Long-chain polyphosphates inhibit SARS-CoV-2 infection by targeting a host receptor and a viral RNA polymerase.
Polyphosphates versus SARS-CoV-2
Long-chain, inorganic polyphosphates (polyPs), which are found in many cells in the blood, have cytoprotective and antiviral activities, particularly against HIV-1 infection. Ferrucci et al. tested the effects of polyPs of various lengths on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in vitro. Molecular docking and binding analyses showed that polyPs bound to the host receptor ACE2, which facilitates viral entry, and a viral RNA polymerase required for replication. Both proteins underwent proteasomal degradation in cells incubated with polyP120, the optimal species tested, resulting in inhibition of SARS-CoV-2 replication and a reduced inflammatory response. Given that polyPs have low toxicity, these results suggest that their potential therapeutic use should be further explored.
Abstract
Inorganic polyphosphates (polyPs) are linear polymers composed of repeated phosphate (PO43−) units linked together by multiple high-energy phosphoanhydride bonds. In addition to being a source of energy, polyPs have cytoprotective and antiviral activities. Here, we investigated the antiviral activities of long-chain polyPs against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. In molecular docking analyses, polyPs interacted with several conserved amino acid residues in angiotensin-converting enzyme 2 (ACE2), the host receptor that facilitates virus entry, and in viral RNA-dependent RNA polymerase (RdRp). ELISA and limited proteolysis assays using nano– LC-MS/MS mapped polyP120 binding to ACE2, and site-directed mutagenesis confirmed interactions between ACE2 and SARS-CoV-2 RdRp and identified the specific amino acid residues involved. PolyP120 enhanced the proteasomal degradation of both ACE2 and RdRp, thus impairing replication of the British B.1.1.7 SARS-CoV-2 variant. We thus tested polyPs for functional interactions with the virus in SARS-CoV-2–infected Vero E6 and Caco2 cells and in primary human nasal epithelial cells. Delivery of a nebulized form of polyP120 reduced the amounts of viral positive-sense genomic and subgenomic RNAs, of RNA transcripts encoding proinflammatory cytokines, and of viral structural proteins, thereby presenting SARS-CoV-2 infection in cells in vitro.
INTRODUCTION
Coronaviruses (CoVs) contain positive-sense, single-stranded RNA (~30 kb). Four major categories have been reported, with alpha-CoVs and beta-CoVs known to infect humans. These viruses replicate in the lower respiratory tract and cause pneumonia, which can be fatal (1, 2). These infections include severe acute respiratory syndrome (SARS-CoV), Middle East respiratory syndrome (MERS-CoV), and SARS-CoV-2. This last CoV belongs to the beta CoV genus (3) and has resulted in pandemic acute respiratory syndrome in humans, known as coronavirus disease 2019 (COVID-19). This can progress to acute respiratory distress syndrome, generally at 8 to 9 days after symptom onset (4). As for the other respiratory CoVs, SARS-CoV-2 is transmitted by respiratory droplets, with fecal-oral transmission also possible (5).
When SARS-CoV-2 infects host cells, it replicates its genomic RNA (gRNA) to produce smaller RNAs known as subgenomic RNAs (sgRNAs). These sgRNAs are mainly used to synthesize the SARS-CoV-2 conserved structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N) (6). The receptor used by SARS-CoV-2 to enter host cells is angiotensin-converting enzyme 2 (ACE2), which is mainly expressed in the lungs (7, 8), together with the cellular serine protease transmembrane serine protease 2 (TMPRSS2) (9). Binding of the virus to ACE2 is enhanced by heparan sulfate on cell surface, which alters the structure of the viral S protein to favor the open conformation of the receptor-binding domain (RBD) (10). The SARS-CoV-2 RNA-dependent RNA polymerase [RdRp, also known as nonstructural protein 12 (nsp12)] is a key component of the viral replication and transcription machinery. RdRp contributes to viral genome mutation, and hence to viral adaptation, and is part of a complex (RdRp/nsp7/nsp8) that is fundamental to transcriptional fidelity (that is, transcription “proofreading”) (2). RdRp thus coordinates the replication of both the viral genome (which is used as the template for replication and transcription) and the shorter sgRNAs, according to the prevailing “leader-to-body” fusion model (11). Note that the RdRp mutation 14408C>T-(P323L) in nsp12 in European strains of SARS-CoV-2 suggests that the proofreading activity has been affected, thus altering SARS-CoV-2 mutation rates (12).
Because of the global spread of COVID-19 and its associated relatively high mortality rate, the development of therapeutic strategies represents an urgent unmet medical need. We therefore investigated the potential activities of long-chain inorganic polyphosphates (polyPs) against SARS-CoV-2 infection. At this time, more efficient viral replication, transmission, and evasion of host defenses might be related to emerging SARS-CoV-2 variants with genomic plasticity, with mutations in S protein, thus emerging around the world (B.1.1.7, B1.351, and P1) (13). These three major variants are the result of selective pressures and adaptation of the virus during prolonged individual infections and the subsequent transmission, and they pose potential challenges for vaccination and antibody-based therapies.
PolyPs are composed of chains of a few to many hundreds of inorganic phosphate (Pi) units. They are ubiquitous, being prevalent in peripheral blood mononuclear cells and erythrocytes (14), and they are found subcellularly in the nucleus, cytoplasm, plasma membrane, and mitochondria (15). PolyPs are involved in blood coagulation (16) and inhibition of the complement pathway (17). They are also involved in chelation of calcium (Ca2+) for bone mineralization (18) and activation of apoptosis (14), and they act as “chaperone-like” (19) and neuronal excitability molecules (20, 21). Note that linear polyPs show cytoprotective and antiviral activities against HIV-1 infection in vitro (22). Furthermore, and of particular relevance here, polyPs (as Pi ×40) substantially inhibit the interaction of the RBD of SARS-CoV-2 S protein with ACE2 in vitro (23).
Here, we showed that long-chain inorganic polyPs had antiviral activities against SARS-CoV-2 in different cellular models, including Vero E6 and Caco2 cells and primary human epithelial cells obtained from a healthy donor by nasal brushing. We demonstrated that intracellularly, polyPs enhanced the proteasome-mediated decrease in the abundance of ACE2 and RdRp through steric hindrance, which resulted in marked reductions in the amounts of SARS-CoV-2 structural sgRNAs and proteins (that is, S, E, M, and N). These activities were then tested in vitro for their effects on replication of virus variants, including the B.1.1.7 variant. Because polyPs are already known not to be toxic (24), these data suggest that the therapeutic use of polyPs to prevent SARS-CoV-2 infections should be explored.
RESULTS
The antiviral effects of long-chain polyPs against SARS-CoV-2 in vitro are dependent on interactions with ACE2 and RdRp
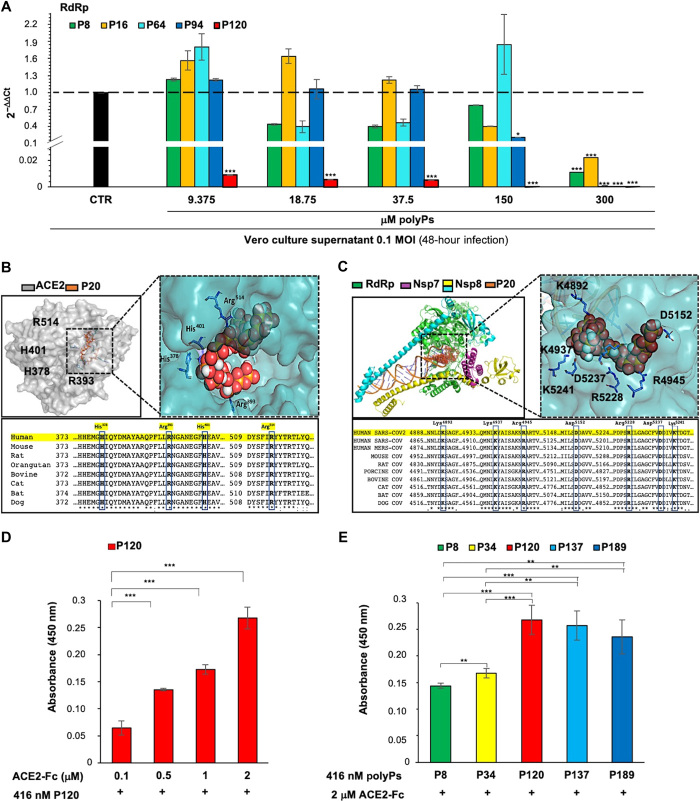
Antiviral activities of linear polyPs with average chain lengths of 15, 34, and 91 Pi residues have been shown in vitro for HIV-infected cells (22). To measure the antiviral effects of polyPs, we used SARS-CoV-2 viral particles obtained from a Korean patient who was positive for COVID-19 (BetaCov/Korea/KCDC03/2020; registered ID: EPI_ISL_407193) to infect Vero E6 cells. At 24 hours after infection, the cells were treated with increasing concentrations of polyPs (9.375 to 300 μM) of different chain lengths (Pi ×8, ×16, ×64, ×94, and ×120). Pi concentrations were calculated on a per-Pi basis, as described by Lorenz et al. (22). After 24 hours of polyP treatment (that is, 48 hours after infection), the viral RNA in the cell culture supernatants was quantified for RdRp expression by quantitative real-time polymerase chain reaction (qRT-PCR; fig. S1A). These data showed that polyP ×120 (polyP120) statistically significantly decreased the abundance of RdRp RNA at all concentrations tested. None of the other polyPs tested here had significant effects at concentrations <300 μM, except for polyP94, which also significantly decreased RdRp RNA abundance at 150 and 300 μM (Fig. 1A). The quantity of viral RNA was also expressed as plaque-forming unit (PFU) equivalents (fig. S1B). Therefore, polyP120 showed better antiviral activity compared with the other polyPs with shorter chain lengths, with a median inhibitory concentration (IC50) of 57.29 μM [coefficient of determination (R2) = 0.957; fig. S1C].
Fig. 1. The antiviral effects of polyPs are due to their binding to ACE2 and RdRp.
(A) Viral RdRp expression was measured by RT-PCR analysis of viral RNA extracted from the culture medium of Vero E6 cells (4 × 105) that were infected with SARS-CoV-2 for 24 hours and then treated with increasing concentrations of polyPs (9.375, 18.75, 37.5, 150, and 300 μM) of different chain lengths (P8, polyP8; P16, polyP16; P64, polyP64; P94, polyP94; P120, polyP120) for an additional 24 hours. The qRT-PCR analysis was performed with primer-probe sets that targeted the RdRp region of SARS-CoV-2 virus. Note that ΔCt was calculated as the difference between the Ct for RdRp expression in SARS-CoV-2–infected cells treated with polyPs and the Ct for RdRp expression in SARS-CoV-2–infected cells without polyPs. The quantity of viral RNA (ct) was also expressed as PFU equivalents (fig. S1B). Data are means ± SD. *P < 0.05 and ***P < 0.001 [by unpaired two-tailed Student’s t test; RdRp versus untreated Vero E6 cells (black column); n = 3 independent experiments per group]. (B) Top: Molecular docking of polyP20 on the SARS-CoV-2 ACE2 domain (PDB structure: 6M0J, chain A). Left: ACE2. The transparent molecular surface is colored according to electrostatic potential, as −10 kT/e (red) to +10 kT/e (blue). The orange sticks represent polyP20. Right: Magnified view of the ACE2 receptor as a cyan transparent surface to indicate the binding interface. Bottom: Alignment analysis of ACE2 protein regions with potential binding sites for polyP20. The amino acid residues mainly responsible for the interactions between ACE2 and polyP20 are shown as blue boxes (His378, Arg393, His401, and Arg514). (C) Top: Molecular docking of polyP20 (P20) on SARS-CoV-2 RdRp (PDB structure: 6 M71). Left: RdRp. The molecular surface is colored according to electrostatic potential, from −10 kT/e (red) to +10kT/e (blue). The red balls represent polyP20. Right: Magnified view of RdRp as a cyan transparent surface to indicate the binding interface. Bottom: Alignment analysis of the RdRp protein (nsp12) region that contains the potential binding sites for polyP20. The amino acid residues mainly responsible for interactions between RdRp and polyP20 are shown in blue boxes (Lys4892, Lys4937, Arg4945, Asp5152, Arg5228, Asp5237, and Arg5241). (D) ELISAs of increasing concentrations (0.1 to 2.0 μM) of human ACE2-Fc chimera protein in 96-well plates coated with 416 nM polyP120. Absorbance at 450 nm was measured after a 1-hour incubation at 25°C. Comparison among the different ACE2-Fc concentrations compared with the lowest concentration (0.1 μM) for binding to polyP120 was evaluated. Data are means ± SD. ***P < 0.001 by unpaired two-tailed Student’s t test; n = 3 independent experiments per group. (E) ELISAs of 2 μM human ACE2-Fc chimera protein and 416 nM polyPs of the indicated chain lengths coated on 96-well plates. Absorbance at 450 nm was measured after a 1-hour incubation at 25°C. Comparison among the different polyPs for the binding to ACE2-Fc was evaluated comparing them to polyP8 and polyP34 (P34) (e.g., polyP120 versus polyP8: P = 9.4 × 10−5; unpaired two-tailed t test, adjusted with the Bonferroni method). Data are means ± SD. **P < 0.01 and ***P < 0.001 by unpaired two-tailed Student’s t test; n = 3 independent experiments per group.
The anti-HIV effects of polyPs are due to binding to both the host cell surface and the virus (22). For this reason, we investigated whether the antiviral activity of polyPs against SARS-CoV-2 could be ascribed to their binding to ACE2 on the host cells and to the viral RdRp. To address these hypotheses, potential binding sites for polyP120 on both ACE2 and viral RdRp were investigated through molecular docking analysis with shorter-chain polyPs, because longer-chain polyPs do not produce reliable docking results. These data showed the potential for binding between ACE2 and the shorter chain polyPs (e.g., polyP20), which was mainly mediated through four amino acid residues of ACE2: His378, Arg393, His401, and Arg514 (Fig. 1B and Table 1), which are conserved across different vertebrates (sequence identity, 63.6%; Fig. 1B). Further docking studies with RdRp in its active replicated form (i.e., as part of the RdRp/nsp7/nsp8 complex; Table 1) illustrated the possible binding modes for polyP20. This binding was mediated through seven RdRp amino acid residues within the RNA binding site of RdRp: Lys4892, Lys4937, Arg4945, Asp5152, Arg5228, Asp5237, and Lys5241 (Fig. 1C and Table 1). These amino acids are conserved in the RdRp proteins of other viruses in the family Coronoviridae (e.g., SARS-CoV and MERS-CoV; Fig. 1C) and potentially in the main families of positive-sense, single-stranded RNA viruses from other hosts (e.g., Nidovirales; see www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=76804).
Table 1. Best scores obtained through the AutoDock Vina docking analysis.
| Receptor | Best score (kcal/mol) |
| RdRp | −5.3 |
| ACE2 | −5.9 |
| RdRp/nsp7/nsp8 | −4.8 ± 0.2 |
The potential interaction between polyPs and ACE2 was evaluated by enzyme-linked immunosorbent assays (ELISAs). Briefly, 416 nM polyP120 was used to coat 96-well plates, and the recombinant ACE2-Fc chimeric protein was added at increasing concentrations (0.1 to 2.0 μM) and allowed to bind for 1 hour at 25°C. Binding was then detected with a horseradish peroxidase (HRP)–conjugated secondary anti-Fc antibody (Fig. 1D). These data showed that ACE2 bound to polyP120 in a dose-dependent manner. As a negative control, the ACE2-Fc chimeric protein was tested under the same conditions on uncoated wells, where no substantial ELISA signal was detected, as expected (table S1). Then, using the maximum concentration of 2 μM ACE2-Fc chimeric protein, we tested other polyPs of different chain lengths, including polyP8, polyP34, polyP120, polyP137, and polyP189 (all at a coating concentration of 416 nM). Together, these data showed that there were greater interactions between the ACE2 protein and the higher–molecular weight polyPs compared with the lower–molecular weight polyPs (Fig. 1E and table S1). In particular, polyP120 showed the greatest binding to ACE2 compared with the lower–molecular weight polyPs in the assay (Fig. 1E).
An additional test was performed to map the regions of ACE2 and polyP120 that interacted with each other by limited proteolysis, using chymotrypsin with the purified ACE2-Fc chimeric protein. The lowest binding with polyP8 (from Fig. 1E) was used as the control in this assay (fig. S1, D and E). This showed that the proteolytic profiles of isolated ACE2 and its complex with polyP8 were comparable (fig. S1E, lanes 1 to 3 and 7 to 9). In contrast, these proteolytic profiles were different from those for the complex between ACE2-Fc and polyP120, with the main difference being a band with electrophoretic mobility of about 50 kDa (fig. S1E, lanes 4 to 6, red arrow). The results from a bottom-up proteomics approach applied to this proteolysis by chymotrypsin showed that all of the peptides identified by nano–liquid chromatography–tandem mass spectrometry (nano LC-MS/MS) were mapped to the C-terminal region (fig. S1F, red, and table S2). The MS data together with the apparent molecular weight of the band of about 50 kDa suggested that the chymotrypsin cleavage site was located upstream of Arg514, within the fragments 510 to 518, which contains several aromatic amino acid residues. This fragment was not detected in the presence of polyP120, which thus suggested that the cleavage site was hidden in the complex formed between ACE2-Fc and polyP120 (fig. S1F and table S2). The binding region suggested by these limited proteolysis experiments was close to Arg514, which was one of the residues included in the binding pocket that was described by the docking calculations (Fig. 1B).
The binding of polyP120 to ACE2 and RdRp promotes their proteasomal degradation
Because a scaffold-like activity has been previously described for polyPs (19), we investigated whether the abundance of the ACE2 protein was modulated by polyPs. Because polyPs can undergo hydrolysis reactions in aqueous solutions and generate monophosphates if they are not stable, we tested phosphate monomers (KH2PO4 form) and double-distilled H2O and showed that they did not modulate ACE2 abundance in Vero E6 cells after 24 hours of treatment (fig. S2A). We then isolated primary human epithelial cells from nasal brushing of a healthy participant (fig. S2B) (25). These cells underwent next-generation sequencing to verify potential nonsynonymous variants able to modulate ACE2 gene expression. These data showed the absence of modifier variants in the ACE2 gene (synonymous_variant, c.2247G>A,p.Val749Val; intron_variant, c.1297+68_1297+69insCTTAT; splice_region_variant&intron_variant, c.439+4G>A; (EVA) - EMBL-EBI; project ID: PRJEB42411; analyses: ERZ1700617).
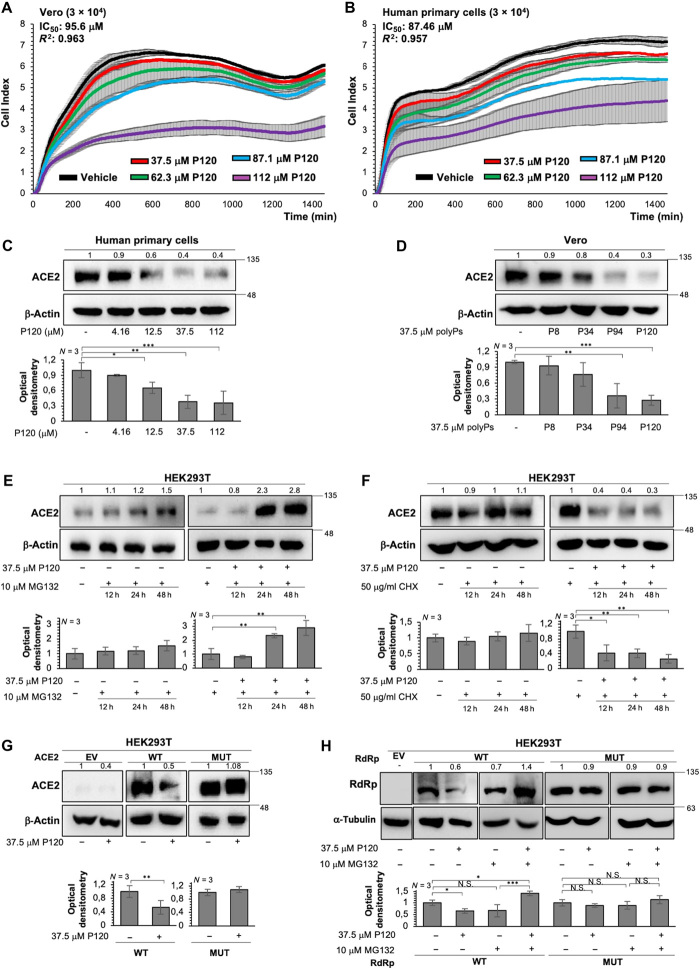
The potential cytotoxicity of different concentrations of polyP120 (37.5 to 112 μM) was evaluated in Vero E6 cells (Fig. 2A) and in the primary human epithelial cells (Fig. 2B), through real-time cell proliferation assays (i.e., cell sensor impedance was measured every 2 min and then expressed as the “cell index”). Double-distilled H2O was used as the vehicle control. The IC50 values for polyP120 in Vero E6 cells and in the primary human epithelial cells were 95.6 μM (R2 = 0.963; Fig. 2A) and 87.46 μM (R2 = 0.957; Fig. 2B), respectively. The lowest concentration of polyP120 used in this assay (37.5 μM) only marginally decreased the proliferation rate in the primary human epithelial cells (although the proliferation curves of both polyP120-treated and vehicle-treated cells had substantially overlapping slopes during early treatment; Fig. 2B, 37.5 μM versus vehicle treatment). Note that this effect was not observed in Vero E6 cells, for which both curves had similar and overlapping slopes (Fig. 2A).
Fig. 2. PolyP120 stimulates the proteasome-mediated degradation of ACE2 and RdRp.
(A and B) Real-time cell proliferation analysis for the cell index (i.e., the cell sensor impedance was expressed every 2 min as a unit called cell index). Vero E6 cells (3 × 104) (A) and primary human epithelial cells from nasal brushing (3 × 104) (B) were plated and treated with the indicated concentrations of polyP120 (4.16 to 112 μM); vehicle-treated cells were the negative control. Impedance was measured every 2 min over 24 hours. The IC50 values were calculated through nonlinear regression analysis performed with GraphPad Prism 9 {[inhibitor] versus response (three parameters)}. Data are means ± SD of three independent experiments. 5′UTR, 5′ untranslated region. (C to H) Representative Western blotting analysis of primary human epithelial cells (C), Vero E6 cells (D), and HEK293T cells (E to H) that were treated as indicated. All experiments were performed in triplicate, and Western blotting was performed with antibodies against the indicated proteins. Double-distilled water was used as vehicle control for all conditions. β-Actin or α-tubulin was used as a loading control. Densitometry analyses were performed with blots from three independent experiments. Data are means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 by unpaired two-tailed Student’s t test; n = 3 independent experiments per group. N.S., not significant. The treatments were as follows. (C) Cells were treated without or with polyP120 at 4.16, 12.5, 37.5, and 112 μM, for 24 hours. (D) Cells treated without or with 37.5 μM polyPs (polyP8, polyP34, polyP94, or polyP120), for 24 hours. Negative controls in (C) and (D) were vehicle-treated cells. (E) Cells were treated without and with 10 μM MG-132 (proteasome inhibitor) alone or in combination with 37.5 μM polyP120 for 12, 24, and 48 hours. Negative controls: untreated cells and MG-132–treated cells. (F) Cells were treated without and with the protein synthesis inhibitor cycloheximide (CHX; 50 μg/ml), alone or in combination with 37.5 μM polyP120, for 12, 24, and 48 hours. Negative controls: untreated cells and CHX-treated cells. (G) Cells were transiently transfected with plasmids expressing WT ACE2 or mutant ACE2 protein (MUT: H378S, R393Q, H401S, and R514Q). Forty-eight hours later, the cells were then treated without or with 37.5 μM polyP120 for a further 24 hours. Negative controls: cells transfected with empty vector (EV) and vehicle-treated cells. For a longer exposure for ACE2 band density acquisition and the densitometric values for the EV condition, see fig. S2G. (H) Cells were transiently transfected with plasmids expressing WT RdRp or mutant RdRp protein (MUT: K4892S, K4937S, R4945Q, R5228Q, and K5241S). Forty-eight hours later, the cells were treated with 10 μM MG-132 alone or in combination with 37.5 μM polyP120 for a further 24 hours. Negative controls: cells transfected with EV, untreated cells, and MG-132–treated cells.
We then treated the primary human epithelial cells with increasing concentrations of polyP120 (4.16 to 112 μM) using double-distilled H2O as vehicle. Western blotting analysis demonstrated that polyP120 reduced the abundance of the ACE2 protein in these cells after 24 hours and in a dose-dependent manner (Fig. 2C). To exclude the possibility that the reduction in ACE2 abundance was due to any antiproliferative toxicity of polyP120, we tested lower concentrations of polyP120 (4.16 to 18.75 μM) on the primary human epithelial cells to determine any modulation of the proliferation rate (fig. S2C) and the amount of ACE2 (fig. S2D) according to the cell index and Western blotting analyses, respectively. This revealed a significant decrease in the ACE2 protein (P = 0.03) in the primary human epithelial cells treated with 18.75 μM polyP120 (fig. S2D), with no effects on the cell proliferation rate (that is, complete overlap of the growth curves at the three polyP120 concentrations, compared with the vehicle control; fig. S2C). These data suggested that the polyP120-induced reduction in ACE2 abundance was not dependent on any antiproliferative effects. With Vero E6 cells, the decrease in ACE2 abundance was dependent on the number of Pi residues in polyPs at 37.5 μM (Fig. 2D). We then tested for reduction of ACE2 abundance by 37.5 μM polyP120 in Vero E6 cells over time, as assessed by Western blotting analysis. This showed time-dependent decreases in ACE2 abundance after addition of 37.5 μM polyP120. A substantial enhancement in ACE2 degradation at 24 and 48 hours after treatment was observed (fig. S2E). Moreover, assays of caspase-3 activity did not show activation of the apoptotic cascade by polyP120 at concentrations up to 112 μM (fig. S2F).
On the basis of these findings, we choose 37.5 μM polyP120 as the best concentration (which was less than the IC50 values) to use for further testing of its effects on ACE2 degradation, given the time of treatment and its prolonged effects (primary human epithelial cells, 60% decrease in ACE2 abundance; Vero E6 cells, 70% decrease in ACE2 abundance) with no apparent toxicity. This concentration was selected to investigate the mechanism of action of polyP120 responsible for its antiviral activity. In human embryonic kidney (HEK) 293T cells treated with 37.5 μM polyP120, we observed greater accumulation of ACE2 at 24 hours after addition of the proteasome inhibitor MG-132 (10 μM) compared with that in the vehicle-treated control cells (Fig. 2E) and the cells treated with MG-132 alone. These data suggested that the polyP120-mediated reduction in ACE2 occurred through the proteasome. We also showed that upon addition of the protein synthesis inhibitor cycloheximide (CHX; 50 μg/ml), polyP120 reduced ACE2 abundance after 12 hours, compared with that in the cells treated with vehicle or CHX alone (Fig. 2F).
To further examine the region of interaction between ACE2 and polyP120 and thus to determine the amino acids responsible for interacting with polyPs, we performed site-directed mutagenesis of the ACE protein (Fig. 1B). HEK293T cells were transfected with plasmids that expressed wild-type (WT) ACE2 or ACE2 that was mutated at various residues, with conversion of the electrically charged (positive) side-chain amino acids [i.e., Arg (R) and His (H)] into polar, uncharged residues [Ser (S) and Gln (Q)]. We generated plasmids that expressed ACE2-encoding complementary DNAs (cDNAs) that contained four mutations, as mutated-(H378S, H401S, R393Q, or R514Q)-ACE2-FLAG_pcDNA3.1(+)-C-DYK, and used these constructs to transfect HEK293T cells. Forty-eight hours later, the cells were treated with 37.5 μM polyP120 for 24 hours. Western blotting analysis showed that polyP120 halved the ACE2 abundance, compared with that in cells transfected with the empty vector, whereas there was no change in the abundance of the mutant ACE2 (Fig. 2G; see also fig. S2G for long exposure).
Because the residues His378, Arg393, and His401 are located in highly structured regions in the binding pocket of the globular ACE2 protein [Protein Data Bank (PDB): 1R42], they were less accessible to partial hydrolysis by chymotrypsin (fig. S1, E and F). For this reason, we performed single mutational analysis of the ACE2 residues that were predicted to bind to polyP120 to dissect further their interactions. We thus generated ACE2-encoding cDNAs with four single mutations in the residues in ACE2 (i.e., H378S, H401S, R393Q, and R514Q) that were predicted to be sites for interaction between ACE2 and polyPs by the molecular docking analysis (Fig. 1B), and these plasmids were used to transfect HEK293T cells. Forty-eight hours later, the cells were treated with 37.5 μM polyP120 for a further 24 hours. Western blotting analysis showed that the single mutated ACE2 proteins (ACE2-H378S, ACE2-H401S, ACE2-R393Q, and ACE2-R514Q) were not reduced in abundance upon treatment with polyP120 (fig. S2H). Similarly, previous assays showed that polyP120-induced ACE2 degradation occurred only with WT ACE2 and not with the mutated ACE2 proteins (MUT: ACE2-H378S-H401S-R393Q-R514Q; Fig. 2G). This suggested that not only Arg514 but also other amino acid residues (i.e., His378, His401, and Arg393) were involved in the binding of ACE2 to polyP120. Together, these data suggested that polyP120 can bind to ACE2 in the region that comprised the residues His378, His401, Arg393, and Arg514, to thereby enhance its proteasome-dependent degradation.
We further tested whether polyP120 also enhanced proteasome-dependent degradation of the RdRp protein overexpressed in HEK293T cells. The cells were transfected with plasmids expressing the WT RdRp protein or RdRp with mutations, whereby amino acid residues with positively charged side chains were mutated to residues with uncharged or polar side chains. A plasmid that contained the RdRp-encoding cDNA with five mutations was generated [mutated-(K4892S, K4937S, R4945Q, R5228Q, and K5241S)-RdRp_pcDNA3.1 (+)-C-HA] and used to transfect HEK293T cells. Forty-eight hours later, the cells were treated with 37.5 μM polyP120 and 10 μM MG-132, alone or in combination, for an additional 24 hours. Western blotting analysis revealed that the abundance of the WT RdRp protein was reduced after 24 hours of treatment with polyP120 alone, whereas it accumulated in the presence of both polyP120 and MG-132 (Fig. 2H). In contrast, the amount of mutated RdRp was not modulated by polyP120 alone or in combination with MG-132 (Fig. 2H). Furthermore, addition of MG-132 alone did not alter RdRp protein abundance after 24 hours (Fig. 2H). Together, these data suggested that both ACE2 and RdRp are modulated intracellularly by polyP120 in a similar manner, which might be through a scaffold-like activity, as was previously reported for polyPs, and thus supported their degradation mediated by the proteasome.
Long-chain polyPs have antiviral actions against different strains of SARS-CoV-2
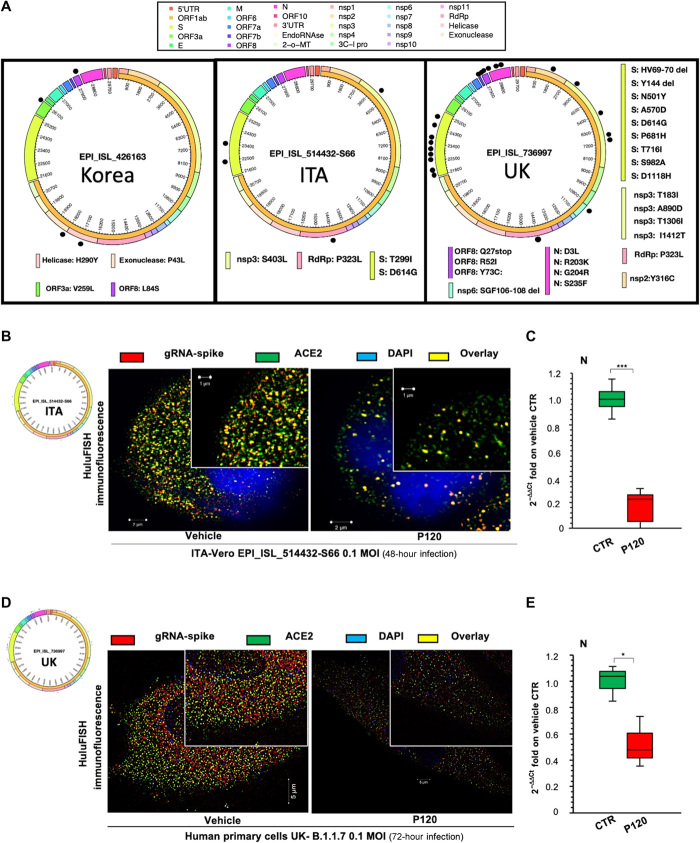
To investigate the antiviral effects of polyP120, we tested its efficacy in experiments with SARS-CoV-2 viral particles isolated from Italian [global initiative on sharing all influenza data (GISAID): code, EPI_ISL_514432-S66; GenBank: MT682732.1] and British (GISAID: code, EPI_ISL_736997) patients. Because different mutations have been identified among these strains (12), we sequenced the viral genomes from the isolated viruses and performed sequence analysis with alignment of the Korean (EPI_ISL_426163), Italian, and British strain sequences. The amino acid differences were plotted as a circular representation of the SARS-CoV-2 genome (with an in-house pipeline; see Materials and Methods) to generate a circus plot (Fig. 3A; for the list of missense variants, see Table 2). Overall, by Nextstrain sequence analysis (https://nextstrain.org/sars-cov-2/), we showed that the Korean strain (EPI_ISL_426163) belonged to the 19B clade, the Italian strain (EPI_ISL_514432-S66) belonged to the 20A clade, and the British strain (EPI_ISL_736997) belonged to the 20I/501Y.V1 (B.1.1.7) clade. These analyses revealed different variants between the isolated 20A clade (Italian) SARS-CoV-2 and the 19A clade (Korean) virus (RdRp: P323L; S: D614G, T299I). The P323L mutation in RdRp was associated with an increased mutation rate, which would appear to be due to an alteration in the proofreading that is finely regulated by the RdRp/nsp7/nsp8 complex (12). The D614G-producing variant in the gene encoding S (26) resulted in faster replication and transmission, thus enhancing the infectivity and competitive fitness of SARS-CoV-2. Furthermore, the 20I/501Y.V1 (B.1.1.7) clade (British variant) contained multiple mutations, including those in the S gene (for H69del, V70del, Y144del, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H; see Table 2). Together, these mutations in the B.1.1.7 variant are implicated in increased transmissibility (up to a 71% increase compared with previous circulating strains of SARS-CoV-2) and an increased risk of death (13, 26).
Fig. 3. polyP120 exerts antiviral action against SARS-CoV-2 variants belonging to 20A and 20I/501Y.V1 (B.1.1.7) clades.
(A) Circos plot showing missense variations among 19A clade (EPI_ISL_426163), 20A clade (EPI_ISL_514432-S66), and 20I/501Y.V1 (B.1.1.7) clade (EPI_ISL_736997) SARS-CoV-2 variants used in this work. Each black dot represents a missense mutation in the viral genomes. (B to D) HuluFISH analyses. (B and D) HuluFISH analysis with a pan–SARS-CoV-2 probe against the S gene (red) coupled to immunofluorescence (IF) staining with an antibody against the ACE2 protein (green). (B) Vero E6 cells were infected with Italian SARS-CoV-2 particles (MOI, 0.1) for 24 hours and then were treated with 37.5 μM polyP120 for additional 24 hours. (D) Primary human epithelial cells from nasal brushing were pretreated with 37.5 μM polyP120 for 1 hour and then infected with Italian SARS-CoV-2 particles (MOI, 0.1) for 72 hours. SARS-CoV-2–infected cells treated with vehicle (CTR) were used as negative controls in (B) and (D). The SIM image was acquired with Elyra 7 and processed with Zeiss ZEN software (blue edition). Magnification, ×63. (C and E) Quantification of mRNA abundance relative to that in the CTR (2−ΔΔCt) of genomic viral RNAs (N gene) from RT-PCR analysis of total RNA extracted from Vero E6 cells infected with Italian SARS-CoV-2 (EPI_ISL_514432-S66) (C) or primary human epithelial cells from nasal brushing infected with B.1.1.7 SARS-CoV-2 (UK) (EPI_ISL_736997) (E). SARS-CoV-2–infected cells treated with vehicle (CTR) were used as controls. Data are means ± SD. *P < 0.05 and ***P < 0.001 by unpaired two-tailed Student’s t test, adjusted with the Bonferroni method; n = 3 independent experiments per group. DAPI, 4′,6-diamidino-2-phenylindole.
Table 2. Sequence alignment analysis for genetic differences within the viral genomes from isolated 19A Korean (EPI_ISL_426163), 20A Italian (EPI_ISL_514432-S66), and 20I/501Y.V1 (B.1.1.7) British UK (EPI_ISL_736997) SARS-CoV-2 strains.
The missense mutations among the SARS-CoV-2 strains used in this study are listed (genomic and amino acid substitutions, and amino acid substitutions according to the nomenclature of the ORF1ab peptides).
| Viral genome |
Genomic
substitution |
Amino acid
substitution |
Amino acid
substitution (ORF1ab peptides) |
|
EPI_ISL_426163
(Korea) |
C17104T |
ORF1ab: H5614Y |
Helicase: H290Y |
| C18167T |
ORF1ab: P5968L |
Exonuclease: P43L |
|
| G26167T | ORF3a: V259L | ORF3a: V259L | |
| T28144C | ORF8: L84S | ORF8: L84S | |
|
EPI_ISL_514432
(Italy) |
C3927T |
ORF1ab: S1221L |
nsp3: S403L |
| C14408T | ORF1ab: P4715L | RdRp: P323L | |
| C22458T | S: T299I | S: T299I | |
| A23403G | S: D614G | S: D614G | |
|
EPI_ISL_736997
(United Kingdom) |
A1752G | ORF1ab: Y496C | nsp2: Y316C |
| C3267T | ORF1ab: T1001I | nsp3: T183I | |
| C5388A |
ORF1ab: A1708D |
nsp3: A890D | |
| C6636T | ORF1ab: T2124I | nsp3: T1306I | |
| T6954C | ORF1ab: I2230T | nsp3: I1412T | |
| C14408T | ORF1ab: P4715L | RdRp: P323L | |
| A23063T | S: N501Y | S: N501Y | |
| C23271A | S: A570D | S: A570D | |
| A23403G | S: D614G | S: D614G | |
| C23604A | S: P681H | S: P681H | |
| C23709T | S: T716I | S: T716I | |
| T24506G | S: S982A | S: S982A | |
| G24914C | S: D1118H | S: D1118H | |
| C27972T | ORF8: Q27stop | ORF8: Q27stop | |
| G28048T | ORF8: R52I | ORF8: R52I | |
| A28111G | ORF8: Y73C | ORF8: Y73C | |
| G28280C | N: D3L | N: D3L | |
| G28881A | N: R203K | N: R203K | |
| G28882A | N: G204R | N: G204R | |
| C28977T | N: S235F | N: S235F |
Here, we verified the antiviral action of polyP120 against these different SARS-CoV-2 strains. For this purpose, Vero E6 cells were infected with the Italian SARS-CoV-2 (EPI_ISL_514432-S66) at a multiplicity of infection (MOI) of 0.1 for 24 hours, which was followed by addition of 37.5 μM polyP120 for a further 24 hours (fig. S3A). Immunofluorescence (IF) coupled to fluorescence in situ hybridization in cells (HuluFISH) technology showed a reduction in both ACE2 on host cells and the viral gRNA-S gene (Fig. 3B and fig. S3B) in Vero E6 cells treated with polyP120, compared with vehicle-treated control cells. These data were confirmed by qRT-PCR analysis that showed statistically significant decreased viral N gene expression in Vero E6 cells infected with Italian SARS-CoV-2 and treated with polyP120 (Fig. 3C). Furthermore, similar results were obtained with the primary human epithelial cells pretreated with 37.5 μM polyP120 for 1 hour and then infected with B.1.1.7 SARS-CoV-2 (EPI_ISL_736997) at an MOI of 0.1 for 72 hours (Fig. 3, D and E, and fig. S3C). Together, these data showed the antiviral efficacy of 37.5 μM polyP120 against the 20A (Fig. 3, B and C) and B.1.1.7 variants as 78% (after 48 hours of infection) and 52% (after 72 hours of infection) inhibition of N gene transcription, respectively (Fig. 3, C and E).
The antiviral action of long-chain polyPs involves the expression of both gRNAs and sgRNAs
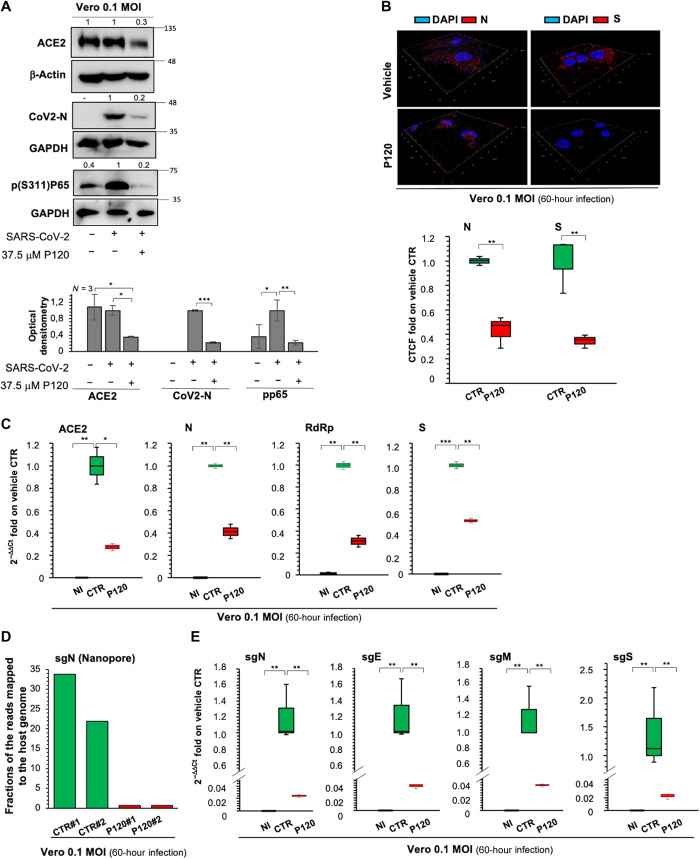
To dissect the antiviral action of polyP120, Vero E6 cells were infected with Italian SARS-CoV-2 (EPI_ISL_514432-S66) at an MOI of 0.1 for 24 hours, which was followed by the addition of 37.5 μM polyP120 for a further 36 hours (fig. S3D). Western blotting (Fig. 4A) and IF analysis (Fig. 4B) showed reduced amounts of the viral N and S proteins in polyP120-treated cells (Fig. 4B and fig. S3E). These data also showed inhibition of the inflammatory nuclear factor κB (NF-κB) pathway in the cells infected with SARS-CoV-2 and treated with polyP120, as demonstrated by reduced amounts of phosphorylated (Ser311)-p65 protein (Fig. 4A).
Fig. 4. Antiviral action of polyP120 in Vero E6 cells.
(A) Top: Representative Western blotting analysis (with antibodies against the indicated proteins) of Vero E6 cells (8 × 105) infected with Italian SARS-CoV-2 viral particles (MOI, 0.1) for 24 hours and then treated with 37.5 μM polyP120 or vehicle for a further 36 hours. All experiments were performed in triplicate. Bottom: Densitometry analysis of the indicated band intensities on blots from three independent experiments. Data are means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 by unpaired two-tailed Student’s t test; n = 3 independent experiments per group. Negative controls: noninfected (NI) cells and SARS-CoV-2–infected cells treated with vehicle (CTR). GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) Representative three-dimensional IF reconstruction (top) and its quantification (bottom) with antibodies against the SARS-CoV-2 N and S proteins for cells treated as described for (A). Magnification, ×63. Fluorescence intensity was measured in each cell and compared with that in the vehicle control. More than 50 cells were counted. Quantification is presented as the corrected total cell fluorescence (CTCF) intensity. (C) Quantification of mRNA abundance relative to that in CTR cells (2−ΔΔCt) for ACE2 and genomic viral RNAs (N, RdRp, and S) from RT-PCR analysis with SYBR Green of total RNA extracted from cells treated as described for (A). Noninfected and SARS-CoV-2–infected cells treated with vehicle (CTR) were used as controls. Data are means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 by unpaired two-tailed Student’s t test, adjusted with the Bonferroni method; n = 3 independent experiments per group. (D) Quantification of direct RNA sequencing reads (through Nanopore technology) assigned to the sgN transcript expressed as fractions of reads mapped to the host genome. Two independent experiments are shown (#1 and #2; SRA data accession: PRJNA688696). (E) Quantification of mRNA abundance relative to that in CTR cells (2−ΔΔCt) of subgenomic (sg) viral RNAs from the RT-PCR analysis with SYBR Green of cells treated as described for (A). Noninfected cells and SARS-CoV-2–infected cells treated with vehicle (CTR) were used as controls. Data are means ± SD. **P < 0.01 by unpaired two-tailed Student’s t test, adjusted with the Bonferroni method; n = 3 independent experiments per group.
In addition, there were reductions in ACE2 abundance at both the protein (Fig. 4A) and mRNA (Fig. 4C) levels upon treatment of SARS-CoV-2–infected cells with polyP120. As observed here in Vero E6 cells and as reported by others (27), ACE2 mRNA abundance was increased in SARS-CoV-2–infected cells compared with that in noninfected cells (Fig. 4C), with a trend toward increased expression with greater viral load (MOI range, 0.001 to 0.01; fig. S3F). This was not seen at the protein level when noninfected Vero E6 cells were compared with SARS-CoV-2–infected cells at an MOI of 0.1 (Fig. 4A), which was consistent with a previous report (27). The antiviral activity of polyP120 was further validated by RT-PCR analysis to determine the extent of expression the viral genes N, RdRp, and S (Fig. 4C). Note that we observed reductions in the abundance of both RdRp RNA (Fig. 4C) and the RdRp protein (Fig. 2H). One study showed that polyPs interfere with the ACE2-S complex by binding to the RBD on the surface of the S protein (23). Note that similar antiviral activities were observed for polyPs of longer chain lengths (e.g., polyP126 and polyP189) on the expression of the viral N gene (fig. S3, G and H). There were no significant differences in the decreased expression of the viral N gene fragments seen for these different chain-length polyPs (fig. S3H).
To further dissect the antiviral activity mediated by polyP120 through RdRp, we focused on its discontinuous transcription, taking into account the four structural sgRNAs (i.e., sgS, sgE, sgM, and sgN). For this purpose, SARS-CoV-2–infected Vero E6 cells (MOI, 0.1) were treated with polyP120 and then analyzed for transcription of these sgRNAs (fig. S3D). Nanopore technology and direct RNA sequencing of the RNAs extracted after the infection of the Vero E6 cells with SARS-CoV-2 showed decreased amounts of the sgN transcripts in the polyP120-treated cells in two independent experiments (Fig. 4D; Sequence Read Archive (SRA)–BioProject, PRJNA688696). Furthermore, through RT-PCR analysis with SYBR Green, we also confirmed the decreased expression of sgN, sgE, sgM, and sgS (Fig. 4E). Together, these data indicated that polyP120 had an antiviral effect on SARS-CoV-2–infected Vero E6 cells through reduced abundance of ACE2 on the host cells, and the consequent reduction in the amounts of both the protein and RNA of the viral factors, including their subgenomic transcripts.
PolyP120 decreases COVID-19–associated release of cytokines in human cell lines
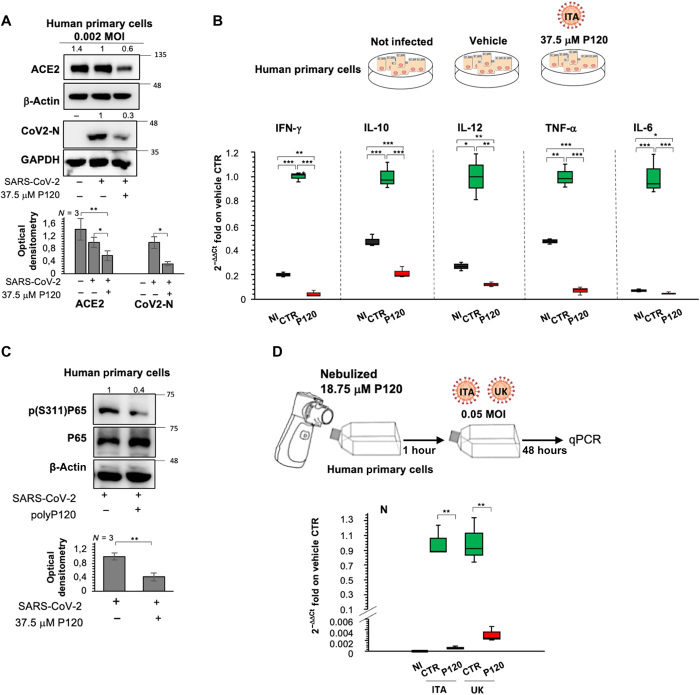
We also tested the effectiveness of polyP120 in SARS-CoV-2–infected primary human epithelial cells (obtained from nasal brushing). Twenty-four hours after infection of primary human epithelial cells with the Italian SARS-CoV-2 (EPI_ISL_514432-S66) (MOI, 0.002), the cells were treated for an additional 36 hours with 37.5 μM polyP120 (fig. S4A). Western blotting analysis showed reductions in the amounts of the ACE2 and N proteins in the polyP120-treated primary human epithelial cells (Fig. 5A). This treatment also resulted in decreased expression of various viral genes (S, RdRp, and N), as well as of ACE2 (fig. S4B).
Fig. 5. Antiviral actions of polyP120 in human cell lines result in decreased cytokine production.
(A) Top: Representative Western blotting analysis (using antibodies against the indicated proteins) of human epithelial cells (4.25 × 103) infected with Italian SARS-CoV-2 (MOI, 0.002) for 24 hours and then treated with 37.5 μM polyP120 or vehicle for a further 36 hours. All experiments were performed in triplicate. Bottom: Densitometry analysis of the indicated band intensities on blots from three independent experiments. Data are means ± SD. *P < 0.05 and **P < 0.01 by unpaired two-tailed Student’s t test; n = 3 independent experiments per group. Negative controls: noninfected cells and SARS-CoV-2–infected cells treated with vehicle (CTR). (B) Quantification of mRNA abundance relative to that in CTR cells (2−ΔΔCt) for IFN-γ, IL-10, IL-12, tumor necrosis factor–α (TNF-α), and IL-6 from RT-PCR analysis with SYBR Green of RNA extracted from cells treated as described for (A). Noninfected cells and SARS-CoV-2–infected cells treated with vehicle (CTR) were used as controls. Data are means ± SD. *P < 0.05, **P < 0.01, and ***P < 0.001 by unpaired two-tailed Student’s t test, adjusted with the Bonferroni method; n = 3 independent experiments per group. (C) Top: Representative Western blotting of the primary human epithelial cells from nasal brushing treated as described for (A) with antibodies against the indicated proteins. All experiments were performed in triplicate. Bottom: Densitometry analysis of the indicated band intensities on blots from three independent experiments. Data are means ± SD. **P < 0.01 by unpaired two-tailed Student’s t test; n = 3 independent experiments per group. (D) Top: Experimental plan. Primary human epithelial cells from nasal brushing were plated in flasks and treated with nebulized 18.75 μM polyP120. After 1 hour, the cells were infected with 20A (EPI_ISL_514432-S66) or 20I/501Y.V1 (B.1.1.7) (UK) (EPI_ISL_736997) SARS-CoV-2 viral particles (MOI, 0.05), and noninfected cells were used as the negative control for infection. After 48 hours, the cells were lysed and their RNA was extracted. Bottom: Quantification of mRNA abundance relative to that in CTR cells (2−ΔΔCt) of N from RT-PCR analysis with SYBR Green. Noninfected cells and SARS-CoV-2–infected cells treated with vehicle (CTR) were used as controls. Data are means ± SD. **P < 0.01 by unpaired two-tailed Student’s t test, adjusted with the Bonferroni method; n = 3 independent experiments per group. qPCR, quantitative PCR.
Patients with COVID-19 show increased circulating amounts of inflammatory cytokines, including interferon-γ (IFN-γ), interleukin-6 (IL-6), IL-10, and IL-12 (28). Because the polyPs have been described to have anti-inflammatory functions (16), we investigated the potential modulation by polyP120 of these cytokines in the primary human epithelial cells infected with SARS-CoV-2 (fig. S4A). In SARS-CoV-2–infected cells after treatment with polyP120, RT-PCR analysis showed significant reductions in the amounts of mRNAs encoding IFN-γ, IL-6, IL-10, IL-12, and tumor necrosis factor–α (Fig. 5B). This anti-inflammatory action of polyP120 might be ascribed to its inhibition of the NF-κB pathway, as was demonstrated here by its inhibition of p65 phosphorylation in Vero E6 cells (Fig. 4A) and in the primary human epithelial cells infected with SARS-CoV-2 (Fig. 5C). To determine whether this reduced expression of genes encoding inflammatory cytokines in polyP120-treated cells was due to the ability of polyP120 to block viral entry or anti-inflammatory effects, we treated the noninfected primary human epithelial cells with 37.5 μM polyP120 for 48 hours. RT-PCR analyses showed reduction in the expression of genes encoding all of the cytokines tested in the polyP120-treated cells (fig. S4C). These data suggested that this anti-inflammatory action of polyP120 was independent of viral infection. Because polyPs are associated with inflammatory pathways that are mainly regulated by NF-κB (29) and mechanistic target of rapamycin kinase (mTOR) (30), we hypothesized that this decreased expression of cytokine-encoding genes can be ascribed to modulation of the inflammatory cascades by polyP120 in autocrine and paracrine manners.
In vitro pretreatment of primary human epithelial cells and colorectal cancer cells with polyP120 results in antiviral activity
To further confirm whether pretreatment with longer-chain polyPs (e.g., polyP120 and polyP126) had any antiviral activities, we pretreated the primary human epithelial cells for 30 min with polyP120 or polyP126 and then infected the cells with SARS-CoV-2 at different viral loads (MOIs, 0.01 and 0.16) for 12 hours, which was followed by analysis of the expression of viral N gene fragments (fig. S4, D to F). Here, polyP126 showed antiviral activity, although that of polyP120 was significantly greater (fig. S4, E and F). Together, these data suggested that polyP120 and polyP126 had antiviral effects in the primary human epithelial cells through inhibition of N gene transcription.
The antiviral actions exerted by pretreatment with polyPs were also confirmed in SARS-CoV-2–infected Caco2 cells, a human colorectal carcinoma cell line. The antiproliferative effect of polyP120 was measured in Caco2 cells, which showed an IC50 value of 68.91 μM (R2, 0.99) and a marginally decreased proliferation rate at 37.5 μM (fig. S5A). The polyP120-treated Caco2 cells (37.5 μM, 24 hours) also showed reduced amounts of ACE2 (fig. S5B). Furthermore, for these SARS-CoV-2–infected cells (36 hours of infection), pretreatment for 8 hours with polyP120 significantly decreased expression of the viral N and RdRp genes (fig. S5, C and D) and reduced the amount of ACE2 mRNA (fig. S5D). The antiviral efficacy of the pretreatment with 37.5 μM polyP120 for 30 min was also demonstrated in SARS-CoV-2–infected Vero E6 cells, which showed reduced expression of viral N gene fragments (fig. S5, E and F). These data suggested that the mechanism of action of polyP120 was effective across different cell types. Together, the data presented here enhance our understanding of the potential use of polyP120 in early phases of SARS-CoV-2 infection, to reduce viral replication and infectivity.
Delivery of polyP120 through a nonambulatory nebulizer system results in antiviral effects
Because of the respiratory failure experienced by patients with COVID-19, new drug formulations that are appropriate for aerosol inhalation, and thus more direct delivery to the lungs, represent an unmet medical need. For this reason, we tested the delivery of polyP120 into primary human epithelial cells through a nebulizer system. Because 18.75 μM polyP120 showed antiviral actions (Fig. 1A) and reduced the abundance of ACE2 (fig. S2D) without any antiproliferative effects (fig. S2C), we used this concentration of polyP120. After removal of the medium from attached primary human epithelial cells from nasal brushing in tissue culture flasks, nebulized 18.75 μM polyP120 was directed into the flasks for 2 min (condensation rate, 1 ml/min) (Fig. 5D and movie S1). After this treatment, the medium was added back to the cells, and the cells were infected with 20A and 20I/501Y.V1 (B.1.1.7) SARS-CoV-2 at an MOI of 0.05 for 48 hours. RT-PCR analysis showed reduced expression of the viral N gene in these polyP120-treated cells (Fig. 5D). Overall, in these cells pretreated with nebulized polyP120 and infected with the 20A and 20I/501Y.V1 (B.1.1.7) SARS-CoV-2 variants, qRT-PCR analysis demonstrated inhibition of N gene transcription of 99.95 and 99.80%, respectively (Fig. 5D). This result indicated that polyP120 delivered in this aerosol formulation at 18.75 μM maintained its antiviral actions against infection by different strains of SARS-CoV-2.
DISCUSSION
Here, we showed that long-chain polyPs (e.g., polyP120; with 120 Pi units) can inhibit the expression of SARS-CoV-2 viral genes (as genomic and subgenomic transcripts). Through their targeting of ACE2 and RdRp, long-chain polyPs impaired replication of the virus in host cells and enhanced proteasome-mediated degradation of ACE2 and RdRp. We also described the mechanisms of action behind this antiviral function of polyPs (Fig. 6). The first mechanism involved time- and dose-dependent proteasome-mediated degradation of ACE2 in the presence of long-chain polyPs. This appeared to be due to binding of polyPs to ACE2, which is likely mediated by four amino acid residues of ACE2 that are also conserved across different species: Arg514, His401, His378, and Arg393. The presence of these conserved amino acids in the human ACE2 protein (Fig. 1B) suggested that inhibition of SARS-CoV-2 infection and replication by polyP120 potentially occurs in other infected species and may be exploited for preventive therapeutic applications.
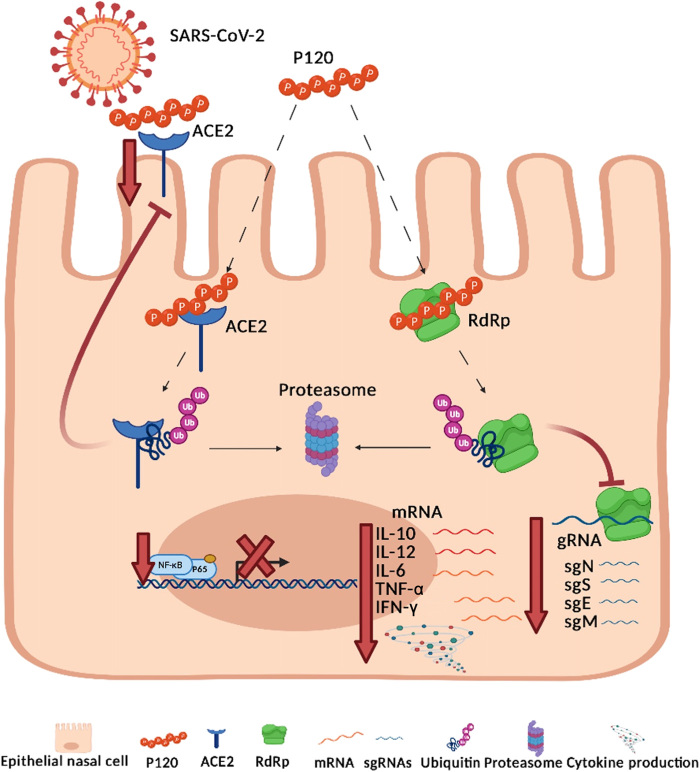
Fig. 6. Antiviral mechanisms of action of polyP120.
Cartoon representation to illustrate our hypothesis for the antiviral actions of polyP120 in primary human epithelial cells. PolyP120 can stimulate the proteasome-mediated degradation of ACE2 and RdRp, thus reducing the expression of the SARS-CoV-2 gRNAs and sgRNAs. Thus, polyP120 can act extracellularly through direct interactions with the host cell ACE2, which is the receptor for the S protein of SARS-CoV-2, and intracellularly through its interactions with the RdRp protein of SARS-CoV-2. PolyP120 also led to the reduced expression of the genes encoding IL-10, IL-12, IL-6, IFN-γ, and TNF-α by decreasing the phosphorylation of the NF-κB protein p65. These cytokines are typically responsible for the cytokine storm observed in patients with COVID-19.
Another study showed that polyPs (as a 40-Pi polymer) can impair the interaction between the S protein RBD and ACE2 in vitro (21). Although these findings need to be further confirmed, especially because of the emerging variants of the S protein (British-UK B1.1.7; B.1.351 and P.1 variants), these data highlight the potential for the use of polyPs in vivo to limit viral entry into human host cells. The results presented here that show that polyP120 inhibits replication of the UK B1.1.7 variant in vitro suggest that therapeutic applications of polyP120 should be explored further. We also showed that SARS-CoV-2–infected cells had reduced ACE2 mRNA abundance after treatment with polyP120. We suggest that this is because the expression of ACE2 is regulated by the transcription factor HNF1 Homeobox A (HNF1 alpha) (31) and epigenetically by Sirtuin 1 (SIRT1) (32). The role of polyPs in transcriptional regulation processes will be investigated in future studies.
Note that in directing any treatment at ACE2 as the receptor for SARS-CoV-2 entry into cells, the use of ACE inhibitors has been discouraged for patients with COVID-19. This is due to the consequent increase in the abundance of ACE2 mRNA, which might then facilitate the engagement and entry of SARS-CoV-2 into cells (33). However, the future use of polyPs might indeed overcome this side effect of the use of ACE inhibitors. Thus, the combination of ACE inhibitors with polyPs can be postulated additionally for the treatment of patients with COVID-19. The second potential mechanism of action for polyPs involves inhibition of RdRp (Fig. 6). PolyPs appear to bind to the SARS-CoV-2 RdRp protein and to drive its proteasome-mediated degradation. This binding appeared to occur through five residues within the RdRp RNA binding site (Lys4892, Lys4937, Arg4945, Arg5228, and Lys5241). PolyP120 appeared to decrease the expression of the main SARS-CoV-2 genes through this mechanism. Note also that transcription of the main SARS-CoV-2 sgRNAs was also impaired (e.g., sgN). Thus, we suggest that polyPs reduce active viral replication (34). This antiviral action was also seen with mutations in the S gene, which enhanced viral infectivity and fitness; this further supports the potential for the use of polyPs to inhibit different emerging variants of SARS-CoV-2. Furthermore, analysis of the BNT162b2 vaccine (Pfizer) showed that immune responses are elicited that can potently neutralize SARS-CoV-2; thus, the emerging variants with mutations in the S gene (e.g., B.1.1.7, B1.135.1, and P1) are likely to be highly resistant to antibody neutralization because of partial escape for humoral immunity, whereas S-specific CD4+ T cell activation is not affected by the mutations in the variants (35–37). Here, we showed that throughout our model of action (Fig. 6), polyP120 reduced the amounts of ACE2 and RdRp through proteasome-mediated degradation. Thus, this antiviral activity is independent of the acquired mutations in the RBD S variants. We thus suggest that patients infected with these emerging variants might benefit from treatment with polyP120 in an aerosol formulation. Furthermore, the amino acid residues in RdRp that are involved in binding to polyPs have been conserved through evolution in different species, and thus, they might also be conserved in other positive-sense, single-stranded RNA viruses from other hosts (e.g., Nidovirales, as listed at www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?taxid=76804). These findings open the way to examine the potential use of polyPs against other viral infections and thus for investigations into other mechanisms of action in which polyPs might be involved.
We note that based on in silico analyses with RASMOT 3D-PRO (available at http://biodev.cea.fr/rasmot3d/) and combined with other bioinformatics tools, including HHpred (https://toolkit.tuebingen.mpg.de/tools/hhpred), HMMScan (www.ebi.ac.uk/Tools/hmmer/search/hmmscan), DALI (http://ekhidna2.biocenter.helsinki.fi/dali/), PSI-Blast, (https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE=Proteins&PROGRAM=blastp&RUN_PSIBLAST=on), and FATCAT (https://fatcat.godziklab.org/fatcat-cgi/cgi/fatcat.pl?-func=search), there were 25 human proteins that contained three-dimensional (3D) motifs of the four specific residues identified in the ACE2 protein (i.e., H378, R393, H401, and R514; see table S3). Among the proteins that we identified, there were three with fully conserved 3D motifs of these amino acids, which we identified by the gene names LIG3, DOT1L, and SOD1. The protein products of the first two of these genes are DNA binding proteins, and thus, they might have stronger affinities for negatively charged molecules, such as polyPs. The third, SOD1, is active in mitochondria, where it destroys free radicals and reactive oxygen species and where its metabolism is regulated by polyPs (38). We will address the hypothesis of a potential modulation of these proteins driven by polyP120 (e.g., via proteasome-dependent degradation) in the near future.
Here, we demonstrated that increasing polyP120 concentrations showed antiproliferative effects on primary human epithelial cells. This action might correlate with the modulation of proliferative pathways (e.g., the mitogen-activated protein kinase cascade through extracellular signal–regulated kinase), which has been reported for polyPs (39). PolyPs are also key modulators of inflammatory pathways, with actions mainly through the NF-κB cascade (29). Here, we showed that through its effects on the NF-κB cascade, polyP120 decreased the abundances of mRNAs encoding the main cytokines (IFN-γ, IL-6, IL-10, and IL-12) that are part of the cytokine storm often seen in patients with COVID-19 (28). The mechanism of action of polyP120 in modulation of NF-κB signaling will be a topic of future studies. Among these cytokines, IL-6 amounts in patients with COVID-19 are substantially increased and are associated with adverse clinical outcomes. Whereas inhibition of IL-6 with tocilizumab appears to be effective and safe in preliminary investigations, the data from ongoing clinical trials need to be evaluated to better define the role of tocilizumab in COVID-19 and its potential for routine clinical application (40). Furthermore, in COVID-19 infections, microcapillary clotting in the lungs generates acute pulmonary embolisms, which impair O2 acquisition by the lungs, thus resulting in insufficient respiratory function. Of importance, low–molecular weight heparin given to patients reduces their pain, improves their clinical status, reduces their IL-6 release, and stabilizes their oxygen exchange (41). At this time, however, it is not clear whether polyPs can suppress the complement protein by the terminal pathway (17) and enhance clot formation. For these reasons, caution should be taken in the treatment of late-stage disease patients with polyPs.
Asymptomatic and symptomatic SARS-CoV-2–infected patients with comorbidities can use immune system components to fight the virus, where memory CD4+ T cells have been shown to have similar numbers and quality between the two groups (42, 43). Preexisting T cell immunity to SARS-CoV-2 might be relevant also because it might influence COVID-19 disease severity. It is possible that patients with high numbers of preexisting memory CD4+ T cells that recognize SARS-CoV-2 can mount a faster and stronger immune response upon exposure to SARS-CoV-2 and thereby limit disease severity. At this time, we showed that polyP120 interferes with the amounts of the cytokines produced. Thus, potential control of immune cell activation, including T cells and memory T cells, can be hypothesized. These will be issues for future studies.
Patients with COVID-19 suffer with early respiratory failure. Thus, new drugs formulated as aerosols might represent a promising therapeutic approach. Here, we showed that the efficacy of polyP120 was maintained when nebulized at the concentration of 18.75 μM for delivery to cells in vitro. PolyP120 still showed antiviral activity against different emerging SARS-CoV-2 strains [i.e., 20A and 20I/501Y.V1 (B.1.1.7) clade emerging variants] in infected primary human epithelial cells. Thus, we envisage potential aerosol delivery of polyP120 for treatment of patients with respiratory failure. The final formulation needs to be determined, although there is a window of opportunity at 18.75 μM for antiviral, noncytotoxic activity. On the basis of this, it will be important to determine a safe dose of polyP120 that might serve in humans for prophylactic use as an anti–SARS-CoV-2 treatment.
In conclusion, our data showed that pretreatment of cells with long-chain polyPs impaired SARS-CoV-2 infection. In this regard, the European panel on Food Additives and Flavourings has already provided information about the safety of phosphates as food additives (e.g., E450–452) (24). To date, among the drugs tested on patients with COVID-19, the safety and antiviral activity of remdesivir (GS-5734) is emerging in two completed phase 3 clinical trials (i.e., NCT04292899 and NCT04292730). Assuming that polyP120 lacks toxicity and given its role in enhancing proteasomal degradation of RdRp as presented here, we hypothesize that polyP120 might be used in clinics in the early phases of COVID-19 disease, alone or in combination with inhibitors of RdRp-mediated RNA replication and transcription. These analyses open a scenario in which polyPs might act through additional mechanisms that are involved in cellular processes that use the pool of available adenosine 5′-triphosphate (produced by a source of Pi) to modulate protein degradation mediated by the proteasome (44). This enhancement by polyPs might provide overall modulation of responses of virus-host interactions and viral replication. These activities might be of importance for preventing infections and progression of SARS-CoV-2 and other viruses, thus underlining the importance of polyPs and their substantial antiviral functions.
MATERIALS AND METHODS
Ethical committee approval
Ethical committee approvals for the COVID-19 samples use in this study were as follows: (i) protocol no. 141/20; date: 10 April 2020, CEINGE TaskForce Covid19; Azienda Ospedaliera Universitaria Federico II, Direzione Sanitaria, protocol no. 000576 of 10 April 2020; (ii) protocol no. 157/20; date: 22 April 2020, GENECOVID, the experimental procedures within the use of SAR-CoV-2 in a biosafety level 3 (BSL3) laboratory were authorized by Ministero della Sanità and Dipartimento Di Medicina Molecolare e Biotecnologie Mediche, Università degli Studi di Napoli Federico II and Azienda Ospedaliera Universitaria Federico II, Direzione Sanitaria protocol no. 0007133 of 08 May 2020; (iii) protocol no. 18/20; date: 10 June 2020, Genetics CEINGE TaskForce Covid19; Azienda Ospedaliera Universitaria Federico II, Direzione Sanitaria protocol no. 000576 of 10 April 2020.
Cell culture
Vero E6 cells and HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; 41966-029, Gibco) with 10% fetal bovine serum (FBS; 10270-106, Gibco), 2 mM l-glutamine (25030-024, Gibco), and 1% penicillin/streptomycin (P0781, Sigma-Aldrich). Freshly isolated human nasal epithelial cells were collected by nasal brushing of healthy donors [as previously described (45)]. These primary human epithelial cells were cultured in PneumaCult (no. 05009; STEMCELL Technologies) with 2 mM l-glutamine (25030-024, Gibco), and 1% penicillin/streptomycin (P0781, Sigma-Aldrich). Caco2 cells were cultured in Eagle’s minimum essential medium (M2279, Sigma-Aldrich) with 10% FBS (10270-106, Gibco), 2 mM l-glutamine (25030-024, Gibco), and 1% penicillin/streptomycin (P0781, Sigma-Aldrich). For proteasome inhibition, the cells were incubated in DMEM, with 10% FBS containing 10 μM MG-132 (M8699, Sigma-Aldrich) for 24 hours. For inhibition of protein synthesis, the cells were incubated in DMEM, with 10% FBS containing CHX (50 μg/ml; C7698, Sigma-Aldrich) for 24 hours. After each treatment, the cells were analyzed by Western blotting. HEK293T cells were transiently transfected with the following plasmid DNA constructs: WT-ACE2-FLAG_pcDNA3.1(+)-C-DYK (GenScript); mutated-(H378S, H401S, R393Q, R514Q)-ACE2-FLAG_pcDNA3.1(+)-C-DYK (GenScript); WT-ACE2- pCEP4-myc (Addgene); mutated-(H378S)-ACE2_pCEP4-myc; mutated-(H401S)-ACE2_pCEP4-myc; mutated-(R393Q)-ACE2_pCEP4-myc; mutated-(R514Q)-ACE2_pCEP4-myc; WT-RdRp_pcDNA3.1(+)-C-HA (GenScript); and mutated-(K4892S, K4937S, R4945Q, R5228Q, K5241S)-RdRp_pcDNA3.1(+)-C-HA (GenScript). Transient transfections were performed with HilyMaxTransfection Reagent (H357-10, Dojindo), according to the manufacturer’s instructions. Forty-eight hours after transfection, the HEK293T cells were treated with polyp120, vehicle, and/or 10 μM MG-132 for an additional 24 hours before being harvested for Western blotting analysis.
In vitro treatments and infections
SARS-CoV-2 was isolated from patient samples as follows: Nasopharyngeal swabs in 2 ml of viral transport medium were collected for molecular diagnosis and frozen. Confirmed PCR-positive specimens were aliquoted and refrozen until virus isolation was initiated. Vero E6 cells were used for virus isolation from nasopharyngeal swabs. Vero E6 cells (8 × 105) were trypsinized and resuspended in DMEM, with 2% FBS in T25 flasks to which 100 μl of the clinical specimen was added. The inoculated cultures were grown in a humidified 37°C incubator with 5% CO2 and observed for cytopathic effects daily. When cytopathic effects were observed (7 days after infection), the cell monolayers were scrapped with the back of a pipette tip. The cell culture supernatant containing the viral particles was aliquoted (100 μl) and immediately frozen at −80°C. Viral lysates were used for total nucleic acid extraction for confirmatory testing and sequencing. Cells were pretreated with polyPs before infection, as follows. For Vero E6 and primary human epithelial cells, these were plated in T25 flasks for treatment with 37.5 μM polyPs (i.e., polyP120 and polyP126). After 30 min (for Vero E6 and primary human epithelial cells), without changing the cell culture medium containing polyPs, these pretreated cells were infected with viral particles that belonged to the 20A and 20I/501Y.V1 (B.1.1.7) clades. Uninfected cells were used as the negative control. After 12, 24, 60, or 72 hours of infection, the cells were lysed, and their RNA was extracted. Vehicle-treated cells were used as the negative control for this treatment. These experiments were performed in a BSL3-authorized laboratory. For Caco2 cells, these were plated in T25 flasks for treatment with 37.5 μM polyP120. After 8 hours, without changing the cell culture medium containing polyP120, these pretreated cells were infected with SARS-CoV-2 viral particles obtained from a frozen swab from an Italian patient positive for COVID-19. Uninfected cells were used as the negative control. After 36 hours, the cells were lysed, and their RNA was extracted. Vehicle-treated cells were used as the negative control for this treatment. These experiments were performed in a BSL3-authorized laboratory. Treatment of cells with polyPs after infection with SARS-CoV-2 viral particles from an Italian patient positive for COVID-19 was performed as follows. Vero E6 or primary human epithelial cells were plated in T75 flasks for infection with viral particles belonging to the 20A and 20I/501Y.V1 (B.1.1.7) clades. Uninfected cells were used as the negative infection control. After 24 hours, the infected cells were treated with polyPs (37.5 μM). Vehicle-treated cells were used as the negative treatment control. After a further 36 hours, the cells were lysed, and their RNA was extracted. These experiments were performed in a BSL3-authorized laboratory. Treatment of cells with polyPs after infection with SARS-CoV-2 viral particles from a Korean patient positive for COVID-19 was performed as follows. Vero E6 cells (4 × 105) were plated in six-well plates for infection with SARS-CoV-2 viral particles, which were obtained from a frozen swab from Korean patients that had tested positive for COVID-19. Uninfected cells were used as the negative infection control. After 24 hours, the infected cells were treated with polyPs of different chain lengths (polyP8, polyP16, polyP64, polyP94, and polyP120) at different concentrations (9.38, 18.75, 37.5, 150, and 300 μM). Vehicle-treated cells were used as the negative treatment control. After a further 24 hours, the cell culture supernatant was used to extract RNA. Analysis by qRT-PCR was performed with primer-probe sets targeting the RdRp region of SARS-CoV-19 virus (AccuPower SARS-CoV-2 Real-Time RT-PCR kits, Bioneer). The quantity of viral RNA cycle threshold (ct) was also expressed as PFU equivalents (fig. S1B). The IC50 for polyP120 was calculated through nonlinear regression analysis performed with GraphPad Prism 9 {[inhibitor] versus response (three parameters)}. N = 3 independent experiments per group. For treatment with nebulized polyP120, primary human epithelial cells (6 × 105) were plated in a T25 flask. The polyP120 (18.75 μM) was dissolved in 2 ml of water and nebulized for 2 min with a nebulizer system (mesh nebulizer; YM–252; input, 5 V/1 A; oscillation frequency, 110 kHz; atomized particles, <5 μM; condensation rate, 1 ml/min). After removal of the medium from the flask, the nebulized polyP120 was directed into the flask. The medium was then replaced after the nebulizing of the polyP120. Those cells treated with nebulized polyP120 were then infected with viral particles belonging to 20A and 20I/501Y.V1 (B.1.1.7) clades, for 48 hours.
RT-PCR assays
N1-3 detection was performed as follows. RNA samples were extracted with TRIzol RNA Isolation Reagent (no. 15596018, Ambion, Thermo Fisher Scientific) according to the manufacturer’s instructions. RT-PCR was performed with quanty COVID-19 kits (ref. RT-25; Clonit; US Food and Drug Administration “in vitro diagnostic”–approved). These kits enable specific quantitative detection of the N1, N2, and N3 fragments (from the SARS-CoV-2 N gene) using differentially labeled target probes. These runs were performed on a PCR machine (CFX96, Bio-Rad) under the following conditions: 25°C for 2 min, 50°C for 15 min, and 95°C for 2 min [95°C for 3 s and 55°C for 30 s (×45 cycles)]. The quantification cycle (Cq) values of N1, N2, and N3 are reported as means ± SD normalized to the internal control detected with the Clonit quanty COVID-19 kits. Specific quantitative detection of ACE2, N1, RdRp, S, sgM, sgE, sgN, and sgS was performed as follows. RNA samples were extracted with TRIzol RNA Isolation Reagent according to the manufacturer’s instructions. Reverse transcription was performed with 5X All-In-One RT MasterMix (catalog no. g486, ABM) according to the manufacturer’s instructions. The cDNA preparation was through the cycling method, as follows: incubation of the complete reaction mix at 25°C for 5 min, at 42°C for 30 min, and at 85°C for 5 min and then held at 4°C. The reverse transcription products (cDNA) were amplified by qRT-PCR using an RT-PCR system (7900, Applied Biosystems, Foster City, CA, USA). The relative expression of the target genes was determined using the 2−ΔΔCt method as the fold increase compared with the vehicle-treated SARS-CoV-2–infected control. The data are presented as means ± SD of the 2−ΔΔCt values of two or three replicates. The target genes were detected with a BrightGreen 2X qRT-PCR MasterMix Low-ROX (catalog no. MasterMix-LR, ABM). ACTB was used as the housekeeping gene (forward primer: 5′-GACCCAGATCATGTTTGAGACCTT-3′; reverse primer: 5′-CCAGAGGCGTACAGGGATAGC-3). The details of the primers used in these assays are provided in table S4.
Viral genome sequencing
For library construction, RNA samples were obtained from Vero E6 cells infected by 19A (Korean), 20A (Italian), or 20I/501Y.V1 (B.1.1.7) (British UK) SARS-CoV-2. After performing quality control, qualified samples proceeded to library construction with TruSeq Stranded Total RNA with Ribo-Zero H/M/R Gold. Samples were sequenced with the Illumina platform (type of read: paired end). To map cDNA fragments obtained from RNA sequencing, NC_045512.2 was used as the reference genome. Circos plot generation was performed as follows. Amino acid changes in the 19A Korean (EPI_ISL_426163), 20A Italian (EPI_ISL_514432-S66), and 20I/501Y.V1 (B.1.1.7) British UK (EPI_ISL_736997) SARS-CoV-2 viral sequences were plotted on a circular representation of the viral genome using a pipeline developed in-house. The procedure took as its input the list of sequence variations identified for each virus strain/variant and built circos plots with the R platform (R Core Team 2019. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; www.R-project.org/) and the package “circlize” (46).
Nanopore RNA direct sequencing
To perform direct sequencing of the RNA with Nanopore technology, we prepared the RNA library using direct RNA sequencing kits (SQK-RNA002, Oxford Nanopore kits), according to the manufacturer’s instructions. Briefly, we used a poly(dT) adapter and SuperScript II Reverse Transcriptase (Thermo Fisher Scientific) to generate RNA-DNA hybrids. Agencourt RNAClean XP magnetic beads (Beckman) were used to purify the RNA-DNA duplexes. Subsequently, RNA-DNA hybrids were ligated to Nanopore sequencing adapters (RMX) with T4 DNA ligase (NEB E6056) before sequencing. After another step of purification using Agencourt RNAClean XP magnetic beads, the library concentration was estimated by Qubit assay (Thermo Fisher Scientific). The Nanopore direct RNA library was then loaded into the flow cell (R9.06) using MinION, for 26 hours. The motor protein RMX pulls the 3′ end of the RNA strand into the Nanopore channel (47). Changes in the ionic current were then detected at each pore by a sensor. Sequence data were validated as follows. Quality control checks on raw sequence data from the Nanopore were performed with NanoPlot v1.28.2 (41) and FastQC v0.11.9 (42). Only reads with a Phred quality score > 7 were used in the following analysis. In this phase, reads were not trimmed or filtered out on the basis of their length (48). Reads from sgRNAs were classified as follows. Reads from Nanopore direct sequencing were initially identified by alignment to a set of reference sequences using Minimap2 version 2.17 (40), with the “long-read spliced alignment” preset. The reference sequences included the SARS-CoV-2 viral genome (GenBank, NC_045512), the Chlorocebus sabaeus host genome (assembly ChlSab 1.1), and the human ribosomal DNA repeat (GenBank, U13369.1). Reads identified as viral were then realigned to the same viral genome sequence with Minimap2, with the following options: -k 8 -w 1 -g 30000 --max-chain-skip 40 -p 0.7 -N 32 --frag=no --splice -G 30000 -C 0 -u n --splice-flank=no -A 1 -B 2 -O 2,24 -E 1,0 -z 400,200 --no-end-flt. Reads were assigned to each canonical sgRNA by selecting those that overlapped with the genomic interval of 54 to 86 (leader), did not overlap with the genomic interval of 100 to 21,546 (ORF1ab), and contained the specific downstream transcription regulatory sequence preceding each open reading frame (ORF). Filtering and counting of mapped reads were performed with SAMtools version 1.9 (http://htslib.org). The sequence alignments were generated with Clustal Omega software (www.uniprot.org/align/). sgRNA quantification was performed by counting the leader sequence that contained reads mapped to the N ORF sequence. RNA amounts from different samples were normalized against the number of reads that mapped to the host genome; only host genome reads longer than 1500 nucleotides were used (i.e., the length of N sgRNA), to compensate for different average read lengths in different samples.
Docking experiments
The polyP molecule (ligand) containing 20 phosphate atoms (polyP20) was built with the tools available within the ZINC data bank web server (49) and then translated into PDB coordinates. The 3D structures of the receptor molecules were downloaded from the PDB. The atom types and charges were assigned to the ligands and receptors with the AutoDock Tools resources. All of the docking experiments were performed with AutoDock Vina (50).
ELISAs to test for binding between ACE2 and polyPs
ELISAs were performed with the human ACE2-Fc chimera protein (GenScript). Flat-bottom, 96-well plates (Thermo Fisher Scientific) were coated with 416 nM of each polyP. After blocking with 5% nonfat dry milk in phosphate-buffered saline (PBS) for 1 hour at 37°C, the plates were incubated with the ACE2-Fc chimeric protein used at increasing concentrations (0.1 to 2 μM) in 3% bovine serum albumin (BSA)–PBS for 1 hour at room temperature with gentle shaking. In parallel assays, the ACE2-Fc chimeric protein was tested under the same conditions on uncoated wells after blocking, as a negative control. The plates were then washed with PBS and incubated with an anti-human HRP-conjugated antibody (AP113P, Sigma-Aldrich, USA) for 1 hour at room temperature with gentle shaking. The remaining steps were performed as previously described (51). Absorbance at 450 nm was measured with a plate reader (EnVision; 2102, PerkinElmer), and the binding values are reported as the means of at least three determinations.
Preparation and characterization of sodium polyPs
Sodium phosphate, monobasic (NaH2PO4 ≥ 99.0%, Sigma-Aldrich) was polymerized in an aluminum crucible in an electric furnace (HQ-DMF3, CORETECH) at 700°C for 1 hour. The molten sodium polyP was then poured onto a copper plate for rapid cooling, and the sodium polyP glass obtained was ground into a powder. In general, the synthesized polyP polymer has a wide range of chain lengths. Fractionation of polyPs was performed by fractional precipitation. The sodium polyP powder (20 g) was dissolved in 200 ml of distilled water (i.e., at 1:10, w/v), the pH was adjusted to 7, and this was left at 25°C for 12 hours. Acetone was then added to this solution of sodium polyP dropwise, until the solution became cloudy because of precipitation of polyPs. The precipitated polyPs were separated by centrifugation at 5.8g for 5 min (1248R, LABOGENE). This fractional precipitation was repeated 25 times, and the polyP fractions that were obtained were dried in a freeze dryer for 48 hours. A gel permeation chromatography–multiangle light-scattering system was used to determine the polyP chain lengths in each of the precipitation fractions. This included a high-performance LC system (LC-20 AD, Shimadzu) and a multiangle light-scattering detector (miniDAWN TREOS II, Wyatt Technology). The gel permeation chromatography column (PL aquagel-OH; 7.5 mm by 50 mm; 8 μm; Agilent Technology) was protected by a guard column (PL aquagel-OH Mixed-M; 7.5 mm by 300 mm; 8 μm; Agilent Technology). The system was run with 150 mM NaCl. The concentration of polyPs in the sample injection was 25 mg/ml, and the injection volume was 50 μl. The temperature of the column was maintained at 30°C. Polymerization of sodium phosphate monobasic took place as follows
The polymer obtained is known as Graham’s salt, which contains polyPs as three mixed molecular structures: linear, cyclic, and branched. Branched polyPs are very labile, and when they dissolve in water, the branching points of these polyPs are hydrolyzed at a high rate, irrespective of pH, and even at room temperature. In contrast, linear polyPs and cyclopolyPs are hydrolyzed very slowly at neutral pH and room temperature. The “half hydrolysis time” for the P─O─P bonds in linear polyPs at pH 7 at 25°C is several years. These synthesized polyPs usually contain very small amounts of cyclopolyPs, and almost all of these have chain lengths of <6 Pi units (52). Here, the minimum chain length obtained for these polyPs was 20, and therefore, when these polyPs were separated by fractional precipitation, cyclopolyPs were not precipitated. After fractionation of polyPs by gel permeation chromatography, their chain lengths (n) were determined according to the following equation
The range of the chain lengths in each fraction was then obtained through the polydispersity index [weight-average molar mass/number-average molar mass (Mw/Mn)], using the Astra software, version 6.1 (Wyatt Technology).
Western blotting analysis
Cells for Western blotting analysis were washed in cold PBS and lysed in cell lysis buffer [20 mM sodium phosphate (pH 7.4), 150 mM NaCl, 10% (v/v) glycerol, 1% (w/v) sodium deoxycholate, 1% (v/v) Triton X-100] supplemented with protease inhibitors (Roche). The resulting cell lysates were cleared by centrifugation at 16,200g for 10 min at room temperature, and the supernatants were removed and assayed for protein concentrations with the Protein Assay Dye Reagent (Bio-Rad). The cell lysates (50 μg) were resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gels of different percentages, which depended on the molecular weights of the proteins of interest. The proteins were then electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Millipore). After 1 hour in blocking solution with 5% (w/v) dry milk fat in PBS or 5% (w/v) BSA (Sigma-Aldrich) in tris-buffered saline [both of which contained 0.02% (v/v) Tween 20] (TBS-Tween), the PVDF membranes were incubated with the required primary antibody overnight at 4°C: anti-ACE2 (1:1000; ab15348), anti–SARS-CoV-2 nucleoprotein (1:250; 35-579, ProSci Inc.), anti–SARS-CoV-2 S (1: 250; ab272504, Abcam), anti-RdRp (1:100; ARG11093, Arigo), anti-p65 (1:1000; sc-372, Santa Cruz Biotechnology), anti–p-(Ser311)p65–NF-κB (1:500; sc-101748. Santa Cruz Biotechnology), anti–glyceraldehyde-3-phosphate dehydrogenase (1:10,000; sc-365062. Santa Cruz Biotechnology), anti–α-tubulin (1:3000; Ab15246, Abcam), or anti–β-actin (1:10,000; A5441, Sigma-Aldrich). The membranes were then incubated with the required secondary antibodies for 1 hour at room temperature: secondary mouse or rabbit HRP-conjugated antibodies (NC 15 27606, ImmunoReagents Inc.), diluted in 5% (w/v) BSA in TBS-Tween or in 5% (w/v) milk fat in PBS-Tween, according to the manufacturer’s instructions. The protein bands were visualized by chemiluminescence detection (Pierce, Thermo Fisher Scientific, IL, USA). Densitometry analysis was performed with ImageJ software. The peak areas of the bands were measured on the densitometry plots, and the relative proportions (percentages) were calculated. Then, the density areas of the peaks were normalized with those of the loading controls, and the ratios for the corresponding controls are presented as fold changes. Western blotting was performed in triplicate. Densitometry analyses shown were derived from three independent experiments.
IF and HuluFISH
Cells (1 × 104) were fixed in 4% paraformaldehyde (PFA) in PBS for 30 min, washed three times with PBS, and permeabilized for 15 min with 0.1% Triton X-100 (215680010, Acros Organics) diluted in PBS. The cells were then washed with PBS and blocked with 3% BSA (A9418, Sigma-Aldrich) in PBS for 1 hour at room temperature. The samples were incubated with the appropriate primary antibodies overnight at 4°C: anti-ACE2 (1:50; ab15348, Abcam), anti–SARS S (1:50; ab272420, Abcam), or anti–SARS-CoV-2 nucleoprotein (1:50; 35-579, ProSci Inc.). After washing twice with PBS, the samples were incubated with the appropriate secondary antibody at room temperature for 1 hour: anti-mouse Alexa Fluor 647 (1:100; ab150115, Abcam) or anti-rabbit Alexa Fluor 647 (1:100; ab150075, Abcam). DNA was stained with 4′,6-diamidino-2-phenylindole (1:1000; no. 62254, Thermo Fisher Scientific). The slides were washed and mounted with coverslips with 50% glycerol (G5150, Sigma-Aldrich). HuluFISH was performed on the pan–SARS-CoV-2 probe against gRNA-S (MetaSystem, Zeiss) of SARS-CoV-2–infected Vero E6 cells previously fixed in 4% PFA in PBS and stained with anti-ACE2 antibody. Microscopy images were obtained with the Elyra 7 platform (Zeiss) with the optical Lattice SIM technology, using the 63× oil immersion objective. Z-stacking imaging, 3D reconstruction, and colocalization scatterplots were performed with the ZEN software (Zeiss, blue edition). Quantification of the fluorescence intensity was performed with ImageJ software (version 1.52r; https://imagej.nih.gov/ij/index.html), and the corrected total cell fluorescence (CTCF) was estimated according to the following equation:
CTCF = integrated density − (area of selected cell × mean background fluorescence).
Real-time cell proliferation assay for cell index
Cellular proliferation rates were determined with the xCELLigence system (Roche). Primary human epithelial cells, Vero E6 cells, and Caco2 cells (3 × 104) were treated with increasing concentrations of polyP120, harvested, and then added to a single well of an xCELLigence E-plate 16. The proliferation rates were determined at 2-min intervals by measuring the impedance changes across the electrodes at the bottoms of the wells. These cell index values were calculated by the xCELLigence RTCA software.
Caspase-3 activity assay
Primary human epithelial cells (3 × 105) were treated with increasing concentrations of polyP120 (4.16 to 112 μM) for 24 hours. Vehicle-treated cells and cells treated with 1 μM staurosporine were used as negative and positive controls, respectively. The cells were then harvested, and apoptosis was evaluated with Caspase-3 Activity Assay kits (no. 5723, Cell Signaling Technology), according to the manufacturer’s instructions. Briefly, the cells were lysed in 30 μl of PathScan lysis buffer (no. 7018, Cell Signaling Technology). Cell lysates (4 μg) were mixed with substrate solution (N-acetyl-Asp-Glu-Val-Asp-7-amino-4-methylcoumarin; also known as Ac-DEVD-AMC Fluorescent Substrate) and incubated at 37°C in the dark for 1 hour. Relative fluorescent units (RFUs) were acquired (excitation, 380 nm; emission, 460 nm) using a multimode plate reader (PerkinElmer).
Limited proteolysis
Limited proteolysis was carried out on human recombinant ACE2-Fc (GenScript) and in parallel on its complexes with polyP120 and polyP8 after preincubation for 2 hours at room temperature in a 10-fold molar excess of polyPs. The experiments were performed directly in shipping buffer [20 mM tris-HCl, 300 mM NaCl, 1 mM ZnCl2, and 10% glycerol (pH 7.4)] using chymotrypsin as the enzymatic probe with an enzyme:substrate ratio of 1:50 (w/w). The digestion reactions were followed as time courses, with volumes withdrawn that contained the equivalent of 4 μg of ACE2-Fc, at 30 min, 1 hour, and 2 hours of proteolysis. The digestion products were resolved on 10% SDS-PAGE gels; an aliquot of 4 μg of intact ACE2-Fc was used as reference. The differential bands displayed in the three samples were cut and in situ hydrolyzed with trypsin. Peptide mixtures were analyzed by nano–LC-MS/MS (Proxeon-nanoEasy II-LTQ Orbitrap XL; Life Technologies, Carlsbad, CA, USA). Peptide fractionation was performed on a C18 capillary reverse-phase column (200 mm, 75 μm by 5 μm) working at a flow rate of 250 nl/min, using a step gradient of eluent B (0.2% formic acid, 95% acetonitrile; LC-MS grade) from 10 to 60% over 69 min and from 60 to 95% over 3 min. The MS data were recorded from 300 mass/charge ratio (m/z) to 1800 m/z, and the MS/MS data were collected in data-dependent acquisition mode: The 10 most abundant ions in each scan were fragmented and the respective tandem mass spectra were recorded. The peptide identification was performed using the Mascot software (Matrix Science, Boston, USA), searching in a database that contained only the sequence of ACE2-Fc. The search was performed by setting cysteine carbamidomethylation as a fixed modification, pyro-Glu of N-terminal Gln and pyro-carbamidomethyl of N-terminal Cys as variable modifications, trypsin as the digestion enzyme, and 10 parts per million as the MS tolerance. The identified peptides were filtered at P < 0.05 and ion scores >20.
Site-directed mutagenesis
Site-directed mutagenesis for the generation of plasmids expressing mutated pCEP4-myc-ACE2 was performed with the following primers: His378: 5′-ATGAGATGGGGTCAATCCAGTAT-3′ (forward) and 5′-ATACTGGATTGACCCCATCTCAT-3′ (reverse); Arg393: 5′-ACCTTTTCTGCTACAAAATGGAGCTAATGA-3′ (forward) and 5′-TCATTAGCTCCATTTTGTAGCAGAAAAGGT-3′ (reverse); His401: 5′-AAGGATTCTCAGAAGCTGTT-3′ (forward) and 5′-AACAGCTTCTGAGAATCCTT-3′ (reverse); Arg514: 5′-CTCATTCATTCAATATTACACAAGGA-3′ (forward) and 5′-TCCTTGTGTAATATTGAATGAATGAG-3′ (reverse). The reactions were performed with PCR kits (KAPA HiFi HotStart; KR0369; v10.17) on a PCR machine under the following conditions: 95°C for 3 min, 98°C for 20 s, 60°C for 15 s, and 72°C for 6 min; these three conditions were cycled 16 times; then 72°C for 7 min. The nonmutated PCR template was removed from the pool of PCR products by incubation with the restriction endonuclease Dpn I [R0176L, New England Biolabs (NEB)] at 37°C for 1 hour and then at 60°C for 20 min. The plasmids were circularized through a ligation reaction with T4 DNA ligase (M0202S) at room temperature for 2 hours before being used to transform 5-α Competent Escherichia coli (High Efficiency, NEB). All of the plasmids were then sequenced to verify the mutations.
Statistical analysis
Statistical significance was defined as P < 0.05 by unpaired two-tailed Student’s t tests. All of the data are given as means ± SD. In the figures, statistical significance is represented as follows: *P < 0.05, **P < 0.01, and ***P < 0.001. For multiple comparisons, the P values were adjusted by the Bonferroni method. The statistical significance of the data was also confirmed by two-tailed Mann-Whitney tests. All of the data from the qRT-PCR assays were analyzed using unpaired two-tailed Student’s t tests, by comparing (i) uninfected versus vehicle control–infected cells (i.e., cells infected and treated with the vehicle as a control for polyPs) and (ii) polyP120-treated cells versus vehicle control–infected cells. ELISA data were analyzed by applying unpaired, two-tailed Student’s t tests to the absorbance values, and comparing these with those obtained with 0.1 μM human ACE2-Fc chimera protein (Fig. 1D) or 416 nM polyP8 or polyP34 (Fig. 1E). The background values were subtracted from the absorbance values under each condition. Caspase-3 activities were analyzed by unpaired two-tailed Student’s t tests by comparing RFU values measured after 1 hour of incubation to those obtained at the experimental starting point (i.e., time zero). The IC50 for polyP120 was calculated by nonlinear regression analysis performed with GraphPad Prism 9 {using [inhibitor] versus response (three parameters)}. All of the experiments were performed in triplicate.
Acknowledgments
We thank C. Berrie for providing professional editing and graphical consultation. We thank D. Schlessinger, Scientist Emeritus Intramural Research Program Ageing, NIH Bethesda, MD, USA, for critical discussion of the manuscript; A. Cometta, Zeiss, Milan, Italy, for assistance with the Zeiss Elyra 7 images; R. Tufano of the Bioinformatic Core for bioinformatic assistance on the SARS-Cov-2 emerging-variants data analyses; and C. Angelini, Istituto per le Applicazioni del Calcolo “Mauro Picone,” Consiglio Nazionale delle Ricerche, Naples, Italy, for helpful discussions and data evaluation on selecting the best statistical analysis tool used. We further thank A. Limone (IZSM Director), P. Forestieri (President, CEINGE), and M. Giustino (CEO, CEINGE) for collaborative support of the program within the Regione Campania Covid19 Taskforce. We thank I. Gentile (Head of the Covid19 Therapy Unit at AOU Federico II) for comments on clinical trials of patients with COVID-19. Funding: This study was supported by the project “CEINGE TaskForce COVID19,” code D64I200003800 by Regione Campania for the fight against Covid-19 (DGR no. 140; 17 March 2020). We further thank for support the Italian Association for Cancer Research (AIRC) Grant IG no. 22129 (M.Z.), the European School of Molecular Medicine for a doctorate program Fellowship (F.A.), Fondazione Celeghin Italiana (M.Z.), and Ministero dell’Università e della Ricerca Italiana (PRIN) grant no. 2017FNZRN3 (M.Z.). We further thank NRF 2018R1A5A2025079–Korean Ministry of Science and ICT (J.-H.C.). Author contributions: V.F. performed qPCR, in vitro treatment, and Western blotting analysis. F.A. performed quantitative IF staining, site-directed mutagenesis, transfections, and Western blotting analysis. R.S. and F.Q. prepared RNA from cell lysates and did qPCR analysis. L.M., F.A., and C.A. performed Western blotting. V.F., F.A., and L.M. performed in vitro treatments on noninfected cells. V.F. and L.M. performed the caspase assays. C.D.L. and M.P. performed ELISAs. M.M. and I.I. performed limited proteolysis experiments. F.A. amplified viral sequences for sequencing analyses. G.B., M.Z., G.F., M.V., S.B., P.C., and B.M.P. performed SARS-CoV-2 infections in a BSL3-authorized laboratory. G. Castaldo and M.C. obtained the primary human epithelial cells by nasal brushing. G.P. and A.B. performed viral sequencing and bioinformatic analyses. L.C. and R.D.M. performed Nanopore sequencing. S.P. and M.B. performed docking experiments. C.T., L.A., and E.C. obtained qualified samples from Italian patients with COVID-19. I.P. and B.I. performed staining on Vero cells. D.-Y.K., K.-S.Y., J.-H.C., and H.-Y.K. performed experiments in the Korean laboratory. G.B. and G. Criscuolo contributed to data acquisition and analysis. G. Criscuolo contributed to the qPCR expression analysis. M.Z., J.-H.C., and H.-Y.K. designed the experiments, critically discussed experimental plans with the authors, and wrote the paper. All authors discussed the results and commented on the paper. Competing interests: H.-Y.K. (50%), M.Z. (25%), and the University Industry Foundation of Yonsei University Health System (25%) are inventors on patent application (KR: 10-2020-0123355; PCT: PCT/KR2020/018981) held/submitted by the Yonsei University Industry Foundation that covers composition including polyP to treat infectious disease or COVID-19 virus. The other authors declare that they have no competing interests. Data and materials availability: Gene sequence accession numbers: Viral sequences used for the experiments were deposited to GISAID (www.epicov.org/) or GenBank (www.ncbi.nlm.nih.gov). GISAID, BetaCov/Korea/KCDC03/2020, registered ID: EPI_ISL_407193 (entire virus sequence); British UK variant (B.1.1.7): EPI_ISL_736997. GenBank: MT682732.1 (SARS-CoV-2 isolate SARS-CoV-2/human/ITA/Naples/2020, complete genome) and MT890669 (SARS-CoV-2 isolate SARS-CoV-2/human/ITA/Naples/2020, complete genome). Whole-exome sequencing of the primary human epithelial nasal-brushing–derived cells is deposited at the European Variation Archive (EVA) (www.ebi.ac.uk/eva/). EMBL-EBI; project ID: PRJEB42411; analyses: ERZ1700617. Direct RNA sequencing by Nanopore Technology for sgRNAs is deposited at the SRA (www.ncbi.nlm.nih.gov/sra). Accession: PRJNA688696 (release date: 5 January 2021) available at www.ncbi.nlm.nih.gov/sra/PRJNA688696. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Tay M. Z., Poh C. M., Renia L., MacAry P. A., Ng L. F. P., The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 20, 363–374 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F. G., Horby P. W., Cao B., Wang C., Remdesivir in adults with severe COVID-19: A randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395, 1569–1578 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen K. G., Rambaut A., Lipkin W. I., Holmes E. C., Garry R. F., The proximal origin of SARS-CoV-2. Nat. Med. 26, 450–452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B., Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D. S. C., Du B., Li L.-j., Zeng G., Yuen K.-Y., Chen R.-c., Tang C.-l., Wang T., Chen P.-y., Xiang J., Li S.-y., Wang J.-l., Liang Z.-j., Peng Y.-x., Wei L., Liu Y., Hu Y.-h., Peng P., Wang J.-m., Liu J.-y., Chen Z., Li G., Zheng Z.-j., Qiu S.-q., Luo J., Ye C.-j., Zhu S.-y., Zhong N.-s.; China Medical Treatment Expert Group for Covid-19 , Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim D., Lee J.-Y., Yang J.-S., Kim J. W., Kim V. N., Chang H., The Architecture of SARS-CoV-2 Transcriptome. Cell 181, 914–921.e10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walls A. C., Park Y.-J., Tortorici M. A., Wall A., McGuire A. T., Veesler D., Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181, 281–292.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wrapp D., Wang N., Corbett K. S., Goldsmith J. A., Hsieh C. L., Abiona O., Graham B. S., McLellan J. S., Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N.-H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S., SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e8 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clausen T. M., Sandoval D. R., Spliid C. B., Pihl J., Perrett H. R., Painter C. D., Narayanan A., Majowicz S. A., Kwong E. M., McVicar R. N., Thacker B. E., Glass C. A., Yang Z., Torres J. L., Golden G. J., Bartels P. L., Porell R. N., Garretson A. F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B. M., Caradonna T. M., Kellman B. P., Martino C., Gordts P. L. S. M., Chanda S. K., Schmidt A. G., Godula K., Leibel S. L., Jose J., Corbett K. D., Ward A. B., Carlin A. F., Esko J. D., SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 183, 1043–1057.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sola I., Almazan F., Zuniga S., Enjuanes L., Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2, 265–288 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., Masciovecchio C., Angeletti S., Ciccozzi M., Gallo R. C., Zella D., Ippodrino R., Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J. Transl. Med. 18, 179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCormick K. D., Jacobs J. L., Mellors J. W., The emerging plasticity of SARS-CoV-2. Science 371, 1306–1308 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Abramov A. Y., Fraley C., Diao C. T., Winkfein R., Colicos M. A., Duchen M. R., French R. J., Pavlov E., Targeted polyphosphatase expression alters mitochondrial metabolism and inhibits calcium-dependent cell death. Proc. Natl. Acad. Sci. U.S.A. 104, 18091–18096 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Angelova P. R., Agrawalla B. K., Elustondo P. A., Gordon J., Shiba T., Abramov A. Y., Chang Y. T., Pavlov E. V., In situ investigation of mammalian inorganic polyphosphate localization using novel selective fluorescent probes JC-D7 and JC-D8. ACS Chem. Biol. 9, 2101–2110 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Müller F., Mutch N. J., Schenk W. A., Smith S. A., Esterl L., Spronk H. M., Schmidbauer S., Gahl W. A., Morrissey J. H., Renné T., Platelet polyphosphates are proinflammatory and procoagulant mediators in vivo. Cell 139, 1143–1156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wat J. M., Foley J. H., Krisinger M. J., Ocariza L. M., Lei V., Wasney G. A., Lameignere E., Strynadka N. C., Smith S. A., Morrissey J. H., Conway E. M., Polyphosphate suppresses complement via the terminal pathway. Blood 123, 768–776 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoac B., Kiffer-Moreira T., Millan J. L., McKee M. D., Polyphosphates inhibit extracellular matrix mineralization in MC3T3-E1 osteoblast cultures. Bone 53, 478–486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie L., Jakob U., Inorganic polyphosphate, a multifunctional polyanionic protein scaffold. J. Biol. Chem. 294, 2180–2190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stotz S. C., Scott L. O., Drummond-Main C., Avchalumov Y., Girotto F., Davidsen J., Gómez-Gárcia M. R., Rho J. M., Pavlov E. V., Colicos M. A., Inorganic polyphosphate regulates neuronal excitability through modulation of voltage-gated channels. Mol. Brain 7, 42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmstrom K. M., Marina N., Baev A. Y., Wood N. W., Gourine A. V., Abramov A. Y., Signalling properties of inorganic polyphosphate in the mammalian brain. Nat. Commun. 4, 1362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorenz B., Leuck J., Kohl D., Muller W. E., Schroder H. C., Anti-HIV-1 activity of inorganic polyphosphates. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 14, 110–118 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Neufurth M., Wang X., Tolba E., Lieberwirth I., Wang S., Schröder H. C., Müller W. E. G., The inorganic polymer, polyphosphate, blocks binding of SARS-CoV-2 spike protein to ACE2 receptor at physiological concentrations. Biochem. Pharmacol. 182, 114215 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.EFSA Panel on Food Additives and Flavourings (FAF), Younes M., Aquilina G., Castle L., Engel K.-H., Fowler P., Fernandez M. J. F., Fürst P., Gürtler R., Husøy T., Mennes W., Moldeus P., Oskarsson A., Shah R., Waalkens-Berendsen I., Wölfle D., Aggett P., Cupisti A., Fortes C., Kuhnle G., Lillegaard I. T., Scotter M., Giarola A., Rincon A., Tard A., Gundert-Remy U., Re-evaluation of phosphoric acid-phosphates - di-, tri- and polyphosphates (E 338-341, E 343, E 450-452) as food additives and the safety of proposed extension of use. EFSA J. 17, e05674 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K. B., Yoshida M., Barnes J. L.; HCA Lung Biological Network , SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 26, 681–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hou Y. J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K. H. III, Leist S. R., Schäfer A., Nakajima N., Takahashi K., Lee R. E., Mascenik T. M., Graham R., Edwards C. E., Tse L. V., Okuda K., Markmann A. J., Bartelt L., de Silva A., Margolis D. M., Boucher R. C., Randell S. H., Suzuki T., Gralinski L. E., Kawaoka Y., Baric R. S., SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zamorano Cuervo N., Grandvaux N., ACE2: Evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. eLife 9, e61390 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M., The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 53, 25–32 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hassanian S. M., Avan A., Ardeshirylajimi A., Inorganic polyphosphate: A key modulator of inflammation. J. Thromb. Haemost. 15, 213–218 (2017). [DOI] [PubMed] [Google Scholar]
- 30.Hassanian S. M., Dinarvand P., Smith S. A., Rezaie A. R., Inorganic polyphosphate elicits pro-inflammatory responses through activation of the mammalian target of rapamycin complexes 1 and 2 in vascular endothelial cells. J. Thromb. Haemost. 13, 860–871 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen K. B., Chhabra K. H., Nguyen V. K., Xia H. J., Lazartigues E., The transcription factor HNF1α induces expression of angiotensin-converting enzyme 2 (ACE2) in pancreatic islets from evolutionarily conserved promoter motifs. BBA Gene Regul. Mech. 1829, 1225–1235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clarke N. E., Belyaev N. D., Lambert D. W., Turner A. J., Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin. Sci. 126, 507–516 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Kai H., Kai M., Interactions of coronaviruses with ACE2, angiotensin II, and RAS inhibitors-lessons from available evidence and insights into COVID-19. Hypertension Res. 43, 648–654 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zollo M., Ferrucci V., Izzo B., Quarantelli F., Domenico C. D., Comegna M., Paolillo C., Amato F., Siciliano R., Castaldo G., Capoluongo E., SARS-CoV-2 subgenomic N (sgN) transcripts in oro-nasopharyngeal swabs correlate with the highest viral load, as evaluated by five different molecular methods. Diagnostics 11, 288 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B. F., Hahn A. S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., Winkler M. S., Schulz S., Jäck H.-M., Jahrsdörfer B., Schrezenmeier H., Müller M., Kleger A., Münch J., Pöhlmann S., SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 184, 2384–2393.e12 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geers D., Shamier M. C., Bogers S., den Hartog G., Gommers L., Nieuwkoop N. N., Schmitz K. S., Rijsbergen L. C., van Osch J. A. T., Dijkhuizen E., Smits G., Comvalius A., van Mourik D., Caniels T. G., van Gils M. J., Sanders R. W., Munnink B. B. O., Molenkamp R., de Jager H. J., Haagmans B. L., de Swart R. L., Koopmans M. P. G., van Binnendijk R. S., de Vries R. D., Kessel C. H. G., SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 6, eabj1750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia-Beltran W. F., Lam E. C., Astudillo M. G., Yang D., Miller T. E., Feldman J., Hauser B. M., Caradonna T. M., Clayton K. L., Nitido A. D., Murali M. R., Alter G., Charles R. C., Dighe A., Branda J. A., Lennerz J. K., Lingwood D., Schmidt A. G., Iafrate A. J., Balazs A. B., COVID-19-neutralizing antibodies predict disease severity and survival. Cell 184, 476–488.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gray M. J., Jakob U., Oxidative stress protection by polyphosphate--new roles for an old player. Curr. Opin. Microbiol. 24, 1–6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han K. Y., Hong B. S., Yoon Y. J., Yoon C. M., Kim Y. K., Kwon Y. G., Gho Y. S., Polyphosphate blocks tumour metastasis via anti-angiogenic activity. Biochem. J. 406, 49–55 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coomes E. A., Haghbayan H., Interleukin-6 in COVID-19: A systematic review and meta-analysis. Rev. Med. Virol. 30, 1–9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costanzo L., Palumbo F. P., Ardita G., Antignani P. L., Arosio E., Failla G.; Italian Society for Vascular Investigation and the Italian Society of Vascular Medicine , Coagulopathy, thromboembolic complications, and the use of heparin in COVID-19 pneumonia. J. Vasc. Surg. Venous Lymphat. Disord. 8, 711–716 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grifoni A., Weiskopf D., Ramirez S. I., Mateus J., Dan J. M., Moderbacher C. R., Rawlings S. A., Sutherland A., Premkumar L., Jadi R. S., Marrama D., de Silva A. M., Frazier A., Carlin A. F., Greenbaum J. A., Peters B., Krammer F., Smith D. M., Crotty S., Sette A., Targets of T Cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 181, 1489–1501.e15 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Z., Yang X., Zhong J., Zhou Y., Tang Z., Zhou H., He J., Mei X., Tang Y., Lin B., Chen Z., McCluskey J., Yang J., Corbett A. J., Ran P., Exposure to SARS-CoV-2 generates T-cell memory in the absence of a detectable viral infection. Nat. Commun. 12, 1724 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tai H. C., Schuman E. M., Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 9, 826–838 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Di Lullo A. M., Scorza M., Amato F., Comegna M., Raia V., Maiuri L., Ilardi G., Cantone E., Castaldo G., Iengo M., An "ex vivo model" contributing to the diagnosis and evaluation of new drugs in cystic fibrosis. Acta Otorhinolaryngol. Ital. 37, 207–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu Z., Gu L., Eils R., Schlesner M., Brors B., circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Bayega A., Fahiminiya S., Oikonomopoulos S., Ragoussis J., Current and future methods for mRNA analysis: A drive toward single molecule sequencing. Methods Mol. Biol. 1783, 209–241 (2018). [DOI] [PubMed] [Google Scholar]
- 48.De Coster W., D’Hert S., Schultz D. T., Cruts M., Van Broeckhoven C., NanoPack: visualizing and processing long-read sequencing data. Bioinformatics 34, 2666–2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sterling T., Irwin J. J., ZINC 15—Ligand Discovery for Everyone. J. Chem. Inf. Model. 55, 2324–2337 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trott O., Olson A. J., AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borriello M., Laccetti P., Terrazzano G., D’Alessio G., De Lorenzo C., A novel fully human antitumour immunoRNase targeting ErbB2-positive tumours. Br. J. Cancer 104, 1716–1723 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulaev I. S., Biochemistry of inorganic polyphosphates. Rev. Physiol. Biochem. Pharmacol. 73, 131–158 (1975). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.