Abstract
With the increased appreciation for the significance of noncoding RNAs (ncRNAs), the present research aimed to determine the role of competing endogenous RNA (ceRNA) in the process of particulate matter (PM) exposure-induced pulmonary damage. Alterations in messenger RNA (RNA), microRNA and long non-coding RNA (lncRNA) profiles of human bronchial epithelial (HBE) cells treated with PM were analyzed by microarray assays. Next, we identified that lncRNA taurine upregulated gene 1 (TUG1) acted as a competing endogenous RNA for microRNA-222–3p (miR-222–3p) and subsequently attenuated the inhibitory effect of miR-222–3p on CUGBP elav-like family member 1 (CELF1). The binding potency among ceRNAs was verified by RNA immunoprecipitation (RIP) assay and dual-luciferase reporter assay. Knockdown of TUG1 attenuated HBE cell apoptosis and cell cycle arrest by downregulation of CELF1 and protein 53 (p53). Further, we confirmed that Tug1/mir-222–3p/CELF1/p53 network aggravated PM-induced airway hyper-reactivity (AHR) in mice. In summary, our novel findings revealed that TUG1 triggered dysfunction of pulmonary cells followed by PM exposure by serving as a sponge for miR-222–3p and thereby upregulating the expression of CELF1and p53.
Keywords: PM, TUG1, CeRNAs, Apoptosis, Airway hyper-reactivity
1. Introduction
Exposure to particulate matter (PM) has become a global threat to public health. In 2017, almost 3 million deaths come as a result of ambient PM pollution directly (Collaborators, 2018). PM readily enters the respiratory pathways (Yang et al., 2020), causing tissue inflammation, oxidative stress, fibrosis and even tumors (Schraufnagel et al., 2019). Since ambient PM was classified as first-class carcinogen, the benefit of reduction in PM emission has been broadly recognized (Bai et al., 2020; Mannucci et al., 2019; Tshala-Katumbay et al., 2015); however, reduction of ambient particle concentration to a harmless level remains a long-term undertaking. Therefore, deciphering the mechanism of PM-induced damage, identifying potential biomarkers and effective treatment strategies are pressing and timely issues.
Airway hyper-reactivity (AHR) is an anomalous condition with excessive airway contraction when the airways respond to stimuli (Caraher et al., 2017; Hao et al., 2003). Increased AHR predicts the development of respiratory symptoms in adults and serves as a major feature of asthma and chronic obstructive pulmonary disease (COPD) that correlates with disease severity (Brutsche et al., 2006). It is well-known that multiple inflammatory cells, cytokines, and injuries to airway epithelia lead to AHR when airway is under stimulation (Busse, 2010). Although airway inflammation is one of the most important mechanisms in causing AHR, the contribution of apoptosis in epithelial cells should not be ignored. Several studies have indicated that TIM-1-expressing natural killer T (NKT) cells responded to apoptotic airway epithelial cells by secreting cytokines, resulting in AHR (Kim et al., 2013; Lee et al., 2010). However, the molecular network of PM-induced epithelial injury remains unclear.
With the development of deep sequencing, nearly 90% of the noncoding sequences of the mammalian genome have been identified (Cech and Steitz, 2014). The majority of these sequences transcribe into noncoding RNAs (ncRNAs), including miRNAs and lncRNAs, which have been certified as vital regulatory factors in a plethora of biological processes. MiRNAs, short noncoding RNA with the length of 18–25 nt, have the ability to regulate the expression of target genes by degrading mRNA or blocking mRNA translation (Cheng et al., 2015). Furthermore, lncRNAs, a class of ncRNAs with much longer length than miRNAs, act as vital multi-dimensional modifiers in the process of translation, post-transcription, and epigenetic modulation (Gao et al., 2020). LncRNAs can competitively sponge target miRNAs and abolish their inhibition of target genes (Ergun and Oztuzcu, 2015). However, the biological function of ceRNAs in response to PM-induced AHR has yet to be reported.
In the present study, we identified a critical ceRNA network and explored its function in PM-induced epithelial injury. The characterization of this network affords the development of new therapeutic strategies for AHR induced by PM exposure.
2. Methods
2.1. Particulate matter (PM)
Standard reference particular matter (SRM 1648a) acquiring from the National Institute of Standards and Technology (NIST, USA) was used in this study. The stock solution of PM was prepared by phosphate buffered saline (PBS) at a concentration of 100 mg/mL. Prior to application, the stock solutions were subjected to ultrasonication (Sonics, USA) for 30 min and immediately diluted with PBS to working concentrations.
2.2. Cell culture
Human bronchial epithelial cell (HBE) was purchased from American Type Culture Collection (ATCC, USA). Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and 1% penicillin (100 U/mL)/streptomycin (100 μg/mL) (PS; Enpromise, China) at 37 °C in 5% carbon dioxide.
Previous studies have suggested phenotype and ncRNA profile alterations in HBE cells in response to PM exposure (100 or 500 μg/mL) for 24 h (Wang et al., 2019; Zhang et al., 2020). Therefore, for the microarray assays, HBE cells were seeded in six-wells plates and treated with equal volume DMEM with 0, 100 or 500 μg/mL PM, which were respectively defined as control, PM (low dose) and PM (high dose) groups for microarray assays. Next, to validate the expression levels of target molecules, HBE cells were treated for 24 h with PM at doses ranging from 0 to 500 μg/mL (including 0, 25, 50, 100, 200 and 500 μg/mL). Three biological replicates were performed.
2.3. Microarray assays
HBE cells treated with 0, 100 and 500 μg/mL PM for 24 h were used for the mRNAs, miRNAs and lncRNAs microarray assays.
For microRNAs, after miRNA labeling and array hybridization, the slides were scanned utilizing the Axon GenePix 4000B microarray scanner (Axon Instruments, CA). Then, scanned images were analyzed by GenePix Pro 6.0 software (Axon Instruments, CA) to extract raw expression data. Duplicated miRNAs were averaged, and those miRNAs with intensities lesser than 30 in all samples were omitted. Normalization was performed in the chosen expression data for consequent analysis.
For the expression of mRNA and lncRNA, microarray was performed using the Arraystar Human lncRNA Array (8 × 15 K, Arraystar) under a contract service with Shanghai Kangchen Technology. Normalization and subsequent processing of raw data were conducted by R packages.
Significant differentially expressed mRNAs, miRNAs or lncRNAs between control and exposure group were determined with the screening criteria (Fold Change ≥ 1.5, P < 0.05). Volcano plots, network diagram or heat maps were depicted using R software (version 3.6.1).
2.4. Construction of ceRNA network and enrichment analysis
Next, the differentially expressed lncRNAs were submitted to online databases ENCORI (http://starbase.sysu.edu.cn/ceRNA.php?source=lncRNA) and lncRNASNP2 (http://bioinfo.life.hust.edu.cn/lncRNASNP/), to identify the hub lncRNAs (hub DE-lncRNAs) potentially binding to miRNAs. Identical miRNAs between target miRNAs and differentially expressed miRNAs (DE-miRNAs) was regarded as hub DE-miRNAs. Next, to predict the molecular interactions between hub DE-miRNAs and differentially expressed mRNAs (DE-mRNAs), the potential binding sites were predicted with the online databases miRWalk (http://mirwalk.umm.uni-heidelberg.de/), TargetScan (http://www.targetscan.org/) and ENCORI (http://starbase.sysu.edu.cn/agoClipRNA.php?source=mRNA). Finally, the predicted consequences were integrated and depicted with Sankey diagram by R software.
2.5. Nuclear, cytoplasmic and total RNA extraction and quantitative real-time PCR (qRT-PCR) analysis
Total RNA of HBE cells and murine lungs was extracted with RNA isolated reagent (Invitrogen, USA) followed by the manufacturer’s instructions. Cytoplasmic and nuclear RNA of HBE cells were separated and purified with the PARIS Kit (Life Technologies, USA) according to the manufacturer’s procedures. Extracted RNA (1000 ng) was utilized to synthesize cDNA with a One Step Prime-Script® miRNA cDNA Synthesis Kit (Takara, Japan) or a One Step Prime-Script® lncRNA cDNA Synthesis Kit (Takara, Japan), respectively. Next, qRT-PCR analysis was performed by applying the SYBR Premix Ex Taq kit (Takara, Japan) on the Quant Studio 6 Flex system (Applied Biosystems, USA). β-actin was used as an internal control for total RNAs and the relative expressions of RNAs were calculated with the 2−ΔΔCt method. And β-actin was also used as a cytoplasmic control, while U6 was treated as a nuclear control when the ratio of cytoplasm to nucleus in the expression level of RNAs was calculated. All of the experiments were performed independently in triplicates. Primer sequences for mRNAs and lncRNAs are listed in Table S4. Bulge-loopTM miRNA qRT-PCR Primer Sets specific for miRNAs were designed and synthesized by RiboBio (Guangzhou, China).
2.6. Antisense oligonucleotides (ASO), short interfering RNA (siRNA), microRNA mimics, and plasmid construction and cell transfection
To knockdown the expression of taurine upregulated gene 1(TUG1) and CUGBP elav-Like family member 1(CELF1), TUG1- or Tug1- targeting ASO (ASO-TUG1, ASO-mmu-Tug1), CELF1-targeting siRNA (si-CELF1) and scrambled negative control (NC) sequences (ASO-NC, ASO-mmu-NC, si-NC) were designed and synthesized by RiboBio (Guangzhou, China). ASO-mmu-Tug1 and ASO-mmu-NC were specially designed for murine experiments in vivo. HBE cells were seeded in six-well plates and transfected with ASOs or siRNAs utilizing Lipofectamine 3000 Transfection Reagent (Invitrogen, USA) combined with Opti-MEM (Gibco, USA) in line with the manufacturer’s protocol.
Mimic-NC and microRNA-222–3p (miR-222–3p) specific mimics (mimic-miR-222–3p) were synthesized from RiboBio (Guangzhou, China). Prior to transfection, the cells were seeded on six-well plates and cultured for 24 h. Then, the cells were transiently transfected with the corresponding oligonucleotides utilizing Lipofectamine 3000.
Twenty-four hours after transfection, RNA was collected and qRT-PCR was performed. All experiments were carried out independently in triplicates. The ASO or siRNA with the highest knockdown efficiency among three candidates was selected for the further research. The sequences of ASO and siRNA are listed in Table S5.
2.7. Western blotting assay
Total proteins were extracted by a total protein extraction kit (Life Technologies, USA) from cells. A total of 20 μg of protein was subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred onto polyvinylidene difluoride (PVDF) membranes, and blocked with 5% skim milk for 1 h. Blots were then incubated with primary antibodies against CELF1 (1: 1000 dilution; Abcam, USA), p53 (1: 1000 dilution; Cell Signaling Technology, USA) or IL13 (1: 1000 dilution; Bioss, China) for 48 h, follow by incubation with HRP-conjugated secondary antibodies for 1 h. GAPDH (1:1000 dilution; Cell Signaling Technology, USA) and β-actin (1:5000 dilution; GenStar, China) were used as a control. The immunoreactive signals were visualized with a Super ECL Plus Kit (Yeasen, China).
2.8. Flowcytometry assay
After transfection and treatment with PM, HBE cells were washed twice with cold PBS. Binding buffer (1×) was added to resuspend cells at a concentration of 106 cells/mL. Then 5 μl FITC Annexin V and 5 μl propidium iodide (PI) were added into 100 μl cells solutions. After incubated for 15 min at 25 °C in the dark, samples were detected on the flow cytometer (BD FACSCalibur Flow Cytometer, USA) at 488 nm.
To detect alterations in cell cycle, HBE cells were fixed with pre-cooled 70% ethanol for 24 h at a concentration of 1 × 105 cells/mL after washed three times with PBS. Then, HBE cells were collected and treated with 100 μl RNase A in 37 °C for 30 min. Samples were incubated with PI at 37 °C in the dark and the cell cycle was evaluated by flow cytometry. All assays were independently performed independently in triplicates.
2.9. Luciferase reporter assays
The overexpression plasmid of miR-222–3p and the control plasmid were obtained from Shanghai Genechem (China). The predicted binding sites of miR-222–3p with wild type CELF1-3′-UTR (WT) were replaced by ACTCGTCCTA for mutagenesis (MUT). The sequences of Renilla luciferase (Rluc) and firefly luciferase (Luc) were constructed in plasmid vectors for reporter fluorescence (Rluc) and internal reference (Luc), respectively. Next, well-constructed reporter plasmids were cotransfected into HBE cells, and the luciferase activity was assessed with a dual-luciferase reporter assay system (Promega, USA). Experiments were conducted independently in triplicates.
2.10. RNA immunoprecipitation (RIP) and MS2-GFP-RIP system
To clarify whether TUG1 interacts the miRNA mediated RNA-induced silencing complex, RIP assay was performed. After washed in PBS, HBE cells were lysed and harvested by AGO2 antibody (5 μg/sample; Abcam, USA) at 4 °C for 16 h, with IgG (5 μg/sample; Cell Signaling Technology, USA) served as a negative control. The coprecipitated RNAs were extracted from the magnetic bead-antibody compounds and assessed by qRT-PCR with specific primers.
In order to define the endogenous binding between TUG1 and miRNAs, full-length TUG1 was synthesized and ligated into the overexpression plasmid pcDNA-12×MS2 (named as pMS2, ORIGENE, USA), named as pMS2-TUG1. Besides, pcDNA-MS2/GFP encoding the MS2-GFP fusion protein which can combine with the MS2 stem ring structure was constructed. Then, 5 μg of pcDNA-MS2/GFP and 5 μg of pMS2-TUG1 plasmid were cotransfected into the corresponding HBE cells. Next, pMS2 was used as the negative control. After incubation for 48 h, cells were lysed and harvested with GFP antibody (5 μg/sample; Abcam, USA). Afterwards, followed by the RIP procedure as mentioned above, the levels of combined RNAs were analyzed. The sequences of primers are provided in Table S4.
Besides, miR-222–3p-Cy3 was also designed and cotransfected with pMS2-TUG1 and pcDNA-MS2/GFP into HBE cells. The ratio of plasmid to siRNA was 1 μg: 30 pmol. Forty-eight h after transfection, the fluorescence signals were measured with a fluorescent microscope (Zeiss, Germany).
2.11. In vivo model
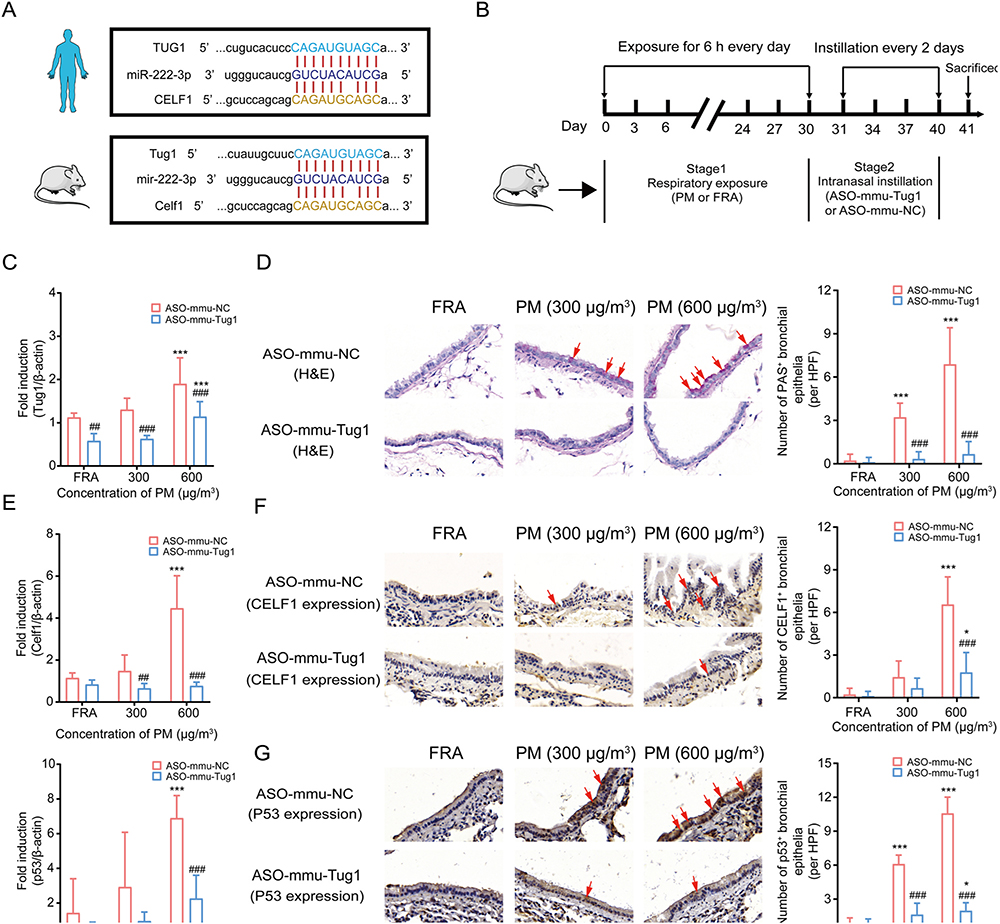
Altogether, 30 male wild type (WT) C57BL/6 mice (20–22 g) purchased from Model Animal Research Center of Nanjing University, China were included in animal models. All the mice were treated humanely in a setting with 12/12 h light/dark light cycles, 24 °C ± 2 °C room temperature with 40–70% relative humidity. Ethical approval of experimental protocols was obtained from Committee on Animal Use and Care of Southeast University. Animal exposure to PM was performed by a dynamic inhalation exposure chamber outfitted with an aerosol generator (Beijing HuiRongHe Technology Co. Ltd., China). Then, 300 μg/m3 and 600 μg/m3 were set as low PM and high PM exposure dose, respectively, according to a previous study (Fu et al., 2019). Briefly, mice were randomly divided into 6 groups (n = 5) and treated in two stages as follows: mice exposed to filtered room air FRA + ASO-mmu-NC (50 nmol/kg), FRA + ASO-mmu-Tug1 (50 nmol/kg); mice exposed to PM (300 μg/m3) + ASO-mmu-NC (50 nmol/kg), PM (300 μg/m3) + ASO-mmu-Tug1 (50 nmol/kg); mice exposed to PM (600 μg/m3) + ASO-mmu-NC (50 nmol/kg), PM (600 μg/m3) + ASO-mmu-Tug1 (50 nmol/kg). For stage one, mice in each group were exposed in whole body dynamic inhalation exposure system for 2 h per day for 30 consecutive days. For stage two, mice were treated with ASO (50 nmol/kg) every 2 days for 10 days by intranasal administration. All mice were sacrificed 24 h after the last treatment.
2.12. Histopathological and immunohistochemistry (IHC) pathological analysis
Half of left murine lung tissues were fixed in 4% paraformaldehyde (PFA) and embedded with paraffin. Paraffin-embedded tissues were serially sectioned for 4-μm thick. Hematoxylin-eosin staining (HE) was performed followed by the protocols as mentioned in the previous study (Li et al., 2020). The number of periodic acid Schiff reaction positive (PAS+) bronchial epithelia were counted in 3 non overlapping HPFs (400× magnification) in the bronchial airway of each section. The HPFs with most positive cells were selected. In each field, only positive cells with the nucleus at the focal plane were counted. The results are presented as arithmetic mean ± standard deviation (SD) of PAS+ cells per HPF. n = 9/group (3 HPF/section × 1 section/mouse × 3 mice/group).
For IHC staining, series of treatments were performed to dewax, rehydrate, antigen retrieval and block the activity of endogenous peroxidase of the tissue. Then the sections were treated with rabbit monoclonal antibody against CELF1 (1:100) (Abcam, USA) and mouse monoclonal antibody against p53 (1:100) (Cell Signaling Technology, USA) at 4 °C overnight, followed by incubation with the biotinylated secondary antibody for 30 min. Negative controls were conducted without primary antibodies. Finally, after stained using peroxidase substrate DAB kit, dehydrated, cleared and sealed with hematoxylin, each section was analyzed under light microscopy by two histologists who are blinded to the experiment design and representative images were photographed. As mentioned above, the number of CELF1 positive (CELF1+) or p53 positive (p53+) bronchial epithelia were counted in 3 non overlapping HPFs (400× magnification) in the bronchial airway of each section. n = 9/group (3 HPF/section × 1 section/mouse × 3 mice/group).
2.13. Immunofluorescence (IF) staining
Paraffin-embedded murine lung tissues were serially sectioned for 4-μm thick. Tissue section were permeabilized in 0.1% Triton X-100 for 10 min and blocked with 5% skim milk for 1 h. After washed, rabbit anti-IL13 (1:100) was stained overnight at 4 °C, followed by 2 μg/mL of goat anti-rabbit IgG secondary antibody conjugated with Alexa Flour 488, and the counter-stained with DAPI. The fluorescence signals were measured with a fluorescent microscope (Zeiss, Germany).
2.14. Statistical analysis
Values are presented as mean ± SD unless indicated otherwise. Statistical analysis was performed using SPSS 20.0 software, and statistical significance was selected at P < 0.05. Unpaired student’s t-test or paired student’s t-test were used for comparison of statistical differences between two groups appropriately; one-way or two-way ANOVA followed by post hoc Bonferroni test was performed to analyze statistical differences between multiple groups. In correlation analyses, Spearman correlation was performed with R packages and a regression line is indicated.
3. Results
3.1. Differentially expressed RNAs in HBE cells followed by PM exposure
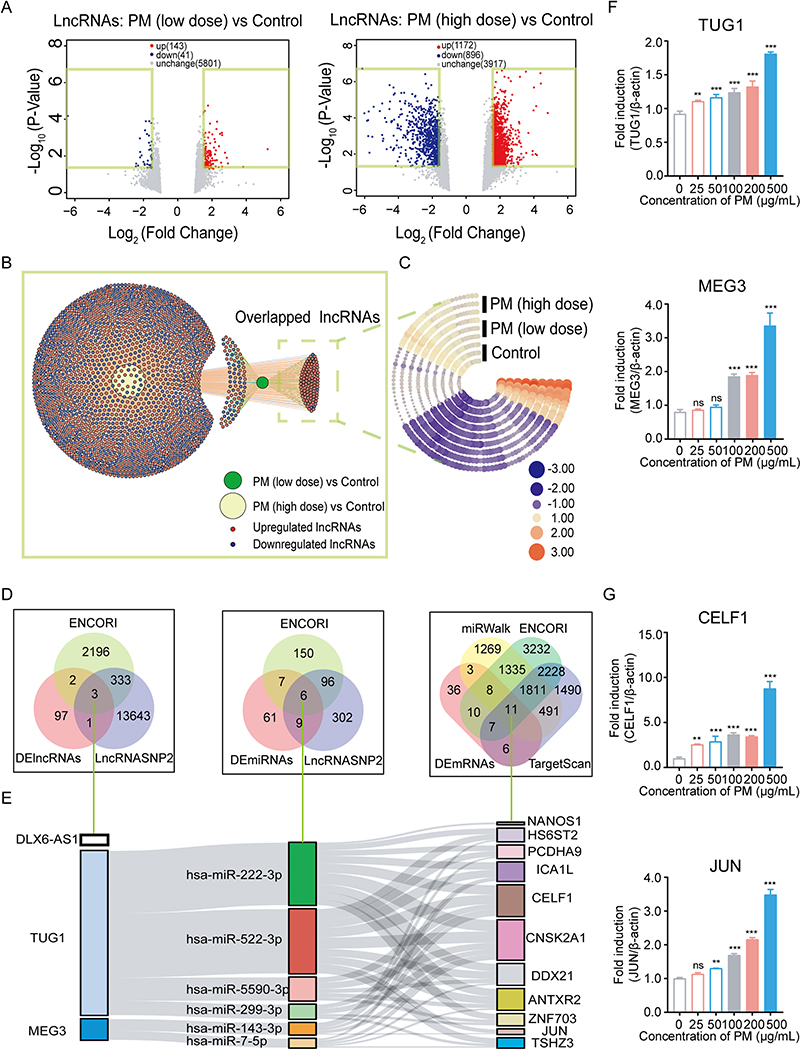
To determine whether ncRNAs act a vital part in PM-induced lesion and identify hub ncRNAs, microarray analysis was conducted with HBE cells exposed to PM. After trimming the raw data, expression profiles of lncRNAs, miRNAs and mRNAs were presented. A total of 143 and 1, 172 lncRNAs were significantly upregulated; 41 and 896 lncRNAs were significantly downregulated in the low- and high-dose PM treated HBE cells, respectively (Fig. 1A). The overlapped significantly altered lncRNAs between low- and high-dose PM exposure groups are shown in Fig. 1B. These overlapped lnRNAs were defined as differentially expressed RNAs (DE-RNAs) induced by PM and depicted as a heat map (Fig. 1C, Table S1).
Fig. 1.
Differentially expressed RNAs in HBE cells followed by PM exposure. (A) The volcano plots revealed differentially expressed lncRNAs in HBE cells between the PM-exposed group and control, with a cut-off as fold change ≥ 1.5 and P-value < 0.05. (B) Network diagram showing the overlapped differentially expressed lncRNAs between PM (low dose) and PM (high dose) groups. Each red or blue point represents a single upregulated or downregulated lncRNA, respectively. (C) Heatmap showing expression levels of overlapped lncRNAs in HBE cells. Each arc represents a cellular sample, each column represents a lncRNA. The color code represents expression levels: red or blue color represent higher or lower expression levels relative to the control, respectively. (D-E) Venn diagram displaying a total of 3 lncRNAs, 6 miRNAs, 11 mRNAs, which were overlapped between differentially expressed RNAs and bioinformatics database. Sankey diagram depicting the interaction between the overlapped differentially expressed RNAs. (F-G) Quantitative RT-PCR analysis of the expression levels of TUG1, MEG3, CELF1 and JUN in HBE cells treated with PM. ** P < 0.01, *** P < 0.001, compared with control.
Notably, 2 and 110 miRNAs were significantly upregulated; 95 and 262 miRNAs were significantly downregulated in the low- and high-dose PM-treated HBE cells, respectively (Fig. S1A). The overlapped miRNAs between the low- and high-dose PM groups are shown in a Venn diagram and heatmap (Fig. S1B and C, Table S2). The significantly altered mRNAs are shown in Fig. S1D–F and Table S3.
Next, to explore the network among the DE-lncRNAs, DE-miRNAs and DE-mRNAs, we identified 3 hub DE-lncRNAs, with the requisite sequence to bind to miRNAs, based on the prediction in ENCORI and lncRNASNP2 databases (Fig. 1D). A total of 3 hub DE-lncRNAs were identified, namely lncRNA DLX6 antisense RNA 1 (DLX6-AS1), maternally expressed 3 (MEG3), and taurine upregulated gene 1 (TUG1).
Next, 6 hub DE-miRNAs were identified after screening the potential binding sites to the 3 hub DE-lncRNAs according to ENCORI and lncRNASNP2 databases. However, there was no predicted miRNAs targeting to DLX6-AS1 in our DE-miRNAs set. Further, 11 hub DE-mRNAs, which can be integrated by 6 hub DE-miRNAs were identified based on DEmRNAs, TargetScan, miRWalk and ENCORI database (Fig. 1D). Ultimately, a lncRNAs-miRNAs-mRNAs ceRNA network in reference to PM-induced DE-RNAs was constructed as presented in Fig. 1E.
qRT-PCR assay was used to validate the expression levels of RNAs in the ceRNA network. TUG1, but not MEG3, were significantly increased in a dose-dependent manner in PM-treated HBE cells (Fig. 1F). At the mRNA level, only CELF1 and JUN showed an increase in a dose-dependent manner, but not the rest 9 hub DE-mRNAs (Fig. 1G and Fig. S2A). Compared to JUN, expression level of CELF1 was significantly increased at a very low dose (25 μg/mL) PM exposure, suggesting that CELF1 was more sensitive to PM exposure than JUN. Notably, a dose-response relationship between PM and inhibitory effects on HBE cells was demonstrated recently, with a dose of 64 μg/mL PM sufficiently potent to cause apoptosis (Zhou et al., 2014). Based on this, 50 and 100 μg/mL PM were chosen as low and high exposure level, respectively, for further experiments. At the protein level, CELF1 was also upregulated with both low- and high-dose PM exposure, compared to the control group (Fig. S2B).
3.2. PM exposure induced apoptosis, cell cycle arrest and activation of p53 in HBE cells
Compared to the control, PM exposure groups showed a significantly higher ratio of apoptotic cells (Fig. S3A). Cell cycle was also measured with a flow cytometry assay. Relative to control, low-dose PM exposure led to significant G1 arrest; and high dose PM exposure led to significant G1 and S arrests in HBE cells (Fig. S3B).
Compared to the control, p53, a critical protein in mediating apoptosis and cell cycle, was significantly induced in PM-treated HBE cells at both the mRNA and protein levels (Fig. S3C and D). These results suggested phenotypic alterations in HBE cells followed by PM exposure; however, their association with the ceRNA network remains to be characterized.
3.3. TUG1 regulated CELF1 by acting as a ceRNA sponge for miR-222–3p
To confirm the interaction between TUG1 and CELF1 or JUN, we transfected ASO-TUG1 in HBE cells followed by PM exposure. ASO-TUG1 #3 displaying the highest knockdown efficiency was chosen for subsequent experiments (Fig. S4). As shown in Fig. 2A, TUG1 knockdown significantly decreased the expression level of CELF1 while increasing JUN expression in HBE cells; thus, CELF1 was deemed a potential target mRNA in the ceRNA network. Next, compared with the ASO-NC group, transfection of ASO-TUG1 significantly decreased CELF1 expression at both mRNA and protein levels in control, 50 and 100 μg/mL PM-treated HBE cells (Fig. 2B and C). These results demonstrated that TUG1 was an up-stream molecule that positively regulated CELF1 expression in HBE cells.
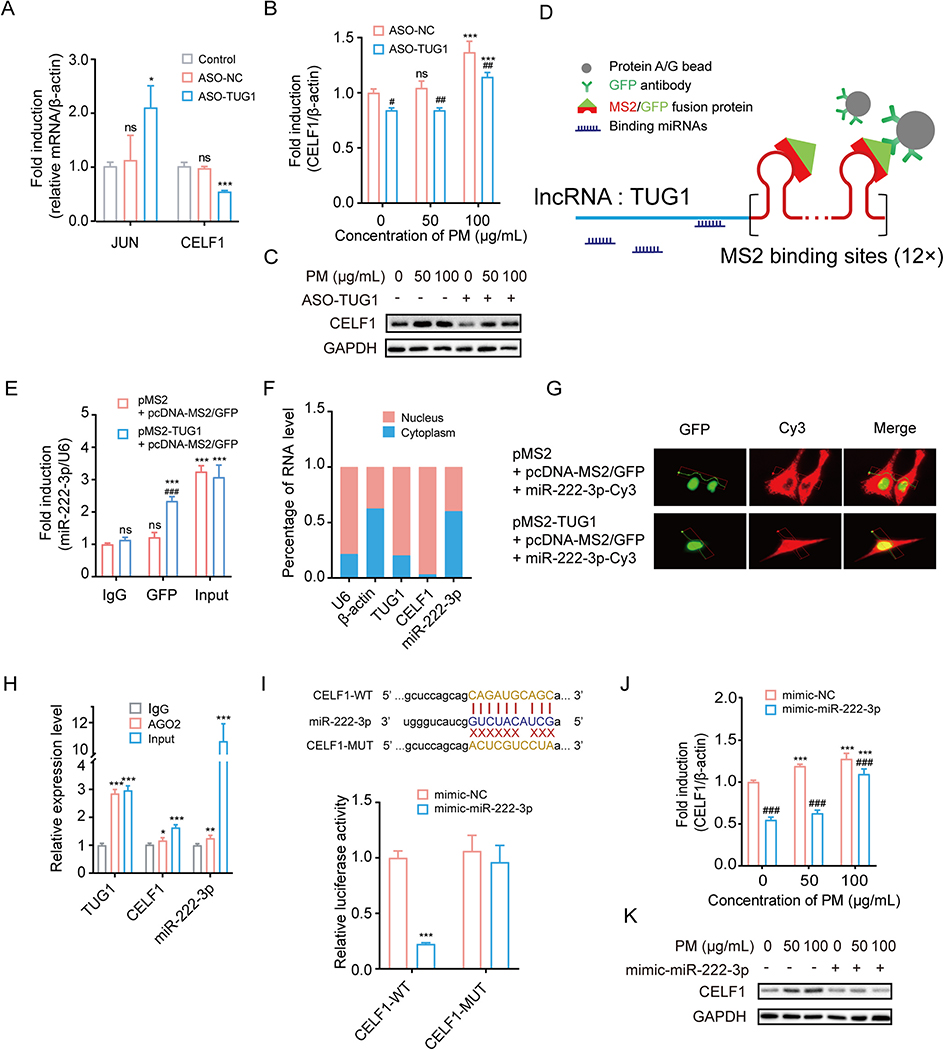
Fig. 2.
TUG1 regulates CELF1 by acting as a ceRNA sponge for miR-222–3p. (A) mRNA expression levels of JUN and CELF1 in HBE cells transfected with ASO-NC or ASO-TUG1. * P < 0.05, *** P < 0.001, compared with control. (B-C) mRNA and protein levels of CELF1 in HBE cells treated with ASO-TUG1 after exposed to PM (0, 50 or 100 μg/mL). *** P < 0.001, compared with control; # P < 0.05, ## P < 0.01, ### P < 0.001, compared with ASO-NC within each group. (D) Schematic diagram of MS2-GFP-RIP system. (E) miR-222–3p levels bound to GFP in HBE cells. *** P < 0.001, compared with IgG; ### P < 0.001, compared with control within each group. (F) Subcellular localization of TUG1, CELF1 and miR-222–3p in HBE cells. (G) Representative images of HBE cells with co-transfection of pMS2-TUG1, pcDNA-MS2/GFP and miR-222–3p-Cy3 or pMS2, pcDNA-MS2/GFP and miR-222–3p-Cy3. Green, GFP; Red, Cy3. (H) TUG1, CELF1 and miR-222–3p levels bound to AGO2 in HBE cells. * P < 0.05; ** P < 0.01; *** P < 0.001, compared with IgG. (I) The predicted binding sites of miR-222–3p on 3′ UTR of CELF1. Luciferase reporter assays validated the binding of miR-222–3p to CELF1. *** P < 0.001, compared with mimic-NC. (J-K) mRNA and protein levels of CELF1 in HBE cells treated with mimic-miR-222–3p after exposed to PM (0, 50 or 100 μg/mL). *** P < 0.001, compared with control; ### P < 0.001, compared with mimic-NC within each group.
To further explore the ceRNA network, we performed MS2-GFP-RIP assay to verify the endogenous binding capacity of TUG1 with miRNAs (Fig. 2D). The results showed that only miR-222–3p was pulled-down and enriched by the GFP antibody (Fig. 2E). miR-299–3p, miR-522–3p and miR-5590–3p were not enriched by the GFP antibody (Fig. S5).
Furthermore, the TUG1 was primarily located in nucleus, while miR-222–3p was predominantly located in the cytoplasm of HBE cells (Fig. 2F). To clarify the potential binding between TUG1 and miR-222–3p, HBE cells were cotransfected with pMS2-TUG1, pcDNA-MS2/GFP and miR-222–3p-Cy3. Of note, HBE cells were set as control group (HBE cells were cotransfected with pMS2, pcDNA-MS2/GFP and miR-222–3p-Cy3) and TUG1 overexpression (OEX) group (HBE cells were cotransfected with pMS2-TUG1, pcDNA-MS2/GFP and miR-222–3p-Cy3). In the control group, we found that the red fluorescence (Cy3-labeled miR-222–3p) was mainly captured in the cytoplasm and there was no co-localization between pcDNA-MS2/GFP and miR-222–3p (Fig. 2G). In contrast, in the TUG1 OEX group, co-localization of green and red fluorescence was detected in the nucleus, suggesting migration of miR-222–3p from cytoplasm to nucleus, and binding to TUG1 (Fig. 2G). Notably, the expression of AGO2 protein in the nucleus has been previously reported supporting the interaction between lncRNA and miRNA in the nucleus (Gagnon et al., 2014; Rashid et al., 2016). Taken together, the AGO2-RIP results suggest that TUG1 and CELF1 were directly associated with AGO2-containing miRNA ribonucleoprotein complexes, which were likely enriched by miR-222–3p (Fig. 2H).
The binding between miR-222–3p and CELF1 was validated by luciferase reporter assays. As Fig. 2I showed, luciferase activity of WT CELF1-3′-UTR luciferase plasmid, but not MUT CELF1-3′-UTR luciferase plasmid, was decreased by miR-222–3p. Further, transfection of miR-222–3p mimic significantly decreased the expression level of CELF1 in HBE cells (Fig. 2J and K). Taken together, our results showed that lncRNA TUG1 positively regulated CELF1 expression by sponging miR-222–3p in HBE cells.
3.4. TUG1-miR-222–3p-CELF1 ceRNA network mediated PM-induced apoptosis and cell cycle arrest in vitro
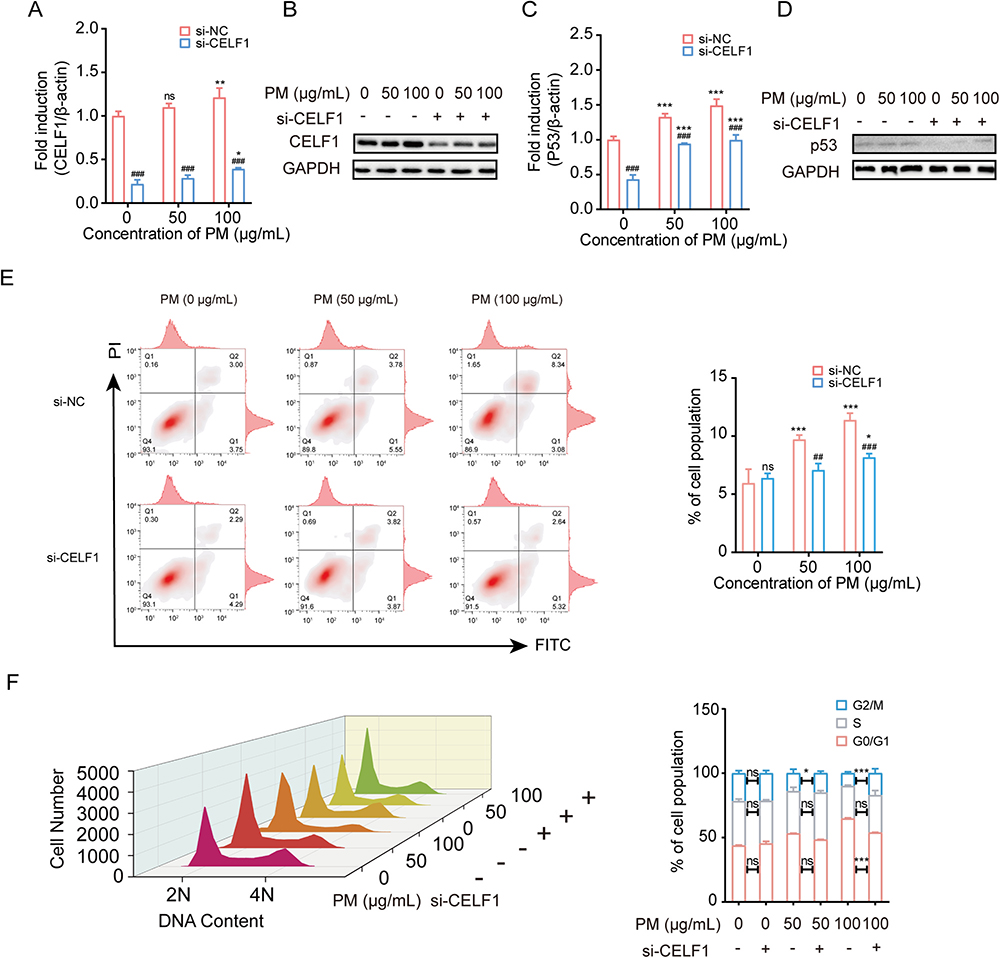
To explore the association between TUG1-miR-222–3p-CELF1 ceRNA network and pulmonary cell injuries, siRNA was transfected to knockdown the expression of CELF1 in HBE cells (Fig. S6). Compared to the si-NC within each group, transfection of si-CELF1 significantly decreased the expression levels of CELF1 in 0, 50 and 100 μg/mL PM-treated HBE cells (Fig. 3A and B). The expression level of p53 was also decreased in HBE cells followed by the si-CELF1 treatment (Fig. 3C and D). The results of flowcytometry assay demonstrated that the proportion of apoptosis and cell cycle arrest were inhibited after decreasing the expression level of CELF1 in HBE cells (Fig. 3E and F).
Fig. 3.
Knockdown CELF1 inhibited PM-induced apoptosis, cell cycle arrest, and activation of P53. (A-B) mRNA and protein levels of CELF1 in HBE cells treated with si-CELF1 after exposed to PM (0, 50 or 100 μg/mL). * P < 0.05, ** P < 0.01, compared with control; ### P < 0.001, compared with si-NC within each group. (C-D) mRNA and protein levels of JUN in HBE cells treated with si-CELF1 after exposed to PM (0, 50 or 100 μg/mL). *** P < 0.001, compared with control; ### P < 0.001, compared with si-NC within each group. (E) Flow cytometric analysis of apoptosis in HBE cells treated with si-CELF1 after exposure to PM (0, 50 or 100 μg/mL). * P < 0.05, *** P < 0.001, compared with control; ## P < 0.01, ### P < 0.001, compared with si-NC within each group. (F) Flow cytometric analysis of cell cycle in HBE cells treated with si-CELF1 after exposure to PM (0, 50 or 100 μg/mL). * P < 0.05, ** P < 0.01, *** P < 0.001, compared with si-NC. The cell cycle is divided into distinct consecutive phases defined as quiescent state and first gap phase (G0/G1), DNA synthesis phase (S), second gap phase and mitosis phase (G2/M). 2 N represents G0/G1 and 4 N represents G2/M while 2–4 N represents S.
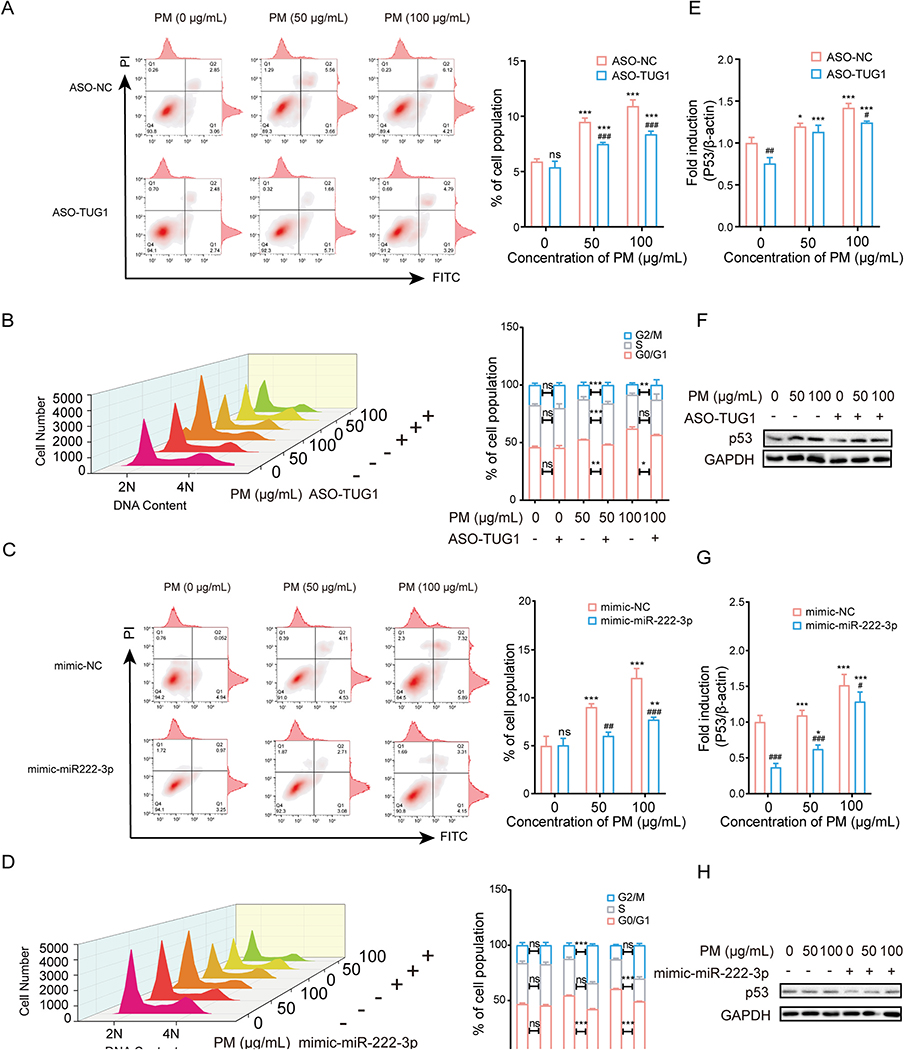
To further explore the role of TUG1 and miR-222–3p in the PM-induced cellular injury, flow cytometric analysis was performed. As expected, ASO-TUG1 or mimic-miR-222–3p treatment effectively inhibited the PM-induced increase in the apoptotic ratio and G1 arrest (Fig. 4A–D). In addition, the expression level of p53 was also inhibited by TUG1 knockdown or miR-222–3p overexpression (Fig. 4E–H). These results suggested that TUG1 knockdown attenuated PM-induced injury by decreasing CELF1/p53 expression levels in vitro.
Fig. 4.
Knockdown TUG1 and overexpressed miR-222–3p inhibited PM-induced apoptosis and cell cycle arrest by decreasing CELF1/P53 in vitro. (A) Flow cytometric analysis of apoptosis in HBE cells treated with ASO-TUG1 after exposure to PM (0, 50 or 100 μg/mL). *** P < 0.001, compared with control; ### P < 0.001, compared with ASO-NC within each group. (B) Flow cytometric analysis of cell cycle in HBE cells treated with ASO-TUG1 after exposure to PM (0, 50 or 100 μg/mL). * P < 0.05, ** P < 0.01, *** P < 0.001, compared with ASO-NC. The cell cycle is divided into distinct consecutive phases defined as quiescent state and first gap phase(G0/G1), DNA synthesis phase (S), second gap phase and mitosis phase (G2/M). 2 N represents G0/G1 and 4 N represents G2/M while 2–4 N represents S. (C) Flow cytometric analysis of apoptosis in HBE cells treated with mimic-miR-222–3p after exposure to PM (0, 50 or 100 μg/mL). ** P < 0.01, *** P < 0.001, compared with control; ## P < 0.01, ### P < 0.001, compared with mimic-NC within each group. (D) Flow cytometric analysis of cell cycle in HBE cells treated with mimic-miR-222–3p after exposure to PM (0, 50 or 100 μg/mL). *** P < 0.001, compared with mimic-NC. (E-F) mRNA and protein levels of p53 in HBE cells treated with ASO-TUG1 after exposed to PM (0, 50 or 100 μg/mL). * P < 0.05, *** P < 0.001, compared with control; # P < 0.05, ## P < 0.01, compared with ASO-NC within each group. (G-H) mRNA and protein levels of p53 in HBE cells treated with mimic-miR-222–3p after exposure to PM (0, 50 or 100 μg/mL). * P < 0.05, *** P < 0.001, compared with control; # P < 0.05, ### P < 0.001, compared with mimic-NC within each group.
3.5. ASO-mmu-Tug1 attenuated PM-induced experimental AHR in murine model through the CELF1/p53 pathway
Based upon sequence alignment analysis between human and mouse, we found that the sequences of TUG1, CELF1 and miR-222–3p were analogous to their murine homologs (Supplementary Files B). Homology diagram of RNAs in the binding sites between human and mouse was depicted in Fig. 5A. To corroborate existing evidence in vitro and in consideration of the vital role of ceRNAs in PM-induced AHR, we exposed mice to PM in the presence or absence of ASO-mmu-Tug1 treatment (Fig. 5B). Intranasally instillation of ASO-mmu-Tug1 can markedly decrease expression of Tug1 in murine lung tissues, which proved the success of establishing Tug1 knockdown in vivo model (Fig. 5C). Airway hypersecretion, serving as an established index of AHR dysfunction, was significantly increased in PM-exposed murine airway epithelia (Fig. 5D). These pathological alterations were alleviated by downregulation of Tug1 (Fig. 5D). Meanwhile, mRNA expression levels of Celf1 and p53 were also suppressed followed by instillation of ASO-mmu-Tug1 (Fig. 5E). Similar changes were also demonstrated at the protein level (Fig. 5F and G).
Fig. 5.
Downregulation of Tug1 rescued PM exposure-induced AHR in mice through the Tug1/Celf1/p53 regulatory axis. (A) Homology diagram of RNAs in the binding sites of ceRNA network between human and mouse. (B) Workflow of murine model treated with ASO-mmu-NC or ASO-mmu-Tug1 after expose to PM (0, 300 or 600 μg/m3). (C) Expression levels of Tug1 in murine lung tissues followed by PM (0, 300 or 600 μg/m3) with ASO-Tug1 treatment. *** P < 0.001, compared with control; ## P < 0.01, ### P < 0.001, compared with ASO-mmu-NC within each group. (n = 5 /group). (D) Representative images of AHR and the number of PAS+ bronchial epithelia per HPF in mice bronchial airway followed by PM (0, 300 or 600 μg/m3) with ASO-mmu-Tug1 treatment. *** P < 0.001, compared with control; ### P < 0.001, compared with ASO-mmu-NC within each group. n = 9/group (3 HPF/section × 1 section/mouse × 3 mice/group); HPF: high-power field at 400 × magnification, scale bar = 50 μm. (E) mRNA expression levels of Celf1 and p53 in murine lung tissues exposed to PM (0, 300 or 600 μg/m3) with ASO-mmu-Tug1 treatment. *** P < 0.001, compared with control; ## P < 0.01, ### P < 0.001, compared with ASO-mmu-NC within each group. (n = 5 /group), (F-G) Expression levels of CELF1 and p53 protein in mice bronchial airway exposed to PM (0, 300 or 600 μg/m3) with ASO-mmu-Tug1 treatment. * P < 0.05, *** P < 0.001, compared with control; # P < 0.05, ### P < 0.001, compared with ASO-mmu-NC within each group. n = 9/group (3 HPF/section × 1 section/mouse × 3 mice/group); HPF: high-power field at 400 × magnification, scale bar = 50 μm.
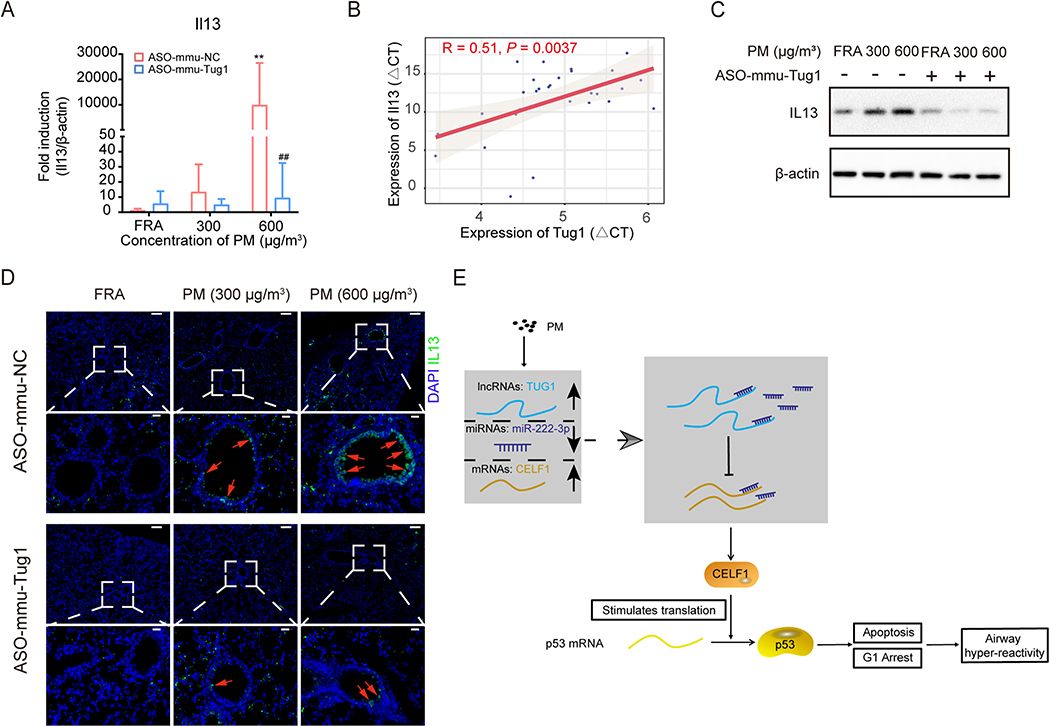
To further evaluate the linkage between TUG1 and AHR in vivo, we addressed the expression of AHR-associated molecules in PM (with or without ASO-mmu-Tug1)-exposed murine lung tissues. The mRNA expression levels of Il13, Il4, Il5, Il33, CclL11, and Muc5ac were significantly increased in 600 μg/m3 PM-exposed lung tissues compared to the control, suggesting activation of AHR hallmarks in vivo (Fig. 6A, Fig. S7A). Furthermore, ASO-mmu-Tug1 treatment significantly inhibited the expression levels of AHR-associated molecules, compared to those in PM-exposed murine lung tissues (Fig. 6A, Fig. S7A). Of note, the expression of Il13, Il5, or Il33 was significantly correlated with the expression of Tug1 (Fig. 6B, Fig. S7B). Given the pivotal role of IL13 for promoting AHR (Walter et al., 2001), we further identified the protein levels of IL13 and found that both the results of WB and IF were consistent with the results of qRT-PCR (Figs. 6C and D). Given these results, we concluded that PM exposure led to AHR in mice, which was mitigated by Tug1 inhibition.
Fig. 6.
ASO-mmu-Tug1 treatment inhibited the expression level of IL13 in vivo.
(A) mRNA expression level of Il13 in murine lung tissues exposed to PM (0, 300 or 600 μg/m3) with ASO-mmu-Tug1 treatment. ** P < 0.01, compared with control; ## P < 0.01, compared with ASO-mmu-NC within each group. (n = 5 /group). (B) Correlation of Tug1 and Il13 mRNA expression level as determined by qRT-PCR in murine lung tissues (n = 30). Spearman’s R and a regression line were indicated, and P < 0.05 was marked in red. (C) The protein expression level of IL13 in murine lung tissues exposed to PM (0, 300 or 600 μg/m3) with ASO-mmu-Tug1 treatment. β-actin was used as a control. (D) Representative images of IL-13 immunohistochemistry staining on the sections of lung tissue from mice exposed to PM (0, 300 or 600 μg/m3) with ASO-mmu-Tug1 treatment. Red arrows highlight IL-13 positive airway epithelia. Scale bars represent 100 μm (upper row), or 20 μm (lower row). (E) Graphical abstract. PM exposure triggered the expression of TUG1 in pulmonary cells. Next, TUG1 indirectly promoted the expression of CELF1 through sponge miR-222–3p, and then CELF1 facilitated the accumulation of p53 protein to mediate apoptosis and cell cycle arrest, eventually leading to AHR.
Taken together, the results suggested a molecular pathway whereby TUG1 promoted the expression of CELF1 through sponge miR-222–3p and then activated p53 pathways to mediate apoptosis and cell cycle arrest in response to PM exposure in bronchial epithelia (Fig. 6E).
4. Discussion
In the present study, we demonstrated that the lncRNA-miRNA-mRNA network plays a critical role in mediating PM exposure-induced AHR. LncRNA TUG1 competes with CELF1 for miR-222–3p binding, therefore abolishing the suppressive effect of miR-222–3p on CELF1. Increased expression levels of TUG1 promoted apoptosis and cell cycle arrest in pulmonary epithelia, resulting in the ensuing AHR in PM-exposed mice.
Existing studies suggest that imbalanced expression of ceRNAs gives rise to altered proliferation and migration (Hua et al., 2020), abnormal autophagy and apoptosis (Zhao et al., 2020), and even tumorigenesis (Huang et al., 2020). Recent studies have also suggested that altered murine ncRNAs may be associated with intra-tracheal instillation with fine PM (Zhong et al., 2019). Consistent with previous studies, our microarray data demonstrated distinctly altered lncRNA, miRNA and mRNA profiles in HBE cells followed by PM exposure, and we have proceeded to identify the TUG1-miR-222–3p-CELF1 ceRNA network as playing a critical role in mediating the effects of PM exposure in HBE cells.
TUG1, is a lncRNA localized to human chromosome 22q12.2, which has been previously associated with COPD (Poulet et al., 2020). Indeed, lncRNA microarray results established that TUG1 was significantly increased in the lung tissue of COPD patients, compared to those from non-COPD (Tang et al., 2016). Another recent study suggested the TUG1-mediated ceRNAs underlying cigarette smoke (CS)- induced chronic inflammation in lungs, which aggravated airway remodeling (Gu et al., 2019). The study highlighted a distinct ceRNAs pathway, namely, TUG1/miR-145–5p/DUSP6, which was involved in COPD. However, we note that this TUG1-involved ceRNAs pathway was associated with airway remodeling, not AHR. Though AHR and airway remodeling are both characteristics inherent to COPD, their development is mediated by distinct mechanisms. In addition, the chemical composition in ambient PM differs from CS, potentially explaining their propensity to trigger distinct molecular responses (Drago et al., 2018).
Mucous hypersecretion, eosinophil accumulation, and increased expression of a set of inflammatory cytokines were characterized as hallmarks for AHR. In terms of inflammatory mediators, Th2 (Boonpiyathad et al., 2019) and eosinophils (Lee et al., 2004) associated cytokines are critical in driving clinical features of AHR. Briefly, IL-4 is a cytokine that induces differentiation of naïve T cells to Th2 cells; IL-5 is responsible for the activation and recruitment of eosinophils to the airways; IL-13 enhances proliferation of IgE-producing B cells and endothelial cells, induces goblet cell hyperplasia and increases the sensitivity of airway to stimuli; IL-33 has been demonstrated to stimulate Th2, basophils and eosinophils to produce Th2-cytokines (Hirose et al., 2017). These cytokines had distinct temporal roles in the development of inflammation and AHR. Indeed, IL4 acts as early phase effector of inflammation, which is consistent with our results that low dose PM-exposure significantly induced IL4 expression, while other cytokines were only induced following high dose PM-exposure.
IL13 plays an essential role in AHR, as suggested by a previous study that IL13−/− mice resisted to allergen-induced AHR (Walter et al., 2001). Induction of IL13 expression in mRNA and protein levels were observed in the PM-exposed murine lung tissues, suggested contribution of PM exposure to AHR. Otherwise, treatment with ASO-mmu-Tug1 significantly inhibited IL13 induction, suggesting that lncRNA TUG1 could be an up-stream trigger of IL13 under the PM exposure. Together with previous studies (Li et al., 2018; Tang et al., 2016), our results emphasized the important role of TUG1 associated with AHR-like lesions both in vivo and in vitro.
The regulatory role of miRNAs in response to air pollution has been studied extensively. Recently, association between the regulation in the expression levels of miRNAs and diverse PM exposure has been summarized (Cheng et al., 2020). Among the PM-related miRNAs, miR-222–3p was the only overlapping miRNA in the PM-exposure groups to different particle sizes. High expression level of miR-222–3p alleviated apoptosis of epithelial cells induced by PM in our study, indicative of a crucial role of miR-222–3p in miRNA related epigenetic mechanisms in PM-induced damage. However, whether miR-222–3p mediated its effects by ceRNAs in PM-induced injury has yet to be determined.
Several studies have documented that miR-222–3p acts an anti-apoptotic effect by suppressing expression of p53 and apoptosis related proteins (Sun et al., 2019; Zhang et al., 2010). Therefore, the expression level of p53 was analyzed in our study, indicating it was activated upon PM treatment and restrained by miR-222–3p mimic application. Furthermore, we showed that silencing TUG1 can also cause p53 restraint. Hence, p53 appears to be a downstream target of the ceRNA network. The vital role of p53 in apoptosis has been broadly acknowledged (Levine and Oren, 2009). It is widely believed that p53 mediates biological functions mainly through its transcriptional activity upon activation in response to cellular stress, such as DNA damage, oncogene activation, or hypoxia (Vousden and Prives, 2009). Interestingly, increased expression of miR-222–3p elicited a decrease in CELF1, while that CELF1 mediated the expression of p53 by expediting its protein translation, leading to sustained G2 arrest and G1 arrest as previously reported (Yan et al., 2019). These observations are consistent with a positive regulatory relationship between CELF1 and p53.
CELF1 is one of the RNA binding proteins that directly interacts with mRNA and elicits mRNA decay (Kim and Gorospe, 2008). However, in some instances, CELF1 also facilitates mRNA translation by mediating the stability or translation rate of specific target transcripts (Chang et al., 2012; Iakova et al., 2004). In this study, after CELF1 knockdown, a correspondingly low expression level of p53 was detected. This regulatory relationship was also demonstrated in intestinal epithelial cells, corroborating in part the findings reported herein (Yan et al., 2019). Furthermore, the silencing of TUG1 and CELF1, as well as the overexpression of miR-222–3p, alleviated to some extent the cycle arrest induced by PM. Overall, our novel findings establish that the TUG1/miR-222–3p /CELF1/p53 network is a critical mediator of PM-induced apoptosis and cell cycle arrest.
5. Conclusion
In summary, the ceRNA network TUG1/miR-222–3p/CELF1 mediates the deleterious effects of airborne PM exposure, triggering the ensuing bronchial hyper-reactivity. Further evaluation of the TUG1-related ceRNA network in larger human populations is required for a better understanding on their expression profiles and mediation of the adverse outcomes caused by exposures to ambient air pollution.
Supplementary Material
Acknowledgments
Funding
This work was financially supported by the State Key Program of the National Natural Science Foundation of China, China (81730088), the National Natural Science Foundation of China, China (81973084, 91943301, 91743112); the National Science Fund for Distinguished Young Scholars, China (82025031); Guangdong Provincial Natural Science Foundation Team Project, China (2018B030312005). MA was supported in part by grants from the National Institute of Environmental Health Sciences (NIEHS), U.S.A. R01ES10563 and R01ES07331.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data Availability
The miRNA, lncRNA and mRNA microarray datasets supporting the conclusions of this article are available in the GEO repository; the accession numbers are GSE134430 and GSE138870, respectively.
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jhazmat.2021.125878.
References
- Bai W, Li Y, Niu Y, Ding Y, Yu X, Zhu B, Duan R, Duan H, Kou C, Li Y, Sun Z, 2020. Association between ambient air pollution and pregnancy complications: a systematic review and meta-analysis of cohort studies. Environ. Res 185, 109471. 10.1016/j.envres.2020.109471. [DOI] [PubMed] [Google Scholar]
- Boonpiyathad T, Sözener ZC, Satitsuksanoa P, Akdis CA, 2019. Immunologic mechanisms in asthma. Semin. Immunol 46, 101333. 10.1016/j.smim.2019.101333. [DOI] [PubMed] [Google Scholar]
- Brutsche MH, Downs SH, Schindler C, Gerbase MW, Schwartz J, Frey M, Russi EW, Ackermann-Liebrich U, Leuenberger P, SAPALDIA T, 2006. Bronchial hyperresponsiveness and the development of asthma and COPD in asymptomatic individuals: SAPALDIA cohort study. Thorax 61, 671–677. 10.1136/thx.2005.052241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busse, 2010. The relationship of airway hyperresponsiveness and airway inflammation: airway hyperresponsiveness in asthma: its measurement and clinical significance. Chest 138, 4S–10S. 10.1378/chest.10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caraher EJ, Kwon S, Haider SH, Crowley G, Lee A, Ebrahim M, Zhang L, Chen LC, Gordon T, Liu M, Prezant DJ, Schmidt AM, Nolan A, 2017. Receptor for advanced glycation end-products and World Trade Center particulate induced lung function loss: a case-cohort study and murine model of acute particulate exposure. PLoS One 12, 0184331. 10.1371/journal.pone.0184331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA, 2014. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 157, 77–94. 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Chang ET, Donahue JM, Xiao L, Cui Y, Rao JN, Turner DJ, Twaddell WS, Wang JY, Battafarano RJ, 2012. The RNA-binding protein CUG-BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. Biochem. J. 446, 113–123. 10.1042/BJ20120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DL, Xiang YY, Ji LJ, Lu XJ, 2015. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol. 36, 479–488. 10.1007/s13277-015-3093-z. [DOI] [PubMed] [Google Scholar]
- Cheng M, Wang B, Yang M, Ma J, Ye Z, Xie L, Zhou M, Chen W, 2020. microRNAs expression in relation to particulate matter exposure: a systematic review. Environ. Pollut 260, 113961. 10.1016/j.envpol.2020.113961. [DOI] [PubMed] [Google Scholar]
- Collaborators, 2018. Global, regional, and national comparative risk assessment of 84 behavioural, environmental and occupational, and metabolic risks or clusters of risks for 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet 392, 1923–1994. 10.1016/S0140-6736(18)32225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago G, Perrino C, Canepari S, Ruggieri S, L’Abbate L, Longo V, Colombo P, Frasca D, Balzan M, Cuttitta G, Scaccianoce G, Piva G, Bucchieri S, Melis M, Viegi G, Cibella F, RESPIRA Collaborative Project, G., Indoor and Outdoor Air Quality and Respiratory Health in Malta and Sicily - RESPIRA Study, G., Balzan M, Bilocca D, Borg C, Montefort S, Zammit C, Bucchieri S, Cibella F, Colombo P, Cuttitta G, Drago G, Ferrante G, L’Abbate L, Grutta S, Longo V, Melis MR, Ruggieri S, Viegi G, Minardi R, Piva G, Ristagno R, Rizzo G, Scaccianoce G, 2018. Relationship between domestic smoking and metals and rare earth elements concentration in indoor PM2.5. Environ. Res 165, 71–80. 10.1016/j.envres.2018.03.026. [DOI] [PubMed] [Google Scholar]
- Ergun S, Oztuzcu S, 2015. Oncocers: ceRNA-mediated cross-talk by sponging miRNAs in oncogenic pathways. Tumour Biol. 36, 3129–3136. 10.1007/s13277-015-3346-x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Lu R, Cui J, Sun H, Yang H, Meng Q, Wu S, Aschner M, Li X, Chen R, 2019. Inhibition of ATP citrate lyase (ACLY) protects airway epithelia from PM2.5-induced epithelial-mesenchymal transition. Ecotoxicol. Environ. Saf 167, 309–316. 10.1016/j.ecoenv.2018.10.033. [DOI] [PubMed] [Google Scholar]
- Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR, 2014. RNAi factors are present and active in human cell nuclei. Cell Rep. 6, 211–221. 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Cai Y, Kapranov P, Xu D, 2020. Reverse-genetics studies of lncRNAs-what we have learnt and paths forward. Genome Biol. 21, 93. 10.1186/s13059-020-01994-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Yuan Y, Wang L, Yang H, Li S, Tang Z, Li Q, 2019. Long non-coding RNA TUG1 promotes airway remodelling by suppressing the miR-145–5p/DUSP6 axis in cigarette smoke-induced COPD. J. Cell. Mol. Med 23, 7200–7209. 10.1111/jcmm.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao M, Comier S, Wang M, Lee JJ, Nel A, 2003. Diesel exhaust particles exert acute effects on airway inflammation and function in murine allergen provocation models. J. Allergy Clin. Immunol 112, 905–914. 10.1016/j.jaci.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Hirose K, Iwata A, Tamachi T, Nakajima H, 2017. Allergic airway inflammation: key players beyond the Th2 cell pathway. Immunol. Rev 278, 145–161. 10.1111/imr.12540. [DOI] [PubMed] [Google Scholar]
- Hua K, Deng X, Hu J, Ji C, Yu Y, Li J, Wang X, Fang L, 2020. Long noncoding RNA HOST2, working as a competitive endogenous RNA, promotes STAT3-mediated cell proliferation and migration via decoying of let-7b in triple-negative breast cancer. J. Exp. Clin. Cancer Res 39, 58. 10.1186/s13046-020-01561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Li X, Ye H, Liu Y, Liang X, Yang C, Hua L, Yan Z, Zhang X, 2020. Long non-coding RNA NCK1-AS1 promotes the tumorigenesis of glioma through sponging microRNA-138–2-3p and activating the TRIM24/Wnt/β-catenin axis. J. Exp. Clin. Cancer Res 39, 63. 10.1186/s13046-020-01567-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakova P, Wang GL, Timchenko L, Michalak M, Pereira-Smith OM, Smith JR, Timchenko NA, 2004. Competition of CUGBP1 and calreticulin for the regulation of p21 translation determines cell fate. EMBO J. 23, 406–417. 10.1038/sj.emboj.7600052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Gorospe M, 2008. GU-rich RNA: expanding CUGBP1 function, broadening mRNA turnover. Mol. Cell 29, 151–152. 10.1016/j.molcel.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Chang YJ, Chuang YT, Lee HH, Kasahara DI, Martin T, Hsu JT, Savage PB, Shore SA, Freeman GJ, Dekruyff RH, Umetsu DT, 2013. T-cell immunoglobulin and mucin domain 1 deficiency eliminates airway hyperreactivity triggered by the recognition of airway cell death. J. Allergy Clin. Immunol 132, 414–425. 10.1016/j.jaci.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HH, Meyer EH, Goya S, Pichavant M, Kim HY, Bu X, Umetsu SE, Jones JC, Savage PB, Iwakura Y, Casasnovas JM, Kaplan G, Freeman GJ, DeKruyff RH, Umetsu DT, 2010. Apoptotic cells activate NKT cells through T cell Ig-like mucin-like-1 resulting in airway hyperreactivity. J. Immunol 185, 5225–5235. 10.4049/jimmunol.1001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Dimina D, Macias MP, Ochkur SI, McGarry MP, O’Neill KR, Protheroe C, Pero R, Nguyen T, Cormier SA, Lenkiewicz E, Colbert D, Rinaldi L, Ackerman SJ, Irvin CG, Lee NA, 2004. Defining a link with asthma in mice congenitally deficient in eosinophils. Science 305, 1773–1776. 10.1126/science.1099472. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Oren M, 2009. The first 30 years of p53: growing ever more complex. Nat. Rev. Cancer 9, 749–758. 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, et al. , 2018. Identification of abnormally expressed lncRNAs induced by PM2.5 in human bronchial epithelial cells. Biosci. Rep 38. 10.1042/BSR20171577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang Y, Li B, Yang H, Cui J, Li X, Zhang X, Sun H, Meng Q, Wu S, Li S, Wang J, Aschner M, Chen R, 2020. Activation of NLRP3 in microglia exacerbates diesel exhaust particles-induced impairment in learning and memory in mice. Environ. Int 136, 105487. 10.1016/j.envint.2020.105487. [DOI] [PubMed] [Google Scholar]
- Mannucci PM, Harari S, Franchini M, 2019. Novel evidence for a greater burden of ambient air pollution on cardiovascular disease. Haematologica 104, 2349–2357. 10.3324/haematol.2019.225086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulet, et al. , 2020. Exosomal long non-coding RNAs in lung diseases. Int. J. Mol. Sci 21. 10.3390/ijms21103580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid F, Shah A, Shan G, 2016. Long non-coding RNAs in the cytoplasm. Genom. Proteom. Bioinform 14, 73–80. 10.1016/j.gpb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraufnagel DE, Balmes JR, Cowl CT, De Matteis S, Jung SH, Mortimer K, Perez-Padilla R, Rice MB, Riojas-Rodriguez H, Sood A, Thurston GD, To T, Vanker A, Wuebbles DJ, 2019. Air pollution and noncommunicable diseases: a review by the Forum of International Respiratory Societies’ Environmental Committee, Part 2: Air Pollution and Organ Systems. Chest 155, 417–426. 10.1016/j.chest.2018.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Zhao X, Ming J, Liu X, Liu D, Jiang C, 2019. Stepwise detection and evaluation reveal miR-10b and miR-222 as a remarkable prognostic pair for glioblastoma. Oncogene 38, 6142–6157. 10.1038/s41388-019-0867-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Shen Z, Guo J, Sun S, 2016. Screening of long non-coding RNA and TUG1 inhibits proliferation with TGF-beta induction in patients with COPD. Int. J. Chronic Obstr. Pulm. Dis 11, 2951–2964. 10.2147/COPD.S109570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tshala-Katumbay D, Mwanza JC, Rohlman DS, Maestre G, Oriá RB, 2015. A global perspective on the influence of environmental exposures on the nervous system. Nature 527, S187–S192. 10.1038/nature16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vousden KH, Prives C, 2009. Blinded by the light: the growing complexity of p53. Cell 137, 413–431. 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- Walter DM, McIntire JJ, Berry G, McKenzie AN, Donaldson DD, DeKruyff RH, Umetsu DT, 2001. Critical role for IL-13 in the development of allergen-induced airway hyperreactivity. J. Immunol 167, 4668–4675. 10.4049/jimmunol.167.8.4668. [DOI] [PubMed] [Google Scholar]
- Wang G, Zheng X, Duan H, Dai Y, Niu Y, Gao J, Chang Z, Song X, Leng S, Tang J, Zheng Y, 2019. High-content analysis of particulate matters-induced oxidative stress and organelle dysfunction in vitro. Toxicol. Vitr 59, 263–274. 10.1016/j.tiv.2019.04.026. [DOI] [PubMed] [Google Scholar]
- Yan JK, Zhang T, Dai LN, Gu BL, Zhu J, Yan WH, Cai W, Wang Y, 2019. CELF1/p53 axis: a sustained antiproliferative signal leading to villus atrophy under total parenteral nutrition. FASEB J. 33, 3378–3391. 10.1096/fj.201801695R. [DOI] [PubMed] [Google Scholar]
- Yang L, Li C, Tang X, 2020. The impact of PM2.5 on the host defense of respiratory system. Front Cell Dev. Biol 8, 91. 10.3389/fcell.2020.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, Kang C, 2010. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int. J. Oncol 37, 1621–1626. 10.3892/ijo_00000816. [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhang Y, Meng Q, Sun H, Wu S, Xu J, Yun J, Yang X, Li B, Zhu H, Xue L, Li X, Chen R, 2020. MicroRNA-382–5p is involved in pulmonary inflammation induced by fine particulate matter exposure. Environ. Pollut 262, 114278. 10.1016/j.envpol.2020.114278. [DOI] [PubMed] [Google Scholar]
- Zhao X, Su L, He X, Zhao B, Miao J, 2020. Long noncoding RNA CA7–4 promotes autophagy and apoptosis via sponging MIR877–3P and MIR5680 in high glucose-induced vascular endothelial cells. Autophagy 16, 70–85 10.1080/15548627.2019.1598750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wang Y, Zhang C, Hu Y, Sun C, Liao J, Wang G, 2019. Identification of long non-coding RNA and circular RNA in mice after intra-tracheal instillation with fine particulate matter. Chemosphere 235, 519–526. 10.1016/j.chemosphere.2019.06.122. [DOI] [PubMed] [Google Scholar]
- Zhou B, Liang G, Qin H, Peng X, Huang J, Li Q, Qing L, Zhang L, Chen L, Ye L, Niu P, Zou Y, 2014. p53-Dependent apoptosis induced in human bronchial epithelial (16-HBE) cells by PM(2.5) sampled from air in Guangzhou, China. Toxicol. Mech. Methods 24, 552–559. 10.3109/15376516.2014.951814. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.