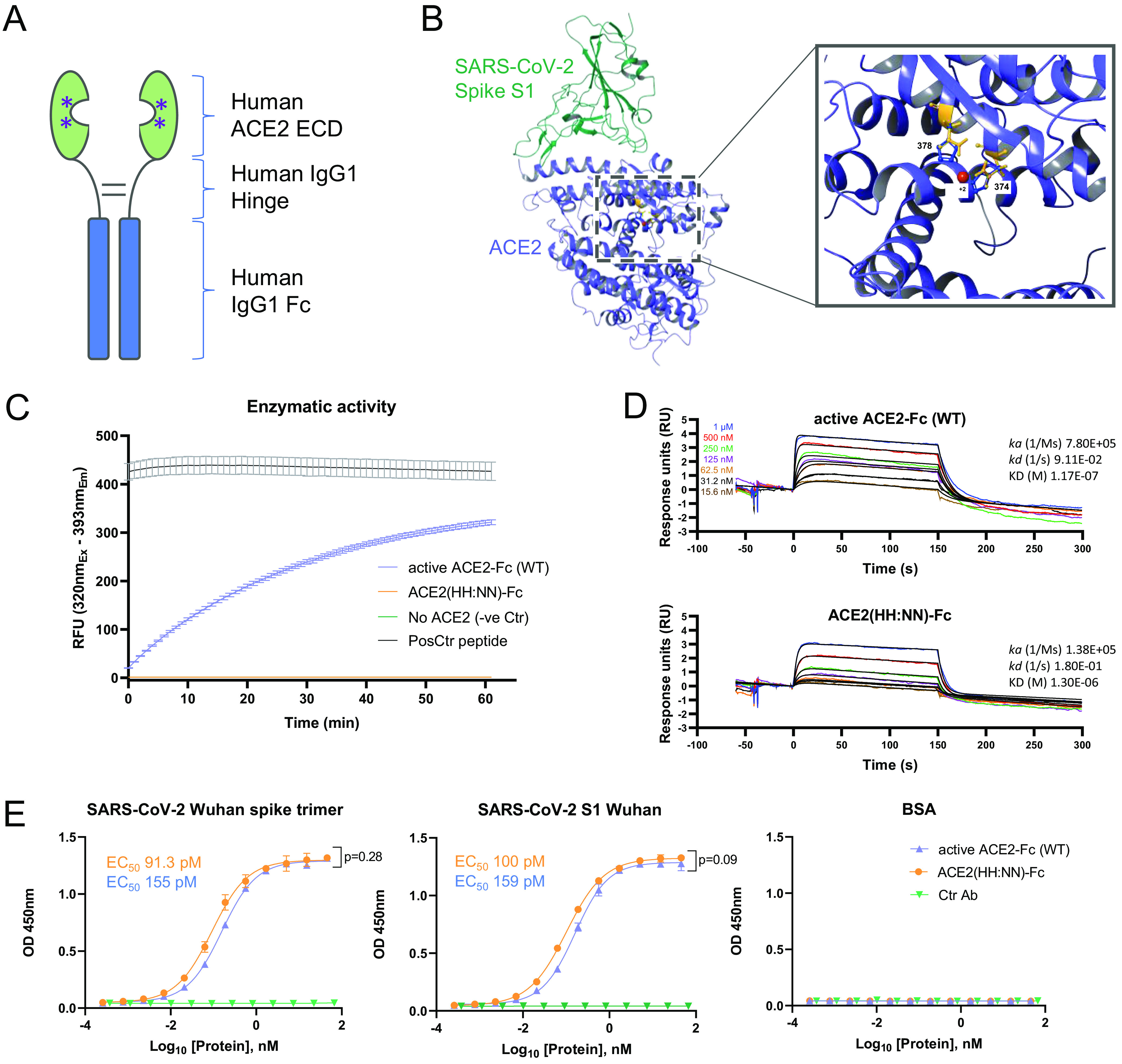
Characterization of ACE2-Fc receptor decoy. (A) Schematic representation of ACE2-Fc molecule with a streamlined antibody-like expression/purification process and biophysical characterization. (B) Three-dimensional (3D) structure of SARS-CoV-2 spike S1 domain (green) in complex with ACE2 (blue) (PDB 6M0J). Inset, zoomed section of the ACE2 catalytic site showing the H374 and H378 residues (blue) in complex with Zn (red), and the H374N and H378N mutations (orange). (C) Enzymatic activity of active (blue) and H374N, H378N mutated (orange) ACE2-Fc using Mca-APK(Dnp) fluorogenic peptide (mean ± SD). (D) Binding kinetics of active (top) and inactive (bottom) ACE2-Fc with Ang II. (E) ELISA of SARS-CoV-2 active spike trimer (left) or S1 domain (center) against WT active and ACE2(HH:NN)-Fc, showing comparable binding capacity. No binding detected with control antigen (right) or negative-control antibody (mean ± SD). EC50, half maximal effective concentration; RFU, relative fluorescent units; OD, optical density. Unpaired t test of AUC (left t = 1.086, df = 24; center t = 1.79, df = 24).
We first set out to confirm inactivation of the ACE2 component. In vitro testing using a fluorogenic substrate for ACE2, Mca-APK(Dnp), showed complete abrogation of enzymatic activity for the ACE2-Fc construct carrying the HH:NN mutation, while the wild-type (WT) active ACE2-Fc molecule was able to efficiently process the peptide (Fig. 2C). Furthermore, the kinetic interaction of both WT and mutated ACE2 domains for their natural substrate Ang II was investigated using SPR. Both constructs interacted with the substrate; however, the mutated ACE2 was characterized by a lower on-rate (ka 7.80E+05 versus 1.38E+05, for active and HH:NN ACE2, respectively) and a higher off-rate (kd 9.11E−02 versus 1.80E−01, for active and HH:NN ACE2, respectively), culminating in a final affinity (KD) of 1.3 μM for the ACE2 HH:NN versus 117 nM for the WT active ACE2 (Fig. 2D).
We next explored whether the ACE2 mutations affected SP binding. Both WT and mutated ACE2 showed comparable binding capacity for recombinant SARS-CoV-2 full S trimer and S1 domain by ELISA (Fig. 2E). SPR measurements of kinetic interaction for the S1 domain of SARS-CoV-2 showed comparable kinetic profiles between active WT and HH:NN ACE2 (Table 1), further suggesting the preservation of an unaltered spike binding domain.
Engineered Fc domain with abrogated FcγR engagement.
To overcome the risk of activating the host immune system, thus exacerbating the hyperinflammatory response often associated with severe COVID-19 development (28), the Fc domain was engineered to remove FcγR interactions. The well-established L234A/L235A (LALA) (29) mutations of the CH2 domain and the LALA combination with P329G (LALA-PG) (30) were introduced in the human IgG1 Fc portion of the ACE2-Fc fusion protein. We first investigated the expression yields of the ACE2(HH:NN) with WT Fc, LALA Fc, and LALA-PG Fc and ACE2 domain activity. All constructs showed comparable expression and purification efficiencies using protein A affinity chromatography (data not shown). Mutations on the Fc domain did not affect the binding capacity of ACE2 for SARS-CoV-2 S protein, and all three versions showed highly comparable dose/response curves to recombinant SARS-CoV-2 S trimer or S1 domain by ELISA (Fig. 3A). Similarly, all three variants were able to bind SupT1 cell lines expressing SARS-CoV-2 S trimer as a transmembrane protein (Fig. 3B), further confirming binding capacity for the glycoprotein in a more physiological environment.
FIG 3.
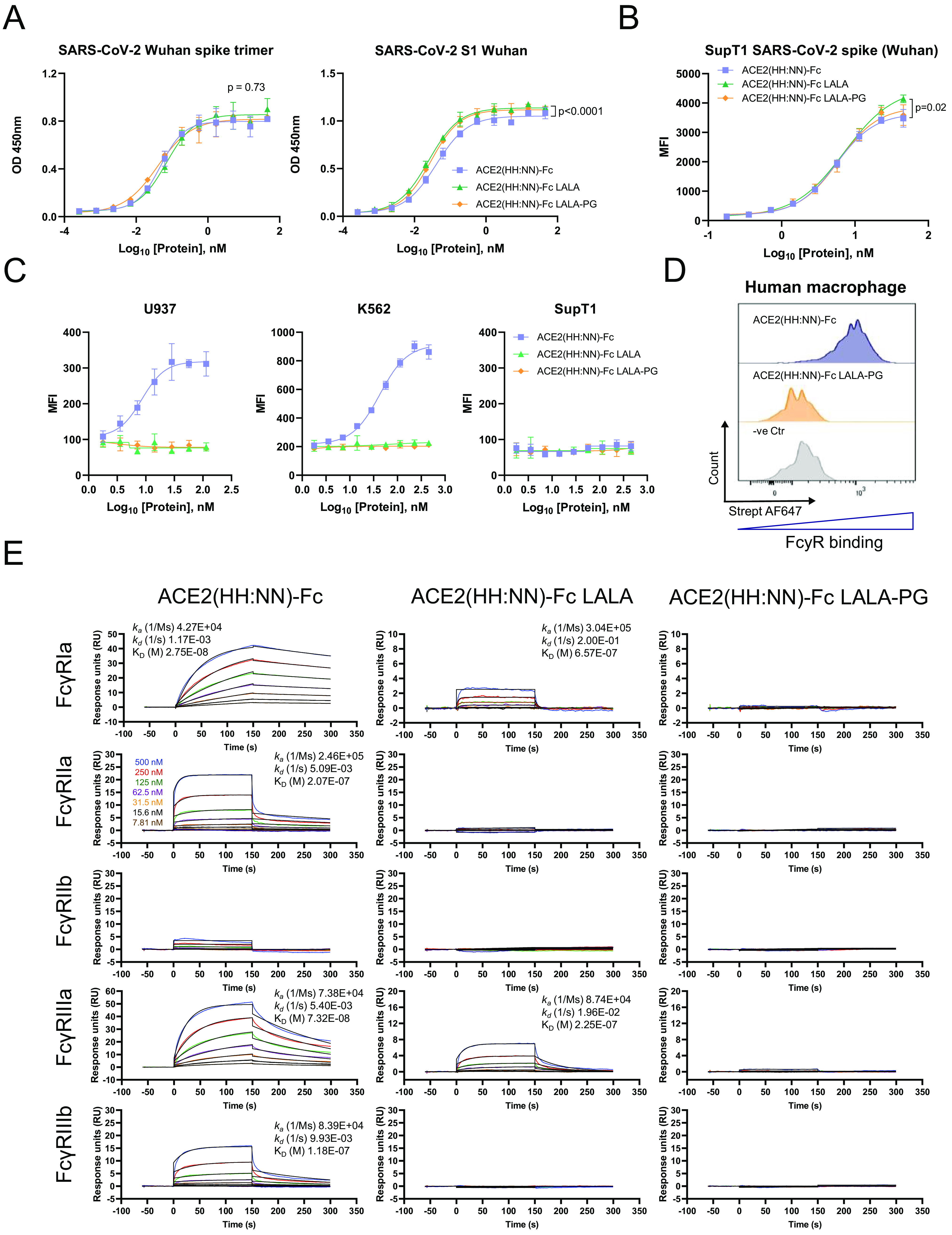
Characterization of Fc effector functions. (A) ELISA of SARS-CoV-2 active spike trimer (left) or S1 domain (right) with ACE2(HH:NN) WT Fc (blue), LALA Fc (green), or LALA-PG Fc (orange), showing comparable binding capacity (mean ± SD). One-way ANOVA of AUC with Tukey’s multiple comparisons (left F = 0.3121, df = 2, 72; right F = 34.17, df = 2, 72, compared to blue). (B) Binding capacity on SupT1 cell line expressing SARS-CoV-2 full-length spike, by flow cytometry with ACE2(HH:NN) WT Fc (blue), LALA Fc (green), or LALA-PG Fc (orange) (mean ± SD). One-way ANOVA of AUC with Tukey’s multiple comparisons (blue versus green, F = 3.986, df = 2, 54). MFI, mean fluorescence intensity. (C) Fc-mediated binding capacity to U937, K562, and SupT1 of ACE2(HH:NN) WT Fc (blue), LALA Fc (green), or LALA-PG Fc (orange), detected with biotinylated SARS-CoV-2 S1 and streptavidin conjugated secondary agent. No binding was detected with ACE2-Fc constructs carrying the LALA or LALA-PG mutations (mean ± SD). (D) Representative flow cytometry of Fc-mediated binding of ACE2(HH:NN) WT Fc (blue) and LALA-PG Fc (orange) on human monocyte-derived M1 macrophages. No binding detected with Fc carrying the LALA-PG mutation (n = 4). (E) SPR binding kinetic of ACE2(HH:NN) WT Fc, LALA Fc, or LALA-PG Fc on human FcγRIa, FcγRIIa, FcγRIIb, FcγRIIIa, and FcγRIIIb. LALA-PG mutations mediated a complete abrogation of FcγR interaction. Sensograms fitted with 1:1 Langmuir binding model.
Next, we investigated the residual interaction of the engineered Fc domains for human FcγRI, FcγRII, and FcγRIII on K562, U937, and SupT1 human cell lines. K562 are reported to express RNA for FcγRIIa and IIIa/b, while U937 express FcγRIa/b, IIa/b, and IIIb (source www.proteinatlas.org, v20.0). A reverse flow cytometry detection assay, using biotinylated SARS-CoV-2 S1 as secondary reagent, demonstrated that the ACE2 construct with WT Fc efficiently bound both K562 and U937 in a dose-dependent manner (Fig. 3C). No binding was detected with either LALA or LALA-PG Fc mutations. SupT1 cells are not reported to express FcγR on the membrane and, consequently, failed to show binding events with the tested molecules. Equally, human M1 polarized monocyte-derived macrophages (MDM) from healthy donors and showed strong interaction with the ACE2 carrying WT Fc, while no detectable engagement was obtained with the LALA-PG Fc mutation (Fig. 3D).
Binding affinities of WT, LALA, and LALA-PG ACE2(HH:NN)-Fc variants for human FcγRs were tested via SPR. ACE2(HH:NN)-Fc showed strong interaction with FcγRIa and IIIa (27.5 nM and 73.2 nM, respectively) and reduced binding affinity for FcγRIIa and IIIb (207 nM and 118 nM, respectively). The LALA mutation still maintained residual binding to the FcγRIa and IIIa (657 nM and 225 nM, respectively) but no detectable binding to the remainder of the receptors. The LALA-PG mutation, however, showed a complete abrogation of FcγR binding, suggesting a more silent immunomodulatory profile (Fig. 3E).
Analysis of ACE2 decoy cross-reactivity and spike binding affinity.
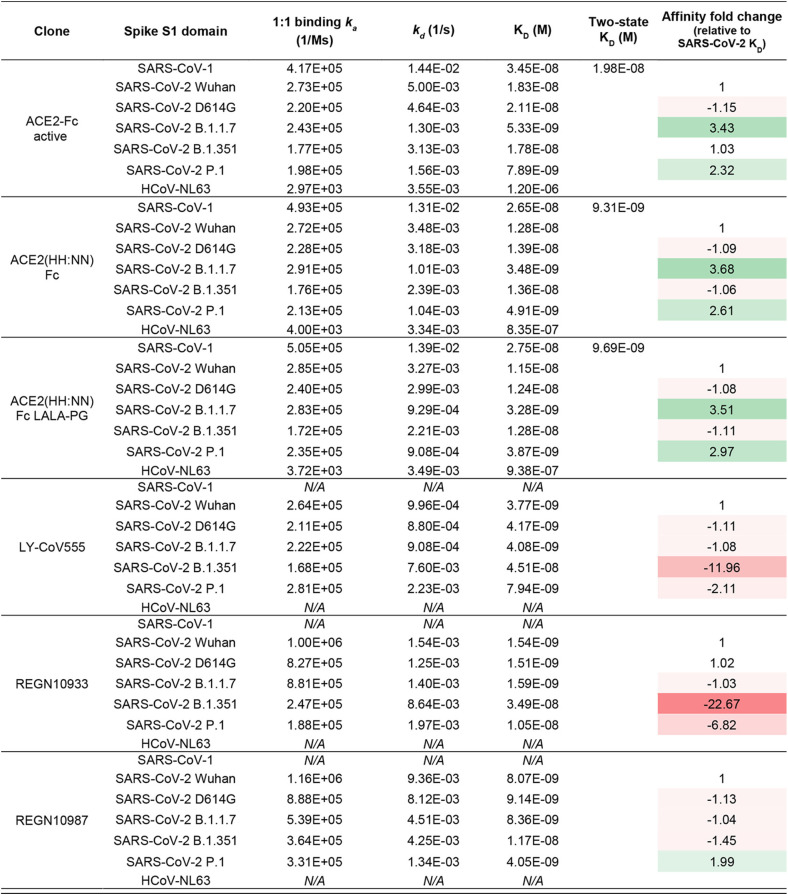
As this receptor decoy has the potential to bind S glycoproteins of viruses that utilize ACE2 as host cell receptor, binding kinetics were generated for the S1 spike domain of SARS-CoV-1, SARS-CoV-2 Wuhan, D614G, B.1.1.7, B.1.351, and P.1 variants, and HCoV-NL63, comparing to the leading anti-SARS-CoV-2 antibodies LY-CoV555 (31), REGN10933, and REGN10987 (32). The ACE2-Fc fusion constructs mediated specific interaction toward all spike proteins tested, while the monoclonal antibodies showed specificity for only the SARS-CoV-2 related S1 (Fig. 4A). The ACE2(HH:NN)-Fc and ACE2(HH:NN)-Fc LALA-PG showed comparable affinities for the tested S1 domains, confirming no effect of the Fc mutations on ACE2 binding (Fig. 4A and Table 1). Similar to the active ACE2-Fc, the inactive ACE2-Fc constructs also displayed an affinity enhanced 3-fold for the SARS-CoV-2 S1 B.1.1.7 and >2.5-fold for P.1. While the monoclonal antibody REGN10987 maintained a similar affinity for the SARS-CoV-2 S1 variants tested, with an almost 2-fold increase for the P.1 S1, the LY-CoV555 and REGN10933 were dramatically affected by the B.1.351 variant with 12- and 23-fold reduction in affinity compared to that of S1 Wuhan, respectively, and with a reduced impact, also by the P.1 variant (Fig. 4A and Table 1).
FIG 4.
SARS-CoV-2 spike binding and neutralization. (A) SPR binding kinetics of ACE2(HH:NN) WT Fc, LALA-PG Fc, LY-CoV555, REGN10933, and REGN10987 against SARS-CoV-1, SARS-CoV-2 variants (Wuhan, D614G, B1.1.7, B.1.351, and P.1), and HCoV-NL63 S1 domains. ACE2(HH:NN) Fc and ACE2(HH:NN) Fc LALA-PG were able to efficiently bind all spike protein tested. All sensograms were fitted with Langmuir 1:1 binding model, except for SARS-CoV-1 S1 kinetics, which were fitted with two-state kinetics. Two-fold serial dilutions starting from 250 nM (500 nM for HCoV-NL63 S1). (B) Cell microarray screening of human cell-membrane proteome with ACE2-Fc (LALA-PG), control Fc (LALA-PG), CTLA4-hFc, or PBS. Depicted is a selection of antigens (key legend on the right panel). ACE2-Fc (LALA-PG) shows strong specific interaction with SARS-CoV-2 spike protein only. See also Table S1. (C) Neutralization assay of authentic SARS-CoV-2 virus with ACE2(HH:NN) WT Fc (blue) and LALA-PG Fc (orange). Both variants show comparable neutralization efficiencies (mean ± SD). Unpaired t test of AUC (t = 1.695, df = 28). (D) Neutralization assay of SARS-CoV-1, SARS-CoV-2 Wuhan, SARS-CoV-2 D614G, SARS-CoV-2 B1.1.7, and SARS-CoV-2 B1.351 pseudotyped vectors with ACE2(HH:NN)-Fc (LALA-PG), LY-CoV-55, REGN10933/REGN10987 cocktail, REGN10933, and REGN10987. Marked decrease of neutralization capacity for SARS-CoV-2 B1.351 detected for LY-CoV555, REGN10933/REGN10987 cocktail, REGN10933, and REGN10987. No loss of neutralization capacity detected for ACE2(HH:NN)-Fc (LALA-PG) receptor decoy (mean ± SD). One-way ANOVA of AUC with Dunnett’s multiple comparisons to blue (F = 369.2, df = 4,88). Bottom right panel, fold change of neutralization capacity based on NT50 values. *, unmeasurable NT50 value.
Binding specificity and cross-reactivity of the ACE2(HH:NN)-Fc LALA-PG construct were assessed using a cell-based protein microarray assay, screening 5,477 full-length plasma membrane and cell surface-tethered human secreted proteins, 371 human heterodimers, and the SARS-CoV-2 S (Table S1). The test construct showed strong specific binding to the target protein SARS-CoV-2 S, while no other interaction was detected across the comprehensive panel of human protein (Fig. 4B). An Fc LALA-PG only construct with the ACE2 domain omitted did not display any interaction with SARS-CoV-2 S or any other target tested. The control fusion protein CTLA4-hFc instead showed strong interaction for its predicted target CD86, and the FcγRIa, due to the presence of a WT IgG1 Fc domain. A secondary anti-human Fc antibody interaction with human IgG3 was detected across all conditions tested (Fig. 4B).
In vitro neutralization of SARS-CoV-2 variants of concern.
We first assessed the neutralization capacity of our decoy receptor against the authentic replication competent SARS-CoV-2. Both ACE2(HH:NN)-Fc and ACE2(HH:NN)-Fc LALA-PG showed comparable neutralization efficiency for authentic SARS-CoV-2 virus in vitro, with half maximal neutralization titer (NT50) values of 5.2 and 4.1 nM, respectively, providing evidence of potent therapeutic activity (Fig. 4C).
Next, to investigate the degree of neutralization efficiency against the SARS-CoV-2 variants of interest, the receptor decoy was tested against the engineered replication-deficient lentiviral vectors pseudotyped with the glycoproteins of SARS-CoV-2 Wuhan, D614G mutation, B.1.1.7, and B.1.351 variants and SARS-CoV-1. The ACE2(HH:NN)-Fc LALA-PG was able to efficiently neutralize SARS-CoV-2, with tight dose-response curves among the SARS-CoV-2 variants, and SARS-CoV-1 (Fig. 4D). Interestingly, the neutralization capacity was slightly improved for the B.1.1.7 and B.1.351 variants compared to that for SARS-CoV-2 Wuhan. The monoclonal antibody LY-CoV555 showed a marked reduction in neutralization capacity for the D614G and B.1.1.7 variants, 3- and 8-fold, respectively, significantly affecting the antibody efficacy, with an almost complete abrogation of neutralization against the B.1.351 variant (Fig. 4C). The 1:1 REGN10933/REGN10987 antibody cocktail was more resilient in its response to the SARS-CoV-2 variants but was characterized by a 4-fold reduction in neutralization for the B.1.351 variant. When the two antibodies constituting the cocktail were analyzed individually, the REGN10933 showed a 3-fold decrease in neutralization capacity for the D614G and B.1.1.7 variants, with a staggering >1,000-fold reduction for the B.1.351 variant, while the REGN10987 showed a 4-fold neutralization reduction for the B.1.1.7 variant and a 10-fold shift for the D614G variant (Fig. 4D).
In vivo neutralization of SARS-CoV-2 in a hamster model of disease.
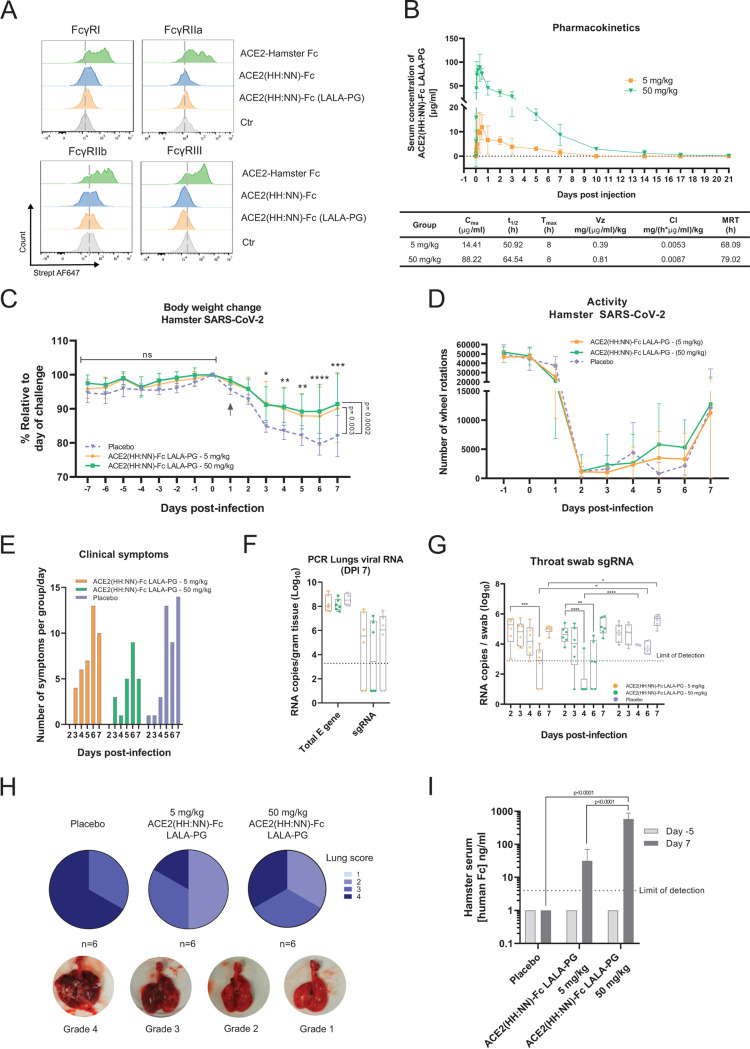
It has been previously reported that hamsters are a relevant small animal model for SARS-CoV-2 infection, reporting symptoms such as reduced body weight and pathological lesions on the lung (33). The hamster FcγRs show an interaction profile with human IgG1 Fc molecules different from that of hamster IgG Fc; nonetheless, the LALA-PG mutation of our construct still showed a complete lack of interaction with hamster FcγRs (Fig. 5A). However, human Fc-tagged molecules are not expected to experience extended circulating half-life due to lack of hamster FcRn engagement. The pharmacokinetics of ACE2(HH:NN)-Fc LALA-PG were assessed in healthy golden Syrian hamsters (Mesocricetus auratus) via intraperitoneal (i.p.) administration. The drug showed a half-life of 64.5 h and detectable levels up to 17 days postinjection for the 50 mg/kg dose (Fig. 5B).
FIG 5.
In vivo SARS-CoV-2 neutralization. (A) Fc-mediated binding capacity of ACE2(HH:NN) WT Fc (blue), LALA-PG Fc (orange), or ACE2(HH:NN) hamster Fc (green) to HEK293T cells expressing hamster FcγRI, FcγRIIa, FcγRIIb, and FcγRIII receptors, detected with biotinylated SARS-CoV-2 S1 and streptavidin conjugated secondary agent. No binding was detected with ACE2(HH:NN)-Fc constructs carrying the LALA-PG mutations on the hamster FcγRs, while limited binding was detected with the ACE2(HH:NN) WT Fc. (B) Syrian hamster serum concentration of i.p. injected ACE2(HH:NN)-Fc LALA-PG at 5 and 50 mg/kg doses, over the course of 28 days (n = 6/group, 3 animals per time point). Cmax, maximum detected concentration; Tmax, peak concentration time; t1/2, half-life; Vz, volume of distribution; Cl, clearance rate; MRT, mean residence time. (C to I) Syrian hamster intranasally challenged with authentic SARS-CoV-2. ACE2(HH:NN)-Fc (LALA-PG) administered i.p. at day 1 postchallenge at 5 mg/kg, 50 mg/kg, or placebo (PBS) (n = 6 per group). (C) Body weight change (%) relative to that on the day of viral inoculation. Day of therapeutic administration marked with arrow. Significant reduction of body weight change relative to that of the placebo, detected for both treatment regimens (mean ± SD). Individual day comparison placebo versus 50 mg/kg dose two-way ANOVA with Sidak’s multiple comparisons compared to placebo group. *, P = 0.01; **, P = 0.004; ***, P = 0.0001; ****, P < 0.0001. One-way ANOVA of AUC with Tukey’s multiple comparisons (F = 9.379, df = 2, 225). (D) Hamster activity monitoring (wheel rotation) showing faster recovery trend at 5 to 6 DPI for the treated groups. (E) Clinical symptoms scoring per group per day, based on fur appearance, nasal/ocular discharge, posture, breathing, activity, and body weight. (F) Total E RNA and subgenomic RNA PCR assay from lung extracts at 7 DPI. Limit of detection 3.28 RNA copies. Samples with undetectable RNA were assigned a value of 1 (mean ± SD). (G) Subgenomic RNA PCR swab test. Limit of detection 2.88 RNA copies; samples with undetectable RNA were assigned a value of 1 (mean ± SD). Two-way ANOVA with Dunnett’s multiple comparisons. (H) Necropsy pathology lung score (categories 1 to 4) showing reduction in lung damage for ACE2(HH:NN)-Fc LALA-PG treated groups. Bottom, representative lung damage for grade scores 1, 2, 3, and 4. (D) Human IgG1 Fc concentration in hamster sera at day −5 and day 7 relative to viral inoculation. Limit of detection 4 ng/ml. Samples with undetectable levels were assigned a value of 1 (mean ± SD). Two-way ANOVA with Sidak’s multiple comparisons (F = 39.2, df = 2, 22).
For in vivo SARS-CoV-2 neutralization, Syrian hamsters were challenged intranasally with 104.5 median tissue culture infectious dose (TCID50) viral inoculum and then dosed 24 h later via i.p. injections of ACE2(HH:NN)-Fc LALA-PG at either 5 mg/kg or 50 mg/kg. Phosphate-buffered saline (PBS) injections were used for the placebo control group. The hamster groups treated with either high or low ACE2(HH:NN)-Fc LALA-PG doses showed a significant protection against body weight loss, with a maximum average weight loss of 11% compared to 21% for the placebo group, relative to the day of viral inoculum (Fig. 5C). All groups showed a drastic reduction in motor activity with a trend for faster recovery in the two treated groups from day 5 post-viral challenge (Fig. 5D). Throat swabs revealed a substantial reduction in viral RNA copies between day 4 and day 6 compared to the viral load in the placebo; several animals showed undetectable levels of RNA between day 3 and day 6 for the high ACE2(HH:NN)-Fc LALA-PG dose and an overall viral load significantly lower than that of the placebo control group (Fig. 5E). Generally, hamsters treated with the ACE2(HH:NN)-Fc (LALA-PG), especially at high dose, showed fewer clinical symptoms of disease, such as ruffled fur, body weight loss, and increased breathing, compared to those in the control group (Fig. 5F). Macroanalysis on lung necropsies (day 7) also showed an overall reduction in lung damage for the ACE2(HH:NN)-Fc LALA-PG-treated groups, characterized by fewer lesions and blood clotting (Fig. 5G). Finally, i.p. administered ACE2(HH:NN)-Fc LALA-PG was still detectable in the hamster sera at day 7, with levels for the high-dose treatment almost 20-fold higher than those for the low-dose treatment (Fig. 5H).
Formulation optimization and streamlined manufacturing of ACE2(HH:NN)-Fc decoy.
To define a suitable formulation considering manufacturing scale-up for clinical application, the well-established antibody formulation buffer 20 mM His (34, 35) was used to solubilize the ACE2(HH:NN)-Fc at a range of pH conditions from 3.5 to 7. The ACE2(HH:NN)-Fc in PBS at pH 7.4 showed good thermal stability with a first unfolding event at 46.1°C, attributed to the unfolding of the ACE2 domain (Fig. 6A). When tested in 20 mM His buffer, the first unfolding event occurred at a Tm between 42.3 and 51.6°C, with the lowest Tm associated with pH 3.5 and the most stable Tm obtained at pH 6.5 (Fig. 6A).
FIG 6.
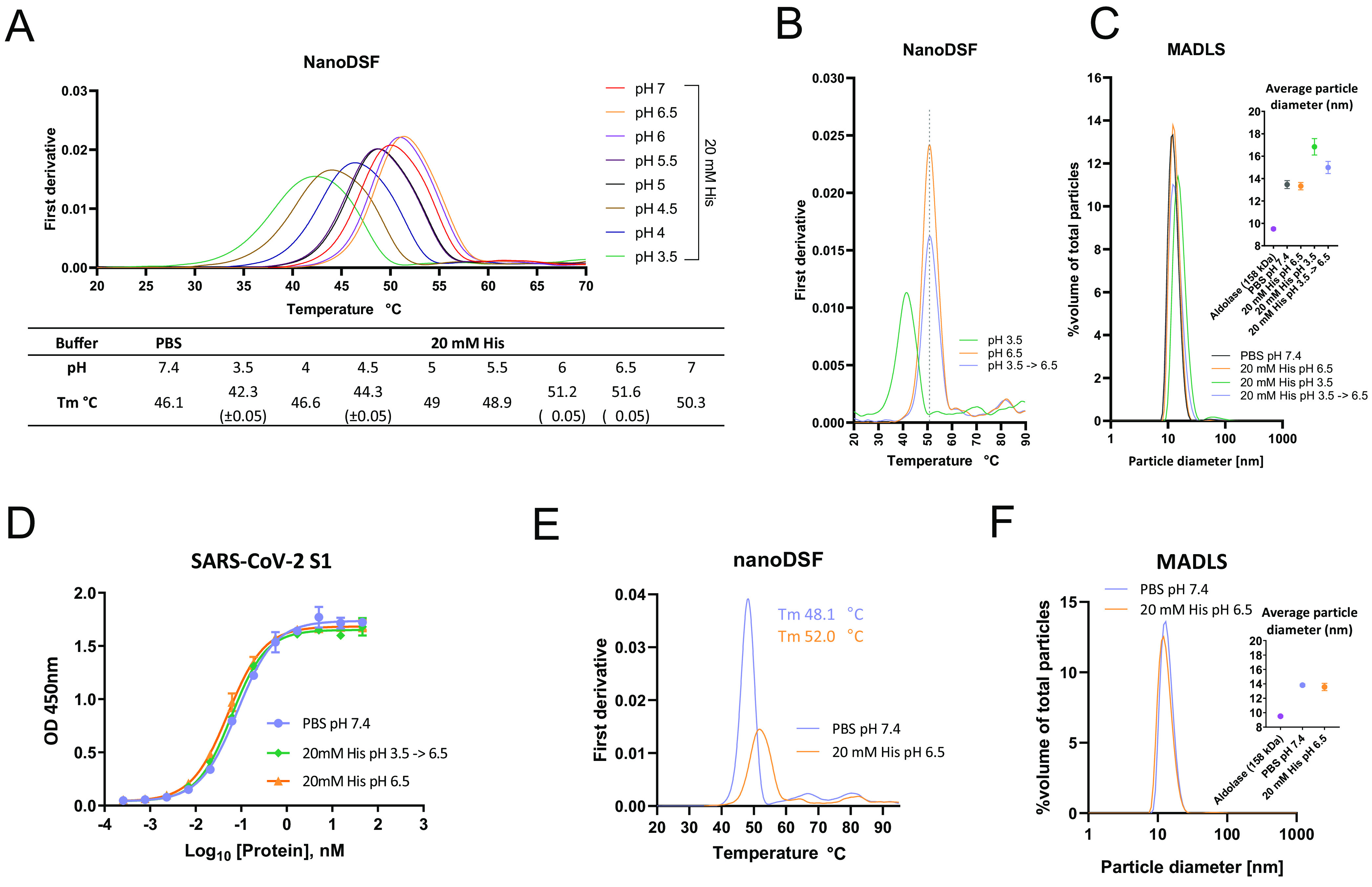
ACE2-Fc formulation optimization. (A) Thermal stability analysis via nanoDSF of ACE2(HH:NN)-Fc in PBS at pH 7.4 or in 20 mM His buffer with pH range of 3.5 to 7. Highest stability obtained with 20 mM His (pH 6.5). (B) Thermal stability of ACE2(HH:NN)-Fc in 20 mM His pH 6.5 (orange) and 3.5 (green) following 2 h of incubation at RT. Full stability could be recovered following buffer exchange of ACE2(HH:NN)-Fc from pH 3.5 to pH 6.5 (blue). (C) Particle size distribution analysis via MADLS of ACE2(HH:NN)-Fc at 1 mg/ml in PBS (pH 7.4), 20 mM His *(pH 3.5, 6.5), or buffer exchanged from pH 3.5 to 6.5. Increase of particle size of sample at pH 3.5 was partially recovered upon buffer exchange in 20 mM His (pH 6.5) (mean ± SD). (D) ACE2(HH:NN)-Fc binding capacity for SARS-CoV-2 S1 in ELISA in PBS (pH 7.4; blue), 20 mM His (pH 6.5; orange), or 20 mM His (pH 3.5) followed by buffer exchange in 20 mM His (pH 6.5; green) (mean ± SD). (E) Thermal stability analysis via nanoDSF of ACE2(HH:NN)-Fc (LALA-PG) in PBS at pH 7.4 (blue) or in 20 mM His (pH 6.5; orange) showing a 3.9°C Tm shift. (F) Particle size distribution analysis via MADLS of ACE2(HH:NN)-Fc (LALA-PG) at 1 mg/ml in PBS (pH 7.4; blue) and 20 mM His (pH 6.5; orange) showing comparable profile (mean ± SD).
A crucial phase during manufacturing of monoclonal antibodies lies in the viral inactivation step, often carried out at low pH (36), which can affect the stability and aggregation state of the proteins in solution. To investigate this, the ACE2(HH:NN)-Fc was exposed to pH 3.5 for 90 min before dialysis in 20 mM His pH 6.5. Thermal stability comparison of ACE2(HH:NN)-Fc at pH 3.5, 6.5, and 3.5 dialyzes to 6.5 showed how the initial instability due to pH 3.5 could efficiently be restored to that of the ACE2(HH:NN)-Fc following dialysis at pH 6.5 (Fig. 6B). The distribution of particles within the solution showed a predominantly monodispersed profile for the ACE2(HH:NN)-Fc in PBS and 20 mM His pH 6.5, with an average diameter of 13.5 and 13.3 nm, respectively, in agreement with a molecule with a predicted molecular weight (MW) of 219 kDa. The suspension in a low pH buffer of 3.5 did not significantly enhance aggregation of ACE2(HH:NN)-Fc (Fig. 6C). Furthermore, the change of buffer from PBS pH 7.4 to 20 mM His pH 6.5 and, crucially, the viral inactivation step at pH 3.5 with subsequent dialysis to pH 6.5 did not affect the capacity of the ACE2(HH:NN)-Fc to bind the SARS-CoV-2 S1 protein, further validating the proposed process (Fig. 6D).
The ACE2(HH:NN)-Fc LALA-PG also showed an increased thermal stability when in 20 mM His pH 6.5 buffer, with Tm moving from 48.1°C to 52.0°C and CH2 CH3 unfolding happening at 64.3°C and 81.8°C, respectively (Fig. 6E). The ACE2(HH:NN)-Fc LALA-PG was also characterized by a monodispersed particle profile with an average diameter size of 13.6 nm in 20 mM His pH 6.5 (Fig. 6F). Finally, the formulation in 20 mM His pH 6.5 of ACE2(HH:NN)-Fc LALA-PG did not alter the SARS-CoV-2 neutralization capacity of the construct (data not shown).
DISCUSSION
We have described the generation of a catalytically inactive ACE2 receptor decoy fused to an engineered human Fc domain with abrogated FcγR engagement, showing optimal biophysical properties and manufacturability. The construct showed strong neutralization potency against several SARS-CoV-2 variants of concern in vitro and evidence of efficacy as a therapeutic administration in a live viral challenge model in vivo.
Monoclonal antibodies developed for the treatment of COVID-19 have shown efficacy in the treatment of early phases of the infection, potentially useful in prophylaxis or as an alternative for people who cannot be vaccinated (37). However, cumulative S mutants may render therapeutic MAbs ineffective. For instance, the variants of concern B.1.351 and P.1 have been shown to affect the neutralization capacity of the approved antibody therapeutics. The LY-CoV555 antibody reported an almost complete abrogation of neutralization, while the antibody cocktail REGN-COV2 showed a severe impairment for one of its components, suggesting preservation of limited therapeutic efficacy (18, 19). By analyzing kinetic of interactions, we determined that the antibodies LY-CoV555, REGN10933, and REGN10987 showed strong binding capacity for the SARS-CoV-2 Wuhan, D614G, and B.1.1.7 S1 domains. Strikingly, the LY-CoV555 and REGN10933 MAbs were strongly impaired in their binding to the B.1.351 variant. Although the final constant domain sequences used in our version of the aforementioned antibodies may vary compared to the clinical products, the variable domains and antibody formats were generated according to published information (31, 32). While not yet present in naturally occurring variants, the single amino acid mutation E406W has recently been shown to be able to escape both antibodies in the REGN-COV2 cocktail (38), further highlighting the weakness of the MAb approach.
“Receptor traps” are an established therapeutic approach, e.g., the anti-TNF Etanercept (20), the VEGF-trap Aflibercept (21), and the CTLA-4-Ig Abatacept (22). Since ACE2, the receptor for SARS-CoV-2, is a type I transmembrane protein with a discrete extracellular domain, ACE2-based receptor decoys may be effective against COVID-19. A theoretical advantage of this approach is resistance to S mutational drift since mutations disrupting interaction with ACE2 would render the virus inactive. ACE2-based therapeutics have been described: A soluble catalytically active human ACE2 showed efficacy in the treatment of a severe COVID-19 patient by reducing plasma viremia (39). Two recent reports have described engineered ACE2 molecules with sub nM affinities for the S glycoprotein (24, 26). Similarly, ACE2-derived inhibitory peptides with improved manufacturability and stability also showed enhanced SARS-CoV-2 neutralization efficacy (25). However, the lack of an Fc domain may affect serum half-life and manufacturing efficiency, and importantly, optimized designs may allow viral mutational escape due to differences with the endogenous receptor. Plain ACE2-Fc fusions were developed against SARS-CoV-1 in 2003 and were also proposed for SARS-CoV-2 (40, 41). Recently, a tetravalent ACE2-Fc was described which showed improved neutralization efficiency compared to that of standard ACE2-Fc formats, without ACE2 domain engineering (42). These ACE2-Fc fusions retained catalytic activity of ACE2 and maintained full Fc effector function.
In contrast to previously reported formats, we developed an ACE2-Fc decoy engineered to be catalytically inactive to prevent systemic activity and with a completely abrogated FcγR interaction to minimize proinflammatory activity. We provided evidence for lack of enzymatic activity on a synthetic substrate while showing that these mutations still maintain reversible engagement with the natural substrate Ang II, avoiding the risk of acting as a substrate sink and offering a safer profile over systemic interference on the renin/angiotensin system. Differently from a previously described ACE2-Fc construct carrying the LALA Fc mutation (43), our design using the LALA-PG mutation shows a complete abrogation of human FcγR engagement while maintaining FcRn interaction to provide extended half-life (30). Although its relevance has not been conclusively determined for SARS-CoV-2, the engineered Fc should alleviate the risk of antibody-dependent enhancement (ADE) of infection as reportedly mediated through FcγRII for SARS-CoV-1 and MERS-CoV (44, 45). A cross-reactivity study against a comprehensive panel of close to 6,000 human-soluble and membrane-bound proteins has highlighted the exquisite specificity of this construct for the target protein. Finally, the introduced mutations to inactivate the ACE2 enzymatic activity, which sit outside the S protein-targeting region, maintained affinity and kinetic interactions equivalent to those of the active receptor for the SARS-CoV-2 S1 variants, minimizing the risk of mutational escape.
Despite the enhanced affinity of the S1 variants B.1.1.7 and P.1 for the ACE2 receptor and the increased infectivity of the pseudotyped vectors displaying the D614G, B.1.1.7, and B.1.351 S glycoproteins, the ACE2(HH:NN)-Fc LALA-PG decoy maintained efficient neutralization capacity toward all SARS-CoV-2 variants tested, showing enhanced potency driven by mutational drift. Paradoxically, the spike mutations enhancing affinity for the ACE2 receptor would improve the neutralization potency of ACE2-based decoys. Additionally, we showed binding capacity to SARS-CoV-1 and HCoV-NL63, alongside neutralization of SARS-CoV-1 pseudovirus, providing evidence for broad-spectrum activity over ACE2-tropic viruses.
Conversely, we have observed a substantial drop in neutralization capacity of the LY-CoV555, the 1:1 REGN10933/REGN10987 antibody cocktail, and the latter’s individual antibodies for the SARS-CoV-2 variants, in line with previous reports (18). The LY-CoV555 and REGN10933 were especially impaired by the B.1.351 variant. We also noticed a generally reduced neutralization capacity for the D614G variant, which is likely due to its enhanced infectious capacity compared to that of the Wuhan strain. However, as the lentiviral pseudotyped system adopted in this work has limitations, results could be confirmed in future studies using the authentic virus.
Despite the lack of FcRn engagement in hamster, the i.p. injected ACE2(HH:NN)-Fc LALA-PG showed detectable serum levels for up to 17 days in vivo and was also able to affect the replication of the authentic SARS-CoV-2 virus in a hamster model, reducing body weight loss and lung damage in infected animals.
In conclusion, we describe detailed in vitro and in vivo characterization of a soluble catalytically inactive ACE2-Fc receptor decoy molecule resistant to spike protein mutation. We also demonstrate that our decoy molecule has the potential for rapid upscale manufacturability. In theory, our decoy should be active against any new ACE2-tropic virus which might emerge in the future. In this phase of the SARS-CoV-2 pandemic where viral variants are exerting pressure over the efficacy of vaccines and monoclonal antibodies, the development of biotherapeutics which are inherently resistant to SARS-CoV-2 mutations may be prudent.
MATERIALS AND METHODS
Cell line generation and maintenance.
HEK-293T cells (ATCC, CRL-11268) were cultured in Iscove’s modified Dulbecco’s medium (IMDM) (Lonza, 12-726F) supplemented with 10% fetal calf serum (FCS; Biosera, FB 1001/500) and 2 mM GlutaMAX (Gibco, 35050061) at 37°C with 5% CO2. Sup-T1 (ATCC, CRL-1942), U937 (ATCC, CRL-1593.2), and K562 (ATCC, CCL-243) were cultured in RPMI 1640 medium (Gibco 21875034) supplemented with 10% fetal calf serum (FCS; Biosera, FB 1001/500) and 2 mM GlutaMAX (Gibco, 35050061) at 37°C with 5% CO2.
Sup-T1 cells were γ-retrovirally transduced to express the S glycoprotein of SARS-CoV-2 Wuhan Hu-1 strain coexpressed with enhanced blue fluorescent protein (eBFP) as a marker gene. Briefly, 5 × 105 cells were incubated with 1 ml of unquantified vector supernatant in the presence of retronectin in non-tissue culture-treated 24-well plate and subjected to spin-inoculation at 1,000 × g for 40 min. Cells were recovered 24 h later by culturing in serum-supplemented RPMI 1640 for two passage before use in experiments.
For the generation of MDM, human monocytes were isolated from blood of healthy donors using Easy Sep human monocyte isolation kit (Stemcell, 19359), according to manufacturer’s recommendations. Monocyte isolation was determined with the following flow cytometry antibody panel after 10 min incubation with anti-human CD32 (StemCell, 18520): allophycocyanin (APC) anti-human CD14 (Biolegend, 301808), PE-Cy7 anti-human CD3 (Biolegend, 344186), AF488 anti-human CD20 (Biolegend, 302316), and live/dead Sytox Blue stain (Invitrogen, S34857). Monocytes were activated by culturing in Immunocult serum-free macrophage differentiation medium (Stemcell, 10961) supplemented with 50 ng/ml macrophage colony-stimulating factor (M-CSF) (Stemcell, 78057). At day 6, cells were supplemented with 50 ng/ml gamma interferon (IFN-γ) (Stemcell, 78020) and 10 ng/ml lipopolysaccharide (LPS) (Sigma, L4391) to stimulate M1 polarization. M1 macrophages were harvested by Accutase dissociation (Stemcell, 07920). The following flow cytometry antibody panel was used to determine monocyte differentiation and M1 polarization after 10 min incubation with anti-human CD32: APC anti-human CD14 (BioLegend, 344186), BV421 anti-human CD80 (BioLegend, 305222), phycoerythrin anti-human CCR7 (BioLegend 353204), APC/Fire750 anti-human CD209 (330116), and 7-aminoactinomycin D (7-AAD) viability staining solution at 5 μl/1 × 106 cells. Samples from both flow staining panels were acquired using the MacsQuant10 instrument (Miltenyi Biotech).
Protein expression, purification, and characterization.
Human ACE2 containing amino acids 18 to 740 (UniProt Q9BYF1) was fused to the human IgG1 hinge and Fc (UniProt P01857). Inactive ACE2 was generated by introducing H374N and H378N mutations. Silent Fc variants were generated with L234A/L235A and L234A/L235A/P329G mutations. Chimeric human inactive ACE2 with hamster Fc fusion was generated using Cricetulus migratorius IgG heavy chain hinge-Fc sequence (GenBank U17166.1). Variable domain sequences for LY-CoV-555 were obtained from published crystal structure PDB 7L3N (31); REGN10933 and REGN10987 sequences were obtained from published crystal structure PDB 6XDG (32). Heavy variable domains were fused to human IgG1 constant chain (UniProt P01857); kappa variables were fused to human kappa constant domain (UniProt P01834); lambda variable was fused to human lambda constant 1 (UniProt P0CG04). All constructs were cloned in an AbVec vector (46). REGN10933/REGN10987 antibody cocktail was generated as a 1:1 mix of REGN10933 and REGN10987. Recombinant Fc-tagged proteins were expressed by transient transfection in ExpiCHO, according to manufacturer’s recommendations (Thermo Fisher, A29133). Supernatant from transfected CHO cells was purified using 1 ml HiTrap MabSelect PrismA (GE Healthcare, 17549851) affinity chromatography with in-line dialysis in PBS via HiTrap 5 ml desalting columns (GE Healthcare, 29048684) using an Akta Pure system (GE Healthcare), following manufacturer’s recommendations.
SARS-CoV-2 S1 domains (amino acids [aa] 1 to 681) from Wuhan (GenBank, QHD43416.1) or including the D614G (11), B.1.1.7 (13), B.1.351 (14), and P.1 (17) mutations were cloned in fusion with a dual 6×His tag using an AbVec vector. Supernatant from Expi293-transfected cells was manually purified using TALON metal affinity chromatography (TaKaRa bio Inc., 635502), according to manufacturer’s recommendations. Purified proteins were buffer exchanged in PBS using Zeba spin desalting columns (Thermo Fisher, 89890).
Purified proteins were analyzed for purity determination via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 4 to 20% gradient gel (Bio-Rad, 4568094), with or without the presence of 2-mercaptoethanol as reducing agent.
Differential scanning fluorimetry.
Thermal stability was determined by nano differential scanning fluorimetry (nanoDSF) on a Prometheus NT.48 instrument (Nanotemper) using first derivative of 350/330 nm ratio to determine the melting temperature (Tm) value. Samples were loaded on a glass capillary and temperature scanned from 20 to 95°C at 1°C/min.
Aggregation and particle size measurement.
Aggregation propensity and average particle size of the test proteins were determined using a Zetasizer Ultra device and ZS Xplorer software (Malvern Panalytical) by MADLS. Samples were loaded into a low-volume quartz cuvette (Malvern Panalytical, ZEN2112) at a concentration of 1 mg/ml. Triplicate measurements were taken for each sample. Particle size of aldolase (158 kDa) was used as reference.
ACE2 enzymatic activity.
Enzymatic activity of active ACE2-Fc (ACRO biosystems, AC2-H5257) and ACE2(HH:NN)-Fc was measured by using Mca-APK(Dnp) (Enzo Life Science, BML-P163) as the substrate in 96-well black microtiter plates. Samples were diluted in reaction buffer (50 mM 4-morpholineethanesulfonic acid [pH 6.5], 300 mM NaCl, 10 μM ZnCl2, and 0.01% Triton X-100) at a concentration of 0.1 μg/ml in the presence of 20 μM Ma-APK(Dnp) or control peptide BML-P127 (Enzo Life Sciences) in a final volume of 100 μl/well. The reaction was performed in triplicate at room temperature for 1 h. Activity was measured as fluorescence intensity at 320 nm/393 nm (Ex/Em) wavelength at 1-min intervals using a Varioskan LUX instrument (Thermo Scientific).
ELISA on spike protein.
Nunc Maxisorp clear 96-well plates were coated with 1 μg/ml (in PBS) of SARS-CoV-2 S trimer (ACRO biosystems, SPN-C52H9), SARS-CoV-2 S1 domain (ACRO biosystems, S1N-C52H3), or bovine serum albumin (BSA) (Sigma, A7906) overnight at 4°C in 50 μl/well. Plates were blocked with PBS 2% BSA for 1h at room temperature (RT). Test proteins were incubated at 45.6 nM concentration with 3-fold serial dilutions for 1 h at RT in PBS 0.5% BSA. Bound Fc-tagged proteins were detected with anti-human horseradish peroxidase (HRP)-conjugated secondary antibodies (Jackson ImmunoResearch, 109-035-088) at 1:3,000 dilution in PBS 0.5% BSA. Incubation was allowed for 1 h at RT. All washes were performed in PBS 0.05% Tween 20. Specific interaction revealed with 1-step TMB (3,3′,5,5′-tetramethylbenzidine) Ultra reagent (Thermo Fisher, 34028) at 45 μl/well and blocked with 45 μl/well of 1 M H2SO4. Plates were acquired on a Varioskan Lux instrument at a wavelength of 450 nm. Data were analyzed with GraphPad Prism 8 (GraphPad software).
Flow cytometry.
For FcγR binding assay on U937, K562, SupT1 cells, and MDM, test constructs were incubated at specified concentrations for 30 min at 4°C to prevent dissociation/internalization. Protein labeled cells were stained with biotinylated SARS-CoV-2 S1 (ACRO biosystems, S1N-C82E8) and detected with streptavidin AF647 (Invitrogen, S21374). Cells were stained with 7-AAD viability staining solution at 5 μl/1 × 106 cells to determine live cells. Stained samples were acquired using a MacsQuant10 instrument (Miltenyi Biotec) and analyzed on FlowJo software (BD).
Binding capacity of ACE2(HH:NN) Fc, LALA Fc, and LALA-PG Fc to SupT1 expressing wild-type SARS-CoV-2 full-length spike was assessed via incubation of test protein at 45.6 nM with 2-fold serial dilutions for 30 min at RT, followed by secondary incubation with anti-human IgG (H+L) AF647 (Invitrogen, A21445) for 20 min at RT in the dark. Cells were stained with 7-AAD viability staining solution at 5 μl/1 × 106 cells to determine live cells and subsequently acquired using MacsQuant10 instrument. Flow cytometry data were analyzed on FlowJo software (BD).
Surface plasmon resonance.
Recombinant active ACE2-Fc (ACRO biosystems, AC2-H5257) and ACE2(HH:NN)-Fc constructs were captured on flow cell 2 of a series S protein A sensor chip (GE Healthcare, 29127555) to a density of 500 response units (RU) using a Biacore 8K instrument (GE Healthcare). HBS-EP+ buffer was used as running buffer in all experimental conditions. Recombinant purified Angiotensin II (Sigma, A9525) at 1 μM with 2-fold serial dilutions, was used as the ‘analyte’ and injected over the flow channels with 150 s contact time and 500 s dissociation.
For SARS-CoV-1 S1 (ACRO biosystems, S1N-S52H5), HCoV-NL63 S1 (SIN-V52H3), SARS-CoV-2 S1 WT (ACRO biosystems, S1N-C52H3), and inhouse expressed SARS-CoV-2 S1 WT, D614G, B.1.1.7, and B.1.351 kinetics, test ACE2-Fc constructs and antibodies were captured to a density of 70 RU or 50 RU, respectively, on a series S protein A sensor chip (GE Healthcare, 29127555) using a Biacore T200 and Biacore 8K instruments (GE Healthcare). HBS-P+ buffer was used as running buffer in all experimental conditions. Recombinant purified spike proteins at known concentrations were used as the “analyte” and injected over the respective flow cells with 150 s contact time and 300 s dissociation.
The binding kinetics to FcγRIa (ACRO biosystems, FCA-H52H1), FcγRIIa (ACRO biosystems, CD1-H5223), FcγRIIb (ACRO biosystems, CDB-H5228), FcγRIIIa (ACRO biosystems, CDA-H5220), and FcγRIIIb (ACRO biosystems, CDB-H5222) were captured to a density of 50 RU (or 150 RU for FcγRIIa, FcγRIIb, and FcγRIIIb) on flow cell 2, 3, or 4 of a series S CM5 chip (GE Healthcare) functionalized with an anti-His capture kit (GE Healthcare) using a Biacore T200 instrument. HBS-EP+ buffer was used as running buffer in all experimental conditions. Purified ACE2(HH:NN)-Fc, ACE2(HH:NN)-Fc LALA, and ACE2(HH:NN)-Fc LALA-PG at a concentration of 500 nM with 2-fold serial dilutions were used as the analyte and injected over the respective flow cells with 150 s contact time and 300 s dissociation.
All experiments were performed at 25°C with a flow rate of 30 μl/ml. Flow cell 1 was unmodified and used for reference subtraction. A “0 concentration” sensogram of buffer alone was used as a double reference subtraction to factor for drift. Data were fit to a 1:1 Langmuir binding model using Biacore insight evaluation software (GE Healthcare). SARS-CoV-1 S1 sensograms were also fit to a two-state kinetic model. Since a capture system was used, a local Rmax parameter was used for the data fitting in each case.
Viral vector production.
Viral vectors were produced by triple transient transfection of HEK-293T cells in 100-mm plates using GeneJuice (Merck, 70967) with a total of 12.5 μg of DNA. γ-retroviral vectors were produced by triple transient transfection of 4.69 μg Peq-Pam plasmid (encoding Moloney GagPol), 3.13 μg of RDF plasmid (encoding RD114 envelope), and 4.69 μg retroviral backbone SFG (47) expressing full-length SARS-CoV-2 S glycoprotein coexpressed with eBFP as the marker gene. Similarly, for lentiviral vector production, cells were transfected with 5.42 μg of pCMV-dR8.74 (encoding lentiviral GagPol), 2.92 μg of envelope plasmid expressing codon-optimized SARS-CoV S glycoproteins with their endoplasmic reticulum (ER) retention signals deleted (deletion of the last 19 amino acids on the carboxy terminus), and 4.17 μg of lentiviral backbone pCCL encoding enhanced green fluorescent protein (eGFP) as transgene driven by internal viral spleen focus-forming virus (SFFV) promoter.
Culturing medium was changed 24 h posttransfection, and vector supernatants were collected 48 h after transfection and processed by centrifugation at 1,000 × g for 10 min at 4°C to remove cellular debris followed by microfiltration using Millex-HV 0.45 μm syringe filter units (Merck, SLHV033RB). Viral supernatants were either kept on ice for further use or frozen down at −80°C for storage.
p24 ELISA.
Physical particles were determined by measuring p24 levels using the QuickTitre lentivirus titer, which quantifies lentivirus-associated HIV rather than free p24 proteins (Cell Biolabs, VPK-107-T). Manufacturer’s protocol was followed, and samples were assayed in triplicates. Briefly, after incubation with kit’s ViraBind reagents and virus inactivation, samples were incubated in microwell plates precoated with anti-p24 antibodies followed by a subsequent incubation with secondary fluorescein isothiocyanate (FITC)-conjugated anti-HIV p24 monoclonal antibody (1:1,000). Subsequently, wells were exposed to HRP-conjugated anti-FITC monoclonal antibody (1:1,000). Plates were acquired on a Varioskan Lux instrument at a wavelength of 450 nm. Data were analyzed with Graph Prism 8 (GraphPad software).
SARS-CoV-2 lentiviral pseudotyped viral vector titration.
Functional infectious viral titers were determined by flow cytometry analysis (BD LSRFortessa X-20 cell analyzer) of transgene expression in transduced HEK-293T cells that were previously engineered to express human ACE2 and TMPRSS2. Experiments were performed in 24-well plates (50,000 cells/well). Serially diluted viral supernatants were added onto seeded cells in the presence of 8 μg/ml Polybrene. Transduction efficiencies were determined 72 h later using BD LSRFortessa X-20 cell analyzer. eGFP expression between 0.5% and 20% was used in the following equation to determine viral titer:
SARS-CoV-2 lentiviral pseudotyped viral vector neutralization assay.
Proteins were serially diluted in PBS to 7 decreasing concentrations ranging from 100 mg/ml to 6.1 ng/ml (4-fold serial dilution). Each antibody dilution was then mixed 1:1 with lentiviral vectors pseudotyped with SARS-CoV S glycoproteins normalized to 1.0 × 105 physical particles of vectors pseudotyped with Wuhan glycoprotein to a final volume of 200 μl and incubated at 37°C for 1 h. Antibody-virus mixtures were then cultured with 3 × 104 HEK-293T cells previously genetically engineered to express human ACE2 and TMPRSS2, in the presence of 8 μg/ml of Polybrene, in 48-well plates with a final volume of 0.5 ml per well. Plates were spin-inoculated at 1,000 × g for 10 min and incubated for 72 h. Viral titers were then quantified by eGFP expression in target cells using BD LSRFortessa X-20 cell analyzer, and infectivity of all fractions was determined as a percentage of viral titers in the PBS only control.
SARS-CoV-2 virus neutralization assay.
Vero cells (ATC-CCL81) cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Sigma, catalog no. D6546) with 10% FCS, 2 mM l-glutamine (Sigma-Aldrich, G7513), and 1% penicillin/streptomycin (Invitrogen, 15140148) were seeded the day prior to infection at 2 × 104 cells per well in a 96-well flat-bottom plate. Serial dilutions of proteins of interest were incubated with 100 TCID50 of SARS-CoV-2 (strain England/02/2020) for 1 h at 37°C, 5% CO2. After careful removal of culturing medium from Vero monolayer, 100 μl of protein-virus mixtures was added to the cells and incubated 1 h at 37°C, 5% CO2. Subsequently, 100 μl of culturing medium with 4% FBS was added to occupied wells, and plates were incubated at 37°C, 5% CO2 for 48 h. After removal of culturing medium, occupied wells were fixed with 4% paraformaldehyde (PFA) in PBS for 1 h at room temperature for viral inactivation, followed by incubation with 0.1% Triton X-100 for 15 min at room temperature for cell permeabilization. Plates were washed with 0.05% vol/vol PBS-Tween and sequentially incubated with mouse anti-SARS-CoV-2 N protein antibody (The Native Antigen Company, MAB12183-100) at 1:500 dilution and HRP-conjugated goat anti-mouse IgG antibody (Jackson ImmunoResearch, 115-035-146) at 1:5,000 dilution in 3% wt/vol milk in 0.05% PBS-Tween. Plates were acquired on a BMG FLUOstar Omega at a wavelength of 450 nm.
Cell microarray test.
A total of 5,477 expression vectors, encoding both ZsGreen1 and a full-length human plasma membrane protein or a cell surface-tethered human secreted protein, and 371 human heterodimers were coarrayed across a microarray slide in duplicate (Table S1). HEK293 cells were used for reverse transfection and expression. Test protein was incubated at 20 μg/ml upon cell fixation. Hits were detected by fluorescent secondary antibody using ImageQuant software (GE Healthcare). An expression vector (pIRES-hEGFR-IRES-ZsGreen1) was spotted in quadruplicate on every slide and used as the transfection control. Assay was performed by Retrogenix Ltd.
In vivo hamster model.
For pharmacokinetics (PK) studies, male LVG golden Syrian hamsters 7 to 9 weeks old (100 to 140 g) were single dosed via i.p. injections of ACE2(HH:NN)-Fc LALA-PG at 5 and 50 mg/kg (n = 6 per group). A total of 200 μl of blood was collected from orbital vein at specific time points: 15 min ± 3 min, 30 min ± 3 min, 1 h ± 5 min, 2 h ± 5 min, 4 h ± 5 min, 8 h ± 5 min, 12 h ± 5 min, 24 h ± 5 min, 48 h ± 5 min, 72 h ± 5 min, 120 h ± 5 min, 168 h ± 5 min, 240 h ± 5 min, 336 h ± 5 min, 408 h ± 5 min, 504 h ± 5 min, 672 h ± 5 min postadministration (n = 3 per time point per group). Samples were incubated at 2 to 8°C for 1 h and centrifuged at 2,000 × g for 10 min at 2 to 8°C. Separated serum was stored at −80°C. ACE2(HH:NN)-Fc LALA-PG concentration was measured via ELISA. Briefly, 96-well plates (Corning Inc., 42592) were coated with anti-human IgG antibody (Sigma-Aldrich, I6260) at 1 μg/ml in 50 μl/well overnight at 4°C. Plates were washed 4 times in PBS 0.05% Tween 20 and blocked with 200 μl/well of blocking buffer (GenScript, DD-PK-009) for 1 h at RT on a shaking platform. Upon wash, plates were incubated with 50 μl/well of test sample diluted 100-fold in assay buffer (GenScript, DD-PK-009) in duplicate for 1 h at RT on a shaking platform. Standard curve was generated using purified ACE2(HH:NN)-Fc LALA-PG at 80 μg/ml with 2-fold serial dilutions in blank hamster serum. Plates were washed as described above and incubated with anti-human Fc HRP-conjugated detection antibody (Jackson ImmunoResearch, 109-035-088) at 1:10,000 dilution in assay buffer for 1 h at RT on a shaking platform. Upon wash, plates were incubated with 100 μl/well of TMB solution (GenScript, DD-PK-009) and blocked with 100 μl/well of stop solution (GenScript, DD-PK-009). Plates were read at 450 nm by using a Multiskan FC plate reader (Thermo Fisher Scientific). Pharmacokinetic parameters were calculated by WinNonlin 8.1 (Certara) with the noncompartmental model (NCA).
Syrian hamsters (Mesocricetus auratus) RjHan:AURA strain, male and females 4 to 10 weeks old, were individually caged in a human biosafety level 3 laboratory. At day 0, animals were challenged by intranasal inoculum of 0.1 ml of authentic SARS-CoV-2 with a dose of 104.5 TCID50 under medetomidine and ketamine sedation. At 1 day post-infection (DPI), animals were treated with i.p. injections of 5 mg/kg or 50 mg/kg ACE2(HH:NN)-Fc LALA-PG or PBS, at equal volumes, and monitored until 7 DPI (group n = 6). Nonterminal blood samples were collected at −5 DPI and terminal blood samples were collected at 7 DPI. For human Fc detection, blood samples were inactivated by incubation at 56°C for 2 h before being stored at −20°C. Detection of residual human Fc in the hamster sera was performed by ELISA, using an anti-human Fc MAb (Sigma-Aldrich, I6260) as capture. The standard curve was generated using purified ACE2(HH:NN)-Fc LALA-PG, passed through the same heat-inactivation step as the serum samples and ranging from 22.8 nM to 22.3 pM via a 2-fold serial dilution. Changes relative to ELISA protocol detailed above: Nunc MaxiSorp clear 96-well plates were coated with 1 μg/ml (in PBS) of anti-human Fc MAb overnight at 4°C in 50 μl/well. Plates were blocked with 200 μl/well of PBS 2% BSA for 1 h at RT. Wash steps were performed 3 times in PBS 0.05% Tween 20. Serum samples were tested at both 1:100 and 1:1,000 dilutions in duplicate. Bound proteins were detected with anti-human HRP-conjugated secondary antibody (Jackson ImmunoResearch, 109-035-088) at 1:3,000 dilution in PBS 0.5% BSA. Specific interaction revealed with 1-step TMB Ultra reagent (Thermo Fisher, 34028) at 45 μl/well and blocked with 45 μl/well of 1 M H2SO4. Plates were acquired on a Varioskan Lux instrument at a wavelength of 450 nm.
Body weight measurements were recorded daily throughout the study. Wheel rotations were counted automatically (4 counts are equal to 1 full rotation) every day between 8:15 and 12:00 a.m. Clinical symptoms were graded depending on severity. Active animals (alert with normal behavior and <15% body weight loss) were assigned a score of 0. Animals with ruffled fur, curled bodies, nasal/ocular discharge, sneezing/coughing, or mild increased respiratory rate or showing reduced activity were assigned a score of 1. Animals inactive and sluggish, showing abdominal breathing or >20% body weight loss were assigned a score of 2. Animals with passive/absent behavior or with dyspnea were assigned a score of 3. The symptoms were summed in a ranked manner, using the calculation N0 × 0 + N1 × 1 + N2 × 2 + N3 × 3 + N4 × 4, where N is the number of animals and the number in subscript indicates the number of symptoms per animal. Throat swabs were collected at 2, 3, 4, 6, and 7 DPI to test for presence of SARS-CoV-2 by qPCR. RNA was isolated using a Direct-zol RNA miniprep kit (Zymo Research, R2056) and subgenomic RNA detected as described previously (48, 49). Postmortem examinations were performed at 7 DPI. For RNA analysis, lung tissues were homogenized by an Ultra-Turrax homogenizer before RNA extraction and analysis by total viral RNA and subgenomic RNA as described above. Macroscopic lung lesions were assessed by a pathologist according to the following scoring scheme: 0, no macroscopical changes; 1, focal discoloration of lung <20%; 2, multifocal discoloration of lung 20 to 50%; 3, multifocal discoloration of lung 50 to 80%; 4, whole lung affected >80%. Animal caretakers and pathology personnel were blinded for the treatment groups. Experiments were performed by Wageningen Bioveterinary Research Division Virology of Wageningen University. Animal work was approved by the Dutch Central Authority for Scientific procedures on Animal (CCD), experimental application 2020.D-0007.016 by the Animal Welfare Body of Wageningen University and Research.
Statistical analysis.
All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software). Specific analysis is detailed in figure legends. A P value <0.05 was considered significant.
ACKNOWLEDGMENTS
This work was funded by the UK Research and Innovation business-led innovation in response to global disruption grant. England/02/2020 isolate of SARS-CoV-2 was kindly provided by Public Health England (PHE), UK. We thank Nadia Oreshkova for helpful discussions and guidance in the in vivo assays.
M.F., S.C.O., and M.P. designed the study. M.F., S.C.O., F.T.I., and R.B. planned and/or performed protein purification and biophysical characterizations. J.S., K.L., K.W., F.P., and C.A. planned and/or performed plasmid design and cloning. L.M., Z.A., and R.K. planned and/or performed lentiviral production and characterization as well as flow cytometry assays. Z.A. and V.B. planned and performed MDM work. L.M., J.S., and P.W. planned and performed cell line development. G.M., E.M.B., and Y.T. planned and/or performed authentic virus neutralization assays. A.K., M.F., and S.C.O. planned in vivo and cross-reactivity studies. M.F. wrote the paper, and all authors reviewed the manuscript.
M.F., L.M., F.T.I., Z.A., K.L., F.P., C.A., P.W., V.B., J.S., P.D., A.K., M.P., and S.C.O., are employees of Autolus Ltd.
Footnotes
Supplemental material is available online only.
Contributor Information
Martin Pule, Email: m.pule@autolus.com.
Tom Gallagher, Loyola University Chicago.
REFERENCES
- 1.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z. 2020. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA 323:1061–1069. 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schett G, Sticherling M, Neurath MF. 2020. COVID-19: risk for cytokine targeting in chronic inflammatory diseases? Nat Rev Immunol 20:271–272. 10.1038/s41577-020-0312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Peñaranda S, Bankamp B, Maher K, Chen M-H, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TCT, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Günther S, Osterhaus ADME, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 6.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, Müller MA, Drosten C, Pöhlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, Kugathasan R, Penn R, Brown JC, Sanchez-David RY, Braga L, Williamson MK, Hassard JA, Staller E, Hanley B, Osborn M, Giacca M, Davidson AD, Matthews DA, Barclay WS. 2020. The furin cleavage site of SARS-CoV-2 spike protein is a key determinant for transmission due to enhanced replication in airway cells. bioRxiv 10.1101/2020.09.30.318311 [DOI] [PubMed]
- 8.Raybould MIJ, Kovaltsuk A, Marks C, Deane CM. 2020. CoV-AbDab: the coronavirus antibody database. Bioinformatics 37:734–735. 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L, Liu W, Yu X, Wu M, Reichert JM, Ho M. 2020. COVID-19 antibody therapeutics tracker: a global online database of antibody therapeutics for the prevention and treatment of COVID-19. Antib Ther 3:205–212. 10.1093/abt/tbaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang J, Wang W, Chen Z, Lu S, Yang F, Bi Z, Bao L, Mo F, Li X, Huang Y, Hong W, Yang Y, Zhao Y, Ye F, Lin S, Deng W, Chen H, Lei H, Zhang Z, Luo M, Gao H, Zheng Y, Gong Y, Jiang X, Xu Y, Lv Q, Li D, Wang M, Li F, Wang S, Wang G, Yu P, Qu Y, Yang L, Deng H, Tong A, Li J, Wang Z, Yang J, Shen G, Zhao Z, Li Y, Luo J, Liu H, Yu W, Yang M, Xu J, Wang J, Li H, Wang H, et al. 2020. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 586:572–577. 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 11.Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, Hengartner N, Giorgi EE, Bhattacharya T, Foley B, Hastie KM, Parker MD, Partridge DG, Evans CM, Freeman TM, de Silva TI, McDanal C, Perez LG, Tang H, Moon-Walker A, Whelan SP, LaBranche CC, Saphire EO, Montefiori DC, Angyal A, Brown RL, Carrilero L, Green LR, Groves DC, Johnson KJ, Keeley AJ, Lindsey BB, Parsons PJ, Raza M, Rowland-Jones S, Smith N, Tucker RM, Wang D, Wyles MD. 2020. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 182:812–827.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hodcroft EB, Zuber M, Nadeau S, Crawford KHD, Bloom JD, Veesler D, Vaughan TG, Comas I, Candelas FG, Stadler T, Neher RA, SeqCOVID-SPAIN consortium. 2020. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv 10.1101/2020.10.25.20219063 [DOI] [Google Scholar]
- 13.Wise J. 2020. Covid-19: new coronavirus variant is identified in UK. BMJ 371:m4857. 10.1136/bmj.m4857. [DOI] [PubMed] [Google Scholar]
- 14.Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N, Mlisana K, von Gottberg A, Walaza S, Allam M, Ismail A, Mohale T, Glass AJ, Engelbrecht S, Van Zyl G, Preiser W, Petruccione F, Sigal A, Hardie D, Marais G, Hsiao M, Korsman S, Davies M-A, Tyers L, Mudau I, York D, Maslo C, Goedhals D, Abrahams S, Laguda-Akingba O, Alisoltani-Dehkordi A, Godzik A, Wibmer CK, Sewell BT, Lourenço J, Alcantara LCJ, Pond SLK, Weaver S, Martin D, Lessells RJ, Bhiman JN, Williamson C, de Oliveira T. 2020. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 10.1101/2020.12.21.20248640 [DOI] [Google Scholar]
- 15.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. 2021. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 29:463–476.e6. 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Naveca F, Nascimento V, Souza V, Corad A, Nascimento F, Silva G, Costa Á, Duarte D, Pessoa K, Gonçalves L, Brandão MJ, Jesus M, Fernandes C, Pinto R, Silva M, Mattos T, Wallau GL, Mendonça Siqueira M, Resende PC, Delatorre E, Gräf T, Bello G. 2021. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585.
- 17.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DDS, Mishra S, Crispim MAE, Sales FCS, Hawryluk I, McCrone JT, Hulswit RJG, Franco LAM, Ramundo MS, de Jesus JG, Andrade PS, Coletti TM, Ferreira GM, Silva CAM, Manuli ER, Pereira RHM, Peixoto PS, Kraemer MUG, Gaburo N, Camilo CDC, Hoeltgebaum H, Souza WM, Rocha EC, de Souza LM, de Pinho MC, Araujo LJT, Malta FSV, de Lima AB, Silva JDP, Zauli DAG, Ferreira ACS, Schnekenberg RP, Laydon DJ, Walker PGT, Schlüter HM, dos Santos ALP, Vidal MS, Del Caro VS, Filho RMF, dos Santos HM, Aguiar RS, Proença-Modena JL, Nelson B, Hay JA, Monod M, Miscouridou X, Coupland H, Sonabend R, Vollmer M, Gandy A, et al. 2021. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 372:815–821. 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, Nair MS, Liu L, Iketani S, Luo Y, Guo Y, Wang M, Yu J, Zhang B, Kwong PD, Graham BS, Mascola JR, Chang JY, Yin MT, Sobieszczyk M, Kyratsous CA, Shapiro L, Sheng Z, Huang Y, Ho DD. 2021. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. bioRxiv 10.1101/2021.01.25.428137 [DOI] [PubMed]
- 19.Wang P, Wang M, Yu J, Cerutti G, Nair MS, Huang Y, Kwong PD, Shapiro L, Ho DD. 2021. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. bioRxiv 10.1101/2021.03.01.433466 [DOI] [PMC free article] [PubMed]
- 20.Weinblatt ME, Kremer JM, Bankhurst AD, Bulpitt KJ, Fleischmann RM, Fox RI, Jackson CG, Lange M, Burge DJ. 1999. A trial of etanercept, a recombinant tumor necrosis factor receptor:Fc fusion protein, in patients with rheumatoid arthritis receiving methotrexate. N Engl J Med 340:253–259. 10.1056/NEJM199901283400401. [DOI] [PubMed] [Google Scholar]
- 21.Holash J, Davis S, Papadopoulos N, Croll SD, Ho L, Russell M, Boland P, Leidich R, Hylton D, Burova E, Ioffe E, Huang T, Radziejewski C, Bailey K, Fandl JP, Daly T, Wiegand SJ, Yancopoulos GD, Rudge JS. 2002. VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc Natl Acad Sci U S A 99:11393–11398. 10.1073/pnas.172398299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Genovese MC, Becker J-C, Schiff M, Luggen M, Sherrer Y, Kremer J, Birbara C, Box J, Natarajan K, Nuamah I, Li T, Aranda R, Hagerty DT, Dougados M. 2005. Abatacept for rheumatoid arthritis refractory to tumor necrosis factor α inhibition. N Engl J Med 353:1114–1123. 10.1056/NEJMoa050524. [DOI] [PubMed] [Google Scholar]
- 23.Khan A, Benthin C, Zeno B, Albertson TE, Boyd J, Christie JD, Hall R, Poirier G, Ronco JJ, Tidswell M, Hardes K, Powley WM, Wright TJ, Siederer SK, Fairman DA, Lipson DA, Bayliffe AI, Lazaar AL. 2017. A pilot clinical trial of recombinant human angiotensin-converting enzyme 2 in acute respiratory distress syndrome. Crit Care 21:234. 10.1186/s13054-017-1823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glasgow A, Glasgow J, Limonta D, Solomon P, Lui I, Zhang Y, Nix MA, Rettko NJ, Zha S, Yamin R, Kao K, Rosenberg OS, Ravetch JV, Wiita AP, Leung KK, Lim SA, Zhou XX, Hobman TC, Kortemme T, Wells JA. 2020. Engineered ACE2 receptor traps potently neutralize SARS-CoV-2. Proc Natl Acad Sci U S A 117:28046–28055. 10.1073/pnas.2016093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linsky TW, Vergara R, Codina N, Nelson JW, Walker MJ, Su W, Barnes CO, Hsiang T-Y, Esser-Nobis K, Yu K, Reneer ZB, Hou YJ, Priya T, Mitsumoto M, Pong A, Lau UY, Mason ML, Chen J, Chen A, Berrocal T, Peng H, Clairmont NS, Castellanos J, Lin Y-R, Josephson-Day A, Baric RS, Fuller DH, Walkey CD, Ross TM, Swanson R, Bjorkman PJ, Gale M, Blancas-Mejia LM, Yen H-L, Silva D-A. 2020. De novo design of potent and resilient hACE2 decoys to neutralize SARS-CoV-2. Science 370:1208–1214. 10.1126/science.abe0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chan KK, Dorosky D, Sharma P, Abbasi SA, Dye JM, Kranz DM, Herbert AS, Procko E. 2020. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369:1261–1265. 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlothauer T, Herter S, Koller CF, Grau-Richards S, Steinhart V, Spick C, Kubbies M, Klein C, Umaña P, Mössner E. 2016. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng Des Sel 29:457–466. 10.1093/protein/gzw040. [DOI] [PubMed] [Google Scholar]
- 28.Manson JJ, Crooks C, Naja M, Ledlie A, Goulden B, Liddle T, Khan E, Mehta P, Martin-Gutierrez L, Waddington KE, Robinson GA, Ribeiro Santos L, McLoughlin E, Snell A, Adeney C, Schim van der Loeff I, Baker KF, Duncan CJA, Hanrath AT, Lendrem BC, De Soyza A, Peng J, J'Bari H, Greenwood M, Hawkins E, Peckham H, Marks M, Rampling T, Luintel A, Williams B, Brown M, Singer M, West J, Jury EC, Collin M, Tattersall RS. 2020. COVID-19-associated hyperinflammation and escalation of patient care: a retrospective longitudinal cohort study. Lancet Rheumatol 2:e594–e602. 10.1016/S2665-9913(20)30275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hezareh M, Hessell AJ, Jensen RC, van de Winkel JGJ, Parren PWHI. 2001. Effector function activities of a panel of mutants of a broadly neutralizing antibody against human immunodeficiency virus type 1. J Virol 75:12161–12168. 10.1128/JVI.75.24.12161-12168.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lo M, Kim HS, Tong RK, Bainbridge TW, Vernes J-M, Zhang Y, Lin YL, Chung S, Dennis MS, Zuchero YJY, Watts RJ, Couch JA, Meng YG, Atwal JK, Brezski RJ, Spiess C, Ernst JA. 2017. Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J Biol Chem 292:3900–3908. 10.1074/jbc.M116.767749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones BE, Brown-Augsburger PL, Corbett KS, Westendorf K, Davies J, Cujec TP, Wiethoff CM, Blackbourne JL, Heinz BA, Foster D, Higgs RE, Balasubramaniam D, Wang L, Bidshahri R, Kraft L, Hwang Y, Žentelis S, Jepson KR, Goya R, Smith MA, Collins DW, Hinshaw SJ, Tycho SA, Pellacani D, Xiang P, Muthuraman K, Sobhanifar S, Piper MH, Triana FJ, Hendle J, Pustilnik A, Adams AC, Berens SJ, Baric RS, Martinez DR, Cross RW, Geisbert TW, Borisevich V, Abiona O, Belli HM, de Vries M, Mohamed A, Dittmann M, Samanovic M, Mulligan MJ, Goldsmith JA, Hsieh C-L, Johnson NV, Wrapp D, McLellan JS, et al. 2020. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv 10.1101/2020.09.30.318972 [DOI]
- 32.Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K, Chung KM, Hermann A, Ullman E, Cruz J, Rafique A, Huang T, Fairhurst J, Libertiny C, Malbec M, Lee W-Y, Welsh R, Farr G, Pennington S, Deshpande D, Cheng J, Watty A, Bouffard P, Babb R, Levenkova N, Chen C, Zhang B, Romero Hernandez A, Saotome K, Zhou Y, Franklin M, Sivapalasingam S, Lye DC, Weston S, Logue J, Haupt R, Frieman M, Chen G, Olson W, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. 2020. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369:1010–1014. 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imai M, Iwatsuki-Horimoto K, Hatta M, Loeber S, Halfmann PJ, Nakajima N, Watanabe T, Ujie M, Takahashi K, Ito M, Yamada S, Fan S, Chiba S, Kuroda M, Guan L, Takada K, Armbrust T, Balogh A, Furusawa Y, Okuda M, Ueki H, Yasuhara A, Sakai-Tagawa Y, Lopes TJS, Kiso M, Yamayoshi S, Kinoshita N, Ohmagari N, Hattori S, Takeda M, Mitsuya H, Krammer F, Suzuki T, Kawaoka Y. 2020. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc Natl Acad Sci U S A 117:16587–16595. 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek Y, Singh N, Arunkumar A, Zydney AL. 2017. Effects of histidine and sucrose on the biophysical properties of a monoclonal antibody. Pharm Res 34:629–639. 10.1007/s11095-016-2092-0. [DOI] [PubMed] [Google Scholar]
- 35.Warne NW. 2011. Development of high concentration protein biopharmaceuticals: the use of platform approaches in formulation development. Eur J Pharm Biopharm 78:208–212. 10.1016/j.ejpb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Mattila J, Clark M, Liu S, Pieracci J, Gervais TR, Wilson E, Galperina O, Li X, Roush D, Zoeller K, Brough H, Simpson-Platre C. 2016. Retrospective evaluation of low-pH viral inactivation and viral filtration data from a multiple company collaboration. PDA J Pharm Sci Technol 70:293–299. 10.5731/pdajpst.2016.006478. [DOI] [PubMed] [Google Scholar]
- 37.Cohen MS. 2021. Monoclonal antibodies to disrupt progression of early Covid-19 infection. N Engl J Med 384:289–291. 10.1056/NEJMe2034495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Starr TN, Greaney AJ, Addetia A, Hannon WW, Choudhary MC, Dingens AS, Li JZ, Bloom JD. 2021. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science 371:850–854. 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zoufaly A, Poglitsch M, Aberle JH, Hoepler W, Seitz T, Traugott M, Grieb A, Pawelka E, Laferl H, Wenisch C, Neuhold S, Haider D, Stiasny K, Bergthaler A, Puchhammer-Stoeckl E, Mirazimi A, Montserrat N, Zhang H, Slutsky AS, Penninger JM. 2020. Human recombinant soluble ACE2 in severe COVID-19. Lancet Respir Med 8:1154–1158. 10.1016/S2213-2600(20)30418-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moore MJ, Dorfman T, Li W, Wong SK, Li Y, Kuhn JH, Coderre J, Vasilieva N, Han Z, Greenough TC, Farzan M, Choe H. 2004. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J Virol 78:10628–10635. 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei C, Fu W, Qian K, Li T, Zhang S, Ding M, Hu S. 2020. Potent neutralization of 2019 novel coronavirus by recombinant ACE2-Ig. bioRxiv 10.1101/2020.02.01.929976 [DOI] [PMC free article] [PubMed]
- 42.Miller A, Leach A, Thomas J, McAndrew C, Bentley E, Mattiuzzo G, John L, Mirazimi A, Harris G, Gamage NN, Carr S, Ali H, Montfort RV, Rabbitts T. 2021. A super-potent tetramerized ACE2 protein displays enhanced neutralization of SARS-CoV-2 virus infection. Sci Rep 11:10617. 10.1038/s41598-021-89957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwanaga N, Cooper L, Rong L, Beddingfield B, Crabtree J, Tripp RA, Qin X, Kolls JK. 2020. Novel ACE2-IgG1 fusions with improved in vitro and in vivo activity against SARS-CoV2. bioRxiv 10.1101/2020.06.15.152157 [DOI] [PMC free article] [PubMed]
- 44.Jaume M, Yip MS, Cheung CY, Leung HL, Li PH, Kien F, Dutry I, Callendret B, Escriou N, Altmeyer R, Nal B, Daeron M, Bruzzone R, Peiris JSM. 2011. Anti-severe acute respiratory syndrome coronavirus spike antibodies trigger infection of human immune cells via a pH- and cysteine protease-independent Fc R pathway. J Virol 85:10582–10597. 10.1128/JVI.00671-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang S-F, Tseng S-P, Yen C-H, Yang J-Y, Tsao C-H, Shen C-W, Chen K-H, Liu F-T, Liu W-T, Chen Y-MA, Huang JC. 2014. Antibody-dependent SARS coronavirus infection is mediated by antibodies against spike proteins. Biochem Biophys Res Commun 451:208–214. 10.1016/j.bbrc.2014.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods 329:112–124. 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Büeler H, Mulligan RC. 1996. Induction of antigen-specific tumor immunity by genetic and cellular vaccines against MAGE: enhanced tumor protection by coexpression of granulocyte-macrophage colony-stimulating factor and B7-1. Mol Med 2:545–555. 10.1007/BF03401639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 25. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, Niemeyer D, Jones TC, Vollmar P, Rothe C, Hoelscher M, Bleicker T, Brünink S, Schneider J, Ehmann R, Zwirglmaier K, Drosten C, Wendtner C. 2020. Virological assessment of hospitalized patients with COVID-2019. Nature 581:465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1<br>. Download JVI.00685-21-s0001.xlsx, XLSX file, 0.08 MB (77.1KB, xlsx)